Abstract
Clostridium perfringens is an anaerobic, gram-positive, spore-forming bacterium that ubiquitously inhabits a wide variety of natural environments including the gastrointestinal tract of humans and animals. C. perfringens is an opportunistic enteropathogen capable of producing at least 20 different toxins in various combinations. Strains of C. perfringens are currently categorized into 7 toxinotypes (A, B, C, D, E, F, and G) based on the presence or absence of 6 typing-toxins (α, β, epsilon, iota, enterotoxin, and netB). Each toxinotype is associated with specific histotoxic and enteric diseases. Spontaneous enteritis due to C. perfringens has been reported in laboratory animals; however, the source of the bacteria was unknown. The Quality Assurance Laboratory (QAL) at the National Institute of Environmental Health Sciences (NIEHS) routinely screens incoming animal feeds for aerobic, enteric pathogens, such as Salmonella spp. and E. coli. Recently, QAL incorporated anaerobic screening of incoming animal feeds. To date, the lab has isolated numerous Clostridium species, including C. perfringens, from 23 lots of natural ingredient laboratory animal diets. Published reports of C. perfringens isolation from laboratory animal feeds could not be found in the literature. Therefore, we performed a toxin profile screen of our isolated strains of C. perfringens using PCR to determine which toxinotypes were present in the laboratory animal diets. Our results showed that most C. perfringens strains we isolated from the laboratory animal feed were toxinotype A with most strains also possessing the theta toxin. Two of the C. perfringens strains also possessed the β toxin. Our results demonstrated the presence of C. perfringens in nonsterile, natural ingredient feeds for laboratory animals which could serve as a source of this opportunistic pathogen.
Abbreviations: C.perfringens, Clostridium perfringens; GI, gastrointestinal; NE, necrotic enteritis
Introduction
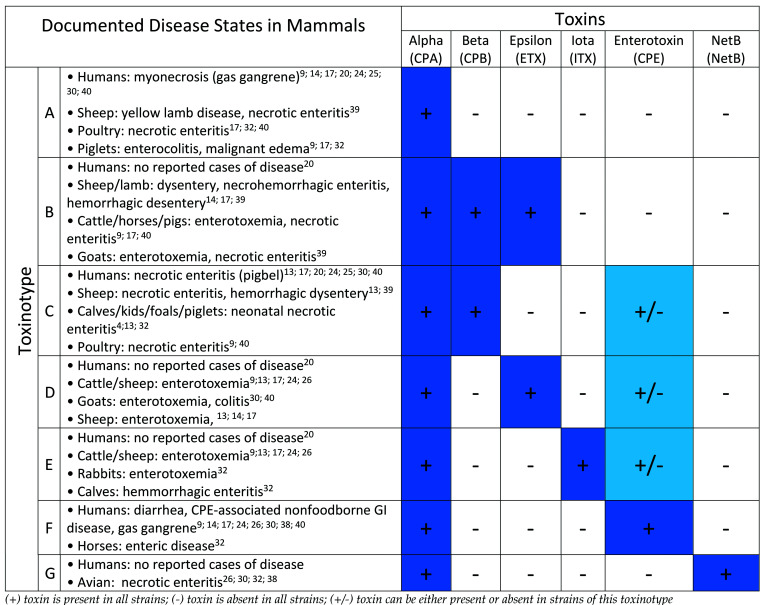
Clostridium perfringens is a well-known and widely dispersed gram-positive, nonmotile, anaerobic bacterium that ubiquitously inhabits most terrestrial and aquatic environments. Like some other members of the Firmicute phylum, its survivability is greatly enhanced by its ability to produce endospores during periods of environmental stress.1,14,24,30 C. perfringens commonly resides in the gastrointestinal (GI) tract of numerous animal species and has been widely reported in a variety of domesticated animal species (Figure 1).
Figure 1.
Toxinotypes and pathogenicity.
C. perfringens is an important human and animal pathogen that causes a wide spectrum of diseases.34 However, not all strains of C. perfringens cause disease in animals or humans, and the presence of C. perfringens in the intestinal tract usually does not lead to illness. C. perfringens typically does not exhibit adherence and invasive properties toward healthy intestinal mucosa, and the development of clinical disease appears to be the result of a complex interaction between host immune status, strain virulence, and other nonspecific factors.34 Host stresses that lead to abnormal gut microbiota appear to be an important predisposing factor to disease development. Gut microbiota disturbances and C. perfringens vulnerability are known to occur from host antibiotic exposure, alterations in feeding regimens, overeating, and dietary changes.7,22,37 C. perfringens is known to produce at least 20 different toxins in varying combinations; these toxins cause a broad range of diseases including necrotic enteritis, gas gangrene, and various enterotoxemia (Figure 1).22,29,34 The presence or absence of 6 particular toxins (“typing toxins”) classifies each strain into 1 of 7 currently recognized toxinotypes (A-G; Figure 1).29 The 6 typing toxins that cause most of the diseases reported include α, β, epsilon, iota toxins (previously defined as major toxins), enterotoxin, and netB (previously defined as minor toxins). Over the last few years, the C. perfringens typing system has been expanded from 5 toxinotypes (A through E) to 7 toxinotypes (A through G). Toxinotype F is a reclassification of enterotoxin-positive (CPE-positive) toxinotype A strains, and toxinotype G strains produce the necrotic enteritis B-like (netB) toxin.29 The nontyping toxin perfringolysin O (theta) is not a main virulence factor for animal disease but was added to the study because it is thought to have a synergistic action with CPA-mediated and ETX-mediated diseases.39,41 Table 1 summarizes the mode of action, biologic activity and gene location (plasmid or chromosome) of each toxin of interest within our study.
Table 1.
Mechanism of action and biologic effects of toxin-encoding genes.*
| Toxin | Gene | Mode of Action | Biologic Activity | Gene Location |
|---|---|---|---|---|
| Alpha (α) | cpa | Phospholipase C (plc) And sphingomyelinase | Cytotoxic, hemolytic, necrotic, smooth muscle contraction, enteritis | Chromosome |
| Beta (β) | cpb | Pore-forming activity | Cytotoxic, dermonecrotic, edema, hemorrhagic dysentery, enteritis | Plasmid |
| Epsilon (ε) | etx | Pore-forming activity and alteration of cellularmembrane permeability | Edema of various organs, smooth muscle contraction, dermonecrotic, enterotoxic, blood pressure elevation | Plasmid |
| Iota (ι) | iap and ibp | Ia-mediated ADP-ribosylation of G-actin and pore-forming activity | Disruption of actin cytoskeleton and cell membrane, inhibition of smooth muscle contraction, necrotic | Plasmid |
| Enterotoxin | cpe | Pore-forming activity | Enterotoxic, cytotoxic, erythema, leakage of water and ions, oncosis | Plasmid or Chromosome |
| Theta (θ) | pfo | Perfringoysin and pore-forming activity | Cholesterol-specific hemolysin, tissue destruction, antiinflammatory | Chromosome |
| NetB | netB | Pore-forming activity | Necrotic enteritis of poultry, hemolytic gut lesions | Plasmid |
Enzyme-linked immunosorbent assays (ELISAs) have been used to toxinotype C. perfringens strains. ELISAs have been traditionally used to detect α toxin (CPA), β toxin (CPB), epsilon toxin (ETX), and enterotoxin (CPE), but no commercially available ELISA kit has been developed that can reliably detect iota toxin (ITX).1,14,39 Furthermore, biochemical tests alone cannot distinguish between different C. perfringens toxinotypes and may overlook samples not actively producing toxins.26 Recent C. perfringens toxinotyping efforts have used traditional polymerase-chain reactions or quantitative polymerase-chain reactions (PCR or qPCR) to successfully determine the presence of toxin-associated genes.22
In 2018, strain isolation and toxinotyping of C. perfringens was reported from agricultural animal feeds manufactured in Serbia.22 To our knowledge, there are no published reports of any Clostridium spp. being isolated from natural ingredient, laboratory animal feeds. In this paper, we report the isolation and toxigenic profile of 29 Clostridium spp. including C. perfringens and 5 other clostridial species (Clostridium baratii, Clostridium beijerinckii, Clostridium bifermentans, Clostridium butyricum, and Clostridium sordellii) from 10 different laboratory animal diets, including both open and closed formulations, obtained from 4 different commercial feed manufacturers. These results demonstrate that opportunistic, pathogenic bacteria, such as C. perfringens, are present in unsterilized, natural ingredient, laboratory animal diets and could act as a source to colonize the GI tract of laboratory species and cause disease, especially in immunocompromised or biologically stressed animals. The presence of C. perfringens in these feeds provides a rationale for feed sterilization before use to avoid the introduction of unwanted, potentially pathogenic organisms that may cause unwanted physiologic effects, disease, or death.
Materials and Methods
Clostridium spp. cultivation from animal feed and initial identification.
Twenty-three separate lots of laboratory animal feed were tested from large, commercial, US-based manufacturers. These included several lots of our standard, open-formula NIH-31 rodent feed produced under contract by a commercial source. We also tested 2 lots of the open formula NIH-07 rodent diet from 2 different manufacturers; the NIH-2004 open-formula swine diet, 4 different natural ingredient, closed-formula rodent diets from 2 different manufacturers; and 1 purified, high-fat, rodent diet. For enrichment of Clostridium spp., approximately 25 g of each feed sample was aseptically placed into 250 mL of thioglycolate broth and incubated at 35 °C for 24 h. After incubation, each thioglycolate broth bottle was briefly mixed and streaked onto blood agar plates (BAPs) using sterile cotton tipped applicators. BAPs were then incubated at 37 °C (98.6 °F) under anaerobic conditions using a generating system (GasPak EZ, BD Diagnostics, Franklin Lakes, NJ) inside an anaerobic chamber. After 24 to 48 h of incubation, each BAP was examined; suspect colonies were isolated onto fresh BAPs and incubated again at 37 °C (98.6 °F) aerobically and anaerobically. Isolates indicating growth only under anaerobic conditions were archived, and a sample of each isolate was shipped to Charles River Laboratories (Wilmington, MA) for identification using matrix associated laser desorption/ionization time of flight mass spectrometry (MALDI-TOF).
DNA purification and quantification.
For each isolate identified as a Clostridium spp. by MALDI-TOF, a loopful of colony biomass was placed inside a 2 mL tube prefilled with approximately 1200 mg of acid washed, 100 µm zirconium beads (Ops Diagnostics, Lebanon, NJ) along with the initial reagents recommended by the tissue kit manufacturer (DNeasy Blood and Tissue Kit, Qiagen, Hilden, Germany) for cultured cells. Next, bead tubes were homogenized using a homogenizer (FastPrep-96, MP Biomedicals, LLC, Santa Ana, CA) at max speed for 3 min. After homogenization, tubes were centrifuged at 8,000 × g for 30 s and incubated at 56 °C (132.8 °F) for 30 min. After incubation, tubes were centrifuged at 13,000 × g for 1 min, and each sample’s supernatant was then transferred to a new DNA/RNA-free 1.5 mL microcentrifuge tube. The kit’s quick-start protocol was then continued from step 3 until purified DNA was eluted into a new DNA/RNA-free 1.5 mL microcentrifuge tube (step 8). Before downstream analysis, purified DNA was quantified fluorometrically DS-11 FX Spectrophotometer/Fluorometer, DeNovix, Wilmington, DE) and an assay kit (dsDNA Broad Range Assay Kit, DeNovix) was used following the manufacturer’s instructions. Total genomic DNA was isolated from a subset of test diets DNeasy PowerMax Soil Kit, Qiagen, Hilden, Germany) to assess our ability to identify C. perfringens directly from feed via PCR. Five (5.0) g of each diet were added to the kit’s 50 mL conical tube along with 0.7 mm garnet beads and 15 mL of the kit’s PowerBead and C1 solutions per kit instructions. The tubes were vortexed for 10 min then incubated at 37 °C (98.6 °F) overnight on a shaking tray. The remainder of the kit protocol was followed, and the total genomic DNA was eluted from the Qiagen column using 5.0 mL of C6 solution (elution buffer). The DNA was precipitated with 0.3 M sodium acetate and 10 mL of isopropyl alcohol, washed with 1.5 mL of 75% ethanol, and resuspended in 1.0 mL of DNAse/RNAse-free water. Purified DNA was quantified fluorometrically DS-11 FX Spectrophotometer/Fluorometer, DeNovix) and a broad-range assay kit (dsDNA Broad Range Assay Kit, DeNovix) was used following the manufacturer’s instructions.
Sequence verification of cultured isolates.
The bacterial identity of each isolate was verified by 16S rRNA gene analysis using the universal bacterial primers 27F (5′-AGAGTTTGATCMTGGCTCAG-3′, M = C/A) and 1492R (5′-GGTTACCTTGTTACGACTT-3′).42 Fifty (50) µl polymerase chain reaction (PCR) mixtures were carried out T100 Thermal Cycler, (Bio-Rad Laboratories, Hercules, CA) using 100 ng of total DNA template along with polymerase reagents (Applied Biosystems, Foster City, CA) according to the kit’s suggested protocol. PCR amplifications were performed as follows: Initial denaturation at 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 1 min with a final 5 min extension period at 72 °C. 16S rRNA gene amplicons were cleansed (QIAquick PCR Purification Kit, Qiagen, Hilden, Germany). Approximately 50 ng of PCR amplicon template was sent to Genewiz (Morrisville, NC) in a premix tube (amplicon + primer) for Sanger sequencing. Sanger sequencing results were trimmed and assembled (CLC Main Workbench 8, Qiagen, Hilden, Germany) using the default settings. Assembled 16S contigs were identified by uploading sequences into the National Center for Biotechnology Information’s (NCBI) Basic Local Alignment Search Tool (BLAST) using the ‘16S ribosomal RNA sequences (bacteria and archaea)’ database and the top BLAST score result.
C. perfringens toxin gene profiling.
To screen isolates for various toxin-associated genes, 50 µl PCR reactions were carried out using 10 ng of sample DNA template and toxin-specific gene primers (Table 2) along with Platinum Taq Polymerase (Invitrogen/ThermoFisher, Carlsbad, CA) in accordance with the manufacturer’s guidelines. PCR cycle parameters were as follows: Initial denaturation at 95 °C for 3 min, followed by 35 cycles each of 95 °C for 30 s, 50 °C to 60 °C for 30 s (Table 2; optimal annealing temperature for each primer set), and 72 °C for 1 min with a final 5 min extension period at 72 °C. Upon completion of PCR, 20 µl of product was run on a 1.2% agarose gel containing a 1× concentration of GelGreen Nucleic Acid Stain (Biotium, Fremont, CA). For added assurance, amplicons were sent to Genewiz (Morrisville, NC) for Sanger sequencing, and trimmed results were matched against NCBI’s BLAST using the ‘nucleotide collection (nr/nt)’ database.
Table 2.
Toxin-specific gene primers
| Toxin Gene | Oligonucleotide Sequence (5′-3′) | Anneal Temp | Amplicon Size |
|---|---|---|---|
| cpa (α toxin)21 | GCTAATGTTACTGCCGTTGA | 50 °C | approximately 324 bp |
| TCTGATACATCGTGTAAG | |||
| plc (α toxin)24 | CCGTTGATAGCGCAGGACA | 54 °C | approximately 219 bp |
| CCCAACTATGACTCATGCTAGCA | |||
| cpb (β toxin)21 | GCGAATATGCTGAATCATCTA | 57 °C | approximately 196 bp |
| GCAGGAACATTAGTATATCTTC | |||
| etx (ε toxin)21 | GCGGTGATATCCATCTATTC | 54 °C | approximately 655 bp |
| CCACTTACTTGTCCTACTAAC | |||
| iap (ι toxin)5 | AATGCCATATCAAAAAATAA | 53 °C | approximately 821 bp |
| TTAGCAAATGCACTCATATT | |||
| cpe (enterotoxin)21 | GGAGATGGTTGGATATTAGG | 56 °C | approximately 233 bp |
| GGACCAGCAGTTGTAGATA | |||
| pfoA (θ toxin)12 | ATCCAACCTATGGAAAAGTTTCTGG | 60 °C | approximately 532 bp |
| CCTCCTAAAACTACTGCTGTGAAGG | |||
| netB (NetB toxin) | GCTGGTRaCTGGAATAAATGC | 56 °C | approximately 384 bp |
| TCGCCATTGAGTAGTTTCCC |
Ra = A/G (nucleotide at this position was modified from Keyburn and colleagues’s original primer version of nucleotide “G”
Identification of C. perfringens directly from feed.
Fifty (50) nanograms of DNA isolated from feed samples were used in a 50 µl total volume PCR reaction using the plc (α toxin) primers listed in Table 2 and Platinum Taq Polymerase. Touchdown PCR cycle parameters were as follows: 95 °C initial denaturation for 3 min; followed by 10 cycles of 95 °C for 30 s, 65 °C for 30 s with a 0.5 °C decrease/cycle, and 72 °C for 30 s; followed by 25 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s; and ending with a single 72 °C extension for 3 min. Twenty µl of each reaction was run on a 2.0% agarose gel containing 1× GelGreen Nucleic Acid Stain (Thomas Scientific, Swedesboro, NJ).
Results
Sequence identification of clostridial isolates.
From the 23 separate lots of laboratory animal feed tested, we isolated and identified 7 different species of Clostridium (C. perfringens, C. bifermentans, C. butyricum, C. baratii, C. beijerinckii, C. sordellii, and C. tertium). Table 3 lists the Clostridium isolates selected for our toxinotyping experiment.
Table 3.
Clostridium sp. strains isolated from natural ingredient laboratory animal feed
| Sample ID | Sample | Feed Manufacturera | Lotb | Institutea | Species |
|---|---|---|---|---|---|
| ATCC 12917 | Positive Control Strain - Toxinotype D | NA | NA | ATCC | C. perfringens |
| ATCC 3626 | Positive Control Strain - Toxinotype B | NA | NA | ATCC | C. perfringens |
| ATCC 8009 | Positive Control Strain - Toxinotype E | NA | NA | ATCC | C. perfringens |
| ATCC 27324 | Positive Control Strain - Toxinotype E | NA | NA | ATCC | C. perfringens |
| Uzal Lab Isolate | Positive Control Strain - Toxinotype G | NA | NA | UC Davis | C. perfringens |
| QA1011-17_F | Diet 1 (NIH-31) | 1 | 1 | NIEHS | C. perfringens |
| QA3081-14_B | Diet 1 (NIH-31) | 1 | 2 | NIEHS | C. perfringens |
| QA1881-14_A | Diet 1 (NIH-31) | 1 | 3 | NIEHS | C. perfringens |
| QA4249-17 | Diet 2 (Closed Formula) | 2 | 1 | NIEHS | C. perfringens |
| QA3578-18_A | Diet 3 (Closed Formula) | 1 | 1 | NIEHS | C. perfringens |
| QA1535-17_A | Diet 3 (Closed Formula) | 1 | 2 | NIEHS | C. perfringens |
| QA411-18_C | Diet 4 (NIH-07) | 1 | 1 | NIEHS | C. perfringens |
| QA1027-18_C | Diet 4 (NIH-07) | 3 | 2 | 2 | C. perfringens |
| QA1025-18_C | Diet 5 (Closed Formula) | 2 | 1 | 2 | C. perfringens |
| QA1026-18_C | Diet 6 (Closed Formula) | 2 | NA | 2 | C. perfringens |
| QA1028-18_C | Diet 7 (NIH-2004 - Swine) | 3 | NA | 2 | C. perfringens |
| QA407-18_A | Diet 8 (Closed Formula) | 2 | NA | 3 | C. perfringens |
| QA408-18_B | Diet 9 (Closed Formula) | 2 | NA | 3 | C. perfringens |
| QA409-18_C1 | Diet 10 (Closed Formula) | 2 | 1 | 3 | C. perfringens |
| QA1787-16_A | Diet 11 (Purified - High-fat) | 4 | NA | NIEHS | C. perfringens |
| QA1011-17_B | Diet 1 (NIH-31) | 1 | 1 | NIEHS | C. bifermentans |
| QA1011-17_C | Diet 1 (NIH-31) | 1 | 1 | NIEHS | C. butyricum |
| QA3081-18_D | Diet 1 (NIH-31) | 1 | 2 | NIEHS | C. butyricum |
| QA4071-14_A | Diet 1 (NIH-31) | 1 | 4 | NIEHS | C. bifermentans |
| QA1024-18_E | Diet 1 (NIH-31) | 1 | 5 | 2 | C. baratii |
| QA3913-17_B | Diet 2 (Closed Formula) | 2 | 2 | NIEHS | C. beijerinckii |
| QA409-18_A | Diet 10 (Closed Formula) | 2 | 1 | 3 | C. sordellii |
| QA1025-18_D | Diet 5 (Closed Formula) | 2 | 1 | 2 | C. bifermentans |
| QA218-16_E | Environmental swab | NA | NA | NIEHS | C. tertium |
Numbers were assigned to commercial feed manufacturers and institutes in place of their name.
Lot numbers assigned solely to indicate separate lots of a given diet.
Toxin gene profiling of C. perfringens isolates.
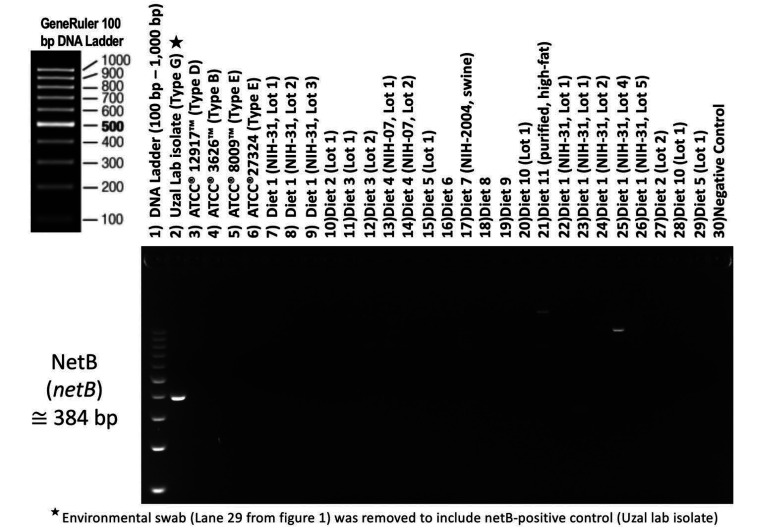
As expected, PCR, gel documentation, and Sanger sequencing verified the presence of toxin genes in all C. perfringens isolates, but not in any other species of Clostridium isolated from laboratory animal feed (Figure 2). All C. perfringens strains isolated from laboratory animal diets possessed the cpa gene encoding the α toxin, and 2 of the isolates possessed the cpb gene encoding the β toxin. No other typing toxin (epsilon, iota, enterotoxin, and NetB) was found in any C. perfringens strain isolated from laboratory animal feed (Figures 2 and 3). Nine of the 15 C. perfringens strains tested (60%) had the pfoA gene encoding the theta toxin, and 1 strain had the cpb2 gene encoding the β2 toxin. Two of our 4 positive control C. perfringens strains used for PCR validation did not correspond completely with the toxin profile certification provided by the vendor in that the expected toxin genes (epsilon and iota) was not amplifiable using any of our referenced toxin primers (Table 4). Amplification of the enterotoxin gene was possible with only 2 of the 4 referenced primer sets tested in our experiment (Table S1 and Figure S1, Figure S2, Figure S3 and Figure S4).
Figure 2.
Toxin gene profile of Clostridium sp. isolates.
Figure 3.
NetB toxin gene profile of Clostridium sp. isolates.
Table 4.
Toxin profile of C. perfringens strains isolated from natural diets
| Lane | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain Source | DNA Ladder | ATCC® 12917™ | ATCC® 3626™ | ATCC® 8009™ | ATCC® 27324™ | Diet 1 | Diet 1 | Diet 1 | Diet 2 | Diet 3 | Diet 3 | Diet 4 | Diet 4 | Diet 5 | Diet 6 | Diet 7 | Diet 8 | Diet 9 | Diet 10 | Diet 11 | ||||||||||
| Toxin Gene | cpa | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ||||||||||
| plc | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | |||||||||||
| cpb | ✹ | ✹ | ✹ | ✹ | ||||||||||||||||||||||||||
| etx | ✹ | ✹ | ||||||||||||||||||||||||||||
| iap | ✹ | ✹ | ||||||||||||||||||||||||||||
| cpe | ✹ | ✹ | ||||||||||||||||||||||||||||
| cpb2 | ✹ | ✹ | ✹ | |||||||||||||||||||||||||||
| pfoA | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | ✹ | |||||||||||||||||
| netB | ||||||||||||||||||||||||||||||
| Toxinotype | F | B | A | E | C | A | A | A | C | A | A | A | A | A | A | A | A | A | A | |||||||||||
| Detected | Not Detected (Excpected) | Detected with 2 of 4 primer sets | ||||||||||||||||||||||||||||
Toxin genes verified to be present by PCR are highlighted in green. Toxin genes documented to be present in positive control strain (ATCC®) but nonamplifiable by PCR using referenced primer sets are highlighted in red. Toxin genes documented to be present in control strain but only amplifiable using some of the referenced primer sets are highlighted in orange.
Identification of C. perfringens directly from feed.
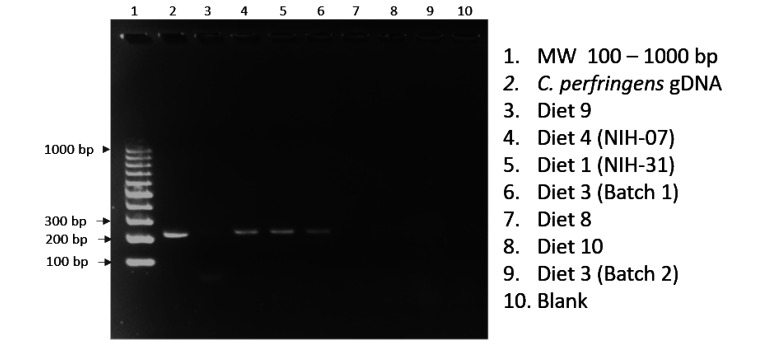
We attempted to identify C. perfringens in animal feed by direct PCR screening for the plc gene (α toxin) in total gDNA isolated from a subset of tested feeds. All feeds tested for direct PCR were confirmed to possess C. perfringens by culture and isolation. Our initial attempts to amplify the gene target directly from feed using the same PCR parameters used on the purified bacterial isolates were unsuccessful. By using a “touchdown” PCR technique, in which a higher initial annealing temperature (65 °C) is used to minimize nonspecific amplifications, we were able to correctly amplify the expected 219 bp amplicon in 4 of the 7 diets tested by direct PCR screening (Figure 4).
Figure 4.
Direct feed analysis for C. perfringens (α toxin – plc gene).
Discussion
Our results established that most of the C. perfringens strains that we isolated from natural ingredient animal feed diets were toxinotype A, which is the basic toxinotype for which accumulation of toxin-encoding plasmids yield other distinctive toxinotypes (one exception to this rule is the enterotoxin gene (cpe) which is not always plasmid-encoded, but can sometimes be located on the chromosome).20 Our results indicated that 2 of the C. perfringens strains isolated from laboratory animal feed possessed the β toxin gene; this toxin is known to be fatal in mice at low concentrations.20,38 Although the 2 β toxin amplicons observed in our animal feed isolates by gel documentation were faint (low PCR amplification), repeated gel screening and Sanger sequencing of these amplicons confirmed them as authentic. With the technique we used to cultivate and isolate C. perfringens strains from nonsterile natural ingredient, laboratory animal feeds (thioglycollate broth to isolation plates), it is difficult to prove isolated colonies used for this study are of a single strain type and multiple strain types may be present (a fraction of the bacteria possessed a plasmid encoding the β toxin gene). Most of the C. perfringens toxin genes are solely plasmid-encoded (for example, cpb, epsilon, itx and netB), and strains are capable of transferring toxin-encoding plasmids to neighboring strains of C. perfringens which is an important element of its pathologic progression.20 C. perfringens strains can carry up to 3 different toxin plasmids with each plasmid encoding up to 3 different toxins.20 The nonspecific amplicons observed on the electrophoresis gel (Figure 2) using our referenced netB primers did not reveal any relevant information about the genetic origin by Sanger sequencing and BLAST analysis. Nine of the 15 C. perfringens strains tested from laboratory animal feed also possessed the theta toxin. Theta toxin has a synergistic action with CPA which can increase necrotizing and lethal effects on animals.12,40 Two of our 4 positive controls did not work as expected (Table 4). The toxin profile certification provided by the vendor (ATCC) did not correlate with the toxin profile identified by our PCR analysis of the epsilon (etx) and iota (iap) genes. For both discrepancies (toxin gene absent in the positive control strain), another positive control sample effectively amplified and confirmed that our referenced gene primers did effectively target the toxin of interest. Multiple referenced primer sets for the etx gene5,41 and iap gene21,29,41 were tested on these positive controls but gene amplification was still unsuccessful (Table S1, Figures S1, S2, S3 and S4). Since the toxin gene missing from each of these positive controls (epsilon and iota) are plasmid-based genes, one possible explanation for the failure to identify them via PCR is that the plasmid responsible for encoding the respective toxin gene was lost due to continual growth on defined media in a laboratory setting outside a GI environment.
Although we were able to demonstrate C. perfringens in laboratory animal diets by direct PCR of total gDNA isolated from feed, our PCR results were not consistent. Lack of consistent PCR amplification among the diets is likely due to the concentration of C. perfringens in the diet and/or PCR inhibitors remaining in the gDNA isolated from the feed. Therefore, further analysis using quantitative PCR and spiking experiments would be necessary to determine the sensitivity/detection limit of our assay, and the possible presence of PCR inhibitors in animal diets. As such, we caution against the use of PCR screening of feed samples as the sole method to screen animal feeds for C. perfringens.
C. perfringens is ubiquitous in the environment, including in the digestive tract of healthy animals; therefore, analyzing the evolution of pathogenic strains is difficult. In addition, host factors including diet, innate immunity, and normal flora greatly impact colonization by foreign bacteria.28,31 Because few environmental bacteria are known to carry these types of toxin gene variants, strain adaptation to acquire such toxins is suited for survival in a GI environment.27 It has been noted that, unlike commensal strains, pathogenic strains of C. perfringens isolated from NE or wound infections show rearrangement of chromosomal regions that include hydrolytic enzymes and toxins, which may confer selective advantages for colonizing GI environments.28,31 Spontaneous disease from C. perfringens is rare in laboratory animals but has been reported most often in female mice nursing large litters with older pups (>14 d of age).8,19,36 The nutritional stress of lactation and an increased intake of high carbohydrate diets may be predisposing factors in these cases, and autoclaving the diet was a way to reduce infections.8,18 A study demonstrated that C3H/HeJ mice, which have innate immune deficiencies due to absence of the toll-like receptor 4 (Tlr4), were more sensitive to experimental infection with C. perfringens as compared with the C3H/HeN strain, which has a normal Tlr4.23 While disease due to C. perfringens has not been seen in the rodent population at NIEHS, immunocompromised and gnotobiotic animals could be more susceptible to colonization with C. perfringens and disease development due to toxins. Therefore, the introduction of C. perfringens from laboratory animal feed could pose a serious risk to studies using these types of rodents.
In addition, because of the bacterium’s ubiquitous nature, almost all food sources, whether animal- or plant-based, can potentially be contaminated with C. perfringens.15 Microbial screening for C. perfringens has limited utility because positive results are common and indicate very little unless extremely high numbers of C. perfringens are enumerated.22 Most sterilization processes, including ionizing radiation, do well at destroying vegetative cells, but do not always effectively eliminate their spores.10 For this reason, it may be beneficial to couple irradiation with microbial screening when using nonautoclavable diets. Surviving spores in sterilized feed can germinate and multiply rapidly.6,10,35 Proper heating is the most reliable method of spore inactivation, but the required temperature and time is dependent on feed properties such as pH, water content, and fat content.6 We test our facility’s feed autoclaves weekly using VERIFY dual-species self-contained biologic indicators (STERIS Life Sciences; Mentor, OH) which contains spores from Geobacillus stearothermophilus and Bacillus atrophaeus. Our facility’s vivarium houses over 50,000 rodents, and we have not documented an animal clinical case or sentinel necropsy that has demonstrated the presence of disease associated with C. perfringens by in-sourced or out-sourced diagnostic testing.
Because the isolation of C. perfringens from laboratory animal diets has not been previously reported, we wanted to develop our own capabilities to rapidly screen incoming diets for C. perfringens and effectively evaluate their toxigenic profile via PCR-based assays. Based on our findings, C. perfringens appears to be a common contaminant of laboratory animal feeds. Almost all environmental isolates identified exclusively fall into the toxinotype A category. The presence of C. perfringens in these laboratory animal diets provides a rationale for feed sterilization before use, and we strongly recommend proper feed sterilization and, if deemed necessary, microbial screening prior to use to prevent exposure of animals to potentially pathogenic strains of C. perfringens that can lead to disease, death or subclinical alterations in physiology that can affect research outcomes.
Supplementary Materials
ATCC’s PCR validation (assay 2 and 3) of toxin genes etx, iap (iA) and cpe.
Validation of etx gene primer sets against positive controls.
Validation of iap (iA) gene primer sets against positive controls.
Validation of cpe gene primer sets against positive controls.
Acknowledgments
We would like to thank Mr. Dennis Barnard, NIH, Division of Veterinary Resources, and Mr. Ned Collins, Alpha-Omega, Inc., US EPA contractor, for assistance, as well as the NIEHS Summer Internship Program for funding a portion of this project. We would also like to express our gratitude to Francisco A. Uzal and Juliann Beingesser, California Animal Health and Food Safety Lab – UC Davis, for providing our lab a netB positive control strain to validate our netB PCR assay.
References
- 1.Albini S, Brodard I, Jaussi A, Wollschlaeger N, Frey J, Miserez R, Abril C. 2008. Real-time multiplex PCR assays for reliable detection of Clostridium perfringens toxin genes in animal isolates. Vet Microbiol 127:179–185. 10.1016/j.vetmic.2007.07.024. PubMed [DOI] [PubMed] [Google Scholar]
- 2.Asha NJ, Wilcox MH. 2002. Laboratory diagnosis of Clostridium perfringens antibiotic-associated diarrhoea. J Med Microbiol 51:891-894. 10.1099/0022-1317-51-10-891 [DOI] [PubMed] [Google Scholar]
- 3.Awad MM, Ellenor DM, Bod RL, Emmins JJ, Rood JI. 2001. Synergistic effects of alpha-toxin and perfringolysin O in Clostridium perfringens-mediated gas gangrene. Infect Immun 69:7904–7910. 10.1128/IAI.69.12.7904-7910.2001. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baums CG, Ulrich S, Gunter A, Goethe R. 2004. Diagnostic multiplex PCR for toxin genotyping of Clostridium perfringens isolates. Vet Microbiol 100:11–16. 10.1016/S0378-1135(03)00126-3. PubMed [DOI] [PubMed] [Google Scholar]
- 5.Braun M, Herholz C, Straub R, Choisat B, Frey J, Nicolet J, Kuhnert P. 2000. Detection of the ADP-ribosyltransferase toxin gene (cdtA) and its activity in Clostridium difficile isolates from Equidae. FEMS Microbiol Lett 184:29-33. 10.1111/j.1574-6968.2000.tb08985.x [DOI] [PubMed] [Google Scholar]
- 6.Clifford WJ, Anellis A. 1975. Radiation resistance of spores of some Clostridium perfringens strains. Appl Microbiol 29:861–863. 10.1128/am.29.6.861-863.1975. PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collier CT, Hofacre CL, Payne AM, Anderson DB, Kaiser P, Mackie RI, Gaskins HR. 2008. Coccidia-induced mucogenesis promotes the onset of necrotic enteritis by supporting Clostridium perfringens growth. Vet Immunol Immunopathol 122:104–115. 10.1016/j.vetimm.2007.10.014. PubMed [DOI] [PubMed] [Google Scholar]
- 8.Dagnaes-Hansen F, Moser JM, Smith-John T, Aarup M. 2010. Sudden death in lactating inbred mice. Lab Anim (NY) 39:205–207. 10.1038/laban0710-205. PubMed [DOI] [PubMed] [Google Scholar]
- 9.Elsify A. 2015. A review of Clostridium perfringens toxins with special reference to Beta 2 toxin. Minufiya Vet J 9:85–100. [Google Scholar]
- 10.European Food Safety Authority. 2005. Opinion of the scientific panel on biological hazards on a request from the commission related to Clostridium spp in foodstuffs. EFSA J 3(4):1-65. 10.2903/j.efsa.2005.199. [DOI] [Google Scholar]
- 11.Feinstein RE, Morris WE, Halldén WA, Hedenqvist P, Lindberg R. 2008. Fatal acute intestinal pseudoobstruction in mice. J Am Assoc Lab Anim Sci 47:58–63. Available at https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2654011. [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher DJ, Fernandez-Miyakawa ME, Sayeed S, Poon R, Adams V, Rood JI, Uzal FA, McClane BA. 2006. Dissecting the contributions of Clostridium perfringens type c toxins to lethality in the mouse intravenous injection model. Infect Immun 74:5200–5210. 10.1128/IAI.00534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fohler S, Klein G, Hoedemaker M, Scheu T, Seyboldt C, Campe A, Jensen KC, Abdulmawjood A. 2016. Diversity of Clostridium perfringens toxin-genotypes from dairy farms. BMC Microbiol 16:199. 10.1186/s12866-016-0812-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hüseyin H, erganiş O, Sayin Z, Aras Z. 2012. Toxinotyping of Clostridium perfringens isolates by ELISA and PCR from lambs suspected of enterotoxemia. Turk J Vet Anim Sci 36: 409–415. [Google Scholar]
- 15.Jost BH, Billington SJ, Trinh HT, Bueschel DM, Songer JG. 2006. Association of genes encoding beta2 toxin and a collagen binding protein in Clostridium perfringens isolates of porcine origin. Vet Microbiol 115:173–182. 10.1016/j.vetmic.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Rubbo AD, Rood JI, Moore R. 2008. NetB, a net toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog 4:e26. 10.1371/journal.ppat.0040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klaasen HLBM, Molkenboer MJCH, Bakker J, Miserez R, Häni H, Frey J, Popoff MR, van den Bosch JF. 1999. Detection of the β2 toxin gene of Clostridium perfringens in diarrhoeic piglets in the Netherlands and Switzerland. FEMS Immunol Med Microbiol 24:325–332. 10.1016/S0928-8244(99)00049-8. [DOI] [PubMed] [Google Scholar]
- 18.Krugner-Higby L, Girard I, Welter J, Gendron A, Rhodes JS, Garland T, Jr. 2006. Clostridial enteropathy in lactating outbred swiss-derived (ICR) mice. J Am Assoc Lab Anim Sci 45:80–87. [PubMed] [Google Scholar]
- 19.Lepp D, Gong J, Songer JG, Boerlin P, Parreira VR, Prescott JF. 2013. Identification of accessory genome regions in poultry Clostridium perfringens isolates carrying the netB plasmid. J Bacteriol 195:1152–1166. 10.1128/JB.01032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Adams V, Bannam TL, Miyamoto K, Gacia JP, Uzal FA, Rood JI, McClane BA. 2013. Toxin plasmids of Clostridium perfringens. Microbiol Mol Biol Rev 77:208–233. 10.1128/MMBR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meer RR, Songer JG. 1997. Multiplex polymerase chain reaction assay for genotyping Clostridium perfringens. Am J Vet Res 58:702–705. [PubMed] [Google Scholar]
- 22.Milanov DS, Petrović TR, Todorović DS, Aleksić NR, Čabarkapa IS. 2018. Toxin genotypes of Clostridium perfringens in animal feed and their role in the ethiology of enterotoxemia in domestic animals. Food Feed Res 45:67–76. 10.5937/FFR1801067M. [DOI] [Google Scholar]
- 23.Myers GS, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, Ren Q, Varga J, Awad MM, Brinkac LM, Daugherty SC, Haft DH, Dodson RJ, Madupu R, Nelson WC, Rosovitz MJ, Sullivan SA, Khouri H, Dimitrov GI, Watkins KL, Mulligan S, Benton J, Radune D, Fisher DJ, Atkins HS, Hiscon T, Jost BH, Billington SJ, Songer JG, McClane BA, Titball RW, Rood JI, Melville SB, Paulsen IT. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res 16:1031–1040. Available at http://www.genome.org/cgi/doi/10.1101/gr.5238106. 10.1101/gr.5238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagpal R, Ogata K, Tsuji H, Matsuda K, Takahashi T, Nomoto K, Suzuki Y, Kawashima K, Nagata S, Yamashiro Y. 2015. Sensitive quantification of Clostridium perfringens in human feces by quantitative real-time PCR targeting α-toxin and enterotoxin genes. BMC Microbiol 15:219. 10.1186/s12866-015-0561-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petit L, Gilbert M, Popoff MR. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol 7:104–110. 10.1016/S0966-842X(98)01430-9. [DOI] [PubMed] [Google Scholar]
- 26.Petit L, Gibert M, Gourch A, Bens M, Vandewalle A, Popoff MR. 2003. Clostridium perfringens epsilon toxin rapidly decreases membrane barrier permeability of polarized MDCK Cells. Cell Microbiol 5:155–164. 10.1046/j.1462-5822.2003.00262.x. [DOI] [PubMed] [Google Scholar]
- 27.Popoff MR, Bouvet P. 2013. Genetic characteristics of toxigenic Clostridia and toxin gene evolution. Toxicon 75:63–89. 10.1016/j.toxicon.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Rakoff-Nahoum S, Medzhitov R. 2008. Innate immune recognition of the indigenous microbial flora. Mucosal Immunol 1 S1:S10–S14. 10.1038/mi.2008.49. [DOI] [PubMed] [Google Scholar]
- 29.Rood JI, Adams V, Lacey J, Lyras D, McClane BA, Melville SB, Moore RJ, Popoff MR, Sarker MR, Songer JG, Uzal FA, Van Immerseel F. 2018. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe 53:5–10. 10.1016/j.anaerobe.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rood JI, Cole ST. 1991. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev 55:621–648. 10.1128/mr.55.4.621-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenstiel P. 2013. Stories of love and hate: innate immunity and host-microbe crosstalk in the intestine. Curr Opin Gastroenterol 29:125–132. 10.1097/MOG.0b013e32835da2c7. [DOI] [PubMed] [Google Scholar]
- 32.Savva CG, Fernandes da Costa SP, Bokori-Brown M, Naylor CE, Ambrose RC, Moss DS, Titball RW, Basak AK. 2013. Molecular architecture and functional analysis of NetB, a pore-forming toxin from Clostridium perfringens. J Biol Chem 288:3512–3522. 10.1074/jbc.M112.430223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawires YS, Songer JG. 2006. Clostridium perfringens: insight into virulence evolution and population structure. Anaerobe 12:23–43. 10.1016/j.anaerobe.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Shrestha A, Uz Uzal FA, McClane BA. 2018. Enterotoxic clostridia: Clostridium perfringens enteric diseases. Microbiol Spectr 6. 10.1128/microbiolspec.GPP3-0003-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smart JL, Roberts TA, Stringer MF, Shah N. 1979. The incidence and serotypes of Clostridium perfringens on beef, pork and lamb carcasses. J Appl Bacteriol 46:377–383. 10.1111/j.1365-2672.1979.tb00834.x. [DOI] [PubMed] [Google Scholar]
- 36.Takehara M, Kobayashi K, Nagahama M. 2021. Toll-like receptor 4 protects against Clostridium perfringens infection in mice. Front Cell Infect Microbiol 11:633440. 10.3389/fcimb.2021.633440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Timbermont L, Haesebrouck F, Ducatelle R, Van Immerseel F. 2011. Necrotic enteritis in broilers: an updated review on the pathogenesis. Avian Pathol 40:341–347. 10.1080/03079457.2011.590967. [DOI] [PubMed] [Google Scholar]
- 38.Uzal FA. 2004. Diagnosis of Clostridium perfringens intestinal infections in sheep and goats. Anaerobe 10:135–143. 10.1016/j.anaerobe.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Uzal FA, Fisher DJ, Saputo J, Sayeed S, McClane BA, Songer G, Trinh HT, Fernandez-Miyakawa ME, Gard S. 2008. Ulcerative enterocolitis in two goats associated with enterotoxin- and beta2 toxin-positive Clostridium perfringens type D. J Vet Diagn Invest 20:668–672. 10.1177/104063870802000526. [DOI] [PubMed] [Google Scholar]
- 40.Uzal FA, Vidal JE, McClane BA, Gurjar AA. 2010. Clostridium perfringens toxins involved in mammalian veterinary diseases. Open Toxinology J 3:24–42. Available at https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3917546. 10.2174/1875414701003020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Asten AJ, van der Wiel CW, Nikolaou G, Houwers D, Gröne A. 2009. A multiplex PCR for toxin typing of Clostridium perfringens. Vet Microbiol 136:411–412. 10.1016/j.vetmic.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 42.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 1991. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697–703. 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ATCC’s PCR validation (assay 2 and 3) of toxin genes etx, iap (iA) and cpe.
Validation of etx gene primer sets against positive controls.
Validation of iap (iA) gene primer sets against positive controls.
Validation of cpe gene primer sets against positive controls.