Abstract
Burkholderia pseudomallei, the causative agent of melioidosis, is intrinsically resistant to a wide range of antimicrobial agents including β-lactams, aminoglycosides, macrolides, and polymyxins. We used Tn5-OT182 to mutagenize B. pseudomallei to identify the genes involved in aminoglycoside resistance. We report here on the identification of AmrAB-OprA, a multidrug efflux system in B. pseudomallei which is specific for both aminoglycoside and macrolide antibiotics. We isolated two transposon mutants, RM101 and RM102, which had 8- to 128-fold increases in their susceptibilities to the aminoglycosides streptomycin, gentamicin, neomycin, tobramycin, kanamycin, and spectinomycin. In addition, both mutants, in contrast to the parent, were susceptible to the macrolides erythromycin and clarithromycin but not to the lincosamide clindamycin. Sequencing of the DNA flanking the transposon insertions revealed a putative operon consisting of a resistance, nodulation, division-type transporter, a membrane fusion protein, an outer membrane protein, and a divergently transcribed regulator protein. Consistent with the presence of an efflux system, both mutants accumulated [3H]dihydrostreptomycin, whereas the parent strain did not. We constructed an amr deletion strain, B. pseudomallei DD503, which was hypersusceptible to aminoglycosides and macrolides and which was used successfully in allelic exchange experiments. These results suggest that an efflux system is a major contributor to the inherent high-level aminoglycoside and macrolide resistance found in B. pseudomallei.
Burkholderia pseudomallei is a motile, gram-negative bacillus found in tropical regions of the world including Southeast Asia, northern Australia, and temperate regions bordering the equator (5, 14). It is the causative agent of melioidosis, a disease which can manifest itself as an acute, subacute, or chronic form of illness (7, 38). The acute form of the disease is a septicemic illness which is often rapidly fatal, despite antibiotic treatment, and which is responsible for considerable morbidity and mortality, especially in northeastern Thailand (7). Subacute melioidosis is described as a prolonged febrile illness with multiorgan involvement, systemic abscess formation, and bacteremia (38). The existence of chronic melioidosis, perhaps the most common form of the disease, becomes evident by the postmortem examination of infected tissues or by activation of other forms of the disease as a result of a traumatic event (35). Infection likely occurs as a result of contact with the organism in soil or water, with entrance into the body via inhalation, via aspiration, or through cuts and abrasions (14, 44). Successful treatment of melioidosis patients is difficult because B. pseudomallei is inherently resistant to a variety of antibiotics including β-lactams, aminoglycosides, macrolides, and polymyxins (14, 17). We initiated the studies described here in an effort to understand the antibiotic resistance mechanisms present in this organism. We report here on the identification of AmrAB-OprA, a unique multidrug efflux system in B. pseudomallei which is specific for both aminoglycoside and macrolide antibiotics. This system shows substantial homology to multidrug efflux systems found in Pseudomonas aeruginosa (16, 30–32), Neisseria gonorrhoeae (13), and Escherichia coli (21, 23), although in terms of antibiotic specificity, the multidrug efflux system in B. pseudomallei appears to have little if any similarity to those in the other organisms.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in these studies are listed in Table 1. Unless noted otherwise, cultures were grown at 37°C on Luria broth (LB) agar (Becton Dickinson, Cockeysville, Md.) or in LB. Antibiotics were used as described previously (15).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strains | ||
| E. coli SM10λpir | SM10 with a λ prophage carrying the gene encoding the π protein; Kmr | 26 |
| B. pseudomallei | ||
| 1026b | Clinical isolate; AGr Tcs | 11 |
| C21 | 1026b derivative; gspD::Tn5-OT182 AGr Tcs | 10 |
| RM101 | 1026b derivative; amrA::Tn5-OT182 AGs Tcr | This study |
| RM102 | 1026b derivative; amrB::Tn5-OT182 AGs Tcr | This study |
| RM104 | 1026b derivative; 1026b::pSK240 Tcr | This study |
| RM102 Smr | RM102 derivative; rpsL (Smr) | This study |
| DD502 | RM102 Smr::pDD502 Sms Kmr Tcr | This study |
| DD503 | DD502 derivative; Δ(amrR-oprA) rpsL (Smr) AGs Tcs | This study |
| Plasmids | ||
| pOT182 | PSUP102(Gm)::Tn5-OT182 Cmr Gmr Apr Tcr | 24 |
| pUCP28T | Broad-host-range vector; IncP OriT; pRO1600 ori; Tpr | 36 |
| p34E | Cassette cloning vector; Apr | 42 |
| pKAS46 | Mobilizable allelic exchange vector; π-dependent R6K replicon; Apr KmrrpsL+ (Sms) | 37 |
| pSKM11 | Mobilizable allelic exchange vector; Tcr Apr | 27 |
| pSKM240 | Mobilizable allelic exchange vector; pSKM11 containing a 240-bp internal Sst1 fragment from amrR; Tcr Apr | This study |
| pBluescript SK | General cloning vector; Apr | Stratagene |
| pAW22 | 14.0-kb NotI fragment from RM101 obtained by self-cloning; Apr Tcr | This study |
| pAW14 | 1.0-kb SalI fragment from RM101 obtained by self-cloning; Apr Tcr | This study |
| pAW31 | 12.1-kb BamHI fragment from RM102 obtained by self-cloning; Apr Tcr | This study |
| pAW32 | 19.3-kb BamHI fragment from RM101 obtained by self-cloning; Apr Tcr | This study |
| pRM105 | pUCP28T containing a 14.7-kb EcoRI fragment from pAW32 | This study |
| pSR20 | pSK-Bluescript containing a 4.0-kb SacI fragment from pAW32 | This study |
| pDD500 | pKAS46 derivative; 2.4-kb SacI-EcoRI fragment from pSR20 containing 3′ end of oprA and 1.4 kb of downstream DNA | This study |
| pDD501 | p34E derivative; 2.1-kb BamHI fragment from pAW22 containing the 3′ end of amrR and 1.5 kb of downstream DNA | This study |
| pDD502 | pDD500 containing a 2.11-kb XbaI fragment from pDD501 (XbaI sites flank the BamHI sites in pDD501) | This study |
r, resistant; s, susceptible; AG, aminoglycoside; Tc, tetracycline; Ap, ampicillin; Sm, streptomycin; Km, kanamycin.
Tn5-OT182 mutagenesis and isolation of aminoglycoside-susceptible strains of B. pseudomallei.
Mutagenesis of B. pseudomallei was performed as described previously (11) with slight modifications in the selection of mutants. Briefly, pOT182 (Table 1) was introduced into B. pseudomallei 1026b via conjugation with E. coli SM10(pOT182). Transposon mutants of B. pseudomallei were selected by plating the conjugation mixtures onto Pseudomonas isolation agar (Difco) containing 50 μg of tetracycline per ml. The colonies that arose on this medium were picked and transfered to LB agar containing tobramycin (50 μg/ml) or streptomycin (50 μg/ml) and to a new Pseudomonas isolation agar-tetracycline plate. Colonies which did not grow on tobramycin- and streptomycin-containing media were characterized further.
Streptomycin uptake assays.
Uptake assays were performed essentially as described by Bryan and Van den Elzen (4). Broth cultures of B. pseudomallei were grown overnight at 37°C at 250 rpm in 3 ml of nutrient broth. When required, the medium contained 50 μg of tetracycline per ml. Overnight cultures were diluted 1:100 in 20 ml of nutrient broth in a 250-ml flask without antibiotic and were allowed to grow to an optical density at 600 nm of 0.4 to 0.6. When the culture was at the proper optical density, 10 ml of the culture was transferred to another flask and was allowed to incubate for another 5 min. Assays were begun by the addition of a mixture of [3H]dihydrostreptomycin (10 Ci/mmol; American Radiolabeled Chemicals Inc., St. Louis, Mo.) and unlabeled streptomycin (Sigma) to a final concentration of 50 μg of streptomycin per ml. The specific activity of streptomycin in the assay was 15 to 20 dpm of streptomycin per ng. Control experiments showed that there was no difference in uptake if dihydrostreptomycin rather than streptomycin was used as a carrier. Samples (1 ml) were removed at various time points and were filtered through a 0.45-μm-pore-size cellulose acetate membrane filter (Sartorius) held in a Millipore filter apparatus. The filters were soaked in a 2.5-mg/ml solution of streptomycin prior to the assay to prevent nonspecific binding of labeled antibiotic. Samples were washed with 15 ml of 3% NaCl. The filters were allowed to air dry and were placed in scintillation vials, and 4 ml of Readysafe liquid scintillation fluid (Beckman) was added. The radioactivity in the samples was counted in a Beckman LS6500 scintillation counter.
DNA sequencing.
DNA was sequenced by the University Core DNA Services (University of Calgary, Calgary, Alberta, Canada) and by ACGT, Inc. (Northbrook, Ill.). The DNA flanking the Tn5-OT182 insertions was isolated by self-cloning (24) and was subcloned into p-Bluescript SK (Stratagene). DNA alignments were performed with Gene Jockey, version 1.2, for the Macintosh, and homology searches were performed with the BLASTX and BLASTP programs, which were provided by the National Center for Biological Information (1).
Antimicrobial agent susceptibility testing.
The susceptibilities of B. pseudomallei strains to antimicrobial agents were determined via the agar dilution method (28) with a multiwell replicator and Mueller-Hinton II agar (Becton Dickinson Microbiology Systems) or by E tests (AB Biodisk, Solna, Sweden) according to the manufacturer’s instructions. The bacteria used for MIC determinations were resuspended in phosphate-buffered saline to a 0.5 McFarland standard.
Animal studies.
The hamster model of acute B. pseudomallei infection has been described elsewhere (3). The 50% lethal dose (LD50) for each of various B. pseudomallei strains was calculated 48 h after infection by the method of Reed and Muench (34).
Construction of pRM105.
To attempt complementation of the aminoglycoside-macrolide-susceptible phenotype of RM102, plasmid pRM105 was constructed by ligating a 14.7-kb EcoRI fragment from pAW32 into the EcoRI site of pUCP28T. The 14.7-kb fragment contained 7.9 kb of Tn5-OT182 DNA originating at the EcoRI site in Tn5-OT182 and 6.8 kb of DNA downstream of the transposon insertion terminating at a EcoRI site located downstream of the oprA gene. The fragment contains all of amrB. The resulting construct was electroporated into E. coli SM10 and was conjugated into RM102 as described previously (11). A single transconjugant, RM102(pRM105), was selected, and the aminoglycoside and macrolide MICs for the transconjugant were determined.
Construction of strain DD503.
B. pseudomallei DD503 (Table 1) was constructed for use in allelic exchange experiments with the vector pKAS46 (37). An Smr derivative of RM102 was selected by plating 100-μl aliquots of an overnight LB culture onto LB agar plates containing 100 μg of streptomycin per ml and 50 μg of tetracycline per ml. One Smr Tcr colony was selected for further studies and was named RM102-Smr. With the exception of Smr, this strain exhibited the same antibiotic susceptibility profile as RM102, which strongly suggested that Smr was due to an rpsL mutation. Plasmid pDD502 (Table 1) was conjugated from SM10λpir to RM102-Smr, and transconjugants were selected on LB agar plates containing 50 μg of tetracycline per ml and 50 μg of kanamycin per ml. One Tcr Kmr colony (RM102-Smr::pDD502) was selected for further studies and was named DD502. Several isolated colonies of DD502 were spread onto separate LB agar plates containing 100 μg of streptomycin per ml to select for transconjugants that excised pKAS46 DNA. A Smr Tcs Kms colony was selected and was named DD503.
PCR amplification of the Δ(amrR-oprA) locus of DD503.
Oligodeoxyribonucleotide primers DD503L (5′-CAGATAGTGCGACGCGGCAA-3′ [nucleotides 311 to 330 of the submitted nucleotide sequence]) and DD503R (5′-TTCGGCGAGCCCCAGTTGAT-3′ [within oprA]) were used to amplify a 575-bp product from B. pseudomallei DD503 chromosomal DNA by PCR with Taq DNA polymerase (Gibco BRL). Dimethyl sulfoxide was added to the PCR mixture at a final concentration of 10%. The conditions for PCR involved a 2-min denaturation step at 97°C, followed by 30 cycles of 97°C for 30 s, 60°C for 30 s, and 72°C for 30 s. The 575-bp PCR product was purified with the Magic PCR Preps DNA Purification System (Promega, Madison, Wis.), and DNA sequencing reactions were initiated with DD503R and DD503L.
β-Galactosidase assays.
β-Galactosidase activity was measured as described by Miller (25). Overnight cultures were diluted in LB and were grown to the early logarithmic phase. Aliquots (1 ml) were removed over the course of several hours, and the β-galactosidase activity was determined.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in the GenBank database under accession no. AF072887.
RESULTS
Isolation of aminoglycoside- and macrolide-susceptible mutants.
Prior studies of the antibiotic susceptibility of B. pseudomallei have demonstrated high levels of resistance to aminoglycoside antibiotics (14, 17). On the basis of these findings, it was hypothesized that an efflux system might be responsible for aminoglycoside resistance in B. pseudomallei. To identify proteins associated with a putative efflux system, B. pseudomallei 1026b was mutagenized with Tn5-OT182, and aminoglycoside-susceptible mutants were isolated and analyzed. Approximately 3,000 transposon mutants were screened for their susceptibilities to streptomycin and tobramycin. We identified two mutants, RM101 and RM102, which were sensitive to both antibiotics. Further examination revealed that both RM101 and RM102 were susceptible to a variety of aminoglycoside antibiotics, with a 64-fold reduction in the MIC of streptomycin, a greater than 16-fold reduction in the MIC of kanamycin, a 32- to 64-fold reduction in the MIC of tobramycin, and a greater than 128-fold reduction in the MIC of gentamicin (Table 2). In addition, both mutants showed increased susceptibility to the macrolide antibiotics erythromycin and clarithromycin but not to clindamycin. The MICs of ampicillin, benzalkonium chloride, ceftazidime, cetrimide, chloramphenicol, ciprofloxacin, nalidixic acid, protomine sulfate, rifampin, sodium dodecyl sulfate, trimethoprim, and polymyxin B for the mutants were not different from those for the parent strain, strain 1026b (data not shown). In addition, strain C21 had the same MIC profile as strain 1026b, indicating that Tn5-OT182 had no effect on susceptibility to antibiotics (data not shown).
TABLE 2.
Susceptibilities of B. pseudomallei strains to antimicrobial agents
| Antimicrobial agent | MIC (μg/ml)
|
||||
|---|---|---|---|---|---|
| 1026b | RM101 | RM102 | RM102 (pRM105) | DD503 | |
| Gentamicin | 256 | 2 | 2 | 96 | 0.75 |
| Kanamycin | 16 | <1 | <1 | 24 | NDa |
| Neomycin | 32 | <4 | <4 | ND | <4 |
| Spectinomycin | >1,064 | 64 | 64 | ND | 64 |
| Streptomycin | 1,064 | 16 | 16 | 512 | >1,024 |
| Tobramycin | 48 | 0.75 | 1.5 | 16 | 0.75 |
| Clarithromycin | >256 | 12 | 16 | 192 | 8 |
| Erythromycin | >256 | 8 | 16 | >256 | 8 |
| Clindamycin | >256 | >256 | >256 | ND | >256 |
ND, not determined.
Streptomycin uptake in RM101 and RM102.
Uptake assays demonstrated that both transposon mutants rapidly accumulated streptomycin, while the parent strain, strain 1026b, did not (Fig. 1). Strain C21, a B. pseudomallei 1026b derivative with a Tn5-OT182 insertion in a type II secretion pathway gene (gspD) (10), did not accumulate streptomycin, indicating that streptomycin accumulation in strains RM101 and RM102 was not a function of the transposon. These results are consistent with the presence of an efflux system which recognizes aminoglycosides. Uptake of streptomycin in parent strain 1026b was not enhanced by the addition of carbonyl cyanide m-chlorophenylhydrazone (CCCP) (100 μM to 5 mM) or arsenate (10 mM) (data not shown).
FIG. 1.
[3H]dihydrostreptomycin (streptomycin) uptake by B. pseudomallei 1026b (open triangles), B. pseudomallei C21 (closed triangles), B. pseudomallei RM101 (open squares), and B. pseudomallei RM102 (closed squares) in nutrient broth. OD600, optical density at 600 nm.
Analysis of DNA flanking transposon insertions in RM101 and RM102.
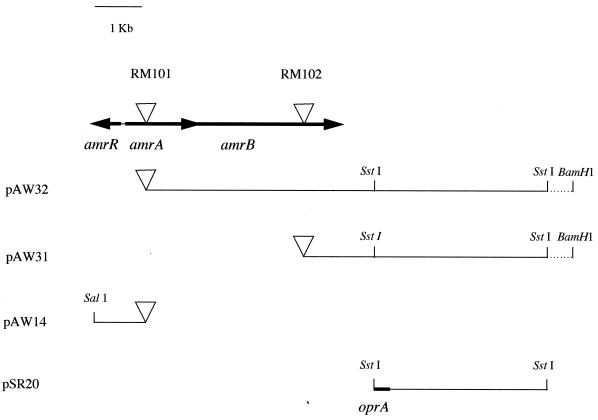
The DNA flanking the transposon insertions in both mutants was isolated by self-cloning (11, 15, 24). Three of the resulting plasmids, pAW14, pAW31, and pAW32, were sequenced, and nucleic acid and protein databases were analyzed for homologous sequences. These analyses revealed that the transposon insertions in RM101 and RM102 were located in two genes whose products showed homology to proteins belonging to the resistance, nodulation, division-type (RND-type), multidrug resistance proteins. We have given the genes involved in aminoglycoside and macrolide resistance in B. pseudomallei the prefix amr (aminoglycoside and macrolide resistance). The plasmids, the locations of the transposon insertions, and the arrangements of the amr genes are presented in Fig. 2. The transposon insertion in RM101 was in an open reading frame 1,199 nucleotides in length. This open reading frame was named amrA. The product of this open reading frame showed strong homology to the P. aeruginosa membrane fusion protein MexC (50% identity). A putative ribosome-binding sequence, GAG, was located 8 to 10 bp upstream of the potential ATG start codon. Analysis of the Tn5-OT182 insertion in RM102 revealed an open reading frame 3,131 nucleotides in length (amrB). The product of this open reading frame showed strong homology to the E. coli AcrD protein (57% identity) and the P. aeruginosa RND-type efflux protein MexB (54% identity). Analysis of the sequence downstream of amrB in pAW32 subclones revealed a gene encoding a potential outer membrane protein (Fig. 2). The partial sequence of this gene indicates a protein with strong similarity to amino acids 90 to 216 of OprJ, an outer membrane protein associated with the mexC-mexD-oprJ efflux operon in P. aeruginosa (31). We were unable to fully sequence the outer membrane gene (tentatively named oprA), presumably because of a secondary structure resulting from the high GC content (68%) of B. pseudomallei DNA (46). In addition, a fourth open reading frame immediately upstream and divergently transcribed from amrA was identified. The product of this gene, named amrR, showed strong homology (41% identity) to MtrR, a protein in N. gonorrhoeae which acts as a transcriptional repressor of the mtrRCDE efflux system and which is arranged in a similar, divergent manner (13). The amrR gene was 564 bp in length and had a putative ribosome-binding sequence, GGAG, located 10 to 13 bp upstream of the ATG start codon.
FIG. 2.
Arrangement of the amr locus in B. pseudonallei 1026b. Triangles indicate Tn5-OT182 insertions within the amr locus and the transposon location relative to the location of the cloned flanking DNA in pAW32, pAW31, and pAW14.
Complementation of RM102.
Plasmid pRM105, which contains 6.8 kb of DNA downstream of the transposon insertion in RM101, was able to restore aminoglycoside and macrolide resistance in susceptible mutant RM102. These results strongly suggest that the aminoglycoside-macrolide-susceptible phenotype in RM102 is due to the transposon insertion in this gene and that the amr operon is responsible for aminoglycoside and macrolide resistance in this organism.
lacZ transcriptional fusion analysis of RM101 and RM102.
Both mutants RM101 and RM102 were lacZ transcriptional fusions, as indicated by sequence analysis and by their high levels of β-galactosidase activity. β-Galactosidase activity increased during growth in liquid cultures for both RM101 and RM102 but was not inducible by the addition of 2 μM streptomycin. Induction of β-galactosidase activity was also examined in RM102 Smr, an rpsL derivative of RM102; however, streptomycin (100 μg/ml) did not induce β-galactosidase activity beyond the level seen in control cultures which did not receive streptomycin (data not shown). In addition, stress conditions (4% ethanol or 0.5% NaCl [22]) did not result in the induction of β-galactosidase activity in these strains (data not shown).
Characterization of B. pseudomallei DD503, an amr locus deletion mutant suitable for allelic exchange.
An amr locus deletion mutant, DD503, was constructed (Table 1). As a result of the amr locus deletion, DD503 displays the same antibiotic resistance and susceptibility profile as RM101 and RM102, thus confirming the effect of the transposon insertions in these strains. The increased susceptibility makes DD503 a useful recipient for allelic exchange experiments and allows the use of many currently available allelic exchange vectors containing the conditionally counterselectable Sms marker (rpsL+), such as pSS1129 and pKAS46 (39, 40). The antibiotic susceptibility of DD503 is similar to those of RM101 and RM102, with the exception that DD503 is Smr Tcs and lacks lacZ because Tn5-OT182 was deleted during construction of the deletion. The nucleotide sequence at the junction of the Δ(amrR-oprA) mutation in DD503 was determined by amplification and sequencing of the DNA of a PCR product generated with an amrR-specific oligodeoxyribonucleotide primer (DD503L) and an oprA-specific primer (DD503R). Although we have not determined the entire sequence of oprA, the sequence of primer DD503R was determined from the partial sequence of a subclone of pAW32 (pSR20) which contained a portion of oprA. The nucleotide sequence of the PCR product confirmed the predicted junction of the Δ(amrR-oprA) mutation extending from the 5′ end of amrR (nucleotide 550) to the oprA sequence determined from pSR20, including a 136-bp portion of the pKAS46 polylinker from XbaI to SacI.
Relative virulence of amr mutants in Syrian hamsters.
The virulence of amr mutants was assessed in a Syrian hamster model of acute B. pseudomallei infection (3). There was no significant difference in the LD50 for parent strain 1026b, RM101, RM102, and DD503. The LD50s for all strains were <10 bacteria at 2 days. In addition, all of the infected hamsters were bacteremic at the time of death. This demonstrates that transposon mutant strains RM101 and RM102 and deletion mutant DD503 remain highly virulent and suggests that the amr locus is probably not a virulence determinant in this animal model of acute B. pseudomallei infection. Similarly, the rpsL mutation (Smr) in DD503 has no effect on virulence in hamsters.
DISCUSSION
Classical mechanisms of drug resistance include enzyme modification of antibiotics, target modification, and active efflux of specific drugs (e.g., tetracycline). Somewhat recently, it has been recognized that bacteria, in addition to the methods mentioned above, use multidrug efflux systems capable of recognizing a wide range of seemingly unrelated compounds. In P. aeruginosa, at least three multidrug efflux systems which contribute to the intrinsic antibiotic resistance found in this organism exist. These systems include the MexAB-OprM system (30), the MexCD-OprJ system (31), and the MexEF-OprN system (16). The three systems overlap in their ability to efflux compounds such as tetracycline, chloramphenicol, and fluoroquinolones but show specificity toward the efflux of β-lactams and newer cephems such as cefepime and cefpirome. In other bacteria, such as N. gonorrhoeae and E. coli, multidrug efflux systems confer resistance to hydrophobic antibiotics, detergents, and dyes (13, 21, 23).
In this study we report on the identification of a multidrug efflux system in B. pseudomallei which is specific for both aminoglycoside and macrolide antibiotics. To our knowledge, this is a unique mechanism of high-level aminoglycoside resistance (9). Inactivation of either amrA or amrB in B. pseudomallei resulted in enhanced susceptibility to aminoglycoside and macrolide antibiotics. Further analysis revealed the presence of genes encoding an outer membrane protein and a putative regulatory protein. Despite the similarity between the system described here and the products of the mex genes in P. aeruginosa, there was no apparent similarity in substrate specificity. Whereas mexAB-oprM and mexCD-oprJ are associated with enhanced resistance to tetracycline, chloramphenicol, fluoroquinolones, and β-lactams, the amr locus in the B. pseudomallei system did not confer resistance to these compounds. That these genes served as an efflux pump was made evident by the accumulation of [3H]dihydostreptomycin in RM101 and RM102. The amr locus also confers resistance to macrolides such as erythromycin but not to lincosamides such as clindamycin. Efflux systems which recognize macrolides but not lincosamides have been described in Streptococcus and Staphylococcus (6, 41, 45). In addition, efflux-mediated macrolide resistance has been reported in gonococci (13), although in gonococci, the system described was not specific for macrolides and the level of resistance mediated by the efflux system was increased only fourfold. In addition, MdfA, a multidrug resistance protein in E. coli, is reported to recognize erythromycin and certain aminoglycosides, although for the E. coli strains that expressed MdfA from multicopy plasmids, only two- to fourfold increases in the MICs of these antibiotics were detected (12).
A hallmark of multidrug efflux systems in gram-negative bacteria is energy-dependent drug exclusion (29). Thus, collapse of a proton gradient across the cell membrane by the addition of CCCP should result in a marked increase in drug accumulation. Surprisingly, however, efflux of streptomycin was not inhibited by the addition of CCCP. However, we have recently sequenced a region of the B. pseudomallei chromosome that encodes a product with strong homology to emrA from E. coli (20), a locus reported to confer resistance to CCCP. We are examining this homolog in B. pseudomallei to determine if this may account for the inability of CCCP to inhibit the efflux capacity of the amr locus.
The homology of the amrR sequence with the sequences of other multidrug efflux system-negative regulators would suggest that the amrR gene product functions in this capacity as well. Construction of amrR knockout mutants should confirm this hypothesis.
At least one substrate for the amr system, streptomycin, was not able to induce β-galactosidase activity in amr-lacZ fusion strains. In addition, stress conditions (4% ethanol or 0.5% NaCl) which have been shown to increase the level of transcription of acrAB (22) did not result in the induction of β-galactosidase activity in these strains, indicating some other means of mediating amr regulation. Further work is required to identify inducers of the amr system.
We have constructed and characterized B. pseudomallei DD503, a strain that should greatly enhance the study of genetic determinants of this organism. The results of the studies with animals reported here demonstrate that DD503 exhibits a virulence capacity that is indistinguishable from that of the parent strain, strain 1026b, which is an essential characteristic for a strain that will be used in allelic exchange experiments involving virulence determinants. We have used DD503, in conjunction with pKAS46, in allelic exchange experiments to introduce defined mutations (antibiotic resistance cassettes and deletions) into the flagellin structural gene (fliC) and into a genetic locus encoding resistance to serum (10). In addition, many broad-host-range cloning vectors and antibiotic resistance cassettes encode resistance to antibiotics such as kanamycin, gentamicin, and erythromycin as selective markers. These vectors and cassettes are not useful as selective markers in B. pseudomallei because it is inherently resistant to these antibiotics by the multidrug resistance pump encoded by the amrAB oprA operon. These selective markers may be used in B. pseudomallei DD503, however, and should further facilitate the genetic analysis of this organism.
ACKNOWLEDGMENTS
This work was funded by the Canadian Bacterial Diseases Network of Centres of Excellence Program. D. DeShazer was the recipient of an Alberta Heritage Foundation for Medical Research Postdoctoral Fellowship.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bianco N, Neshat S, Poole K. Conservation of the multidrug resistance efflux gene oprM in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1997;41:853–856. doi: 10.1128/aac.41.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett P J, DeShazer D, Woods D E. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol Infect. 1997;118:137–148. doi: 10.1017/s095026889600739x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryan L E, Van den Elzen H M. Streptomycin accumulation in susceptible and resistant strains of Escherichia coli and Pseudomonas aeruginosa. J Antibiot. 1976;28:696–703. doi: 10.1128/aac.9.6.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaowagul W, White N J, Dance D A, Wattanagoon Y, Naigowit P, Davis T M, Looareesuwan S, Pitakwatchara N. Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J Infect Dis. 1989;159:890–899. doi: 10.1093/infdis/159.5.890. [DOI] [PubMed] [Google Scholar]
- 6.Clancy J, Petitpas J, Dib-Hajj F, Yaun W, Cronan M, Kamath A V, Bergeron J, Retsema J A. Molecular cloning and functional analysis of a novel macrolide-resistance determinant, mefA, from Streptococcus pyogenes. Mol Microbiol. 1996;22:867–879. doi: 10.1046/j.1365-2958.1996.01521.x. [DOI] [PubMed] [Google Scholar]
- 7.Dance D A B. Melioidosis: the tip of the iceberg? Clin Microbiol Rev. 1991;4:52–60. doi: 10.1128/cmr.4.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dance D A B. Melioidosis. In: Cook G C, editor. Manson’s tropical diseases. 20th ed. London, England: The W. B. Saunders Co. Ltd.; 1996. pp. 925–930. [Google Scholar]
- 9.Davies J, Wright G D. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997;5:234–240. doi: 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- 10.DeShazer, D., P. J. Brett, and D. E. Woods. Unpublished data.
- 11.DeShazer D, Brett P J, Carlyon R, Woods D E. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J Bacteriol. 1997;179:2116–2125. doi: 10.1128/jb.179.7.2116-2125.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagman K E, Pan W, Spratt B G, Balthazar J T, Judd R C, Shafer W M. Resistance of Neisseria gonorrhoeae to antimicrobial hydrophobic agents is modulated by the mtrRCDE efflux system. Microbiology. 1995;141:611–622. doi: 10.1099/13500872-141-3-611. [DOI] [PubMed] [Google Scholar]
- 14.Howe C, Sampath A, Spotnitz M. The pseudomallei group: a review. J Infect Dis. 1971;124:598–606. doi: 10.1093/infdis/124.6.598. [DOI] [PubMed] [Google Scholar]
- 15.Jones A L, DeShazer D, Woods D E. Identification and characterization of a two-component regulatory system involved in invasion of eukaryotic cells and heavy-metal resistance in Burkholderia pseudomallei. Infect Immun. 1997;65:4972–4977. doi: 10.1128/iai.65.12.4972-4977.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohler T, Michea-Hamzehpour M, Henze U, Gotoh N, Kocjancic Curty L, Pechere J-C. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Micro. 1997;23:345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 17.Leelarasaimee A, Bovornkitti S. Melioidosis: review and update. Rev Infect Dis. 1989;11:413–425. doi: 10.1093/clinids/11.3.413. [DOI] [PubMed] [Google Scholar]
- 18.Li X-Z, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: resistance to tetracycline, chloamphenicol, and norfloxacin. Antimicrob Agents Chemother. 1994;38:1732–1741. doi: 10.1128/aac.38.8.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li X-Z, Ma D, Livermore D M, Nikaido H. Role of efflux pump(s) in intrinsic resistance of Pseudomonas aeruginosa: active efflux as a contributing factor to β-lactam resistance. Antimicrob Agents Chemother. 1994;38:1742–1752. doi: 10.1128/aac.38.8.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomovskaya O, Lewis K. emr, an Escherichia coli locus for multidrug resistance. Proc Natl Acad Sci USA. 1992;89:8938–8942. doi: 10.1073/pnas.89.19.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma D, Cook D N, Alberti M, Pon M G, Nikaido H, Hearst J E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J Bacteriol. 1993;175:6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma D, Cook D N, Alberti M, Pon M G, Nikaido H, Hearst J E. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol Microbiol. 1995;16:45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x. [DOI] [PubMed] [Google Scholar]
- 23.Ma D, Alberti M, Lynch C, Nikaido H, Hearst J E. In the regulation of the acrAB genes of Escherichia coli by global stress signals, the local repressor AcrR plays a modulating role. Mol Microbiol. 1996;19:101–112. doi: 10.1046/j.1365-2958.1996.357881.x. [DOI] [PubMed] [Google Scholar]
- 24.Merriman T R, Lamont I L. Construction and use of a self-cloning promoter probe vector for gram-negative bacteria. Gene. 1993;126:17–23. doi: 10.1016/0378-1119(93)90585-q. [DOI] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mongkolsuk S, Rabibhadana S, Vattanaviboon P, Loprasert S. Generalized and mobilizable positive-selection cloning vectors. Gene. 1994;143:145–146. doi: 10.1016/0378-1119(94)90620-3. [DOI] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 2nd ed. Approved standard M7-T2. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 29.Nikaido H. Multidrug efflux pumps of gram-negative bacteria. J Bacteriol. 1996;178:5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poole K, Tetro K, Zhao Q, Neshat S, Heinrichs D E, Bianco N. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, Yamasaki T, Neshat S, Yamagishi J-I, Li X-Z, Nishino T. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21:713–724. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 32.Poole K, Krebs K, McNally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement or an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puthucheary S D, Parasakthi N, Lee M K. Septicemic melioidosis: a review of 50 cases from Malaysia. Trans R Soc Trop Med Hyg. 1992;86:683–685. doi: 10.1016/0035-9203(92)90191-e. [DOI] [PubMed] [Google Scholar]
- 34.Reed L J, Muench H. A simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 35.Sanford J P. Pseudomonas species (including melidosis and glanders) In: Mandell G L, Douglas R G Jr, Bennett J E, editors. Principles and practice of infectious diseases. 3rd ed. New York, N.Y: Churchill Livingstone; 1990. pp. 1692–1696. [Google Scholar]
- 36.Schweizer H P, Klassen T, Hoang T. Improved methods for gene analysis and expression in Pseudomonas spp. In: Nakazawa T, Furukawa K, Haas D, Silver S, editors. Molecular biology of pseudomonads. Washington, D.C: American Society for Microbiology; 1996. pp. 229–237. [Google Scholar]
- 37.Skorupski K, Taylor R K. Positive selection vectors for allelic exchange. Gene. 1996;169:47–52. doi: 10.1016/0378-1119(95)00793-8. [DOI] [PubMed] [Google Scholar]
- 38.Smith C J, Allen J C, Embi M N, Othman O, Razak N, Ismail G. Human melioidosis: an emerging medical problem. MIRCEN J Appl Microbiol Biotechnol. 1987;3:343–366. [Google Scholar]
- 39.Stibitz S. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 1994;235:458–465. doi: 10.1016/0076-6879(94)35161-9. [DOI] [PubMed] [Google Scholar]
- 40.Stibitz S, Black W, Falkow S. The construction of a cloning vector designed for gene replacement in Bordetella pertussis. Gene. 1986;50:133–140. doi: 10.1016/0378-1119(86)90318-5. [DOI] [PubMed] [Google Scholar]
- 41.Sutcliff J, Tait-Kamradt A, Wondrack L. Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob Agents Chemother. 1996;40:1817–1824. doi: 10.1128/aac.40.8.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsang T, Copeland V, Bowden G T. A set of cassette cloning vectors for rapid and versatile adaption of restriction fragments. BioTechniques. 1991;10:330. [PubMed] [Google Scholar]
- 43.Weinberg A N. Unusual bacterial pneumonias. In: Pennington J E, editor. Respiratory infections: diagnosis and management. 2nd ed. New York, N.Y: Raven Press; 1989. pp. 411–415. [Google Scholar]
- 44.Whitmore A, Krishnaswami C S. An account of the discovery of a hitherto undescribed infective disease occurring amoung the population of Rangoon. Indian Med Gazette. 1912;47:262–267. [PMC free article] [PubMed] [Google Scholar]
- 45.Wondrack L, Massa M, Yang B V, Sutcliffe J. Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob Agents Chemother. 1996;40:992–998. doi: 10.1128/aac.40.4.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]