Abstract
Cytochrome P450 2B6 (CYP2B6) is a human enzyme important in chemical detoxification, steroid and fatty acid metabolism that is primarily hepatic. Therefore, induction or inhibition of CYP2B6 may perturb endo- and xenobiotic metabolism and cause adverse reactions. Recent research indicates that mice lacking Cyp2b enzymes are obese with liver steatosis [1] (Heintz et al., J Nutr Biochem, 70:125–137, 2019). Current work is underway to determine the role of CYP2B6 in obesity and fatty acid metabolism, and CYP2B6 fluorescent inhibition assays were used to determine the IC50s of multiple industrial chemicals, pesticides, bile acids, steroids, and fatty acids. In many cases, inhibition of CYP3A4 was also performed in comparison because CYP3A4 is the most abundant hepatic detoxification CYP and therefore by abundance alone may also play a key role in the chemical's metabolism. Further, using the ratio of comparative potency of these compounds for CYP2B6 and CYP3A4, specificity can be estimated for these CYP2B6 inhibitors. These results indicate strong preferential inhibition (greater than 10-fold) of CYP2B6 and include lithocholic acid, arachidonic acid, atrazine, chlorpyrifos, endosulfan, parathion, and nonylphenol. Estradiol was a strong preferential inhibitor of CYP3A4. Other screened CYP2B6 inhibitors include triclosan, ticlopidine, jet fuel, docosahexaenoic acid, linoleic acid, linolenic acid, oleic acid, lithocholic acid, butylate, hexachlorocyclohexane, vinclozolin, pentachlorophenol, metalachlor, butylate, diazinon, avermectin, tribufos, ticlopidine, and bisphenol A. Documentation of xenobiotic and endobiotic inhibition by these CYPs is necessary for proper modeling of the effects of diet, chemical exposure or even mixtures on drug metabolism and potential adverse reactions.
Keywords: CYP2B6, CYP3A4, Enzyme inhibition, Lipids, Adverse drug reactions, Mixtures
Specifications Table
| Subject | Endocrinology and Metabolism |
| Specific subject area | Inhibition of CYP2B6 and CYP3A4 by endobiotics and xenobiotics |
| Type of data | Table Graph |
| How data were acquired | Gen5 fluorescent microplate reader (Synergy H1 Hybrid Reader, BioTek, Winooski, Vermont, USA). Data was stored in Excel and analyzed in GraphPad Prism 7.0 (San Diego, CA USA) |
| Data format | Raw Analyzed |
| Parameters for data collection | CYP2B6 and CYP3A4 transfected baculosomes in Vivid® CYP450 Blue and Red screening kits from Life Technologies (Carlsbad, CA) were exposed to chemical inhibitors. |
| Description of data collection | Fluorescent data was collected from a Gen5 microplate reader in kinetic mode during the performance of CYP2B6 and CYP3A4 Vivid P450 assays in the absence and presence of a number of potential CYP2B6 and CYP3A4 inhibitors. Collected data was analyzed on GraphPad Prism 7.0 to determine IC50s and Hillslopes. |
| Data source location | Biological Sciences, Clemson University, Clemson, SC 29,634 |
| Data accessibility | Public repository Repository name: Mendeley Data Data identification number: doi: 10.17632/pfgg68yd2y.5 Direct URL to data: https://data.mendeley.com/datasets/pfgg68yd2y/5 |
| Related research article | None; not at this time. |
Value of the Data
-
•
Inhibition of crucial detoxification Cytochrome P450s such as cytochrome P450 2B6 (CYP2B6) and cytochrome P450 3A4 (CYP3A4) are important for our understanding of potential CYP substrates and adverse drug reactions.
-
•
Pharmacologists, toxicologists, and endocrinologists that study endobiotic or xenobiotic (i.e., drug, pesticide) metabolism may benefit from this data.
-
•
Enclosed is a list of CYP2B6 and CYP3A4 inhibitors, their IC50s (Inhibitory Concentration that reduces activity 50%), and Hillslopes that could be used to estimate other putative xenobiotic and endobiotic inhibitors or compare inhibition between CYPs.
-
•
These data might be used to estimate how chemicals interact and increase toxicity in mixture models [2], and determine the potential for adverse drug reactions
-
•
The data provided in this manuscript could be used to determine the potential for cholestatic disease or test whether diets high in unsaturated fats might interact with chemical exposures and cause adverse reactions.
1. Data Description
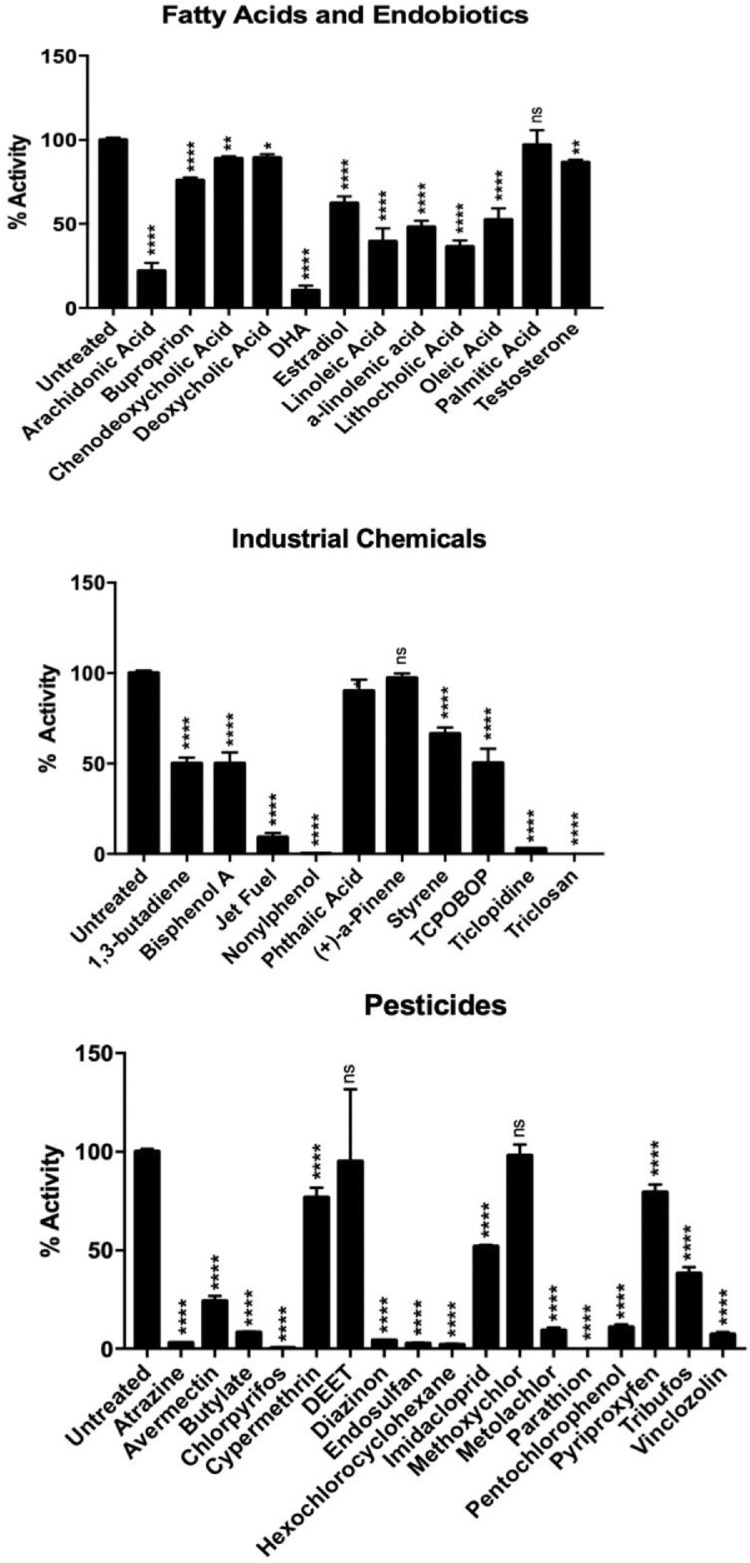
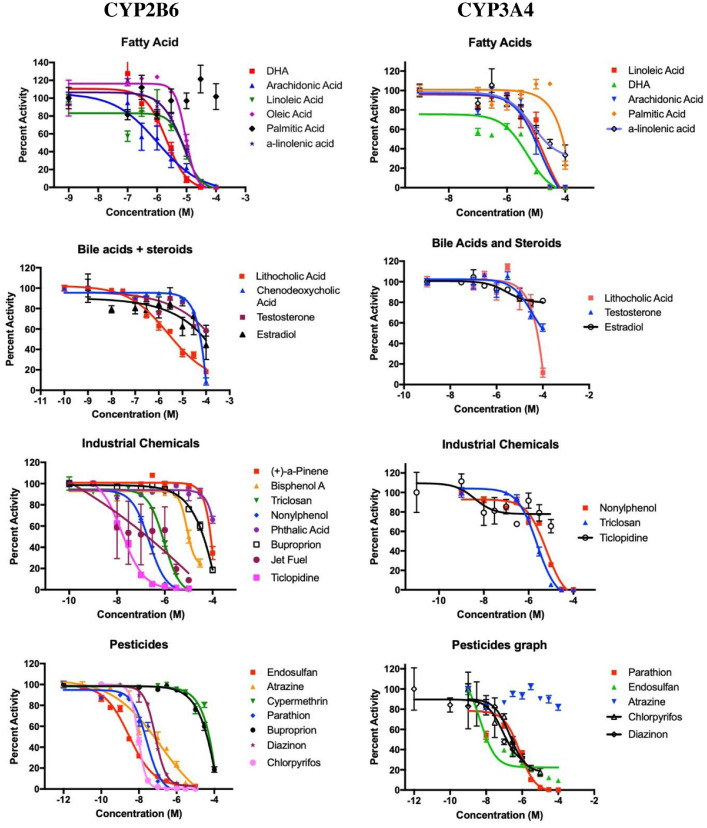
Inhibition of CYP2B6 by a number of chemicals, including fatty acids, bile acids, steroids, pharmaceuticals, industrial chemicals, and pesticides was quantified. Fig. 1 shows the results of initial screening of potential CYP2B6 inhibitors at 10 µM concentrations except the constitutive androstane receptor (CAR) activator, 1,4-Bis(3,5-Dichloro-2-pyridinyloxy)benzene (TCPOBOP) [3], which was screened at 1 µM. Fig. 2 shows the results of concentration-dependent inhibition of CYP2B6 and CYP3A4 by select strong CYP2B6 inhibitors. Relevant data such as IC50’s and Hillslopes for CYP2B6 and CYP3A4 mediated inhibition are provided in Table 1. Inhibition ratios comparing CYP2B6 and CYP3A4 inhibition are also provided. Raw and relative data from initial screening studies, as well as raw and normalized data for determining IC50’s, Hillslopes, and 95% Confidence Intervals are provided in Supplementary File 1.
Fig. 1.
Screening of CYP2B6 inhibition by endo- and xenobiotics. Percent of control activity from chemically-exposed CYP2B6-transfected baculosomes. All exposures were performed at 10 µM with the exception of TCPOBOP, which was exposed at 1 µM. (A) Fatty acids and other endobiotics, (B) industrial chemicals and pharmaceuticals, and (C) pesticides. Statistical differences in absorbance were determined by ANOVA followed by fisher's LSD as the post-hoc test. (n = 2–14) *indicates p-value < 0.05; **indicates p-value < 0.01; ***indicates p-value < 0.001; ****indicates p-value < 0.0001.
Fig. 2.
Concentration-dependent inhibition of CYP2B6 and CYP3A4. CYP2B6 or CYP3A4 containing baculosomes were exposed to fatty acids, bile acids, steroids, pesticides, industrial chemicals or pharmaceuticals at increasing concentrations. IC50’s and Hillslopes were determined as described in the materials and methods using GraphPad Prism 7.0 (n = 3–12).
Table 1.
IC50 and 95% confidence intervals (CI) of select CYP2B6 and CYP3A4 inhibitors.
| CYP2B6 |
CYP3A4 |
||||||
|---|---|---|---|---|---|---|---|
| Chemical | IC50(µM) | 95% CI (µM) | Hillslope | IC50 (µM) | 95% CI (µM) | Hillslope | IC50 Ratio1 |
| Fatty acids | |||||||
| Arachidonic acid | 1.04 | 0.20–5.35 | −0.59 | 13.7 | 7.21–21.62 | −4.08 | + 13.2 |
| Docosahexaenoic acid | 2.06 | 1.30–3.27 | −1.22 | 5.08 | 2.66–9.69 | −5.0 | + 2.47 |
| α-Linolenic acid | 7.32 | 5.91–9.30 | −1.45 | 4.04 | 0.427–38.3 | −0.701 | − 2.36 |
| Linoleic acid | 9.19 | 5.23–16.1 | −0.69 | 18.3 | 10.10–33.10 | −4.46 | + 1.99 |
| Oleic acid | 9.30 | 7.18–12.10 | −2.45 | np | np | np | np |
| Palmitic acid | nd | nd | nd | 19,890 | nd | nd | nd |
| Bile acids and steroids | |||||||
| Chenodeoxycholic acid | 49.0 | 48.3–62.8 | −1.32 | np | np | np | np |
| Lithocholic acid | 2.47 | 0.924–6.61 | −0.51 | 3233 | 3.5E-6–2.9E7 | −3.26 | +1313 |
| Testosterone | 90.2 | 64.4–126.4 | −0.32 | 33.9 | 12.53–91.52 | −4.73 | −2.66 |
| Estradiol | 42.2 | 24.8–71.6 | −0.33 | 2.78 | 0.617–12.5 | −0.903 | −15.2 |
| Pesticides | |||||||
| Endosulfan | 0.0030 | 0.0024–0.0037 | −0.66 | 0.00460 | 0.0025–0.0086 | −0.261 | +1.53 |
| Parathion | 0.026 | 0.021–0.032 | −1.13 | 0.729 | 0.369–1.439 | −0.094 | +28.03 |
| Atrazine | 0.144 | 0.022–0.952 | −0.34 | nd | nd | nd | +++ |
| Cypermethrin | 6040 | nd | −0.79 | np | np | np | np |
| Chlorpyrifos | 0.0092 | 0.0087–0.0097 | −2.09 | 0.280 | 0.0050–1.57 −0.507 | +30.4 | |
| Diazinon | 0.0754 | 0.0675–0.0840 | −1.51 | 0.0665 | 0.022–0.199 | −2.52 | −1.13 |
| Other chemicals | |||||||
| (+)-α-Pinene | 292 | 2.0E-12–4.2E16 | −1.74 | np | np | np | np |
| Buproprion | 50.7 | 2.74–9.4E4 | −0.70 | np | np | np | np |
| Ticlopidine | 0.0196 | 0.0174–0.0219 | −0.95 | nd | nd | nd | nd |
| Phthalic acid | 1170 | nd | −1.71 | np | np | np | np |
| Bisphenol A | 8.44 | 6.50–10.90 | −2.65 | np | np | np | np |
| Nonylphenol | 0.221 | 0.185–0.263 | −1.21 | 6.25 | 4.43–8.81 | −1.02 | + 28.30 |
| Triclosan | 0.944 | 0.693–1.28 | −1.30 | 2.39 | 1.80–3.17 | −1.46 | + 2.53 |
| Jet fuel | 0.171 | 0.062–0.458 | −0.08 | np | np | np | np |
Ratio: is the ratio of the IC50 values from the two CYPs. + indicates greater affinity for CYP2B6 and – indicates greater affinity for CYP3A4.
np: Assay not performed
nd: Parameters were not determined because of poor inhibition.
All chemicals with an IC50 below 5.0 µM for CYP2B6 were also investigated for CYP3A4 inhibition.
Statistics determined using GraphPad Prism 7.0 as described in materials and methods (San Diego, CA).
Supplementary data File 1 is an excel spread sheet that includes: (1) The percent inhibition from screening multiple chemicals used to produce the graphs for Fig. 1. (2) The raw data from the CYP2B6 and CYP3A4 fluorescent inhibition assays on separate pages, and (3) the normalized data for the CYP2B6 and CYP3A4 fluorescent inhibition assays used to produce the graphs in Fig. 2 and determine the Hillslopes and IC50s recorded in Table 1 (available in Mendeley; doi: 10.17632/pfgg68yd2y.4 for all data used and unused and Mendeley; doi: 10.17632/pfgg68yd2y.5 for all used data).
2. Experimental Design, Materials and Methods
2.1. CYP2B6/CYP3A4 inhibition
CYP2B6- and CYP3A4-transfected baculosomes from Vivid® CYP450 Blue and Red screening kits, respectively, were obtained from Life Technologies (Carlsbad, CA, USA). Potential inhibitors (and substrates) were screened as described in the Vivid® CYP450 Screening Kits User Guide. Decreased fluorescence due to chemical inhibition was measured on a Gen5 microplate reader (Synergy H1 Hybrid Reader, BioTek, Winooski, Vermont, USA) at 415/460 nm excitation/emission (CYP2B6) and 550/590 excitation/emission (CYP3A4) at automatic read intervals of 50 s for a 30 min duration in kinetic assay mode. Results were exported into Microsoft Excel, where initial fluorescence was subtracted from fluorescence within the linear part of the curve to calculate change over time and averaged between replicates.
First, screening of several potential CYP2B6 inhibitors was performed using 10 µM concentrations of all chemicals except TCPOBOP, which was screened at 1 µM (n = 2–14). Fatty acids and other endobiotics, pesticides, pharmaceuticals, and industrial chemicals were screened for their inhibitory effects CYP2B6 (Fig. 1). Several of the CYP2B6 inhibitors were further evaluated in concentration-dependent studies to confirm inhibition and determine IC50s and Hillslopes (Fig. 2; Table 1). Strong inhibitors were considered chemicals with IC50’s below 5 µM for CYP2B6. CYP2B6 is not expressed as highly as CYP3A4, the liver's most abundant CYP [4,5]; therefore, we compared the IC50’s of CYP2B6 and CYP3A4 for chemicals with IC50’s below 5 µM for CYP2B6 (Fig. 2)(n = 3–12). Table 1 provides the IC50’s and Hillslopes for CYP2B6 and CYP3A4 mediated inhibition. Inhibition ratios comparing CYP2B6 and CYP3A4 inhibition are also provided. Raw and relative data used to determine significance, IC50’s, Hillslopes, and 95% Confidence Intervals is provided in Supplementary File 1.
2.2. Statistics
Data are presented as mean ± standard error of the mean (SEM). Statistical significance was determined by one-way ANOVA followed by Fisher's LSD as the post-hoc test. Statistical analysis was performed with GraphPad Prism 7.0 (San Diego, CA, USA) with a p-value < 0.05 considered statistically significant. Decreased fluorescence due to chemical inhibition was quantified on a Gen5 microplate reader (Synergy H1 Hybrid Reader, BioTek, Winooski, Vermont, USA) at 415/460 nm excitation/emission at 30 s intervals for 30 min in kinetic assay mode in accordance with manufacturer's protocol. IC50 values were determined as described previously by us using GraphPad Prism 7.0 (Graphpad Software, San Diego, CA, USA) [6,7]. Briefly, chemical concentrations were log10 transformed, sigmoidal concentration-response curves were fit with a variable slope model with least squares ordinary fit. Confidence intervals were produced assuming asymmetrical distribution as recommended by GraphPad.
Ethics Statement
No data was collected from animal experimentation, human subjects, or social media platforms for this study.
CRediT authorship contribution statement
Emily M. Olack: Methodology, Investigation, Visualization, Writing – original draft. Melissa M. Heintz: Investigation. William S. Baldwin: Conceptualization, Writing – review & editing, Data curation, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have or could be perceived to have influenced the work reported in this article.
Acknowledgments
Funding was provided by National Institutes of Health (NIH) grants R15ES017321. The authors would like to thank Dr. Elizabeth Litoff for her technical help.
References
- 1.Heintz M.M., Kumar R., Rutledge M.M., Baldwin W.S. CYP2B-null male mice are susceptible to diet-induced obesity and perturbations in lipid homeostasis. J. Nutr. Biochem. 2019;70:125–137. doi: 10.1016/j.jnutbio.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rider C.V., LeBlanc G.A. An integrated addition and interaction model for assessing toxicity of chemical mixtures. Toxicol. Sci. 2005;87:520–528. doi: 10.1093/toxsci/kfi247. [DOI] [PubMed] [Google Scholar]
- 3.Tzameli I., Pissios P., Schuetz E.G., Moore D.D. The xenobiotic compound 1,4-bis[2-(3,5-dichloropyridyloxy)]benzene is an agonist ligand for the nuclear receptor car. Mol. Cell. Biol. 2000;20:2951–2958. doi: 10.1128/mcb.20.9.2951-2958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H., Tompkins L.M. CYP2B6: new insights into a historically overlooked cytochrome P450 isozyme. Curr. Drug Metab. 2008;9:598–610. doi: 10.2174/138920008785821710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin W.S., Roling J.A. A concentration addition model for the activation of the constitutive androstane receptor by xenobiotic mixtures. Toxicol. Sci. 2009;107:93–105. doi: 10.1093/toxsci/kfn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmidt A.M., Sengupta N., Noorai R.E., Saski C.A., Baldwin W.S. RNA sequencing indicates that atrazine induces multiple detoxification genes in daphnia magna and this is a potential sources of its mixtures interactions with other chemicals. Chemosphere. 2017;189:699–708. doi: 10.1016/j.chemosphere.2017.09.107. [DOI] [PMC free article] [PubMed] [Google Scholar]