Summary
Major histocompatibility complex (MHC) tetramers can work as diagnostic tools to identify antigen-specific T cells in immunological research and monitoring. Here, we provide a general protocol for the production of MHC tetramer. We obtain highly pure N-terminal His-tagged HLA-A2 α chain and β2-microglobulin (β2m) to fold a monomer with a photocleavable peptide, which can exchange with an HLA-A2 presented peptide derived from influenza A virus. Further those monomers compose tetramer to stain antigen-specific CD8+ T cells.
For complete details on the use and execution of this protocol, please refer to Xiao C.C. et al. (2021).
Subject areas: Biotechnology and bioengineering, Cell Biology, Immunology, Molecular Biology, Protein Biochemistry, Protein expression and purification
Graphical abstract
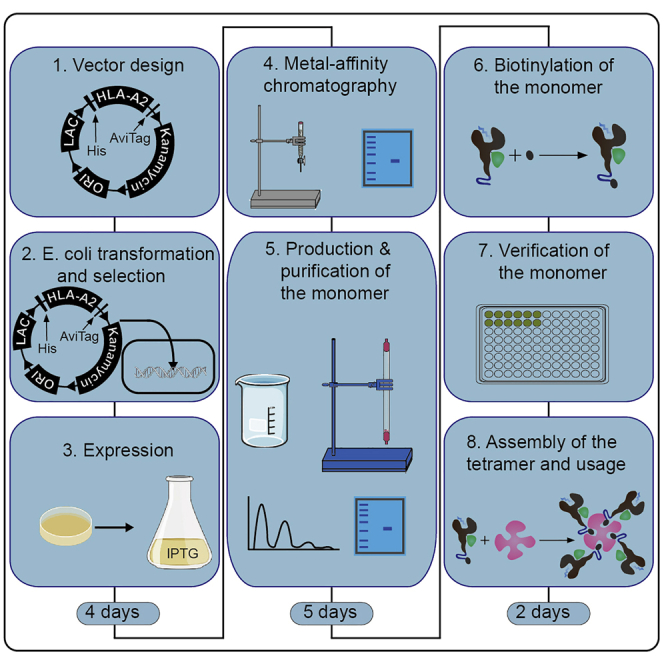
Highlights
-
•
Protocol for the production of HLA-A2 α chain and β2m
-
•
Rapid approach to produce a large number of antigen-specific HLA-A2 tetramers
-
•
The detection Influenza A virus associated antigen-specific CD8+ T cells from PBMCs
Major histocompatibility complex (MHC) tetramers can work as diagnostic tools to identify antigen-specific T cells in immunological research and monitoring. Here, we provide a general protocol for the production of MHC tetramer. We obtain highly pure N-terminal His-tagged HLA-A2 α chain and β2-microglobulin (β2m) to fold a monomer with a photocleavable peptide, which can exchange with an HLA-A2 presented peptide derived from influenza A virus. Further those monomers compose tetramer to stain antigen-specific CD8+ T cells.
Before you begin
Cloning human leukocyte antigen heavy chain and light chain (HLA-A2α chain and β2m) DNA coding sequence into pET28a plasmid
Timing: n/a
-
1.Use molecular biology technique of choice to clone the DNA sequence encoding the open reading frame of the HLA-A2 α chain and β2m in an expression vector for bacteria with the following characteristics:
-
a.N-terminal poly-histidine tag (6×His);
-
b.C-terminal AviTag;
-
c.Resistance to kanamycin
-
d.IPTG-inducible expression using the regulatory sequences of a commercially available plasmid.
-
a.
For example, our designed optimized encoding sequences of HLA-A2 α chain and β2m were respectively cloned in into the pET28a expression plasmid.
Note: We recommend propagating and maintaining the plasmid encoding the HLA-A2 α chain and β2m inserts in an adequate Escherichia coli strain, such as BL21 or DH5α. BL21 is used for protein expression, DH5α is used for plasmid amplification. Please check the construct is in frame by sequencing the vector. It is worth noting that only the HLA-A2 α chain has a C-terminal AviTag.
Obtaining HLA-A2 α chain and β2M-expressing E. coli
Timing: 2 days
-
2.
The plasmid containing HLA-A2 α chain and β2m insert are transform by heat shock method. Add 1 μL of the 100 ng/μL plasmid containing HLA-A2 α chain and β2m into 100 μL of CaCl2-competent E. coli strain BL21. Incubate on ice for 30 min. Then, place the tube in a 42°C water bath for 1 min, immediately followed by 2 min-incubation on ice. Add 1 mL of Luria-Bertani (LB, Table 1) and incubate for 1 h at 37°C whilst shaking at 220 rpm. In parallel, also transform bacteria with the empty plasmid as a transformation control(Prato C.A. et al., 2020).
Note: The plasmid was obtained from a plasmid minipreparation. Judge the quality plasmid by measuring its optical density (OD). Make sure that OD260:OD280 is between 1.8 and 2.0 while the OD260:OD230 is between 2.0 and 2.5.
-
3.
Seed 100 μL of transformed bacteria in LB agar plates with 10 μg/mL kanamycin for 16 h at 37°C.
Note: Please use the appropriate antibiotics for your plasmid.
-
4.
Testing for HLA expressing in at least three colonies.
-
5.
Pilot expression experiment
Collect an aliquot to test, and the rest of the sample is going forward to the next step in this protocol.-
a.Inoculate a single E. coli colony into 6 mL of LB with 10 μg/mL kanamycin in a 10 mL-conical tube.
-
b.Grow at 37°C whilst shaking at 220 rpm.
-
c.Induce with IPTG (final concentration: 1 mM) between an OD of 0.6–0.8.
-
d.Incubate for another 4 h at 23°C whilst shaking at 220 rpm.Optional: Save 1 mL aliquot before and after IPTG addition, centrifuge at 12,000 g and 4°C for 1 min. Then discard the supernatant and add 100 μL gel loading buffer to the pellet. Subsequently, degenerate the sample for 10 min at 95°C. This sample can be refrigerated. Follow with SDS-PAGE electrophoresis to determine whether the protein has expressed.
-
e.Centrifuge the bacterial expression culture for 10 min at 4,000 g and 4°C, then discard the supernatant.
-
f.Resuspend the pellet in 3 mL of ultrasonic liquid 1 (pH8, as described in Table 2).
-
g.Lyse the resuspended bacterial pellet and perform ultrasound by ultrasonic cell crusher (key resources table) for 10 min on ice.
-
h.Centrifuge a 200 μL aliquot for 20 min at 12,000 g and 4°C.
-
i.Save 75 μL supernatant with 25 μL gel loading buffer in a separate Eppendorf (EP) tube at 4°C and resuspend the pellet in 100 μL of gel loading buffer.
-
j.Analyze an aliquot from the resuspended pellet (10 μL), the supernatant (10 μL), and the low molecular weight (LMW) protein ladder (10 μL) by 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE.). Stain the gel with Coomassie blue, using the protocol provided by the manufacturer (https://www.beyotime.com/), verify the presence of a differential band of the expected size in the induced sample obtained from the bacterial culture of insert-containing clone. If different colonies were screened, decide which clone yields the best expression of the recombinant protein in the soluble fraction (Figure 1).
-
a.
-
6.Glycerol stocks
-
a.According to the result of step 4j, mix gently 750 μL of the reserve culture (point 4b) with 250 μL of sterile 40% glycerol.
-
b.Freeze immediately at –80°C.
-
c.For each HLA-A2 α chain and β2m expression and purification, inoculate the bacteria from –80°C culture stocks LB agar plate supplemented with 10 μg/mL kanamycin.
-
a.
Note: HLA-A2 α chain and β2m-transformed bacteria glycerol stocks can be stocked for at least two years at –80°C. Longer than that period, reduced protein expression yields maybe experienced and a new E. coli transformation should be used.
Table 1.
Culture broth
| Reagent | Final concentration | Amount |
|---|---|---|
| Luria Bertani Broth (LB) | ||
| Tryptone | 10 g/L | 10 g |
| Yeast extract | 5 g/L | 5 g |
| NaCl | 5 g/L | 5 g |
| Total | n/a | 1 L |
| Kanamycin | ||
| Kanamycin | 100 mg/mL | 1 g |
| Total | n/a | 10 mL |
Storage: 18°C–25°C.
Prepare a stock solution of 100 mg/mL in deionized water (1,000× solution). Filter-sterilize through a 0.22 μm syringe filter. Store the stock solution in aliquots at –20°C. To prepare media, add kanamycin to a final concentration of 100 μg/mL to the cooled broth.
Table 2.
Buffers
| Reagent | Final concentration (mM) | Amount |
|---|---|---|
| Ultrasonic liquid | ||
| Ultrasonic liquid 1: Ordinary wash buffer (pH 8.0) | ||
| Tris-base | 20 | 2.42 g |
| NaCl | 500 | 29.22 g |
| EDTA | 1 | 0.29 g |
| Triton X-100 | 3% (add before use) | n/a |
| Total | n/a | 1 L |
| Ultrasonic liquid 2: 2 M urea wash buffer (pH 8.0) | ||
| Tris-base | 20 | 2.42 g |
| NaCl | 500 | 29.22 g |
| EDTA | 1 | 0.29 g |
| Urea | 2,000 | 120.12 |
| Triton X-100 | 3% (add before use) | n/a |
| β-Mercaptoethanol | 10 (add before use) | n/a |
| Total | n/a | 1 L |
| Storage: 18°C–25°C | ||
| Nickel column loading buffer (pH 8.0) | ||
| Tris-base | 17.17 | 2.42 g |
| NaCl | 500 | 29.22 g |
| Urea | 8,000 | 480.48 g |
| Imidazole | 10 | 0.68 g |
| β-Mercaptoethanol | 10 (add before use) | n/a |
| Total | n/a | 1 L |
| Storage: 18°C–25°C | ||
| Nickel column cleaning buffer (pH 8.0) | ||
| Tris-base | 20 | 2.42 g |
| NaCl | 500 | 29.22 g |
| Urea | 8,000 | 480.48 g |
| Imidazole | 40 | 2.72 g |
| β-Mercaptoethanol | 10 (add before use) | n/a |
| Total | n/a | 1 L |
| Storage: 18°C–25°C | ||
| Nickel column eluting buffer (pH 8.0) | ||
| Tris-base | 20 | 2.42 g |
| NaCl | 500 | 29.22 g |
| Urea | 8,000 | 480.48 g |
| Imidazole | 150 | 10.21 g |
| β-Mercaptoethanol | 10 (add before use) | n/a |
| Total | n/a | 1 L |
| Storage: 18°C–25°C | ||
| Refolding buffer (pH 8.0) | ||
| Tris-base | 100 | 12.11 g |
| EDTA | 5 | 1.46 g |
| Urea | 5,000 | 300.30 g |
| L-arginine | 400 | 69.68 g |
| Reduced glutathione | 5 (add before use) | n/a |
| Oxidized glutathione | 0.5 (add before use) | n/a |
| Total | n/a | 1 L |
| Storage: 18°C–25°C | ||
| Dialyzate (pH 8.0) | ||
| Tris-base | 25 | 15.14 |
| Total | n/a | 5 L |
| Storage: 2°C–8°C | ||
| 10× dilution buffer (pH 8.0) | ||
| Tris-base | 500 | 30.28 g |
| BSA | 1% | 5 g |
| Tween 20 | 0.2% | 1 mL |
| NaCl | 1,000 | 29.22 g |
| Total | n/a | 500 mL |
| Storage: 2°C–8°C | ||
| 10× substrate buffer (pH 4.0) | ||
| Citric Acid | 59 | 1.13 g |
| Tri-Sodium Citrate dihydrate | 41 | 1.21 g |
| Total | n/a | 100 mL |
| Storage: 2°C–8°C | ||
| 100× Hydrogen peroxide stock solution | ||
| H2O2 | 0.6% (v/v) | 10 mL (3% H2O2) |
| Total | n/a | 50 mL |
| Storage: 2°C–8°C | ||
| Stop solution | ||
| Oxalic acid dehydrate | 2% (w/v) | 2 g |
| Total | n/a | 100 mL |
| Storage: 18°C–25°C | ||
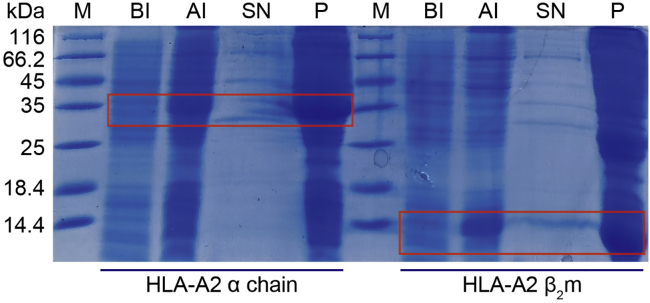
Figure 1.
Analysis of recombinant HLA-A2 α chain and β2m expression by Coomassie Blue-Stained SDS-PAGE
A distinct band of 35.9 kDa and 14 kDa can be observed for insoluble fractions (35.9 kDa and 14 kDa bands are circled with a red rectangle in the picture). BI, before IPTG induce. AI, after IPTG induce. SN, supernatant after ultrasound centrifugation. P, pellet after ultrasound centrifugation. M, molecular weight marker.
HLA-A2 α chain and β2m expression in E.coli
Timing: 2 days
-
7.Starter culture
-
a.Transfer a single colony of transformed bacteria to a sterile 10 mL-conical tube containing 6 mL of LB with 10 μg/mL kanamycin.
-
b.Incubate for 16 h at 37°C while shaking at 220 rpm.
-
a.
-
8.Induction of protein expression
-
a.Expand the starter culture by 1/100 dilution into a sterile 2 L-conical flask containing 500 mL of LB plus 10 μg/mL kanamycin.
-
b.Incubate at 37°C whilst shaking at 220 rpm until 0.6 < OD600 < 0.8
-
c.Add IPTG to a final concentration of 1 mM and incubate for 4 h at 23°C whilst shaking at 220 rpm.
-
a.
-
9.Collection of bacteria
-
a.Centrifuge the culture for 10 min at 4,000 g and 4°C.
-
b.Discard the supernatant and freeze the bacterial pellet at –80°C.
-
a.
Pause point: Bacterial pellets can be stored directly at –80°C for three months. It’s recommended that resuspend the bacterial pellet in a sucrose-based resuspension buffer prior to freezing for longer storage.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| APC anti-human CD8a Antibody | BioLegend | Cat#301049 |
| Purified anti-human CD28 | BioLegend | Cat#302934 |
| HRP anti-human β2-microglobulin antibody | BioLegend | Cat#280303 |
| Bacterial strains | ||
| DH5α | N/A | N/A |
| E. coli BL21 | N/A | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS ) | Merck | Cat#A800764 |
| BeyoPure Ultrapure Water | Beyotime | Cat#ST876 |
| Biotin (50 mM) | Invitrogen | Cat#2110450 |
| Bovine serum albumin (BSA) | Solarbio | Cat#A8020 |
| Citric Acid | Sigma-Aldrich | Cat#C2404 |
| Dialysis Membranes | Solarbio | Cat#YA1045 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat#D2650 |
| DL-Dithiothreitol (DTT) | Beyotime | Cat#ST-043 |
| EDTA ph8.0 (0.5 M) | KeyGen BioTECH | Cat#KGR0084 |
| ELISA Wash Buffer 20× concentrate | BioLegend | Cat#421601 |
| ELISA Coating Buffer (5×) | BioLegend | Cat#421701 |
| Glutathione oxidized | Sangon biotech | Cat#A610228-0001 |
| Glutathione reduced | Sangon biotech | Cat#A600229-0005 |
| High Affinity Ni-NTA Resin | GenScript | Cat#L00250 |
| IL-2 | SL Pharm | N/A |
| Iscove’s Modified Dulbecco’s Medium (IMDM) | HyClone | Cat#SH30228.01 |
| Isopropyl β-D-thiogalactoside (IPTG) | Meilunbio | Cat#MB3026 |
| Kanamycin | Beyotime | Cat#ST101 |
| Nunc™ MaxiSorp™ ELISA Plates, Uncoated | BioLegend | Cat#423501 |
| NuPAGE LDS Sample Buffer (4×) | Invitrogen | Cat#2197595 |
| PE streptavidin | BioLegend | Cat#405203 |
| Protein Marker (14.4–116 kDa) | Beyotime | Cat#P0062 |
| SDS-PAGE Gel Quick Preparation Kit | Beyotime | Cat#P0012AC |
| SDS-PAGE Protein Sample Loading Buffer (1×, Odorless) | Beyotime | Cat#P0287 |
| Streptavidin solution at 1 mg/mL | BioLegend | Cat#280302 |
| Tri-Sodium Citrate dihydrate | Sigma-Aldrich | Cat#S1804 |
| Tryptone | OXOID | Cat#LP0042 |
| Yeast extract | OXOID | Cat#LP0021 |
| EBV EBNA4416-424 IVTDFSVIK peptide | GenScript | Customized |
| Influenza A MP58-66 GILGFVFTL peptide (GIL) | GenScript | Customized |
| Influenza A PB1413–421 NMLSTVLGV (F1) | GenScript | Customized |
| Influenza A PA46–54 FMYSDFHFI (F1) | GenScript | Customized |
| Influenza A PA86–96 RTMAWTVVNSI (F3) | GenScript | Customized |
| Influenza A NP275-283 CLPACVYGL (F4) | GenScript | Customized |
| Influenza A NP372-381 AMDSNTLEL (F5) | GenScript | Customized |
| Influenza A NP383-392 SRYWAIRTR (F6) | GenScript | Customized |
| Influenza A NP458-466 FQGRGVFEL (F7) | GenScript | Customized |
| Influenza A NS1122–130 AIMDKNIIL (F8) | GenScript | Customized |
| Influenza A NA231-239 CVNGSCFTV (F9) | GenScript | Customized |
| Influenza A MP57-68 KGILGFVFTLTV (F10) | GenScript | Customized |
| Influenza A HA344-353 GLFGAIAGFI (F11) | GenScript | Customized |
| KILGFVFJV (phosphosensitive peptide) | GenScript | Customized |
| Critical commercial assays | ||
| Bradford Protein Assay Kit | Beyotime | Cat#P0006-1 |
| Biotin-Protein Ligase Kit | GeneCopoeia | Cat#BI001 |
| EasySepTM Human CD8+T Cell isolation Kit | STEMCELL Technologies | Cat#17853 |
| Experimental models: Cell lines | ||
| T2A2 | Dr. Anna Gil | N/A |
| Software and algorithms | ||
| FlowJo software version 10.7 | FlowJo LLC | https://www.flowjo.com/ |
| Prism version 8 | GraphPad | https://www.graphpad.com/l |
| Adobe Illustrator version 2020 | Adobe Illustrator | https://www.adobe.com/ |
| Other | ||
| Amicon® Ultra | Millipore | C7715 |
| Chromatography instrument | Shhuxi | MC99-2 |
| HiPrep 16/60 Sephacryl S-200 HR | Cytiva | 17116601 |
| Ultrasonic cell crusher | Jingxin | JY92-IIN |
| Ultrasonic cleaning equipment | Kschaosheng | KQ5200DE |
Materials and equipment
Preparing and storing columns
Nickel-Column
The bed volume of Nickel-Column we used is 6 mL, which can be modified according to the practical situation to adapt the different production occasion.
-
•Before using:
-
○On the first time, pack 6 mL of Nickel-Agarose resin into a column with a 10×mL bed volume.
-
○Before each use wash with 10×bed volumes of filtered water and equilibrate with 10×bed volumes of Nickel Loading Buffer.
-
○
-
•After using:
-
○Wash with 5×bed volumes of Nickel Eluting Buffer at the flow rate of 1 mL/min.
-
○Wash with 10×bed volumes of filtered water at the flow rate of 1 mL/min.
-
○Store in filtered water containing 20% ethanol at 2°C–8°C.
-
○
Hiprep 16/60 sephacryl S-200 high resolution
The bed volume of Hiprep 16/60 Sephacryl S-200 High Resolution is 120 mL.
-
•Before using:
-
○Use a 0.22 μm filter to suck and filter the pure water. Then place the filtered water in an ultrasonic cleaning equipment (see key resources table), and ultrasonicate for 10 min to remove bubbles.
-
○Before connecting the column to a chromatography system, start the pump to remove all air form the system, particularly in the tubing and valves.
-
○Before each use wash the column with 0.5×bed volumes (60 mL) of filtered and degassed water and equilibrate with 2×bed volumes of 25 mM Tris-HCl (pH 8.0) at the flow rate of 1 mL/min.
-
○
-
•After using:
-
○Wash with 1×bed volumes of filtered water at the flow rate of 1 mL/min and store in filtered and degassed water containing 20% ethanol 2°C–8°C.
-
○
Step-by-step method details
This protocol describes in detail the purification of HLA-A2 α chain and β2m.
Protein purification
Timing: 1 days
In this part of the protocol, bacterial cells are lysed, and the pellet is disrupted by sonication using ultrasonic fluid. Then, inclusion bodies are dissolved in denaturing loading buffer overnight at 4°C, which is subjected to a Ni2+-affinity chromatography taking advantage of the His-tag presence in the recombinant protein. Subsequently, the HLA-A2 α chain and β2m are refolded in the Refolding Buffer followed by dialysis. Finally, the product is concentrated by ultrafiltration and then purified by the size exclusion chromatography using a Sephacryl S-200 column.
-
1.Cell Lysis
-
a.Unfreeze the bacteria pellet at 20°C–24°C until it starts to melt followed by weighing the cell pellet. Transfer immediately to ice.
 CRITICAL: Maintain the sample and buffer on ice.
CRITICAL: Maintain the sample and buffer on ice. -
b.According to the cell pellet’ weight, resuspend and spin the pellet in 10 mL/g of Ordinary Wash Buffer (pH8, as described in Table 2) and sonicate by soaking the probe of sonicator into the sample, setting the power at 400 watts. Perform 100 cycles of 10 min, alternating the tubes and keeping the samples on ice between each sonication cycle.
-
c.Centrifuge the solution at 12,000 g for 20 min at 4°C.
-
d.Discard the supernatant and repeat step b to c twice.
-
e.Discard the supernatant and repeat step b to c five times after replacing the Ordinary Wash buffer in step b with a 2 M Urea Wash Buffer (pH8, as described in Table 2).
-
f.Discard the supernatant and repeat step b to c twice after replacing the Ordinary Wash buffer in step b with a 25 mM Tris-HCl.
-
g.Add denaturation solution, and place a magnetic stir bar into it and stir overnight at 2°C–8°C.
-
h.Centrifuge the solution at 12,000 g for 20 min at 4°C.
-
i.Filter the supernatant through a 0.22 μm-pore membrane with a syringe.
 CRITICAL: Every sonicator is different and may require optimization.
CRITICAL: Every sonicator is different and may require optimization.
-
a.
-
2.Metal-affinity chromatography
 CRITICAL: All buffers must be filtered through a 0.22 μm-pore membrane.
CRITICAL: All buffers must be filtered through a 0.22 μm-pore membrane.-
a.Load the sample into the pre-equilibrated Nickel Column at a 1 mL/min rate.
-
b.Wash the column with 15×bed column of Nickel Column Clean Buffer as fraction.
-
c.Elute the protein of interest with 6×bed volume of Nickel Column Elute Buffer as fraction.
-
d.Collect 6 number of aliquots of 10 mL (as show in Figures 2A and 2B: mark fraction 0–5).
-
e.Take a sample of 60 μL to test for the presence of the protein of interested.
-
f.Analyze aliquots of the fractions and the LMW protein ladder (10 μL) by 15% SDS-PAGE. Stain the gel with Coomassie blue, using the protocol provided by the manufacturer, and verify the presence of a differential band of the expected size in the induced insert-containing clone (Figures 2A and 2B).
-
g.Pool the fraction 1 and 2 (from step 2d) and determine the absorbance at 280 nm in a nanodrop using an aliquot of the protein containing fractions to determine the concentration of the protein of interest. Use the Nickel Column Elute Buffer as a blank.
-
a.
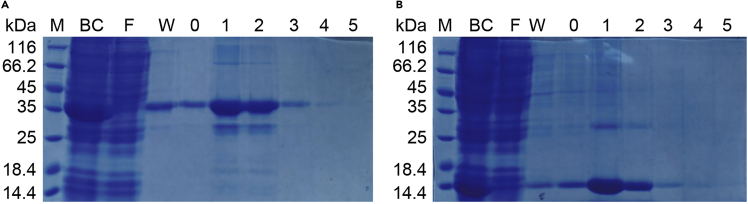
Figure 2.
Purification of HLA-A2 α chain and β2m
(A and B) Aliquots, HLA-A2 α chain (A) / β2m (B), from metal-affinity chromatography eluted fraction (0–5) were solved analyzed by 15% SDS-PAGE and stained with Coomassie blue. Fractions 1 and 2 were pooled for subsequent protein refolding. M, molecular weight marker. BC: before column chromatography. W, wash. F, flow-through.
Protein preparation
Timing: N/a
This part of the protocol serves to assess the purity of the recombinant protein and its activity. Also, proper storage conditions to preserve HLA-A2 α chain and β2m integrity and function are described here.
-
3.
Protein storage
Add 0.2% phenylmethanesulfonyl fluoride (PMSF) to inhibit protein degradation and store the protein for up to 2 months at –80°C.
CRITICAL: Distribute the protein into several tubes and freeze-thaw only once. Once thawed, protein can be preserved at 4°C and used up within a week.
-
4.
Protein analysis
Analyze the purity of HLA-A2 α chain and β2m by a 15% SDS-PAGE. Continue to generate the HLA-A2 α chain and β2m and peptide monomer and check integrity of the refolded protein by Enzyme Linked Immunosorbent Assay (ELISA).
Production of the monomer
Timing: 4 days
A monomer is a combination of the HLA-A2 α chain and β2m with a peptide that can be either photosensitive or not. The photosensitive peptide is used for the exchange assay.
-
5.
Refolding of HLA-A2 α chain and β2m plus peptide, followed by purification.
The peptides are antigen-specific peptide that were collected from Immune Epitope Database and Analysis Resource (IEDB: http://www.iedb.org).-
a.Mix HLA-A2 α chain, β2m and peptide in a 1:2:10 molar ratio and dilute into 40 mL refolding solution. Place a magnetic stir bar into the solution and stir gently at 2°C–8°C for 12 h.
-
b.Dialyze the folding mixture against 25 mM Tris-HCl (pH8.0) for 72 h at 4°C. Replace the dialysate 4–5 times during this period.
-
a.
-
6.Protein ultrafiltration
-
a.Rinse the ultrafiltration tube (Amicon® Ultra) with filtered water.
-
b.Add 10 mL filtered water, and centrifuge the ultrafiltration at 3,000 g for 10 min at 4°C.
-
c.Discard the lower layer of liquid and residual liquid on the upper layer.
-
d.Add 10 mL of sample (if there are more samples, add in multiple times and centrifuge under the same conditions) and centrifuge at 3,000 g for 15 min at 4°C.
-
e.Properly extend the centrifugation time until the upper liquid volume of the ultrafiltration tube is about 1 mL.
-
f.Collect the sample and place it in a new EP tube.
-
g.Temporarily store the sample at 4°C and wait for purification by the Sephacryl S-200 column.
-
a.
-
7.Purify with the Sephacryl S-200 column
-
a.Turn on chromatography instrument (MC99-2, see key resources table) 1 h earlier.
-
b.Carefully immerse the inlet tube of the Sephacryl S-200 column in the sample.
-
c.Start the pump at the flow rate of 0.5 mL/min.
-
d.When the sample is almost exhausted, quickly turn off the pump to avoid air bubbles generate.
-
e.Gently put the inlet tubing into the cold filtered and degassed 25 mM Tris-HCl (pH8.0).
-
f.Start the pump at the flow rate of 0.5 mL/min.
-
g.Change the flow rate to 1 mL/min after 10 min.
-
h.The first peak will appear in the displayer after 50 min or so and the second peak is our target protein. You would receive four peaks (Figure 3A).
-
i.Collect all peak and take a sample of 60 μL to test for the presence of the protein of interested (Figure 3B).
-
a.
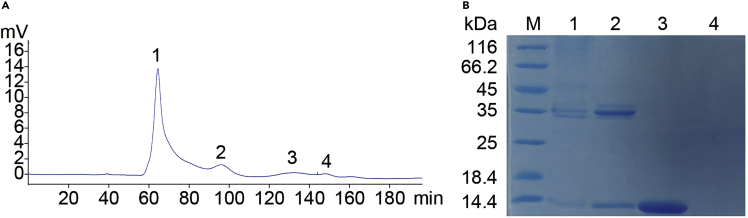
Figure 3.
The refolded protein purified by gel filtration chromatography
(A) Four aliquots collected from the Chromatography instrument. The number 1–4 represents the first to fourth peak collected.
(B) The four aliquots were solved by 15% SDS-PAGE and stained with Coomassie blue.
Biotinylation of monomer and refolding verification
Timing: 1 h Streptavidin has a strong affinity for biotin. The HLA-A2 α chain contains an AviTag, which can be attached to biotin. The biotinylated monomer can connect with PE-conjugated streptavidin to stain antigen-specific CD8+ T cells.
-
8.
The monomer is biotinylated by using the protocol provided by the manufacturer (https://www.genecopoeia.com/product/avitag-cdnaclone-reagent/). Biotin-Protein Ligase kit (key resources table). Fresh biotinylation solution is prepared as described in Table 3. Allow the biotinylation reaction to proceed at 30°C for 30–40 min.
CRITICAL: The biotinylation solution must be prepared freshly.
Pause point: Use immediately, or store the biotinylated monomer solution for up to one week at 4°C. For longer storage, add BSA (0.5% final concentration) and PMSF (0.2 mM final concentration) and divide into several aliquots in 0.2 mL tubes. The biotinylated monomer (MW 50.9 kDa) with a concentration of approximately 1 mg/mL can be stored for up to 1 year at –80°C.
CRITICAL: The monomer combined with a photosensitive peptide (key resources table) is required to be protected from light. Photosensitive peptides are sensitive to light. Please avoid light, otherwise it will affect the subsequent peptide exchange effect.
Table 3.
Biotinylation reaction system
| Reagent | Final concentration | Amount |
|---|---|---|
| 10× Biotin Ligase Buffer A | 1× | 10 μL |
| 10× Biotin Ligase Buffer B | 1× | 10 μL |
| Biotin Ligase | 6.4 ng/μL | 0.68 μL |
| Purified protein | 40 μM | 10 nmol |
| Tris-HCl (pH8.0) | n/a | Up to 100 μL |
Monomer reconstruction confirmation by ELISA
Only when the HLA-A2 α chain, β2m and peptide are combined together, the monomer can be stabilized. The HRP-conjugated antibody is aimed at β2m, and the biotinylated HLA-A2 α chain is bound to the plate coated with streptavidin, so the ELISA signal can only be detected if it is correctly folded into a monomer.
-
9.
Use ELISA to verify whether the monomer has been biotinylated.
Prepare the relevant reagents required for ELISA in advance.-
a.Coat a wells plate with 100 μL streptavidin solution (2 μg/mL). Seal the plate and incubate overnight (16 h–18 h) at room temperature (18°C–25°C).
-
b.Discard the coating solution and wash the plate 3 times with at least 300 μL Wash Buffer per well and remove residual buffer by firmly tapping plate upside down on absorbent paper.
-
c.To block non-specific binding and reduce background, add 300 μL of 1× Dilution Buffer to all wells.
-
d.Seal the plate and incubate it for 30 min at room temperature (18°C–25°C).
-
e.Dilute the sample 1,200-fold in 1×Dilution Buffer. Mix thoroughly.
-
f.Discard the Dilution Buffer from the plate and blot the residual buffer.
-
g.Pipette 100 μL of 1×Dilution buffer (blank) or 100 μL sample into the appropriate wells.
-
h.Seal the plates and incubate for 1 h at 37°C.
-
i.Discard the liquid from the wells and wash 3 times with 300 μL of wash buffer per well.
-
j.Dilute concentration HRP-conjugated antibody to 0.3 μg/mL in 1× Dilution Buffer and add 100 μL of diluted HRP-conjugated β2m antibody per well.
-
k.Seal the plates and incubate for 1 h at 37°C.
-
l.Discard the liquid from the wells and wash 3 times with 300 μL of wash buffer per well.
-
m.Add 100 μL of substrate solution.
-
n.Incubate for 8 min at room temperature (18°C–25°C) in the dark whilst shaking at 450 rpm.
-
o.Add 50 μL of stop solution to each well and read the results at 405 nm on an ELISA reader within 30 min (Sample results are demonstrated in Figures 4A and 4B)
 CRITICAL: Keep an eye on the color development to make sure the reaction is stopped before saturation.
CRITICAL: Keep an eye on the color development to make sure the reaction is stopped before saturation.
-
a.
-
10.
Use the ELISA method to verify whether the photosensitive monomer has been biotinylated (Figures 4A and 4B) and its exchange efficiency (Figures 5A and 5B). Here, we regard the GILGFVFTL peptide as control and define its exchange efficiency with photosensitive peptide as 100%. The exchange efficiency is the percentage of their ratio of OD values corresponding to the antigen-peptide and the GILGFVFTL-peptide(Rodenko B. et al., 2006).
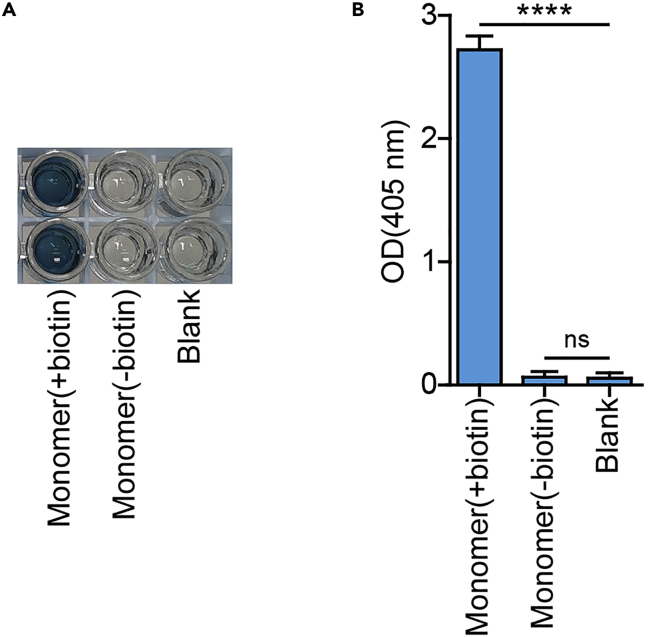
Figure 4.
Verification of the monomer by ELISA
(A) The real shot of ELISA result.
(B) The statistical results of (A). The “Blank” is “1× Dilution Buffer”. Monomer(+biotin): the monomer has been biotinylated; Monomer(-biotin): the monomer has not been biotinylated.Data are shown as mean ± SD (n = 3); ∗∗∗∗ P < 0.0001; ns, not significant.
Figure 5.
The ELISA results showing the photosensitive monomer has been biotinylated and its exchange efficiency
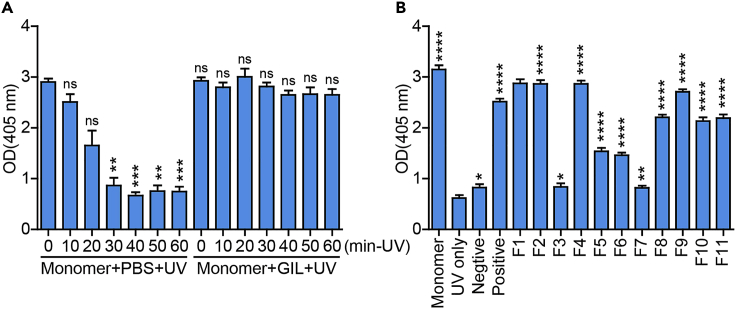
(A) Photosensitive monomer disrupture over time with or without the positive control peptide (GILGFVFTL) after UV irradiation and incubation in the dark.
(B) A photosensitive monomer exchanged with different peptides, showing different binding abilities. UV-only: replace peptide with PBS and irradiate with UV. Negative: EBV antigen peptide (IVTDFSVIK). Positive: the influenza A virus antigen peptide (GILGFVFTL). F1-11: the monomer exchange with the influenza A virus associated peptides (more information see key resources table). All the group in (A) and (B) vs “Monomer +PBS + UV-0min” and “UV only” respectively. Data are shown as mean ± SD (n = 3); ∗ P < 0.05, ∗∗ P < 0.01, ∗∗∗ P < 0.001, ∗∗∗∗ P < 0.0001; ns, not significant.
Tetramer reconstruction and validation
Timing: 7 days
CRITICAL: Bring tetramers, peptides and PE-conjugated streptavidinstreptavidin to 0°C by putting them on ice.
Photosensitive peptide is sensitive to the UV 365 nm. The monomer loaded with a photosensitive peptide offers the flexibility of exchanging the photosensitive peptide to other antigenic peptides of interest before assembling tetramers for flow cytometry analysis.
-
11.Production of antigen specific HLA-A2 tetramers.
-
a.Dilute 1 mg/mL stock solutions of peptides of choice to 100 μg/mL. Mix by pipetting. If the monomer is combined with a photosensitive peptide, to dilute the stock solution to 200 μg/mL
 CRITICAL: If the monomer is not loaded with a photosensitive peptide, proceed directly to step g. If monomer is combined with a photosensitive peptide, proceed to b in order to generate the peptide exchange and test for its efficiency.
CRITICAL: If the monomer is not loaded with a photosensitive peptide, proceed directly to step g. If monomer is combined with a photosensitive peptide, proceed to b in order to generate the peptide exchange and test for its efficiency. -
b.Dilute 10 mM stock solutions of peptides of choice to 400 μM by mixing 2 μL of peptide stock solution with 48 μL PBS, and keep on ice.
-
c.Add 20 μL diluted peptide (400 μM) and 20 μL photosensitive-peptide monomer (0.2 mg/mL) into 96-well V bottom plate. Mix by pipetting up and down.
-
d.Put the plate on ice and illuminate with UV light for 40 min (the distance of the UV lamp to the samples should be 2–5 cm and the time of UV irradiation is selected from the ELISA results shown in Figure 5A).
-
e.Seal the plate and incubate it for 30 min at 37°C in the dark.
-
f.Evaluate the efficiency of the peptide exchange by ELISA (Figure 5B).
-
g.Transfer 30 μL of peptide-exchange monomer (from step 11e) into a 0.2 mL tube, then add 3.3 μL of 0.2 mg/mL PE-conjugated streptavidin, mixing by pipetting up-and-down. Incubate on ice in the dark for 30 min. This is enough for about 15 tests.
-
h.During the incubation, prepare a blocking solution by adding 1.6 μL 50 mM D-Biotin to 198.4 μL PBS, mixing by vortexing. After the incubation, add 2.4 μL of blocking solution and pipette up-and-down to stop the reaction.
-
i.Incubate the tubes at 2°C–8°C overnight.
 CRITICAL: Antigen specific HLA-A2 tetramers can be stored at 2°C–8°C for 1 week, protected from light.
CRITICAL: Antigen specific HLA-A2 tetramers can be stored at 2°C–8°C for 1 week, protected from light.
-
a.
-
12.
Pretreatment of T2A2 cells
T2A2 cells are overexpression HLA-A2 and can be loaded with HLA-A2 antigen to stimulate CD8+T cell. Mitomycin C contributes to the inhibition of DNA synthesis. Carboxy fluorescein succinimidyl ester (CFSE) is a fluorescent dye with cellular permeability and it can persist in cells for 24 h after labeling, which help to distinguish T2A2 cells(Xiao C.C. et al., 2021).-
a.Harvest 2–4 × 108 T2A2 cells from T-75 cell culture flasks.
-
b.Centrifuge for 5 min at 400 g and 4°C.
-
c.Discard the supernatant and resuspend cells in complete Iscove’s Modified Dulbecco’s Medium (IMDM) to a final concentration of approximately 1–2 × 106 cells/mL.
-
d.Transfer the T2A2 cells in 2 mL/well into the 6-well plate with mitomycin C (20 μg/mL) and incubate at 37°C with 5% CO2 for 30 min.
-
e.Wash twice with PBS and centrifuge for 5 min at 400 g and 4°C.
-
f.Discard the supernatant and resuspend cells with 1 mL PBS containing 5 μM CFSE.
-
g.The CFSE labeled T2A2 cells are incubated at 37°C with 5% CO2 for 30 min and then washed twice with PBS.
-
h.Discard the supernatant and the cell concentration is adjusted to 5 cells/mL in serum-free IMDM.
-
i.T2A2 cells are seeded into 96-well plates with 105 cells in 200 μL per well, and then incubated with peptides at a final concentration of 20 μM at 37°C for 4 h, respectively.
-
a.
-
13.CD8+ T cells staining
-
a.Use the EasySepTM human CD8 positive selection kit (with antibodies recognizing the CD8a surface marker) to pre-sort the CD8+ T cells from peripheral blood mononuclear cells (PBMCs) of HLA-A2+ healthy donors.
-
b.Harvest peptide loaded T2A2s from step 24 and mix all cells together.
-
c.Centrifuge cells at 400 g for 5 min at 4°C.
-
d.Discard the supernatant and resuspend cells in primary T medium to a final concentration of approximately 2.5 × 106 cells/mL.
-
e.Add 2.5 × 105 peptide-loaded T2A2 cells to 96-well plates and co-culture with 2.5 × 105 CD8+ T cells plus anti-human CD28 (1 μg/mL) and IL-2 (50 IU/mL).
-
f.Supplement with 50 IU/mL IL-2 and 20 μM peptide every two days.
-
g.7 days later, perform tetramer staining analysis.
-
h.Collect the cells from 96-well plates and wash cells with 1×PBS containing 1% BSA.
-
i.Centrifuge for 5 min at 600 g and 4°C.
-
j.Stain with 10 μL antigen specific tetramer (from 11i) for 1 h at room temperature and then add 2 μL APC labeled human CD8+ (clone T8) for 1 h on ice.
-
a.
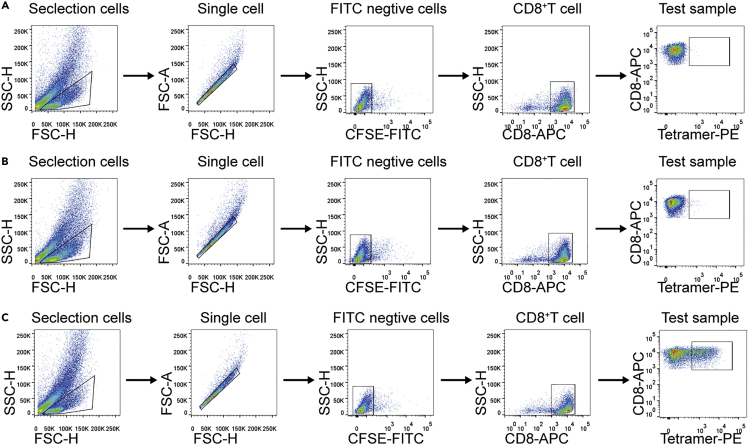
Perform Flow Cytometric Analysis after washing once. Visualization of representative flow cytometry data and gating strategy are shown in Figures 6A–6C.
Figure 6.
Visualization of representative flow cytometry data and gating strategy
(A) The cells only stain with anti-CD8 antibody as a fluorescence minus one (FMO) control.
(B) Tetramer with irrelevant peptide, EBV antigen peptide (IVTDFSVIK), staining of human CD8+ T cells.
(C) Influenza A virus peptide (GILGFVFTL) associated tetramer staining of human CD8+ T cells.
Expected outcomes
After purification of Metal-affinity chromatography, the elution protein is usually observed in fractions 1–2 and about 60 mg of total protein is obtained in those fractions.
After Sephacryl S-200 column purification, the eluted peak is found mostly in fractions 2 and about 20 mg of total protein is generally obtained in those fractions.
As an example, an SDS-PAGE resolution of Metal-affinity chromatography and Sephacryl S-200 column purification-eluted fractions are shown respectively in Figures 2 and 3.
The expected photosensitive monomer biotinylation and exchange efficiency are depicted respectively in Figures 4 and 5.
Limitations
One of the limitations is the use of sorted CD8+ T cells rather than total PBMCs. Due to the lower proportion of T cells in PBMCs, the proportion of activated specific CD8+ T cells was even lower. Furthermore, while our current study focuses on HLA-A2 presented peptides derived from the influenza A virus, other HLA class I allotype-restricted CD8+ T cell-specific peptides and other infectious disease-associated tetramers should be included for a broader exploration, which will be addressed in future studies.
Troubleshooting
Problem 1
Low yield of the recombinant HLA-A2 α chain and β2m after purification.
Potential solution
Adjust the concentration of IPTG to improve the induction of protein expression.
Make sure that the Nickel-Column is correctly regenerated and suitable for use. If necessary, recharge the Nickel-Column following the manufacturer’s instructions (https://www.genscript.com.cn/).
Problem 2
Protein precipitation occurs during dialysis and biotinylation.
Potential solution
To avoid precipitation, maintain the protein in a folding solution at a concentration below 2 mg/mL.
During the process of biotinylation, maintain the process of biotinylation at a Tris-HCl (pH8.0) system. The pH value of Tris-HCl system cannot be lower than 8.0.
Problem 3
Light-sensitive monomers have low exchange efficiency with other peptides.
Potential solution
Explore the most suitable UV irradiation time as shown in Figure 5A.
Resource availability
Lead contact
Further information and requests for resources and regents should be directed to and will be fulfilled by the lead contact, Guobing Chen (guobingchen@jnu.edu.cn)
Materials availability
The corresponding tetramers may be obtained from the research group of Guobing Chen, Jinan University, China.
Acknowledgments
This work was supported by grants from the National Key Research and Development Program of China (2018YFC2002003), the Natural Science Foundation of China (U1801285, 81971301,82074133, and 92169102), Guangzhou Planned Project of Science and Technology (201904010111, 202002020039), Zhuhai Planned Project of Science and Technology (ZH22036302200067PWC), and the Initial Supporting Foundation of Jinan University.
Author contributions
Ye J.Z. and Chen G.B. conceived and wrote the protocol. Ye J.Z., Lei W., and Xiao C.C. performed the experiments. Gao J., Li X.R., Su X.M., and Li W.X. assisted with experiments; Ye J.Z., Wang P.C., and Chen G.B. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Zhenyou Jiang, Email: tjzhy@jnu.edu.cn.
Guobing Chen, Email: guobingchen@jnu.edu.cn.
Data and code availability
No new code or data were generated as part of this study.
References
- Prato C.A., Carabelli J., Cattaneo V., Campetella O., Tribulatti M.V. Purification of recombinant galectins expressed in bacteria. STAR Protoc. 2020;1:100204. doi: 10.1016/j.xpro.2020.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenko B., Toebes M., Hardrup S.R., Van Esch W.J., Molenaar A.M., Schumacher T.N., Ovaa H. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nat. Protoc. 2006;1:1120. doi: 10.1038/nprot.2006.121. [DOI] [PubMed] [Google Scholar]
- Xiao C., Qiu C., Deng J., Ye J., Gao L., Su J., Luo O.J., Wang P., Chen G. Optimization of antigen-specific CD8+ T cell activation conditions for infectious diseases including COVID-19. STAR Protoc. 2021;2:100789. doi: 10.1016/j.xpro.2021.100789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new code or data were generated as part of this study.