Significance
Genes on sex chromosomes (i.e. human chX) are regulated differently in males and females to balance gene expression levels between sexes (XY vs. XX). This sex-specific regulation is called dosage compensation (DC). DC is achieved by altering the shape and compaction of sex chromosomes specifically in one sex. In this study, we use Oligopaints to examine DC in silkworms. This study visualizes this phenomenon in a species with ZW sex chromosomes, which evolved independently of XY. Our data support a long-standing model for how DC mechanisms evolved across species, and we show potential similarity between DC in silkworms and nematodes, suggesting that this type of DC may have emerged multiple independent times throughout evolution.
Keywords: dosage compensation, 3D genome organization, chromosome biology, X-inactivation, lepidoptera
Abstract
Interphase chromatin is organized precisely to facilitate accurate gene expression. The structure–function relationship of chromatin is epitomized in sex chromosome dosage compensation (DC), where sex-linked gene expression is balanced between males and females via sex-specific alterations to three-dimensional chromatin structure. Studies in ZW-bearing species suggest that DC is absent or incomplete in most lineages except butterflies and moths, where male (ZZ) Z chromosome (chZ) expression is reduced by half to equal females (ZW). However, whether one chZ is inactivated (as in mammals) or both are partially repressed (as in Caenorhabditis elegans) is unclear. Using Oligopaints in the silkworm, Bombyx mori, we visualize autosomes and chZ in somatic cells from both sexes. We find that B. mori chromosomes are highly compact relative to Drosophila. We show that in B. mori males, both chZs are similar in size and shape and are more compact than autosomes or the female chZ after DC establishment, suggesting both male chZs are partially and equally downregulated. We also find that in the early stages of DC in females, chZ chromatin becomes more accessible and Z-linked expression increases. Concomitant with these changes, the female chZ repositions toward the nuclear center, revealing nonsequencing-based support for Ohno’s hypothesis. These studies visualizing interphase genome organization and chZ structure in Lepidoptera uncover intriguing similarities between DC in B. mori and C. elegans, despite these lineages harboring evolutionarily distinct sex chromosomes (ZW/XY), suggesting a possible role for holocentricity in DC mechanisms.
The interphase genome is a complex network of three-dimensional (3D) chromatin interactions. Individual chromosomes occupy discrete regions of the nucleus, termed chromosome territories (CTs), that are nonrandomly organized in interphase (1). Chromosome size can influence CT position, with smaller chromosomes being more central in human nuclei (2), and studies from flies to humans have shown that gene-poor CTs are positioned closer to the nuclear periphery than gene-rich CTs (3–5). In agreement, both genomics and imaging-based approaches have revealed that the nuclear periphery is dense with repressive heterochromatin, while the nuclear interior is more permissive to active transcription (6–10). Additionally, repositioning genes away from the nuclear periphery is associated with increased transcription (11–14).
At the subchromosomal level, chromatin is further organized into clusters of short-range interactions and chromatin loops. These intrachromosomal interactions can result in specific long-range chromosome folding patterns that may be species- or context-specific (5). This intricate 3D genome structure at both the whole- and subchromosome levels is essential for properly regulating gene expression in different developmental and tissue-specific contexts (15–19).
One prime example of how 3D chromosome architecture facilitates accurate gene expression is sex chromosome dosage compensation (DC). DC classically refers to the normalization of expression between sex chromosomes and autosomes, while sex chromosome dosage balance is the process of equalizing gene-expression levels of sex-linked genes between homogametic (XX or ZZ) and heterogametic (XY or ZW) sexes (20, 21). Here, we refer to both processes collectively as DC. Studies in diverse eukaryotic species have revealed an array of epigenetic mechanisms for establishing DC, all of which are associated with sex-specific alterations to the 3D structure of the sex chromosomes (22–28). For example, in mammals where X-linked expression is reduced in females (XX) by inactivation of a single X chromosome (29), the inactive chX is more peripheral in the nucleus than the active chX (30). Furthermore, the inactive chX adopts a unique, highly compact 3D configuration (known as a Barr body) compared to the active chX (31, 32).
One of the first models for the evolution of DC was proposed by Susumo Ohno. Ohno predicted that mechanisms would evolve to not only down-regulate X-linked expression in the homogametic sex (XX) to maintain balanced chX expression between the sexes, but also to up-regulate X-linked expression in both sexes to balance chX expression with autosomal expression (33). This two-step model for achieving DC is known as Ohno’s hypothesis, and it remains the prevailing model for the evolution of DC. Yet, studies both supporting and refuting particularly the latter part of Ohno’s hypothesis exist, leaving this model still widely debated (34–38).
Interestingly, the few existing studies in species with female heterogametry (ZW sex chromosomes) have suggested that DC mechanisms may be absent or incomplete in these lineages (39–47). However, previous expression-based studies demonstrated that Lepidopteran insects are an exception to this rule, where Z-linked gene expression in males (ZZ) is repressed to match the gene expression of the single chZ in females (ZW) (20, 48–53). In the silkworm moth Bombyx mori, DC has been shown to occur in late embryonic development (48). Yet whether one chZ is completely silenced (similar to X-inactivation in mammals) or both are partially repressed [similar to Caenorhabditis elegans (54)] in Lepidopteran DC remains unclear. Furthermore, how 3D genome organization may more broadly be linked to transcriptional regulation in Lepidoptera is unknown, as the 3D organization of chromosomes in interphase has never been visualized in any moth or butterfly.
Here, we use Oligopaint DNA FISH to visualize autosomes and the Z sex chromosome in male and female B. mori embryonic and larval cells. We reveal that B. mori chromosomes form distinct CTs, with both CT volume and position in the nucleus being influenced by both genomic size and gene density. Additionally, we show that gene-poor chromosomes harbor more robust long-range cis interactions than their gene-rich counterparts. We further show that both copies of chZ in males are compact and highly similar in volume, shape, and chromosome folding patterns. These findings suggest a mechanism in which both chZs in males are partially down-regulated to achieve DC, similar to DC in C. elegans (54, 55). Furthermore, in support of Ohno’s hypothesis, we find that chZ in females is repositioned away from the nuclear edge after DC is initiated, and this central positioning of the chZ correlates with an increase in Z-linked chromatin accessibility and an increase in Z-linked gene expression in females. This result is again similar to findings in C. elegans (56), further supporting a model in which DC in Lepidopteran insects and nematodes may be established via similar mechanisms. Interestingly, both C. elegans and B. mori harbor holocentric chromosomes (where centromeres form all along the length of the chromosome), raising the possibility that the holocentric chromosome structure may influence DC mechanisms. Together, these findings illustrate that DC is achieved in B. mori by both partially repressing both copies of chZ in males and up-regulating Z-linked genes in females, and reveal nonsequencing-based evidence for Ohno’s theory of the two-step mechanism of DC.
Results and Discussion
B. mori Interphase Chromosomes Are Highly Compact.
Before specifically interrogating chZ organization in B. mori, we needed to establish basic principles of interphase chromatin organization in this species. We began by measuring nuclear volume in B. mori embryonic cells using DAPI staining, which suggested that B. mori nuclei may be extremely compact for their DNA content. The B. mori genome is over three times as large as the Drosophila melanogaster genome (57, 58), and therefore, we expected B. mori nuclei to be three times as large. However, our measurements of nuclear volume revealed instead that B. mori diploid nuclei are similar in volume to Drosophila diploid nuclei (SI Appendix, Fig. S1A). This finding suggests that chromatin compaction may be more robust in moths than in flies. Interestingly, when we compared nuclear volumes across a broader range of species, we found that Drosophila may be more of an outlier than Bombyx, with fly nuclei appearing to be larger than expected for their genomic content (SI Appendix, Fig. S1 B–D). However, we also observed that species with larger genomes and more complex tissues show more variation in nuclear volume (SI Appendix, Fig. S1C), making these broad comparisons difficult to interpret.
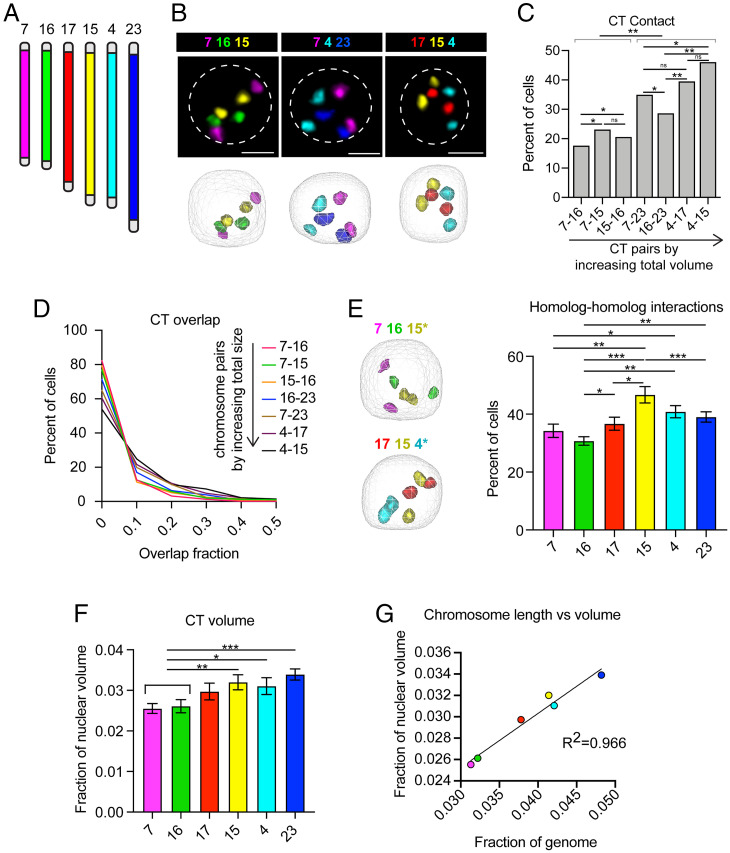
Still, we wanted to use what is known about Drosophila genome organization as a baseline for comparison to Bombyx. Our nuclear volume measurements suggest that chromatin organization in Bombyx and Drosophila are likely distinct. One possibility is that B. mori interphase CTs are more intermixed than fly CTs, allowing for a larger genome to occupy a smaller nuclear space. Supporting this hypothesis, the Condensin II complex that is required for CT separation in flies is reportedly incomplete in B. mori (5, 59). Thus, to measure CT intermixing, we labeled interphase chromosomes in B. mori embryonic nuclei with whole-chromosome Oligopaints for six autosomes (chromosomes 4, 7, 15, 16, 17, and 23) (SI Appendix, Table S1) of the 28 B. mori chromosomes (60, 61) (Fig. 1A). This assay clearly revealed the formation of spatially distinct CTs (Fig. 1B), with the majority of cells (50 to 80%) harboring no overlap at all between any given representative CT pair (Fig. 1 C and D). Also, contact between homologous chromosome copies occurs at comparable rates to heterologous contacts for two chromosomes of similar size (Fig. 1 C and E and SI Appendix, Fig. S1E), supporting previous findings that somatic homolog pairing is absent in moths (59, 61). These results suggest that CT size is the main factor dictating CT–CT contacts in B. mori. However, size cannot fully explain all observed trans interactions, as chromosome 7 (ch7) has more heterologous and homologous contacts than ch16 (Fig. 1 C and E), despite these two chromosomes being similar in size. Together, these assays demonstrate that B. mori harbor spatially distinct CTs and that high levels of CT intermixing cannot explain the small size of B. mori nuclei.
Fig. 1.
B. mori interphase chromosomes are compact and nonrandomly organized. (A) Schematic of whole-chromosome Oligopaints used. Chromosomes and chromosome pairs are ordered by size (small to large) in all figure panels. (B, Upper) Representative Oligopaint DNA FISH images showing B. mori CTs. (Scale bar, 2.5 µm.) Dotted line indicates approximate nuclear edge. (Lower) Three-dimensional rendering of cells from TANGO. (C) Bar graph showing the absolute contact frequency for all replicates (including at least three biological replicates per chromosome pair). Statistics: Fisher’s exact test comparing contact versus no contact. *P = 0.003–0.0003; **P < 0.0003. (D) Histogram showing CT overlap as a fraction of the smaller chromosome (for example, 15–16 overlap fraction is the fraction of ch16 colocalizing with ch15). All pairwise comparisons are statistically significant (Mann–Whitney U test, P < 0.0001). (E, Left) Representative 3D rendering of nuclei, as in B (Lower), showing homolog–homolog interactions of the chromosome pair indicated with an asterisk. (Right) Quantification of homolog–homolog interactions shown as the fraction of cells harboring any voxel overlap between the two homologs of a single chromosome (indicated on the x axis). Bars show the average between biological replicates. Error bars show SEM. Statistics = unpaired t tests. *P < 0.05, **P < 0.01, ***P < 0.001. (F) Quantification of CT volume. Bars show average CT volume as fraction of nuclear volume across biological replicates. Statistics: unpaired t tests. *P = 0.035, **P = 0.008, ***P = 0.0006. (G) Dot plot showing chromosome genomic length relative to total genome size (x axis) versus CT volume relative to nuclear volume (y axis). R2 = 0.966. For all data shown, at least three biological replicates (embryos) were analyzed for each chromosome and at least 100 cells per embryo. Error bars show SEM.
Another possible explanation for the small size of B. mori nuclei is that chromatin within CTs may be more tightly packaged in B. mori than in flies, leading to smaller CTs and overall reduced nuclear volumes. We therefore measured CT volumes, which illustrated that on average, each chromosome occupies only 2 to 4% of the nucleus (Fig. 1F). This amount is precisely the percent of the B. mori genome each chromosome encompasses, and thus the genomic length of chromosomes scales nearly perfectly with chromosome volume (Fig. 1G). Still, the volume of B. mori chromosomes is significantly smaller than that of fly chromosomes of similar genomic size (SI Appendix, Fig. S1F). These findings suggest that cis compaction is more robust in B. mori than in Drosophila, but basic principles of large-scale nuclear organization, such as CT formation, are largely conserved in this species.
B. mori Autosomes Are Tightly Folded.
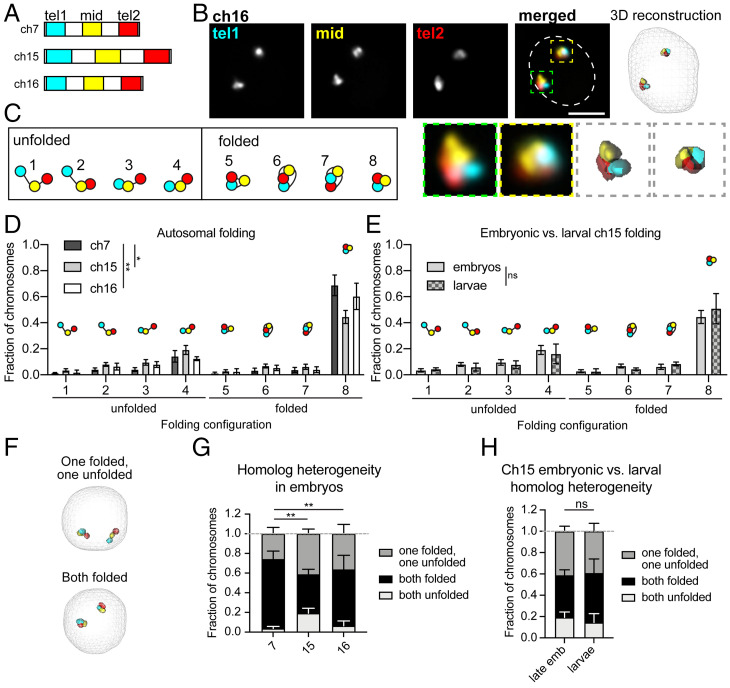
Our findings regarding B. mori and fly chromosome volumes suggest that subchromosomal compaction is more robust in moths than flies. As previous studies in flies illustrated that within CTs, chromosomes can have preferential folding configurations (5), we reasoned that one way to extensively compact chromosomes would be to fold them into tighter configurations. To test this hypothesis and to broadly assess long-range cis interactions in B. mori nuclei, we used Oligopaints to label ∼3-Mb stripes at both telomeres (tel1 and tel2) and in the center (mid) of ch7, ch15, and ch16 (Fig. 2 A and B and SI Appendix, Table S2). This strategy allowed us to trace chromosomes in 3D, and folding configurations were quantified based on contact patterns between each of the three stripes. Eight possible folding configurations can be detected with this assay based on the presence or absence of contact between the three stripes (Fig. 2C) (5). Among these configurations, the four in which the two telomeres are not in contact were classified as “unfolded” configurations, and the four in which the telomeres are in contact were classified as “folded” (Fig. 2C). This assay revealed that in both embryonic and larval nuclei, B. mori chromosomes tend to adopt a single folded configuration where all three domains are in contact with each other (Fig. 2 D and E). Ch7, which is small and gene-poor, is found in this completely folded configuration at even higher frequencies (∼75%). Importantly, this folding pattern is dramatically more compact than the folding patterns observed in fly cells, where only 30% of chromosomes are in a configuration with all stripes in contact (SI Appendix, Fig. S1G) (5).
Fig. 2.
B. mori autosomes are tightly folded. (A) Schematic of stripe Oligopaints. Each chromosome is divided into five equal domains. The first (tel1), middle (mid), and last (tel2) domains are labeled as indicated. (B, Upper Left) Representative Oligopaint DNA FISH images showing stripe paints for ch16. (Scale bar, 2.5 µm.) Dotted line indicates approximate nuclear edge. (Upper Right) Three-dimensional rendering of cell on the left. (Lower Left) Zoom of individual chromosomes labeled with Oligopaints. (Lower Right) Zoom of 3D rendering. (C) Schematic of eight possible folding configurations detected by this assay. Configurations with telomere–telomere contact (cyan and red) are categorized as folded, while configurations without telomere–telomere contact are classified as unfolded. (D) Quantification of folding for ch7 (dark gray), ch15 (gray), and ch16 (white) in embryos. As each nucleus has two chromosome copies, two data points were obtained per nucleus. The fraction of chromosomes in a given configuration (noted on the x axis and in the imbedded schematics) is shown on the y axis. Statistics: χ2 test. *P = 0.01, **P = 0.003. (E) Quantification of folding for ch15 in late embryos and larvae. Statistics: χ2 test. (F) Representative 3D rendering of nuclei, as in B, showing a cell with homologs in different folding configurations (Upper) or the same configuration (Lower). (G) Quantification of homolog heterogeneity in embryos. Bars represent average between biological replicates and error bars represent SEM. Statistics: χ2 test. **P = 0.002–0.009. (H) Quantification of homolog heterogeneity for ch15 in embryos and larvae. Statistics: χ2 test. For all graphs, bars show the average of at least three biological replicates with at least 100 cells analyzed from each replicate. Error bars represent SEM.
Notably, for all three examined B. mori chromosomes, both homologous copies are folded similarly in 60 to 75% of nuclei (Fig. 2 F and G). These low levels of intranuclear homolog heterogeneity are in contrast to findings from human cells, which showed high levels of intranuclear homolog heterogeneity in interphase (62). When considering only the 60 to 75% of cells with similarly folded homologs, the majority harbor two folded copies (Fig. 2 F and G). Overall, this extensive chromosome folding may at least partially explain why B. mori nuclei are so much smaller than their fly counterparts. Taken together, our autosomal Oligopaint assays reveal basic principles of interphase nuclear organization in B. mori.
ChZ Is More Compact in Males than in Females.
As DC in all studied species has been associated with sex-specific alterations to the 3D structure of the sex chromosomes, we next examined the 3D organization of the B. mori chZ sex chromosome by FISH to interrogate possible structural changes associated with DC. In B. mori, a transcriptome study found that Z-linked gene expression is reduced in males (ZZ) to match the expression of the single chZ in females (ZW), and this equalization is complete by late embryonic development (48). However, whether one chZ is completely silenced (as in mammals) or both are partially and equally repressed (as in C. elegans) in moth DC is unknown. In agreement with previous studies in Lepidoptera (63), we saw no cytological evidence of Barr body formation in male (ZZ) nuclei, suggesting complete Z-inactivation may not be occurring (SI Appendix, Fig. S2A). However, we noted that nuclei from both males and females appear to be completely devoid of dense heterochromatin based on a lack of DAPI-bright signal (SI Appendix, Fig. S2A). In agreement with this observation, previous ChIP-seq (chromatin immunoprecipitation followed by sequencing) studies revealed only minimal H3K9me2/3 genome-wide in B. mori-cultured cells (64). Therefore, we reasoned that it is possible that complete Z-inactivation could occur even in the absence of a cytological Barr body.
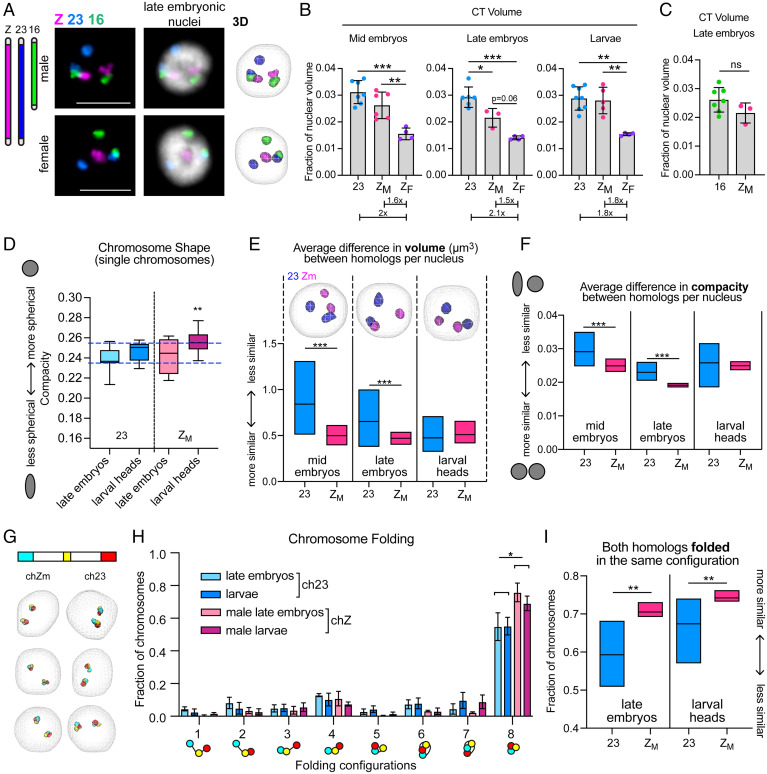
Irrespective of Barr body formation, we reasoned that if Z-inactivation occurs in B. mori, one male chZ homolog would be significantly more compact than the other copy by DNA FISH. Visualizing chZ by FISH would also reveal whether, instead, both chZ homologs in males are similar in size but are more compact than the single chZ in females, indicating partial repression of both chZ copies in males. To this end, we used Oligopaints for the B. mori chZ that were similar in design to those used to label autosomes (61). As a control, we used ch23, which is a similar genomic size and gene density to chZ (Fig. 3A and SI Appendix, Table S1). We examined chZ organization at three distinct developmental timepoints: a mid-embryonic stage (before DC is complete), a late-embryonic stage (when complete DC is achieved), and a late larval stage (fifth instar, at which DC should be stable) (48). At each of these time points, we measured CT volume to look for differences between the male and female chZ relative to ch23. CT volume measurements before and after DC is complete revealed that total chZ volume per nucleus in males is less than that of total ch23 volume, with this difference being statistically significant only after DC is complete (Fig. 3B). After DC is complete in late male embryos, both chZ copies together occupy only ∼2% of the nuclear volume, while ch23 occupies ∼3% (Fig. 3B and SI Appendix, Fig. S2B). Instead, chZ is more similar in volume to smaller ch16, which occupies ∼2.5% of the nucleus (Fig. 3C and SI Appendix, Fig. S2B). This significantly reduced volume of chZ compared to the autosomes suggests that chZ is more compact than expected for a chromosome of its size and gene density in males. Additionally, both before and after DC is complete in embryos, the total volume of both chZ copies in males is only 1.6× greater than the volume of the single chZ in females (not 2× greater), suggesting that chZ in females is not as compact as the chZs in males (Fig. 3B). However, chZ in females is still slightly smaller than expected compared to the autosomes (SI Appendix, Fig. S2B). This result is highly reminiscent of FISH-based assays in C. elegans showing that each chX in hermaphrodite worms is more compact than the single chX in males (25, 28).
Fig. 3.
Male Z chromosomes are similar in size and shape, suggesting partial repression of both homologs. (A, Left) Schematic of whole chromosome Oligopaints. (Right) Representative FISH images showing B. mori CTs for chZ, 16, and 23. Nucleus from male late embryo (ZZ, Upper) and nucleus from female late embryo (ZW, Lower) are shown. (Scale bar, 5 µm.) (Right) Three-dimensional rendering of cells. (B) Quantification of total CT volume per nucleus at different developmental stages. Each dot represents the average of a biological replicate (embryo or larva). Bars represent average between biological replicates, and error bars represent SD. Below, average ch23 or chZM value is normalized to average ZF value. For all data in A and B, at least 100 cells were analyzed per replicate. Error bars represent SEM. ZF = chZ in female nuclei; ZM = chZ in male nuclei. Statistics: unpaired t tests. *P < 0.05, **P < 0.01, ***P < 0.001. (C) Quantification of chZM (20.7 Mb) total volume compared to ch16 (14.3 Mb) volume in late embryos. Bars represent average CT volume as a fraction of nuclear volume between replicates. Error bars represent SEM. (D) Tukey box and whisker plot showing quantification of chromosome shape (compacity; y axis) for individual chromosomes at the indicated time points. Midline: median. Statistics: Welch’s t tests. **P < 0.01. Blue dashed lines indicate the shape range of ch23. (E, Upper) Three-dimensional rendering of male nuclei showing ch23 (blue) or chZ (pink) at indicated developmental time points. (Lower) Quantification of interhomolog differences in volume (µm3) for ch23 (blue) or chZ (pink) in male embryos or larvae. Midline: mean. Statistics: Mann–Whitney U test. ***P < 0.0001. (F) Quantification of interhomolog differences in shape as measured by compacity (ratio between volume and surface area of object, where 1 = perfect sphere) for ch23 (blue) or chZ (pink) in male embryos or larvae. Midline: mean. Statistics: Mann–Whitney U test. ***P < 0.0001. (G, Upper) Schematic of stripe Oligopaints for ch23 and chZ; 3-Mb domains are painted at both telomeres (cyan, stripe 1; red, stripe 3) and a 1.5-Mb domain is painted in the middle of the chromosome (yellow, stripe 2). (Lower) Representative 3D renderings of male nuclei with stripe paints for chZ (Left) and ch23 (Right). (H) Quantification of chromosome folding for ch23 (blue) and chZ (pink) in embryos and larvae. Bars represent the average of at least three biological replicates with at least 100 cells analyzed from each replicate. Error bars represent SD. *P = 0.01; Multiple t tests. (I) Quantification of the fraction of cells with both homologs for either ch23 (blue) or chZ (pink) folded in the same configuration based on stripe paint assay. Midline: mean. Statistics: unpaired t test. Late embryos **P = 0.003; larval heads **P = 0.005.
We next measured the volume of chZ compared to ch23 in fifth-instar larvae. Interestingly, by this stage, we found that the total volume of chZ is equal to that of ch23 in males, and the volume of the single chZ in females is half this volume (Fig. 3 B, Right), indicating that similar levels of compaction are occurring for chZ and autosomes in both males and females by this developmental stage (when DC should be stable) (48). However, the hypercompaction of male chZ is not entirely lost by the larval stage. We next measured the shape of the chromosomes, which can also be a metric for compaction as chromosomes become more linear and less spherical when they decondense (5). We observed that male chZ is significantly more spherical at the larval stage than in late embryos, while ch23 remains largely unchanged (Fig. 3D). This finding suggests that male chZ is still compact compared to ch23 at the larval stage, but a different mechanism of compaction may be occurring at this later time point.
Both Male chZ Homologs Are Similar in Size, Shape, and Folding Configuration.
While our measurements of total chZ volume per nucleus support the repression of Z-linked genes in male late embryos, whether both chZ copies are partially down-regulated or one chZ is completely inactivated in males is still unclear. To address this question, we measured the differences in volume and shape between the two chZ homologs and between the two ch23 homologs within individual male nuclei (defined here as intranuclear difference). If both male chZs are partially and equally repressed, we would expect both chZ copies to be similar in volume and shape. However, if one chZ is inactivated, the two chZ copies would be different in volume and shape while the two ch23 copies would be more similar to each other.
Measuring the intranuclear volume difference for chZ and ch23 revealed that in both mid and late embryos, the two chZ copies in males are significantly more similar in volume than the two ch23 copies (Fig. 3E and SI Appendix, Fig. S2C), suggesting that the two chZ copies are similarly compacted. In agreement with this finding, measuring the intranuclear difference in shape illustrated that both chZs are significantly more similar in shape compared to ch23 at both embryonic stages (Fig. 3F and SI Appendix, Fig. S2D). These results suggest that the two chZ homologs are more similar to each other both before and after DC is thought to be complete in mid and late embryos, respectively. Curiously, these differences between chZ and ch23 seem to subside by the larval stage (Fig. 3 E and F and SI Appendix, Fig. S2 C and D), where chZ and ch23 homologs are equally similar, supporting the idea that a distinct compaction mechanism facilitates DC at this developmental stage. Furthermore, at all time points, ch23 shows much higher variance for intranuclear size and shape than chZ, indicating not only higher homolog–homolog variability, but higher cell-to-cell variability for ch23. Similarly high variance was observed for the small, gene-rich ch16 (SI Appendix, Fig. S2E), suggesting that this variability may be exhibited by all autosomes. The finding that the two chZs in males are highly similar in volume and shape after DC is complete supports a model of DC in which expression from both chZs is partially and equally reduced.
To further investigate chZ compaction and organization, we examined the folding configurations of chZ and ch23 after DC is complete in late embryos and larvae by measuring contact patterns between subchromosomal stripes, as previously described for autosomes (Fig. 2). This assay revealed that on average in a population of cells, the chZs in males are more often completely folded (with contact among all three stripes) than ch23 (Fig. 3 G and H). Next, we measured intranuclear folding heterogeneity by quantifying how often the two homologs in the same nucleus were either both unfolded or both folded for chZ and ch23. We reasoned that if both chZ copies are equally repressed, the two homologs would harbor similar folding patterns in most nuclei, whereas if only one chZ is inactivated, there would be higher intranuclear heterogeneity. Indeed, we found that chZ homologs in males are significantly more often in the same configuration compared to ch23, and these results are similar for both time points after DC is complete (Fig. 3I).
Taken together, these data suggest that Z-linked repression in B. mori males is achieved by partially repressing both chZ copies and not by entirely inactivating one copy. This type of DC is reminiscent of DC in C. elegans, despite these two lineages bearing evolutionarily distinct ZW and XY sex chromosomes. Intriguingly, like B. mori and all Lepidoptera, C. elegans harbor holocentric chromosomes in somatic cells. Whether or not this holocentric configuration inhibits the inactivation of an entire chromosome, or perhaps facilitates partial repression of both sex chromosomes in the homogametic sex, remains to be explored. Additionally, whether Condensin-like complexes are involved in DC mechanisms in Bombyx (as in C. elegans DC) is yet to be tested.
Female chZ Undergoes Changes in Nuclear Position at the Early Stages of DC.
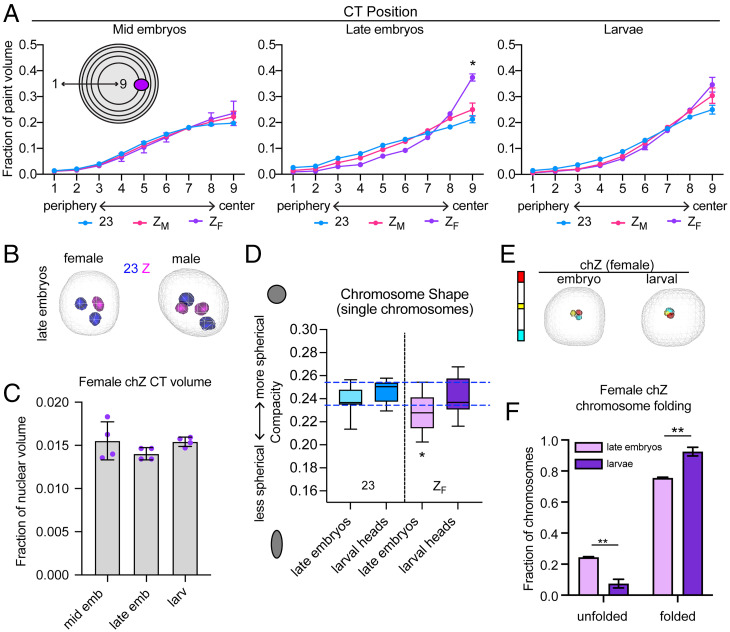
We next wanted to determine whether there are any differences in the nuclear position of chZ in males versus females. To address this question, we measured the position of autosomes and chZ in the nucleus using a shell analysis. In this assay, nuclei are divided into nine concentric shells of equal volume and the total paint volume in each shell is measured (Fig. 4A). Shell analysis of the autosomes revealed that gene-poor chromosomes (like ch23) are preferentially located at the nuclear periphery in late embryos (SI Appendix, Fig. S3A). This observation is in agreement with data on CT position in other species (3–5) and in agreement with the fact that chromatin at the nuclear periphery is broadly repressed (6–10). Interestingly, we also observed that small chromosomes are at the nuclear periphery more often than larger chromosomes, even when they are gene-rich (SI Appendix, Fig. S3A). This finding is distinct from what has been previously shown in humans, where smaller chromosomes tend to be more central in the nucleus (2).
Fig. 4.
Female chZ shifts toward the nuclear center and undergoes changes in shape in late embryos. (A) Quantification of CT position. Nuclei are divided into nine shells of equal volume, with shell 1 being most peripheral and shell 9 being most central. The 3D volume of each chromosome paint in each nuclear shell is quantified. Shown are averages and SE across at least three biological replicates (embryos or larvae), where at least 100 cells were measured per replicate. Statistics: χ2 test. *P = 0.03. (B) Three-dimensional rendering of female (Left) or male (Right) nuclei showing ch23 (blue) or chZ (pink) in late embryos. (C) Bar graph showing chZF volume as a fraction of nuclear volume in mid embryos (Left), late embryos (Center), and larval heads (Right). Each dot represents the average of a biological replicate. (D) Tukey box-and-whisker plot showing quantification of chromosome shape (compacity; y axis) for individual chromosomes at the indicated time points. Midline: median. Statistics: Welch’s t tests. *P < 0.05. Blue dashed lines indicate the shape range of ch23. (E, Left) Schematic of stripe Oligopaints for female chZ. (Right) Three-dimensional rendering of stripe paints in female late embryos or larvae. (F) Quantification of fraction of chZF in unfolded (Left) or folded (Right) configuration in late embryos (lavender) and larvae (purple). Statistics: Fisher’s exact test comparing unfolded versus folded chromosomes in embryos and larvae. **P < 0.01.
We next tested whether chZ is positioned similarly to ch23, which is similar in both size and gene density, and found no significant differences in their nuclear positions in males at any developmental stage (Fig. 4 A and B and SI Appendix, Fig. S3B). Furthermore, we calculated the intranuclear difference in position for the two chZ and ch23 copies in males by measuring the distance of individual homologs from the nuclear periphery. Again, no significant difference was observed between chZ and ch23 (SI Appendix, Fig. S3C). Strikingly, in females, we observed a substantial difference in the nuclear position of chZ relative to ch23. We found that in late embryos, when DC is reportedly complete (48), the female chZ is repositioned to the center of the nucleus (Fig. 4 A and B and SI Appendix, Fig. S3B). This shift toward the nuclear center persists into the larval stage but to a lesser extent. A similar, more modest shift toward the center was observed for chZ in males, but this was not significantly different from ch23, as described earlier in this paragraph. Notably, the single chX in C. elegans males is also preferentially positioned in the nucleus after DC, although in this case the male chX is positioned close to the nuclear periphery and interacts with nuclear pore proteins (28, 65).
Female chZ Is More Linear at the Early Stages of DC.
We wondered whether this shift in position of female chZ to the center of the nucleus corresponds to decompaction and increased gene expression, since the nuclear interior is generally more permissive to active transcription. Additionally, an increase in Z-linked expression in females would directly support Ohno’s hypothesis (33). To test whether changes in chromosome compaction occur for female chZ, we revisited our earlier volume measurements and found no change in volume for chZ in females at any developmental stage (Fig. 4C). However, differences in chromosome shape can alternatively indicate loss of chromatin compaction (5). Thus, we measured the shape of female chZ after DC onset (late embryos and larvae) and compared it to our earlier chromosome shape data from males (Fig. 3D). This analysis revealed that female chZ in late embryos is significantly less spherical than both ch23 and male chZs in late embryos (SI Appendix, Fig. S3D) as well as female chZ in larvae (Fig. 4D). Together, these results suggest that female chZ is decondensed specifically in late embryos, consistent with possible up-regulation of female chZ during this time point.
To further interrogate whether female chZ decondenses in late embryos, we measured chromosome folding of female chZ after DC onset using stripe paints. We found that chZ is significantly more often unfolded in late embryos compared to larvae (Fig. 4 D and E). This unfolded configuration of chZ in late female embryos is consistent with its decompaction, elongation, and shift in nuclear position. Together with our earlier results, these findings suggest that compaction of female chZ and decompaction of male chZ at the onset of DC may be modulated by distinct molecular mechanisms. Male chZ repression is largely facilitated by local compaction and reduction in volume while female decompaction hinges more on changes in folding and long-range cis interactions.
ChZ Chromatin Is More Accessible in Females than in Males, Specifically at the Late Embryonic Stage.
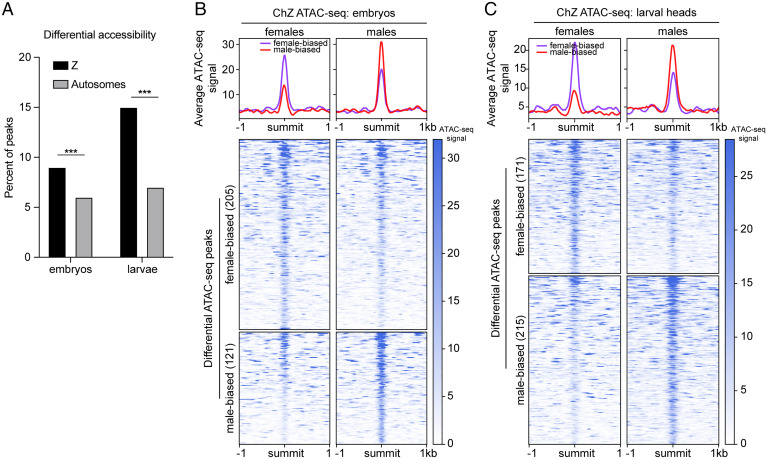
Next, we wanted to determine whether we could detect differences between male and female chZ chromatin at higher resolution after DC is complete. To this end, we performed assay for transposase accessible chromatin using sequencing (ATAC-seq) (66) to measure sex-specific chromatin accessibility at base pair resolution in late embryos and larvae (48). To account for the difference in copy number, chZ was analyzed separately from the autosomes. On the autosomes, we identified 106,235 ATAC peaks in late embryos and 164,727 ATAC peaks in larval heads. Of those, only 5,847 (5.5%) peaks were differentially accessible between male and female late embryos and 11,143 (6.7%) in larvae. On chZ, we identified 3,792 total peaks in late embryos and 3,028 in larvae. Of those, 326 (8.6%) showed differential accessibility between male and female late embryos and 386 (12.7%) in larvae (SI Appendix, Fig. S4). Therefore, a significant difference was observed between chZ and autosomes in the percent of peaks that are differentially accessible in both late embryos and larvae, demonstrating differences in the chromatin landscape specifically on chZ consistent with DC (Fig. 5A).
Fig. 5.
ATAC-seq reveals female chZ is more accessible than male chZ in late embryos. (A) Bar graph showing the percent of ATAC-seq peaks with significant differential accessibility (P < 0.05) between males and females on chZ (black) and autosomes (gray) in embryos (Left) and larvae (Right). Significantly more peaks are differentially accessible on chZ at both time points (***P < 0.0001; two-tailed Fishers exact test comparing embryo Z to A, Left, or larval Z to A, Right). (B and C, Upper) Average ATAC-seq signal across differentially accessible ATAC-seq peaks between males and females on chZ in late embryos (B) and larvae (C). Average male signal is shown in red and female signal is in purple. (Lower) Heatmaps of signal at individual differential peaks in embryos (B) and larvae (C) are shown. The number of differential peaks is indicated in parentheses. Female-biased peaks are shown in the top map and male-biased peaks in the bottom map. Signal in females is shown on the left. Signal in males is shown on the right. Peaks are sorted by decreasing signal.
Further analysis of the 326 differentially accessible peaks on chZ in late embryos revealed that there are significantly more sites with higher accessibility in females than males on chZ (205 female-biased peaks versus 121 male-biased peaks) (Fig. 5B). When analyzing the 386 differential peaks in larvae, we observed a shift toward increased accessibility in males, with slightly more of the differential ATAC-seq peaks being male-biased on chZ (171 female-biased peaks and 215 male-biased peaks) (Fig. 5C). Together, these results support our FISH-based findings that chZ in late female embryos undergoes decompaction and that this decompaction is transient and only occurs during the late embryonic stage.
Changes in Female chZ Chromatin Accessibility and Nuclear Position Correlate with Changes in Z-Linked Transcription.
To determine if the increased accessibility of chZ in late female embryos and the shifted nuclear position by FISH correspond to changes in Z-linked expression, we reanalyzed existing sex-specific RNA-sequencing (RNA-seq) data from B. mori mid embryos (before DC), late embryos (DC established), and larvae (DC stable) (48). Previously, these data were used to conclude that the male chZ is repressed to establish DC by the late embryonic stage (48). Here, we wanted to focus specifically on the female chZ and investigate whether female Z-linked expression is up-regulated when DC is first established in late embryos. In our analysis, we included both genic and intergenic transcripts, and any loci that were not expressed were excluded (defined as fragments per kilobase of transcript per million mapped reads [FPKM] < 0.01) (Methods).
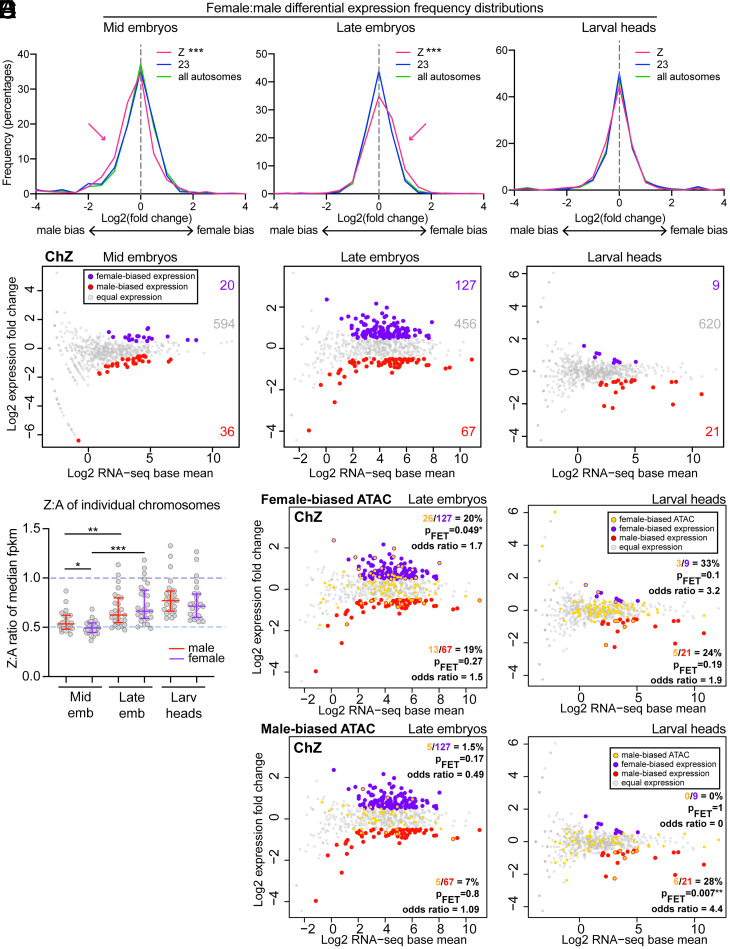
In agreement with the previous analyses of these data, we find that overall, chZ is repressed in males, resulting in approximately equal Z-linked transcription between males and females. This DC between male and female chZ expression is not yet complete in mid embryos, where we find significantly male-biased expression on chZ (but not ch23 or other autosomes) (Fig. 6 A and B and SI Appendix, Figs. S5–S9). However, median chZ expression in both males and females is approximately half the median autosomal expression at this stage, resulting in median Z:A ratios of 0.53 and 0.49 in males and females, respectively (Fig. 6C and SI Appendix, Fig. S5). Since females harbor only a single chZ chromosome, a median Z:A ratio of 0.5 is expected. However, males harbor two chZ copies, so a median ratio of 1 would be expected in males if chZ were not repressed. Together, these findings indicate that, while still incomplete, the process of establishing DC has already begun in mid embryos. These findings support our FISH data showing that the two male chZ homologs are already highly similar in size and shape in mid embryos (Fig. 3 D and E).
Fig. 6.
RNA-seq reveals male chZ repression by the mid-embryonic stage and robust up-regulation of female chZ at the late-embryonic stage. (A) Frequency histograms showing male versus female differential RNA-seq signal for all expressed loci (genes + intergenes) on chZ (pink), ch23 (blue), or all autosomes (green): (Left) mid embryos, (Center) late embryos, (Right) larval heads. ***P < 0.0001, Mann–Whitney U test. (B) MA plots showing differential expression on chZ between male and female mid embryos (Left), late embryos (Center), and larval heads (Right). Loci with significantly differential expression [q < 0.01 and fold-change (log2) > ±0.5] are shown in red (male-biased) or purple (female-biased). Numbers of differential or equally expressed loci are indicated to the right. (C) Dot plot showing median Z:A expression (FPKM) per autosome (each dot indicates the ratio for Z versus a single autosome). Midlines are median ratios. Upper and lower bars indicate interquartile ranges. Dashed line at 0.5 indicates reduced expression of male Z compared to autosomes, while dashed line at 1 indicates equal expression of Z and autosomes. *P < 0.05, **P < 0.01, ***P < 0.001; Mann–Whitney U test. (D and E) MA plots labeling genes associated with differential ATAC-seq peaks in yellow on top of RNA-seq results (female-biased ATAC-seq in D, male-biased ATAC-seq in E). MA plots for late embryos are on the left, larval heads on the right. Two-sided Fisher’s exact test was used to calculate P values and odds ratios.
In late embryos, Z-linked transcription increases compared to the autosomes in both males and females, resulting in median Z:A expression ratios of 0.62 in males and 0.66 in females (Fig. 6C and SI Appendix, Fig. S5). This increase in chZ-linked expression is most pronounced in females, resulting in significantly female-biased Z-linked transcription in late embryos despite females having half the allelic copy number (Fig. 6 A–C). This hyperexpression of female chZ is concurrent with the shift of female chZ toward the nuclear interior observed by FISH (Fig. 4A) and in agreement with our ATAC-seq results showing more female-biased accessibility on chZ at this time point (Fig. 5 A and B). Importantly, female-biased expression is limited to chZ and was not observed for ch23 or the autosomes overall (SI Appendix, Figs. S5–S9). While it is possible that this female-biased chZ expression in late embryos could reflect female-specific developmental genes being activated at this time, we do not believe that this is the case since we find that many genes are slightly (but significantly) female-biased rather than a few genes being extremely female-biased (Fig. 6 A and B).
At the larval stage, Z-linked expression is not significantly biased between sexes (Fig. 6A), and Z:A expression ratios are closer to 1 (0.77 and 0.71 in males and females, respectively) (Fig. 6C). This finding suggests more equalized expression between male and female chZ and also between chZ and autosomes in larvae. These results are consistent with both our ATAC-seq data showing that chZ accessibility is not substantially sex-biased at this stage (Fig. 5C) and our FISH data showing fewer differences in the 3D shape and size of chZ compared to ch23 in larvae (Fig. 3 B, E, and F).
Taken together, our RNA-seq analyses suggest that Z-linked repression in males may occur earlier than previously appreciated. We observed the most dramatic levels of repression in mid embryos and then a modest but significant increase in Z-linked expression in both males and females throughout development (Fig. 6C and SI Appendix, Figs. S5 and S6). This repression of sex chromosomes in the homogametic sex in conjunction with sex chromosome activation in both males and females directly supports Ohno’s hypothesis of the evolution of DC (33).
Increased Z-Linked Transcription in Late Female Embryos Is Directly Associated with Changes in Chromatin Accessibility.
Finally, we wanted to determine if the genes that are up-regulated in female late embryos by RNA-seq are associated with the female-biased ATAC-seq peaks observed at this time point. To this end, we mapped our differential ATAC-seq peaks to their closest genes (or intergenes) and tested whether these correspond to loci with female-biased expression. In late embryos, genes associated with female-biased ATAC-seq peaks are indeed significantly enriched for female-biased expression (Fig. 6D and SI Appendix, Figs. S10 and S11), whereas this enrichment was not observed for males (Fig. 6E and SI Appendix, Fig. S11). These results suggest a functional relationship between chromatin accessibility and transcription on the female chZ and supports our hypothesis that the female chZ is decompacted and repositioned (Fig. 4) for active gene expression during this stage. On the other hand, in larvae, very few differentially expressed genes were detected on chZ between males and females. Nevertheless, we found that male-biased accessibility is enriched for male-biased expression on chZ in larvae (Fig. 6E). These few genes with male-biased expression in larvae may be indicative of high levels of spermatogenesis occurring at this developmental stage, since the Z chromosome is enriched for genes involved in this pathway (67). Altogether, these results support a model in which female chZ is transiently decompacted and up-regulated in late embryos.
Conclusions
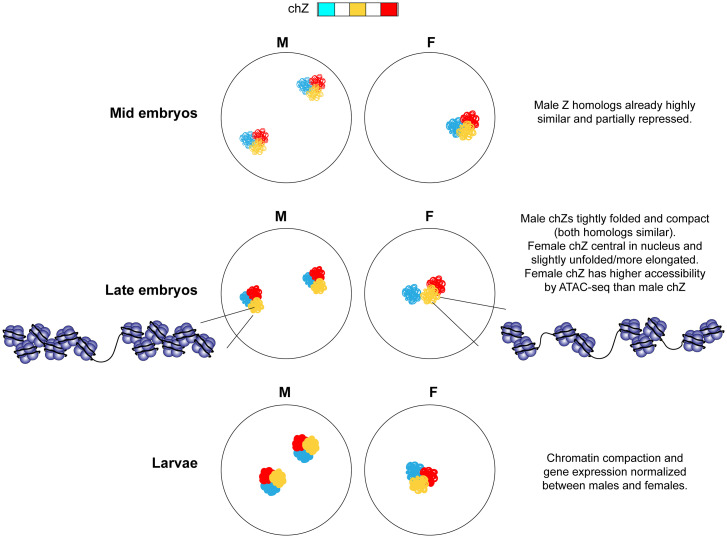
In this study, we use Oligopaints to visualize both autosomes and chZ in female and male B. mori at different stages of development before and after DC establishment. We show that B. mori nuclei are highly compact overall and that chromosomes are spatially partitioned into distinct CTs. These CTs occupy preferential positions in the nucleus that follow both conserved and divergent rules of organization, with small and gene-poor chromosomes being most peripheral. We show that both chZ copies in male silkworms are similarly condensed and folded, supporting a model where Z-linked expression is normalized by partially and equally repressing both chZ copies in males. By combining these FISH assays with genomics, we demonstrate that in fact, this repression of male chZ likely occurs before the mid-embryonic stage, earlier in development than previously appreciated. In late embryos, we find transcriptional up-regulation of chZ in both sexes, but this up-regulation is more pronounced in females. This up-regulation is concomitant with chZ repositioning in 3D nuclear space and chZ decompaction, observed both at the whole chromosome level by FISH and at higher-resolution ATAC-seq (Fig. 7). These findings directly support Ohno’s hypothesis for the evolution of DC (33). Our Oligopaint FISH results importantly reveal nonsequencing-based evidence in support of this long-standing model, and link increased transcription to changes in the 3D position and organization of chromosomes in Lepidoptera.
Fig. 7.
Summary and model. F, female; M, male.
Our studies revealed striking similarities between DC in moths and C. elegans, despite these two lineages harboring evolutionarily distinct sex chromosomes. Whether the holocentric chromosome structure can explain this possible convergent evolution remains unclear. For example, the large number of centromeres interspersed along the length of the chromosomes in holocentric species might disfavor chromosome-wide up-regulation in the heterogametic sex (as seen in monocentric flies) (68, 69), which could increase the likelihood of aberrant up-regulation of transposable elements and other centromeric-proximal sequences. Additionally, repression of an entire chromosome in the homogametic sex (such as in mammalian X-inactivation) could be incompatible with the holocentric chromosome structure, as many more loci need to remain accessible for kinetochore formation during cell division than in a monocentric species. Finally, while DC studies in other holocentric species have suggested that histone modifications (63) or specialized Condensin complexes (70) can regulate sex chromosome-linked expression, the molecular mechanisms regulating DC and overall nuclear organization in B. mori remain subjects for future interrogation.
Methods
B. mori Strains.
Moth embryos were obtained from Carolina Biological, Coastal Silkworms, Mulberry Farms, or were freshly laid in the laboratory from moths derived from embryos from these sources. Some larvae were obtained from Rainbow Mealworms. Embryos were kept in diapause at 4 °C for 3 mo to 1 y. For rearing, embryos were transferred to 25–28 °C, and larvae were fed fresh mulberry leaves or powdered mulberry chow (Carolina Biological).
B. mori Staging and Sexing.
B. mori embryonic and larval stages were defined as follows: mid embryos = during embryonic diapause (at 4 °C less than 1 mo); late embryos = postdiapause (at 25–28 °C 7 to 10 d after removal from 4 °C, before hatching); larvae = fifth instar (∼3 in long and after cessation of eating). Single embryos or single larval heads were used for slides involving Z chromosome analyses, and the number of Z chromosomes was used to sex the embryos. Larvae were sexed during dissection based on gonad morphology. For details on slide preparation, see SI Appendix, Extended Methods.
Drosophila Nuclear Analyses and Oligopaint Data.
Fly images used for analyses of CTs and chromosome folding were from previously published studies (5, 71). For nuclei analysis in pupal neurons, pupal brains were dissected, and neurons were isolated by FACS as previously described (72). Neurons were then settled on Poly-l-lysine–coated slides for 30 min in Schneider’s media, fixed with 4% PFA for 10 min, washed with PBS, stained with DAPI DNA stain, and mounted with Prolong Diamond (Invitrogen). Methods for nuclear analyses for other species can be found in the SI Appendix, Extended Methods.
B. mori Oligopaint Design and Synthesis.
Oligopaint libraries were previously published (61) and designed as previously described using the Oligominer pipeline (73). Coordinates and chromosome information for all paints can be found in SI Appendix, Tables S1 and S2. Information on paint multiplexing can be found in SI Appendix, Extended Methods.
Oligopaint DNA FISH in B. mori Cells.
Oligopaint synthesis and FISH were performed as previously described (5, 74). Briefly, after fixation slides were washed in 0.1% Triton X-100 in PBS (PBS-T0.1%) then permeabilized with PBS-T0.5%. Cells were predenatured in 2× SSCT/50% formamide at 92 °C for 2 min and then at 60 °C for 20 min. Primary Oligopaint probes in hybridization buffer were then added to slides and sealed under a coverslip before being denatured at 92 °C for 2.5 min. Slides were incubated overnight at 37 °C. Slides were then washing in 2× SSCT before applying secondary probes containing fluorophores. Slides were incubated at 37 °C for 2 h before washing with 2× SSCT, staining with DAPI, and mounting. A detailed version of the FISH protocol can be found in the SI Appendix, Extended Methods.
Imaging, Quantification, and Data Analysis.
Images were acquired on a Leica DMi6000 widefield fluorescence microscope using an HCX PL APO 63×/1.40–0.60 Oil objective or HCX PL APO 100×/1.40–0.70 Oil objective (Leica), and Leica DFC9000 sCMOS Monochrome Camera. DAPI, CY3, CY5, and FITC filter cubes were used for image acquisition. All images were processed using the LasX software and Huygens deconvolution software, and tiffs were created in ImageJ. For quantification, deconvolved images were segmented and measured using a modified version of the TANGO 3D-segmentation plug-in for ImageJ (75), using either the ‘Hysteresis’ or ‘Spot Detector 3D’ algorithms. Statistical analyses were performed using Prism 9 software by GraphPad. Figures were assembled in Adobe Illustrator.
RNA-Seq Analysis.
Raw RNA-seq data were downloaded from BioProjectID: PRJNA388026. Briefly, adaptors and low-quality bases were trimmed, the filtered reads were mapped to an updated version of the Ensembl (2013) B. mori (ASM15162v1) reference genome using bowtie v2.4.1 (76). The GTF file was downloaded from Silkbase (2016 gene models). Raw read counts were normalized by FPKM. Genes that were not expressed in any samples (raw FPKM < 0.01 in all samples) were removed from analyses. Differential analyses were performed using edgeR (77). More detailed methods can be found in SI Appendix, Extended Methods.
ATAC-Seq Sample Preparation and Data Analysis.
ATAC-seq was performed using the ATAC-seq Kit from Active Motif (cat#53150) according to the manufacturer’s instructions. ATAC-seq libraries were sequenced using the NovaSeq 6000 SP, PE 150 bp with 300 cycles (Illumina) at the National Heart, Lung, and Blood Institute Sequencing and Genomics core, and analyzed as previously described (78). Briefly, sequencing reads were trimmed for adapters and aligned to an updated version of the Ensembl (2013) B. mori (ASM15162v1) reference genome with bowtie2 (76). For proper normalization, reads from chZ and autosomes were analyzed separately. Peaks were called using MACS2 (79), and differential accessibility was called with the R package DiffBind v2.6.6 (80) and associated with the closest gene to generate MA plots integrating RNA-seq data. The ATAC-seq data have been deposited in the Gene Expression Omnibus (GEO) database under accession number GSE191164. Further details on ATAC-seq methodology can be found in SI Appendix, Extended Methods.
Supplementary Material
Acknowledgments
We thank the McJunkin and Kimmel laboratories for providing C. elegans and Dictyostelium, respectively; members of the E.P.L., Drinnenberg, and Joyce laboratories, as well as Jamie Walters, Brian Oliver, and Jeannie Lee for critical reading of the manuscript and helpful discussions; Catherine McManus for assistance with rearing and staging C. elegans; and Saumya Keremane for assistance preparing mouse eye slides. This work was funded by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH (Grant DK015602 to E.P.L.) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (Grant 1K99HD104851 to L.F.R). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113374119/-/DCSupplemental.
Data Availability
The ATAC-seq data have been deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo), accession number GSE191164 (81). All other study data are included in the main text and/or SI Appendix. Previously published data were used for this work (48).
References
- 1.Cremer T., Cremer M., Chromosome territories. Cold Spring Harb. Perspect. Biol. 2, a003889 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun H. B., Shen J., Yokota H., Size-dependent positioning of human chromosomes in interphase nuclei. Biophys. J. 79, 184–190 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritz A. J., Sehgal N., Pliss A., Xu J., Berezney R., Chromosome territories and the global regulation of the genome. Genes Chromosomes Cancer 58, 407–426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parada L., Misteli T., Chromosome positioning in the interphase nucleus. Trends Cell Biol. 12, 425–432 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Rosin L. F., Nguyen S. C., Joyce E. F., Condensin II drives large-scale folding and spatial partitioning of interphase chromosomes in Drosophila nuclei. PLoS Genet. 14, e1007393 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guelen L., et al. , Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Pickersgill H., et al. , Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38, 1005–1014 (2006). [DOI] [PubMed] [Google Scholar]
- 8.van Steensel B., Belmont A. S., Lamina-associated domains: Links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Steensel B., Henikoff S., Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 18, 424–428 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Towbin B. D., et al. , Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150, 934–947 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Briand N., Collas P., Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 21, 85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vigouroux C., et al. , Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J. Cell Sci. 114, 4459–4468 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Worman H. J., Nuclear lamins and laminopathies. J. Pathol. 226, 316–325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrai C., de Castro I. J., Lavitas L., Chotalia M., Pombo A., Gene positioning. Cold Spring Harb. Perspect. Biol. 2, a000588 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blackwood E. M., Kadonaga J. T., Going the distance: A current view of enhancer action. Science 281, 60–63 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Chepelev I., Wei G., Wangsa D., Tang Q., Zhao K., Characterization of genome-wide enhancer-promoter interactions reveals co-expression of interacting genes and modes of higher order chromatin organization. Cell Res. 22, 490–503 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciabrelli F., Cavalli G., Chromatin-driven behavior of topologically associating domains. J. Mol. Biol. 427, 608–625 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Cullen K. E., Kladde M. P., Seyfred M. A., Interaction between transcription regulatory regions of prolactin chromatin. Science 261, 203–206 (1993). [DOI] [PubMed] [Google Scholar]
- 19.Kleinjan D. A., van Heyningen V., Long-range control of gene expression: Emerging mechanisms and disruption in disease. Am. J. Hum. Genet. 76, 8–32 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu L., Walters J. R., Knipple D. C., Conserved patterns of sex chromosome dosage compensation in the lepidoptera (WZ/ZZ): Insights from a moth neo-Z chromosome. Genome Biol. Evol. 9, 802–816 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marín I., Siegal M. L., Baker B. S., The evolution of dosage-compensation mechanisms. BioEssays 22, 1106–1114 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Jordan W. III, Rieder L. E., Larschan E., Diverse genome topologies characterize dosage compensation across species. Trends Genet. 35, 308–315 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ercan S., Mechanisms of X chromosome dosage compensation. J Genomics 3, 1–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimaud C., Becker P. B., The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes Dev. 23, 2490–2495 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau A. C., Nabeshima K., Csankovszki G., The C. elegans dosage compensation complex mediates interphase X chromosome compaction. Epigenetics Chromatin 7, 31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Splinter E., et al. , The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 25, 1371–1383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow J. C., Heard E., Nuclear organization and dosage compensation. Cold Spring Harb. Perspect. Biol. 2, a000604 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma R., et al. , Differential spatial and structural organization of the X chromosome underlies dosage compensation in C. elegans. Genes Dev. 28, 2591–2596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyon M. F., Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190, 372–373 (1961). [DOI] [PubMed] [Google Scholar]
- 30.Ohno S., Kaplan W. D., Kinosita R., Somatic association of the positively heteropycnotic X-chromosomes in female mice (Mus musculus). Exp. Cell Res. 15, 616–618 (1958). [DOI] [PubMed] [Google Scholar]
- 31.Giorgetti L., et al. , Structural organization of the inactive X chromosome in the mouse. Nature 535, 575–579 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barr M. L., Bertram E. G., A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163, 676–677 (1949). [DOI] [PubMed] [Google Scholar]
- 33.Ohno S., Sex Chromosomes and Sex-Linked Genes (Springer Science & Business Media, 1967). [Google Scholar]
- 34.Albritton S. E., et al. , Sex-biased gene expression and evolution of the X chromosome in nematodes. Genetics 197, 865–883 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng X., et al. , Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat. Genet. 43, 1179–1185 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng X., Berletch J. B., Nguyen D. K., Disteche C. M., X chromosome regulation: Diverse patterns in development, tissues and disease. Nat. Rev. Genet. 15, 367–378 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veitia R. A., Veyrunes F., Bottani S., Birchler J. A., X chromosome inactivation and active X upregulation in therian mammals: Facts, questions, and hypotheses. J. Mol. Cell Biol. 7, 2–11 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Wheeler B. S., et al. , Chromosome-wide mechanisms to decouple gene expression from gene dose during sex-chromosome evolution. eLife 5, e17365 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ellegren H., et al. , Faced with inequality: Chicken do not have a general dosage compensation of sex-linked genes. BMC Biol. 5, 40 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itoh Y., et al. , Dosage compensation is less effective in birds than in mammals. J. Biol. 6, 2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh Y., et al. , Sex bias and dosage compensation in the zebra finch versus chicken genomes: General and specialized patterns among birds. Genome Res. 20, 512–518 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vicoso B., Bachtrog D., Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol. Evol. 3, 230–235 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolf J. B., Bryk J., General lack of global dosage compensation in ZZ/ZW systems? Broadening the perspective with RNA-seq. BMC Genomics 12, 91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Julien P., et al. , Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 10, e1001328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uebbing S., Künstner A., Mäkinen H., Ellegren H., Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome Biol. Evol. 5, 1555–1566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vicoso B., Emerson J. J., Zektser Y., Mahajan S., Bachtrog D., Comparative sex chromosome genomics in snakes: Differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11, e1001643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen S., et al. , Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46, 253–260 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Gopinath G., Srikeerthana K., Tomar A., Sekhar S. M. C., Arunkumar K. P., RNA sequencing reveals a complete but an unconventional type of dosage compensation in the domestic silkworm Bombyx mori. R. Soc. Open Sci. 4, 170261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huylmans A. K., Macon A., Vicoso B., Global dosage compensation is ubiquitous in lepidoptera, but counteracted by the masculinization of the Z chromosome. Mol. Biol. Evol. 34, 2637–2649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kiuchi T., et al. , A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 509, 633–636 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Smith G., Chen Y.-R., Blissard G. W., Briscoe A. D., Complete dosage compensation and sex-biased gene expression in the moth Manduca sexta. Genome Biol. Evol. 6, 526–537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walters J. R., Hardcastle T. J., Getting a full dose? Reconsidering sex chromosome dosage compensation in the silkworm, Bombyx mori. Genome Biol. Evol. 3, 491–504 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walters J. R., Hardcastle T. J., Jiggins C. D., Sex chromosome dosage compensation in heliconius butterflies: Global yet still incomplete? Genome Biol. Evol. 7, 2545–2559 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer B. J., Casson L. P., Caenorhabditis elegans compensates for the difference in X chromosome dosage between the sexes by regulating transcript levels. Cell 47, 871–881 (1986). [DOI] [PubMed] [Google Scholar]
- 55.Ercan S., Lieb J. D., C. elegans dosage compensation: A window into mechanisms of domain-scale gene regulation. Chromosome Res. 17, 215–227 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Gupta V., et al. , Global analysis of X-chromosome dosage compensation. J. Biol. 5, 3 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mita K., et al. , The genome sequence of silkworm, Bombyx mori. DNA Res. 11, 27–35 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Xia Q., et al. ; Biology Analysis Group, A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306, 1937–1940 (2004). [DOI] [PubMed] [Google Scholar]
- 59.King T. D., et al. , Recurrent losses and rapid evolution of the condensin II complex in insects. Mol. Biol. Evol. 36, 2195–2204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawamoto M., et al. , High-quality genome assembly of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 107, 53–62 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Rosin L. F., Gil Jr. J., Drinnenberg I. A., Lei E. P., Oligopaint DNA FISH reveals telomere-based meiotic pairing dynamics in the silkworm, Bombyx mori. PLoS Genetics 17, e1009700 (2021). [DOI] [PMC free article] [PubMed]
- 62.Finn E. H., et al. , Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176, 1502–1515.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gu L., et al. , Dichotomy of dosage compensation along the neo Z chromosome of the monarch butterfly. Curr. Biol. 29, 4071–4077.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Senaratne A. P., et al. , Formation of the CenH3-deficient holocentromere in Lepidoptera avoids active chromatin. Curr. Biol. 31, 173–181.e7 (2020). [DOI] [PubMed] [Google Scholar]
- 65.Sharma R., Meister P., Linking dosage compensation and X chromosome nuclear organization in C. elegans. Nucleus 6, 266–272 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buenrostro J., Wu B., Chang H., Greenleaf W., ATAC-seq: A method for assaying chromatin accessibility genome-wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arunkumar K. P., Mita K., Nagaraju J., The silkworm Z chromosome is enriched in testis-specific genes. Genetics 182, 493–501 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mukherjee A. S., Beermann W., Synthesis of ribonucleic acid by the X-chromosomes of Drosophila melanogaster and the problem of dosage compensation. Nature 207, 785–786 (1965). [DOI] [PubMed] [Google Scholar]
- 69.Park Y., Kuroda M. I., Epigenetic aspects of X-chromosome dosage compensation. Science 293, 1083–1085 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Chuang P. T., Albertson D. G., Meyer B. J., DPY-27: A chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell 79, 459–474 (1994). [DOI] [PubMed] [Google Scholar]
- 71.Bag I., et al. , M1BP cooperates with CP190 to activate transcription at TAD borders and promote chromatin insulator activity. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/2020.10.27.357533v4. [DOI] [PMC free article] [PubMed]
- 72.Chen D., Dale R. K., Lei E. P., Shep regulates Drosophila neuronal remodeling by controlling transcription of its chromatin targets. Development 145, dev154047 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Beliveau B. J., et al. , OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl. Acad. Sci. U.S.A. 115, E2183–E2192 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen S. C., Joyce E. F., Programmable chromosome painting with oligopaints. Methods Mol. Biol. 2038, 167–180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ollion J., Cochennec J., Loll F., Escudé C., Boudier T., TANGO: A generic tool for high-throughput 3D image analysis for studying nuclear organization. Bioinformatics 29, 1840–1841 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Langmead B., Salzberg S., Fast gapped-read alignment with Bowtie 2. Nat. Meth, 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen D., McManus C. E., Radmanesh B., Matzat L. H., Lei E. P., Temporal inhibition of chromatin looping and enhancer accessibility during neuronal remodeling. Nat. Commun. 12, 6366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stark R., Brown G., DiffBind: Differential binding analysis of ChIP-Seq peak data. R package version 100. http://bioconductor.org/packages/release/bioc/vignettes/DiffBind/inst/doc/DiffBind.pdf (2011).
- 81.L. F. Rosin, D. Chen, Y. Chen, E. P. Lei, Sex-specific ATAC-seq data from B. mori embryos and larvae. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE191164. Deposited 17 December 2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ATAC-seq data have been deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo), accession number GSE191164 (81). All other study data are included in the main text and/or SI Appendix. Previously published data were used for this work (48).