Graphical abstract
Keywords: Fatty acid desaturase, Machine learning, Haplotype, Protein structure, Docking, Oleic/linoleic acid ratio
Abstract
Fatty Acid Desaturase 2 (FAD2), a key enzyme in the fatty acid biosynthesis pathway, is involved in the desaturation and conversion of oleic acid to linoleic acid. Therefore, it plays a crucial role in oleic/linoleic acid ratio and the quality of olive oil. DNA sequencing of 19 FAD2 genes from a set of olive oil varieties revealed several single-nucleotide polymorphisms (SNPs) and highlighted associations between some of the SNPs and saturated fatty acids contents. This was further confirmed by SNP-interaction and machine learning approach. Haplotype diversity analysis led to the discovery of three highly polymorphic SNPs and four haplotypes harboring differential oleic/linoleic acid ratios. Moreover, a combination of molecular modeling and docking experiments allowed a deeper and better understanding of the structure–function relationship of the FAD2 enzyme. Sequence patterns and variations involved in the regulation of the FAD2 activity were also identified. Furthermore, S82C and H213N substitutions in OeFAD2 make the Oueslati variety more interesting in terms of fatty acid profile and oleic acid level.
1. Introduction
One of the most ancient domesticated species, a native of the middle east, highly popular and appreciated for its delicious drupe and healthy fruit oil, Olea europaea L. is an emblematic evergreen tree cultivated all over the Mediterranean basin [1]. The Food and Agriculture Organization (FAO) had declared 2021 as the international year of fruits and vegetables, aiming to raise awareness about balanced and nutritious food and a healthy diet [2], [3]. Indeed, unhealthy diets and malnutrition are among the top-ten risk factors for disease globally [3]. Food and diet are part of one’s culture and identity, which change over time with a preference for a more convenient lifestyle. However, the consumption of junk food containing highly processed and degraded unhealthy vegetable oil is increasing due to various reasons. Olive oil is considered a healthy oil and its consumption is strongly recommended due to its several benefits. Benefits of olive oil consumption include balanced growth and development, better mental health, healthy heart, lower cardiovascular diseases and cancer risks, lower diabetes and obesity risks, and improved immunity.
Although olive oil is consumed worldwide, the Mediterranean basin is its principal zone of production [4], mainly Spain, Greece, Tunisia, and Italy, with respectively 1790, 327, 278, and 277 thousand tons of Virgin Olive Oil (VOO) produced in 2018 [5]. The world olive oil market size is expected to reach $4.79 Billion by 2025 [6]. Besides, olive oil price and demand depend on several factors, mainly its quality and geographic origin [7]. Moreover, consumers’ choices are guided by the origin and category of the olive oil [8], and the main worry of consumers is purity of the oil [9]. The quality of olive oil is determined mainly by its acidity, expressed as oleic acid content. The Extra Virgin Olive Oil (EVOO) is considered as the best quality olive oil (<0.8 g/100 g) [10]. Consequently, the level of acidity represents the main parameter of reference, and oleic acid content is a crucial factor in determining the category of olive oil.
The biosynthesis of fatty acids, particularly oleic acid, is a complex pathway that spans several cellular compartments and where different substrates, precursors, and enzymes intervene. Fatty acid synthesis, as for oleic acid, starts with sugar as the backbone precursor [11]. The carbon comes from leaf through translocation of photosynthates as well as the olive drupe itself since it is a photosynthetic organ [11]. The biosynthesis of fatty acids proceeds through a series of enzymatic reactions, including several enzymes, namely fatty-acid synthase, desaturase, transacylase, esterase, transferase, and carboxylase [12]. Unfortunately, being complex, this pathway has not been completely characterized yet, nor have its individual enzymes revealed. Therefore, much research still needs to be performed to fully understand the mechanism of fatty acids biosynthesis in Olea europaea. Knowing the characteristics and the specificities of these enzymes and their corresponding genes is a pre-requisite for genetic improvement of olive oil quality. In particular, the Fatty Acid Desaturase 2 (FAD2) enzyme is involved in the desaturation and conversion of oleic acid (C18:1) to linoleic acid (C18:2). It therefore plays a key role in oleic/linoleic acid ratio in olive oil [13], [14].
Breeding for improved olive oil quality could be realized by monitoring the oleic/linoleic acid ratio. Indeed, the development of several oilseeds with decreased linoleic acid and/or increased oleic acid contents has been achieved through FAD2 manipulation using different biotechnological approaches [15], [16], [17], [18], [19]. The FAD2 gene family comprises several members, sometimes including isoforms, displaying distinct or similar patterns of expression, and involved in fatty acid metabolism and stress response [20], [21], [22]. Recently, in Olea europaea subsp. europaea, two sequences of microsomal FAD2 genes (OeFAD2-1 and OeFAD2-2) were characterized [23]. Given that OeFAD2-2 is more implicated and responsible for oleic acid desaturation in the mesocarp [11], [24], [25], [26], [27], it would be a pertinent candidate gene to focus on for genetic improvement of olive oil quality.
Therefore, given that olive tree biodiversity constitutes the primary source for breeding programs aiming at improving the oil quality [28], and since Tunisia comes after EU in terms of production and exportation, with a rich and varied olive germplasm constituted by hundreds of cultivars and varieties; our study focused on the identification through gene sequencing, molecular characterization, Single Nucleotide Polymorphism (SNP) discovery, docking studies and structure modeling of OeFAD2-2 in 19 Tunisian olive varieties. Our findings are expected to provide a deeper insight into the biochemistry and molecular basis of olive oil constituents and quality. This, in turn, may contribute to the production of olive oil with a higher nutritional value by increasing its oleic/linoleic ratio.
2. Materials and methods
2.1. Plant material and olive oil extraction
A total of 19 olive tree varieties were chosen and used in this study (Supplementary Table 1). They were selected from different regions of Tunisia, from north to south. Two trees were considered for each variety, and for which olive oil was obtained across the steps of stoned olives grinding (2.5 Kg), followed by a mechanical oil extraction. Therefore, olive oil was obtained following the standard procedure commonly used in olive mills, which includes a milling step, followed by kneading step for 30 min at 25 °C, centrifugation for 3 min at 2000g, and finally natural decantation. All samples were kept at 4 °C until fatty acid composition determination and DNA extraction.
2.2. Determination of fatty acid composition
The composition of fatty acids of olive oil samples was determined according to the analytical method and procedure recommended by the European Union Commission Regulation 2568/91 [29]. As a prerequisite, fatty acids were converted into methyl esters to make them suitable and detectable via CPG analysis. Fatty acid methyl esters (FAMEs) were prepared through a reaction of cold trans-esterification. FAMEs were obtained by mixing the olive oil vigorously in hexane (0.2 g in 3 ml) with 0.4 ml of methanolic solution of KOH (2 N), and then analyzed by gas chromatography (Shimadzu Gas Chromatograph, Kyoto, Japan) equipped with a flame ionization detector and a fused silica capillary column (30 m length × 0.32 mm internal diameter × 0.25 μm film thickness). 1 μL was used as the injection volume, and the nitrogen was used as carrier gas with 1 ml/min flow rate. Both detection and injection temperatures were set at 220 °C, whereas 180 °C was the oven temperature. Based on their retention times, ten fatty acids (C16:0, C16:1, C17:0, C17:1, C18:0, C18:1, C18:2, C18:3, C20:0 and C20:1) could be identified. The obtained values of C18:1, C18:2, and C18:3 were indicated in Supplementary Table 2.
2.3. DNA extraction
DNA was extracted from olive using the QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany) and following the manufacturer’s instructions with slight modifications, according to Ben Ayed et al. [30]. Thereafter, DNA detection and quantification were performed with a SpectroFluorometer (Tecan GENIOS Plus, Männedorf, Switzerland) following Hoechst H33258 dye incorporation. A serial dilution of Lamda DNA (D150A Promega, Wisconsin, USA) served for standard calibration. DNAs extracted from olive samples were dissolved in TE buffer and stored at −20 °C until further PCR amplification and sequencing.
2.4. Primer design, PCR amplification and sequencing of OeFAD2-2
PCR primers (FAD2F: 5′ GAATTGAAGGGCGAGCAGT 3′ and FAD2R: 5′ TGGAATGTAATGCAAACACTGA 3′) were designed using the Primer3 program [31] based on the 5′- and 3′- UTR regions of the OeFAD2 mRNA sequence available at NCBI (http://www.ncbi.nlm.nih.gov) (accession number AY083163.1) and the OeFAD2 gene was PCR amplified from the 19 olive varieties. PCRs were performed in 30 µL reaction volumes consisting of 100 ng of olive genomic DNA, 2.5 µL of 25 mM MgC12, 2 μL of 10 mM dNTPs, 1 μL of each primer (10 μM), 1.25U of GoTaq Flexi DNA polymerase (Promega, Wisconsin, USA), 10 µL of GoTaq buffer (5X) and 3.25 μL of distilled water. PCR amplifications were performed on a 96-well Verity Thermal Cycler (Applied Biosystems, Massachusetts, USA) with a starting denaturation at 95 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 57 °C for 30 s, 72 °C for 30 s, with a final elongation at 72 °C for 10 min. PCR products from each variety were visualized by agarose gel electrophoresis, allowing to check amplification efficiency and ensure amplicon specificity and characteristics. PCR products were purified by passing through WizardR SVGel and PCR Clean-Up System purification columns (Promega, Wisconsin, USA), and sequenced in triplicate from either end using the same primers as used in PCR amplification and analyzed on the 3130XL Genetic Analyzer (Applied Biosystems, Massachusetts, USA) for sequence determination.
2.5. Sequence analysis and SNP marker discovery
FAD2 gene was sequenced from 19 Tunisian olive oil varieties having different fatty acid compositions. DNA polymorphisms in the OeFAD2 gene sequence of the 19 olive varieties were discovered by sequence alignment using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) [32] to determine the positions and types of SNPs. The outputs from the sequencer were visually inspected to confirm the possible heterozygous sequence. The potential SNPs were resequenced to minimize false positives due to sequencing artifacts. Likewise, the deduced amino acid sequences of the 19 FAD2 genes were aligned to determine conserved motifs in each sequence.
2.6. Haplotype analysis
DNA polymorphism and haplotype variation were determined within all gene sequences using DnaSP v5 [33]. The latter allowed extensive analyses and measures, namely number of polymorphic sites, haplotype number, and haplotype diversity (Hd). The haplotype analysis and the construction of the haplotype block structure were assessed using Haploview v4.2 [34]. We took into account a minimum Lewontin’s D’ value of 0.8 to design the haplotype block, solid spine of the linkage disequilibrium algorithm. The estimated values of the linkage disequilibrium were based on those of the frequency of haplotype, which was determined without genetic information on parents using the algorithm and based on the methods of the Haploview v4.2 program. Moreover, according to Gabriel et al. [35], any breed whose D’ values had an upper limit of > 0.98 and a lower limit of > 0.7 at a 95% confidence interval corresponded to strong linkage disequilibrium. The haplotype block was selected as a domain for which the confidence interval between the pairwise SNP exceeded 95%.
2.7. Statistical methods
The descriptive statistical analysis of the background data on quantitative variables was performed using R packages. Genotyping quality check and association analysis of alleles were done using the “Genetics” package of R software [36]. Genotype-phenotype association analyses using logistic regression assuming different genetic models-dominant, codominant, recessive, over dominant, and log-additive were performed using the “SNPassoc” R package [37]. The relationship between the fatty acids composition and SNPs was analyzed and determined through various statistical tools and techniques. One-way ANOVA and the t-test were applied for the fatty acids composition to assess the significant differences between the means of variety groups for each SNP. A variance multi-way analysis was performed to examine the association of the studied SNPs simultaneously with fatty acids composition. Binary logistic regression was used to test the associations of the discovered SNPs with fatty acids composition separately. All analyses were performed using R program. Two-sided P-values < 0.05 were considered as statistically significant. Machine learning analysis was carried out by Bayesian network through two constraint-based machine learning (ML) algorithms (Grow-shrink and Incremental Association) and the Pearson’s correlation conditional independence test. The Bayesian network learned via Constraint-based methods served to generate nodes and arcs. Each generated node was characterized by Gaussian distribution. This analysis aimed to study the interactions between unsaturated fatty acid contents (C18:1, C18:2 and C18:3) and the discovered SNPs on the one hand and between SNPs on the other hand. Machine learning analysis was performed using the “bnlearn” R package.
2.8. Cluster analysis
Two-way hierarchical clustering of the olive oil varieties and the studied parameters (oleic acid (C18:1), linoleic acid (C18:2), linolenic acid (C18:3), and the 15 newly identified SNPs) were performed using the “Heatmap” R package [38]. The phylogenetic analyses based on the 19 FAD2 gene sequences were realized using MEGA v11 (https://www.megasoftware.net/) [39] using the Neighbor-Joining method.
2.9. Bioinformatics analysis
Multiple sequence alignment of FAD2 sequences was performed using ClustalW2 with default parameters [40]. The TMHMM server (http://www.cbs.dtu.dk/services/TMHMM) [41] was used to predict transmembrane regions.
2.10. Three-dimensional protein modeling
Three-dimensional (3D) protein models were predicted and proposed for the four desaturases. Homology modeling was performed using the (PS)2-v2 Protein Structure Prediction Server (http://ps2v2.life.nctu.edu.tw/) [42], [43], where the reported crystal structure of a mammalian stearoyl-CoA desaturase (PDB ID 4YMK) was used as a template (42% identity and 98.7% alignment with the targeted protein sequence). The four low-energy modeled structures were further validated by performing Ramachandran plot analysis (model structure with the maximum number of amino acids in allowed regions) using the PROCHECK suite of programs [44]. The models were viewed using The PyMol Molecular Visualization System (https://pymol.org/2/) [45].
2.11. Protein docking
Protein docking was performed using GRAMM-X online server (http://vakser.compbio.ku.edu/resources/gramm/grammx/) [46]. The Arabidopsis ferredoxin structure (PDB ID 4ZHO) was used as the ligand protein. The structure provided by the NCBI data bank was prepared for docking using Discovery Studio 4.1 Visualizer (https://discover.3ds.com/discovery-studio-visualizer-download) [47]. The 4ZHO molecule was opened using Discovery Studio; after removing water molecules and other bound ligand molecules, the file was saved as a PDB file. The Discovery Studio software was also used to visualize hydrophobic surfaces of predicted models. Finally, Verify3D allowed the validation of the obtained models (http://nihserver.mbi.ucla.edu/Verify_3D/) [48]. Molecular modeling and docking analyses were combined to study the structure–function relationship of the poorly structurally characterized integral membranes of FAD2. The findings allowed us to investigate the role of each substitution within FAD2 gene sequences of four olive varieties.
3. Results and discussion
3.1. FAD2 gene sequencing, SNP discovery, and molecular characterization
To discover a large number of genic SNP markers across the olive tree genome, we sequenced the FAD2 gene from 19 olive oil varieties. All the obtained FAD2 sequences were submitted to NCBI GenBank (accession numbers are provided in Supplementary Table 3). FAD2 sequences of the studied olive varieties revealed intronless genes (IG) and were identified as mono-exonic genes containing full-length ORFs [49]. FAD2 CDS is made of 1152 bases and encodes a protein of 383 amino acids. The multiple sequence alignment of the FAD2 nucleotide sequences showed 15 variations (Table 1), of which only six corresponded to non-synonymous substitutions in the FAD2 protein sequence, namely (Q26H, S82C, H213N, E286G, H309R, and G341W) in all 19 varieties, representing new haplotypes and protein isoforms. The average nucleotide diversity for the FAD2 gene in the coding region was estimated to be the observed genetic diversity (π) = 0.00438 and the expected diversity (θw: per-site Watterson’s theta) = 0.00339. Given that the two values π and θw were different, the nucleotide polymorphism of the FAD2 gene in coding regions does not fit into the neutral and silent mutation hypothesis. Furthermore, the values of Tajima’s D (D = 1.1035), Fu and Li’s D* statistic test (D* = 1.2431), and Fu and Li's F* statistic test (F* = 1.3947) were also not significant (P > 0.1) for this gene; therefore, confirming our previous hypothesis. Among the total SNPs, substitution patterns and rates were estimated according to the Tamura and Nei model [50]. Transition and transversion SNPs were 12.632 and 6.185, respectively. The transition–transversion ratio (ts/tv) was found to be 2.042. Fifteen candidate SNPs located in the FAD2 gene were further statistically validated to determine the genotypic and allelic frequencies, heterozygosity, minor allele frequency (MAF), and Hardy-Weinberg equilibrium (HWE). Table 2 shows seven monomorphic loci (SNP87, SNP140, SNP307, SNP593, SNP755, SNP988 and SNP1083) in olive FAD2 gene. As all individuals have the same form (genotype) at the monomorphic sites, they were excluded from the genetic association analysis, because they do not provide information.
Table 1.
Details of the SNPs detected in the FAD2 gene from the 19 studied olive oil varieties.
| Olive oil varieties / SNP | SNP87 | SNP140 | SNP206 | SNP307 | SNP593 | SNP699 | SNP755 | SNP776 | SNP788 | SNP902 | SNP919 | SNP988 | SNP1083 | SNP1181 | SNP1237 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Besbessi | A | A | T | C | C | C | T | G | C | T | G | G | T | C | G |
| JemriB | A | A | T | C | C | C | T | G | C | T | G | G | T | C | G |
| JemriD | A | A | T | C | C | C | T | G | C | T | G | G | T | C | G |
| Jarboui | A | A | T | C | C | C | T | G | C | T | G | G | G | T | G |
| Zalmati | G | A | T | C | C | C | C | G | G | A | A | A | G | T | G |
| ChemlSfax | A | A | T | C | C | C | C | G | G | A | A | A | G | T | G |
| Meski | A | A | T | C | C | C | C | G | G | A | A | A | G | T | G |
| Arbequina | A | A | T | C | C | C | C | G | G | A | A | G | G | C | G |
| Tounsi | G | T | C | G | T | A | T | A | C | T | G | G | T | C | C |
| Zarrazi | A | T | C | G | T | A | T | A | C | T | G | G | T | C | C |
| Fakhari | A | T | C | G | C | A | T | A | C | T | G | G | T | C | C |
| Oueslati | A | T | C | G | C | A | T | A | C | T | G | G | T | C | C |
| ElHorr | A | T | C | G | C | C | T | A | C | T | G | G | T | C | C |
| Chemcheli | A | T | C | G | C | C | T | A | C | T | G | G | T | C | G |
| ChemlTat | G | T | C | C | C | C | T | A | C | T | G | G | T | C | G |
| SahliMG | G | T | C | C | C | C | T | A | C | T | G | G | T | C | G |
| Toffehi | A | T | T | C | C | C | T | A | C | T | G | G | T | C | G |
| Chetoui | A | T | T | C | C | C | T | A | C | T | G | G | T | C | G |
| Fougi | A | A | T | C | C | C | T | A | C | T | G | G | T | C | G |
Table 2.
Characteristics of the identified SNPs.
| SNPs | Ho | He | HWE (p value) | MAF | alleles |
|---|---|---|---|---|---|
| SNP87 | 0.474 | 0.411 | 1.0000 | 0.289 | A: G |
| SNP140 | 0.526 | 0.499 | 1.0000 | 0.474 | T: A |
| SNP206 | 0.632 | 0.432 | 0.1525 | 0.316 | T: C |
| SNP307 | 0.474 | 0.494 | 1.0000 | 0.447 | G: C |
| SNP593 | 0.421 | 0.432 | 1.0000 | 0.316 | C: T |
| SNP699 | 0.579 | 0.500 | 0.9103 | 0.5 | A: A |
| SNP755 | 0.474 | 0.478 | 1.0000 | 0.395 | T: C |
| SNP776 | 0.368 | 0.5 | 0.4152 | 0.5 | A: A |
| SNP788 | 0.789 | 0.494 | 0.0369 | 0.447 | G: C |
| SNP902 | 0.316 | 0.465 | 0.2971 | 0.368 | T: A |
| SNP919 | 0.368 | 0.478 | 0.5172 | 0.395 | G: A |
| SNP988 | 0.526 | 0.488 | 1.0000 | 0.421 | G: A |
| SNP1083 | 0.526 | 0.488 | 1.0000 | 0.421 | T: G |
| SNP1181 | 0.263 | 0.411 | 0.2491 | 0.289 | C: T |
| SNP1237 | 0.632 | 0.432 | 0.1525 | 0.316 | G: C |
Ho: Observed Heterozygosity; He: Expected Heterozygosity; MAF: Minor allele frequency; HWE: Hardy-Weinberg equilibrium.
3.2. Genotype-phenotype correlation and SNP-SNP interaction
3.2.1. Association statistics for virgin olive oil quality and genetic variants in FAD2 gene
Associations between mono and polyunsaturated fatty acid composition and genotypes of 15 SNPs in the FAD2 gene were investigated. Significant results for each genetic model are summarized in Table 3. By assuming each genetic model, SNPs over the correction threshold line with the lowest Bayesian Information Criteria (BIC), Akaike Information Criteria (AIC), and P values were identified.
Table 3.
Genetic control for 14 SNPs associated with oleic acid (C18:1) content.
| SNP | Model | N | Genotype | Mean | SE | P | AIC | BIC |
|---|---|---|---|---|---|---|---|---|
| SNP87 | Dominant | 9 | AA | 64.66 | 1.94 | 0.3 | 122.9 | 125.7 |
| 10 | AG-GG | |||||||
| SNP140 | Recessive | 15 | T/T-A/T | 67.8 | 1.23 | 0.005 | 115.1 | 117.9 |
| 4 | AA | 59.62 | 1.53 | |||||
| SNP206 | – | 7 | TT | 61.13 | 1.32 | 9e-04 | 111.4 | 114.2 |
| 12 | TC | 68.97 | 1.27 | |||||
| SNP307 | Additive | < 0.0001 | 85.1 | 87.9 | ||||
| SNP593 | Additive | 0.014 | 117.1 | 119.9 | ||||
| SNP699 | Additive | < 0.0001 | 82.5 | 85.4 | ||||
| SNP755 | Recessive | 16 | T/T-T/C | 67.7 | 1.1 | 0.001 | 111.8 | 114.6 |
| 3 | CC | 57.43 | 0.22 | |||||
| SNP776 | Additive | 0.0017 | 112.8 | 115.7 | ||||
| SNP788 | Dominant | 3 | GG | 57.43 | 0.22 | 0.001 | 111.8 | 114.6 |
| 16 | C/G-C/C | 67.7 | 1.1 | |||||
| SNP902 | Additive | 1e-04 | 106.2 | 109.1 | ||||
| SNP919 | Additive | < 0.0001 | 91.8 | 94.6 | ||||
| SNP988 | Additive | < 0.0001 | 90.2 | 93 | ||||
| SNP1083 | Additive | <0.0001 | 99.9 | 102.8 | ||||
| SNP1181 | Additive | 1e-04 | 106.4 | 109.3 |
N: number of olive oil varieties; SE: standard deviation; P: P-value; AIC: Akaike Information Criteria; BIC: Bayesian Information Criteria.
3.2.1.1. Interaction between SNPs of FAD2 gene and C18:1 content
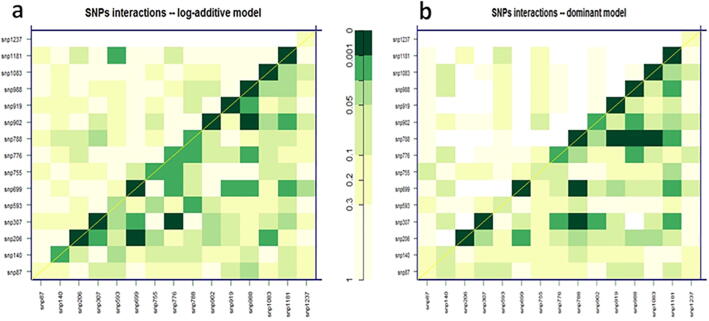
Table 3 shows the mean, standard deviation (SE), difference, and significant values of the first 14 SNPs related to C18:1 for the model harboring the lowest P-value, AIC and BIC. The five following SNPs, SNP307, SNP699, SNP919, SNP988, and SNP1083, displayed the lowest P-value (P < 0.0001) and significant associations under the additive model. In addition, these five SNPs reported significant values under three additional models (i.e., dominant, recessive, and codominant). However, the best fit for genotypic variation of the oleic acid content analyzed for these markers is the additive model based on AIC and BIC values. Fig. 1 displays the interaction analysis plots drawn using all the studied SNP markers with the highest significant or lower significant effects (P < 0.001) on C18:1. Each plot contains the P values obtained from different likelihood ratio tests. As shown by the diagonal in Fig. 1a, a considerable number of SNP markers displayed epistasis with high and moderate significance levels. In fact, not all the interactions exhibited high P values, and the color intensity of some cases on the diagonal is weak. SNP206, SNP307, SNP699, SNP902, SNP919, SNP988, SNP1083 and SNP1181 showed the highest significant interactions. Additionally, another group of SNP pairs composed of SNP206-SNP699, SNP307-SNP776, and SNP902-SNP988, also exhibited significant interaction..
Fig. 1.
Interaction analysis for significantly associated markers with C18:1 (a) and C18:2 (b) rates. Increasing intensity of green color corresponds to an increasing level of interaction between SNPs. The same color indicates similar levels of statistical significance between SNP cases. P-values for the interaction (epistasis) log-likelihood ratio test are found in the upper triangle of the matrix. In contrast, those from the likelihood ratio test (LRT) compare the two-SNP additive likelihood to the best of the single-SNP models are in the lower part. P values from LRT for the crude effect of each SNP are written on the diagonal. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2.1.2. Interaction between SNPs of FAD2 gene and C18:2 content
Table 4 shows the mean, standard deviation (SE), and the difference and significance values of the first 14 SNPs related to C18:2 for the model with the lowest P-value, AIC and BIC. The following five SNPs, SNP307, SNP699, SNP919, SNP988, and SNP1083, had the lowest P-value (P < 0.0001) and significant associations under the additive model. In addition, these five SNPs provided significant values under three additional models (i.e., dominant, recessive, and codominant). However, the best fit for genotypic variation of the linoleic acid content analyzed for these markers is the additive model based on AIC and BIC values. As shown by the diagonal in Fig. 1b, there is a considerable number of SNP markers demonstrating epistasis with high and moderate significance levels. However, not all interactions exhibited high P-values, and the color intensity of some points on the diagonal is low. The SNP206, SNP307, SNP699, SNP 788, SNP919, SNP988, SNP1083 and SNP1181 showed the highest significant interactions. Additionally, another group of SNP pairs composed of SNP307-SNP788, SNP307-SNP788, SNP919-SNP788, SNP988-SNP788, and SNP1083-SNP788, also exhibited significant interaction.
Table 4.
Genetic control for 14 SNPs associated with linoleic acid (C18:2) level.
| Marker | Model | N | Genotype | Mean | SE | P | AIC | BIC |
|---|---|---|---|---|---|---|---|---|
| SNP87 | Additive | 0.18 | 108.4 | 111.2 | ||||
| SNP140 | Recessive | 15 | T/T-A/T | 12.91 | 1.01 | 0.094 | 107.3 | 110.1 |
| 4 | AA | 16.57 | 1.06 | |||||
| SNP206 | – | |||||||
| SNP307 | Additive | 1e-04 | 93.4 | 96.2 | ||||
| SNP593 | Recessive | 17 | C/C–C/T | 14.22 | 0.9 | 0.077 | 106.9 | 109.7 |
| 2 | TT | 9.1 | 1.9 | |||||
| SNP699 | Additive | 6e-04 | 96.8 | 99.7 | ||||
| SNP755 | Additive | 0.0042 | 101.1 | 103.9 | ||||
| SNP776 | Additive | 0.044 | 105.8 | 108.6 | ||||
| SNP788 | Dominant | 3 | GG | 19.1 | 1.07 | 0.0046 | 101.2 | 104.1 |
| 16 | C/G-C/C | 12.66 | 0.82 | |||||
| SNP902 | Recessive | 15 | T/T-T/A | 12.44 | 0.84 | 0.0035 | 100.7 | 103.5 |
| 4 | AA | 18.32 | 1.08 | |||||
| SNP919 | Additive | 5e-04 | 96.6 | 99.5 | ||||
| SNP988 | Additive | 1e-04 | 92.9 | 95.7 | ||||
| SNP1083 | Additive | 0.0024 | 99.9 | 102.8 | ||||
| SNP1181 | Additive | 4e-04 | 96.3 | 99.1 |
N: number of olive oil varieties; SE: standard deviation; P: P-value; AIC: Akaike Information Criteria; BIC: Bayesian Information Criteria.
3.2.1.3. Interaction among SNPs of the FAD2 gene and both C18:1 and C18:2 fatty acid levels
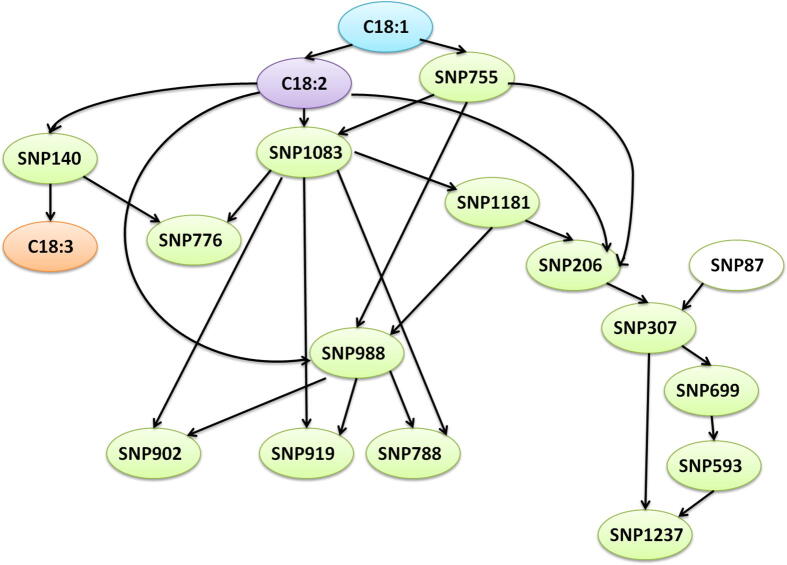
Correlations between the levels of two fatty acids (C18:1 and C18:2) and the discovered SNPs were studied by the Bayesian network (BN) through two constraint-based machine learning (ML) algorithms (Grow-shrink and Incremental Association) and the Pearson’s correlation conditional independence test. The Grow-shrink machine learning algorithm based on the Grow-Shrink (GS) Markov Blanket detection algorithm [51] used a structure learning algorithm. The Incremental Association is based on the Incremental Association Markov Blanket (IAMB) algorithm [52], which is based on a two-phase selection scheme (a forward selection followed by an attempt to remove false positives). These two algorithms learn the network structure by analyzing the probabilistic relations entailed by the Markov property of Bayesian networks with conditional independence tests and then constructing a graph that satisfies the corresponding d-separation statements. The resulting models are often interpreted as causal models even when learned from observational data. The output of this analysis gives graphical models where nodes represent random variables and arrows represent probabilistic dependencies between them. The BN is a directed acyclic graph (DAG). The latter defines a factorization of the joint probability distribution of nodes (variables: in our cases SNPs and C18:1, C18:2, and C18:3) called a global probability distribution. The form of the factorization is given by the Markov property of Bayesian networks.
As shown in Fig. 2, 18 nodes and 27 directed arcs were generated by the analysis. Highly significant associations were obtained between C18:1 and the SNPs (Table 5): SNP140 (r = 0.812, p < 0.001), SNP206 (r = 0.803, p < 0.001), SNP307 (r = 0.772, p < 0.001), SNP593 (r = −0.546, p = 0.016), SNP699 (r = −0.736, p < 0.001), SNP755 (r = −0.763, p < 0.001), SNP776 (r = −0.802, p < 0.001), SNP788 (r = −0.763, p < 0.001), SNP902 (r = 0.763, p < 0.001), SNP919 (r = 0.763, p < 0.001), SNP988 (r = 0.691, p = 0.001), SNP1083 (r = −0.772, p < 0.001), SNP1181 (r = 0.689, p = 0.001) and SNP1237 (r = −0.629, p = 0.004). Moreover, the C18:2 fatty acid levels showed highly significant associations with the identified SNPs (Table 5): SNP140 (r = −0.674, p = 0.002), SNP206 (r = −0.804, p < 0.001), SNP307 (r = −0.634, p = 0.004), SNP699 (r = 0.506, p = 0.027), SNP755 (r = 0.634, p = 0.004), SNP776 (r = 0.701, p = 0.001), SNP788 (r = 0.634, p = 0.004), SNP902 (r = −0.634, p = 0.004), SNP919 (r = −0.634, p = 0.004), SNP988 (r = −0.620, p = 0.005), SNP1083 (r = 0.784, p < 0.001), SNP1181 (r = −0.767, p < 0.001) and SNP1237 (r = 0.564, p = 0.012). On the other hand, we found a negative, highly significant association between C18:1 and C18:2 (r = −0.837, p < 0.001). There was no interaction between SNP87 and the fatty acid or the other novel identified SNPs. However, we confirmed correlations between SNP307-SNP699 (r = −0.760, p < 0.001), SNP919-SNP988 (r = 0.839, p < 0.001), SNP919-SNP1083 (r = −0.864, p < 0.001), SNP1083-SNP1181 (r = −0.864, p < 0.001), SNP1083-SNP755 (r = 0.864, p < 0.001) and SNP776-SNP140 (r = −0.899, p < 0.001).
Fig. 2.
Machine learning analysis carried out by Bayesian network based on Grow-shrink learning algorithm and the Pearson’s correlation conditional independence test. The analysis generated 18 nodes and 27 directed arcs. C18:1, C18:2, and C18:3 nodes are indicated by three different colors. All SNP nodes are colored green, except SNP87 in white for not being correlated with the oleic (C18:1) and linoleic (C18:2) fatty acid levels. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 5.
Pearson’s correlation coefficients and p-values of the association results between C18:1, C18:2, and the discovered SNPs.
| SNP | C18:1 |
C18:2 |
||
|---|---|---|---|---|
| r | p | r | p | |
| SNP87 | 0.100 | 0.684 | −0.349 | 0.144 |
| SNP140 | 0.812 | < 0.001 | −0.674 | 0.002 |
| SNP206 | 0.803 | < 0.001 | −0.804 | < 0.001 |
| SNP307 | 0.772 | < 0.001 | −0.634 | 0.004 |
| SNP593 | − 0.546 | 0.016 | 0.415 | 0.077 |
| SNP699 | −0.736 | < 0.001 | 0.506 | 0.027 |
| SNP755 | −0.763 | < 0.001 | 0.634 | 0.004 |
| SNP776 | −0.802 | < 0.001 | 0.701 | 0.001 |
| SNP788 | −0.763 | < 0.001 | 0.634 | 0.004 |
| SNP902 | 0.763 | < 0.001 | −0.634 | 0.004 |
| SNP919 | 0.763 | < 0.001 | −0.634 | 0.004 |
| SNP988 | 0.691 | 0.001 | −0.620 | 0.005 |
| SNP1083 | −0.772 | < 0.001 | 0.784 | < 0.001 |
| SNP1181 | 0.689 | 0.001 | −0.767 | < 0.001 |
| SNP1237 | −0.629 | 0.004 | 0.564 | 0.012 |
r: Pearson’s correlation coefficient; p: p-value.
These two ML algorithms allowed a deeper description of the genotypes (SNPs) and phenotypes (unsaturated fatty acids, particularly oleic and linoleic) interaction. In fact, the combination of these two algorithms allowed us to detect interactions between all SNPs and C18:1 and C18:2 contents. Compared to other conventional statistical methods, ML algorithms have had a spectacular success in specific plant application areas such as plant genetics and plant breeding. The ML algorithms can perform genotype-phenotype prediction analyses to better understand the link between phenotypic variation and corresponding variation at the genotype level in a more efficient and faster way. The eventual applications of these approaches are to anticipate genomic regions that can be modified to produce the desired phenotype (olive oil varieties with high C18:1 level). However, effective high-throughput genotyping systems and deep biometrics ML methods can provide researchers and breeders with reliable tools to evaluate these choices to genetic selection in breeding programs. As a result, ML research and development may contribute to solving olive oil quality and food security problems within constantly changing settings such as the era of the worrisome climate and environmental change.
Currently, researchers and breeders take advantage of the reliable ML models to support decision-making with rigorous ML algorithms in a range of areas in plant research, which are tailored to consider prior knowledge on the problem to be solved [53]. Indeed, an ensemble machine learning model was developed for Alfalfa yield prediction [54], for predicting the performance of wastewater treatment plants [55], and for modeling and optimizing in plant tissue culture [56], [57]. Other ML algorithms were also used, such as Multilayer perceptron (MLP) being applied for drought tolerance prediction [58], Support vector machine (SVM) for gene selection in cancer classification [59], and plant cell and tissue culture [57] area. In the recent years and near future, the deep learning (DL) method, which is a type of machine learning (ML) approach that is a subfield of artificial intelligence (AI), will provide powerful tools for researchers, allowing interpretation of their decisions in the plant genetics field, especially models prediction and biochemical data interpretation to improve our understanding of plant biology in rapidly changing environments. In fact, due to its ability to combine different kinds of inputs and exploit all the available collected data, DL prediction models could become an efficient method for synthetic biology by learning to automatically generate new DNA sequences and new proteins with desirable properties and also to select the best candidate individuals in breeding programs.
3.2.2. SNP-SNP interaction and FAD2 haplotype analysis
The nucleotide and deduced amino acid sequences of this desaturase gene in the 19 olive varieties were reported earlier. Among the 19 varieties, 15 variable sites of FAD2 polymorphism were found, which resulted in 14 haplotypes with a haplotype diversity (Hd) value of 0.965. Indeed, most varieties had their distinct haplotypes, except the haplotypes 1, 4, 12, and 13 that were shared by three (Besbessi, JemriD, JemriB), two (Chemlali Sfax and Meski), two (Chemlali Tataouine and Sahli mgargeb), and two (Toffehi and Chetoui) varieties, respectively. As mentioned above, five polymorphic SNPs (SNP307, SNP699, SNP919, SNP988, and SNP1083) were found and revealed a highly significant difference in the level of the unsaturated fatty acids C18:1 and C18:2. We studied the important genetic function by analyzing the linkage disequilibrium (LD). To perform haplotype analysis for polymorphisms in the FAD2 gene, we used the Haploview software that provided a single haplotype block including the five SNPs (Supplementary Fig. 1) and having an average Lewontin’s D’ = 0.88. The interaction between the five SNPs generated eight different haplotypes (Supplementary Table 4). On the other hand and according to Gabriel et al. [35], the lower limit of a 95% confidence interval exceeded 0.7, and the status of linkage disequilibrium was reached. Fig. 3 shows only three SNPs (SNP 307, SNP 699 and SNP 919) with a strong degree of linkage disequilibrium, and forming a single block with an average of Lewontin’s D′ = 1.0 (D′ = 1.0 value demonstrates the strongest linkage disequilibrium between two given polymorphisms). Hence, these three SNPs located in the FAD2 gene have direct linkage disequilibrium and indicate the involvement of a multitude of variations rather than a single SNP.
Fig. 3.
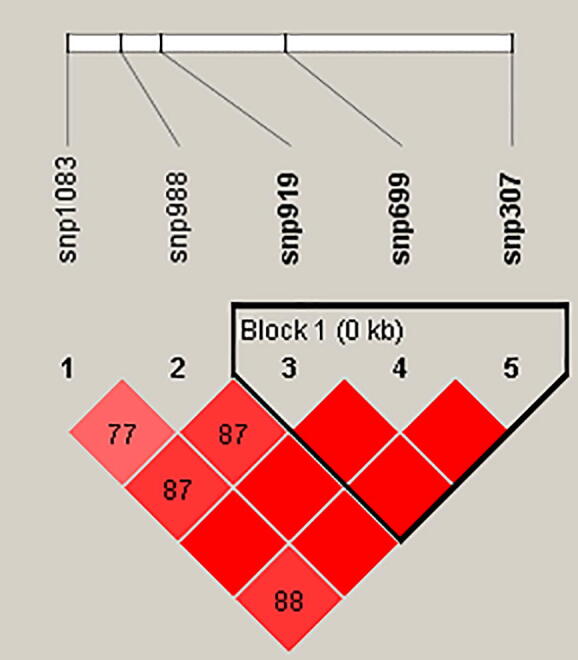
SNP within the FAD2 gene and linkage disequilibrium among the three SNP pairs in FAD2. The color code on the Haploview plot follow the standard color scheme: red (|D’|<1, |D’|=1, LOD ≥ 2). Numbers in cells are D’ value. However, the D’ values of 1.0 are not shown (empty). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The distributions of the four haplotypes and frequencies are presented in Table 6, where the four haplotypes demonstrated a significant difference in the levels of C18:1 and C18:2. For which, the Gsnp307- Asnp699-Gsnp919 haplotype (frequency = 0.5) was significantly associated with a high C18:1 and low C18:2 levels, corresponding to Oueslati olive variety; whereas the Csnp307- Csnp699-Asnp919 haplotype (frequency = 0.395) corresponds to Chemlali Sfax olive variety. The other two haplotypes (Gsnp307- Csnp699-Gsnp919 and Csnp307- Csnp699-Gsnp919) (frequency = 0.053) concerned the olive varieties, which have a fatty acid profile with medium C18:1 and C18:2 contents such as Chetoui variety. The finding of our results confirmed that the different haplotypes had direct effects on C18:1 and C18:2 levels. When the Gsnp307- Asnp699-Gsnp919 haplotype was changed to the Csnp307- Csnp699-Asnp919 type in the FAD2 gene, the proportion of the monounsaturated fatty acid C18:1 was decreased; whereas the proportion of the polyunsaturated fatty acid C18:2 increased. As shown in this study, the content of the oleic unsaturated fatty acid C18:1 appears higher when we obtained the functions of the SNPs located on the FAD2 gene in olive oil as follow: snp307 G > C, snp699 A > C and snp919 G > A.
Table 6.
Haplotype frequencies based on the interaction between five SNPs located in the FAD2 gene.
| Haplotypes Block 1 |
Haplotype frequency |
Phenotype |
Variety Model |
|
|---|---|---|---|---|
| C18:1 | C18:2 | |||
| Gsnp307- Asnp699-Gsnp919 | 0.5 | 74.5 | 10.7 | Oueslati |
| Csnp307- Csnp699-Asnp919 | 0.395 | 57 | 19 | Chemlali Sfax |
| Gsnp307- Csnp699-Gsnp919 | 0.053 | 68.2 | 11.9 | Chemcheli |
| Csnp307- Csnp699-Gsnp919 | 0.053 | 66.5 | 17.5 | Chetoui |
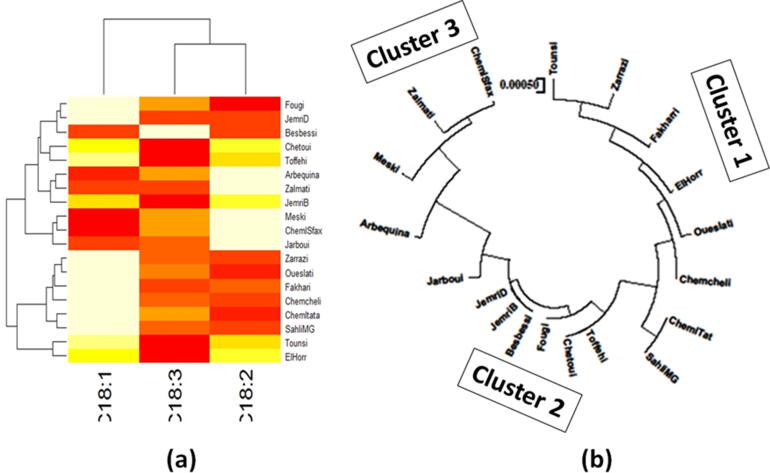
3.3. Two-way hierarchical cluster analysis
Two-way hierarchical cluster analysis was performed to assess the relationship between the olive oil FAD2 sequences and the studied C18:1, C18:2, and C18:3 fatty acid compositions to identify which of these parameters are the most important (Fig. 4a). The first dimension allowed classifying olive oil varieties according to the various studied fatty acid parameters. Three clusters were defined, where the first presented olive oil varieties with high C18:1 and low C18:2 levels, which are the Fougi, Jemri D, Zarrazi, Oueslati, Fakhari, Chemcheli, Chemlali Tataouine, and Sahli mgargeb. The second cluster contains olive oil varieties with medium C18:1 and C18:2 such as, Chetoui, Toffehi, JemriB, Tounsi, and Elhorr. The third cluster represents olive oil varieties with lower C18:1 and high C18:2 levels like Besbessi, Arbequina, Zalmati, Meski, Chemlali Sfax, and Jarboui. This classification of the studied olive oil varieties into three clusters was confirmed by the phylogenetic tree generated by the Neighbor-Joining (NJ) method (Fig. 4b). Based on this cluster analysis, we selected four olive oil varieties: Oueslati and Chemcheli, representing the first cluster, and Chetoui and Chemlali Sfax as a representative of clusters 2 and 3, respectively.
Fig. 4.
(a) Two-way hierarchical clustering of olive oil varieties and the various traits (oleic acid content, linoleic acid content, linolenic acid content, and the identified SNPs. Colors show the proportion of the bands present at every designated band location. Color indicates relative concentration: white, increased; red, decreased; from red to white: gradually increasing Concentration. (b) phylogenetic tree based on Neighbor Joining (NJ) method. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Structure prediction and analysis of desaturase in four olive varieties
3.4.1. Sequence alignment
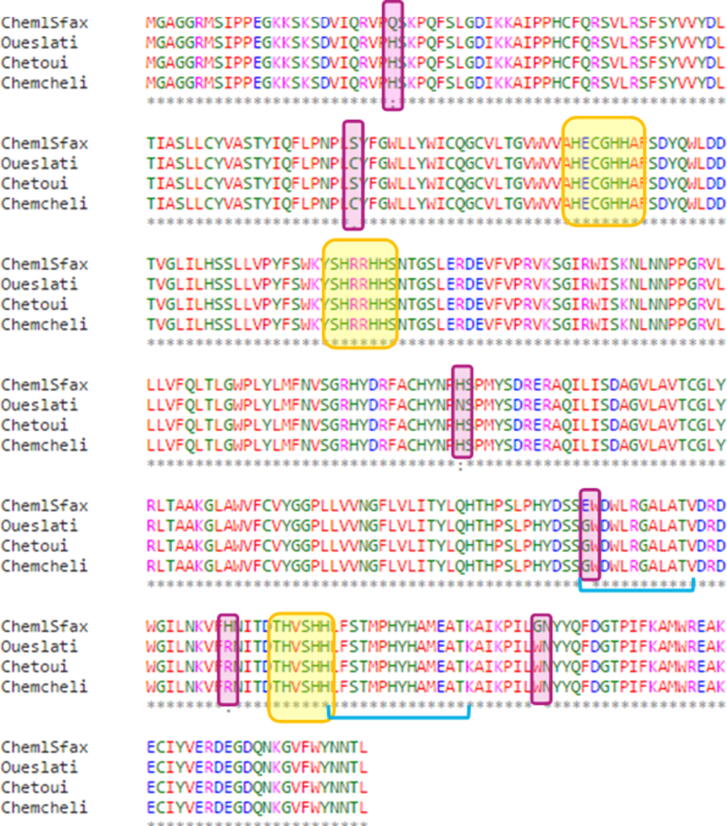
The 3D structure prediction and modeling of the FAD2 proteins of these four olive oil varieties was performed. Alignment was performed to check differences among the four chosen sequences and to identify and localize conserved regions with functional implications. The results of alignment show six positions of changed amino acids (aa) as follows: Q26H, S82C, H213N, E286G, H309R and G341W (Fig. 5). In previous alignment studies of plants, FAD2 showed several conserved motifs in FAD2 sequences [60]. Among others, three conserved histidine motifs are demonstrated to be directly involved in the enzyme activity. In our case, and as shown in Fig. 5, the three histidine boxes are conserved, and none of the changed residues in the four olive sequences was located in these boxes. However, the authors described two strongly conserved sequences among Δ-12 oleate desaturases, EWDWLRGALAT (position 286) and LFSTMPHYHAMEAT (position 320) located before and after the third histidine box, respectively. Interestingly, the substitution E286G was found in the first conserved motif and is suspected of having an important role in enzyme activity or interaction.
Fig. 5.
Multiple sequence alignment of FAD2 sequences showing the catalytic histidine boxes (yellow), the differences between the four sequences (purple), and the highly conserved region underlined in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4.2. Structure prediction
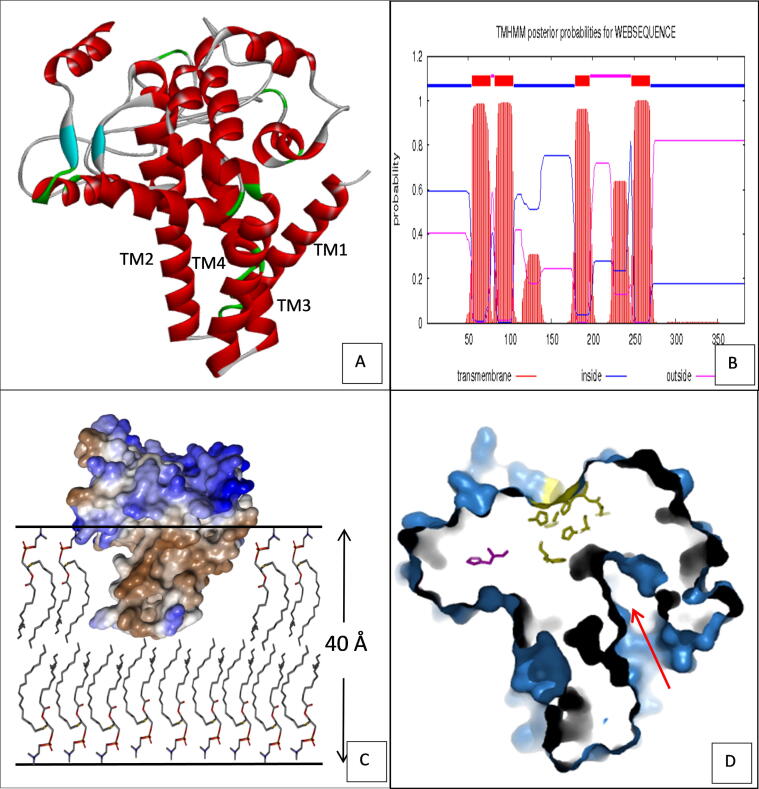
The 3D structures of FAD2 proteins were predicted by homology modeling, based on the 42% sequence identity with a mammalian stearoyl-CoA desaturase (PDB ID 4YMK). The quality of the produced models was assessed by Ramchandran plot analysis. The latter shows 97.2% of the residues located in the allowed Φ and Ψ dihedral angles, while only 2.8% of the residues were outliers. All the FAD2 generated models consisted of 13 α-helices, among which, four longer α-helices (TM1, TM2, TM3 and TM4) with 29, 23, 21 and 20 residues, respectively were present, which together displayed a conical shape (Fig. 6A). These helices are most likely organized in transmembrane domains, since they match with the large red peaks obtained by the TMHMM server (Fig. 6B). To confirm this observation, a hydrophobicity surface representation was generated. As shown in Fig. 6C, the four transmembrane helices are highly hydrophobic (shown in brown). At the same time, the rest of the protein is polar and solvent accessible, proving its cytosolic side location in the plasma membrane. Considering that the membrane thickness is about 40 Å [61] and that the length of the transmembrane moiety is about 20 Å, the protein could be anchored in only one layer of the membrane and considered as a peripheral protein (Fig. 6C).
Fig. 6.
(A) 3D protein models of FAD2 (helix regions are shown in red and β-sheet in blue). (B) Predicted transmembrane domains in FAD2 by TMHMM server (CBS; Denmark). Red color boxes represent the putative transmembrane domains, blue and pink color lines indicate the inner and outer membranes of the cell respectively. (C) Surface representation of FAD2 model anchored in the lipid bilayer (brown color indicated hydrophobic surface while blue color showed solvent accessible surface). (D) Slabbed surface representation of FAD2 in the same orientation as in (C) showing the substrate pocket (red arrow) and the catalytic histidine in yellow color. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The models also involved a C-terminus anti-parallel β-sheet located at the top of the solvent accessible domain. It should be noted that, the generated models are truncated in the N and C terminus region (a total of 78 residues). Besides, these regions are also truncated in the template structure. The predicted structure showed the presence of the three conserved Histidine-boxes, namely H1 (H105, H109 and H110), H2 (H141, H144 and H145) and H3 (H315, H318 and H319). These boxes are located almost at the centre of the cytosolic moiety in contact with the catalytic pocket and are involved in the reaction mechanism by maintaining two metal ions implicated in electron transfer during desaturation [62]. According to the obtained models, FAD2 could be part of a distinct integral membrane proteins (IMP) class since it cannot strictly be placed into either the αIMP or βIMP classes [63]. In fact, FAD2 acts on the lipophilic substrates and has a soluble homolog (Acyl-ACP desaturase). Secondly, as shown in Fig. 6D, the active site cavity pointed directly to the bilayer, implying that the substrate channel is buried in the edge of the transmembrane domain and could access its substrates by direct recruitment from the bilayer. The FAD2 reaction product could also be released in the membrane by dint of the intimate relationship between its membrane-binding domain and active site.
3.4.3. Interpretation of structural differences
As explained above, the sequence alignment of the four varieties showed six substitutions. Nevertheless, in structural analysis, only five could be studied since the first one (Q26H) is situated in the truncated N terminal region. To determine the possible implications of these substitutions in different fatty acid profiles in terms of C18:1 and C18:2 levels, the generated models were analyzed, and no drastic structural changes were observed when the four FAD2 models were superimposed. Nevertheless, careful inspection of the changed amino acid positions showed that four of them are placed in the cytosolic domain, and only one is in the tip of the transmembrane domain. Interestingly, no substitutions were observed near the catalytic cavity. Docking experiments with plant-type ferredoxin protein were conducted to find a plausible explanation of the changes that occurred in the cytosolic domain, since it was demonstrated that in plant FAD2, the electrons needed for the desaturation reaction are obtained from ferredoxin protein [64].
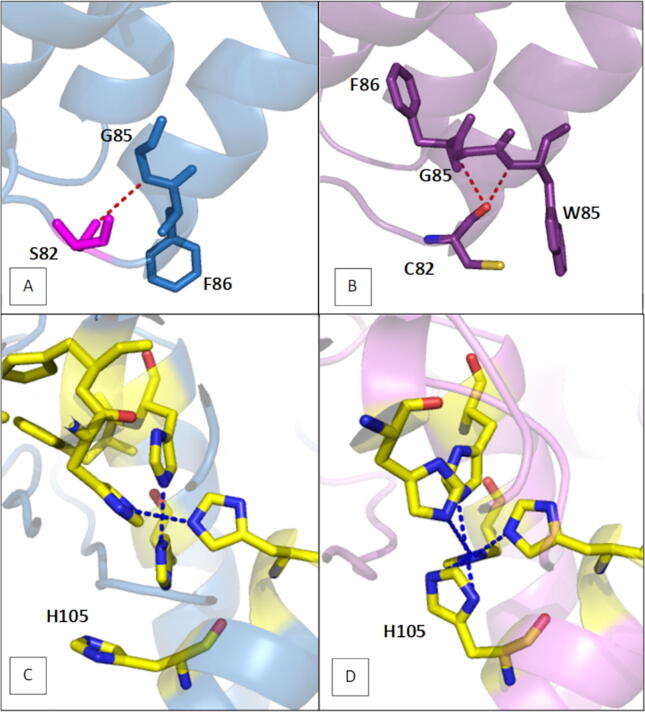
3.4.4. S82C substitution
As shown in Fig. 7A, the S82C substitution is located at the end of the hydrophobic cone and, more precisely in the turn separating TM1 and TM2 helices. Interestingly, on the cytosolic side, TM2 protrudes a helical turn out of the membrane and provides three histidine (H1 box) coordinating the dimetal active site. Polar contact analysis showed that due to its radical OH, S82 established one hydrogen bond with G85 (Fig. 7A), while C82 established two hydrogen bonds with W86 and G85 (Fig. 7B). Furthermore, a rotation of the TM2 helix was observed when comparing models of the two different varieties (S82-models and C82-models). In fact, in variety models displaying a serine residue, the F86 points to the membrane layer (Fig. 7A), while in variety models with a cysteine residue, the F86 points to the interior cone (Fig. 7B). The consequences of this rotation in the H1 histidine conformation were notable. Remarkably, H105 in S82-models was not involved in the first metal coordination (Fig. 7C), while in C82-models, it participates directly in maintaining the ion for the catalytic reaction (Fig. 7D). It has been demonstrated that weakly bound metals lead to accelerating the turnover reaction of the enzyme [65], [66]. Accordingly, S82-FAD2 would be more effective than C82-FAD2 in desaturating C18:1. This result is in accordance with the fatty acid profile of each variety, where Chemcheli and Oueslati varieties (C82-FAD2) displayed lower linoleic acid level than Chemleli and Chetoui varieties (S82-FAD2).
Fig. 7.
(A, B) Close up view of position 82 in S82-model and C82-model respectively, (C, D) Close up view of catalytic histidine involved in one metal binding in S82-model and C82-model respectively. Residues are shown as sticks, H-bonds are shown in red dotted lines, and metal coordinates are represented in blue dotted lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4.5. H213N substitution
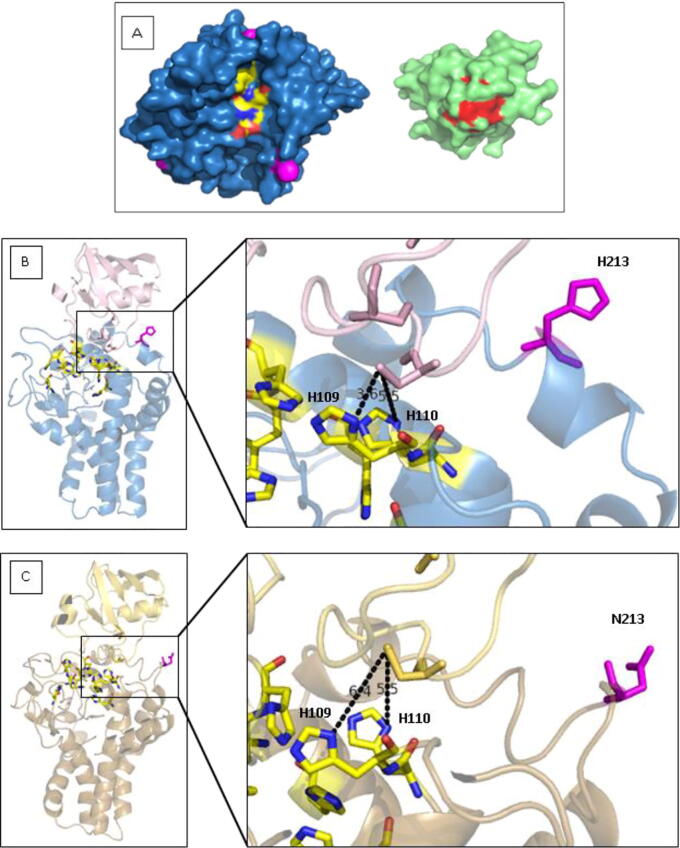
Determination of the H213N effect was of great interest as N213 was found only in the Oueslati variety, which was the most interesting in terms of fatty acid composition. The model analysis showed that the presence of H213 allows the formation of a short helix at the protein surface, while in the case of N213 models, the helix was replaced by a large loop, which means a reduction of the rigidity and stability in this region. Examination of the FAD2 structure shows that the dimetal centre could be accessible from the cytoplasm via a groove at the top of the solvent-accessible domain (Fig. 8A). In the docked models with ferredoxin, the active centre of the latter lies along this groove allowing the placement of the electron donor group within 10 Å from the nearest catalytic histidine centre. This distance is acceptable for electron transfer between biological redox centres [67], [68]. As shown in Fig. 8B, the helix comprising H213 is directly implicated in the surface interaction with ferredoxin and especially with the loop carrying the [2Fe-2S] cluster. In fact, H213 was suspected of playing an important role in the correct positioning and orientation of ferredoxin so that the [2Fe-2S] active centre, coordinated by the S side chains of four highly conserved cysteine residues, will be separated by a short distance to the dimetal centre of the receptor enzyme and could easily release electrons. As opposite, the presence of N213 could hamper the correct placement of ferredoxin and consequently reduce the catalytic activity. The differences in ferredoxin positioning between Ouselati and Chemlali docked models was determined in terms of distances between the lowest cysteine coordinating the [2Fe-2S] ferredoxin active centre and the two nearest catalytic histidine from the FAD2 molecule, which are implicated in the electron transfer and are candidates to form the first electron transfer interface (H109 and H110 from H1 box). In the case of the Chemlali variety model, the distances C39-H109/C39-H110 were respectively 3.6 Å and 5.5 Å (Fig. 8B), whereas, for the Oueslati model, these distances were 6.4 Å and 5.5 Å, respectively (Fig. 8C). Therefore, the electron transfer would be reduced and catalytic activity as well for Oueslati FAD2.
Fig. 8.
(A) Surface presentation of plant-type ferredoxin (left) and FAD2 (right) oriented with their proposed interaction surfaces towards the viewer. (B) Proposed model of the complex between ferredoxin and Chemlali Sfax variety FAD2 (Ferredoxin is colored in pale pink and FAD2 in blue). (C) Docked model of ferredoxin with Oueslati variety FAD2 (Ferredoxin is colored in pale yellow and FAD2 in light brown). The inset shows a closer view of the proposed interaction region between the Histidine boxes shown as yellow sticks and the last Cysteine residue in the ferredoxin protein. Substituted residues are shown as magenta sticks, and distances are shown in black dotted lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4.6. E286G, H309R and G341W substitution
These three changes were analyzed together since all of them exist as E, H, and G in Chemlali Sfax variety and as G, R, and W, respectively in the other three varieties (Oueslati, Chetoui, and Chemcheli). It should be noted that Chemlali Sfax exhibited the highest level of C18:2. Firstly, E286G is a very interesting substitution since it is the first residue in a highly conserved sequence in plant FAD2. This sequence corresponds to a long loop relating to TM4 and a short amphipathic helix (namely AH7) located at the solvent accessible domain in a horizontal conformation and was described to provide interactions between the cytosolic domain and the lipid bilayer [62]. The loop involving E286 seems to be very close to the cytosolic domain in the Chemlali Sfax variety model. Polar contact analysis shows that E286 was not implicated in H-bond formation. However, G286 in all other models was implicated in hydrogen contact, the loop is so much larger, and it seems to be very flexible out of the cytosolic domain. The inspection of the ferredoxin contact surface in G286 models showed that the big loop is directly involved in molecular contact with ferredoxin residues. In contrast, in the E286 model, the loop was retracted, likely hiding the H3 histidine box. A previous study on the ferredoxin mechanism demonstrated that once the electrons are released, the enzyme is broken down from the binding site of the enzyme to begin the catalytic cycle.
For this reason, the interaction surface between ferredoxin and its receptor enzymes cannot be too complimentary. In case of an affinity increase between the two proteins, the activity of the enzyme would be strongly reduced [64]. In our case, less interaction with ferredoxin leads to increasing the catalytic activity of FAD2, suggesting more linoleic acid production. Accordingly, the Chemlali Sfax variety would have the highest amount of C18:2, which is in agreement with fatty acid profile presented in Supplementary Table 4. On the other hand, the inspection of H309R and G341W substitutions allowed us to make conclusions concerning the flexibility of the third histidine box (H3) helix. The latter is located between these two changes. In fact, G341 could provide some suppleness to the helix involving H3, while H309 could be a part of the second coordination shell around the dimetal centre. H309 residue in olive FAD2 sequence corresponds to E291 in mouse stearoyl-CoA desaturase, which was demonstrated to play a role in the enzyme reaction mechanism by coordinating a catalytic histidine residue [62].
4. Conclusions
The FAD2 gene from 19 Tunisian olive oil varieties was sequenced to identify SNPs related to oleic/linoleic acid levels. Fifteen polymorphic variations in FAD2 were identified among the 19 olive oil varieties. Five SNPs were found to be significant, among which eight different haplotypes could be distinguished through interaction studies. Through in-depth analysis, three SNPs showed a strong degree of linkage disequilibrium, with four haplotypes exhibiting high oleic acid level and low linoleic acid level in Oueslati and Chemlali Sfax varieties, and medium levels of both oleic and linoleic acids in Chetoui. Molecular modeling and docking analyses were combined to study the structure–function relationship of the poorly structurally characterized integral membrane protein FAD2. The findings allowed us to investigate the role of each substitution among the FAD2 sequences of the four olive varieties. The results showed the vital role of these genetic substitutions in reducing or enhancing the FAD2 activity to obtain more or less oleic acid content, respectively. The study also highlighted the importance of S82C and H213N substitutions that make the Oueslati variety more promising in terms of fatty acid profile and oleic acid content.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Authors are thankful to Researchers Supporting Project Number RSP/2021/45, King Saud University Riyadh, Saudi Arabia for financial support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.02.028.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Agricultural and Food Biotechnology of Olea europaea and Stone Fruits | Innocenzo Muzzalupo | download n.d. https://b-ok.africa/book/2484983/604968 (accessed January 12, 2021).
- 2.International Year of Fruits and Vegetables. Food Agric Organ U N n.d. http://www.fao.org/fruits-vegetables-2021/en/ (accessed January 19, 2021).
- 3.FAO. Fruit and vegetables – your dietary essentials: The International Year of Fruits and Vegetables, 2021, background paper. Rome, Italy: FAO; 2020. doi:10.4060/cb2395en Also Available in:Spanish.
- 4.Antonialli F., Mesquita D.L., Valadares G.C., de Rezende D.C., de Oliveira A.F. Olive oil consumption: a preliminary study on Brazilian consumers. Br Food J. 2018;120:1412–1429. doi: 10.1108/BFJ-03-2017-0166. [DOI] [Google Scholar]
- 5.FAOSTAT n.d. http://www.fao.org/faostat/fr/#data/QD (accessed January 27, 2021).
- 6.Olive Oil Market Research Report: Market size, Industry outlook, Market Forecast, Demand Analysis, Market Share, Market Report 2020-2025 n.d. https://www.industryarc.com/Report/16677/olive-oil-market.html (accessed January 22, 2021).
- 7.Price analysis of extra virgin olive oil | Emerald Insight n.d. https://www.emerald.com/insight/content/doi/10.1108/BFJ-03-2019-0186/full/html (accessed January 27, 2021).
- 8.Torres-Ruiz F.J., Marano-Marcolini C., Lopez-Zafra E. In search of a consumer-focused food classification system. An experimental heuristic approach to differentiate degrees of quality. Food Res Int. 2018;108:440–454. doi: 10.1016/j.foodres.2018.03.067. [DOI] [PubMed] [Google Scholar]
- 9.Chousou C., Tsakiridou E., Mattas K. Valuing consumer perceptions of olive oil authenticity. J Int Food Agribus Mark. 2018;30:1–16. doi: 10.1080/08974438.2017.1382418. [DOI] [Google Scholar]
- 10.IOC STANDARDS, METHODS AND GUIDES. Int Olive Counc n.d. https://www.internationaloliveoil.org/what-we-do/chemistry-standardisation-unit/standards-and-methods/ (accessed January 27, 2021).
- 11.Aparicio-Ruiz R., Harwood J., editors. Handbook of Olive Oil: Analysis and Properties. 2nd ed. Springer US; 2013. [DOI] [Google Scholar]
- 12.Conde C., Delrot S., Gerós H. Physiological, biochemical and molecular changes occurring during olive development and ripening. J Plant Physiol. 2008;165:1545–1562. doi: 10.1016/j.jplph.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 13.Ben Ayed R., Ennouri K., Ben-Hlima H., Slim S., Hanana M., Mzid R., et al. Identification and characterization of single nucleotide polymorphism markers in FADS2 gene associated with olive oil fatty acids composition. Lipids Health Dis. 2017;16 doi: 10.1186/s12944-017-0530-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hernández M.L., Sicardo M.D., Belaj A., Martínez-Rivas J.M. The Oleic/Linoleic Acid Ratio in Olive (Olea europaea L.) Fruit Mesocarp Is Mainly Controlled by OeFAD2-2 and OeFAD2-5 genes together with the different specificity of extraplastidial acyltransferase enzymes. Front Plant Sci. 2021;12:345. doi: 10.3389/fpls.2021.653997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janick J. John Wiley & Sons; 2008. Plant breeding reviews. [Google Scholar]
- 16.Jarvis B.A., Romsdahl T.B., McGinn M.G., Nazarenus T.J., Cahoon E.B., Chapman K.D., et al. CRISPR/Cas9-Induced fad2 and rod1 Mutations Stacked With fae1 Confer High Oleic Acid Seed Oil in Pennycress (Thlaspi arvense L.) Front Plant Sci. 2021:12. doi: 10.3389/fpls.2021.652319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lakhssassi N., Zhou Z., Liu S., Colantonio V., AbuGhazaleh A., Meksem K. Characterization of the FAD2 Gene Family in Soybean Reveals the Limitations of Gel-Based TILLING in Genes with High Copy Number. Front Plant Sci. 2017:8. doi: 10.3389/fpls.2017.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CRISPR-Cas9 mediated targeted disruption of FAD2–2 microsomal omega-6 desaturase in soybean (Glycine max.L) | BMC Biotechnology | Full Text n.d. https://bmcbiotechnol.biomedcentral.com/articles/10.1186/s12896-019-0501-2 (accessed February 24, 2021).
- 19.Cahoon E.B., Schmid K.M. Adv. Plant Biochem. Mol. Biol. Pergamon; 2008. Metabolic Engineering of the Content and Fatty Acid Composition of Vegetable Oils; pp. 161–200. [DOI] [Google Scholar]
- 20.Lee M.W., Padilla C.S., Gupta C., Galla A., Pereira A., Li J., et al. The FATTY ACID DESATURASE2 family in tomato contributes to primary metabolism and stress responses. Plant Physiol. 2020;182:1083–1099. doi: 10.1104/pp.19.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He M., Qin C.-X., Wang X., Ding N.-Z. Plant unsaturated fatty acids: biosynthesis and regulation. Front Plant Sci. 2020:11. doi: 10.3389/fpls.2020.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luisa Hernández M., Dolores Sicardo M., Arjona P.M., Martínez-Rivas J.M. Specialized Functions of Olive FAD2 gene family members related to fruit development and the abiotic stress response. Plant Cell Physiol. 2020;61:427–441. doi: 10.1093/pcp/pcz208. [DOI] [PubMed] [Google Scholar]
- 23.Contreras C., Mariotti R., Mousavi S., Baldoni L., Guerrero C., Roka L., et al. Characterization and validation of olive FAD and SAD gene families: expression analysis in different tissues and during fruit development. Mol Biol Rep. 2020;47:4345–4355. doi: 10.1007/s11033-020-05554-9. [DOI] [PubMed] [Google Scholar]
- 24.Herńndez M.L., Padilla M.N., Mancha M., Martínez-Rivas J.M. Expression analysis identifies FAD2-2 as the olive oleate desaturase gene mainly responsible for the linoleic acid content in virgin olive oil. J Agric Food Chem. 2009;57:6199–6206. doi: 10.1021/jf900678z. [DOI] [PubMed] [Google Scholar]
- 25.Salimonti A, Zelasco S, Lombardo L, Perri E, Micali S. MUTATION SCANNING OF TWO OLEATE DESATURASE (FAD2) ENCODING GENES IN Olea europaea, L. BY HIGH RESOLUTION MELTING. 2011.
- 26.Salimonti A., Carbone F., Romano E., Pellegrino M., Benincasa C., Micali S., et al. Association Study of the 5′UTR Intron of the FAD2-2 Gene With Oleic and Linoleic Acid Content in Olea europaea L. Front Plant Sci. 2020:11. doi: 10.3389/fpls.2020.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luisa Hernández M., Dolores Sicardo M., Arjona P.M., Martínez-Rivas J.M. Specialized functions of olive FAD2 gene family members related to fruit development and the abiotic stress response. Plant Cell Physiol. 2020;61:427–441. doi: 10.1093/pcp/pcz208. [DOI] [PubMed] [Google Scholar]
- 28.Zelasco S., Carbone F., Lombardo L., Salimonti A. In: Olives Olive Oil Health Dis. Prev. 2nd Ed. Preedy V.R., Watson R.R., editors. Academic Press; San Diego: 2021. Chapter 3 - Olive tree genetics, genomics, and transcriptomics for the olive oil quality improvement; pp. 27–49. [DOI] [Google Scholar]
- 29.Commission Regulation (EEC) No 2568/91 of 11 July 1991 on the characteristics of olive oil and olive-residue oil and on the relevant methods of analysis. Httpswebarchivenationalarchivesgovukeu-Exithttpseur-Lexeuropaeulegal-ContentENTXTuriCELEX01991R2568-20081001 n.d. https://www.legislation.gov.uk/eur/1991/2568/2008-10-01 (accessed May 10, 2021).
- 30.Ayed R.B., Grati-Kamoun N., Moreau F., Rebaï A. Comparative study of microsatellite profiles of DNA from oil and leaves of two Tunisian olive cultivars. Eur Food Res Technol. 2009;229:757–762. doi: 10.1007/s00217-009-1111-3. [DOI] [Google Scholar]
- 31.Rozen S., Skaletsky H. In: Bioinforma. Methods Protoc. Misener S., Krawetz S.A., editors. Humana Press; Totowa, NJ: 1999. Primer3 on the WWW for General Users and for Biologist Programmers; pp. 365–386. [DOI] [Google Scholar]
- 32.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Librado P., Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinforma Oxf Engl. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 34.Barrett JC. Haploview: Visualization and Analysis of SNP Genotype Data. Cold Spring Harb Protoc 2009;2009:pdb.ip71-pdb.ip71. doi:10.1101/pdb.ip71. [DOI] [PubMed]
- 35.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 36.Warnes G. Population Genetics; 2021. Gorjanc with contributions from G, Leisch F, Man and M. genetics. [Google Scholar]
- 37.González J.R., Armengol L., Solé X., Guinó E., Mercader J.M., Estivill X., et al. SNPassoc: an R package to perform whole genome association studies. Bioinforma Oxf Engl. 2007;23:644–645. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 38.heatmap.plus.package: Heatmap with more sensible behavior in heatmap.plus: Heatmap with more sensible behavior. n.d. https://rdrr.io/cran/heatmap.plus/man/heatmap.plus.package.html (accessed September 29, 2021).
- 39.Home n.d. https://www.megasoftware.net/ (accessed September 29, 2021).
- 40.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 42.Chen C.-C., Hwang J.-K., Yang J.-M. (PS)2: protein structure prediction server. Nucleic Acids Res. 2006;34:W152–W157. doi: 10.1093/nar/gkl187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen C.-C., Hwang J.-K., Yang J.-M. (PS)2–v2: template-based protein structure prediction server. BMC Bioinf. 2009;10:366. doi: 10.1186/1471-2105-10-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 45.PyMOL | pymol.org n.d. https://pymol.org/2/ (accessed September 29, 2021).
- 46.Tovchigrechko A., Vakser I.A. GRAMM-X public web server for protein–protein docking. Nucleic Acids Res. 2006;34:W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Systèmes D. Free Download: BIOVIA Discovery Studio Visualizer. Dassault Systèmes 2020. https://discover.3ds.com/discovery-studio-visualizer-download (accessed September 29, 2021).
- 48.Eisenberg D, Lüthy R, Bowie JU. [20] VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol., vol. 277, Academic Press; 1997, p. 396–404. 10.1016/S0076-6879(97)77022-8. [DOI] [PubMed]
- 49.Khadake R., Khonde V., Mhaske V., Ranjekar P., Harsulkar A. Functional and bioinformatic characterisation of sequence variants of Fad3 gene from flax. J Sci Food Agric. 2011;91:2689–2696. doi: 10.1002/jsfa.4515. [DOI] [PubMed] [Google Scholar]
- 50.TN93 model (Tamura-Nei, 93) n.d. http://www.bioinf.man.ac.uk/resources/phase/manual/node69.html (accessed May 14, 2021).
- 51.Margaritis D.□(2003). Learning Bayesian Network Model Structure from Data. Ph.D. thesis, School of Computer Science, Carnegie-Mellon University, Pittsburgh, PA. Available as Technical Report CMU-CS-03-153.
- 52.Tsamardinos I., Aliferis C., Statnikov A. American Association for Artificial Intelligence, FLAIRS; 2003. Algorithms for Large Scale Markov Blanket Discovery; pp. 376–381. [Google Scholar]
- 53.Ben Ayed R., Hanana M. Artificial intelligence to improve the food and agriculture sector. J Food Qual. 2021;2021 doi: 10.1155/2021/5584754. [DOI] [Google Scholar]
- 54.Feng L., Zhang Z., Ma Y., Du Q., Williams P., Drewry J., et al. Alfalfa yield prediction using UAV-based hyperspectral imagery and ensemble learning. Remote Sens. 2020;12:2028. doi: 10.3390/rs12122028. [DOI] [Google Scholar]
- 55.Nourani V., Elkiran G., Abba S.I. Wastewater treatment plant performance analysis using artificial intelligence - an ensemble approach. Water Sci Technol J Int Assoc Water Pollut Res. 2018;78:2064–2076. doi: 10.2166/wst.2018.477. [DOI] [PubMed] [Google Scholar]
- 56.Hesami M., Naderi R., Tohidfar M., Yoosefzadeh-Najafabadi M. Application of Adaptive Neuro-Fuzzy Inference System-Non-dominated Sorting Genetic Algorithm-II (ANFIS-NSGAII) for Modeling and Optimizing Somatic Embryogenesis of Chrysanthemum. Front Plant Sci. 2019:10. doi: 10.3389/fpls.2019.00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hesami M., Jones A.M.P. Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl Microbiol Biotechnol. 2020;104:9449–9485. doi: 10.1007/s00253-020-10888-2. [DOI] [PubMed] [Google Scholar]
- 58.Etminan A., Pour-Aboughadareh A., Mohammadi R., Shooshtari L., Yousefiazarkhanian M., Moradkhani H. Determining the best drought tolerance indices using Artificial Neural Network (ANN): insight into application of intelligent agriculture in agronomy and plant breeding. Cereal Res Commun. 2019;47:170–181. doi: 10.1556/0806.46.2018.057. [DOI] [Google Scholar]
- 59.Duan K.-B., Rajapakse J., Wang H., Azuaje F. Multiple SVM-RFE for gene selection in cancer classification with expression data. IEEE Trans Nanobioscience. 2005;4:228–234. doi: 10.1109/TNB.2005.853657. [DOI] [PubMed] [Google Scholar]
- 60.Dehghan Nayeri F., Yarizade K. Bioinformatics study of delta-12 fatty acid desaturase 2 (FAD2) gene in oilseeds. Mol Biol Rep. 2014;41:5077–5087. doi: 10.1007/s11033-014-3373-5. [DOI] [PubMed] [Google Scholar]
- 61.Rawicz W., Olbrich K.C., McIntosh T., Needham D., Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bai Y., McCoy J.G., Levin E.J., Sobrado P., Rajashankar K.R., Fox B.G., et al. X-ray structure of a mammalian stearoyl-CoA desaturase. Nature. 2015;524:252–256. doi: 10.1038/nature14549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bracey M.H., Cravatt B.F., Stevens R.C. Structural commonalities among integral membrane enzymes. FEBS Lett. 2004;567:159–165. doi: 10.1016/j.febslet.2004.04.084. [DOI] [PubMed] [Google Scholar]
- 64.Hanke G., Mulo P. Plant type ferredoxins and ferredoxin-dependent metabolism. Plant Cell Environ. 2013;36:1071–1084. doi: 10.1111/pce.12046. [DOI] [PubMed] [Google Scholar]
- 65.Harding M.M. Geometry of metal-ligand interactions in proteins. Acta Crystallogr D Biol Crystallogr. 2001;57:401–411. doi: 10.1107/s0907444900019168. [DOI] [PubMed] [Google Scholar]
- 66.Harding M.M. Small revisions to predicted distances around metal sites in proteins. Acta Crystallogr D Biol Crystallogr. 2006;62:678–682. doi: 10.1107/S0907444906014594. [DOI] [PubMed] [Google Scholar]
- 67.Page C.C., Moser C.C., Chen X., Dutton P.L. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 68.Moser C.C., Page C.C., Chen X., Dutton P.L. In: Phototrophic Prokaryotes. Peschek G.A., Löffelhardt W., Schmetterer G., editors. Springer US; Boston, MA: 1999. Intraprotein Electron Transfer; pp. 105–111. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.