Summary
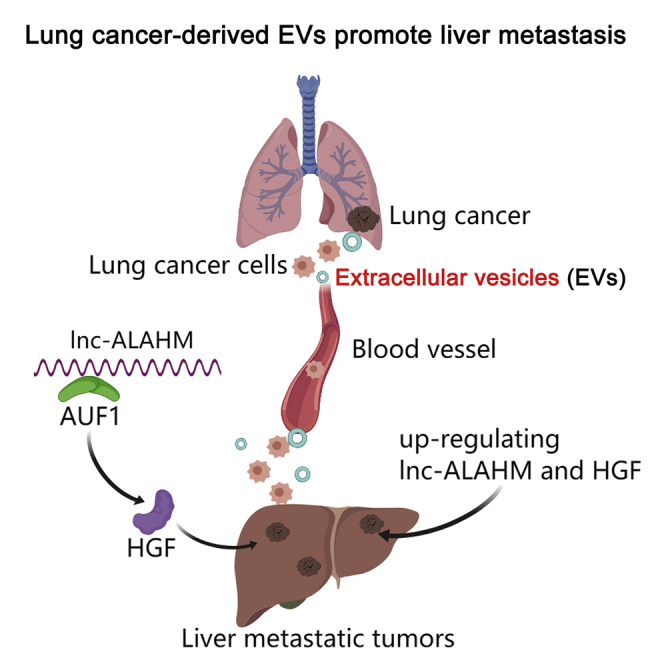
Tumor-derived extracellular vesicles (EVs) are involved in tumor metastasis. Highly enriched lncRNA-ALAHM was identified from serum EVs of lung adenocarcinoma (LUAD) patients with liver metastasis by high-throughput sequencing. A mouse model of in situ lung cancer was used to determine the effect of ALAHM in LUAD cell EVs on liver metastasis. The effects of ALAHM on hepatocyte paracrine HGF as well as proliferation, invasion, and migration of LUAD cells were observed in vitro. As results, ALAHM expression in LUAD cell EVs was significantly increased. LUAD-cell-derived EVs overexpressing ALAHM significantly promoted lung cancer liver metastasis in model mice. ALAHM of LUAD cell EVs also promotes hepatocyte parasecretion of HGF by binding with AUF1 and increases the proliferation, invasion, and migration of LUAD cells. Thus, LUAD-cell-derived EVs containing ALAHM causes increasing HGF and promoting liver metastasis of LUAD cells.
Subject areas: Microenvironment, Molecular biology, Cancer
Graphical abstract
Highlights
-
•
Higher lnc-ALAHM was identified from serum EVs of LUAD patients with liver metastasis
-
•
ALAHM of LUAD-cell-derived EVs promotes liver metastasis by increasing HGF
-
•
ALAHM of LUAD-cell-derived EVs promotes hepatocyte secretion of HGF by binding AUF1
Microenvironment; Molecular biology; Cancer
Introduction
Lung cancer is still one of the most common malignant tumors with high incidence and mortality in the world (Siegel et al., 2021). It is classified into two subtypes: small-cell lung carcinoma (SCLC) and non-small-cell lung carcinoma (NSCLC), and NSCLC is more prevalent than another. Most patients with advanced non-small-cell lung cancer (NSCLC) die within 18 months of diagnosis, with an overall five-year survival rate of only 15%. Lung cancer metastasis is the main cause of patient death (Wood et al., 2014). The liver is a common metastatic organ of NSCLC, as well as an organ prone to various metastatic tumors (Brodt, 2016). There is a long incubation period from the primary tumor to a diagnosis of liver metastasis. Patients with early liver metastasis of lung cancer show almost no symptoms (Tamura et al., 2015), and metastatic cells of NSCLC are fully adapted to survive and proliferate at sites of liver metastasis. Moreover, many patients with NSCLC liver metastases are already at an advanced stage when they are diagnosed, and the survival time is significantly shortened; in fact, most patients died within 6 months (Riihimäki et al., 2014). Furthermore, patients with NSCLC liver metastasis frequently have metastasis to other organs (Yang et al., 2019). This confounding factor is the main reason for the short survival and high mortality of patients with NSCLC liver metastasis.
At present, treatment methods for liver metastasis of lung cancer are mainly divided into either surgical or nonsurgical treatment, both of which have certain limitations including poor long-term efficacy, high recurrence rate, and strict requirements on the location and size of the tumor (Pan et al., 2017). Most patients with advanced metastatic lung cancer are incurable under current therapeutic schemes. Therefore, it is of great significance for the early diagnosis and treatment of liver metastasis of lung cancer to deeply understand the molecular mechanism underlying liver metastasis of lung cancer and to seek and develop new diagnostic molecular markers and effective therapeutic targets.
In recent years, the study of NSCLC metastasis has gradually focused on the tumor microenvironment (TME) (Zhang et al., 2021). TME is the soil for tumor occurrence and development containing many components such as immune cells, inflammatory mediators, and substances secreted by tumor cells, which together affect the growth, invasion, and metastasis of tumor cells (Saxena and Singh, 2021). Intercellular communication in the TME is mainly accomplished by exosomes (Xu et al., 2018), which has been substituted by extracellular vesicles (EVs) according to the 2018 guidelines for studies on extracellular vesicles and exosomes (Théry et al., 2018). Therefore, in this paper, we substituted EVs for exosomes in our study.
Endogenous EVs such as exosomes are a class of extracellular vesicles with a diameter of 30–100 nm that carry small molecules, including proteins, nucleic acids, and lipids, into target cells by endocytosis (Gebeyehu et al., 2021; Xie et al., 2019). A complex EVs communication network between tumor cells and nontumor cells is involved in every step of the whole process from tumor genesis and growth to tumor cell diffusion and metastasis (Meehan and Vella, 2016). Tumor-cell-derived EVs not only exist in large quantities in the microenvironment around primary tumors to promote tumor cell proliferation or immune escape to support tumor growth but also enter microvessels and reach distal organs to form a microenvironment suitable for tumor cell growth to promote tumor metastasis (Wang et al., 2018). At present, a large number of studies have confirmed that lung-cancer-derived EVs are involved in lung cancer metastasis through various pathways, and they are expected to become therapeutic targets and with predictive value in diagnosis, prognosis, and anti-tumor drug resistance (Jiang et al., 2021; Yin et al., 2021; Taverna et al., 2017).
Researchers have devoted significant efforts over the years to reveal the regulating mechanism of EVs in tumor development and identify and develop new and promising biomarkers and therapeutic targets for clinical use. However, studies on EVs in tumor cell metastasis to specific organs, especially liver metastasis, remain lacking. Therefore, we investigated the role of EVs in liver metastasis in lung adenocarcinoma (LUAD) and found that LUAD-cell-derived EVs expresses lncRNA ALAHM and participates in promoting liver metastasis.
Result
Serum-circulating EV-specific lncRNAs are associated with liver metastasis of LUAD
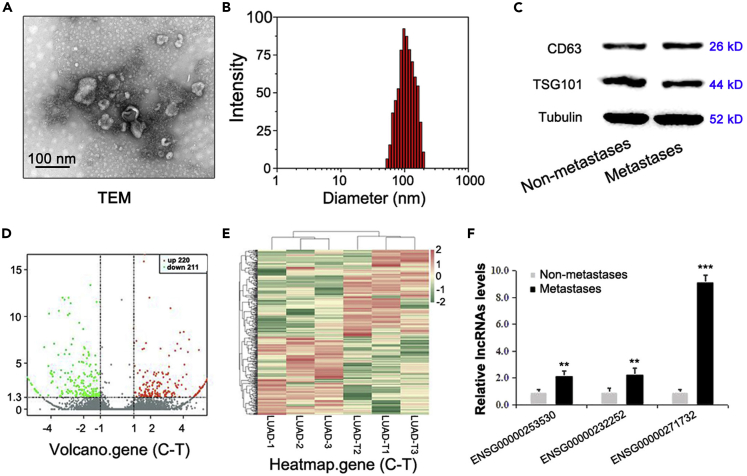
Serum EVs were isolated from patients with LUAD and liver metastasis (without metastasis to other organs) and patients with LUAD without metastasis (three vs. three) (refer to Table 1 for patient details). The morphology, size, and concentration of EVs were determined using TEM and NTA, and Western blotting was used to detect the expression of the EV marker proteins TSG101 and CD63 with the reference "housekeeping" gene tubulin (Representative pictures are shown in Figures 1A–1C).
Table 1.
Clinical characteristics of lung adenocarcinoma patients
| Clinical factors | None metastasis (n = 3) | Liver metastasis (n = 3) |
|---|---|---|
| Age (years) | 65.3 (mean) | 62.7 (mean) |
| Gender (Male/Femle) | 1/2 | 1/2 |
| Smoking (Y/N) | 2/1 | 1/2 |
| Primary (Single or Multiple) | S | S |
| Tumor diameter (cm) | 3.26 (mean) | 3.41 (mean) |
| Metastatic (Single or Multiple) | 0 | 1/2 (S/M) |
| Other organ metastasis | None | None |
| Histology of other cancers | None | None |
Figure 1.
Identification of specific lncRNAs related to liver metastasis in serum EVs of patients with LUAD
After serum EVs were isolated from LUAD patients with or without liver metastasis (three vs. three), EVs samples were detected using TEM and NTA, and the marker proteins TSG101 and CD63 were detected by western blotting. The level of tubulin was used as an internal control. The representative pictures are shown in (A–C). EVs samples underwent transcriptome sequencing, and statistically significant DEGs were identified and are shown in a volcano plot (D) and heatmap (E). The top three lncRNAs in EVs samples were identified, and real-time RT-PCR was used to evaluate their differential expression in EVs of LUAD patients accompanying or without liver metastasis (ten versus ten), of which the one with the highest overexpression was named ALAHM (F). Bar graphs represent ratios plotted as mean ± SD of three separate experiments. Compared with the control group, ∗∗p < 0.01 and ∗∗∗p < 0.001.
The raw sequencing data for the six patients' samples have been uploaded as Sequence Read Archive (SRA) submission (Accession: SUB10791255) through NCBI. A volcano plot (Figure 1D) and heatmap of differentially expressed genes (DEGs) (Figure 1E) were used to analyze the overall distribution of significant DEGs among samples. A list of DEGs of ncRNAs is shown in Table S1.
LncRNAs with significantly increased expression in serum EV samples of LUAD patients with liver metastasis versus those without metastasis were screened using high-throughput sequencing data and the following metrics: original signal value ≥20 and FC ≥ 1.5. As a result, lncRNA ENSG00000271732, ENSG00000253530, and ENSG00000232252 were identified in clinical serum EV samples (n = 20) and validated by real-time RT-PCR (The patients information are shown in Table 2) (All the primers sequences for genes are shown in Table 3). Our results showed that lncRNA ENSG00000271732 (ENST00000607700.1; Location (hg38): chr1:16617391-16617729; Sequence Ontology term: lincRNA; Transcript size: 339 bp) had the greatest difference (Figure 1F), and because of its possible role in liver metastasis of LUAD, it was named lncRNA ALAHM for associated lung adenocarcinoma with hepatic metastasis (abbreviated as lnc-ALAHM or ALAHM).
Table 2.
Clinical characteristics of lung adenocarcinoma patients
| Clinical factors | None metastasis (n = 10) | Liver metastasis (n = 10) |
|---|---|---|
| Age (years) | 68.8 (mean) | 70.9 (mean) |
| Gender (Male/Female) | 5/5 | 5/5 |
| Smoking (Y/N) | 4/6 | 3/7 |
| Primary (Single or Multiple) | S | S |
| Tumor diameter (cm) | 3.14 (mean) | 3.36 (mean) |
| Metastatic (Single or Multiple) | 0 | 7/3 (S/M) |
| Other organ metastasis | None | None |
| Histology of other cancers | None | None |
Table 3.
Primer sequences for lncRNAs, HGF, and GAPDH for qRT-PCR
| Primer name | Primer sequence (5–3′) |
|---|---|
| ENSG00000271732-Fwd | TTCCGAAGGATTGGGTCT |
| ENSG00000271732-Rev | ACACAGAATCCCTCCTGGAAG |
| ENSG00000253530-Fwd | GCAGGTCCGCCAGTCAAA |
| ENSG00000253530-Rev | ACAAGAAGTGCCAGGAGAGAG |
| ENSG00000232252-Fwd | ACACCCAGTTGAAGAAAT |
| ENSG00000232252-Rev | AGCAATCCATCAAAAGATAAA |
| HGF-Fwd | GCTATCGGGGTAAAGACCTACA |
| HGF-Rev | CGTAGCGTACCTCTGGATTGC |
| GAPDH-Fwd | GAAGGTGAAGGTCGGAGTC |
| GAPDH-Rev | GAAGATGGTGATGGGATTTC |
ALAHM is highly expressed in EVs secreted by LUAD cells and uptaken by normal liver cells
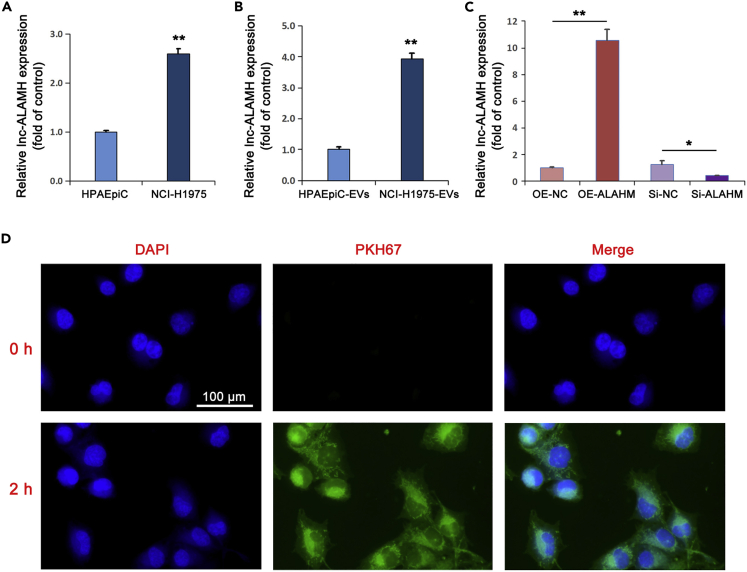
Real-time RT-PCR was used to detect the differential expression of ALAHM in lung cancer NCI-H1975 cells and normal alveolar epithelial HPAEpiC cells. The expression level of ALAHM in NCI-H1975 cells was significantly upregulated than the level in HPAEpiC cells (Figure 2A). EVs of NCI-H1975 and HPAEpiC cells were isolated and detected by TEM (Figure S1). The expression levels of ALAHM were determined by real-time RT-PCR. Similarly, we found increased expression of ALAHM in EVs of NCI-H1975 cells than HPAEpiC cells (Figure 2B). Next, ALAHM overexpression and knockdown NCI-H1975 cell lines were constructed by lentivirus and siRNA, respectively, and ALAHM expression levels in EVs secreted by cells were detected by real-time RT-PCR. Significantly increased expression of ALAHM was found in cells from the overexpression group than cells from the control group, whereas the level was significantly decreased in cells from the knockdown group (Figure 2C). Next, NCI-H1975-cell-derived EVs were labeled with PKH67 and co-cultured with normal liver L02 cells for 2 h and observed by fluorescence microscopy to determine whether uptake of EVs occurred by liver cells (Figure 2D).
Figure 2.
Detection of ALAHM expression in EVs of LUAD cells and EV uptake of normal liver cells
Real-time RT-PCR was used to confirm expression differences in expression levels of ALAHM and EVs-ALAHM in HPAEpiC and NCI-H1975 cells, respectively (A and B). After overexpression or knockdown of ALAHM in NCI-H1975 cells, expression levels of EVs-ALAHM were detected by real-time RT-PCR (C). EVs derived from NCI-H1975 cells were co-cultured with L02 cells. EVs uptake by L02 cells was observed under a fluorescence microscope and images were taken (D). Bar graphs represent ratios plotted as mean ± SD of three separate experiments. Compared with the control group, ∗p < 0.05 and ∗∗p < 0.01.
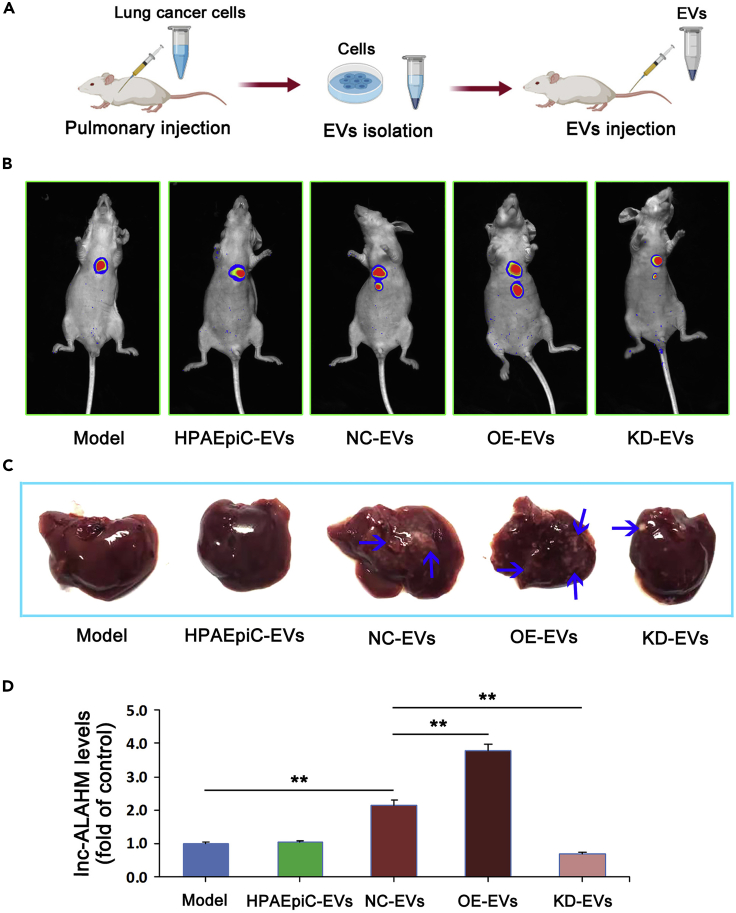
Animal experiments verified that EVs-ALAHM promotes liver metastasis of lung cancer cells
A mouse model of in situ lung cancer was established, and normal lung epithelial HPAEpiC cell EVs (HPAEpiC-EVs), NCI-H1975 cell EVs (NC-EVs), ALAHM-overexpressing NCI-H1975 cell EVs (OE-EVs), and ALAHM knockdown NCI-H1975 cell EVs (KD-EVs) were injected, followed by detection 4 weeks after modeling. The primer sequences for pshR-ALAHM and siRNA-ALAHM are listed in Table 4. An overview of the mouse model experiments is shown in Figure 3A. For live imaging in small animals, we used Fusion FX.EDGE chemiluminescence imaging (VILBER Co., Ltd., France) to detect the formation of in situ lung cancer and liver metastases. Compared with the model (control) group, HPAEpiC-EVs did not promote liver metastasis of lung cancer, whereas lung-cancer-cell-derived EVs (NC-EVs) promoted liver metastasis. Remarkably, OE-EVs administration further promoted liver metastasis of lung cancer, whereas KD-EVs inhibited lung cancer liver metastasis (Figure 3B).
Table 4.
Primer sequences for pshR-ALAHM and siRNA-ALAHM
| Primer name | Primer sequence (5–3′) |
|---|---|
| ShR-ALAHM-F | GATATCCTTCCAGGAGGGATTCTGTG |
| ShR-ALAHM-R | TGATTTATTCCATTCCAAAACTCGAG |
| siRNA-ALAHM | AACATGTGTTAGCCTGATATTTA |
| Negative Control | TGCGCTAGGCCTCGGTTGC |
Figure 3.
Mouse model of in situ lung cancer and EVs-ALAHM promotion of liver metastasis in vivo|
An overview of the mouse model experiments (A). Small-animal imaging detected the formation of in situ lung cancer and liver metastases (B). Representative images are shown. The red fluorescent protein (RFP)-expressing areas are tumors of in situ lung cancer and liver metastases. After livers were removed from model mice, metastasis formation of liver gross specimens was observed and photographed. Metastatic foci are shown by blue arrows (C). The expression of ALAHM in liver tissue was detected by real-time RT-PCR (D). Bar graphs represent ratios plotted as mean ± SD of three separate experiments. Compared with the control group, ∗∗p < 0.01.
The liver metastasis formation was evaluated after liver removal from model mice at the end of the experiment (Figure 3C). We found no significant liver metastases in the model and HPAEpiC-EVs groups. Liver metastasis was observed in the NC-EVs group, but the number of metastases was lower than in the OE-EVs group. More than one significantly fully differentiated liver metastasis was found in mice from the OE-EVs group, whereas no fully differentiated liver metastasis was found in the KD-EVs group.
Real-time RT-PCR was used to detect the expression of ALAHM in liver tissue specimens of mice from different groups. Compared with the model group, there was no difference in the ALAHM expression level in the HPAEpiC-EVs group, whereas ALAHM expression was significantly upregulated in the NC-EVs group and even higher in the OE-EVs group. The expression of ALAHM in the KD-EVs group was significantly downregulated compared with the NC-EVs and OE-EVs groups (Figure 3D).
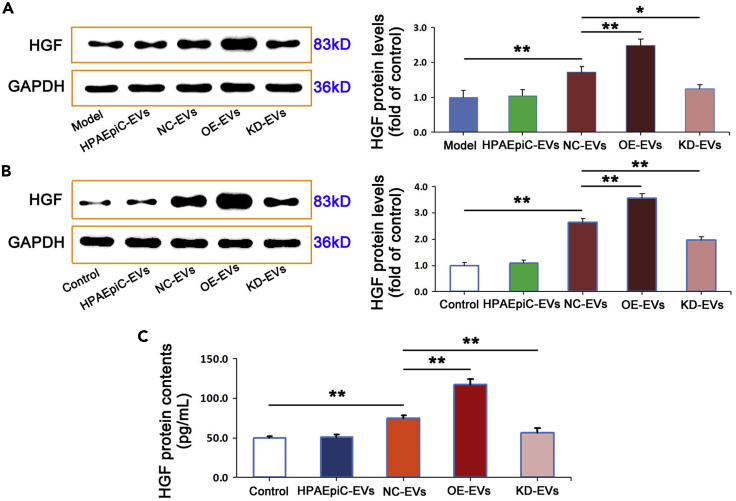
EVs-ALAHM promotes hepatocyte parasecretion of HGF and the proliferation, migration, and invasion of lung cancer cells
To further study the mechanism underlying EVs-ALAHM in liver metastasis of lung cancer, HGF expression in liver tissue specimens of nude mice was detected by western blot. There was no difference in HGF protein expression levels between the model group and HPAEpiC-EVs. It was found that significantly increased HGF levels in mice from the OE-EVs group, with lower but still significantly high levels of expression in mice from the NC-EVs group. In the KD-EVs group, HGF protein expression was significantly downregulated (Figure 4A).
Figure 4.
EVs-ALAHM promoted HGF parasecretion by liver cells
Western blotting was used to detect protein levels of HGF in liver tissue specimens of model mice (A). After L02 cells were co-incubated with different EVs extracts, HGF levels in L02 cells were measured by western blotting (B) and in the supernatant of cell culture medium by ELISA (C). GAPDH was used as a loading control. Representative bands or images are shown. For immunohistochemical results, brown staining indicates strong protein expression and blue staining indicates weak. Ratios were plotted as mean ± SD of three separate experiments. Compared with the control group, ∗p < 0.05 and ∗∗p < 0.01.
Next, the expression levels of HGF were detected by western blotting in liver L02 cells after co-incubation with HPAEpiC cell EVs and NCI-H1975 cell EVs with overexpression or knockdown ALAHM. Compared with the model group, there was no difference in the expression level of HGF in the HPAEpiC-EVs group. However, HGF protein expression was significantly upregulated in the NC-EVs group and further increased in the OE-EVs group. In contrast, the level of HGF in the KD-EVs group was significantly downregulated compared with that in the NC-EVs and OE-EVs group (Figure 4B).
In addition, we evaluated the expression of hepatocellular paracrine HGF after EVs co-incubation with L02 hepatocytes using ELISA and found that the level of HGF in the supernatant of L02 cells did not change following co-incubation with HPAEpiC-EVs. In contrast, HGF levels were significantly increased in the NC-EVs and OE-EVs groups, with a greater expression found in the OE-EVs group. We also found that co-incubation with KD-EVs significantly downregulated the expression of HGF in the supernatant of L02 cells (Figure 4C).
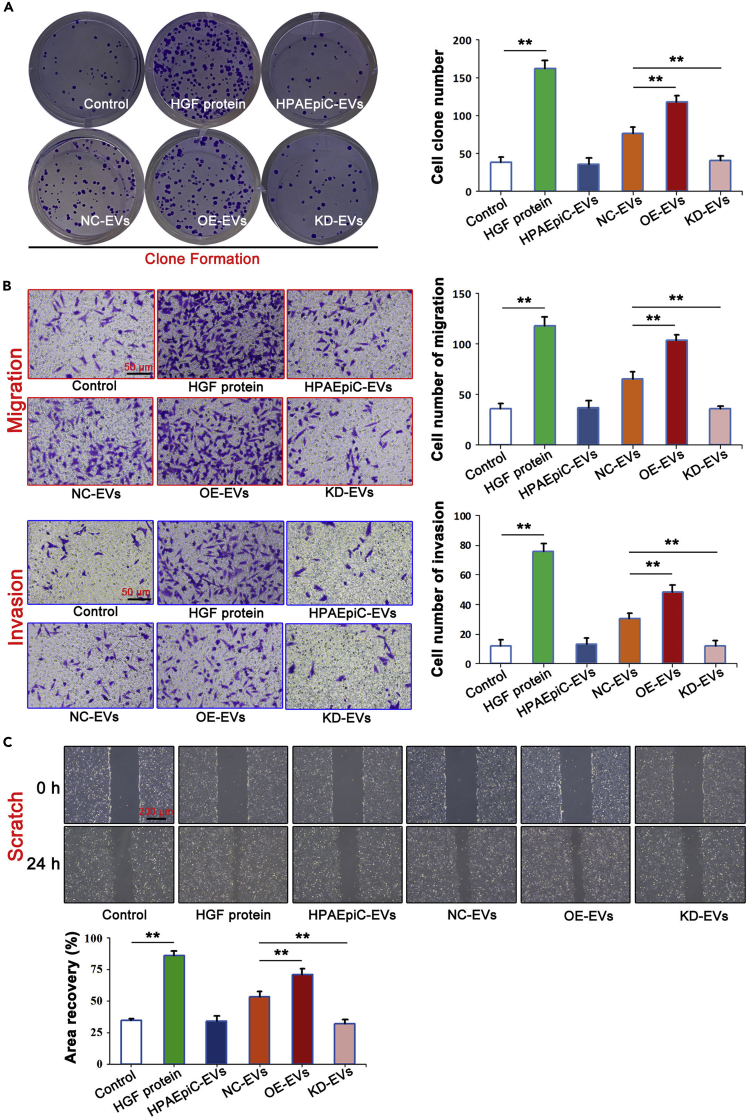
To confirm that EVs-ALAHM of NCI-H1975 cells causes hepatocyte para-secretion of HGF, and thus, promotes the proliferation, migration, and invasion of lung cancer cells, NCI-H1975 cells were co-cultured with HGF, HPAEpiC-EVs, OE-EVs, or KD-EVs, and cell proliferation was detected by clone formation (Figure 5A). Compared with the control group, HGF protein significantly increased the proliferation of NCI-H1975 cells, whereas there was no difference in the HPAEpiC-EVs group. The proliferation ability of NCI-H1975 cells in the NC-EVs group was significantly upregulated, with even greater upregulation in the OE-EVs group, whereas in the KD-EVs group, the proliferation ability of NCI-H1975 cells was significantly downregulated.
Figure 5.
EVs-ALAHM enhanced proliferation, migration, and invasion of lung cancer cells
NCI-H1975 cells were co-cultured with HGF or different EVs isolation, and the proliferation of NCI-H1975 cells was detected by clone formation (A). Transwell assay was used to detect differences in migration and invasion abilities of NCI-H1975 cells (B), and scratch testing was used to observe the migration abilities of NCI-H1975 cells under a light microscope (C). Bar graphs represent ratios plotted as mean ± SD of three separate experiments. Compared with the control group, ∗∗p < 0.01.
Finally, the Transwell assay and Scratch assay were used to evaluate the migration and invasion abilities of NCI-H1975 cells (Figures 5B and 5C). The results showed that HGF protein significantly increased the migration and invasion abilities of NCI-H1975 cells. Regarding invasiveness, no change was found in the HPAEpiC-EVs group, but the invasion ability of NCI-H1975 cells in the NC-EVs and OE-EVs groups was significantly increased, with greater invasiveness found in the OE-EVs group. The invasion ability of NCI-H1975 cells was significantly decreased in the KD-EVs group. The results of the Scratch assay were the same as those of the Transwell assay. HGF treatment significantly increased the migration of NCI-H1975 cells, and the migration of cells was significantly increased in the OE-EVs group, but decreased in the KD-EVs group.
ALAHM promotes HGF expression by binding transcription factor AUF1
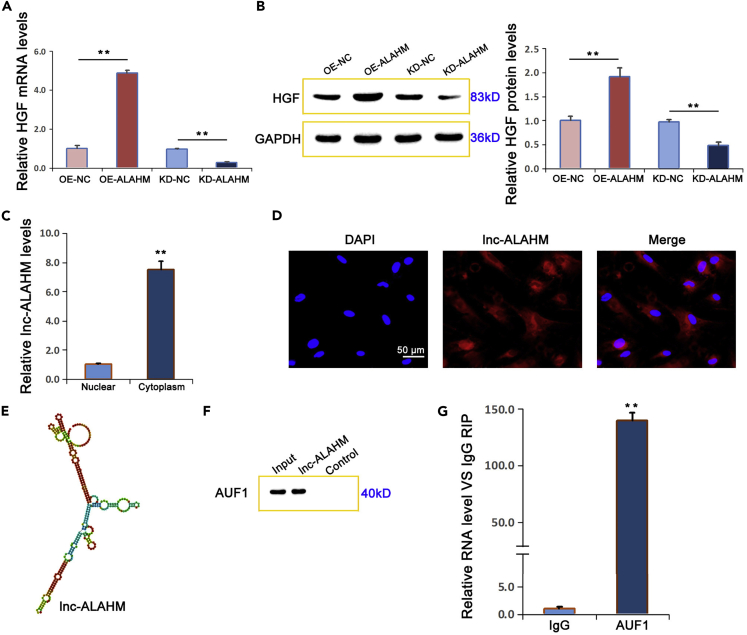
Using lentiviral-mediated overexpression and siRNA knockdown, we investigated how changes in ALAHM expression affected HGF gene and protein expression in L02 cells. Based on real-time RT-PCR and western blotting, we determined that the mRNA and protein expression levels of HGF were significantly increased by overexpressing ALAHM in L02 cells and significantly decreased by ALAHM knockdown (Figures 6A and 6B). Using real-time RT-PCR to detect expression differences in ALAHM in the cytoplasm and nucleus of L02 cells, we found that the expression level of ALAHM in the cytoplasm was significantly higher than that in the nucleus (Figure 6C). Further, the localization of ALAHM in L02 cells was evaluated using RNA FISH, and the results showed that expression was localized in the cytoplasm (Figure 6D).
Figure 6.
ALAHM increased HGF expression by binding to the transcription factor AUF1
After overexpression or knockdown of ALAHM in L02 cells, HGF expression levels of mRNA and protein were detected by real-time RT-PCR and western blot, respectively (A and B). The differential expression of ALAHM in the cytoplasm and nucleus of L02 cells was detected by real-time RT-PCR, which showed that ALAHM was preferentially localized in the cytoplasm (C). RNA FISH was performed to verify the localization of ALAHM in L02 cells and confirmed that expression was mainly localized in the cytoplasm (D). A structure diagram of ALAHM is shown in (E), which was obtained from RNAfold Web Server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The results of RNA pull-down and RIP experiments are shown in (F) and (G), respectively. Bar graphs represent ratios plotted as mean ± SD of three separate experiments. Compared with the control group, ∗∗p < 0.01.
By using catRAPID (http://s.tartaglialab.com/page/catrapid_omics2_group), according to the secondary structure of ALAHM (obtained from RNAfold Web Server, http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) (Figure 6E) and the prediction and analysis results of lncRNA binding to protein, it was speculated that this lncRNA readily binds to RNA-binding protein including F-box protein AUF1, which was one predicted highly combined protein. Furthermore, according to a previous report (Moore et al., 2014), ALAHM may promote HGF protein translation by binding to transcription factor AUF1. Using RNA pull-down and RIP assay experiments, we determined that ALAHM binds to AUF1 (Figures 6F and 6G).
Discussion
EVs are an important means of material transfer and information exchange between cells. A large number of studies have shown that EVs are involved in the malignant progression of various tumors, including lung cancer (Li et al., 2021; Shushkova et al., 2019). Secreting cells selectively load proteins, RNA, DNA, and other substances into EVs through various mechanisms, and EVs also provide a relatively stable environment for their contents (Kai et al., 2018). Tumor cells use EVs to change the physiological state of recipient cells and adapt to the microenvironment. Tumor-derived EVs promote the occurrence and development of tumors by promoting epithelial mesenchymal transformation of tumor cells, angiogenesis, cancer-associated fibroblast transformation, immune escape, and premetastatic niche formation (Weidle et al., 2017; Alipoor et al., 2018). In recent years, the function of lncRNAs derived from tumor EVs has been extensively studied, and multiple lncRNAs have been shown to be putative as biomarkers for tumor diagnosis and prognosis assessment (Xu et al., 2020; Abbastabar et al., 2020; Yan et al., 2020). The biological functions and molecular mechanisms of relevant lncRNAs have also been studied further.
The liver is a common metastatic site of lung cancer. As a better mechanistic understanding of liver metastasis of lung cancer may provide new effective biomarkers and therapeutic targets for clinical practice, we used high-throughput sequencing technology to identify differentially expressed lncRNAs in serum EVs of patients with LUAD and liver metastasis from those without metastasis. Subsequently, the top three serum EVs-derived lncRNAs from these patients were verified using real-time RT-PCR. Our verification results demonstrated that lncRNA ENSG00000271732 was the most significantly enriched of these three lncRNAs from serum EVs of patients with LUAD and liver metastasis, which has not been reported in the present studies. It was named lncRNA ALAHM based on its potential role in liver metastasis of LUAD.
To investigate the role and mechanism of ALAHM in liver metastasis of LUAD, we first verified that the expression levels of ALAHM in LUAD NCI-H1975 cells and its EVs were significantly higher than in normal alveolar epithelial HPAEpiC cells. By constructing NCI-H1975 cell lines with altered ALAHM expression, EVs were isolated from modified cells and used to treat normal L02 hepatocytes. Our results showed that NCI-H1975-cell-derived EVs are uptaken by L02 cells and regulated the expression of ALAHM in L02 cells. A nude mouse model of in situ lung cancer was then established using NCI-H1975 cells, and we found that NCI-H1975 cells overexpressing ALAHM not only significantly upregulated ALAHM expression in liver tissue but also significantly promoted liver metastasis of lung cancer in nude mice.
Based on these results, we hypothesized that lung-cancer cell-derived EVs transport ALAHM to liver tissue and consequently, promote the expression of ALAHM in liver cells and liver metastasis of lung cancer cells. We next asked how an ALAHM-mediated TME plays a role in promoting the growth and metastasis of lung cancer cells and how liver cells with abnormal ALAHM expression recruit lung cancer cells to gather and promote liver metastasis. A better understanding of the specific mechanism was the focus of our in-depth study.
HGF is a paracrine cytokine specifically expressed in the liver and is closely related to liver metastasis of various malignant tumors (Yao et al., 2019); in fact, serum HGF is upregulated in many kinds of tumors. Further, the HGF signaling pathway is activated by binding to specific receptors in the TME and participates in tumor invasion and migration, thereby promoting the growth of metastatic tumors. Thus, HGF is a potential biomarker for prognosis (Spina et al., 2015; Ahmed et al., 2016). Studies have found that increased HGF parasecretion is an important factor in attracting tumor cells to gather in liver tissue and form metastases (Zhang et al., 2017).
In the present study, we found that NCI-H1975-cell-derived EVs overexpressing ALAHM significantly increased HGF protein levels in liver tissue of model nude mice. In addition, EVs of NCI-H1975 cells overexpressing ALAHM significantly increased HGF protein expression in liver cells in vitro and HGF protein level in the supernatant of liver cells. These results indicate that EVs-ALAHM of NCI-H1975 cells promotes HGF paracrine secretion in hepatocytes.
According to the in silico structure of ALAHM, we speculated that ALAHM readily binds to RNA-binding protein. AUF1 is one of the best characterized AU RNA-binding proteins that posttranscriptionally regulates gene expression and participates in tumor development. AUF1 expression is upregulated in hepatocellular carcinoma, and overexpressed AUF1 is significantly correlated with tumor size and stage and patient prognosis (Yang et al., 2014). According to the functions of AUF1 (Moore et al., 2014), it was speculated that ALAHM may promote the translation of HGF by binding AUF1. RNA pull-down and RIP experiments verified that ALAHM binds to AUF1 in this study. Our findings indicate that ALAHM promotes HGF translation and paracrine secretion through the recruitment of RNA-binding protein AUF1, thus promoting liver metastasis of LUAD cells.
In this study, we have identified specific lncRNA ALAHM highly enriched in serum EVs of LUAD patients with liver metastasis through gene sequencing. Subsequently, we confirmed that EVs derived from LUAD cells overexpressing ALAHM promoted liver metastasis and upregulated the expression of HGF protein in liver tissues by employing a nude mouse model of LUAD in situ. In vitro studies further revealed that ALAHM in EVs from LUAD cells facilitated paracrine secretion of HGF in hepatocytes by binding the RNA binding protein AUF1, thus enhancing the proliferation and metastasis of LUAD cells. This study provides new insight into the mechanism of EVs derived from LUAD promoting liver metastasis from the perspective of TME.
Conclusions
Collectively, the findings of this study revealed that the lung-cancer cell-derived EVs-ALAHM recruits the RNA-binding protein AUF1 to promote hepatocyte parasecretion of HGF, activates the HGF signaling pathway, forms a TME conducive to lung cancer cell growth and proliferation, and promotes liver metastasis of lung cancer. Our study provides a scientific basis for clarifying the molecular mechanism of ALAHM in the role of lung cancer EVs toward promoting liver metastasis of lung cancer.
Limitations of the study
This study is only a preliminary exploration on the mechanism of the unreported lncRNA in lung cancer EVs promoting liver metastasis. The number of patients with LUAD involved in this study was limited. The mechanism of lung-cancer-cell-derived EVs and the role of lncRNA ALAHM in promoting liver metastasis of lung cancer need in-depth and extensive research.
Abbreviations
AUF1 AU-binding factor 1
DEGs differentially expressed genes
ELISA Enzyme-linked immunosorbent assay
EPF Purification Filter
EVs extracellular vesicles
FBS Fetal bovine serum
FC fold change
FISH fluorescence in situ hybridization
GAPDH glyceraldehyde 3-phosphate dehydrogenase
HGF hepatic growth factor
HPAEpiC human pulmonary alveolar epithelial cells
lncRNA Long noncoding RNAs
LUAD lung adenocarcinoma
NSCLC non-small cell lung cancer
PBS phosphate-buffered saline
RFP red fluorescent protein
RIP RNA-binding protein immunoprecipitation
RNA-seq RNA sequencing
RT-PCR reverse transcription polymerase chain reaction
TEM transmission electron microscopy
TME tumor microenvironment
Ethical approval and consent to participate
This study was approved by the Medical Ethics Committee and Ethics Review Committee for Animal Research of the Tianjin Union Medical Center.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit monoclonal anti-HGF | Abcam | RRID:ab178395 |
| Rabbit monoclonal anti-CD63 | Abcam | RRID:ab134045 |
| Rabbit monoclonal anti-TSG101 | Abcam | RRID:ab125011 |
| Mouse monoclonal anti-Tubulin | Abcam | RRID:ab7291 |
| Rabbit monoclonal anti-AUF1 | Abcam | RRID:ab259895 |
| Mouse monoclonal anti-GAPDH | Abcam | RRID:ab8245 |
| HRP-goat anti-rabbit IgG | Thermo Scientific | RRID:A16104 |
| Biological samples | ||
| Patients serum (gene sequencing detection) | TUMC & Tianjin Chest Hospital | N/A |
| Patients serum (lncRNAs screening and validation) | TUMC & Tianjin Chest Hospital | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Primers | GenePharma Inc. | Syntheses |
| Experimental models: Cell lines | ||
| Human:NCI-H1975 | ATCC | CBP60121 |
| Human:HPAEpiC | Sciencell | 3200 |
| Human:L02 | ATCC | CBP60224 |
| Experimental models: Mice | ||
| Balb/c nude | Charles River Labs | SCXK 2020-0013 |
| Oligonucleotides | ||
| siRNA (targeting ALAHM) | GenePharma Inc. | Syntheses |
| siRNA NC | GenePharma Inc. | Syntheses |
| Recombinant DNA | ||
| pcDNA3.1 plasmid | GenePharma Inc. | Syntheses |
| Software and algorithms | ||
| ImageJ | VILBER Co., Ltd. | FUSION FX EDGE SPECTRA |
| Graphpad Prism 6.02 | GraphPad Software Co., Ltd. | N/A |
| TRANSFAC database | Biostars | https://www.biostars.org/p/69086/ |
| catRAPID | Biostars | http://s.tartaglialab.com/page/catrapid_omics2_group |
| SPSS 20.0 | SPSS Inc. | N/A |
| Other | ||
| RNA FISH probe | Ruibo Biotechnology | RRID:C10910 |
| Human:ELISA kit | Abcam | RRID:ab267631 |
| Cytoplasmic and Nuclear RNA Purification kit | AmyJet Scientific | RRID:NGB-21000 |
| SYBR qPCR Master Mix | Vazyme Biotech | RRID:Q511 |
| HiScript II Q Select RT SuperMix | Vazyme Biotech | RRID:R222-01 |
| RIPA lysate | Fount BioTech | RRID:HC1235 |
| DNA Marker | Thermo Fisher Scientific | RRID:SM1332 |
| BCA Protein Assay kit | Thermo Fisher Scientific | RRID:23227 |
| Dimethylsulfoxide | Sigma-Aldrich | RRID:D8418 |
| Fetal bovine serum | Gibco | RRID:12484010 |
| Trypsin-EDTA | Gibco | RRID:25200072 |
| DMEM | Gibco | RRID:72400047 |
| Database: Sequence data | This paper | NCBI:SUB10791255 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Dr. Chunyang Jiang (chunyangjiang@126.com).
Materials availability
This study did not generate new unique reagents. Primers used are provided in Tables 3 and 4, and available upon request to the corresponding author.
Experimental model and subject details
Animal model
The purpose is to establishment of an in situ lung cancer mouse model. Thirty female nude mice aged 3 weeks (body weight: 18–22 g) were ad libitum access to food and water. Mice were randomly assigned to one of the following five groups: (1) lung cancer group; (2) lung cancer + HPAEpiC EVs group; (3) lung cancer + NCI-H1975-NC EVs group; (4) lung cancer + NCI-H1975-ALAHM OE group; and (5) lung cancer + NCI-H1975-ALAHM KD group.
After feeding for 2 weeks, mice were anesthetized by intraperitoneal injection of sodium thiopental (0.05 mg/kg body weight). They were then supine fixed on an operating table, and the anterior chest wall was disinfected with alcohol. A 5-mm incision was made about 1.5 cm above the costal arch in front of the left axillary line. After exposing the chest wall, a 100-μL suspension of stable red fluorescent protein (RFP)-expressing NCI-H1975 cells (1 × 107) was injected into the left lung of each mouse using an insulin injection needle for about 3 min. After injection, the needle remained for 5 s, and the incision was sutured after needle extraction. After cell injection for 4 weeks, 50 μg of EVs was injected into the tail vein of mice from all groups every week for 4 weeks. Mice were fed normally, and tumor growth both in the lung and liver was observed (by using Fusion FX.EDGE chemiluminescence imaging (VILBER Co., Ltd., France)). Upon liver tumor formation, mice were sacrificed by cervical dislocation, the abdominal cavity was opened, and the liver was excised for testing. The experimental protocol was approved by the Ethics Review Committee for Animal Research at Tianjin Union Medical Center (TUMC).
Patients and samples
Serum EVs for gene sequencing detection were isolated from three LUAD patients with liver metastasis (no metastasis to other organs) and three LUAD patients without metastasis (i.e., three vs. three). Serum EVs for lncRNAs screening and validation were isolated from ten LUAD patients with liver metastasis (no metastasis to other organs) and ten LUAD patients without metastasis (i.e., ten vs. ten). All patients were first-time inpatients who had not received any treatment after diagnosis at the TUMC and the Tianjin Chest Hospital between October 2019 and April 2020. Fasting peripheral blood samples (10 mL in an EDTA tube) were taken from each patient in the morning for serum extraction. The clinical and pathological details of these patients are shown in Table 1. This study section was approved by the Medical Ethics Committee of the TUMC.
Cell lines
The human LUAD NCI-H1975 cell line and human normal hepatocyte L02 cell line were purchased from ATCC (Manassas, VA, USA). The human pulmonary alveolar epithelial cells (HPAEpiC) cell line were purchased from Sciencell (Carlsbad, CA, USA).
Method details
Reagents
Except for main reagents have already indicated in key resources table or detailed in methods, all other reagents used were analytical grade laboratory chemicals from standard commercial suppliers.
EVs isolation
Serum samples or collected cell suspensions were centrifuged at 3,000 g for 10 min at 4°C to remove cell debris. The supernatant was then centrifuged at 10,000 g for 20 min at 4°C to remove impurities. The supernatant was diluted with 1× phosphate-buffered saline (PBS), PureExo Solution was added, and the mixture was mixed in a vortex oscillator for 1 min before being held at 4°C for 2 h. The solution was then centrifuged at 10,000 g for 60 min at 4°C and the supernatant was discarded. The centrifugal precipitate was resuspended with PBS, transferred to a new tube, and centrifuged at 12,000 g for 2 min at 4°C. The resulting supernatant, which was rich in EV particles, was retained.
EVs purification
Coarse EV particles were transferred into the upper chamber of an EVs Purification Filter (EPF) column and centrifuged at 3,000 × g for 10 min at 4°C. After centrifugation, the liquid at the bottom of the EPF column was collected, which contained purified EV particles. The purified EVs were retained in a final volume of 50–100 μL and stored at −80°C for subsequent experiments.
Transmission electron microscopy (TEM) detection of EVs
After EVs were thawed and resuspended, an equal volume (50–100 μL) of 4% PFA was added. A 5-μL aliquot of the EVs suspension was added to sample loading Formvar carbon-coated copper grids. After placing the Formvar film on the copper mesh, a drop of PBS was used to clean the surface. The copper grids were immersed in 50 μL of a 1% glutaraldehyde solution for 5 min, and then washed with ddH2O. Next, the copper grids were placed in a solution of uranium dioxalate, followed by methylcellulose droplets on ice for 10 min. Afterward, the grids were placed on a stainless steel ring, and the excess liquid was removed. Finally, the copper grids were observed under a transmission electron microscope (80 kV) (Philips CM200, FEI Inc., Netherlands) and electron micrographs were taken.
Nanoparticle-tracking analysis (NTA) for quantification detection
The sample pool was cleaned with deionized water, the instrument was calibrated with polystyrene microspheres (110 nm), and the sample pool was cleaned again with 1× PBS (Biological Industries, Israel). After diluting EV samples with 1× PBS, the concentration and particle size of EVs were detected using an automated EVs fluorescence detection and analysis system (NanoSight NS300) (Malvern Instruments Co., Ltd., UK).
EVs transcriptome sequencing detection
EV samples were sent to Guangzhou Ruibo Biotechnology Co., Ltd. for EVs transcriptome sequencing. After raw sequencing data were obtained, low-quality reads were processed, and the sequencing quality was evaluated to obtain clean data. Clean data were compared with the reference genome using the BWT algorithm (FM index) in HISAT2 and BAM files were obtained. Gene Ontology analysis and Kyoto Encyclopedia of Genes and Genomes biopathway enrichment analysis were performed to investigate differentially expressed genes (DEGs). For detected lncRNAs, expression levels were calculated and differences in expression were analyzed. We also determined the corresponding ORF, protein domain, coding potential, secondary structure prediction, and protein family. We used the R package DEGseq, applying different multiples of fold change (FC) such as log2FC > 1 and a significant level (Q value < 0.05), to identify DEGs from RNA-seq data.
Real-time quantitative RT-PCR
Trizol was added to the collected EVs or cells and incubated, followed by the addition of chloroform for 2 min and centrifugation at 12,000 × g for 15 min at 4°C. The supernatant was transferred to a new tube and chloroform was added. The mixture was then centrifuged again and total RNA was transferred to another tube with isopropyl alcohol, mixed, and then centrifuged at 15,000 × g for 10 min at 4°C. The supernatant was discarded and 75% ethanol was added to wash the RNA precipitate. After centrifugation, the supernatant was discarded, and the RNA pellet was air dried for 3 min. Finally, RNase free water was added to dissolve the RNA precipitate. Using deionized water as control, the OD260 value and RNA concentration were measured. Reverse transcription was performed using a Vazyme Reverse Transcription kit according to the manufacturer’s instructions. GAPDH was used as a housekeeping gene for normalization. Primer sequences for lncRNAs, HGF and GAPDH are listed in Table 3. The real-time RT-PCR was performed using an ABI Prism 7700 Sequence Detection System (Applied Biosystems, USA). The cycle threshold (Ct) data were collected by setting the corrected threshold and the relative quantification was measured by the 2-△△Ct method.
Western blot
Lysates of isolated and purified EVs or collected cells were obtained using ice-cold lysis buffer and protein concentrations were determined using a BCA protein quantification kit. According to the protein concentration of each sample, calculated the required volume of the same protein amount of each sample. Protein samples were loaded with 2 × loading buffer and denaturated at 95°C for 5 min. Denaturated protein samples were loaded for SDS-PAGE electrophoresis (Bio-Rad Japan Laboratories Ltd. Co., Tokyo, Japan), which was performed at 80 V and 100 mA for 50 min. Proteins were transferred onto a membrane by wet electroblotting. Next, the membrane was sealed in TBST solution with 5% skim milk, and then washed with TBST. Specific primary antibodies were added and incubated. After a TBST wash, the secondary antibody was added and incubated, followed by another wash with TBST. Chemiluminescence was performed and photographs were taken using an ECL imaging system (Tanon, Shanghai, China).
Cell immunofluorescence assay
L02 cell cultures were treated with 0.25% trypsin and transferred to a 24-well cell culture plate. After cells were adherent, PKH67-labeled EVs derived from NCI-H1975 cells were added to each well. After incubation at 37°C and 5% CO2 for 2 h, cell slides were taken out. Each slide was fixed for 1 h using 500 μL of 4% paraformaldehyde and washed three times with PBS for 5 min each time. After air dried, the slides were immersed three times in PBS (3 min each), and the PBS was sucked dry by absorbent paper. The slides were added with goat serum and sealed at room temperature for 30 min. After removal of the blocking solution, primary antibody was added to each slide and incubated. Next, the slides were washed three times using PBST for 3 min each time, absorbent paper to drain excess liquid from the slides, followed by the addition of a diluted fluorescent secondary antibody and incubation. After immersion cleaning three times with PBST for 3 min each time, DAPI was added and slides were incubated for 5 min. PBST was then used to remove the excess DAPI. Slides were sealed using a sealant containing an anti-fluorescence quenching agent and then observed and images taken under a fluorescence microscope (Olympus Optical Co., Ltd., Tokyo, Japan).
Enzyme-linked immunosorbent assay (ELISA)
HGF expression was detected using ELISA in L02 cells from the following groups: (1) control group: L02 cells; (2) HPAEpiC group: L02 cells + HPAEpiC EVs; (3) NC group: L02 cells + NCI-H1975-NC EVs; (4) overexpression (OE) group: L02 cells + NCI-H1975-ALAHM-OE EVs; and (5) knockdown (KD) group: L02 cells + NCI-H1975-ALAHM-KD EVs. The primer sequences for pshR-ALAHM and siRNA-ALAHM are listed in Table 4. L02 cells were cultured at 37°C and 5% CO2 for 24 h. The cells were then collected, lysed by sonication, and the supernatant was collected. For ELISA, standard solutions and sample supernatants were added to wells. Next, 100 μL of HRP-labeled antibody was added to sample and standard wells, which were then sealed and incubated. After discarding the liquid and following a wash, 50-μL substrate was added to each well and incubated, followed by the addition of 50 μL of Stop Solution. The OD values of each group were determined at 450 nm using a SpectraMax 190 microplate reader (Molecular Devices).
Clone formation assay
L02 cells were divided into the following six experimental groups according to different treatment methods: (1) Control group: L02 cells; (2) L02 cells + HGF (2 μg/mL); (3) L02 cells + HPAEpiC EVs; (4) L02 cells + NCI-H1975-NC EVs; (5) OE group: L02 cells + NCI-H1975-ALAHM-OE EVs; and (6) KD group: L02 cells + NCI-H1975-ALAHM-KD EVs (2 μg/mL of EVs for each group). Cells with different treatments at logarithmic growth stage were collected by digestion and centrifuged at 1,000 rpm for 5 min. The cell precipitates were suspended again for cell counting, and the cell concentration was diluted to 1 × 103/mL. 200 μL cell suspension was inoculated into culture dish (50-200 cells per well), and the medium was supplemented to 10 mL. Shake the culture dish to disperse the cells evenly and put them into a cell incubator for routine culture for 2-3 weeks. When visible clones appeared, the culture was terminated and the culture medium was discarded. Cells were washed twice with PBS at room temperature. After discard PBS, cells were fixed in 4% paraformaldehyde for 15 min at room temperature, then abandoned the fixative. Cells were added with crystal violet stain for 20 min, rinsed in running water and air dried. The cellular clone formation was observed and photographed.
Transwell assay
The scratch assay was performed on cells from the same experimental groups that underwent Clone formation assay. L02 cells were cultured to logarithmic growth stage (the confluence of cells reached 80 percent) and then trypsinized. Cells were suspended in serum-free medium and counted, and the cell concentration was adjusted to 2 × 105/mL. We added 800 μL of medium containing 10% serum to the lower compartment of a 24-well Transwell cell culture plate, and 150 μL of cell suspension was added into the upper compartment. After culturing the plate for 24 h, the upper chamber was carefully removed using tweezers. The liquid in the upper chamber was removed and the chamber was allowed to dry. Next, 800-μL methanol was added to the upper chamber and fixed at 24°C for 30 min. The upper chamber was removed, and the fixative of the upper chamber was dried by filter paper. The fixative of the upper chamber was transferred to a well containing 800 μL of 1% crystal violet dye and stained for 20 min at room temperature. After gently rinsing and soaking with sterile water, the upper chamber was removed and the remaining liquid was absorbed. The cells on the membrane surface at the bottom of the upper chamber were carefully wiped with a wet swab, and a random field of view was observed under a microscope and photographed.
Scratch assay
The scratch assay was performed on cells from the same experimental groups that underwent Clone formation assay. After more than 80% confluence in T25 culture flasks, L02 cells of the different groups underwent scratch assay. Once the fusion rate of treated cells reached 70%–80%, a pipette tip was used to draw a line across the culture plate to disrupt cells. The culture flask was then gently rinsed with sterile PBS three times to ensure that all dislodged cells were removed. Serum-free medium was then added to the culture, and the same visual field was photographed at 0, 24, and 48 h.
RNA fluorescence in situ hybridization (FISH)
A RNA FISH probe was designed by Guangzhou Ruibo Biotechnology Co., Ltd., and used with a Ribo™ Fluorescent In Situ Hybridization kit according to manufacturer’s instructions. L02 cells were inoculated into 24-well plates containing cell slides at a concentration of 5 × 104 cells/mL and cultured for 24 h. The culture medium was discarded, cells were washed with PBS, and then fixed with 4% paraformaldehyde at room temperature for 10 min. The fixation solution was discarded and cells were washed with PBS, followed by the addition of 1-mL PBS containing 0.5% Triton X-100 and placed on ice for 5 min. After another PBS wash, 200 μL of pre-hybridization solution was added to each well, which was then sealed and incubated at 37°C for 30 min. The pre-hybridization solution was discarded and 100 μL of hybridization solution containing the probe was added. Hybridization was carried out overnight at 37°C in the dark. After successive washes with plain hybridization solution and PBS, cells were then stained using DAPI. After a final PBS wash, slides were removed from the plate and fixed with sealing solution for fluorescence detection.
RNA pull-down assay
For each experimental group, cultured L02 cells were trypsinized and washed twice with PBS to collect the cell pellet. Cells were resuspended in PBS containing 0.1% Triton X and underwent three cycles of freeze-thaw using liquid nitrogen, followed by centrifugation at 12,000 rpm for 10 min at 4°C. The supernatant was retained and Biotin-labeled RNA probe was added and mixed. Using a 1.5 mL tube, 20 μL of magnetic beads was added to 400 μL of washing buffer, mixed, and magnetically separated. This wash was performed again before the addition of the beads to the probe-supernatant mixture. Rinse twice with rinse solution, followed by magnetic separation, and repeat twice. Next, 1× protein loading buffer was added, followed by denaturation at 95°C for 5 min. The supernatant was stored at −20°C. Finally, protein expression was detected using Western blot.
RNA-binding protein immunoprecipitation (RIP) assay
For RIP assay, the experimental groups consisted of Input, IgG, and ALAD groups treated with 0, 10, or 100 μM Pb for 48 h. For each group of L02 cells, 30 μL of Protein A/G PLUS-Agarose was mixed with 5 μL of ALAD antibody, followed by gentle rotation at 4°C for 6 h to prepare the antibody mixture. After treatment, cells were removed from the medium and fixed by UV irradiation. After washing the cells twice with PBS, cell pellets were collected and cells were lysed using ice-cold lysis buffer. After centrifugation, the supernatant was added to the antibody mixture and incubated overnight with gentle mixing at 4°C. Next, the mixture was centrifuged and the supernatant was discarded. The agarose beads were washed twice with a low-salt rinse solution; the supernatant was discarded after centrifugation of each wash. The beads were then washed twice with a high-salt bleaching solution, discarding the supernatant after each centrifugation. Next, agarose beads were suspended in 50 μL of proteinase K solution and digested at 55°C for 10 min. Total RNA was extracted using Trizol and real-time RT-PCR was performed to detect RNA expression.
Quantification and statistical analysis
All data were analyzed using two-way univariate analysis of variance followed by Tukey’s (equal variances assumed or homogeneity of variance after variable transformation) or Dunnett’s T3 (equal variances not assumed after variable transformation justification) post-hoc test between groups using the Statistical Package for Social Sciences software version 20.0 (SPSS Inc., Chicago, IL, USA). Results are expressed as mean ± S.E.M. All tests were two sided. Values of p < 0.05 were considered significant.
Acknowledgments
This work was supported by the Natural Science Foundation of Tianjin-Municipal Science and Technology Commission (CN) (Grant Number: 20JCYBJC01030) and Social Development Science and Technology Project of TaiZhou City (Grant Number: 21ywb99). We deeply thank the language editor Justin F. of Enago English language service Co., Ltd.
Author contributions
C.Y.J. conceived and designed this study. C.Y.J., X.L.H., and S.J.Y. supervised and directed the project. All authors carried out the experiments, analyzed the experimental data, and wrote the paper. C.Y.J. mainly presented the tables and the figures. C.Y.J. selected and prepared all clinical specimens. All authors reviewed and agreed the manuscript.
Declaration of interests
The authors have declared that no competing interests exist.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103984.
Contributor Information
Chunyang Jiang, Email: chunyangjiang@126.com.
Shijing Yue, Email: shijingyue@nankai.edu.cn.
Xiaoli Hu, Email: 752591421@qq.com.
Supplemental information
After gained raw gene sequencing data, different expression of ncRNAs in serum EVs between LUAD patients with liver metastasis and without metastasis (three vs. three) were analyzed by DE-Gseq procedure (|log2 (FoldChange)| > 1) (Qvalue <0.05).
Data and code availability
The data reported in this paper will be shared upon request to the lead corresponding author (chunyangjiang@126.com). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abbastabar M., Sarfi M., Golestani A., Karimi A., Pourmand G., Khalili E. Tumor-derived urinary exosomal long non-coding RNAs as diagnostic biomarkers for bladder cancer. EXCLI. J. 2020;19:301–310. doi: 10.17179/excli2019-1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M., Kumar G., Moussa M., Wang Y., Rozenblum N., Galun E., Goldberg S.N. Hepatic radiofrequency ablation-induced stimulation of distant tumor growth is suppressed by c-Met inhibition. Radiology. 2016;279:150080. doi: 10.1148/radiol.2015150080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipoor S.D., Mortaz E., Varahram M., Movassaghi M., Kraneveld A.D., Garssen J., Adcock I.M. The potential biomarkers and immunological effects of tumor-derived exosomes in lung cancer. Front. Immunol. 2018;9:819. doi: 10.3389/fimmu.2018.00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodt P. Role of the microenvironment in liver metastasis: from pre- to prometastatic niches. Clin. Cancer Res. 2016;22:5971–5982. doi: 10.1158/1078-0432.CCR-16-0460. [DOI] [PubMed] [Google Scholar]
- Gebeyehu A., Kommineni N., Meckes D.G., Jr., Sachdeva M.S. Role of exosomes for delivery of chemotherapeutic drugs. Crit. Rev. Ther. Drug Carrier Syst. 2021;38:53–97. doi: 10.1615/CritRevTherDrugCarrierSyst.2021036301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Zhang N., Hu X., Wang H. Tumor-associated exosomes promote lung cancer metastasis through multiple mechanisms. Mol. Cancer. 2021;20:117. doi: 10.1186/s12943-021-01411-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai K., Dittmar R.L., Sen S. Secretory microRNAs as biomarkers of cancer. Semin. Cell Dev. Biol. 2018;78:22–36. doi: 10.1016/j.semcdb.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Li Z.B., Chen X., Yi X.J. Tumor promoting effects of exosomal microRNA-210 derived from lung cancer cells on lung cancer through the RUNX3/PI3K/AKT signaling pathway axis. J. Biol. Regul. Homeost. Agents. 2021;35:473–484. doi: 10.23812/20-570-A. [DOI] [PubMed] [Google Scholar]
- Meehan K., Vella L.J. The contribution of tumour-derived exosomes to the hallmarks of cancer. Crit. Rev. Clin. Lab. Sci. 2016;53:121–131. doi: 10.3109/10408363.2015.1092496. [DOI] [PubMed] [Google Scholar]
- Moore A.E., Chenette D.M., Larkin L.C., Schneider R.J. Physiological networks and disease functions of RNA-binding protein AUF1. Wiley Interdiscip. Rev. RNA. 2014;5:549–564. doi: 10.1002/wrna.1230. [DOI] [PubMed] [Google Scholar]
- Pan T., Xie Q.K., Lv N., Li X.S., Mu L.W., Wu P.H., Zhao M. Percutaneous CT-guided radiofrequency ablation for lymph node oligometastases from hepatocellular carcinoma: a propensity score-matching analysis. Radiology. 2017;282:259–270. doi: 10.1148/radiol.2016151807. [DOI] [PubMed] [Google Scholar]
- Riihimäki M., Hemminki A., Fallah M., Thomsen H., Sundquist K., Sundquist J., Hemminki K. Metastatic sites and survival in lung cancer. Lung Cancer. 2014;86:78–84. doi: 10.1016/j.lungcan.2014.07.020. [DOI] [PubMed] [Google Scholar]
- Saxena S., Singh R.K. Chemokines orchestrate tumor cells and the microenvironment to achieve metastatic heterogeneity. Cancer Metastasis Rev. 2021;40:447–476. doi: 10.1007/s10555-021-09970-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shushkova N.A., Novikova S.E., Zgoda V.G. Exosomes of malignant tumors: prospects of omiсs diagnostics. Biomed. Khim. 2019;65:457–467. doi: 10.18097/PBMC20196506457. [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- Spina A., De Pasquale V., Cerulo G., Cocchiaro P., Della Morte R., Avallone L., Pavone L.M. HGF/c-MET Axis in tumor microenvironment, and metastasis formation. Biomedicines. 2015;3:71–88. doi: 10.3390/biomedicines3010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T., Kurishima K., Nakazawa K., Kagohashi K., Ishikawa H., Satoh H., Hizawa N. Specific organ metastases and survival in metastatic non-small-cell lung cancer. Mol. Clin. Oncol. 2015;3:217–221. doi: 10.3892/mco.2014.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S., Pucci M., Giallombardo M., Di Bella M.A., Santarpia M., Reclusa P., Gil-Bazo I., Rolfo C., Alessandro R. Amphiregulin contained in NSCLC-exosomes induces osteoclast differentiation through the activation of EGFR pathway. Sci. Rep. 2017;7:3170. doi: 10.1038/s41598-017-03460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Song X., Liu L., Niu L., Wang X., Song X., Xie L. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 2018;109:1701–1709. doi: 10.1111/cas.13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidle U.H., Birzele F., Kollmorgen G., Rüger R. The multiple roles of exosomes in metastasis. Cancer Genomics Proteomics. 2017;14:1–15. doi: 10.21873/cgp.20015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood S.L., Pernemalm M., Crosbie P.A., Whetton A.D. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat. Rev. 2014;40:558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- Xie Y., Dang W., Zhang S., Yue W., Yang L., Zhai X., Yan Q., Lu J. The role of exosomal noncoding RNAs in cancer. Mol. Cancer. 2019;18:37. doi: 10.1186/s12943-019-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Dong X., Chen Y., Wang X. Serum exosomal hnRNPH1 mRNA as a novel marker for hepatocellular carcinoma. Clin. Chem. Lab. Med. 2018;56:479–484. doi: 10.1515/cclm-2017-0327. [DOI] [PubMed] [Google Scholar]
- Xu H., Zhou J., Tang J., Min X., Yi T., Zhao J., Ren Y. Identification of serum exosomal lncRNA MIAT as a novel diagnostic and prognostic biomarker for gastric cancer. J. Clin. Lab. Anal. 2020;34:e23323. doi: 10.1002/jcla.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Peng A., Wang B., Gusdon A.M., Sun X., Jiang G., Zhang P. The prognostic impact of lymph node metastasis in patients with non-small cell lung cancer and distant organ metastasis. Clin. Exp. Metastasis. 2019;36:457–466. doi: 10.1007/s10585-019-09985-y. [DOI] [PubMed] [Google Scholar]
- Yang Y., Kang P., Gao J., Xu C., Wang S., Jin H., Li Y., Liu W., Wu X. AU-binding factor 1 expression was correlated with metadherin expression and progression of hepatocellular carcinoma. Tumour Biol. 2014;35:2747–2751. doi: 10.1007/s13277-013-1362-2. [DOI] [PubMed] [Google Scholar]
- Yan S., Du L., Jiang X., Duan W., Li J., Xie Y., Zhan Y., Zhang S., Wang L., Li S., et al. Evaluation of serum exosomal lncRNAs as diagnostic and prognostic biomarkers for esophageal squamous cell carcinoma. Cancer Manag. Res. 2020;12:9753–9763. doi: 10.2147/CMAR.S250971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J.F., Li X.J., Yan L.K., He S., Zheng J.B., Wang X.R., Zhou P.H., Zhang L., Wei G.B., Sun X.J. Role of HGF/c-Met in the treatment of colorectal cancer with liver metastasis. J. Biochem. Mol. Tox. 2019;33:e22316. doi: 10.1002/jbt.22316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Liu X., Shao X., Feng T., Xu J., Wang Q., Hua S. The role of exosomes in lung cancer metastasis and clinical applications: an updated review. J. Transl. Med. 2021;19:312. doi: 10.1186/s12967-021-02985-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Deng T., Liu R., Bai M., Zhou L., Wang X., Li S., Wang X., Yang H., Li J., et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 2017;8:15016. doi: 10.1038/ncomms15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Shi X., Zhao H., Jia X., Yang Y. Identification and validation of a tumor microenvironment-related gene signature for prognostic prediction in advanced-stage non-small-cell lung cancer. Biomed. Res. Int. 2021;2021:8864436. doi: 10.1155/2021/8864436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
After gained raw gene sequencing data, different expression of ncRNAs in serum EVs between LUAD patients with liver metastasis and without metastasis (three vs. three) were analyzed by DE-Gseq procedure (|log2 (FoldChange)| > 1) (Qvalue <0.05).
Data Availability Statement
The data reported in this paper will be shared upon request to the lead corresponding author (chunyangjiang@126.com). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.