Abstract
The activity of vancomycin and teicoplanin combined with gentamicin was investigated in vitro against strains of Enterococcus faecalis resistant to vancomycin and susceptible to teicoplanin (VanB type) and against mutants that had acquired resistance to teicoplanin by three different mechanisms. In vitro, gentamicin selected mutants with two- to sixfold increases in the level of resistance to this antibiotic at frequencies of 10−6 to 10−7. Teicoplanin selected teicoplanin-resistant mutants at similar frequencies. Both mutations were required to abolish the activity of the gentamicin-teicoplanin combination. As expected, simultaneous acquisition of the two types of mutations was not observed. In therapy with gentamicin or teicoplanin alone, each selected mutants in three of seven rabbits with aortic endocarditis due to VanB-type E. faecalis BM4275. The vancomycin-gentamicin combination selected mutants that were resistant to gentamicin and to the combination. In contrast, the teicoplanin-gentamicin regimen prevented the emergence of mutants resistant to one or both components of the combination. These results suggest that two mutations are also required to suppress the in vivo activity of the teicoplanin-gentamicin combination.
Acquired resistance to glycopeptides in enterococci is due to production of peptidoglycan precursors ending in the depsipeptide d-alanyl-d-lactate (d-Ala-d-Lac), instead of the dipeptide d-Ala-d-Ala present in susceptible bacteria (4, 30, 31). The substitution prevents formation of complexes between glycopeptides and peptidoglycan precursors at the cell surface that are responsible for inhibition of cell wall synthesis (8, 22). Acquired glycopeptide resistance by this mechanism is conferred by two classes of genetic elements (vanA or vanB) that encode a dehydrogenase (VanH or VanHB) and a ligase (VanA or VanB) for synthesis of d-Ala-d-Lac (3, 8, 10). Each element also encodes a d,d-dipeptidase (VanX or VanXB) that limits synthesis of precursors containing the target of glycopeptides by hydrolyzing d-Ala-d-Ala produced by the host Ddl d-Ala:d-Ala ligase (23, 33).
Synthesis of the resistance proteins is regulated by two-component regulatory systems composed of a membrane-bound kinase that senses the presence of glycopeptides in the external medium (VanS or VanSB) and a cytoplasmic response regulator (VanR or VanRB) that activates a promoter for transcription of the resistance genes (1, 3, 6, 10, 15, 32). The sensor is thought to control the activity of the response regulator positively by phosphorylation and negatively by dephosphorylation. According to this model, the sensor acts as a kinase in inducing conditions leading to phosphorylation of the response regulator and promoter activation. In the absence of glycopeptides, the sensor acts as a phosphatase and prevents accumulation of the phosphorylated form of the response regulator.
The VanS sensor mediates induction in response to vancomycin and teicoplanin, leading to inducible expression of resistance to high levels of both glycopeptides (VanA phenotype) (2). In contrast, the majority of enterococci harboring vanB-type gene clusters are inducibly resistant to various levels of vancomycin but remain susceptible to teicoplanin since the VanSB sensor triggers induction only in response to vancomycin (VanB phenotype) (2, 6, 10, 21). Teicoplanin-resistant mutants of VanB-type strains obtained in vitro harbor mutations in vanSB that are responsible for various alterations in the regulation of the resistance genes. The mutants are inducibly or constitutively resistant to vancomycin and teicoplanin (Vmr Ter) or are heterogeneously resistant to these antibiotics (VmHet TeHet) (6, 7, 13).
Emergence of teicoplanin resistance in VanB-type strains has also been observed in a patient (14) and in rabbits with experimental endocarditis treated with vancomycin or teicoplanin (5). In the latter model, the combination of gentamicin and teicoplanin was efficient since it reduced the number of surviving bacteria in the vegetations and prevented emergence of mutants resistant to teicoplanin (5). We undertook the present study to identify the conditions of emergence of mutants resistant to one or both components of the glycopeptide-gentamicin combination in VanB-type strains, in vitro and in the rabbit model of aortic endocarditis.
MATERIALS AND METHODS
Bacterial strains and media.
Enterococcus faecalis JH2-2 (Vms Tes) is susceptible to glycopeptides and β-lactams and is intrinsically resistant to low levels of aminoglycosides (16). E. faecalis BM4281 (Vmr Tes) and BM4275 (Vmr Tes) harbor a 250-kb chromosomal genetic element conferring VanB-type resistance and were obtained by conjugal transfer of vancomycin resistance from clinical isolate Enterococcus faecium BM4120 (Vmr Tes) to JH2-2 (20). E. faecalis BM4309 (Vmr Ter), BM4310 (heterogeneously resistant to vancomycin and teicoplanin [VmHet TeHet]), and BM4307 (vancomycin dependent [Vmd]) are spontaneous in vitro mutants of BM4281 (5). Cultures and antibiotic susceptibility testing were performed in brain heart infusion (BHI) broth or agar (Difco Laboratories, Detroit, Mich.) at 37°C.
Amplification, cloning, and sequencing.
The vanSB and ddl genes were amplified by PCR with Pfu polymerase (Stratagene, La Jolla, Calif.) and oligodeoxyribonucleotide primers (Unité de Chimie Organique, Institut Pasteur, Paris, France) as described previously (7). The amplified genes were cloned into pUC18 and sequenced by the dideoxy chain termination method with T7 DNA polymerase (T7 sequencing kit; Pharmacia, Uppsala, Sweden) and [α-35S]dATP (Amersham Radiochemical Centre, Amersham, England).
Regulation of VanXB d,d-dipeptidase synthesis.
Bacteria were grown to an optical density at 600 nm of 0.7 in broth containing or not containing vancomycin or teicoplanin, treated with lysozyme, and lysed by sonication (2). The lysate was centrifuged at 100,000 × g for 45 min at 4°C, and the supernatant was assayed for d,d-dipeptidase activity by using d-Ala-d-Ala as a substrate and d-amino acid oxidase coupled to peroxidase for the indicator reactions (2).
In vitro susceptibility testing and selection of mutants.
Antibiograms were performed by disk-agar diffusion with disks containing 30 μg of vancomycin, 30 μg of teicoplanin, or 500 μg of gentamicin (Sanofi Diagnostics Pasteur, Marnes-la-Coquette, France). The MICs of vancomycin (Eli Lilly & Co., Saint-Cloud, France), teicoplanin (Marion Merrell Dow, Levallois-Perret, France), and gentamicin (Unilabo, Levallois Perret, France) were determined by the method of Steers et al. (28) with 105 CFU per spot on BHI agar after 24 h of incubation. The MICs of gentamicin were determined by using ca. 1.25-fold dilution of the antibiotic (i.e., 10, 8, 6.4, 5, 4, etc., μg/ml) instead of the usual 2-fold dilution.
For time-kill curves, exponentially growing E. faecalis bacteria were diluted in glass tubes containing 10 ml of broth to obtain 5 × 107 CFU/ml and incubated with vancomycin (10 μg/ml), teicoplanin (10 μg/ml), or gentamicin (4 μg/ml) alone or in combination. Aliquots taken after 0, 3, 6, and 24 h of incubation were plated on agar to enumerate the surviving bacteria. Aliquots taken at 24 h were also plated on agar containing the same antibiotics at the same concentrations as in broth to number the mutants resistant to the gentamicin-glycopeptide combinations, and the plates were incubated for 48 h. As previously shown, antibiotic carryover does not cause a diminution in counts of surviving bacteria (5, 11). The MICs of gentamicin for bacteria recovered from the selective media were determined.
Selection of spontaneous gentamicin-resistant mutants was investigated by plating 0.1 ml of an overnight culture on agar containing or not containing gentamicin at twice the MIC. Mutation frequencies were determined by dividing the number of CFU obtained on selective media by the number of CFU obtained on media devoid of antibiotic after 48 h of incubation.
Experimental endocarditis.
Polyethylene catheters were inserted through the right carotid artery into the left ventricle of New Zealand White rabbits (2.0 to 2.5 kg) (11). Twenty-four hours after catheter insertion, the rabbits were inoculated by the ear vein with approximately 5 × 108 CFU of E. faecalis BM4275 (Vmr Tes) in 1 ml of 0.9% NaCl. The catheter was left in place throughout the experiment. Forty-eight hours after inoculation, animals received intramusculary twice daily for 5 days vancomycin (50 mg/kg of body weight), teicoplanin (20 mg/kg after a loading dose of 40 mg/kg), gentamicin (3 mg/kg), or combinations of gentamicin plus vancomycin or teicoplanin. These regimens were previously shown to lead to peak and trough antibiotic serum levels, respectively, of 57 ± 5.5 and 7.0 ± 1.5 μg/ml for vancomycin, 63 ± 23 and 25 ± 10 μg/ml for teicoplanin, and 7.5 ± 2.0 and <0.2 μg/ml for gentamicin (5). Control animals were left untreated. Animals were killed by intravenous injection of pentobarbital. At the time of sacrifice, the heart was removed and the chambers on the left side were examined to confirm vegetative endocarditis. All vegetations from single rabbits were excised, pooled, weighed, and homogenized in 1 ml of sterile distilled water. Vegetation homogenates were plated on agar to count surviving bacteria and on agar containing teicoplanin (16 μg/ml) or gentamicin at twice the MIC to enumerate mutants after 48 h of incubation. The MICs of gentamicin were determined for bacteria recovered from the plate containing this antibiotic. The phenotype of the mutants selected on teicoplanin was determined by disk-agar diffusion.
Statistics.
Variance analysis followed by the Fisher test for multiple comparisons was used to compare the bacterial counts in vegetations from animals treated with various regimens (27). Comparisons of bacterial counts from animals with or without mutants and treated with the same regimen were performed by the Mann-Whitney U test. The Fisher exact test was used for comparisons of the proportions of animals with vegetations retaining resistant mutants. A P value of <0.05 was considered significant.
RESULTS AND DISCUSSION
Properties of the teicoplanin-resistant mutants.
We previously reported the in vitro selection of spontaneous mutants of E. faecalis BM4281 (Vmr Tes), in particular BM4309, which was homogeneously resistant to vancomycin and teicoplanin (Vmr Ter), BM4310, which was heterogeneously resistant to these antibiotics (VmHet TeHet), and BM4307, which required vancomycin for growth (vancomycin-dependent phenotype [Vmd]) (5). In the present study, we characterized these mutants for the presence of mutations in the vanSB sensor and ddl d-Ala:d-Ala ligase genes (Table 1). Regulation of the glycopeptide resistance genes was also studied by determination of VanXB d,d-dipeptidase activity in crude extracts from bacteria grown under various inducing conditions.
TABLE 1.
Properties of E. faecalis strains
| Strain | Phenotypea | Regulationb | Amino acid substitution in:
|
MIC (μg/ml)c
|
Gentr mutantsd
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| VanSB | Ddl | Vm | Te | Gent | Gent + Vm 10 | Gent + Te 10 | Frequency | MIC range (μg/ml) | |||
| JH2-2 | Vms Tes | NAe | NA | None | 2 | 1 | 16 | NA | NA | 10−7 | 40–64 |
| BM4281 | Vmr Tes | Vmi | None | None | 128 | 1 | 5 | 2 | NA | 10−6 | 13–32 |
| BM4275 | Vmr Tes | Vmi | None | None | 128 | 1 | 16 | 0.5 | NA | 10−7 | 25–64 |
| BM4309 | Vmr Ter | Vmi Tei | E221-G | None | 1,024 | 1,024 | 5 | 2 | 1 | 10−6 | 10–32 |
| BM4310 | VmHet TeHet | Vmi Tei | Q425-stop | None | 512 | 256 | 5 | 1 | <0.25 | 10−6 | 20–40 |
| BM4307 | Vmd | Vmi | None | E203-stop | 1,024 | NA | NA | 2 | NA | NA | NA |
| BM4360 | Vmr Ter | Constitutive | Q31-stop | E203-stop | 1,024 | 1,024 | 5 | 2 | 2 | 10−6 | 13–32 |
Vm, vancomycin; Te, teicoplanin; s, susceptible; r, resistant; Het, heterogeneous; d, dependent.
Vmi, inducible by Vm; Tei, inducible by Te.
MICs of gentamicin were determined in agar containing 10 μg of vancomycin per ml (Gent + Vm 10) or 10 μg of teicoplanin per ml (Gent + Te 10).
Mutants were selected on agar containing gentamicin (Gent) at twice the MIC. The MICs of gentamicin were determined for a minimum of eight mutants for each strain.
NA, not applicable.
E. faecalis BM4309 (Vmr Ter) was inducibly resistant to vancomycin and teicoplanin and harbored a mutation (GAG to GGG) in codon 221 of vanSB which led to a glutamic acid (E) to glycine (G) substitution in the glycopeptide sensor domain of VanSB. The E221-G substitution altered the specificity of the VanSB sensor, since it allowed induction by teicoplanin.
E. faecalis BM4310 (VmHet TeHet) was heterogeneously resistant to vancomycin and teicoplanin. This phenotype is characterized by the presence of small colonies in the inhibition zones of glycopeptides (5). BM4310 (VmHet TeHet) produced a truncated sensor due to a mutation in vanSB that converted the CAA codon specifying glutamine (Q) at position 425 to a TAA translation stop codon. Synthesis of VanXB was inducible by vancomycin and teicoplanin, as previously reported for similar VmHet TeHet mutants harboring nonsense mutations in vanSB (7).
E. faecalis BM4307 (Vmd) required vancomycin for growth and was highly resistant to vancomycin. This vancomycin-dependent mutant did not produce a functional host d-Ala:d-Ala ligase, since a mutation in ddl converted the GAA codon specifying glutamic acid (E) at position 203 to a TAA stop codon. Previous analyses established that the VanB d-Ala:d-Lac ligase is essential for peptidoglycan synthesis in ddl null mutants (7, 12, 24, 29). Vancomycin is required for growth of the mutants since VanB is produced only under inducing conditions (7, 12, 24). VanB-type strains produce small amounts of peptidoglycan precursors ending in the d-Ala-d-Ala target of glycopeptides, in addition to precursors ending in d-Ala-d-Lac, leading to inhibition of cell wall synthesis by high concentrations of vancomycin (2). The ddl null mutations increase the level of vancomycin resistance since they abolish synthesis of d-Ala-d-Ala by the host ligase and prevent production of peptidoglycan precursors containing the target of glycopeptides (7, 12, 24). Thus, the ddl E203-stop mutation accounts for high-level vancomycin resistance and vancomycin dependence of BM4307 (Vmd).
E. faecalis BM4360 (Vmr Ter) was obtained by plating BM4307 (Vmd) on teicoplanin (16 μg/ml). The mutant was constitutively resistant to vancomycin and teicoplanin and produced a truncated sensor following conversion of the CAG codon specifying glutamine (Q) at position 31 to a TAG stop codon. The vanSB Q31-stop mutation abolished the vancomycin requirement for growth since the VanB ligase was produced both in the presence and in the absence of glycopeptides in the culture medium.
In vitro activity of gentamicin.
BM4281 (Vmr Tes) and the teicoplanin-resistant mutants BM4309 (Vmr Ter), BM4310 (VmHet TeHet), and BM4360 (Vmr Ter) were significantly more susceptible to gentamicin (MIC = 5 μg/ml) than the parental strain JH2-2 (MIC = 16 μg/ml) (Table 1). Two lines of evidence indicate that expression of the glycopeptide resistance genes was not responsible for increased susceptibility to gentamicin. VanB-type BM4281 (Vmr Tes) was more susceptible to gentamicin than JH2-2 although the resistance genes were tightly regulated in BM4281, as indicated by the absence of d-Ala-d-Lac-ending peptidoglycan precursors and of VanXB d,d-dipeptidase activity in the absence of induction (2). The MICs of gentamicin were variable in 15 independent transconjugants obtained by mating between VanB-type clinical isolate E. faecalis BM4120 (Vmr Tes) and JH2-2 (20, 21). The MIC of gentamicin was 5 μg/ml for 5 transconjugants, including BM4281 (Vmr Tes), and 16 μg/ml for JH2-2 and the remaining 10 transconjugants, such as BM4275 (Vmr Tes) (Table 1 and data not shown). Thus, increased activity of gentamicin may not be due to acquisition of the vanB gene cluster, since it did not depend upon expression of the glycopeptide resistance genes and was not detected in all of the transconjugants that had received the same vanB cluster. Chromosomal markers were recently reported to be cotransferred with conjugative elements in E. faecalis (7), suggesting that certain VanB transconjugants may have acquired genetic information responsible for increased activity of gentamicin in addition to the vanB element.
MICs of gentamicin determined in the presence of glycopeptides.
The MICs of gentamicin for BM4281 (Vmr Tes) (5 μg/ml) and BM4275 (Vmr Tes) (16 μg/ml) were reduced 2.5- and 32-fold, respectively, when 10 μg of vancomycin per ml was added to the medium (Table 1). Vancomycin or teicoplanin at 10 μg/ml decreased by at least 2.5-fold the MICs of gentamicin against the teicoplanin-resistant mutants derived from BM4281 (Vmr Tes). Thus, the gentamicin-glycopeptide combinations were more active than gentamicin alone in spite of resistance to the glycopeptide present in the combination (Table 1).
Selection of spontaneous gentamicin-resistant mutants.
Mutants resistant to gentamicin were obtained at frequencies of 10−6 to 10−7 on agar containing gentamicin at concentrations of twice the MICs (Table 1). The MIC of gentamicin was increased two- to eightfold and the phenotype was stable after three serial subcultures in the absence of the antibiotic. The aspects of the colonies of the mutants and of the parental strains were similar. The generation times of JH2-2 and of two gentamicin-resistant derivatives of this strain were indistinguishable. Thus, acquisition of gentamicin resistance was not associated with a severe growth defect.
The frequencies in obtaining mutants were similar for JH2-2 (Vms Tes), for transconjugants BM4281 (Vmr Tes) and BM4275 (Vmr Tes), and for the teicoplanin-resistant derivatives of BM4281 (Vmr Tes). Thus, selection of mutants with increased resistance to gentamicin was obtained in JH2-2 and in VanB strains with low (MIC = 5 μg/ml) or moderate (MIC = 16 μg/ml) intrinsic resistance to this antibiotic.
In vitro bactericidal activity of glycopeptide-gentamicin combinations.
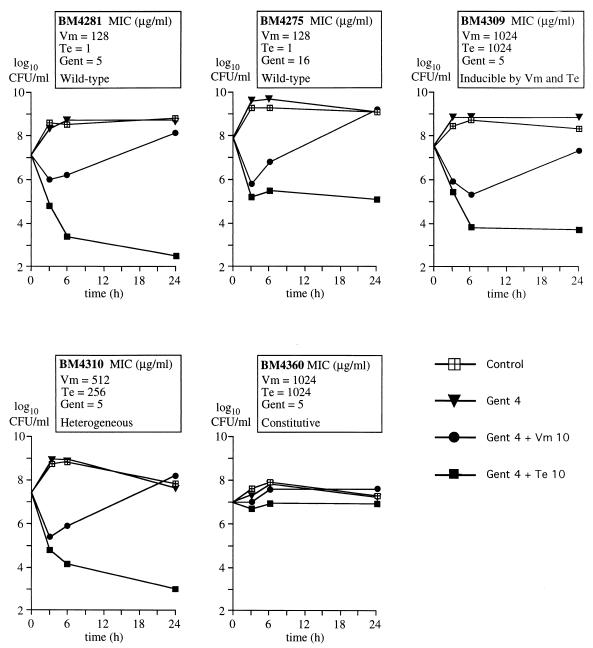
The gentamicin-vancomycin combination initially killed BM4281 (Vmr Tes) and BM4275 (Vmr Tes), but the number of CFU increased after 6 h (Fig. 1), leading to an overall increase in the number of CFU after 24 h of incubation (Table 2). Bacteria incubated with the combination for 24 h formed colonies on agar containing gentamicin and vancomycin (Table 2). The MICs of gentamicin against these bacteria were increased two- to sixfold in comparison with BM4281 (Vmr Tes) and BM4275 (Vmr Tes) (data not shown). Thus, the gentamicin-vancomycin combination selected mutants with increased resistance to gentamicin that were also resistant to the combination. Of note, an initial phase of killing followed by growth was previously observed for VanB-type strains incubated in the presence of vancomycin and streptomycin (19, 25), suggesting that mutants could also be selected by this combination.
FIG. 1.
Time-kill curves. Bacteria were incubated in BHI broth in the absence of antibiotic (control) or in broth containing 4 μg of gentamicin per ml (Gent 4), 4 μg of gentamicin per ml and 10 μg of vancomycin per ml (Gent 4 + Vm 10), or 4 μg of gentamicin per ml and 10 μg of teicoplanin per ml (Gent 4 + Te 10). Surviving bacteria were enumerated on agar plates after 0, 3, 6, and 24 h of incubation at 37°C.
TABLE 2.
Bactericidal activity of gentamicin associated with a glycopeptide and selection of mutants resistant to the combinationsa
| Strain | Phenotypeb | Regulationc | Vancomycin plus gentamicin
|
Teicoplanin plus gentamicin
|
||
|---|---|---|---|---|---|---|
| Variation of log10 CFU in 24 h (mean ± SD) | Resistant mutantsd | Variation of log10 CFU in 24 h (mean ± SD) | Resistant mutantsd | |||
| JH2-2 | Vms Tes | NA | −(1.7 ± 1.4) | − | −(2.6 ± 1.0) | − |
| BM4281 | Vmr Tes | Vmi | +(0.3 ± 0.9) | + | −(4.6 ± 0.5) | − |
| BM4275 | Vmr Tes | Vmi | +(0.7 ± 0.8) | + | −(2.2 ± 1.6) | − |
| BM4309 | Vmr Ter | Vmi Tei | −(0.6 ± 1.3) | + | −(3.4 ± 1.3) | + |
| BM4310 | VmHet TeHet | Vmi Tei | +(0.8 ± 0.2) | + | −(4.0 ± 1.9) | + |
| BM4360 | Vmr Ter | Constitutive | +(0.3 ± 0.1) | + | +(0.6 ± 0.6) | + |
Bacteria (5 × 107 CFU/ml) were inoculated in broth containing gentamicin (4 μg/ml) and vancomycin (10 μg/ml) or teicoplanin (10 μg/ml) and incubated at 37°C for 24 h. Surviving bacteria and mutants were enumerated on agar or on agar containing the same antibiotics at the same concentrations.
Vm, vancomycin; Te, teicoplanin; s, susceptible; r, resistant; Het, heterogeneous.
NA, not applicable; Vmi, inducible by Vm; Tei, inducible by Te.
+, presence of mutants; −, absence of mutants.
The combination of gentamicin and teicoplanin was active against BM4281 (Vmr Tes) and, to a lesser extent, against BM4275 (Vmr Tes), producing 4.6 ± 0.5 and 2.2 ± 1.6 log10 reductions in the initial inoculum after 24 h of incubation, respectively (Fig. 1 and Table 2). Mutants resistant to the combination were not detected by plating the surviving bacteria on agar containing gentamicin and teicoplanin (Table 2).
The mutants of BM4281, BM4309 (Vmr Ter), and BM4310 (VmHet TeHet), which were inducibly resistant to vancomycin and teicoplanin, were initially killed by vancomycin and gentamicin, but the number of CFU increased after 6 h (Fig. 1 and Table 2). BM4309 (Vmr Ter) and BM4310 (VmHet TeHet) were killed by the teicoplanin-gentamicin combination despite high-level resistance to teicoplanin. The gentamicin-glycopeptide combinations did not kill BM4360 (Vmr Ter), which was constitutively resistant to vancomycin and teicoplanin.
Derivatives of BM4309 (Vmr Ter), BM4310 (VmHet TeHet), and BM4360 (Vmr Ter) resistant to the glycopeptide-gentamicin combinations were detected among surviving bacteria after a 24-h incubation period (Table 2). These derivatives were more resistant to gentamicin than the parental strains.
In summary, selection of gentamicin-resistant mutants by the combinations occurred if the strain was resistant to the glycopeptide present in the combination. In contrast, teicoplanin and gentamicin did not select mutants of BM4281 (Vmr Tes) and BM4275 (Vmr Tes) that were susceptible to teicoplanin. The combination of gentamicin and teicoplanin killed the inducible mutants BM4309 (Vmr Ter) and BM4310 (VmHet TeHet) and was bacteriostatic against constitutive mutant BM4360 (Vmr Ter) (Fig. 1; Tables 1 and 2). Thus, acquisition of teicoplanin resistance by E. faecalis BM4281 (Vmr Tes) was not sufficient to totally suppress the in vitro activity of the gentamicin-teicoplanin combination. A second mutation increasing the level of gentamicin resistance was required for growth in the presence of gentamicin and teicoplanin. Spontaneous teicoplanin-resistant mutants of VanB-type strains were obtained at a frequency of 10−7 (5, 7). Spontaneous gentamicin-resistant mutants of BM4275, BM4281, and their derivatives were obtained at frequencies of 10−6 to 10−7 (Table 1). Simultaneous acquisition of the two types of mutations was not observed. As expected for two rare events, the mutations conferring teicoplanin and gentamicin resistance were obtained only in two steps.
Experimental endocarditis.
Various 5-day antibiotic regimens were tested in the rabbit model of endocarditis against E. faecalis BM4275 (Vmr Tes), which did not display increased susceptibility to gentamicin in comparison to JH2-2 (Vms Tes) (Table 1). The efficacy of glycopeptides and gentamicin, alone or in combination, was analyzed in terms both of reduction of bacterial counts in the vegetations and of selection of mutants resistant to these antibiotics (Fig. 2).
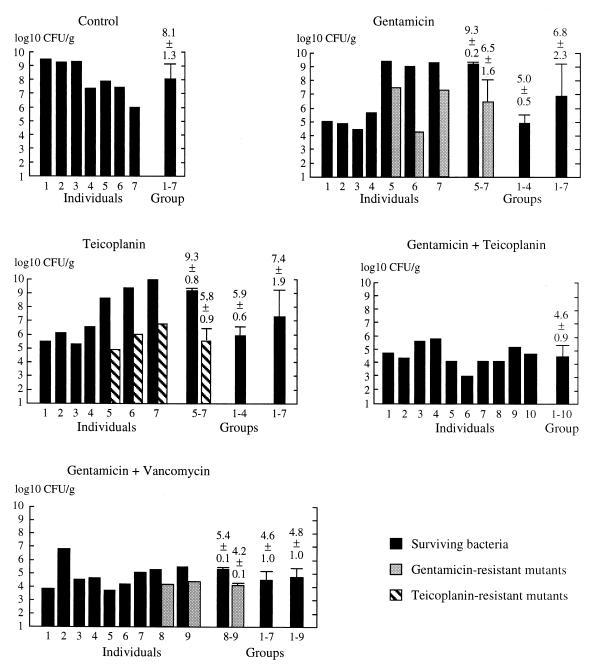
FIG. 2.
Efficacy of various antibiotic regimens for treatment of experimental endocarditis due to E. faecalis BM4275 (Vmr Tes). Vegetation homogenates were plated on agar to enumerate surviving bacteria and on agar containing teicoplanin (16 μg/ml) or gentamicin (32 μg/ml) to enumerate mutants resistant to teicoplanin or gentamicin, respectively. Log10 CFU per gram of vegetation are shown for individual rabbits. For the gentamicin, teicoplanin, and gentamicin-plus-vancomycin regimens, animals were assigned to two groups based on the presence or absence of mutants. The mean ± standard deviation of log10 CFU per gram was calculated for the two groups and for all rabbits.
The gentamicin regimen selected gentamicin-resistant mutants of BM4275 (Vmr Tes) in three (rabbits 5, 6, and 7) of seven rabbits. The mutants were two- to fivefold more resistant to gentamicin than was BM4275 (Vmr Tes) (data not shown). The total number of bacteria per gram of vegetation was significantly lower for the group of four animals that did not contain mutants (rabbits 1, 2, 3, and 4) than for the remaining three animals (5.0 ± 0.5 versus 9.3 ± 0.2 [P < 0.05]) or the control animals (5.0 ± 0.5 versus 8.1 ± 1.3 [P < 0.05]). The results obtained for the latter two groups were not different (P > 0.05). Overall, the gentamicin regimen did not significantly reduce bacterial titers in vegetations in the entire group of seven animals in comparison with control animals (6.8 ± 2.3 versus 8.1 ± 1.3 [P > 0.05]).
As previously described (5), the teicoplanin regimen selected mutants resistant to this antibiotic in three of seven animals. Both Vmr Ter and VmHet TeHet mutants were detected in rabbits 6 and 7. Heterogeneous mutants were present in rabbit 5. The total number of bacteria per gram of vegetation was significantly lower for the group of four animals that did not harbor mutants than for the remaining animals (5.9 ± 0.6 versus 9.3 ± 0.8 [P < 0.05]) or control animals (5.9 ± 0.6 versus 8.1 ± 1.3 [P < 0.05]). Overall, the teicoplanin regimen did not significantly reduce the number of bacteria in the vegetations (7.4 ± 1.9 versus 8.1 ± 1.3).
Each rabbit was inoculated with 5 × 108 CFU, which included approximately 50 mutants since spontaneous mutants resistant to gentamicin or teicoplanin were obtained in vitro at a frequency of approximately 10−7 (Table 1). The number of mutants in the cardiac vegetations of individual rabbits ranged from 1,400 to 800,000 at the end of therapy. Thus, the number of mutants increased 28- to 16,000-fold during therapy with gentamicin or teicoplanin alone.
The combinations of gentamicin plus teicoplanin and gentamicin plus vancomycin were similarly active against BM4275 (Vmr Tes), leading to 3.5 or 3.3 log10 reductions in the number of bacteria per gram of vegetation (P < 0.05 [versus control]). No mutants resistant to gentamicin or teicoplanin were detected in rabbits treated with the gentamicin-teicoplanin combination. In contrast, gentamicin-resistant mutants were detected in the vegetations of two of nine rabbits treated with gentamicin and vancomycin. Statistically, the proportion of animals containing mutants resistant to gentamicin or teicoplanin was significantly lower in the 10 animals treated with the teicoplanin-gentamicin combination than in the 14 animals treated with gentamicin or teicoplanin alone (0 of 10 versus 6 of 14 [P = 0.04]). Thus, the teicoplanin-gentamicin combination was the only regimen effective in preventing the emergence of resistant mutants in vivo.
Conclusions.
Combination of a glycopeptide with an aminoglycoside is the reference treatment for severe enterococcal infections if the patient is allergic to β-lactams or if the strain is highly resistant to the latter antibiotics (18, 31). Efficacy of the combination is abolished, both in vitro and in animal models, by high-level resistance to vancomycin and teicoplanin mediated by gene clusters related to vanA (9, 17, 26). In contrast, combination of an aminoglycoside and teicoplanin may be efficient against VanB-type strains since the bacteria remain susceptible to teicoplanin. Indeed, combinations of teicoplanin and gentamicin (5) or streptomycin (19) were reported to be active against VanB-type enterococci in vitro and in experimental endocarditis, leading to the sterilization of cardiac vegetations in 25% (5) and 75% (19) of rabbits.
Mutants of E. faecalis BM4275 (Vmr Tes) highly resistant to teicoplanin were recovered from the cardiac vegetations of rabbits treated with teicoplanin, indicating that emergence of teicoplanin resistance under treatment with this glycopeptide may be responsible for therapeutic failure (Fig. 2). However, combination of teicoplanin and gentamicin prevented emergence of such mutants. In order to investigate the basis for the lack of emergence of mutants, the in vitro activity of the gentamicin-teicoplanin combination was determined against mutants of E. faecalis BM4281 (Vmr Tes) that were resistant to high levels of teicoplanin by three different mechanisms (Table 1). None of the corresponding mutations totally abolished the in vitro activity of the gentamicin-teicoplanin combination (Fig. 1; Table 2). A second mutation increasing the intrinsic level of gentamicin resistance was required for growth in the presence of the combination. It is therefore likely that teicoplanin-resistant mutants of VanB-type strains do not emerge under treatment with gentamicin and teicoplanin, since growth of such mutants would be inhibited by the combination in the vegetations. Therapy with gentamicin alone selected mutants resistant to this antibiotic (Fig. 2). However, simultaneous acquisition of two mutations conferring teicoplanin and gentamicin resistance was not observed in the animals, as expected for the combination of two rare events (Fig. 2). In agreement with this notion, the vancomycin-gentamicin regimen selected mutants of BM4275 (Vmr Tes) that were resistant to the combination. In this case, a single mutation conferring gentamicin resistance was sufficient, since the strain was already resistant to vancomycin. These observations imply that sequential treatments with the vancomycin-gentamicin and teicoplanin-gentamicin combinations may lead to the selection of teicoplanin- and gentamicin-resistant mutants in two steps. Thus, prior treatment with the vancomycin-gentamicin combination may compromise the efficacy of the gentamicin-teicoplanin combination.
ACKNOWLEDGMENTS
A.L. was supported by the Fondation pour la Recherche Médicale, and M.B. was supported by Programa Praxis XXI of the Fundaçao para a Ciência e Tecnologia and by the Programa Gulbenkian de Doutoramento em Biologia e Medicina, Portugal. This work was supported in part by Marion Merrell Dow Europe and by a Bristol-Myers Squibb unrestricted Biomedical Research Grant in Infectious Diseases.
REFERENCES
- 1.Arthur M, Depardieu F, Gerbaud G, Galimand M, Leclercq R, Courvalin P. The VanS sensor negatively controls VanR-mediated transcriptional activation of glycopeptide resistance genes of Tn1546 and related elements in the absence of induction. J Bacteriol. 1997;179:97–106. doi: 10.1128/jb.179.1.97-106.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Depardieu F, Reynolds P, Courvalin P. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol. 1996;21:33–44. doi: 10.1046/j.1365-2958.1996.00617.x. [DOI] [PubMed] [Google Scholar]
- 3.Arthur M, Molinas C, Courvalin P. The VanS-VanR two-component regulatory system controls synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1992;174:2582–2591. doi: 10.1128/jb.174.8.2582-2591.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur M, Reynolds P, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. doi: 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 5.Aslangul E, Baptista M, Fantin B, Depardieu F, Arthur M, Courvalin P, Carbon C. Selection of glycopeptide-resistant mutants of VanB-type Enterococcus faecalis BM4281 in vitro and in experimental endocarditis. J Infect Dis. 1997;175:598–605. doi: 10.1093/infdis/175.3.598. [DOI] [PubMed] [Google Scholar]
- 6.Baptista M, Depardieu F, Courvalin P, Arthur M. Specificity of induction of glycopeptide resistance genes in Enterococcus faecalis. Antimicrob Agents Chemother. 1996;40:2291–2295. doi: 10.1128/aac.40.10.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baptista M, Depardieu F, Reynolds P, Courvalin P, Arthur M. Mutations leading to increased levels of resistance to glycopeptide antibiotics in VanB-type enterococci. Mol Microbiol. 1997;25:93–105. doi: 10.1046/j.1365-2958.1997.4401812.x. [DOI] [PubMed] [Google Scholar]
- 8.Bugg T D H, Wright G D, Dutka-Malen S, Arthur M, Courvalin P, Walsh C T. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. 1991;30:10408–10415. doi: 10.1021/bi00107a007. [DOI] [PubMed] [Google Scholar]
- 9.Caron F, Carbon C, Gutmann L. Triple-combination penicillin-vancomycin-gentamicin for experimental endocarditis caused by a moderately penicillin- and highly glycopeptide-resistant isolate of Enterococcus faecium. J Infect Dis. 1991;164:888–893. doi: 10.1093/infdis/164.5.888. [DOI] [PubMed] [Google Scholar]
- 10.Evers S, Courvalin P. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J Bacteriol. 1996;178:1302–1309. doi: 10.1128/jb.178.5.1302-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fantin B, Leclercq R, Arthur M, Duval J, Carbon C. Influence of low-level resistance to vancomycin on efficacy of teicoplanin and vancomycin for treatment of experimental endocarditis due to Enterococcus faecium. Antimicrob Agents Chemother. 1991;35:1570–1575. doi: 10.1128/aac.35.8.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraimow H S, Jungkind D L, Lander D W, Delso D R, Dean J L. Urinary tract infection with an Enterococcus faecalis isolate that requires vancomycin for growth. Ann Intern Med. 1994;121:22–26. doi: 10.7326/0003-4819-121-1-199407010-00004. [DOI] [PubMed] [Google Scholar]
- 13.Gutmann L, Billot-Klein D, Al-Obeid S, Klare I, Francoual S, Collatz E, van Heijenoort J. Inducible carboxypeptidase activity in vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1992;36:77–80. doi: 10.1128/aac.36.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayden M K, Trenholme G M, Schultz J E, Sahm D F. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J Infect Dis. 1993;167:1224–1227. doi: 10.1093/infdis/167.5.1224. [DOI] [PubMed] [Google Scholar]
- 15.Holman T R, Wu Z, Wanner B L, Walsh C T. Identification of the DNA-binding site for the phosphorylated VanR protein required for vancomycin resistance in Enterococcus faecium. Biochemistry. 1994;33:4625–4631. doi: 10.1021/bi00181a024. [DOI] [PubMed] [Google Scholar]
- 16.Jacob A E, Hobbs S J. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J Bacteriol. 1974;117:360–372. doi: 10.1128/jb.117.2.360-372.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leclercq R, Bingen E, Su Q H, Lambert-Zechovski N, Courvalin P, Duval J. Effects of combinations of β-lactams, daptomycin, gentamicin, and glycopeptides against glycopeptide-resistant enterococci. Antimicrob Agents Chemother. 1991;35:92–98. doi: 10.1128/aac.35.1.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray B E. The life and times of the enterococcus. Clin Microbiol Rev. 1990;3:46–65. doi: 10.1128/cmr.3.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolau D P, Marangos M N, Nightingale C H, Patel K B, Cooper B W, Quintiliani R, Jr, Courvalin P, Quintiliani R. Efficacy of vancomycin and teicoplanin alone and in combination with streptomycin in experimental, low-level vancomycin-resistant, VanB-type Enterococcus faecalis endocarditis. Antimicrob Agents Chemother. 1996;40:55–60. doi: 10.1128/aac.40.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quintiliani R, Jr, Courvalin P. Conjugal transfer of the vancomycin resistance determinant vanB between enterococci involves the movement of large genetic elements from chromosome to chromosome. FEMS Microbiol Lett. 1994;119:359–364. doi: 10.1111/j.1574-6968.1994.tb06913.x. [DOI] [PubMed] [Google Scholar]
- 21.Quintiliani R, Jr, Evers S, Courvalin P. The vanB gene confers various levels of self-transferable resistance to vancomycin in enterococci. J Infect Dis. 1993;167:1220–1223. doi: 10.1093/infdis/167.5.1220. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds P E. Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur J Clin Microbiol Infect Dis. 1989;8:943–950. doi: 10.1007/BF01967563. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds P E, Depardieu F, Dutka-Malen S, Arthur M, Courvalin P. Glycopeptide resistance mediated by enterococcal transposon Tn1546 requires production of VanX for hydrolysis of d-alanyl-d-alanine. Mol Microbiol. 1994;13:1065–1070. doi: 10.1111/j.1365-2958.1994.tb00497.x. [DOI] [PubMed] [Google Scholar]
- 24.Rosato A, Pierre J, Billot-Klein D, Buu-Hoi A, Gutmann L. Inducible and constitutive expression of resistance to glycopeptides and vancomycin dependence in glycopeptide-resistant Enterococcus avium. Antimicrob Agents Chemother. 1995;39:830–833. doi: 10.1128/aac.39.4.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sahm D F, Kissinger J, Gilmore M S, Murray P R, Mulder R, Solliday J, Clarke B. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shlaes D M, Etter L, Gutmann L. Synergistic killing of vancomycin-resistant enterococci of classes A, B, and C by combinations of vancomycin, penicillin, and gentamicin. Antimicrob Agents Chemother. 1991;35:776–779. doi: 10.1128/aac.35.4.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steel R G D, Torrie J H. Principles and procedures of statistics: a biometrical approach. New York, N.Y: McGraw-Hill; 1980. pp. 172–194. [Google Scholar]
- 28.Steers E, Foltz E L, Graves B S, Riden J. An inocula replicating apparatus for routine testing of bacterial susceptibility to antibiotics. Antibiot Chemother (Basel) 1959;9:307–311. [PubMed] [Google Scholar]
- 29.Van Bambeke F, Chauvel M, Reynolds P E, Fraimow H S, Courvalin P. Vancomycin-dependent Enterococcus faecalis clinical isolates and revertant mutants. Antimicrob Agents Chemother. 1999;43:41–47. doi: 10.1128/aac.43.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walsh C, Fisher S, Park I S, Prahalad M, Wu Z. Bacterial resistance to vancomycin: five genes and one missing hydrogen bond tell the story. Chem Biol. 1996;3:21–28. doi: 10.1016/s1074-5521(96)90079-4. [DOI] [PubMed] [Google Scholar]
- 31.Woodford N, Johnson A P, Morrison D, Speller D C E. Current perspectives on glycopeptide resistance. Clin Microbiol Rev. 1995;8:585–615. doi: 10.1128/cmr.8.4.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright G D, Holman T R, Walsh C T. Purification and characterization of VanR and the cytosolic domain of VanS: a two-component regulatory system required for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1993;32:5057–5063. doi: 10.1021/bi00070a013. [DOI] [PubMed] [Google Scholar]
- 33.Wu Z, Wright G D, Walsh C T. Overexpression, purification and characterization of VanX, a d,d-dipeptidase which is essential for vancomycin resistance in Enterococcus faecium BM4147. Biochemistry. 1995;34:2455–2463. doi: 10.1021/bi00008a008. [DOI] [PubMed] [Google Scholar]