Abstract
Pyrazinamide (PZA), an analog of nicotinamide, is a prodrug that requires conversion to the bactericidal compound pyrazinoic acid (POA) by the bacterial pyrazinamidase (PZase) activity of nicotinamidase to show activity against Mycobacterium tuberculosis. Mutations leading to a loss of PZase activity cause PZA resistance in M. tuberculosis. M. kansasii is naturally resistant to PZA and has reduced PZase activity along with an apparently detectable nicotinamidase activity. The role of the reduction in PZase activity in the natural PZA resistance of M. kansasii is unknown. The MICs of PZA and POA for M. kansasii were determined to be 500 and 125 μg/ml, respectively. Using [14C]PZA and [14C]nicotinamide, we found that M. kansasii had about 5-fold-less PZase activity and about 25-fold-less nicotinamidase activity than M. tuberculosis. The M. kansasii pncA gene was cloned on a 1.8-kb BamHI DNA fragment, using M. avium pncA probe. Sequence analysis showed that the M. kansasii pncA gene encoded a protein with homology to its counterparts from M. tuberculosis (69.9%), M. avium (65.6%), and Escherichia coli (28.5%). Transformation of naturally PZA-resistant M. bovis BCG with M. kansasii pncA conferred partial PZA susceptibility. Transformation of M. kansasii with M. avium pncA caused functional expression of PZase and high-level susceptibility to PZA, indicating that the natural PZA resistance in M. kansasii results from a reduced PZase activity. Like M. tuberculosis, M. kansasii accumulated POA in the cells at an acidic pH; however, due to its highly active POA efflux pump, the naturally PZA-resistant species M. smegmatis did not. These findings suggest the existence of a weak POA efflux mechanism in M. kansasii.
Mycobacterium kansasii is a slow-growing opportunistic human pathogen that often causes pulmonary lesions similar to those caused by M. tuberculosis in immunocompromised patients, such as those with AIDS (24). Unlike M. tuberculosis, M. kansasii is naturally resistant to the antituberculosis drug pyrazinamide (PZA), an analog of nicotinamide, at 50 μg/ml, a concentration at which M. tuberculosis is susceptible (4). In M. tuberculosis, the susceptibility to PZA correlates with the presence of a single enzyme, nicotinamidase, that also has pyrazinamidase (PZase) activity (8, 9, 11, 22). We recently identified the M. tuberculosis PZase/nicotinamidase gene (pncA) (13) and showed that mutation of pncA is a major mechanism of PZA resistance in M. tuberculosis (14, 17). M. bovis and M. bovis BCG, whose genomes are almost identical to that of M. tuberculosis, are naturally resistant to PZA. We have demonstrated that the natural PZA resistance of M. bovis is due to a single point mutation of C to G at nucleotide position 169 of the M. bovis pncA gene, causing an amino acid substitution of aspartate for histidine at amino acid position 57 compared with the M. tuberculosis pncA sequence (13, 14). In this sense, M. bovis strains can be regarded as a special case of PZA-resistant M. tuberculosis. In contrast, the natural PZA resistance in nontuberculous mycobacteria such as M. smegmatis and M. avium is not caused by a defective PZase as in M. tuberculosis with acquired PZA resistance, since these mycobacteria have significant PZase activity (5, 20). Natural PZA resistance, at least in the case of M. smegmatis, is due to a highly active efflux mechanism that rapidly extrudes pyrazinoic acid (POA), the active form of PZA, from the cell after conversion of PZA by the bacterial PZase (27).
The basis for the natural resistance of M. kansasii to PZA is unknown. M. kansasii is known to have reduced PZase activity along with a detectable nicotinamidase activity (5, 20). The role of the reduced PZase activity in the natural PZA resistance of M. kansasii is uncertain. In this study using radioactive [14C]PZA and [14C]nicotinamide, we found that M. kansasii, while having a reasonable amount of nicotinamidase activity, had a very low level of PZase activity that was undetectable by a conventional PZase assay (23). In addition, we cloned the M. kansasii PZase gene (pncA) and showed that it could partially restore PZA susceptibility in naturally PZA-resistant BCG. Furthermore, transformation of M. kansasii with M. avium pncA (known to restore complete susceptibility to BCG) conferred a high degree of susceptibility to PZA in M. kansasii. These results suggest that the natural resistance of M. kansasii to PZA is due to a deficient PZase activity of its nicotinamidase enzyme.
MATERIALS AND METHODS
Growth of mycobacteria and isolation of genomic DNA.
Mycobacterial strains were grown in 7H9 liquid medium with albumin-dextrose-catalase enrichment (Difco) at 37°C for about 2 weeks for M. kansasii ATCC type strain 12478 and for 3 to 4 weeks for M. tuberculosis or M. bovis BCG-Pasteur. Mycobacterial genomic DNA was isolated as described previously (25).
PZA and POA susceptibility testing.
A PZA stock solution (25 mg/ml) was prepared in water. POA was dissolved in dimethyl sulfoxide (1) at a concentration of 100 mg/ml. The susceptibility of M. kansasii, M. kansasii transformants, and BCG transformants to PZA or POA was determined on 7H11 agar plates of acidic pH (pH 5.5) (10, 21) containing 31, 62.5, 125, 250, 500, or 1,000 μg of PZA or POA/ml. Two dilutions (10−2 and 10−4) of early-stationary-phase mycobacterial cultures diluted in 7H9 liquid medium were plated onto the 7H11 plates, which were then incubated at 37°C for about 10 to 14 days for M. kansasii or for 21 to 28 days for BCG. The MICs were determined by the proportional method (indirect test) (6), which is based on determining the lowest concentration of PZA or POA at which bacterial growth (CFU) is inhibited by >99%.
Preparation of protein extracts and determination of protein concentrations.
Protein extracts from M. tuberculosis H37Ra and M. kansasii ATCC type strain 12478 were prepared from late-log-phase cultures by sonication. Briefly, 50-ml cultures were washed with 50 mM potassium phosphate buffer (pH 7.4) twice, and cell pellets were resuspended in 0.4 ml of the same buffer. The concentrated cell suspension was sonicated on ice for 3 min and then centrifuged at 10,000 rpm for 20 min. The protein concentrations of the supernatant fractions (cell lysates) were determined by the Bradford method, using a Bio-Rad protein estimation kit according to the manufacturer’s protocol. The lysates were then used for enzyme activity determinations as described below.
Determination of PZase and nicotinamidase activities.
[14C]carbonyl-PZA ([14C]PZA; 52 mCi/mmol) was provided by the National Institutes of Health (NIH) AIDS Reagents Program, Rockville, Md. [14C]carbonyl-nicotinamide ([14C]NAm; 45.4 mCi/mmol) was purchased from Sigma Chemical Co. To determine the relative PZase and nicotinamidase activities in the bacterial cells, 2 μCi of [14C]PZA or [14C]NAm was added to an equal amount (225 μg of protein) of M. kansasii or M. tuberculosis protein extract in 50 mM potassium phosphate buffer (pH 7.4) in a volume of 100 μl and the mixture was incubated at 37°C for various time periods. Following incubation, 4-μl volumes of the radioactive extracts were spotted onto a 0.25-mm-thick silica G gel 60 thin-layer chromatography (TLC) plate (Whatman), and the plate was air dried and then developed in a solvent system consisting of butanol and 10% ammonium hydroxide (5:1). Following chromatography, the plate was air dried and exposed to X-ray film for autoradiography. The area on the TLC plate where radioactive spots were located, as determined by aligning the autoradiograph with the TLC plate, was cut out and subjected to scintillation counting. The amount of PZase or nicotinamidase in the protein extract was defined and calculated according to the method of Tanigawa et al. (19). One unit of PZase or nicotinamidase was defined as the amount of enzyme required to produce 1 nmol of POA or nicotinic acid (NA) per h.
Southern blot analysis.
Southern blot analysis of mycobacterial genomic DNA was performed as described previously (25). Briefly, genomic DNA from various mycobacterial species was isolated, digested with BamHI, and run on a 0.8% agarose gel. Genomic-DNA fragments from the gel were transferred to a nylon membrane by vacuum blotting, and the membrane was fixed by UV irradiation. The DNA probe for the Southern blotting analysis was prepared by a PCR approach, using primers that were derived from the M. avium pncA gene. The forward primer (5′GCATCAACGCCTACCTGGAC3′) was taken from bp 87 to 107 and the reverse primer (5′TGCACCAGCACCCGGGTGGT3′) was taken from bp 474 to 455 of the M. avium pncA coding sequence (GenBank accession no. U80820) (18). The 391-bp PCR fragment was labeled with [32P]dCTP by using a Random Primer DNA Labeling System (GIBCO BRL) in accordance with the manufacturer’s protocol. The blot was probed with the [32P]dCTP-labeled PCR fragment, washed under low-stringency conditions, and subjected to autoradiography.
Cloning of the M. kansasii pncA gene.
M. kansasii genomic DNA digested with BamHI produced a 1.8-kb fragment that hybridized with the M. avium pncA gene (see Results). The M. kansasii pncA gene was cloned after screening a partial genomic DNA library made by cloning 1.5- to 2-kb BamHI genomic-DNA fragments into pUC19, using a [32P]dCTP-labeled 391-bp PCR fragment from the M. avium pncA gene (18) as a probe, by colony hybridization as described previously (25). A pncA-positive plasmid clone containing the M. kansasii pncA gene was isolated and subjected to DNA sequence analysis as described below. The standard molecular cloning techniques were carried out as described by Sambrook et al. (12).
DNA sequence analysis.
The complete M. kansasii pncA gene sequence was determined for both strands from the 1.8-kb BamHI-pUC19 construct by primer walking in an automatic DNA sequencer (ABI model 377; Applied Biosystems) at the Johns Hopkins University Genetic Core Facility. A multiple-sequence alignment of the M. kansasii, M. avium, M. tuberculosis, and E. coli PncA sequences was performed by the Clustal method, using MegAlign/DNASTAR software.
Transformation of mycobacteria.
The pncA plasmid constructs used for transformation of M. bovis BCG and M. kansasii were made as follows. The 1.8-kb BamHI fragment containing the M. kansasii pncA gene was cloned into the hygromycin resistance-encoding mycobacterial shuttle vector p16R1 (3) as described elsewhere (12). The M. kansasii pncA gene’s start codon is located 584 bp downstream from the 5′-end BamHI site of the 1.8-kb BamHI fragment (see Results) and should contain its own promoter for expression in BCG. The p16R1–1.8-kb M. kansasii pncA construct and the same vector harboring the M. tuberculosis pncA gene on a 3.2-kb EcoRI-PstI fragment (13), along with the vector control, were transformed by electroporation into the naturally PZA-resistant M. bovis BCG as described previously (26). After being subjected to electroporation and incubation at 37°C overnight, the transformed BCG cells were plated on 7H11 agar plates containing 50 μg of hygromycin/ml and incubated at 37°C for 3 to 4 weeks. The M. avium pncA gene, on a 1.6-kb BamHI fragment, was similarly cloned into p16R1, and the p16R1-M. avium pncA construct, along with the p16R1 vector control, was electroporated into M. kansasii as described above. The transformed M. kansasii cells were plated on 7H11 plates containing 100 μg of hygromycin/ml, and the plates were incubated at 37°C for 2 weeks.
[14C]POA accumulation by mycobacteria.
[14C]POA was prepared via [14C]PZA conversion by PZase from an M. smegmatis protein extract. The conversion of [14C]PZA to [14C]POA was complete as judged by TLC. [14C]POA was added at 1 μCi/ml to comparable numbers of bacterial cells (about 2 × 109 each) prepared from late-log-phase cultures of M. kansasii, M. tuberculosis H37Ra, and M. smegmatis at both pH 5.0 and pH 7.0. The radioactive bacterial-cell suspensions were incubated at 37°C for various periods of time, ranging from 0.5 to 6 h. At each time point, portions (100 μl) of bacterial suspension were removed, and the bacterial cells were washed with phosphate-buffered saline onto nitrocellulose membranes (0.45-μm pores) by filtration with the aid of a vacuum pump. The radioactivity associated with bacterial cells on the membranes was determined by scintillation counting.
Nucleotide sequence accession number.
The coding sequence of the M. kansasii pncA gene has been deposited in the GenBank database under accession no. AF002663.
RESULTS
MICs of PZA and POA for M. kansasii.
Previous studies have shown that M. kansasii is resistant to PZA at concentrations higher than 50 μg/ml (4), a concentration at which M. tuberculosis is susceptible (10). However, the exact MIC of PZA for M. kansasii was not reported. In this study, the MIC of PZA for M. kansasii was found to be about 500 μg/ml. The MIC of POA (the active derivative of PZA) for M. kansasii was about 125 μg/ml. As controls, M. bovis BCG and M. tuberculosis H37Ra were both found to have similar POA MICs of about 62 μg/ml.
Relative nicotinamidase and PZase activities of the M. kansasii PncA enzyme.
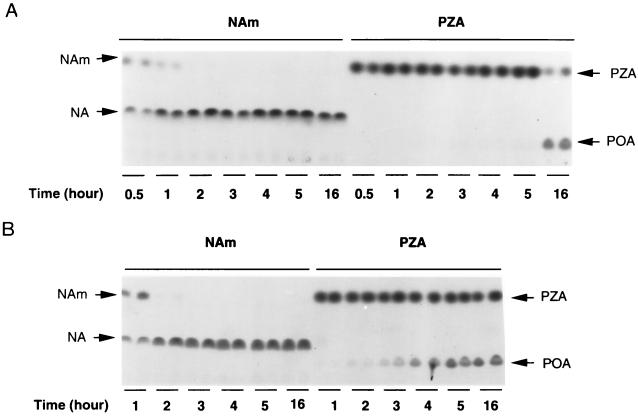
By the conventional Wayne agar method (23), M. kansasii has been shown to have nicotinamidase activity but no detectable PZase activity (5, 20), but some PZase activity was detectable by a more sensitive high-performance liquid chromatography method (16). We compared the relative PZase and nicotinamidase activities of M. kansasii by using [14C]PZA and [14C]NAm, with M. tuberculosis as a control. As shown in Fig. 1A, M. kansasii converted virtually all [14C]NAm to NA at 2 h, whereas conversion of [14C]PZA to POA was hardly seen even by 5 h. However, there was definite conversion of PZA to POA by M. kansasii at 16 h. In contrast, while M. tuberculosis H37Ra converted [14C]NAm to NA by 1 h, M. tuberculosis had much stronger PZase activity, since it converted PZA to POA at 1 to 2 h (Fig. 1B). PZA alone, in the absence of PZase/nicotinamidase (as a control), did not degrade spontaneously into POA even after incubation at 37°C for several days (data not shown). It is worth noting that when [14C]PZA was added to M. tuberculosis or M. kansasii, only two spots were seen on TLC plates; one was PZA itself, and the other was POA. The identities of the spots were confirmed by running “cold” PZA and POA as controls in parallel with samples containing “hot” PZA and POA by TLC, after which the chromatogram was then examined under UV light. Cold PZA and POA gave fluorescence spots, which were marked with pencil, and the TLC plate was then subjected to autoradiography. Under the given set of experimental conditions, no radioactive compounds except PZA or POA were seen.
FIG. 1.
Comparison of nicotinamide and PZA conversions by the PncAs of M. kansasii and M. tuberculosis. [14C]NAm and [14C]PZA were added to M. kansasii (A) and M. tuberculosis H37Ra (B) cultures, and the degrees of conversion of NAm to NA and of PZA to POA were monitored at various time points by TLC. Duplicate samples were taken at each time point. The radioactive NAm and PZA and their derivatives, NA and POA, are indicated by arrows.
To determine the relative amount of nicotinamidase and PZase activities in M. kansasii more quantitatively, we used a more sensitive radioactive assay to measure the enzyme activities in the M. kansasii protein extract, employing [14C]PZA and [14C]NAm, with an equal amount of M. tuberculosis H37Ra protein extract being utilized as a control. The specific activity of the M. kansasii nicotinamidase was about 18-fold higher than its PZase activity (means ± standard deviations, 0.1 ± 0.03 and 1.8 ± 0.12 U/mg of protein, respectively), whereas the M. tuberculosis nicotinamidase activity was about 94-fold higher than its PZase activity (47.4 ± 5.3 and 0.5 ± 0.04 U/mg of protein, respectively). On the other hand, the M. kansasii PZase activity was about 5-fold less than that of the M. tuberculosis PZase, and the M. kansasii nicotinamidase activity was about 25-fold less than that of the M. tuberculosis enzyme.
Cloning and sequence analysis of the M. kansasii pncA gene.
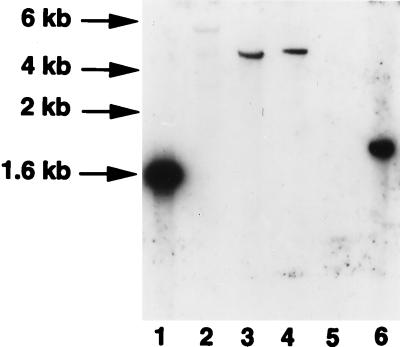
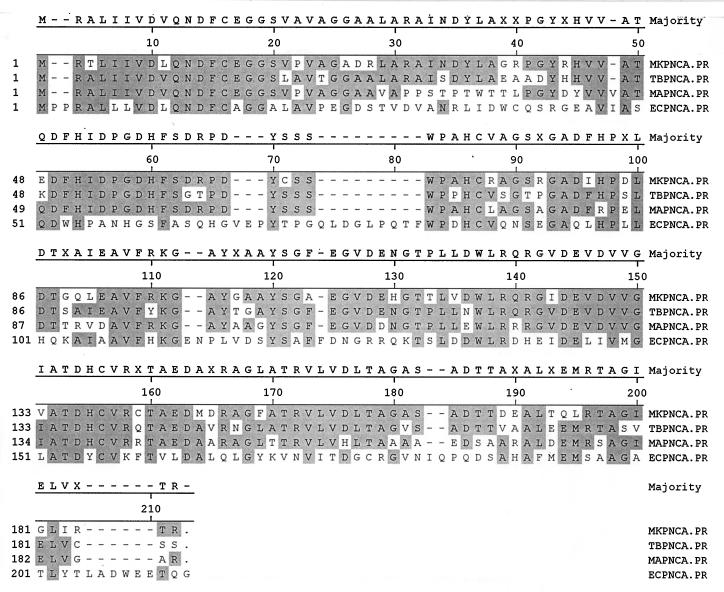
Southern blotting analysis of M. kansasii genomic DNA indicated that a 1.8-kb BamHI fragment hybridized with the 391-bp PCR fragment from the M. avium pncA gene (Fig. 2, lane 6). To clone the M. kansasii pncA gene, a partial genomic-DNA library was constructed by cloning 1.5- to 2-kb BamHI genomic-DNA fragments of M. kansasii into pUC19, and the library was screened with the M. avium pncA gene probe. A positive plasmid clone containing the 1.8-kb BamHI fragment was identified and sequenced. The complete M. kansasii pncA gene was found to be 561 bp long and to encode a protein of about 19.8 kDa (GenBank accession no. AF002663). The M. kansasii PncA showed 69.9, 65.6, and 28.5% amino acid identity with its counterparts from M. tuberculosis, M. avium, and E. coli, respectively (Fig. 3).
FIG. 2.
Determination of the presence of pncA homologues in mycobacterial species by Southern blot analysis. Genomic DNA from various mycobacterial species was digested with BamHI and subjected to hybridization with a [32P]dCTP-labeled M. avium pncA probe. Lanes: 1, M. avium; 2, M. smegmatis; 3, M. tuberculosis H37Rv; 4, M. bovis BCG; 5, M. fortuitum; 6, M. kansasii.
FIG. 3.
Comparison of the sequence of M. kansasii PncA (MKPNCA.PR) with those of its counterparts from M. tuberculosis, M. avium, and E. coli (TBPNCA.PR, MAPNCA.PR, and ECPNCA.PR, respectively). Sequence alignment was performed by the Clustal method. Highlighted areas indicate identical amino acid residues of the PncA enzymes. Accession numbers for the pncA genes of M. kansasii, M. avium, M. tuberculosis, and E. coli are AF002663, U80820, U59967, and P21369, respectively.
Transformation of BCG with the M. kansasii pncA gene partially restored PZA susceptibility.
BCG is naturally resistant to PZA (MIC > 500 μg/ml) due to the presence of a single point mutation in the pncA gene (13). To assess the relative contribution of the pncA genes of M. kansasii and BCG to PZA susceptibility, we transformed BCG with the M. kansasii pncA gene on the 1.8-kb BamHI fragment cloned into p16R1 and also with the M. tuberculosis pncA construct as a positive control. While the M. tuberculosis pncA gene conferred full susceptibility to PZA in BCG (MIC, ca. 31 to 62 μg/ml), the M. kansasii pncA gene could only partially restore PZA susceptibility to BCG (MIC, ca. 125 μg/ml) (Table 1). This indicates that the M. kansasii PncA enzyme indeed has some level of PZase activity, which can enhance the conversion of PZA to active POA, and thus is involved in partial restoration of PZA susceptibility to BCG. Transformation with the pncA gene from either M. tuberculosis or M. kansasii did not alter the POA MIC for BCG (Table 1).
TABLE 1.
MICs of PZA and POA for mycobacterial strains
| Strain | MIC (μg/ml) of:
|
|
|---|---|---|
| PZA | POA | |
| M. kansasii | 500 | 125 |
| M. tuberculosis H37Ra | 31–62 | 62 |
| M. bovis BCG | >500 | 62 |
| BCG + p16R1 vector control | >500 | 62 |
| BCG + p16R1-M. kansasii pncA | 125 | 62 |
| BCG + p16R1-M. tuberculosis pncA | 31–62 | 62 |
| M. kansasii + p16R1 vector control | 500 | 125 |
| M. kansasii + p16R1-M. avium pncA | 31–62 | 125 |
Transformation of M. kansasii with M. avium pncA conferred a high degree of PZA susceptibility.
M. avium pncA has previously been shown to confer to BCG a level of PZA susceptibility similar to that of the PZA-susceptible bacterium M. tuberculosis, indicating that M. avium has a functional PZase/nicotinamidase enzyme capable of potentiating PZA action. To determine whether the natural PZA resistance of M. kansasii is due to the reduced PZase activity of its nicotinamidase enzyme, we transformed M. kansasii (ATCC type strain) with the M. avium pncA gene in the hygromycin vector p16R1. Transformation of the M. kansasii with M. avium pncA caused functional overexpression of the M. avium PZase enzyme, as revealed by an increased conversion of [14C]PZA to [14C]POA compared to that of the vector control (data not shown). The overexpression of the M. avium PZase enzyme rendered the M. kansasii strain more susceptible to PZA (MIC, 31 to 62 μg/ml [a concentration to which the susceptible species M. tuberculosis is sensitive]). This suggests that the natural PZA resistance of M. kansasii (at least in the case of the ATCC type strain) results from its reduced PZase activity, which does not result in efficient conversion of the prodrug PZA to the bactericidal compound POA.
Weak POA efflux activity in M. kansasii.
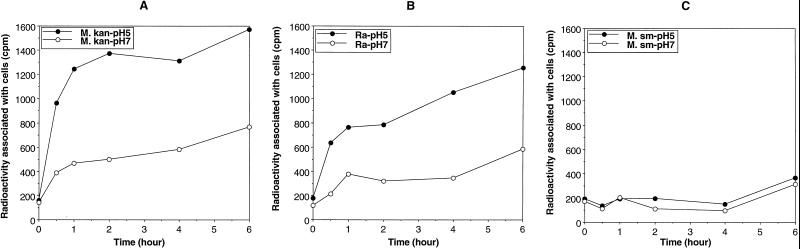
The natural PZA resistance of fast-growing M. smegmatis correlates with a highly active POA efflux mechanism which does not allow POA to accumulate in cells of this species at an acidic pH (27). In contrast, the PZA-susceptible species M. tuberculosis has been found to have a much weaker POA efflux mechanism, as revealed by the increasing accumulation of POA by this organism at an acidic pH (5.0 to 5.5) (27). To determine the potential POA efflux activity in M. kansasii, we compared the POA accumulation patterns of M. tuberculosis, M. smegmatis, and M. kansasii at pH 5.0 and pH 7.0. As shown in Fig. 4, M. kansasii behaved like the susceptible species M. tuberculosis in accumulating POA at pH 5.0 (Fig. 4A and B); in contrast, M. smegmatis cells did not accumulate a significant amount of POA even at an acidic pH (5.0) (Fig. 4C).
FIG. 4.
Comparison of [14C]POA accumulations in M. kansasii (M. kan) (A), M. tuberculosis H37Ra (Ra) (B), and M. smegmatis (M. sm) (C) at an acidic pH (5.0) and at neutral pH (7.0). Closed circles and open circles represent the amounts of radioactivity associated with bacterial cells at pH 5.0 and pH 7.0, respectively. This experiment was repeated at least three times, and the results of a representative experiment are shown here.
DISCUSSION
M. kansasii is known to have a deficient PZase activity in conventional PZase testing by the Wayne agar method, despite having an apparently normal nicotinamidase activity (5, 20). In this study, using a more sensitive radioactive method involving the use of [14C]PZA as a substrate, we showed that M. kansasii definitely has a weak PZase activity that is undetectable by the Wayne method. This finding is in agreement with the results of Speirs et al., who found weak PZase activity in M. kansasii by a sensitive high-performance liquid chromatography analytical method (16). The M. kansasii nicotinamidase activity was about 18-fold higher than its PZase activity, whereas the M. tuberculosis nicotinamidase activity was 94-fold higher than its PZase activity. The PZase activity of M. kansasii was about fivefold lower than that of the M. tuberculosis enzyme. To identify the basis for the separation of PZase activity from nicotinamidase activity of M. kansasii PncA and to assess the role of the reduced PZase activity in the natural resistance of M. kansasii to PZA, we cloned the pncA gene from M. kansasii. Sequence analysis showed that the M. kansasii PncA exhibited a high degree of homology to the M. tuberculosis (69.9%) and M. avium (65.6%) enzymes, which have both PZase and nicotinamidase activities (Fig. 3). However, sequence comparisons did not allow direct identification of the amino acid residues responsible for the weak PZase activity of the M. kansasii nicotinamidase enzyme, because there are several residues in the M. kansasii PncA that are different from the corresponding residues of other mycobacterial PncAs (Fig. 3). The physiological role of the nicotinamidase enzyme (PncA) is to degrade nicotinamide to NA, which can be recycled to NAD via the Preiss-Handler pathway of the pyridine nucleotide cycle in most prokaryotes (2). Because PZase converts PZA to the bactericidal compound POA, the amount of PZase activity, by affecting the rate of conversion of PZA to POA, would be important in determining the susceptibility of mycobacteria to PZA. The presence as well as the amount of PZase activity of the nicotinamidase enzymes from various bacterial species appears to be purely coincidental, because PZA, as an analog of nicotinamide, is an artificial compound that does not exist in nature. It so happens that M. tuberculosis has a nicotinamidase enzyme with a reasonable amount of PZase activity, whereas M. kansasii has a nicotinamidase enzyme with much weaker PZase activity.
Because defective PZase activity caused by pncA mutations correlates with PZA resistance in M. tuberculosis (13, 15, 17), we determined whether the reduced level of PZase activity in M. kansasii is responsible for its natural resistance to PZA. Transformation of M. kansasii with the M. avium pncA gene caused overexpression of PZase and concomitantly conferred a high level of susceptibility to PZA (MIC, 31 to 62 μg/ml); in a previous study, the same M. avium pncA construct was shown to make BCG fully susceptible to PZA (18). This indicates that the reduced PZase activity of M. kansasii is the cause of its natural PZA resistance. On the other hand, transformation of BCG with M. kansasii pncA made BCG more susceptible to PZA (MIC, 125 μg/ml); however, the degree of susceptibility to PZA was not as high as that achieved by transformation with the M. tuberculosis pncA gene (MIC, 31 to 62 μg/ml). This suggests that while M. kansasii PncA has some PZase activity, its PZase activity is lower than that of the M. tuberculosis PncA enzyme, a conclusion also supported by the enzyme assays using [14C]PZA (see Results).
In addition to the level of PZase activity, the activity of efflux pumps (7) could also affect the susceptibility of mycobacteria to PZA. We have recently shown that the natural PZA resistance of M. smegmatis (MIC, >2,000 μg/ml) is not due to a defective PZase but rather is attributable to a highly active efflux pump that extrudes POA from the cell very rapidly (27). Likewise, the natural PZA resistance of M. avium (MIC, 500 μg/ml) also appears to involve an active efflux pump with an efficiency in between those of M. smegmatis and M. tuberculosis (unpublished observation), since the M. avium pncA gene, when transformed into BCG, completely restored PZA susceptibility (18), indicating that the M. avium PZase is fully functional and the natural PZA resistance of M. avium is not due to an inability of its PZase to convert PZA to POA. In this study, M. kansasii, like the PZA-susceptible species M. tuberculosis, could also accumulate POA in the cells at an acidic pH (5.0) (Fig. 4A). The accumulation of POA at an acidic pH, along with the observation that the M. avium pncA gene can confer a high degree of PZA susceptibility to M. kansasii, indicates that M. kansasii has a weak POA efflux mechanism that is unlikely to contribute to its natural PZA resistance. We conclude that the natural PZA resistance of M. kansasii is due to the somewhat-deficient PZase activity of its nicotinamidase enzyme. Site-directed mutagenesis and comparative structural and enzymatic analyses are needed to determine the amino acid residues in PncA that underlie the weak PZase activity of the M. kansasii nicotinamidase.
ACKNOWLEDGMENTS
This work was supported by research grants from the American Lung Association, Potts Memorial Foundation, and the NIH (RO1 AI40584 [to Y.Z.]).
We thank Diane Griffin and Barbara Laughon for encouragement, Salman Siddiqi for M. kansasii strains, and the NIH AIDS Reagents Program for [14C]PZA. We also thank the reviewers for helpful suggestions.
REFERENCES
- 1.Cynamon M H, Gimi R, Gyenes F, Sharpe C A, Bergermann K E, Han H J, Gregor L B, Rapolu R, Luciano G, Welch J T. Pyrazinoic acid esters with broad spectrum in vitro antimycobacterial activity. J Med Chem. 1995;38:3902–3907. doi: 10.1021/jm00020a003. [DOI] [PubMed] [Google Scholar]
- 2.Foster J W, Moat A G. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev. 1980;44:83–105. doi: 10.1128/mr.44.1.83-105.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garbe T, Barathi J, Barnini S, Zhang Y, Abou-Zeid C, Tang D, Mukherjee R, Young D. Transformation of mycobacterial species using hygromycin resistance as selectable marker. Microbiology. 1994;140:133–138. doi: 10.1099/13500872-140-1-133. [DOI] [PubMed] [Google Scholar]
- 4.Good R C, Silcox V A, Kilburn J O, Plikaytis B D. Identification and drug susceptibility test results for Mycobacterium spp. Clin Microbiol Newslett. 1985;7:133–136. [Google Scholar]
- 5.Helbecque D M, Handzel V, Eidus L. Simple amidase test for identification of mycobacteria. J Clin Microbiol. 1975;1:50–53. doi: 10.1128/jcm.1.1.50-53.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inderlied C B, Nash K A. Antimycobacterial agents: in vitro susceptibility testing, spectra of activity, mechanism of action and resistance, and assays for activity in biological fluids. In: Lorian V, editor. Antibiotics in laboratory medicine. 4th ed. Baltimore, Md: Williams & Wilkins; 1996. pp. 127–175. [Google Scholar]
- 7.Jarlier V, Nikaido H. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol Lett. 1994;123:11–18. doi: 10.1111/j.1574-6968.1994.tb07194.x. [DOI] [PubMed] [Google Scholar]
- 8.Konno K, Feldman F M, McDermott W. Pyrazinamide susceptibility and amidase activity of tubercle bacilli. Am Rev Respir Dis. 1967;95:461–469. doi: 10.1164/arrd.1967.95.3.461. [DOI] [PubMed] [Google Scholar]
- 9.McClatchy J K, Tsang A Y, Cernich M S. Use of pyrazinamidase activity in Mycobacterium tuberculosis as a rapid method for determination of pyrazinamide susceptibility. Antimicrob Agents Chemother. 1981;20:556–557. doi: 10.1128/aac.20.4.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDermott W, Tompsett R. Activation of pyrazinamide and nicotinamide in acidic environment in vitro. Am Rev Tuberc. 1954;70:748–754. doi: 10.1164/art.1954.70.4.748. [DOI] [PubMed] [Google Scholar]
- 11.Miller M A, Thibert L, Desjardins F, Siddiqi S H, Dascal A. Testing of susceptibility of Mycobacterium tuberculosis to pyrazinamide: comparison of Bactec method with pyrazinamidase assay. J Clin Microbiol. 1995;33:2468–2470. doi: 10.1128/jcm.33.9.2468-2470.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 13.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 14.Scorpio A, Collins D, Whipple D, Cave D, Bates J, Zhang Y. Rapid differentiation of bovine and human tubercle bacilli based on a characteristic mutation in the bovine pyrazinamidase gene. J Clin Microbiol. 1997;35:106–110. doi: 10.1128/jcm.35.1.106-110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scorpio A, Lindholm-Levy P, Heifets L, Gilman R, Siddiqi S, Cynamon M, Zhang Y. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:540–543. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Speirs R J, Welch J T, Cynamon M H. Activity of n-propyl pyrazinoate against pyrazinamide-resistant Mycobacterium tuberculosis: investigations into mechanism of action of and mechanism of resistance to pyrazinamide. Antimicrob Agents Chemother. 1995;39:1269–1271. doi: 10.1128/aac.39.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sreevatsan S, Pan X, Zhang Y, Kreiswirth B N, Musser J M. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41:636–640. doi: 10.1128/aac.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Z H, Scorpio A, Zhang Y. The pncA gene from the naturally pyrazinamide-resistant Mycobacterium avium encodes pyrazinamidase and confers pyrazinamide susceptibility to resistant M. tuberculosis complex organisms. Microbiology. 1997;143:3367–3373. doi: 10.1099/00221287-143-10-3367. [DOI] [PubMed] [Google Scholar]
- 19.Tanigawa Y, Shimoyama M, Ueda I. Nicotinamide deamidase from Flavobacterium peregrinum. Methods Enzymol. 1980;66:132–136. doi: 10.1016/0076-6879(80)66450-7. [DOI] [PubMed] [Google Scholar]
- 20.Tarnok I, Rohrscheidt E. Biochemical background of some enzymatic tests used for the differentiation of mycobacteria. Tubercle. 1976;57:145–150. doi: 10.1016/0041-3879(76)90052-0. [DOI] [PubMed] [Google Scholar]
- 21.Tarshis M S, Weed W A. Lack of significant in vitro sensitivity of Mycobacterium tuberculosis to pyrazinamide on three different solid media. Am Rev Tuberc. 1953;67:391–395. doi: 10.1164/art.1953.67.3.391. [DOI] [PubMed] [Google Scholar]
- 22.Trivedi S S, Desai S G. Pyrazinamidase activity of Mycobacterium tuberculosis—a test of sensitivity to pyrazinamide. Tubercle. 1987;68:221–224. doi: 10.1016/0041-3879(87)90058-4. [DOI] [PubMed] [Google Scholar]
- 23.Wayne L G. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am Rev Respir Dis. 1974;109:147–151. doi: 10.1164/arrd.1974.109.1.147. [DOI] [PubMed] [Google Scholar]
- 24.Witzig R S, Fazal B A, Mera R M, Mushatt D M, Dejace P M, Greer D L, Hyslop N E. Clinical manifestations and implications of coinfection with Mycobacterium kansasii and human immunodeficiency virus type 1. Clin Infect Dis. 1995;21:77–85. doi: 10.1093/clinids/21.1.77. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Garcia M J, Lathigra R, Allen B, Moreno C, van Embden J D A, Young D. Alterations in the superoxide dismutase gene of an isoniazid-resistant strain of Mycobacterium tuberculosis. Infect Immun. 1992;60:2160–2165. doi: 10.1128/iai.60.6.2160-2165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Garbe T, Young D. Transformation with katG restores isoniazid sensitivity in Mycobacterium tuberculosis isolates resistant to a range of drug concentrations. Mol Microbiol. 1993;8:521–524. doi: 10.1111/j.1365-2958.1993.tb01596.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, Y., A. Scorpio, H. Nikaido, and Z. Sun. Role of acid pH and deficient efflux of pyrazinoic acid in the unique susceptibility of Mycobacterium tuberculosis to the front-line tuberculosis drug pyrazinamide. Submitted for publication. [DOI] [PMC free article] [PubMed]