Summary
With the rapid emergence and spread of SARS-CoV-2 variants, development of vaccines with broad and potent protectivity has become a global priority. Here, we designed a lipid nanoparticle-encapsulated, nucleoside-unmodified mRNA (mRNA-LNP) vaccine encoding the trimerized receptor-binding domain (RBD trimer) and showed its robust capability in inducing broad and protective immune responses against wild-type and major variants of concern (VOCs) in the mouse model of SARS-CoV-2 infection. The protectivity was correlated with RBD-specific B cell responses especially the long-lived plasma B cells in bone marrow, strong ability in triggering BCR clustering, and downstream signaling. Monoclonal antibodies isolated from vaccinated animals demonstrated broad and potent neutralizing activity against VOCs tested. Structure analysis of one representative antibody identified a novel epitope with a high degree of conservation among different variants. Collectively, these results demonstrate that the RBD trimer mRNA vaccine serves as a promising vaccine candidate against SARS-CoV-2 variants and beyond.
Subject areas: Health sciences, Virology, Immunology
Graphical abstract
Highlights
-
•
A mRNA vaccine encoding the RBD trimer of wild-type SARS-CoV-2 was designed and studied
-
•
The vaccine elicited strong RBD-specific memory and plasma B cell responses
-
•
The vaccine induced broadly serum and monoclonal neutralizing antibodies in mice
-
•
The vaccine induced strong and protective immunity against major SARS-CoV-2 variants
Health sciences; Virology; Immunology
Introduction
The current vaccines that were developed by targeting the prototype SARS-CoV-2 strain identified during the early phase of the pandemic demonstrated reduced serum neutralizing activity in vitro and attenuated protection against SARS-CoV-2 variants in humans (Abdool Karim and de Oliveira, 2021; Chen et al., 2021; Corti et al., 2021; Garcia-Beltran et al., 2021; Harvey et al., 2021; Madhi et al., 2021; Planas et al., 2021; Wang et al., 2021a; Wang et al., 2021b; Wang et al., 2021c; Wibmer et al., 2021; Zhou et al., 2021). While these vaccines have been designed and formulated in various modalities, they share one common feature in that they mimic and preserve the native pre-fusion conformation of the spike protein, thus are expected to induce the most relevant and effective immune response against SARS-CoV-2 (Dai and Gao, 2021; Wrapp et al., 2020). The incredible levels of safety and efficacy demonstrated by these vaccines in clinical trials and ongoing vaccine rollout clearly support this shared overall strategy. However, such commonality also predisposes the vaccines to similar escape mechanisms when it comes to SARS-CoV-2 variants. For example, the reduced level of protection provided against these variants is largely attributed to deletions in the N3 and N5 loops that collectively make up the so-called antigenic “supersite” in the N-terminal domain (NTD) and the constellation of substitutions in the receptor-binding domain, such as K417N/T, L452R, E484K/Q, and N501Y (Barnes et al., 2020; Chen et al., 2021; Chi et al., 2020; Corti et al., 2021; Garcia-Beltran et al., 2021; Harvey et al., 2021; McCallum et al., 2021; Planas et al., 2021; Suryadevara et al., 2021; Wang et al., 2021a, 2021b, 2021c; Wibmer et al., 2021; Zhou et al., 2021). Many of these mutations are shared by the B.1.1.7 variant (alpha) that was initially identified in the United Kingdom, the B.1.351 variant (beta) in South Africa, the P1 variant (gamma) in Brazil, and the B.1.617 variant (delta) in India (Abdool Karim and de Oliveira, 2021; Harvey et al., 2021). Fortunately, by screening hundreds and thousands of monoclonal antibodies from convalescent or vaccinated individuals, a small number of broad and potent neutralizing antibodies directed to RBD were identified capable of neutralizing all variants identified thus far (Corti et al., 2021; Harvey et al., 2021; Tortorici et al., 2020; Wang et al., 2021b, 2021c). These neutralizing antibodies target various spatially distributed epitopes on RBD and may contribute to the residual serum neutralizing activity against SARS-CoV-2 variants of concern in these individuals (Chen et al., 2021; Planas et al., 2021; Wang et al., 2021c; Wibmer et al., 2021; Zhou et al., 2021). More importantly, these neutralizing antibodies can be substantially boosted by mRNA vaccines, particularly in convalescent individuals (Stamatatos et al., 2021; Wang et al., 2021c). In contrast, antibodies to regions beyond the RBD, such as the NTD and S2, are inherently weak or completely lose their neutralization to SARS-CoV-2 variants (Corti et al., 2021; Harvey et al., 2021; Liu et al., 2020; McCallum et al., 2021; Robbiani et al., 2020; Rogers et al., 2020; Suryadevara et al., 2021; Zhou et al., 2021; Zost et al., 2020). These results suggest the existence of highly conserved and vulnerable regions within the RBD that could be precisely targeted for the development of next-generation vaccines capable of inducing broad and protective immunity against SARS-CoV-2 variants.
Results
The development of an RBD trimer mRNA vaccine to elicit a potent and durable antibody response
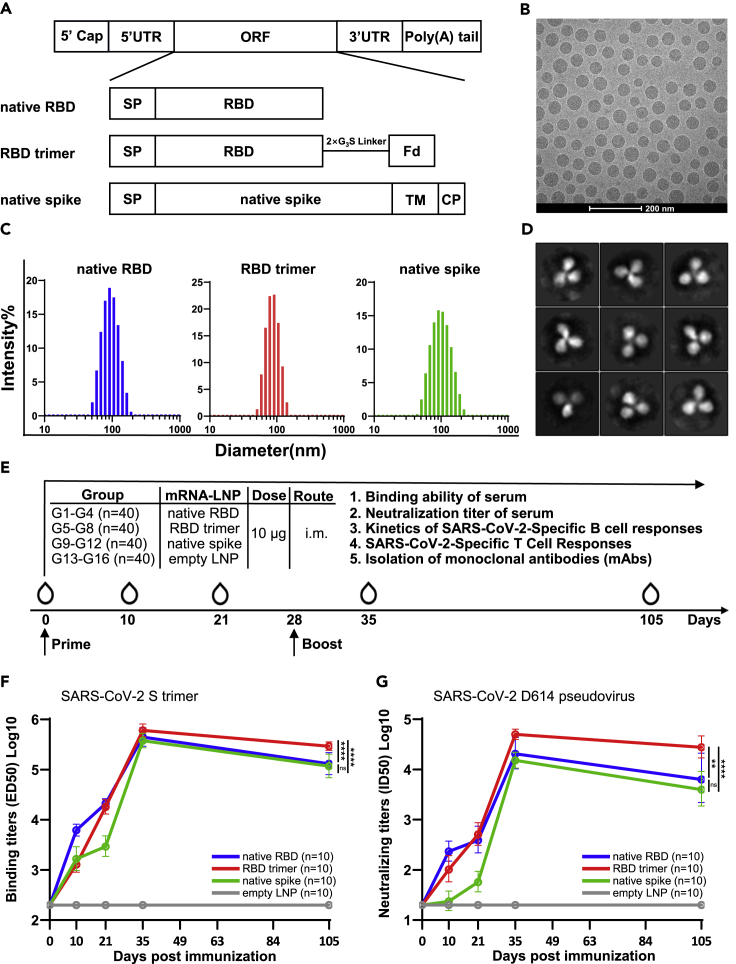
To capitalize on the conserved and vulnerable regions within the RBD that are potentially capable of inducing broad and effective neutralizing antibodies against SARS-CoV-2 variants, we designed a lipid nanoparticle-encapsulated and nucleoside-unmodified mRNA (mRNA-LNP) vaccine encoding the trimerized RBD (RBD trimer). RBD was trimerized via genetic fusion to the foldon trimerization domain, and compared with that of expressing secreted, monomeric version of the native receptor-binding domain (native RBD) and native membrane-anchored full-length spike (native spike) of the prototype SARS-CoV-2 strain (Wuhan Hu-1) (Figure 1A). The RBD sequence used for RBD trimer was 199 amino acids in length, for native RBD was 223 amino acids in length, while the native spike was 1,260 amino acids in length. Quality control tests of these mRNA-LNPs by cryo-TEM imaging and dynamic light-scattering measurements revealed an average particle size between 75 and 110 nm (Figures 1B and 1C). The encapsulation efficiency of three mRNA-LNP vaccines was all more than 95%. Two-dimensional average of recombinant RBD trimers produced by 293F cells bound to soluble ACE2 confirmed the expected trimeric morphology (Figure 1D). A total of 160 female BALB/c mice aged 6–8 weeks were randomly distributed into 16 groups (G1 to G16, n = 10 per group). Of which, four groups each were intramuscularly vaccinated on day 0 and 28 with either the native RBD (G1–G4), the RBD trimer (G5–G8), the native spike (G9–G12), or the empty LNP (G13–G16). On day 10, 21, 35, and 105 after initial immunization, one group each from the native RBD, the RBD trimer, the native spike, and the empty LNP-vaccinated animals were sacrificed for sample collections and evaluation. The RBD trimer vaccine induced the most potent and durable binding and neutralizing antibody response up to 105 days after the initial immunization, followed by the native RBD vaccine, and then the native spike vaccine (Figures 1E–1G). This enhanced immunogenicity was reflected by reciprocal serum binding (ED50) and neutralization (ID50) to a pseudovirus carrying the original strain identified in Wuhan, China (WT D614), particularly following boost immunization (Figures 1F and 1G).
Figure 1.
Design and characterization of mRNA vaccines
(A) Design and construction of three mRNA vaccines expressing native receptor-binding domain (RBD), trimeric RBD with a foldon-trimerization tag (RBD Trimer), or native full-length spike (Native Spike) of SARS-CoV-2. SP: signal peptide; TM: transmembrane domain; CP: cytoplastic domain.
(B) Quality control of LNP-formulated mRNA vaccines through cryo-TEM imaging and (C) dynamic light-scattering measurement. In (B), a scale bar of 200 nm is indicated.
(D) Two-dimensional average of recombinant RBD trimers produced in 293F cells bound to soluble ACE2 and analyzed by Cryo-EM. Box edge, 30.4 nm.
(E) Schedule for immunization and evaluation of immunogenicity in BALB/c mice (n = 160). Mice were immunized and monitored for serum binding and neutralizing antibodies, kinetics of SARS-CoV-2-specific B and T cell responses in spleen, inguinal lymph nodes, and bone marrow, and for isolating monoclonal antibodies from RBD-trimer-immunized animals. The exact date of prime and boost vaccination and sample collections are indicated. On day 10, 21, 35, and 105 after initial immunization, one group of 10 animals from each of the native RBD, the RBD trimer, the native spike, and the empty LNP-vaccinated animals were sacrificed for sample collections and evaluation.
(F and G) Comparison of serum binding (ED50) and neutralizing (IC50) activities among the three mRNA vaccines vaccinated animals. Binding assay was conducted against SARS-CoV-2 S trimer while neutralizing activity was against pseudovirus bearing wild-type spike derived from original strain identified in Wuhan, China (Wu-1 D614). All data are presented as the geometric means together with the standard deviations from at least two independent experiments. Statistical significance was analyzed using unpaired Students’ t-test (two-tailed) (ns, not significant; ∗∗p ≤ 0.01; ∗∗∗∗p ≤ 0.0001).
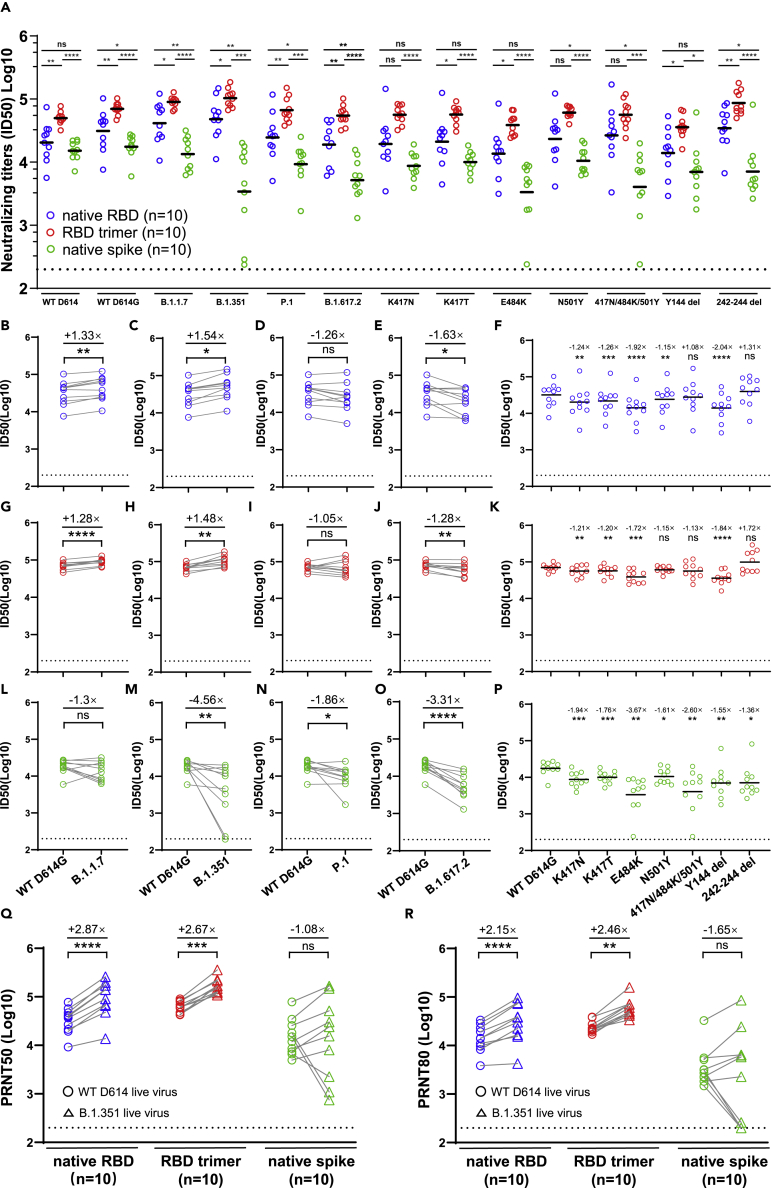
The RBD trimer mRNA vaccine induces broad and potent neutralizing antibody activity against major SARS-CoV-2 variants
The RBD trimer vaccine also induced the most broad and potent serum neutralizing activity against a panel of SARS-CoV-2 variants (B.1.1.7, B.1.351, P.1, and B.1.617.2) and mutant pseudoviruses carrying the key single or triple substitutions derived from the variants (Figures 2A and S1). The overall geometric mean serum ID50 against this panel of viruses was 61,240.6 ± 1.57 for the RBD trimer vaccine, 25,046.5 ± 2.41 for the native RBD vaccine, and 8099.3 ± 2.62 for the native spike vaccine. When compared to the WT D614G pseudovirus, the RBD trimer and native RBD vaccines demonstrated an uninfluenced ID50 to B.1.1.7, B.1.351, P1, and a slight reduction for the B.1.617.2 (Figures 2B–2E and 2G–2J). These broad neutralizing activities correlated with mutant pseudoviruses carrying either single N501Y or triple 417N/484K/501Y mutations (Figure 2K). Interestingly, there was a significant improvement in neutralization against B.1.351 variant, with a 1.52-fold improvement for the RBD trimer vaccine (Figure 2H) and a 1.55-fold improvement for the native RBD vaccine (Figure 2C). These improvements may be related to enhance neutralizing activity to the △242-△244 mutant pseudovirus, with a 1.72-fold increase for the RBD trimer vaccine and a 1.31-fold increase for the native RBD vaccine. It is worth noting that the single △144 mutant pseudoviruses reduced plasma neutralizing activity induced by RBD trimer and native RBD vaccines (Figures 2K and 2P). It was also found that the potency of some RBD-directed monoclonal antibodies was also affected by NTD deletion including △144 and △242-△244 (Wang et al., 2021a, 2021b). In contrast, the native spike vaccine demonstrated reduced activity to all the variants of concern. The loss of plasma neutralizing activity could be largely attributed to either single E484K or triple 417N/484K/501Y substitutions (Figure 2L–2P). A similar trend was also observed against live WT D614 and variant B.1.351 (Figures 2Q, 2R, and S1). These results suggest that the RBD trimer vaccine, and to a lesser extent, the native RBD vaccine, could modulate antibody responses that were less affected by the existing mutations identified in the SARS-CoV-2 variants. The responses elicited by the native spike vaccine, however, were more sensitive to mutations in the variants, similar to the effect seen in convalescent and vaccinated individuals (Chen et al., 2021; Garcia-Beltran et al., 2021; Planas et al., 2021; Wang et al., 2021a, 2021b, 2021c; Wibmer et al., 2021; Zhou et al., 2021).
Figure 2.
Superior antibody response against SARS-CoV-2 variants induced by RBD trimer
(A–P) (A) Serum neutralizing antibody induced by native RBD, RBD trimer, and native spike mRNA vaccines to pseudoviruses carrying full-length, single, or triple mutant spike of SARS-CoV-2 variants. Colour scheme for native RBD (blue), RBD trimer (red), and native spike (green) are consistent throughout the manuscript. Fold change in reciprocal serum neutralization (IC50) from the native RBD (B–F), RBD trimer (G–K), and native spike (L–P) mRNA vaccines immunized mice against variants B.1.1.7, B.1.351, P1, and B.1.617.2, as well as single or triple mutant pseudoviruses, relative to the WT D614G pseudovirus.
(Q and R) Fold change in reciprocal serum neutralization (PRNT50 and PRNT80) in the three mRNA vaccines immunized mice against live B.1.351 variant, relative to the WT D614 live virus. The tested serum samples were collected at week 5 after initial immunization. All data are presented as the original and/or with indicated means. The fold change in ID50, PRNT50, or PRNT80 between mutant and WT D614 virus are shown by overall average at the top in (B–R). The symbol “+” indicates an increase while the symbol “-” a decrease in neutralization. Statistical significance was analyzed using unpaired Students’ t-test (two-tailed) in (A) and paired Students’ t-test (two-tailed) in (B-R) (ns, not significant; ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001).
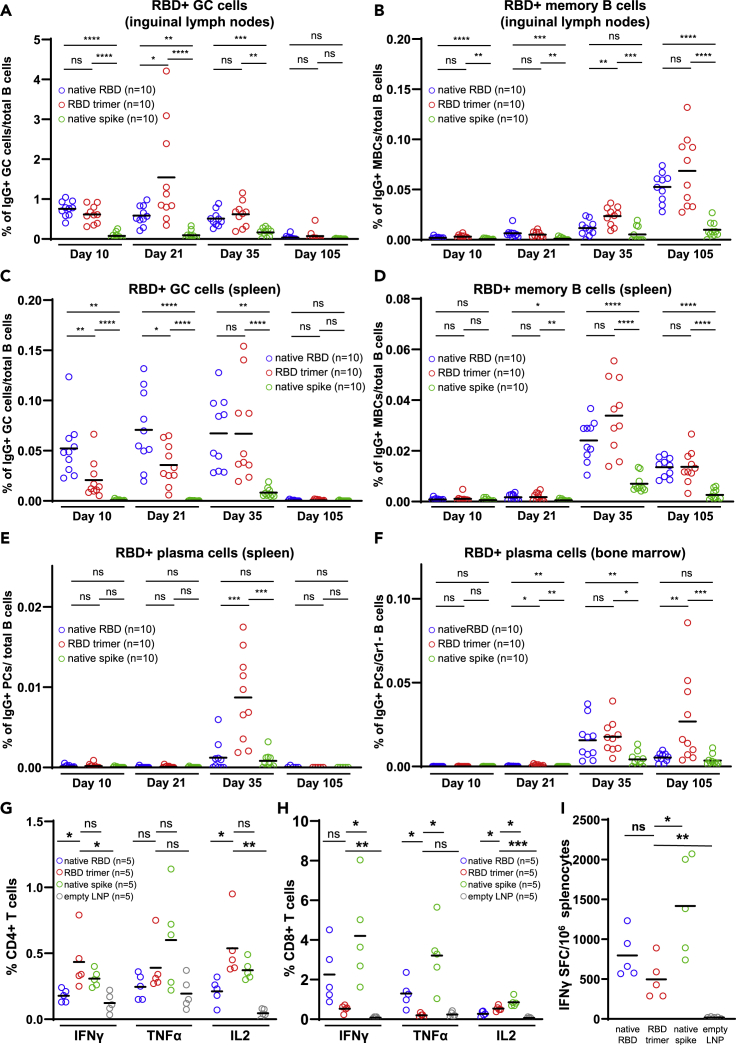
The RBD trimer mRNA vaccine elicits strong antigen-specific germinal center, memory, and long-lived plasma B cell responses
Successful vaccines rely on the generation, development, and maintenance of germinal center B cells (GCBCs), memory B cells (MBCs), and long-lived plasma cells (LLPCs), in a coordinated and spatiotemporal manner. To investigate the cellular basis underlying the ability of the RBD trimer mRNA vaccine to induce broad and potent neutralizing antibody responses, we monitored and compared the kinetics of RBD-specific IgG+ GCBCs, MBCs, and/or LLPCs in inguinal lymph nodes, spleen, and bone marrow among the three groups of fully immunized mice. In the draining inguinal lymph node, we found that RBD trimer induced the strongest GCBCs, peaking at about 1.55 % on day 21 after initial immunization. It waned down to 0.62% by day 35 and to 0.07% by day 105, despite a boosting dose on day 28 (Figure 3A). Furthermore, the RBD trimer vaccine was able to expand MBCs from barely detectable on days 10 and 21 to 0.024% by day 35 and to 0.069% by day 105. A booster dose on day 28 had a clear augmentative effect on MBCs (Figure 3B). The native RBD vaccine was relatively weaker and the native spike vaccine was the weakest with regards to inducing GCBCs and MBCs responses during the same period (Figures 3A and 3B). In the spleen, the RBD trimer and native RBD vaccines appeared to elicit similar kinetics for GCBCs and MBCs, although GCBCs rose substantially slower in the RBD trimer mRNA group than in the native RBD group (Figures 3C and 3D). The boosting dose on day 28 had comparable impact on both GCBCs and MBCs in the two groups of animals, and reached the comparably high levels by day 35, this was then followed by a substantial reduction by day 105 (Figures 3C and 3D). In contrast, the native spike vaccine demonstrated significantly weaker B cell responses throughout the study period.
Figure 3.
RBD trimer mRNA vaccine elicits superior antigen-specific germinal center, memory, and long-lived plasma B cell responses
(A–H) Kinetics of RBD-specific IgG+ GC cells and memory B cells in inguinal lymph nodes (A and B) or in spleen (C and D). Kinetics of RBD-specific IgG+ plasma cells in spleen (E) or in bone marrow (F). Intracellular expression of IFN-γ, TNF-α, and IL-2 in CD4+ (G) and CD8+ T (H) cells 5 week after first immunization.
(I) ELISPOT analysis of IFN-γ-positive splenocytes, samples were collected at week 5 after initial immunization. All data are presented as the original with indicated means. Statistical significance was analyzed using unpaired Students’ t-test (two-tailed) (ns, not significant; ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001; ∗∗∗∗p ≤ 0.0001).
The most striking observation was that the RBD trimer mRNA vaccine induced the strongest RBD-specific plasma B cells in the spleen and in the bone marrow after the booster dose. The highest plasma B cell response in the spleen was observed on day 35, when the trimer group outperformed the native RBD group by a wide margin, and splenic response waned down to a similar level by day 105 (Figure 3E). RBD-specific bone marrow plasma cells became detectable on day 35 and continued to rise by day 105 (Figure 3F). Importantly, at that point, the RBD trimer group attained a much higher level of RBD-specific plasma cells in the bone marrow. Although the precise origin of this superiority of the RBD trimer mRNA vaccine is currently unknown, the fact that the difference appeared after the boost suggests memory B cells induced by the RBD trimer mRNA vaccine immunization may be functionally superior.
Next, we found that all three vaccines were able to induce strong S protein-specific T cell responses on day 35 after the initial immunization. Interestingly, the RBD trimer vaccine tended to elicit more antigen-specific polyfunctional CD4 T cells than CD8 T cells expressing type 1 (Th1) immune response cytokines such as interferon gamma (IFNγ), tumor necrosis factor (TNF)-α, and interleukin (IL)-2 (Figures 3G and 3H). This biased CD4 helper response may help to explain the broad and potent neutralizing antibody response observed in animals that were immunized with the RBD trimer mRNA vaccine (see above). In contrast, the native spike vaccine tended to trigger a higher CD8 T cell response than the RBD trimer mRNA vaccine, as determined by intracellular cytokine staining and enzyme-linked immunospot assay (ELISPOT) (Figures 3H and 3I).
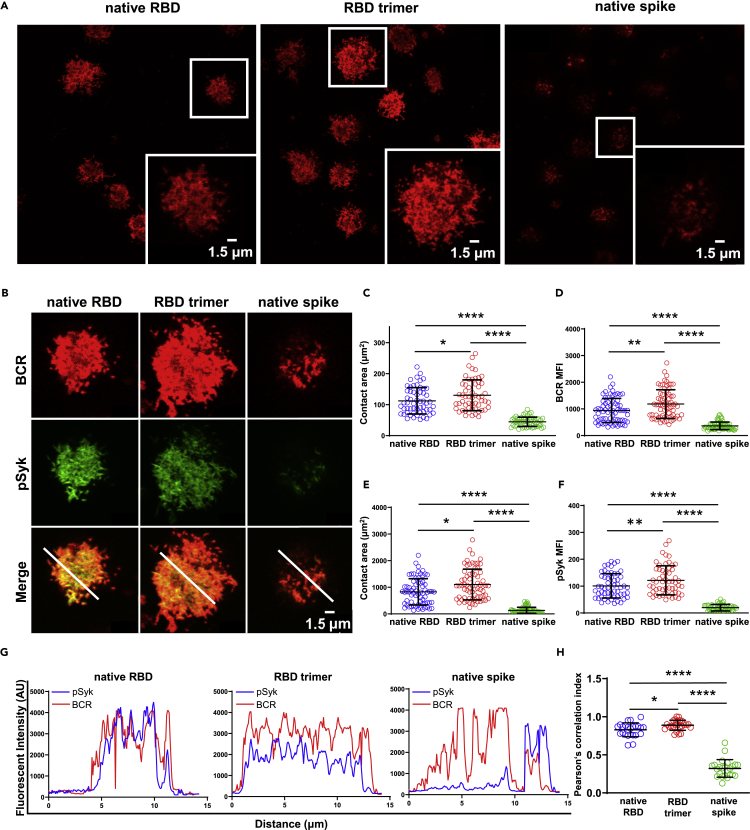
RBD trimer triggers the strongest surface BCR clustering and downstream signaling in B cells
We compared the three immunogens with regards to their ability to activate B cells by triggering SARS-CoV-2-specific B cell receptor (BCR) clustering and downstream signaling; these represent the critical steps for B cells to differentiate into antibody-producing plasma cells. To this end, we first engineered the J558L cells to express human IgG-BCRs with variable regions of the heavy and light chain from a SARS-CoV-2-specific human monoclonal antibody P2B-2F6 (J558L-P2B-2F6 B cells) (Liu et al., 2010). P2B-2F6 was a strong neutralizing antibody isolated by our group from a SARS-CoV-2 convalescent patient that can bind RBD in both “up” and “down” conformations (Ju et al., 2020). The J558L-P2B-2F6 B cells were loaded onto glass coverslips containing the recombinant native RBD, RBD trimer, or native spike in the same molar concentration for 10 min at 37°C. The native spike used here contained the 2P mutations at positions 986 and 987 and GSAS linker to help stabilize the spike in its prefusion conformation (Wrapp et al., 2020). The formation and accumulation of BCR microclusters on the antigen contact area were examined using laser scanning confocal microscopy as previously reported (Liu et al., 2010). Figure 4A shows that the RBD trimer was clearly the most potent, followed by the native RBD and the native spike, in terms of triggering the formation of P2B-2F6-BCR microclusters within B cell immunological synapses, as determined by the contact area on the antigen-containing surface and the mean fluorescence intensity of the P2B-2F6-BCR micro-clusters (Figures 4B, 4C, and 4D). Next, we measured the signaling molecule downstream of P2B-2F6-BCR, phosphorylated spleen tyrosine kinase (pSyk), as this represents the direct outcome of BCR aggregation and has therefore been widely used as the downstream marker for B cell activation (Liu et al., 2010). Consistent with its effect on P2B-2F6-BCR microclusters, RBD trimer exhibited the strongest effect in triggering the recruitment of pSyk into immunological synapses, as demonstrated by the accumulation and significant colocalization with P2B-2F6-BCR (Figures 4B, 4E, and 4F). This was further confirmed by analysis of the spatial distribution of microclusters between P2B-2F6-BCR and pSyk via the fluorescent intensity of colocalization and Pearson correlation index (Figures 4G and 4H). These results indicate that RBD trimer has the best ability to activate B cells through P2B-2F6-BCR clustering and downstream signaling.
Figure 4.
RBD trimer is the most potent in triggering BCR signaling
(A) Synaptic accumulation of BCRs triggered by native RBD, RBD trimer, and native spike on the surface of the J558L B cell line expressing SARS-CoV-2 antibody 2F6-IgG-BCRs captured by a confocal microscopy. The magnified insets (lower right corner) show the highlighted cells in the original image within the white boxes. The BCR molecules are labelled in red, and the scale bar in the inset window represents 1.5 μm.
(B) Synaptic recruitment of phosphorylated Syk (pSyk) to the contact area of a single J558L B cell after BCR accumulation by native RBD, RBD trimer, and native spike captured by a confocal microscopy. The BCRs and pSyk molecules are respectively labelled in red and green, and their merged images are also shown. The scale bar represents 1.5 μm.
(C–F) Statistical analysis for the contact area and mean fluorescence intensity (MFI) of BCRs (C and D) and pSky (E and F) recruited in the immunological synapses on the surface of J558L-2F6-IgG-BCRs B cells. Each dot in the plot represents the data of one cell with indicated means and standard deviation.
(G) Fluorescence intensity profiles from (B) showing degrees of co-localization between BCR and pSyk on J558L-2F6-IgG-BCRs B cells after engagement with native RBD, RBD trimer, and native spike.
(H) Pearson correlation index showing the spatial distribution between BCR and pSky microclusters on J558L-2F6-IgG-BCRs B cells after engagement with native RBD, RBD trimer, and native spike. Each dot represents an individual measurement from a single B cell. Bars indicate the mean values and standard deviations from at least three independent experiments. Statistical significance was analyzed using unpaired Students’ t-test (two-tailed) (∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001).
The RBD trimer mRNA vaccine induces broad and potent monoclonal antibodies against SARS-CoV-2 variants
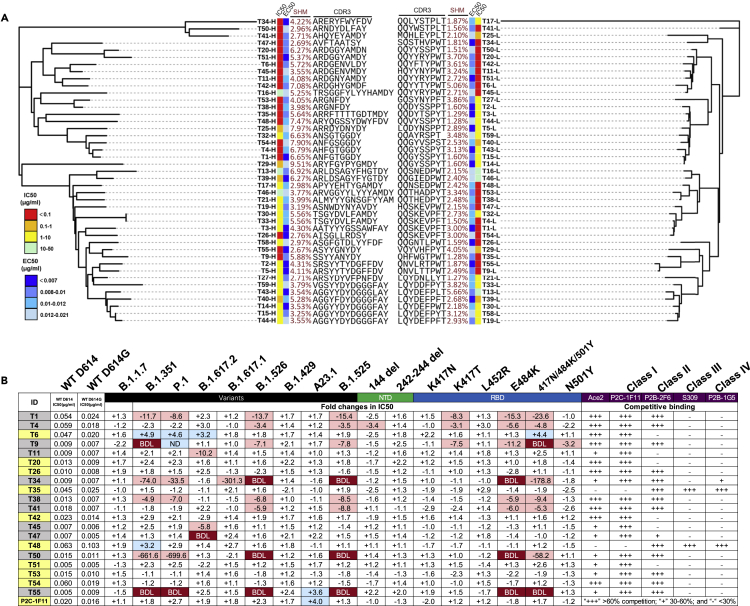
To identify the potential mechanisms that contribute to the broad and potent serum neutralizing activity of the RBD trimer mRNA vaccine, we isolated monoclonal antibodies (mAbs) from five of animals on day 105 after immunization with the RBD trimer vaccine. Spike trimer-specific single B cells were sorted from a suspension of splenocytes into 96-well plates. IgG heavy and light chain variable regions from each well were then amplified by nested PCR, followed by cloning and expression of full-length IgG1 mouse antibodies, as previously described (von Boehmer et al., 2016). Out of total 296 sorted single B cells, we were able to amplify, and express 231 paired IgG heavy and light chain variable regions. Of these, we randomly selected 43 antibodies and determined their relative binding abilities to the recombinant RBD trimer by enzyme-linked immunosorbent assays (ELISAs) and neutralizing activity against WT D614 pseudovirus. Figure 5A shows the genetic relatedness of the 43 antibodies for both heavy and light chain variable regions, and their associated neutralizing (IC50) and binding (EC50) activities, along with their somatic hypermutation (SHM) and CDR3 sequences. The 43 antibodies were scattered across branches without apparent clustering, suggesting that dominant clonal expansion had not occurred during the immunization process even though some antibodies had identical CDR3 sequences in their heavy and light variable regions. Only two antibodies (T30 and T33) shared the same heavy chain variable sequences, although their light chain variable sequences were different. The IC50 and EC50 of these antibodies varied considerably, as indicated by various colors with range of activities. Interestingly, the majority of the potent neutralizing antibodies highlighted in red tended to cluster in one major group on the heavy chain tree while segregated into three relatively small groups on the light chain tree. This matching pattern may suggest that the potent neutralizing activity was attributed more to the heavy chain than to the light chain. Furthermore, the IC50 was not associated with the EC50 in any of the antibodies, thus indicating that binding activity per se does not translate into neutralizing activity (Figure S3A). The degree of SHM also varied among the antibodies and no correlation was detected with the IC50 nor EC50 (Figures S3B, S3C, S3D, and S3E). Notably, no cross neutralization nor cross binding was detected against SARS-CoV or the MERS-CoV spike in the context of pseudoviruses or when expressed on the surface of 293T cells (data not shown).
Figure 5.
RBD trimer induces dominant broad and potent monoclonal antibodies against SARS-CoV-2 variants
(A) Unrooted neighbor-joining tree depicting the genetic relationship among 40 monoclonal antibodies. The left panel is derived from the heavy chain variable region while the right panel from the light chain variable region. Antibody names, and their corresponding IC50, EC50, degree of somatic hyper mutation, and CDR3 sequences are indicated at the tip of the branches. The IC50 was measured against SARS-CoV-2 WT D614 pseudovirus and the EC50 was calculated against recombinant RBD trimer in the ELISA assay.
(B) IC50 and their fold changes against SARS-CoV-2 variants, as well as single or triple mutant pseudoviruses, relative to that of WT D614G. The symbol “+” indicates increased while “-” decreased potency. Those highlighted in blue indicates increased potency at least threefold whereas those in red decreased potency at least threefold. Those in white change less than threefold. BDL, referring to Below Detection Limit, indicates the highest concentration (2 μg/mL) of mAbs failed to reach 50% neutralization. Results were calculated from three independent experiments. Epitope mapping through competitive binding with ACE2 and various well-defined monoclonal antibodies (P2C-1F11, P2B-2F6, S309, and P2B-1G5) was also shown. “+++” indicates >60% competition, while “+” 30%–60%, and “-” <30%. Results are representatives of two independent experiments.
Next, we evaluated the breadth and potency of the top 20 most potent neutralizing mAbs against nine major SARS-CoV-2 variants and eight mutant pseudoviruses carrying single or triple mutations in the NTD and RBD derived from these variants (Figures 5B and S4). As shown in the first column of Figures 5B, 9 out of 20 mAbs (45%), highlighted in yellow, demonstrated broad and potent neutralizing activity against the entire panel of variants and their single or triple mutants. When compared to WT D614G, these mAbs showed either comparable activity or enhanced activity against the major variants. In particular, like that observed for immune serum, many mAbs demonstrated improved neutralizing activity largely due to 242–244del mutation in the NTD (Figure 5B). Furthermore, mAb T6 was able to exert at least a 3-fold increase in neutralization to 3 of the 4 variants of concern, thus suggesting that its epitope might have become more exposed in some of these variants. Examination of the enhancement patterns across the single and triple mutant pseudoviruses further showed that the 242–244del in NTD, K417N/T, and 417N/484K/501Y in RBD were all potential contributors although other substitutions cannot be ruled out. BLI experiments showed that 7 of the 9 mAbs exhibited variable levels of competition with soluble ACE2 and all had strong competition with class 1 mAb P2C-1F11, a broad and potent human mAb isolated from a convalescent individual who was included in a Phase III clinical trial against SARS-CoV-2 infection (Ge et al., 2021; Ju et al., 2020). Some of these mAbs, such as T26, T35, T48, T53, and T54 also competed with class 2 mAb P2B-2F6 (Figure 5B). The remaining 2 of these 9 mAbs failed to compete with P2C-1F11 but did compete with those from class 2 mAb P2B-2F6, class 3 mAb S309, and class 4 mAb P2B-1G5 (Figure 5B).
In contrast, the remaining 11 of the 20 mAbs (55%), highlighted in gray in the first column of Figure 5B, demonstrated reduced levels of activity, or a complete loss of activity, against the testing variants. These effects appeared to be confined to four variants (B.1.351, P.1, B.1.526, and B.1.525) and were largely attributed to the E484K and 417N/484K/501Y mutations, and to a lesser extent, the K417T mutation (Figure 5B). This is in perfect agreement with the E484K mutation shared by these four variants (Corti et al., 2021; Harvey et al., 2021). BLI experiments confirmed their strong competition with typical class 2 mAb P2B-2F6 as well as class 1 mAb P2C-1F11, although their ability to compete with soluble ACE2 was variable. Collectively, these results indicate that the RBD trimer vaccine was able to induce a high proportion of broad and potent mAbs and is likely to contribute to the broad and potent serum neutralizing activity observed in the immunized animals (Figure 2).
Structural basis for the broad and potent T6 antibody against SARS-CoV-2 variants
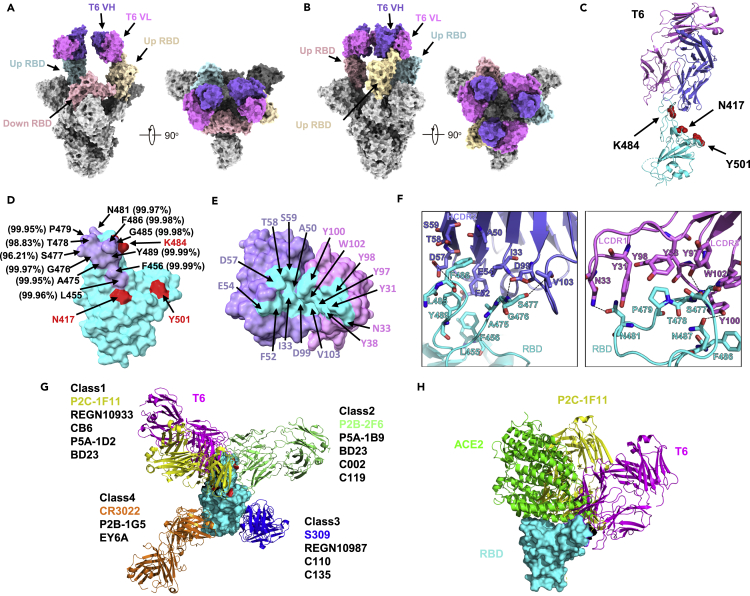
To reveal the molecular basis for broad and potent neutralizing mAbs, we selected T6 with great broad and potency against SARS-CoV-2 and its variants as a representative and determined the cryo-EM structure of its Fab in complex with the spike trimer derived from variant B.1.351 initially identified in South Africa. Data derived from cryo-EM allowed us to build two models for the binding of the T6 Fab to the B.1.351 spike trimer. One model revealed two T6 Fabs bound to two RBDs in the “up” conformation at 3.2 Å resolution. The other model revealed was three T6 Fabs bound to all three “up” RBDs at 3.4 Å resolution (Figures 6A, 6B, S7 and Table S1). In both models, the CL and CH1 of the T6 Fab were not built due to weak densities. As the T6 Fab is only bound to the “up” RBD, it follows that the T6 epitope must be buried when RBD is in the “down” conformation in the spike trimer.
Figure 6.
Cryo-EM and Crystal structure of mAb T6 with spike trimer or RBD of SARS-CoV-2 variant B.1.351
(A) Cryo-EM structure of the soluble, recombinant spike trimer of B.1.351(SA) strain in complex with two variable regions of mAb T6 at 3.2 Å resolution. VH, variable region of heavy chain; VL, variable region of light chain.
(B) Cryo-EM structure of the soluble, recombinant spike trimer of B.1.351(SA) strain in complex with three variable regions of mAb T6 at 3.3 Å resolution.
(C) T6/RBD-3M crystal structure. T6 light chain, heavy chain, and RBD-3M are colored in violet, purple, and cyan, respectively. The three RBD-3M mutated residues (N417, K484, and Y501) are shown as red spheres.
(D) The epitope of mAb T6. The epitope residues containing within 4 Å are indicated in purple. The Conservation of amino acids are labelled beside, the data are collected from GISAID database.
(E) The paratope of mAb T6. The paratope residues containing within 4 Å are indicated in cyan and binding residues within are indicated in either purple or violet, depending on their origin from heavy or light chain Fab.
(F) Interactions between mAb T6 Fab and SARS-CoV-2 RBD-3M. The hydrogen bonds are represented by dashed black lines.
(G) Structural classification of SARS-CoV-2 anti-RBD antibodies. The first one of each class is represented and superposed onto T6/RBD structure. The model of P2C-1F11(PDB:7CDI) is colored in yellow, T6 in pink, P2B-2F6(PDB:7CZX) in light green, S309(PDB:6WPT) in blue, CR3022(PDB:6ZLR) in orange, and SARS-CoV-2 RBD in cyan surface. The three RBD-3M mutated residues (N417, K484, and Y501) are shown as red surface.
(H) The crystal structure of the SARS-CoV-2 RBD/ACE2/P2B-1F11 complex is superimposed onto the T6/RBD-3M crystal structure. RBD is shown in cyan surface, T6 in pink, P2B-1F11 in yellow, and ACE2 in green cartoon.
We further determined the crystal structure of the T6 Fab bound to the RBD of B.1.351 carrying the three signature mutations K417N/E484K/N501Y (RBD-3M) at 2.89 Å resolution (Figure 6C and Table S2). The T6 Fab buries a surface area of 661.4 Å2 on the RBD. Of this, 358.8 Å2 was composed of the heavy chain and 302.6 Å2 by the light chain (Figure 6D). A total of 16 residues from the T6 Fab, and 11 residues from the RBD, are involved in the extensive interactions at the binding interface (Figures 6D and 6E). Of these, RBD residues S477, T478, N481, N487, and Y489 formed hydrogen bonds with the T6 Fab (Figure 6F). The mutated residues N417, K484, and Y501, shared among several SARS-CoV-2 variants were not located within the epitope of the T6 Fab, although N417 and K484 were both located in a proximate position, Y501 has located some distance away (Figure 6D). The L452R mutation, found in several SARS-CoV-2 variants, was not evident in the T6 epitope. Furthermore, structural alignments with four major classes of RBD antibodies indicated that the T6 antibody is a new member of the family of class 1 antibodies although with rather different angle of approach to RBD (Figure 6G) (Barnes et al., 2020). The epitope specificity of the T6 antibody overlapped with that of P2C-1F11, a typical class 1 mAb with a broad and potent neutralizing activity against all variants identified so far (Ge et al., 2021; Wang et al., 2021b). Class 1 antibodies generally exhibit substantial steric clashes with ACE2 when binding to the RBD (Barnes et al., 2020). T6 was no exception, thus indicating that the mechanism underlying its neutralization ability was to compete with ACE2 to bind to the RBD, thereby disrupting the very first step of viral entry (Figure 6H). Finally, analysis of T6 epitope residues in the GISAID database revealed a high degree of conservation, thus providing a sequence and structural basis for its broad and potent neutralizing activity against a wide range of SARS-CoV-2 variants (Figure 6D).
The RBD trimer mRNA vaccine protects K18-hACE2 mice from death after infection with live SARS-CoV-2 WT D614, B.1.351, and B.1.617.2 variants
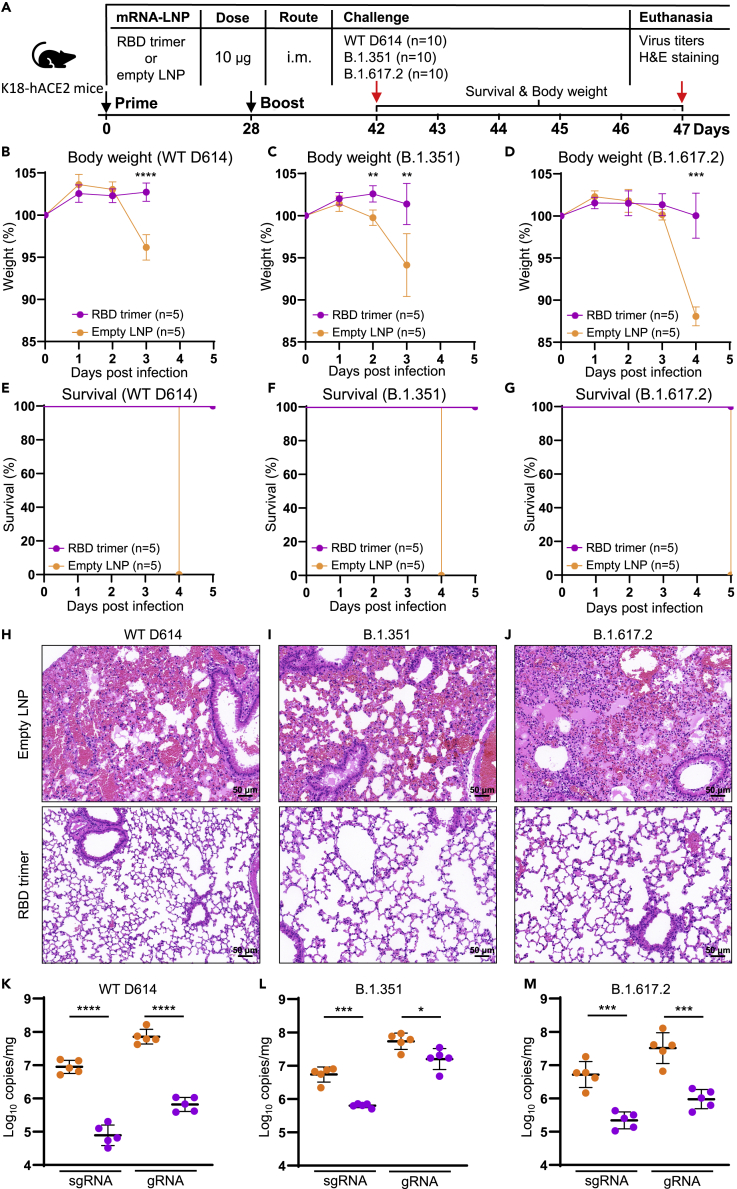
To evaluate the protective potential of RBD trimer mRNA vaccine, we used a well-established K18-hACE2 mouse model of SARS-CoV-2 infection (McCray et al., 2007). A total of 30 mice were divided into three groups (n = 10) and each was immunized with either 10 μg of RBD trimer mRNA vaccine (n = 5 per group) or empty LNP as control (n = 5 per group) on day 0 and 28 (Figure 7A). Two weeks after the second immunization, all animals in the three groups were challenged with 103 plaque-forming units (PFU) of live SARS-CoV-2 WT D614, B.1.351, or B.1.617.2. The mice were monitored for body weight changes and survival during ensuing 5 days. Lung samples were collected at death for viral loads determination and histopathological observation. As shown in Figures 7B–7G, RBD trimer vaccinated animals maintained relatively stable body weight and 100% survival. By contrast, control animals suffered substantial loss of body weight (Figures 7B–7D) and succumbed to infection within 5 days post infection (dpi) (Figures 7E–7G). Among the three live viruses, B.1.351 appeared to be the most pathogenic and resulted in significant weight loss starting from 2 days post infection (dpi) and death on 4 dpi, followed by WT D614 from 3 dpi and 4 dpi, and then by B.1.617.2 from 4 dpi and 5 dpi (Figures 7B–7D), suggesting infection with different viruses could result in different rate of disease progression. Furthermore, histopathological analysis on the lung sections stained with haematoxylin and eosin (H&E) showed that the RBD-trimer-vaccinated mice maintained normal lung structure. By contrast, the control animals showed severe interstitial pneumonia, as evidenced by the infiltration of neutrophil and lymphocyte, edema, and focal presence of hemorrhages in a proportion of alveoli (Figures 7H–7J). All three viral strains lead to similar histopathologic features, consistent with the inflammation induced by SARS-CoV-2 infection. In terms of viral load in lungs, WT D614-infected control animals had 9.6×106 copies/mg for sgRNA and 8.0×107 copies/mg for gRNA, which were about two-logs higher than that in the vaccinated animals (9.6×104 copies/mg for sgRNA and 7.2 ×105 copies/mg for gRNA). Similarly, B.1.351- and B.1.617.2-infected control animals demonstrated significantly higher levels of viral load than the RBD-trimer-vaccinated animals (Figure 7K–7M). These results strongly indicate that the RBD trimer mRNA vaccine can elicit broad and potent protection against live SARS-CoV-2 variants tested.
Figure 7.
RBD trimer mRNA vaccine protects K18-hACE2 mice from death after infection with live SARS-CoV-2 and its variants
(A–G) (A) Timeline for vaccination, challenge with live SARS-CoV-2 WT D614, B.1.351, or B.1.617.2 and monitoring for body weight changes and survival in the ensuing 5 days, and euthanasia up to 5 dpi. Mice were challenged with 103 PFU live SARS-CoV-2 WT D614, B.1.351, or B.1.617.2 via IN route. Body weight relative to day 0 (B–D) and survival percentage (E–G) were recorded daily until death up to 5 dpi.
(H–J) H&E staining of lung sections from immunized or control group. Scale bars, 50 μm. Each image is representative of each group.
(K–M) Viral gRNA and sgRNA copies of N gene in lung tissue samples were quantified by droplet digital PCR (TargetingOne, China). All the samples were collected at death or at 5 dpi.
Discussion
With the rapid emergence and spread of SARS-CoV-2 variants, the development of vaccines with broad and potent protectivity has become a global priority. However, the majority of vaccines that are currently approved and actively being rolled out across the world were based on the prototype SARS-CoV-2 sequences and structural properties of the spike trimer, therefore rendering them to common escape mutations in the spike sequence (Abdool Karim and de Oliveira, 2021; Corti et al., 2021; Dai and Gao, 2021; Harvey et al., 2021; Wrapp et al., 2020). Here, we show that an mRNA-LNP vaccine expressing a structurally designed RBD trimer can induce broad and potent serum neutralizing antibodies against the major SARS-CoV-2 variants, thus bypassing common escape mutations to spike-based vaccine modalities. The exact mechanisms underlying these observations have yet to be elucidated; however, we did identify strong RBD-specific B cell responses, including GCBCs, MBCs, and bone marrow LLPCs, that may all contribute to such a strong antibody response. At the cellular level, the enhanced ability of the RBD trimer to trigger the formation of BCR micro-clusters, and activate BCR downstream signalling, may promote the development of MBCs that are more capable of differentiating into plasma cells in the spleen and LLPCs in the bone marrow, as demonstrated in the present study. The successful generation of MBCs and LLPCs is critical for vaccine protection. This is because MBCs can initiate rapid recall responses while LLPCs can facilitate protection via the persistent production of antigen-specific antibodies (Sallusto et al., 2010; Weisel and Shlomchik, 2017). Furthermore, a substantial proportion of mAbs from animals immunized with our RBD trimer vaccine demonstrated broad and potent neutralizing activity against the major SARS-CoV-2 variants indicated. Cryo-EM and crystal structure analysis of one representative antibody revealed a novel epitope that not only excluded the escape mutations but was also highly conserved with the other SARS-CoV-2 variants. These broad and potent neutralizing mAbs provide a molecular basis for the broad and potent serum neutralizing activity of animals that had been immunized with the RBD trimer vaccine.
More importantly, RBD trimer mRNA vaccine induced protective immunity against high-dose challenge with live wild-type, beta, and delta variants in a mouse model of SARS-CoV-2 infection. Vaccinated animals demonstrated reduced lung pathology and substantial decreases in viral sgRNA and gRNA copies in the lungs, relative to the control animals.
Interestingly, we found a clear inverse correlation between T cell and antibody responses among the three mRNA vaccines tested. The highest antibody response induced by RBD trimer mRNA vaccine was correlated with the lowest CD8+ T cells and ELISPOT positive cells. In contrast, the lowest antibody response induced by native spike mRNA vaccine was mirrored by the highest levels of CD8+ T cells and ELISPOT positive cells. While we are uncertain about exact underlying mechanisms, we believe the non-modified nucleosides used in our mRNA vaccines might have played some roles in triggering a PRR-driven inflammatory response that led to the robust CD8+ T cell stimulation in our study. This is consistent with report by Rauch et al(Rauch, 2021) where non-modified mRNA developed by CureVac induced even higher levels of multifunctional IFN-γ/TNF-positive CD8+ T cells (approximately 4%–10%) in splenocytes of immunized BALB/c mice. By contrast, pseudouridine-modified mRNA is weaker in triggering inflammatory response, hence lower levels of antigen-specific CD8+ T cell response (approximately 1% or less) as demonstrated by Corbett et al(Corbett, 2020); Kariko et al(Karikó, 2005); Kariko et al(Karikó, 2008); Vogel et al(Vogel, 2021). However, this comparison is only a ballpark figure without taking into account of specific mouse species (B6C3F1/J vs. BALB/C), immunization dose (1 vs. 10 μg), structural modification of antigen (S-2P vs. native), and length of the antigen sequence (RBD vs. native spike). In our study, the spike sequence was the longest in length (1,260 residues), followed by mid-length native RBD (223 residues), and shortest trimer RBD (199 residues). In this regard, the length of the antigen or the number of non-modified nucleosides contained in the antigen sequence must quantitatively affect the levels of PRR-driven inflammatory response, leading to various levels of CD8+ T cell response. Following this logic, it is possible that the weakest inflammatory response induced by the shortest RBD trimer vaccine allowed the expression and induction of antibody response to the highest levels among the three mRNA vaccines tested. Alternatively, apart from the levels of expression, the structural and conformational features of antigen were also expected to make significant difference in activating B cell receptor and downstream signaling for development of memory B cells critical for primary and recall responses. Indeed, our results provided strong evidence at molecular and cellular levels that the RBD timer mRNA vaccine is superior to native RBD and native spike vaccines in generating memory and long-lived bone marrow plasma B cells as well as in stimulating BCR and downstream signaling. Collectively, our results indicate that the RBD trimer mRNA vaccine can modulate antibody response toward more conserved regions on the RBD, thereby overcoming the escape mutations that attenuate the efficacy of the current panel of vaccines in mouse models.
Of note, we recognized that the results obtained in mice may not necessarily be predictive of what in higher mammals such as NHP and humans. The BALB/c and K18-hACE2 mice may be inherently insensitive to SARS-CoV-2 variants, perhaps due to their distinctive genetic makeup and immune system from that of NHP and human. Unfortunately, the NHP and human studies are beyond the scope of this study. Nevertheless, our results suggest that structurally distinction of our RBD trimer must at least play some role in generating broad and protective immune responses in mice. It would certainly be exciting to see how the trimeric RBD will be performing in NHP and humans in the near future.
Limitation of the study
Our study was performed exclusively in mice which may not necessarily represent those in NHP and humans. The BALB/c and K18-hACE2 mice, used in immunization and protection experiments, may be inherently insensitive to SARS-CoV-2 variants and the levels of immunity and protection may rely on additional immunological parameters on top of those studied here. Future studies in NPH and humans would be highly desirable to verify and validate these results. In addition, it would be ideal to use regulatory agency approved mRNA vaccines such as mRNA-1273 and BNT162b2 as controls when studying our RBD trimer candidate. However, they were not available in China and therefore proper comparation with these spike-based mRNA could not be conducted. Moreover, given the focus of our study and limited experimental resources, we only tested the protectivity of RBD trimer vaccine against authentic WT D614, B.1.351, and B.1.617.2 variants in K18-hACE2 mouse model.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| P2C-1F11 | Ju et al., 2020 | RRID: AB_2313773 |
| P2B-1G5 | Ju et al., 2020 | RRID: AB_2313773 |
| P2B-2F6 | Ju et al., 2020 | RRID: AB_2313773 |
| S309 | Pinto et al., 2020 | RRID: AB_2313773 |
| PerCP Cy5.5 anti-IgD | BD Biosciences | Cat#564273; RRID: AB_2313773 |
| PE-Cy7 anti-FAS | BD Biosciences | Cat#557653; RRID: AB_2313773 |
| FITC anti-IgG3 | BD Biosciences | Cat#553403; RRID: AB_2313773 |
| EF450 anti-GL7 | eBioscience | Cat#48-5902-82; RRID: AB_2313773 |
| Allophycocyanin (APC)-Cy7 anti-B220 | Biolegend | Cat#103224; RRID: AB_2313773 |
| BV510 anti-CD138 | Biolegend | Cat#142521; RRID: AB_2313773 |
| FITC anti-IgG1 | Biolegend | Cat#406605; RRID: AB_2313773 |
| FITC anti-IgG2b | Biolegend | Cat#406705; RRID: AB_2313773 |
| BV421 anti-IgM | Biolegend | Cat#406532; RRID: AB_2313773 |
| streptavidin-PE | Biolegend | Cat#405203; RRID: AB_2313773 |
| FITC anti-IgG2a | Biolegend | Cat#407105; RRID: AB_2313773 |
| PerCP Cy5.5 anti-Gr1 | Biolegend | Cat#108428; RRID: AB_2313773 |
| CD4-FITC | Biolegend | Cat#100406; RRID: AB_2313773 |
| CD8-PE/Cyanine7 | BD Biosciences | Cat#522877; RRID: AB_2313773 |
| CD19-APC/Cyanine7 | BD Biosciences | Cat#557655; RRID: AB_2313773 |
| INFγ-BV421 | BD Biosciences | Cat#563376; RRID: AB_2313773 |
| TNFα-APC | BioLegend | Cat#506308; RRID: AB_2313773 |
| IL2-PE | BioLegend | Cat#503808; RRID: AB_2313773 |
| Alexa Fluor 647 anti-mouse Fc | Jackson ImmunoResearch Laboratory | Cat#109-607-008; RRID: AB_2313773 |
| Goat non-specific IgG | Jackson ImmunoResearch Laboratory | Cat#005-000-003; RRID: AB_2313773 |
| Alexa Fluor 488 anti-mouse pSyk | Jackson ImmunoResearch Laboratory | Cat#560081; RRID: AB_2313773 |
| Anti-Mouse IgG (H+L), HRP Conjugate | Promega | Cat#W4021; RRID: AB_2313773 |
| rabbit anti-SARS-CoV-N IgG | Sino Biological, Inc. | Cat#40143-R019; RRID: AB_2313773 |
| HRP-conjugated goat anti-rabbit IgG antibody | Jackson ImmunoResearch | Cat# 111-035-003; RRID: AB_2313773 |
| Bacterial and virus strains | ||
| SARS-CoV-2/WH-09/human/2020/CHN | GenBank | |
| SARS-CoV-2/ B.1.1.7 | GISAID | EPI_ISL_601443 |
| SARS-CoV-2/ B.1.351 | GISAID | EPI_ISL_700450 |
| SARS-CoV-2/ P.1 | GISAID | EPI_ISL_792681 |
| SARS-CoV-2/ B.1.617.1 | GISAID | EPI_ISL_1384866 |
| SARS-CoV-2/ B.1.617.2 | GISAID | EPI_ISL_1534938 |
| SARS-CoV-2/ B.1.429 | GISAID | EPI_ISL_2922315 |
| SARS-CoV-2/ B.1.525 | GISAID | EPI_ISL_2885901 |
| SARS-CoV-2/ B.1.526 | GISAID | EPI_ISL_2922249 |
| SARS-CoV-2/ B.1.526 | GISAID | EPI_ISL_2922249 |
| SARS-CoV-2/A23.1 | GISAID | EPI_ISL_2690464 |
| Per site variation v20210517 | GISAID | https://cov.lanl.gov/content/index |
| Chemicals, peptides, and recombinant proteins | ||
| Polyethyleneimine | Polysciences | Cat#24765-1 |
| Trypsin | Macgene | Cat#CC017 |
| Fetal bovine serum | GIBCO | Cat#16000-044 |
| Papain | Sigma-Aldrich | Cat#9001-73-4 |
| SARSr-CoV-2 Spike glycoprotein peptide pool | Genscript | Cat#RP30027 |
| ACK Lysing buffer | Lonza | Cat#10-548E |
| Brefeldin A | Biolegend | Cat#420601 |
| Cellfectin II Reagents | Thermo Fisher | Cat#10362100 |
| Recombinant RNase Inhibitor | TaKaRa | Cat#2313B |
| Critical commercial assays | ||
| QuikChange Lightning Site-Directed Mutagenesis Kit |
Agilent | Cat#210519 |
| Bright-GloTM Luciferase Assay Buffer | Promega | Cat#E264B |
| Bright-GloTM Luciferase Assay Substrate | Promega | Cat#E263B |
| Zombie Yellow™ Fixable Viability Kit | Biolegend | Cat#423103 |
| Mouse IFN-g ELISpot Kit | MabTech | Cat#3321-4AST |
| RNeasy Plus Mini Kit | QIAGEN | Cat#74134 |
| COVID-19 digital PCR detection kit | TargetingOne | Cat#13441 |
| KAPA HiFi HotStart ReadyMix | KAPA Biosystems | Cat#Kk2631 |
| VAHTS DNA Clean Beads | Vazyme | Cat#N411-01 |
| Soluble TMB Kit | CoWin Biosciences | Cat#CW0050 |
| KPL TrueBlue peroxidase substrate | Seracare Life Sciences Inc. | Cat# 5510-0030 |
| BD Cytofix/Cytoper | BD Biosciences | Cat#554714 |
| Strep-Tactin® resin | IBA-lifesciences | Cat#2-1201-002 |
| Ni-NTA Sepharose | GE Healthcare | Cat#17-5318-01 |
| Protein A Sepharose | GE Healthcare | Cat#17127902 |
| Protein G Sepharose | Genscript | Cat# Z02007 |
| Anti-Penta-HIS (HIS1K) Biosensors | Sartorius | Cat#18-5120 |
| Deposited data | ||
| Structure of B.1.351 spike trimer and two T6 Fab | This paper | PDB: 7FJN |
| Structure of B.1.351 spike trimer and three T6 Fab | This paper | PDB: 7FJO |
| Structure of B.1.351 RBD and T6 Fab | This paper | PDB: 7FJS |
| Experimental models: Cell lines | ||
| Human: HEK293T | ATCC | CRL-3216 |
| Human: FreeStyle 293F | Thermo Fisher | R79007 |
| HeLa expressing human ACE2 | Dr. Qiang Ding’s lab | Liu et al., 2021 |
| J558L cells | ATCC | TIB-6 |
| Experimental models: Organisms/strains | ||
| Mouse: BALB/c | Charles river | N/A |
| Mouse: B6/JGpt-H11em1Cin(K18-hACE2)/Gpt | Gempharmatech Co., Ltd | N/A |
| Oligonucleotides | ||
| IS primer: 5’-AAGCAGTGGTATCAACGCAGAGT-3’ |
This paper | N/A |
| IG outer primer: 5’-ATGTCGTTCATACTCGTCCTTGGT-3’ |
This paper | N/A |
| IG outer primer: 5’-GCAGGAGACAGACTCTTC TCCA-3’ |
This paper | N/A |
| IG outer primer: 5’-TCAGCACGGGACAAACTCTTCT-3’ |
This paper | N/A |
| IG outer primer: 5’-GCAGGAGACAGACTCTTCTCCA-3’ |
This paper | N/A |
| IG outer primer: 5’-AACTGGCTGCTCATGGTGT-3’ |
This paper | N/A |
| IG outer primer: 5’-TGGTGCAAGTGTGGTTGAGGT-3’ |
This paper | N/A |
| IG outer primer: 5’-TGGTCACTTGGCTGGTGGTG-3’ |
This paper | N/A |
| IG outer primer: 5’-CACTTGGCAGGTGAACTGTTTTCT-3’ |
This paper | N/A |
| IG outer primer: 5’-AACCTTCAAGGATGCTCTTGGGA-3’ |
This paper | N/A |
| IG outer primer: 5’-GGACAGGGATCCAGAGTTCCA-3’ |
This paper | N/A |
| IG outer primer: 5’-AGGTGACGGTCTGACTTGGC-3’ |
This paper | N/A |
| IG outer primer: 5’-GCTGGACAGGGCTCCATAGTT-3’ |
This paper | N/A |
| IG outer primer: 5’-GGCACCTTGTCCAATCATGTTCC-3’ |
This paper | N/A |
| IG inner primer: 5’-GAAGCACACGACTGAGGCAC-3’ |
This paper | N/A |
| IG inner primer: 5’-TACACACCAGTGTGGCCTT-3’ |
This paper | N/A |
| IG inner primer: 5’-CAGGCCACTGTCACACCACT-3’ |
This paper | N/A |
| IG inner primer: 5’-CAGGTCACATTCATCGTGCCG-3’ |
This paper | N/A |
| IG inner primer: 5’-GAGGCCAGCACAGTGACCT-3’ |
This paper | N/A |
| IG inner primer: 5’-GCAGGGAAGTTCACAGTGCT-3’ |
This paper | N/A |
| IG inner primer: 5’-CTGTTTGAGATCAGTTTGCCATCCT-3’ |
This paper | N/A |
| IG inner primer: 5’-TGCGAGGTGGCTAGGTACTTG-3’ |
This paper | N/A |
| IG inner primer: 5’-CCCTTGACCAGGCATCC-3’ |
This paper | N/A |
| IG inner primer: 5’-AGGTCACGGAGGAACCAGTTG-3’ |
This paper | N/A |
| IG inner primer: 5’-GGCATCCCAGTGTCACCGA-3’ |
This paper | N/A |
| IG inner primer: 5’-AGAAGATCCACTTCACCTTGAAC-3’ |
This paper | N/A |
| Recombinant DNA | ||
| pcDNA3.1 | Thermo Fisher | Cat#V79020 |
| pFastBac-Dual vector | GIBCO | Cat#10712024 |
| pNL4-3- luc -R-E | NIH-AIDS Reagent Program | Cat#3418 |
| Software and algorithms | ||
| Graphpad Prism 8 | GraphPad | www.graphpad.com |
| FlowJo 10 software | FlowJo | https://www.flowjo.com/ |
| PHASER (CCP4 Program Suite) | McCoy et al., 2007 | http://www.phaser.cimr.cam.ac.uk/index.php/Phaser_Crystallographic_Software |
| COOT | Emsley et al., 2004 | http://www2.mrc-lmb.cam.ac.uk/Personal/pemsley/coot/ |
| PHENIX | Adams et al., 2010 | http://www.phenix-online.org/ |
| PyMOL | Janson et al., 2017 | https://pymol.org/2/ |
| Image J | NIH | https://imagej.nih.gov |
| UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera/ |
| UCSF ChimeraX-1.1.1 | Pettersen et al. 2021 | https://www.cgl.ucsf.edu/chimerax/ |
| Motion Correction | Zheng et al. 2017 | http://msg.ucsf.edu/em/software/index.html |
| GCTF v.1.18 | Zhang, 2016 | https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/ |
| Gautomatch | The MRC Laboratory of Molecular Biology | https://www2.mrc-lmb.cam.ac.uk/download/gautomatch-056/ |
| RELION-3.1 | Zivanov et al. 2020 | https://www2.mrc-lmb.cam.ac.uk/relion/index.php?title=Main_Page |
Resource availability
Lead contact
Further information and requests for resources and primary data should be directed to and will be fulfilled by the Lead Contact, Linqi Zhang (Zhanglinqi@mail.tsinghua.edu.cn).
Materials availability
All unique/stable reagents generated in this study are available from the Lead Contact with a completed Material Transfer Agreement.
Experimental model and subject details
Mice
BALB/c mice aged 6 weeks were purchased from Charles River Laboratories. B6/JGpt-H11em1Cin(K18-hACE2)/Gpt mice aged 6 weeks were purchased from Gempharmatech Co., Ltd. All animal experiments were carried out in strict compliance with the Guide for the Care and Use of Laboratory Animals of the People’s Republic of China and approved by the Committee on the Ethics of Animal Experiments of Tsinghua University. Mouse immunization and characterization were conducted in the animal facility of Tsinghua University.
Cell lines
HEK293T cells (ATCC, CRL-3216), HeLa cells expressing ACE2 orthologs (kindly provided by Dr. Qiang Ding) (Liu et al., 2021)were maintained at 37°C in 5% CO2 in Dulbecco’s minimal essential medium (DMEM) containing 10% (v/v) heat-inactivated fetal bovine serum (FBS) and 100 U/mL of penicillin–streptomycin. FreeStyle 293F cells (Thermo Fisher Scientific, R79007) were maintained at 37°C in 5% CO2 in SMM 293-TII (Sinobiological).
Method details
Design and synthesis of lipid-nanoparticle encapsulation of the mRNA vaccines
The expression vectors encoding the native RBD (Arg319–Phe541), the native full-length SARS-CoV-2 spike (Wuhan-Hu-1, GenBank: MN908947.3), and the RBD trimer (Asn331–Lys529) fused with two G3S linkers and a foldon sequence (GYIPEAPRDGQAYVRKDGEWVLLSTFLG) at the C-terminus, were constructed and verified by sequencing. The mRNA was transcribed from the linearized DNA templates using T7 RNA polymerase, and onto which the 5′ and -3′ untranslated regions and a poly-A tail were added. Lipid-nanoparticle (LNP) formulation of mRNA was performed as previously reported (Zhang et al., 2020). Briefly, lipids were dissolved in ethanol containing an ionizable lipid, 1, 2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and PEG-lipid with molar ratios of 50:10:38.5:1.5. The lipid mixture was combined with 20 mM citrate buffer (pH4.0) containing mRNA at a ratio of 1:2 through a T-mixer. Formulations were then diafiltrated against 10× volume of PBS (pH7.4) through a tangential-flow filtratio (TFF) membrane with 100 kD molecular weight cut-offs (Sartorius Stedim Biotech), concentrated to desired concentrations, passed through a 0.22 mm filter, and stored at 2–8°C until use. All formulations were tested for particle size, distribution, RNA concentration and encapsulation.
Quality control of mRNA-LNP by electron microscopy and dynamic light scattering
mRNA-LNP (3 μL) was deposited on a holey carbon grid that was glow-discharged (Quantifoil R1.2/1.3) and vitrificated using a Vitrobot Mark IV (Thermo Fisher Scientific) instrument. Cryo-EM imaging was conducted on a Talos F200C Equipped with a Ceta 4k x 4k camera, operated at 200 kV accelerating voltage. The size of mRNA-LNP was measured using dynamic light scattering (DLS) on a Malvern Zetasizer Nano-ZS (Malvern). Samples were irradiated with red laser (l = 632.8 nm) and scattered light were detected at a backscattering angle of 173°. Results were analyzed using the software (Zetasizer V7.13). Encapsulation of mRNA in the LNP was measured using a Quant-iTTM RiboGreenTM RNA Assay Kit according to the manufacturer’s instructions. Samples were excited at 480 nm and fluorescence intensity was measured at 520 nm, using a SpectraMax iD3 (Molecular Devices).
Mouse immunization and sample collection
A total of 160 female BALB/c mice aged 6–8 weeks were randomly distributed into 16 groups (n = 10 per group). Every 4 groups were intramuscularly vaccinated with two doses of 10 ug mRNA with 28 days interval encoding either the native RBD, the RBD trimer, the native spike, or the empty LNP. The empty LNP was used as the background control. Prior to immunization, serum samples were collected from all 16 groups of animals. On day 10, 21, 35, and 105 after initial immunization, a group of 10 animals from each mRNA vaccinated animals were sacrificed to collect serum, spleens, inguinal lymph nodes, and bone marrow samples. The serum was heat-inactivated at 56°C for 30 min, and stored at −80°C before analysis for SARS-CoV-2 specific antibodies. Spleens, inguinal lymph nodes, and bone marrow were mashed in MACS buffer (PBS supplemented with 1% FBS and 5 mM EDTA) on ice and filtered through a 70 μm cell strainer. The contaminated red blood cells (RBCs) were removed by ACK Lysing buffer (Lonza) and the remaining lymphocytes were washed and resuspended in cold MACS buffer for immediate analysis of various B and T cell populations using flow cytometry.
Serum and antibody binding measured by ELISA
Serum samples were serially diluted and added to 96-well plates pre-coated with purified recombinant SARS-CoV-2 spike trimer produced in HEK 293F cells (100 ng/well). After incubating at 37°C for 1 h, the plates were washed three times with phosphate-buffered saline containing 0.5% Tween 20 (PBST). The secondary horseradish peroxidase (HRP)-conjugated antibody against mouse IgG (1:4000, Promega, USA) was then added and incubated further at 37°C for 1 h. The samples were washed again 3 times with PBST before the substrate TMB (3′,3′,5′,5′, -tetramethyl benzidine) was added. The reaction was stopped by adding 1M H2SO4 and absorbance at 450 nm was measured using an ELISA plate reader. The ED50 value was calculated based on binding curves drawn in Prism 8.0 software (GraphPad Inc., USA). When measuring binding of monoclonal antibodies, we used the 96-well plates pre-coated with purified recombinant RBD trimer produced in HEK 293F cells (100 ng/well) and anti-mouse immunoglobulin G (IgG)-HRP (Promega) as the secondary antibody while the rest of experimental procedures remained the same as described above.
Construction of expression vectors encoding the wild-type and variant spike of SARS-CoV-2
The wildtype and variant spike genes were synthesized, cloned into the pcDNA3.1 expression vector (Invitrogen, USA), and verified by sequencing (Genwiz Inc.), as we previously reported (Wang et al., 2021b). The wildtype used throughout the analysis was derived from the prototype Wuhan-Hu-1 strain (GenBank: MN908947.3) (WT D614). The variant D614G was constructed on the backbone of WT D614 by site directed mutagenesis (Agilent 210519). The variant B.1.1.7 (GISAID: EPI_ISL_601443) was constructed with total of 9 mutations including 69–70del, 144del, N501Y, A570D, D614G, P681H, T716I, S982A and D1118H. The variant B.1.351 (GISAID: EPI_ISL_700450) was constructed with 10 mutations including L18F, D80A, D215G, 242–244del, S305T, K417N, E484K, N501Y, D614G and A701V. The variant P.1 (GISAID: EPI_ISL_792681) was constructed with 12 mutations including L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I and V1176F. The variant B.1.617.1 (GISAID: EPI_ISL_1384866) was constructed with 8 mutations including T95I, G142D, E154L, L452R, E484Q, D614G, P681R, N1071H. The variant B.1.617.2 (GISAID: EPI_ISL_1534938) was constructed with 9 mutations including T19R, 156–157del, R158G, L452R, T478K, D614G, P681R, D950N. The variant B.1.429 (GISAID: EPI_ISL_2922315) was constructed with 4 mutations including S13I, W152C, L452R, D614G. The variant B.1.525 (GISAID: EPI_ISL_2885901) was constructed with 8 mutations including Q52R, A67V, 69–70del, Y144 del, E484K, D614G, Q677H, F888L. B.1.526 (GISAID: EPI_ISL_2922249) was constructed with 6 mutations including L5F, T95I, D253G, E484K, D614G, A701Y. A23.1 (GISAID: EPI_ISL_2690464) was constructed with 4 mutations including F157L, V367F, Q613H, P681R. The single mutations identified from the GISAID database were introduced into the pcDNA3.1 vector encoding WT D614G using QuickChange site-directed mutagenesis (Agilent 210519) (Wang et al., 2021b). Schematic of variant spike of SARS-CoV-2 used in this study are shown in Figure S1.
Serum and antibody neutralization measured by pseudovirus and live SARS-CoV-2
Neutralizing activity of the immune sera and mAbs were determined using SARS-CoV-2 pseudovirus and live virus neutralization assays as previously reported (Ju et al., 2020; Wang et al., 2021b). The pseudovirus was generated by co-transfection of HEK293T cells with the HIV backbone expressing firefly luciferase (pNL43R-E-luciferase) and pcDNA3.1 (Invitrogen, USA) encoding the wildtype Wuhan-Hu-1 (GenBank: MN908947.3) and variants. After 48 h, the cell supernatant containing the pseudovirus was collected, measured for infectivity, and stored at −80C until further use. For measuring neutralizing activity, serum or mAbs were serially diluted 3-fold in the 96-well cell culture plates before SARS-CoV-2 pseudovirus was added. After incubation at 37°C for 1 h, approximately 1.5 × 104 HeLa-ACE2 cells were added into the mixture and incubated further at 37°C for 60 h before cell lysis for measuring luciferase-activity. The ID50 values were calculated based on the relative light units (Bright-Glo Luciferase Assay Vector System, Promega, USA) using Prism 8.0 (GraphPad Software Inc., USA).
For measuring neutralizing activity against live SARS-CoV-2, we performed plaque reduction neutralization test (PRNT) in a certified Biosafety level 3 laboratory against wildtype D614 and B.1.351 variant as we previously reported (Ju et al., 2020). In brief, serial dilutions of serum samples were mixed with live virus and incubated for 1 h at 37°C. The mixtures were then transferred to 96-well plates seeded with Vero E6 cells and incubated for 1 h at 37°C. After changing the medium, the plates were incubated at 37°C for an additional 24 h. The cells were then fixed, permeabilized, and incubated with cross-reactive rabbit anti-SARS-CoV-N IgG (Sino Biological, Inc., China) for 1 h at room temperature before adding an HRP-conjugated goat anti-rabbit IgG antibody (Jackson ImmunoResearch, USA). The reactions were developed using KPL TrueBlue peroxidase substrate (Seracare Life Sciences Inc., USA). The number of SARS-CoV-2 foci was quantified using an EliSpot reader (Cellular Technology Ltd. USA).
Flow cytometry analysis of B cell populations in vaccinated animals
To phenotype various B cell populations by flow cytometry, single cell suspension was stained with indicated antibodies in MACS buffer (PBS supplemented with 1% FBS and 5 mM EDTA). Recombinant RBD used to identify RBD-specific B cells contained the C-terminal Strep-tag. Dead cells and non-singlet events were typically excluded from analysis based on staining of Zombie Yellow (Biolegend) and characteristics of forward and side scatters. All cytometric data were collected on an Aurora (Cytek Biosciences) and analyzed with the FlowJo software (TreeStar).
Germinal center B cells /Memory B cells: Cell suspension was incubated with 500 nM recombinant RBD on ice for 30 min, and washed extensively before subjected to staining with a cocktail of fluorescent antibodies for additional 30 min. The antibodies included PerCP Cy5.5 anti-IgD, PE-Cy7 anti-FAS, FITC anti-IgG3 from BD Biosciences; EF450 anti-GL7 from eBioscience; allophycocyanin (APC)-Cy7 anti-B220, BV510 anti-CD138, FITC anti-IgG1, FITC anti-IgG2b, BV421 anti-IgM, streptavidin-PE, FITC anti-IgG2a from Biolegend. Cell suspension was washed extensively with MACS buffer before analyzed by flow cytometry.
Plasma cells: Cell suspension was permeabilized and incubated with 500 nM RBD on ice for 30 min and washed extensively before stained by the following antibody cocktail for additional 30 min. The antibody cocktail contained allophycocyanin (APC)-Cy7 anti-B220, BV510 anti-CD138, FITC anti-IgG1, FITC anti-IgG2b, BV421 anti-IgM, PerCP Cy5.5 anti-Gr1 (only for bone marrow plasma cell), streptavidin-PE, FITC anti-IgG2a from Biolegend; FITC anti-IgG3 from BD Biosciences. Cell suspension was washed extensively with MACS buffer prior to data acquisition by flow cytometry.
The gating strategies of flow cytometry are shown in Figures S5A–S5C.
T cell responses in vaccinated animals analyzed by ELISPOT and intracellular cytokine staining
T cell responses in the vaccinated mice were assessed using the IFN-γ pre-coated ELISPOT kit according to manufacturer’s protocol (MabTech, Sweden). Splenocytes from immunized mice were collected and stimulated with a peptide pool covering the SARS-CoV-2 spike protein (GenScript, USA) at a concentration of 2 μg/mL of each peptide. Phorbol myristate acetate/ionomycin was used as a positive control while RPMI 1640 medium as a negative control. After incubation at 37°C for 28 h, the plates were washed extensively before a biotinylated anti-mouse IFN-γ antibody was added to each well and incubated further for 2 h at room temperature. The substrate AEC was added and the spots in each well were read using the automated ELISPOT reader (AID, USA). The number of spot-forming units (SFUs) per 1,000,000 cells was calculated and presented.
For intracellular cytokine staining, approximately 1,000,000 mouse splenocytes were stimulated with the same SARS-CoV-2 S peptide pool as above (2 μg/mL of each peptide) and brefeldin A (GolgiPlug; BD, USA) for 6 h at 37°C in 5% CO2. Following two washes with PBS, the splenocytes were permeabilized and stained with the fluorescently conjugated antibodies including CD4-FITC (BioLegend), CD8-PE/Cyanine7 (BD Biosciences), CD19-APC/Cyanine7 (BD Biosciences), INFγ-BV421 (BD Biosciences), TNFα-APC (BioLegend), and IL2-PE (BioLegend). Dead cells were stained using the Zombie Yellow Fixable Viability Kit (BioLegend). The cytometric data were collected using Aurora (Cytek Biosciences) and analyzed with the FlowJo software (TreeStar). The gating strategies of flow cytometry are shown in Figure S5D.
Triggering 2F6-BCR signaling by recombinant RBD, RBD trimer and spike trimer
J558L B cell line expressing SARS-CoV-2 antibody 2F6-IgG-BCRs on the cell membrane (J558L-2F6-IgG-BCRs B cells) were generated accordingly to our previously published report (Liu et al., 2010). Specifically, the VH and VL of P2B-2F6 were respectively subcloned to mouse IgG1-backbone and Igκ –backbone on the pHAGE vector and transfected into J558L cell line through electroporation using Amaxa Nucleofector (Lonza). After recovered and maintained in RPMI-1640 medium for about 24 h. J558L-2F6-IgG-BCRs B cells were stained with the Fab anti-mouse Fc fragment conjugated with Alexa Fluor 647 (Jackson ImmunoResearch Laboratory) and loaded onto the prepared glass coverslips coated with native RBD, RBD trimer and spike trimer. The J558L-2F6-IgG-BCRs B cells were allowed to react with antigens for 10 min at 37°C, fixed by 4% paraformaldehyde (PFA), and imaged by an Olympus FLUOVIEW FV1000 confocal laser scanning microscope equipped with 4 lasers (405, 473, 557, and 635 nm) for fluorescence excitation. For intracellular immunofluorescence staining of the pSyk, the J558L-2F6-IgG-BCRs B cells were fixation with 4% PFA for 30 min and permeabilized with 0.2% Triton X-100 for 20 min at room temperature. The fixed and permeabilized cells were then blocked with 100 μg/mL goat non-specific IgG (Jackson Immuno Research Laboratory) for 1 h at room temperature before stained with the anti-mouse pSyk Fab conjugated with Alexa Fluor 488 (Jackson ImmunoResearch Laboratory). After extensive wash, the cells were ready for imaging as stated above. The contact area with antigen containing surface and mean fluorescence intensity (MFI) of accumulated BCRs recruited to the immunological synapse were analyzed by Image J (NIH, USA).
Recombinant protein expression and purification
Recombinant wildtype RBD, RBD trimer, human receptor ACE2 peptidase domain, and spike trimer of WT D614 and variant B.1.351 were produced by transient transfection of expression vectors into 293F cells with polyethyleneimine (PEI) at a density of 2.0–2.5 × 106/mL. Specifically, the recombinant proteins contained the C-terminal Strep-tag®II (8 amino acids, WSHPQFEK). Approximately 600 μg of expression plasmid in 25 mL of Opti-MEM transfection medium (Life Technologies, CA) was mixed with 2.4 mL of PEI (1.0 mg/mL) in 25 mL of Opti-MEM. After incubation for 15 min, the DNA-PEI complex was added to 600 mL 293F cells. Culture supernatants were collected 4 days after transfection, centrifuged at 4,000 rpm for 15 min, and filtered using 0.45-mm filters. The recombinant proteins were captured from the supernatants using a Strep-Tactin® resin (IBA-lifesciences). The bound proteins were eluted with 10 mL elution buffer containing 100 mM Tris/HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 2.5 mM desthiobiotin. The eluted proteins were purified by SEC on a Superdex 200 Increase 10/300 GL column (GE Healthcare).
Recombinant wildtype RBD, RBD with 417N-484K-501Y mutations (RBD-3M) used for BLI assay and crystallization were expressed using the Bac-to-Bac Baculovirus System (Invitrogen) as previously described (Lan et al., 2020). Specifically, SARS-CoV-2 RBD (residues Arg319 to Lys529) containing the gp67 secretion signal peptide and a C-terminal 6×his tag was inserted into pFastBac-Dual vectors (Invitrogen) and transformed into DH10 Bac component cells. The recombinant bacmid was extracted and further transfected into Hi5 cells using Cellfectin II Reagents (Invitrogen). The recombinant viruses were harvested from the transfected supernatant and amplified to generate high-titer virus stock. Viruses were then used to infect Hi5 cells for protein expression. Secreted RBDs were harvested from the supernatant, captured by Ni-NTA Sepharose (GE Healthcare), and purified by SEC on a Superdex 200 Increase 10/300 GL column (GE Healthcare).
Isolation of RBD-specific single B cells by FACS
Spike-specific single B cells were sorted as previously described (Ju et al., 2020). In brief, single splenocyte suspension from mice vaccinated with RBD trimer mRNA-LNP were collected and incubated with an antibody and SARS-CoV-2 spike trimer cocktail for identification of spike-specific B cells. The cocktail consisted of CD138-BV510, B220-APC-Cy7, GL7-EF450, IgG1-FITC, IgG2a-FITC, IgG2b-FITC, IgD- PercpCy5.5, Streptavidin-PE, and the recombinant spike trimer with Strep-tag®II as described above. Dead cells were stained using the Zombie Yellow Fixable Viability Kit (BioLegend, USA). The spike-specific single B cells were gated as B220+GL7-Spike+IgD-IgG+ and sorted into 96-well PCR plates containing 19ul 0.2% triton-100(sigma) and 1ul Recombinant RNase Inhibitor (TaKaRa). Plates were then snap-frozen on dry ice and stored at −80°C until RT reaction. All cytometric data were collected on an Aria III cytometer (BD Biosciences) and analyzed with the FlowJo software (TreeStar). The gating strategies of flow cytometry are shown in Figure S6.
Single B cell PCR, cloning, and expression of mAbs
Reverse transcription and cDNA amplification for single B cell was performed using standard smart-seq2 protocol (Picelli et al., 2014). Then the purified cDNA of each cell was used for the following BCR target enrichment by a 2-step nest PCR strategy. Target enrichment 1 reaction was performed by mixing ∼5ng cDNA with 25 μL KAPA HiFi HotStart ReadyMix (KAPA Biosystems), 0.1ul IS primer (Sangon), 4.8ul IG outer primer mix (a mix of 12 IGH/L/K constant region outer primers, 10 μM in total, 10/12 μM each; Sangon), and H2O to a total volume of 50ul, and amplified with the following thermo profile (98°C 45 s, 11 cycles of (98°C 20 s, 67°C 30 s, 72°C 1 min), 72°C 1 min). The PCR product was then purified with 0.8× VAHTS DNA Clean Beads (Vazyme). Target enrichment 2 reaction was then performed by mixing 20.1 μL last-step product with 25ul KAPA HiFi HotStart ReadyMix(KAPA Biosystems), 0.1 μL IS primer (10uM; Sangon), 4.8 μL IG inner primer mix (a mix of 12 IGH/L/K constant region inner primers, 10 μM in total, 10/12 μM each; Sangon), followed by the thermo profile (98°C 45 s, 11 cycles of (98°C 20 s, 67°C 30 s, 72°C 1 min), 72°C 1 min), and a double-sided size selection with 0.5× and 0.3× VAHTS DNA Clean Beads (Vazyme). The IgG heavy and light chain variable genes were amplified by nested PCR and cloned into expression vectors to produce full mouse IgG1 antibodies. The primer sequences used are listed as follow:
IS primer
5′-AAGCAGTGGTATCAACGCAGAGT-3’
IG outer primer mix
5′-TCAGCACGGGACAAACTCTTCT-3’
5′-GCAGGAGACAGACTCTTCTCCA-3’
5′-AACTGGCTGCTCATGGTGT-3’
5′-TGGTGCAAGTGTGGTTGAGGT-3’
5′-TGGTCACTTGGCTGGTGGTG-3’
5′-CACTTGGCAGGTGAACTGTTTTCT-3’
5′-AACCTTCAAGGATGCTCTTGGGA-3’
5′-GGACAGGGATCCAGAGTTCCA-3’
5′-AGGTGACGGTCTGACTTGGC-3’
5′-GCTGGACAGGGCTCCATAGTT-3’
5′-GGCACCTTGTCCAATCATGTTCC-3’
5′-ATGTCGTTCATACTCGTCCTTGGT-3’
IG inner primer mix
5′-TACACACCAGTGTGGCCTT-3’
5′-CAGGCCACTGTCACACCACT-3’
5′-CAGGTCACATTCATCGTGCCG-3’
5′-GAGGCCAGCACAGTGACCT-3’
5′-GCAGGGAAGTTCACAGTGCT-3’
5′-CTGTTTGAGATCAGTTTGCCATCCT-3’
5′-TGCGAGGTGGCTAGGTACTTG-3’
5′-CCCTTGACCAGGCATCC-3’
5′-AGGTCACGGAGGAACCAGTTG-3’
5′-GGCATCCCAGTGTCACCGA-3’
5′-AGAAGATCCACTTCACCTTGAAC-3’
5′-GAAGCACACGACTGAGGCAC-3′
Production of mAbs and fab
P2C-1F11, P2B-2F6, and P2B-1G5 are potent neutralizing mAbs initially isolated from SARS-CoV-2 infected patients by our group and previously published (Ju et al., 2020). S309, an antibody published by other groups, was synthesized according to the sequences released in Protein Data Bank (PDB) (Pinto et al., 2020). Antibody production was conducted by co-transfection of the heavy and light chain expression vectors into HEK 293F cells using polyethyleneimine (Sigma). After 96 h, antibodies secreted into the supernatant were captured by Protein A-Sepharose (GE Healthcare) and eluted by solution buffer (Glycine pH 3.0). After further purification by SEC on a Superdex 200 Increase 10/300 GL column (GE Healthcare), antibody concentration was determined by nanodrop 2000 Spectrophotometer (Thermo Scientific). All the mouse mAbs isolated from vaccinated mice with RBD trimer mRNA-LNP were capture by Protein G-Sepharose (Genscript) and purified as illustrated above.
For Fab production, the sequence encoding the mAb T6 VL and VH were separately cloned into the backbone of antibody expression vectors containing the constant regions of human IgG1. The chimeric antibody T6 was expressed in HEK 293F cells by transient transfection and purified by affinity chromatography using Protein A Sepharose (GE Healthcare) and SEC on Superdex 200 Increase 10/300 GL column (GE Healthcare). The purified chimeric mAb T6 was exchanged into phosphate-buffered saline (PBS), and digested with papain protease (Sigma) over night at 4°C. The digested antibody was then passed through Protein A Sepharose to remove the Fc fragment. The flow through containing the unbound Fab was further purified using a Superdex 200 Increase 10/300 GL column (GE Healthcare).
Competitive binding of isolated mAbs with ACE2 or class 1–4 mAbs measured by biolayer interferometry
(BLI) All BLI was carried out on an OctetRED96 (Sartorius) using Anti-Penta-HIS (HIS1K) Biosensors (Sartorius) to immobilize the recombinant native RBD with a 6×his tag. Assays were carried out in 96-well format in black plates (Greiner). For ACE2 or mAb competition studies, the experiment followed a five-step sequential assay: Baseline (PBS, 60 s); immobilization of the recombinant RBD (10 μg/mL, 120 s); Baseline2 (kinetic buffer, 60 s); mAb1 binding (100 nM mAb1 in the kinetic buffer, 250 s); ACE2 or mAb2 binding (100 nM ACE2 or mAb2 in the kinetic buffer, 250 s) in the presence of the mAb1 (100 nM in the kinetic buffer). Kinetic buffer is PBS supplemented with 0.1% BSA and 0.02% tween 20. The level of competition was estimated by the ratios between mAb1 signals in the presence over absence of ACE2 or of mAb2.
Cryo-EM sample preparation and data collection
The spike trimer of variant B.1.351 and T6 Fab, and the wildtype RBD trimer and the human receptor ACE2 peptidase domain, were all mixed at a molar ratio of 1: 3.6, incubated for 30 min at 4°C and purified by SEC on Superdex 200 Increase 10/300 GL column (GE Healthcare). The purified complex concentrated to approximately 2.2 mg/mL in TBS buffer (20 mM Tris pH 8.0 and 150 mM NaCl) was applied to glow-discharged holey carbon grids (Quantifoil grid, Au 300 mesh, R1.2/1.3). The grids were then blotted 2s and plunge-frozen into liquid ethane using Vitrobot Mark IV (Thermo Fisher Scientific). Images for complex were recorded using FEI Titan Krios microscope (Thermo Fisher Scientific) operating at 300 kV with a Gatan K3 Summit direct electron detector (Gatan Inc.) at Tsinghua University. The automated software (AutoEMation2) (Lei and Frank, 2005) was used to collect 4,534 movies for complex in super-resolution mode at a nominal magnification of 64,000× and at a defocus range between −1.5 and −1.8 μm. Each movie has a total accumulate exposure of 50 e−/Å2 fractionated in 32 frames of 175 ms exposure. The final image was binned 2-fold to a pixel size of 1.0979 Å.
Cryo-EM data processing
Motion Correction (MotionCor2 v.1.2.6) (Zheng et al., 2017), CTF-estimation (GCTF v.1.18) (Zhang, 2016) and non-templated particle picking (Gautomatch v.0.56) were automatically executed by TsingTitan.py program. Sequential data processing was carried out on RELION-3.1 (Zivanov et al., 2020). Initially, ∼1,491,000 particles were subjected to 2D classification. After three additional 2D classification, the best selected 1,156,670 particles were applied for initial model and 3D classification. A subset of 286,245 particle images from state 1 (2 RBD up and 1 RBD down) and 258,463 particle images from state 2 (3 RBD up) were further subject to 3D auto-refine and post-processing. The final resolution for state 1 and state 2 are 3.25 Å and 3.34 Å, respectively. The interface between spike protein and T6 Fab was subject to focused refinement with mask on the region of the RBD–Fab complex to improve the map quality. The 306,208 good particles after 3D classified focused on RBD–Fab complex were selected to focused refinement and post-processing with final resolution 4.39 Å. The resolution was estimated with the gold-standard Fourier shell correlation 0.143 criterion.
Model building and refinement
The initial model of complex of spike trimer and T6 Fab were generated using the models (PDB 7A97 and 7A98) and fit into the map using UCSF Chimera v.1.15 (Pettersen et al., 2004). Manual model rebuilding was carried out using Coot v.0.9.2 (Emsley and Cowtan, 2004) and refined with PHENIX v.1.18.2 (Adams et al., 2010) real-space refinement. The quality of the final model was analyzed with PHENIX v.1.18.2 (Adams et al., 2010). All structural figures were generated using PyMOL 2.0 (Janson et al., 2017) and ChimeraX-1.1.1 (Pettersen et al., 2021).
Crystallization and data collection
T6 Fab was mixed with RBD carrying K417N-E484K-N501Y mutations (RBD-3M) at a molar ratio of 1:1.2, incubated for 1 h at 4°C, and purified by gel-filtration chromatography. The purified complex was concentrated to 10 mg/mL in HBS buffer (10 mM HEPES, pH 7.2, 150 mM NaCl) for crystallization. Screening trials were performed at 18°C. The sitting drop vapor diffusion method was used by mixing 0.15 μL of protein with 0.15 μL of reservoir solution. Crystals of RBD-3M-Fab complex were successfully obtained in 2M ammonium sulfate and 5% v/v 2-propanol. Diffraction data were collected at the BL18U beamline of the Shanghai Synchrotron Research Facility (SSRF) and processed with HKL2000.
Crystal structural determination and refinement
Crystal structure was determined by the molecular replacement method using PHASER (CCP4 Program Suite) (McCoy et al., 2007). The T6 Fab cryo-EM structures were used as search models. Subsequent model building and refinement were performed using Coot v.0.9.2 40and PHENIX v.1.18.2 (Adams et al., 2010), respectively. All structural figures were generated using PyMOL 2.0 and ChimeraX-1.1.1 (Pettersen et al., 2021).
Immunization and challenge of K18-hACE2 transgenic mice with live SARS-CoV-2 and variants
Eight-week-old-male K18-hACE2 transgenic mice were obtained from the GemPharmatech Co., Ltd. Three groups of mice (n = 10 per group) were immunized with either 10 μg RBD trimer mRNA vaccine (n = 5) or empty LNP as control (n = 5) on day 0 and 28 via the IM route in a volume of 100 μL. Two weeks after second immunization (day 42), animals were transferred to the BSL-3 animal facility and intranasally challenged with 103 PFU of live SARS-CoV-2 WT D614, B.1.351 or B.1.617.2 variants in a volume of 50 μL DMEM. The body weight and survival percentage of the mice were monitored daily after infection until death upto 5 dpi. Lung tissues were collected for viral loads determination or immediately fixed in 10% PBS-buffered formalin for histopathological observation. All the sample were collected at death or at 5 dpi. Viral gRNA and sgRNA in the lung tissues were measured by droplet digital PCR (Yu et al., 2020), using a COVID-19 digital PCR detection kit (TargetingOne, China). The kit allows the detection of the ORF1ab gene, N gene, and a positive reference gene. The limit of detection is 100 copies/mL. For histopathological analysis, lung tissues were collected and fixed in 10% neutral buffered formalin, embedded in paraffin, and sectioned (5 μm) for standard hematoxylin and eosin staining.
Quantification and statistical analysis
The technical and independent experiment replicates were indicated in the figure legends. Half-maximal effective concentration (EC50) of mAb or dilutions (ED50) of serum were calculated by the equation of four-parameter dose log (agonist) response using Graphpad Prism 8.0. Half-maximal inhibitory concentration (IC50) of mAb or dilutions (ID50) of serum were calculated by the equation of four-parameter dose inhibition response using Graphpad Prism 8.0. The overall fold changes of mutant pseudovirus relative to WT D614G in neutralization of mouse serum and monoclonal antibodies were calculated by the comparison of geometric mean of the ID50 value. Statistical evaluations of statistical significance were performed employing Student’s unpaired or paired t test using GraphPad Prism 8 software, ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001 and ∗∗∗∗p < 0.0001.
Acknowledgments
This study was supported by the National Key Plan for Scientific Research and Development of China (2021YFC0864500, 2020YFC0848800 and 2020YFC0849900), the National Natural Science Foundation (92169205, 81530065, 91442127, and 32000661), the Beijing Municipal Science and Technology Commission (D171100000517 and Z201100005420019), and the Science and Technology Innovation Committee of Shenzhen Municipality (202002073000002). It is also supported by funds from the COVID-19 Science and Technology Project of Beijing Hospitals Authority (YGZX-C1), the Beijing Advanced Innovation Center for Structural Biology, the Tsinghua University Scientific Research Program (20201080053 and 2020Z99CFG004), the Tencent Foundation, the Shuidi Foundation, the TH Capital and the National Science Fund for Distinguished Young Scholars (82025022), and the China Postdoctoral Science Foundation (2020T130062ZX). The funders had no role in study design, data collection, analysis, and interpretation or writing of the report. We thank the SSRF BL17U1 beamline for data collection and processing. Graphical abstract was created with BioRender.com.
Author contributions
L.Z., H.Q., B.Y., X.W., and W.L. conceived and designed the study. Q.L., Y.W., S.Z., J.S., and W.S. performed most of the experiments. Y.J., P.G., and H.Y. conducted the synthesis of mRNA vaccine. Q.L., Y.L., and M.L. carried out the animal experiment. L.C. and Z.Z. performed the neutralization assay of live virus. R.W., R.Z., P.C., Q.Z., and X.S. provided SARS-CoV-2 pseudovirus and its variants. Q.L., Y.W., Y.L., and Z.Y. conducted the analysis of T cell and B cell. Q.L. and W.S. achieved the imaging of BCR triggering. J.L. and J.W. provided assistance with single B cell PCR. Q.L., S.Z., and Y.R produced the RBD, spike, and Fab recombinant proteins and solved the crystal structure of the fab-RBD complex and the cryo-EM structure of the fab-spike complex. Q.L. and J.S. performed the challenge experiment of K18-hACE2 mouse. Q.L., Y.W., S.Z., and W.S. had full access to the data in the study, generated figures and tables, and took responsibility for the integrity and accuracy of the data presentation. H.Q. and L.Z. wrote the manuscript. All authors reviewed and approved the final version of the manuscript.
Declaration of interests
Patent applications have been filed on SARS-CoV-2 RBD trimer (patent application number: 202011294589.2; patent applicants: Tsinghua University). L.Z., Q.L., Y.L., M.L., and X.S. are the inventors.
Published: April 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104043.
Contributor Information
Wanli Liu, Email: liulab@tsinghua.edu.cn.
Xinquan Wang, Email: xinquanwang@mail.tsinghua.edu.cn.
Bo Ying, Email: bo.ying@abogenbio.com.
Jincun Zhao, Email: zhaojincun@gird.cn.
Hai Qi, Email: qihai@tsinghua.edu.cn.
Linqi Zhang, Email: zhanglinqi@mail.tsinghua.edu.cn.
Supplemental information
Data and code availability
The coordinates and structure factors files for T6 and SARS-CoV-2 B.1.351 RBD or spike complexes have been deposited at Protein Data Bank (http://www.rcsb.org) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants — clinical, public health, and vaccine implications. New Engl. J. Med. 2021;384:1866–1868. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett Kizzmekia, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020 doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D., Purcell L.A., Snell G., Veesler D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell. 2021;184:3086–3108. doi: 10.1016/j.cell.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge J., Wang R., Ju B., Zhang Q., Sun J., Chen P., Zhang S., Tian Y., Shan S., Cheng L., et al. Antibody neutralization of SARS-CoV-2 through ACE2 receptor mimicry. Nat. Commun. 2021;12:250. doi: 10.1038/s41467-020-20501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Consortium C.-G.U., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson G., Zhang C., Prado M.G., Paiardini A. PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics. 2017;33:444–446. doi: 10.1093/bioinformatics/btw638. [DOI] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Karikó Katalin, et al. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005 doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Karikó Katalin, et al. Incorporation of Pseudouridine Into mRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Molecular Therapy. 2008 doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Lei J., Frank J. Automated acquisition of cryo-electron micrographs for single particle reconstruction on an FEI Tecnai electron microscope. J. Struct. Biol. 2005;150:69–80. doi: 10.1016/j.jsb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu W., Meckel T., Tolar P., Sohn H.W., Pierce S.K. Intrinsic properties of immunoglobulin IgG1 isotype-switched B cell receptors promote microclustering and the initiation of signaling. Immunity. 2010;32:778–789. doi: 10.1016/j.immuni.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Hu G., Wang Y., Ren W., Zhao X., Ji F., Zhu Y., Feng F., Gong M., Ju X., et al. Functional and genetic analysis of viral receptor ACE2 orthologs reveals a broad potential host range of SARS-CoV-2. Proc. Natl. Acad. Sci. U S A. 2021;118 doi: 10.1073/pnas.2025373118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., et al. Efficacy of the ChAdOx1 nCoV-19 covid-19 vaccine against the B.1.351 variant. N. Engl. J. Med. 2021;384:1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Meng E.C., Couch G.S., Croll T.I., Morris J.H., Ferrin T.E. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30:70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Bjorklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Pinto D., Park Y.J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- Rauch Susanne, et al. mRNA-based SARS-CoV-2 vaccine candidate CVnCoV induces high levels of virus-neutralising antibodies and mediates protection in rodents. NPJ Vaccines. 2021 doi: 10.1038/s41541-021-00311-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.-t., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A., Araki K., Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatatos L., Czartoski J., Wan Y.-H., Homad L.J., Rubin V., Glantz H., Neradilek M., Seydoux E., Jennewein M.F., MacCamy A.J., et al. mRNA vaccination boosts cross-variant neutralizing antibodies elicited by SARS-CoV-2 infection. Science. 2021;372:1413–1418. doi: 10.1126/science.abg9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184:2316–2331.e5. doi: 10.1016/j.cell.2021.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel Annette, et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- von Boehmer L., Liu C., Ackerman S., Gitlin A.D., Wang Q., Gazumyan A., Nussenzweig M.C. Sequencing and cloning of antigen-specific antibodies from mouse memory B cells. Nat. Protoc. 2016;11:1908–1923. doi: 10.1038/nprot.2016.102. [DOI] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang R., Zhang Q., Ge J., Ren W., Zhang R., Lan J., Ju B., Su B., Yu F., Chen P., et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54:1611–1621.e5. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisel F., Shlomchik M. Memory B cells of mice and humans. Annu. Rev. Immunol. 2017;35:255–284. doi: 10.1146/annurev-immunol-041015-055531. [DOI] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K., et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., Gao G., Wang S., Ma C., Xie R., et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.N., Li X.F., Deng Y.Q., Zhao H., Huang Y.J., Yang G., Huang W.J., Gao P., Zhou C., Zhang R.R., et al. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182:1271–1283.e16. doi: 10.1016/j.cell.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361.e6. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Scheres S.H.W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ. 2020;7:253–267. doi: 10.1107/S2052252520000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schafer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors files for T6 and SARS-CoV-2 B.1.351 RBD or spike complexes have been deposited at Protein Data Bank (http://www.rcsb.org) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.