Abstract
Background
Homologous recombination deficiency (HRD) is a phenotype that is characterized by the inability of a cell to effectively repair DNA double-strand breaks using the homologous recombination repair (HRR) pathway. Loss-of-function genes involved in this pathway can sensitize tumors to poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) inhibitors and platinum-based chemotherapy, which target the destruction of cancer cells by working in concert with HRD through synthetic lethality. However, to identify patients with these tumors, it is vital to understand how to best measure homologous repair (HR) status and to characterize the level of alignment in these measurements across different diagnostic platforms. A key current challenge is that there is no standardized method to define, measure, and report HR status using diagnostics in the clinical setting.
Methods
Friends of Cancer Research convened a consortium of project partners from key healthcare sectors to address concerns about the lack of consistency in the way HRD is defined and methods for measuring HR status.
Results
This publication provides findings from the group’s discussions that identified opportunities to align the definition of HRD and the parameters that contribute to the determination of HR status. The consortium proposed recommendations and best practices to benefit the broader cancer community.
Conclusion
Overall, this publication provides additional perspectives for scientist, physician, laboratory, and patient communities to contextualize the definition of HRD and various platforms that are used to measure HRD in tumors.
Keywords: homologous recombination, poly(ADP-ribose) polymerase inhibitors, BRCA1, BRCA2, biomarkers, tumor, DNA repair
This article reports findings of a consortium of project partners from key healthcare sectors that was convened to address concerns about the lack of consistency in the definition of homologous recombination deficiency and methods for measuring homologous repair status.
Implications for Practice.
Analyzing deficiencies in the homologous recombination repair (HRR) machinery becomes increasingly important to identify patients responding to poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) inhibitors. Ovarian, breast, pancreatic, and prostate cancer are at the forefront of this development, but other cancer types will likely follow. Clinically, homologous recombination deficiency (HRD) is broadly defined, ranging from deleterious mutations in single HRR genes (BRCA1/2 and non-BRCA) to complex genomic scars. As it currently stands, assays that determine HR status may not agree on status calls, which can be problematic for the utility of these assays in the clinic. Our work provides an overview of the diagnostic landscape of HRD including a conceptual framework and definitions which will support molecular tumor boards and clinical decision making.
Introduction
Genomic instability is one of the most common underlying aspects of tumorigenesis, and defective DNA repair is described as a hallmark of cancer.1 Homologous recombination repair (HRR) is a DNA repair pathway that acts on DNA double-strand breaks and interstrand cross-links (ICL).2 A deficiency in the HRR pathway has been associated with several tumor types including breast, ovarian, prostate, and pancreatic cancers (Fig. 1) and has been termed homologous recombination deficiency (HRD), whereas tumors that are not HRD are termed homologous recombination proficient (HRP).3,4 The presence of HRD can make tumors more sensitive to ICL-inducing platinum-based therapies and poly(adenosine diphosphate [ADP]–ribose) polymerase (PARP) inhibitors (PARPi).5,6 Adenosine diphosphate-ribose polymerase inhibitors work via synthetic lethality; blocking base excision repair with PARPi results in an accumulation of DNA single-strand breaks and replication fork collapse resulting in DNA double-strand breaks that cannot be repaired by the HRR pathway if HRR is deficient.7,8 Homologous recombination deficiency is a predictive biomarker for treatment with PARPi in ovarian cancer based on patient outcomes in randomized controlled phase III trials.9-12 Additionally, HRD is a positive prognostic marker for both progression-free survival and overall survival.13 Diagnostics developers have created tests to determine homologous recombination (HR) status and aid in treatment decisions; however, these assays may differ in what they measure and may lead to discordant results that can be problematic for prescribing oncologists. Patients are offered treatment at an emotionally difficult time and discordance between assays makes the decision of diagnostic test and therapy selection more challenging due to uncertainty.
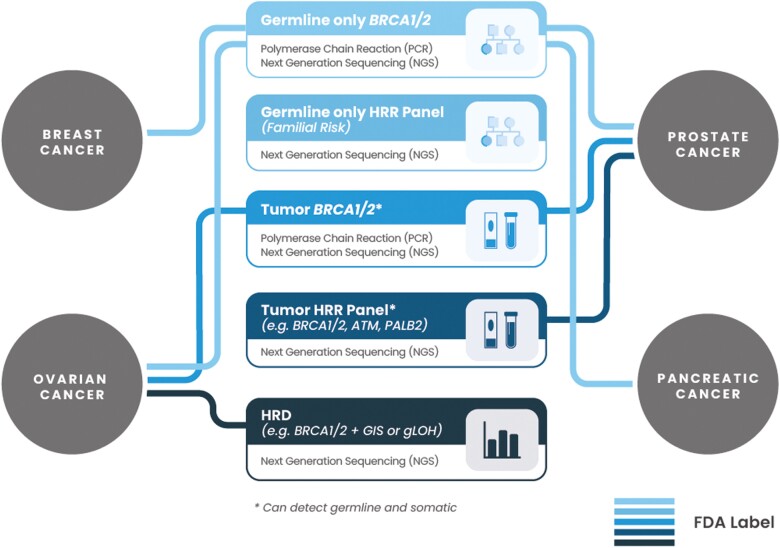
Figure 1.
Present-day landscape of FDA-approved diagnostic tools for PARPi treatments. Not necessarily applicable for all PARPi and indications. Currently approved diagnostic tools can utilize different sample types: tissue, blood, or plasma (refer to Table 1 for details).
The HRD phenotype of sensitivity to platinum-based therapies and PARPi is associated with the HRD genotype defined by impairment in genes involved in the HRR pathway (“causes”) and/or genomic scarring/instability (“consequences”; Fig. 2).4,14 The BRCA1 and BRCA2 genes play prominent roles in the HRR pathway and impaired BRCA gene function is the most studied mechanism in tumor cells among the potential causes that results in HRD. Germline and somatic mutations, as well as epigenetic modifications in BRCA1 and BRCA2, have been consistently associated with an HRD phenotype in breast, ovarian, pancreatic, and prostate cancer,15-17 and have been deemed archetypal in the determination of an HRD phenotype.18 Other HRR pathway genes associated with an HRD phenotype include genes such as ATM, PALB2, RAD51, and others.3 Either genetic or epigenetic alterations in these genes or some combination underlie the HRD phenotype in various cancer types19 including ovarian,16,20,21 endometrial,22 breast,23-25 prostate,26 and pancreatic cancer.27 The association between these genes and an HRD phenotype may be less consistent than BRCA1 and BRCA2 and may vary by the tumor’s tissue of origin. Due to the lack of understanding of the clinical implications of the mutations within HRR pathway genes, more studies to investigate the role of these genes in HRD phenotype in various cancer types are needed.28-30
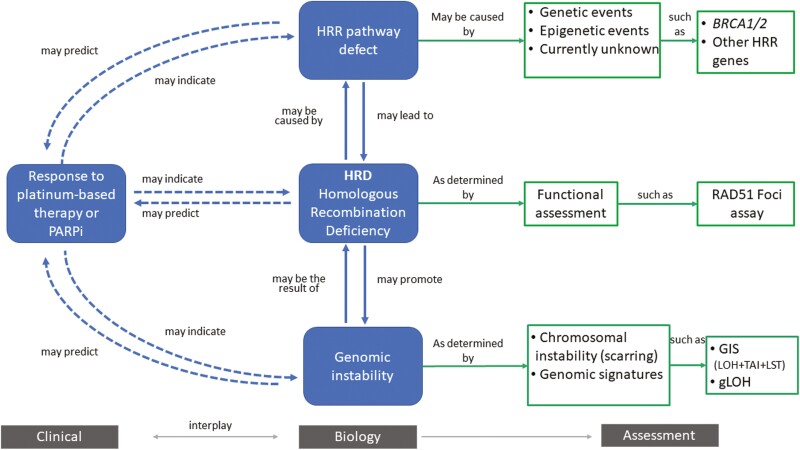
Figure 2.
Overview of homologous recombination deficiency (HRD). Homologous recombination deficiency is a phenotype that is characterized by the inability of a cell to effectively repair double-strand DNA breaks using the homologous recombination repair (HRR) pathway. Alterations in these genes have been deemed “causes” of HRD (eg, genetic events and epigenetic events). This can result in an impaired HRR pathway, which can be deemed “consequences,” and assessed by probing the genome for evidence of genomic instability (eg, chromosomal instability and other genomic signatures).
Testing for the consequences of an impaired HRR pathway is performed by probing the genome for evidence of genomic abnormalities. Several studies in breast and ovarian cancer have identified genomic patterns or signatures of instability associated with an HRD phenotype. These signatures of instability can include genomic patterns of loss of heterozygosity (gLOH), which are regions of intermediate size (>15 MB and < whole chromosome),31 number of telomeric imbalances (TAI), which are the number of regions with allelic imbalance which extend to the sub-telomere but not cross the centromere,32 and large-scale transitions (LST), which are chromosome breaks (translocations, inversions, or deletions).33 These approaches evaluate the presence of HRD-related genomic signatures (often referred to as scars) that are thought to be a consequence of error-prone DNA repair through alternative pathways (eg, non homologous end joining [NHEJ]).
Studies have demonstrated the predictive value of assays to determine the HRD phenotype by evaluating response to platinum-based therapies and PARPi in the context of breast and ovarian cancer.4,14 Various multi-omic studies have investigated how a combination of the above patterns and additional genomic and transcriptomic signatures are associated with an HRD phenotype.5,34-36 A number of studies continue to evaluate genomic instability in breast and ovarian cancer as well as additional cancer types, which could lead to refinements in its use.28 Additional approaches, such as the detection of RAD51 foci, may enable a functional assessment of HR status after a cell’s exposure to a DNA damaging agent. This approach requires multiple slides per patient which must be annotated by a trained professional. Recently, it has been shown that assessment of basal levels of RAD51 foci are possible in clinical samples and appear to show a high degree of correlation with PARPi response. The clinical validity as well as the practicality and implementation for routine clinical use is under investigation.37-39
To date, FDA has approved several companion or complementary diagnostics to facilitate tumor selection for PARPi treatments based on HR status (Table 1). Two of these (FoundationOne CDx and the Myriad myChoice CDx test) assess chromosomal instability to select patients with ovarian cancer that may benefit from an FDA-approved therapy. These assays incorporate both the causes and consequences of HRR impairment, whereas other FDA-approved assays only detect potential causes of HRR impairment without assessing consequences (eg, BRACAnalysis CDx, FoundationOne Liquid CDx, and FoundationOne CDx). It is important to note that while there are broad differences in the approach to test for HRD (causes vs consequences), there are also differences in the assays themselves that need to be considered. In addition to the FDA approved companion diagnostics, other sequencing or single-nucleotide polymorphism (SNP)–based platforms are being evaluated to measure genomic instability. Additional commercial and lab-developed assays that utilize tissue and blood to measure HR status are also available.
Table 1.
Companion diagnostics approved for selection of PARPi.
| Cancer | Assay | Sample type | Therapy | Indication | Trial |
|---|---|---|---|---|---|
| Ovarian | BRACAnalysis CDx | Blood | Olaparib | For the maintenance treatment of adult patients with deleterious or suspected deleterious germline or somatic BRCA-mutated advanced epithelial ovarian, fallopian tube or primary peritoneal cancer who are in complete or partial response to first-line platinum-based chemotherapy | SOLO1 Study |
| BRACAnalysis CDx | Blood | Olaparib | For the treatment of adult patients with deleterious or suspected deleterious germline BRCA-mutated (gBRCAm) advanced ovarian cancer who have been treated with 3 or more prior lines of chemotherapy | Study 19 Study 42 | |
| FoundationOne CDx | Tumor | Olaparib | For the maintenance treatment of adult patients with deleterious or suspected deleterious germline or somatic BRCA-mutated advanced epithelial ovarian, fallopian tube or primary peritoneal cancer who are in complete or partial response to first-line platinum-based chemotherapy | SOLO1 Study | |
| Myriad myChoice CDx testa | Tumor | Olaparib | For the maintenance treatment of adult patients with deleterious or suspected deleterious germline or somatic BRCA-mutated advanced epithelial ovarian, fallopian tube or primary peritoneal cancer who are in complete or partial response to first-line platinum-based chemotherapy | SOLO1 Study | |
| Myriad myChoice CDx testa | Tumor | Olaparib + Bevacizumab | For the maintenance treatment of adult patients with advanced epithelial ovarian, fallopian tube, or primary peritoneal cancer who are in complete or partial response to first-line platinum-based chemotherapy and whose cancer is associated with HRD-positive status defined by either: 1) a deleterious or suspected deleterious BRCA mutation, and/or 2) genomic instability | PAOLA-1 Study | |
| BRACAnalysis CDx | Blood | Rucaparib | For the treatment of adult patients with a deleterious BRCA mutation (germline and/or somatic)-associated epithelial ovarian, fallopian tube, or primary peritoneal cancer who have been treated with 2 or more chemotherapies | ARIEL2 Study | |
| FoundationOne CDx | Tumor | Rucaparib | For the treatment of adult patients with a deleterious BRCA mutation (germline and/or somatic)-associated epithelial ovarian, fallopian tube, or primary peritoneal cancer who have been treated with 2 or more chemotherapies | ARIEL3 Study | |
| FoundationOne Liquid CDx | Blood | Rucaparib | For the treatment of adult patients with a deleterious BRCA mutation (germline and/or somatic)-associated epithelial ovarian, fallopian tube, or primary peritoneal cancer who have been treated with 2 or more chemotherapies | ARIEL2 Study | |
| Myriad myChoice CDx testa | Tumor | Niraparib | For the treatment of adult patients with advanced ovarian, fallopian tube, or primary peritoneal cancer who have been treated with 3 or more prior chemotherapy regimens and whose cancer is associated with HRD-positive status defined by either: (1) a deleterious or suspected deleterious BRCA mutation or (2) genomic instability and who have progressed more than 6 months after response to the last platinum-based chemotherapy | QUADRA Study | |
| Prostate | BRACAnalysis CDx | Blood | Olaparib | For the treatment of adult patients with deleterious or suspected deleterious germline or somatic homologous recombination repair (HRR) gene-mutated metastatic castration-resistant prostate cancer (mCRPC) who have progressed following prior treatment with enzalutamide or abiraterone | PROfound Study |
| FoundationOne CDx | Tumor | Olaparib | For the treatment of adult patients with deleterious or suspected deleterious germline or somatic homologous recombination repair (HRR) gene-mutated metastatic castration-resistant prostate cancer (mCRPC) who have progressed following prior treatment with enzalutamide or abiraterone | PROfound Study | |
| FoundationOne Liquid CDx | Plasma | Olaparib | For the treatment of adult patients with deleterious or suspected deleterious germline or somatic homologous recombination repair (HRR) gene-mutated metastatic castration-resistant prostate cancer (mCRPC) who have progressed following prior treatment with enzalutamide or abiraterone | PROfound Study | |
| Breast | BRACAnalysis CDx | Blood | Olaparib | For the treatment of adult patients with deleterious or suspected deleterious gBRCAm, HER2-negative metastatic breast cancer who have been treated with chemotherapy in the neoadjuvant, adjuvant, or metastatic setting. Patients with hormone receptor (HR)-positive breast cancer should have been treated with a prior endocrine therapy or be considered inappropriate for endocrine therapy | OlympiAD Study |
| BRACAnalysis CDx | Blood | Talazoparib | For the treatment of adult patients with deleterious or suspected deleterious germline BRCA-mutated (gBRCAm) HER2-negative locally advanced or metastatic breast cancer | EMBRACA Study | |
| Pancreatic | BRACAnalysis CDx | Blood | Olaparib | For the maintenance treatment of adult patients with deleterious or suspected deleterious gBRCAm metastatic pancreatic adenocarcinoma whose disease has not progressed on at least 16 weeks of a first-line platinum-based chemotherapy regimen | POLO Study |
Assesses genome-wide characteristics (potentially consequences of HRR impairment). Currently other companion diagnostic claims select patients by assessing genetic mutations (potentially causes of HRR impairment). Labels were accessed on FDA’s website and are current as of June 2021.
There is currently only partial agreement on which parameters contribute to determining the HR status of a sample and what combination of molecular measures are necessary to classify tumors as HRD.40 Additionally, assays may use different cutoffs across tumor types, within tumor types, or for drugs within a similar class.41 For example, a clinical trial on the PARPi veliparib used a cutoff of 33 using the Myriad myChoice CDx test to define HR status in patients with high grade serous ovarian cancer10 while other trials on niraparib9 and olaparib12 used a cutoff of 42. As the application of HR status is investigated in different cancer types, it is important to improve clarity regarding the way HRD is defined, measured, and reported. Addressing this lack of clarity can help optimize the use of this complex biomarker for the selection of patients for therapies targeting the DNA repair pathway, such as PARPi, and identify the elements used to define HR status that should be considered to best achieve consistent results.
Materials and Methods
Friends of Cancer Research convened a group of stakeholders from industry, academia, and government. We hosted bimonthly calls for 4 months with diverse stakeholders to discuss the best way to approach harmonizing HR status measurements using diagnostic assays. We set out to characterize how HRD is currently defined, measured, and used with regards to assays that measure HR status, and, ultimately, to propose common language and recommendations to improve consistency around the use of HR status as a biomarker. We reviewed literature associated with phase III trials, including those that led to FDA approvals, and current guidelines from the American Society of Clinical Oncology (ASCO) and National Comprehensive Cancer Network (NCCN) regarding the use of assays to measure HR status, investigated FDA labels of currently approved PARPi and reports on FDA-approved companion diagnostics validated to assess HR status, and discussed current laboratory and clinical practices.
Results
Assessment of current practices helped to answer what HRD is, how it is currently measured, and how assays currently assess HR status. During routine clinical use, HR status is assessed by measuring either evidence for potential causes of HRD indirectly (eg, genetic mutations) or potential consequence of deficiency in the HRR pathway (eg, genomic instability, mutational signatures; Fig. 2).
Definitions of HRD are Heterogeneous
The definition of HRD varies widely among the scientific and medical communities. Various terms have historically been used in the literature to describe HRD including BRCA-ness, BRCA-like, and genomic scarring.42-44 Additionally, HRD is a complex biomarker whose definition may need refinement based on growing biological and clinical understanding. Defining HRD in terms of specific genetic mutations and/or the success of a PARPi may be a too narrow approach. Homologous recombination deficiency should not be solely defined by response to any one therapy, given that recent studies have shown that HRD has both a positive prognostic value in ovarian and other cancers and predictive value for PARPi and platinum therapy. Additionally, testing capabilities may evolve to better assess HR status with a functional assay.2,38
Approaches for Assessing HR Status Vary Across Current FDA-Approved Companion Diagnostics
Given that HR status can inform treatment decisions and help predict improved outcomes for certain patients, it is essential to understand processes through which these decisions are made and identify best practices to maximize future benefit of HR status determination. To better understand how HR status is currently determined, we reviewed the labels of FDA approved companion diagnostics that test HR status for use with a PARPi. Companion diagnostics are medical devices regulated by FDA to support safe and effective use of a corresponding therapeutic. While “companion diagnostic” as a construct exists in a limited capacity in non-US markets, the clinical and analytical validation conferred by FDA review continues to inform global diagnostic use. In Europe, the European Union In Vitro Diagnostics Regulation emulates FDA’s consideration of companion diagnostic regulation and is increasing review rigor by requiring validation studies.
We reviewed the FDA labels of Lynparza (olaparib), Zejula (niraparib), and Rubraca (rucaparib), PARPi that have used HRD as selection biomarkers in frontline and recurrent epithelial ovarian, fallopian tube, or primary peritoneal cancers. The language on the olaparib and niraparib labels is mostly consistent and defines HRD by either (1) a deleterious or suspected deleterious BRCA mutation and/or (2) genomic instability. Olaparib and niraparib use the FDA-approved diagnostic test the Myriad myChoice CDx test, which determines HR status by assessing the mutation status of BRCA1/2 and/or genomic instability—measured by the evaluation of a combination of molecular measures to derive a genomic instability (gLOH + TAI + LST; the Myriad myChoice CDx test Summary of Safety and Effectiveness Data). PARPi therapies may vary in the requirement of a companion diagnostic based on the clinical evidence for the therapeutic and depending on the indication. The rucaparib label does not mention HRD in the label but refers to patients with a BRCA1/2 mutation (germline or somatic)-associated epithelial ovarian, fallopian tube, or primary peritoneal cancer based on an FDA-approved companion diagnostic for this therapy. Rucaparib uses the Foundation Medicine test Foundation Focus CDx BRCA LOH (now part of a broader FoundationOne CDx panel), which determines BRCA1/2 mutation status as per the companion diagnostic claim, and determines HR status, which is defined by the mutation status of BRCA1/2 and/or genomic instability as measured by gLOH.
In summary, the FDA-approved companion diagnostics test for somewhat different components, which may result in different HR status calls and subsequently different treatment decisions. Given that genomic instability patterns arising from HRD can look different in various tissue types, the assay used to assess HR status should be validated using samples from the intended use population being studied. Within an individual assay, cutoffs for determining HR status may differ by tissue type which requires further transparency around how thresholds are determined and whether these differences are due to chance, clinical trial approach, or biology.
Utilization of HR Status for Clinical Decision-Making Would Benefit from Addressing Uncertainties
Review of the literature, ASCO and NCCN guidelines, and expert discussions suggest uncertainty and inconsistency in how to use assays that measure HR status in the clinic, which could potentially drive low adoption for clinical decision making.45 Because assays that measure HR status can identify cancers that are more likely to respond to PARPi therapies and predict positive outcomes, it is important to address the sources of uncertainty to enable clinicians to use these new diagnostic tools and provide their patients with optimal care.
We identified several areas which may lead to this lack of clarity including inconsistent reporting of HR status between studies and clinical trials, misaligned definitions of HR testing (eg, ‘mutation of HRR genes’ interchanged with ‘HR status’), complexity in the order of testing (eg, germline vs tumor, specific genes vs gene panel), interchangeability of tissue and plasma-based approaches, family history, and cancer type that warrant further investigation.
Discussion
Based on findings, we propose several considerations to bring better alignment in the field for use of HR assays in the clinic.
Use consistent language when describing HRD and align on a foundational definition that allows for a dynamic evolution of the term as HRD knowledge grows. We propose using the following definition: HRD is a phenotype that is characterized by the inability of a cell to effectively repair DNA double-strand breaks using the HRR pathway. Additionally, stakeholders should use the terminology “HRD” and “HRP” to define the presence or absence of an HRD phenotype, respectively.
Adopt a minimum set of requirements for the determination of HR status, the details on how HR status was measured, and clearly report the type of test used should be clearly reported in publications. Greater clarity should be given to whether measures of both cause and consequence are needed to inform the determination of HR status in different contexts (Fig. 2). Evidence of consequence alone may not always be indicative of PARPi sensitivity due to the possibility of reversion mutations41; however, co-occurrence of consequence with cause can potentially support novel loss-of-function mutations.
Additionally, defining mutation status and zygosity of BRCA1/2 (in the context of ovarian cancer) and genomic instability status to then determine HR status is complex, and we recommend transparency and standardization of the type of information used to determine BRCA1/2 mutation status (ie, pathogenicity status of the variant), genomic instability (ie, molecular measures and parameters used to develop a score, value of continuous variable-if any), and criterion for cutoff selection. As we move beyond ovarian cancer, as well as investigate the role other HRR genes play in the cause of HRD, including different patterns of genomic instability exhibited in different cancer types, this additional information will be key to ensure consistency in results obtained across tests that determine HR status. Given that different tests may be used to determine HR status, and each uses different approaches for their determination, publications should include the name of the test used to determine HR status and specific clinical thresholds for level of deficiency (eg, HRD as measured by <assay name> and defined as <features evaluated and cutpoint(s)>).
Conduct studies to identify and assess sources of discordance among assays that assess HR status and identify sources of variability to inform optimal use of these assays for clinical decision making. This can be accomplished through a study that assesses concordance of HR status across assays. Additionally, it would be beneficial to create a clinician-targeted survey to identify major barriers to the understanding or use of HR status as a decision-making factor to determine treatment approaches.
Encourage all testing labs to report a minimum set of elements important for interpreting the clinical report in line with FDA reporting requirements for current FDA approved assays assessing HR status and contextualize clinical meaning to assist clinicians and patients with decision-making. Test developers should report whether the test is tumor-type dependent, what genomic findings were identified (as is being done), and genomic instability/scarring scores (with thresholds as per drug/companion diagnostic approval for that cancer type).
Conclusion
Biomarkers such as HR status play a critical role in treatment decisions for patients with cancer. It is therefore of utmost importance to build consensus on how to define HRD and the methodology for assessing HR status to promote alignment and optimal use of this biomarker to identify patients who would benefit from PARPi therapy. This publication provides findings from the group’s discussions that encourages diagnostic developers to consider the parameters that contribute to the determination of HR status. Perspectives captured in this manuscript complement notable academic efforts by professional societies, such as the European Society for Medical Oncology to assess methods for HRD testing as well as planned activities and surveys being conduct by the Association for Molecular Pathology, Association of Community Cancer Centers, American Society for Clinical Oncology and College of American and Pathologists to assess current clinical practice and provide evidence-based subject matter expert recommendations regarding best practices and performance characteristics of clinical HRD molecular methods. We created recommendations and proposed best practices for industry stakeholders to benefit the entire cancer community that support alignment efforts and can evolve as biological and clinical advancements emerge to support robust use among oncologists and ensure assays enable the best possible care for patients.
Contributor Information
Mark D Stewart, Friends of Cancer Research, Washington, DC, USA.
Diana Merino Vega, Friends of Cancer Research, Washington, DC, USA.
Rebecca C Arend, Division of Gynecologic Oncology, University of Alabama at Birmingham, Birmingam, AL, USA.
Jonathan F Baden, Translational Medicine, Bristol Myers Squibb, New York, NY, USA.
Olena Barbash, Oncology Experimental Medicine Unit, GlaxoSmithKline, Philadelphia, PA, USA.
Nike Beaubier, Tempus Labs, Inc., Chicago, IL, USA.
Grace Collins, Friends of Cancer Research, Washington, DC, USA.
Tim French, Global Medical Affairs, Diagnostics, AstraZeneca, Cambridge, UK.
Negar Ghahramani, Molecular Genetic Pathology Regional Laboratory, SCPMG Regional Reference Laboratories, Los Angeles, CA, USA.
Patsy Hinson, Independent Cancer Research Patient Advocate, Charlotte, NC, USA.
Petar Jelinic, Early Clinical Oncology, Merck & Co., Inc., Kenilworth, NJ, USA.
Matthew J Marton, Early Clinical Oncology, Merck & Co., Inc., Kenilworth, NJ, USA.
Kimberly McGregor, Cancer Genomics Research Group, Foundation Medicine, Cambridge, MA, USA.
Jerod Parsons, Tempus Labs, Inc., Chicago, IL, USA.
Lakshman Ramamurthy, Global Regulatory Affairs, GlaxoSmithKline, Washington, DC, USA.
Mark Sausen, Translational Medicine, Bristol Myers Squibb, New York, NY, USA.
Ethan S Sokol, Cancer Genomics Research Group, Foundation Medicine, Cambridge, MA, USA.
Albrecht Stenzinger, Institute of Pathology, University Hospital Heidelberg, Heidelberg, Germany.
Hillary Stires, Friends of Cancer Research, Washington, DC, USA.
Kirsten M Timms, Myriad Genetics, Inc., Salt Lake City, UT, USA.
Diana Turco, Myriad Genetics, Inc., Salt Lake City, UT, USA.
Iris Wang, Global Precision Medicine, Novartis Pharmaceuticals Corporation, New York, NY, USA.
J Andrew Williams, Precision Medicine & Biosamples, AstraZeneca, Cambridge, UK.
Elaine Wong-Ho, Clinical Sequencing Division, Thermo Fisher Scientific, San Francisco, CA, USA.
Jeff Allen, Friends of Cancer Research, Washington, DC, USA.
Conflict of Interest
Rebecca C. Arend: AstraZeneca, Clovis Oncology, GlaxoSmithKline, KIYATEC, Leap Therapeutics, Merck & Co, Sutro Biopharma, VBL Therapeutics (SAB), Caris Life Sciences (C/A); Jonathan F. Baden: Bristol Myers Squibb (E, OI), Johnson & Johnson (OI); Olena Barbash: GlaxoSmithKline (E); Lakshman Ramamurthy: GlaxoSmithKline (E); Nike Beaubier: Tempus Labs, Inc. (E); Jerod Parsons: Tempus Labs, Inc. (E); Tim French: AstraZeneca (E, OI); Petar Jelinic: Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., (E, OI); Matthew J. Marton: Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., (E, OI); Kimberly McGregor: Foundation Medicine (E), Roche (OI); Ethan S. Sokol: Foundation Medicine (E), Roche (OI); Mark Sausen: Bristol Myers Squibb (E, OI); Albrecht Stenzinger: Aignostics, AstraZeneca, AGCT, Bayer, BMS, Eli Lilly, Illumina, Incyte, Janssen, MSD, Novartis, Pfizer, Roche, Seattle Genetics, Takeda, Thermo Fisher (SAB), Bayer, BMS, Chugai, Incyte (RF); Kirsten M. Timms: Myriad Genetics Inc. (E, OI); Diana Turco: Myriad Genetics Inc. (E, OI); J. Andrew Williams: AstraZeneca (E); Iris Wang: Novartis Pharmaceuticals Corporation (E, OI); Elaine Wong-Ho: Thermo Fisher Scientific (E). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) research funding; (E) employment; (ET) expert testimony; (H) honoraria received; (OI) ownership interests; (IP) intellectual property rights/inventor/patent holder; (SAB) scientific advisory board.
Author Contributions
Conception/design: M.D.S., D.M.V., R.C.A., J.F.B., O.B., N.B., T.F., N.G., P.H., P.J., M.J.M., K.M., J.P., L.R., M.S., E.S.S., A.S., H.S., K.M.T., D.T., I.W., J.A.W., E.W.-H., J.A.
Data analysis and interpretation: M.D.S., D.M.V., R.C.A., J.F.B., O.B., N.B., T.F., N.G., P.H., P.J., M.J.M., K.M., J.P., L.R., M.S., E.S. S., A.S., H.S., K.M.T., D.T., I.W., J.A.W., E.W.-H., J.A. Manuscript writing: M.D.S., D.M.V., R.C.A., J.F.B., O.B., N.B., T.F., N.G., P.H., P.J., M.J.M., K.M., J.P., L.R., M.S., E.S. S., A.S., H.S., K.M.T., D.T., I.W., J.A.W., E.W.-H., J.A. Final approval of manuscript: M.D.S., D.M.V., R.C. A., J.F.B., O.B., N.B., T.F., N.G., P.H., P.J., M.J.M., K.M., J.P., L.R., M.S., E.S.S., A.S., H.S., K.M.T., D.T., I.W., J.A.W., E.W.-H., J.A.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Funding
This work was supported by Friends of Cancer Research.
References
- 1. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646-674. [DOI] [PubMed] [Google Scholar]
- 2. Miller RE, Leary A, Scott CL, et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann Oncol. 2020;31(12):1606-1622. [DOI] [PubMed] [Google Scholar]
- 3. Heeke AL, Pishvaian MJ, Lynce F, et al. Prevalence of Homologous recombination–related gene mutations across multiple cancer types. JCO Precis Oncol. 2018;1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marquard AM, Eklund AC, Joshi T, et al. Pan-cancer analysis of genomic scar signatures associated with homologous recombination deficiency suggests novel indications for existing cancer drugs. Biomark Res. 2015;3:1-1–0.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoppe MM, Sundar R, Tan DSP, Jeyasekharan AD. Biomarkers for homologous recombination deficiency in cancer. J Natl Cancer Inst. 2018;110(7):704-713. [DOI] [PubMed] [Google Scholar]
- 6. Lord CJ, Ashworth A. PARP inhibitors: the first synthetic lethal targeted therapy Europe PMC Funders Group. Science 2017;355:1152-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iglehart JD, Silver DP. Synthetic lethality—a new direction in cancer-drug development. 2009;361:189-191. 10.1056/NEJMe0903044. [DOI] [PubMed] [Google Scholar]
- 8. Banerjee S, Gonzalez-Martin A, Harter P, et al. First-line PARP inhibitors in ovarian cancer: summary of an ESMO Open—Cancer Horizons round-table discussion. ESMO Open. 2020;5(6):e001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. González-Martín A, Pothuri B, Vergote I, et al. ; PRIMA/ENGOT-OV26/GOG-3012 Investigators . Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2019;381(25):2391-2402. [DOI] [PubMed] [Google Scholar]
- 10. Coleman RL, Fleming GF, Brady MF, et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med. 2019;381(25):2403-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moore KN, Secord AA, Geller MA, et al. Niraparib monotherapy for late-line treatment of ovarian cancer (QUADRA): a multicentre, open-label, single-arm, phase 2 trial. Lancet Oncol. 2019;20(5):636-648. [DOI] [PubMed] [Google Scholar]
- 12. Ray-Coquard I, Pautier P, Pignata S, et al. ; PAOLA-1 Investigators . Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med. 2019;381(25):2416-2428. [DOI] [PubMed] [Google Scholar]
- 13. How JA, Jazaeri AA, Fellman B, et al. Modification of homologous recombination deficiency score threshold and association with long-term survival in epithelial ovarian cancer. Cancers. 2021;13(5):1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sokol ES, Pavlick D, Khiabanian H, et al. Pan-cancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by genome-wide loss of heterozygosity. JCO Precis Oncol. 2020;4:442-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vollebergh MA, Lips EH, Nederlof PM, et al. Genomic patterns resembling BRCA1- and BRCA2-mutated breast cancers predict benefit of intensified carboplatin-based chemotherapy. Breast Cancer Res. 2014;16(3):R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frey MK, Pothuri B. Homologous recombination deficiency (HRD) testing in ovarian cancer clinical practice: a review of the literature. Gynecol Oncol Res Pract. 2017;4:1-1–1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nguyen L, Martens J, Van Hoeck A, et al. Pan-cancer landscape of homologous recombination deficiency. Nat Commun. 2020;11(5584):1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prakash R, Zhang Y, Feng W, Jasin M. Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb Perspect Biol. 2015;7(4):a016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heeke AL, Baker T, Lynce F, et al. Prevalence of homologous recombination deficiency among all tumor types. J Clin Oncol. 2017;35:1502. [Google Scholar]
- 20. da Cunha Colombo Bonadio RR, Fogace RN, Miranda VC, Diz MDPE. Homologous recombination deficiency in ovarian cancer: a review of its epidemiology and management. Clinics (Sao Paulo). 2018;73(suppl 1):1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mirza MR, Monk BJ, Herrstedt J, et al. ; ENGOT-OV16/NOVA Investigators . Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375(22):2154-2164. [DOI] [PubMed] [Google Scholar]
- 22. de Jonge MM, Auguste A, van Wijk LM, et al. Frequent homologous recombination deficiency in high-grade endometrial carcinomas. Clin Cancer Res. 2019;25(3):1087-1097. [DOI] [PubMed] [Google Scholar]
- 23. den Brok WD, Schrader KA, Sun S, et al. Homologous recombination deficiency in breast cancer: a clinical review. JCO Precis Oncol. 2017;1:1-13. [DOI] [PubMed] [Google Scholar]
- 24. Zhao EY, Shen Y, Pleasance E, et al. Homologous recombination deficiency and platinum-based therapy outcomes in advanced breast cancer. Clin Cancer Res. 2017;23(24):7521-7530. [DOI] [PubMed] [Google Scholar]
- 25. Melinda LT, Kirsten MT, Julia R, et al. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016;22:3764-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Felice D, Dove Press P. Defective DNA repair mechanisms in prostate cancer: impact of olaparib. Drug Des Dev Ther. 2017:547-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Waddell N, Pajic M, Patch AM, et al. ; Australian Pancreatic Cancer Genome Initiative . Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518(7540):495-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lotan TL, Kaur HB, Salles DC, et al. Homologous recombination deficiency (HRD) score in germline BRCA2- versus ATM-altered prostate cancer. Mod Pathol. 2021;34:1185-1193. 10.1038/s41379-020-00731-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18(1):99-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Norquist BM, Brady MF, Harrell MI, et al. Mutations in homologous recombination genes and outcomes in ovarian carcinoma patients in GOG 218: an NRG Oncology/Gynecologic Oncology Group Study. Clin Cancer Res. 2018;24(4):777-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abkevich V, Timms KM, Hennessy BT, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 2012;107(10):1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Birkbak NJ, Wang ZC, Kim JY, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012;2(4):366-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Popova T, Manié E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res. 2012;72(21):5454-5462. [DOI] [PubMed] [Google Scholar]
- 34. Meijer TG, Verkaik NS, Sieuwerts AM, et al. Functional ex vivo assay reveals homologous recombination deficiency in breast cancer beyond BRCA gene defects. Clin Cancer Res. 2018;24(24):6277-6287. [DOI] [PubMed] [Google Scholar]
- 35. Sztupinszki Z, Diossy M, Krzystanek M, et al. Detection of molecular signatures of homologous recombination deficiency in prostate cancer with or without BRCA1/2 mutations. Clin Cancer Res. 2020;26(11):2673-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Davies H, Glodzik D, Morganella S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23(4):517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Malihi PD, Graf RP, Rodriguez A, et al. Single-cell circulating tumor cell analysis reveals genomic instability as a distinctive feature of aggressive prostate cancer. Clin Cancer Res. 2020;26(15):4143-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Castroviejo-Bermejo M, Cruz C, Llop-Guevara A, et al. A RAD 51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol Med. 2018;10:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. van Wijk LM, Vermeulen S, Meijers M, et al. The recap test rapidly and reliably identifies homologous recombination-deficient ovarian carcinomas. Cancers. 2020;12:1-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Eckstein M, Bloom KJ, Riccelli P, et al. HRD in ovarian cancer: defined today, evolving for the future. 2020;38:e18052. 10.1200/JCO.2020.38.15_suppl.e18052. [DOI] [Google Scholar]
- 41. Takaya H, Nakai H, Takamatsu S, Mandai M, Matsumura N. Homologous recombination deficiency status-based classification of high-grade serous ovarian carcinoma. Sci Rep. 2020;10(1):1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Turner N, Tutt A, Ashworth A. Hallmarks of “BRCAness” in sporadic cancers. Nat Rev Cancer. 2004;4:814-819. [DOI] [PubMed] [Google Scholar]
- 43. Tan DS, Rothermundt C, Thomas K, et al. “BRCAness” syndrome in ovarian cancer: a case-control study describing the clinical features and outcome of patients with epithelial ovarian cancer associated with BRCA1 and BRCA2 mutations. J Clin Oncol. 2008;26(34):5530-5536. [DOI] [PubMed] [Google Scholar]
- 44. Konstantinopoulos PA, Spentzos D, Karlan BY, et al. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J Clin Oncol. 2010;28(22):3555-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stover EH, Fuh K, Konstantinopoulos PA, Matulonis UA, Liu JF. Clinical assays for assessment of homologous recombination DNA repair deficiency. Gynecol Oncol. 2020;159(3):887-898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.