Abstract
Computational toxicology is central to the current transformation occurring in toxicology and chemical risk assessment. There is a need for more efficient use of existing data to characterize human toxicological response data for environmental chemicals in the US and Europe. The Adverse Outcome Pathway (AOP) framework helps to organize existing mechanistic information and contributes to what is currently being described as New Approach Methodologies (NAMs). AOP knowledge and data are currently submitted directly by users and stored in the AOP-Wiki (https://aopwiki.org/). Automatic and systematic parsing of AOP-Wiki data is challenging, so we have created the EPA Adverse Outcome Pathway Database. The AOP-DB, developed by the US EPA to assist in the biological and mechanistic characterization of AOP data, provides a broad, systems-level overview of the biological context of AOPs. Here we describe the recent semantic mapping efforts for the AOP-DB, and how this process facilitates the integration of AOP-DB data with other toxicologically relevant datasets through a use case example.
Keywords: semantic web, adverse outcome pathway, toxcast assays, disease, pathway, ontological mapping
Introduction
There is a need for more efficient use of existing data through improved data integration and compatibility of data structures to characterize human toxicological response data for environmental chemicals. Assessors in the US are moving towards the use of existing mechanistic data (in vitro and in silico) that provide insights into adverse outcomes in humans (National Research Council (NRC), 2007; National Research Council (NRC), 2009; National Research Council (NRC), 2010; (National Research Council (NRC), 2017), and reduced animal testing (Wheeler, 2019). The Adverse Outcome Pathway (AOP) framework helps to organize existing mechanistic information and contributes to what is currently being described as New Approach Methodologies (NAMs) (Thomas et al., 2019). The US EPA Adverse Outcome Pathway-Database (AOP-DB) is a decision support tool for risk assessors, developed by the EPA’s Center for Public Health and Environmental Assessment, which contributes to NAMs (e.g., computational toxicology tools) used for the Toxic Substances Control Act (Public Law 114–182, 2016). The AOP-DB has been made available through the Office of Science Management as a public EPA database since November 2021. Pertinent AOP-DB data is currently integrated with the CompTox Chemicals Dashboard (https://comptox.epa.gov/dashboard/chemical_lists/AOPSTRESSORS), which maps the Distributed Structure-Searchable Toxicity records to the most current list of AOP-DB stressors.
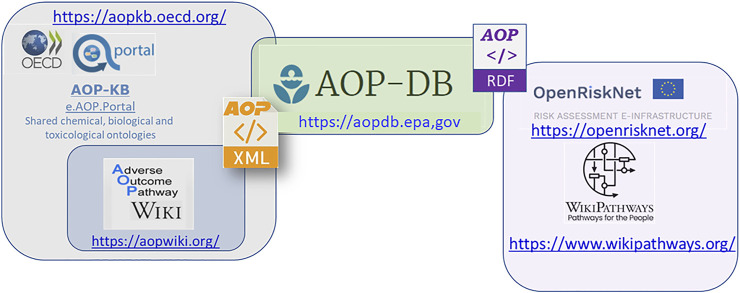
The AOP-DB integrates AOP content to help users characterize AOPs from the OECD-funded AOP-KB (https://aopkb.oecd.org/index.html) effort, where the AOP-Wiki (https://aopwiki.org/) is the primary repository for direct user submission of AOP information to the AOP-KB. Because the AOP-Wiki data is challenging to parse in its current format (Ives et al., 2017; Martens et al., 2018), the AOP-DB was developed to assist in automating and organizing AOP data, as well as integrating with publicly available datasets to allow biological and mechanistic characterization of AOPs and provide a systems-level overview of the biological context of AOPs (Mortensen et al., 2018; Pittman et al., 2018). Recent updates to AOP-DB in version 2 (Mortensen, 2021; Mortensen et al., 2021) include 280 AOPs (1,111 kEs) from the AOP-Wiki XML. The semantic mapping of AOP-DB data, described herein, extends AOP capabilities to users through the incorporation of the Research Description Framework (RDF), which creates additional ontological linkages and improves capabilities for computational analyses (Figure 1). These tools are useful to AOP users trying to retrieve information for AOP development or to understand and characterize existing AOPs. Here we describe the recent semantic mapping efforts for the AOP-DB, and how this process integrates AOP-DB data with other toxicologically relevant datasets.
FIGURE 1.
The OECD funded AOP-KB currently support the AOP-Wiki. The EPA AOP-DB, currently slated as a third-party tool for integration with the AOPKB 2.0, automatically and programmatically pulls AOP data from the AOP-KB XML, and extends AOP capabilities to users with semantic resources like WikiPathways and the OpenRiskNet e-infrastructure that incorporate the Research Description Framework (RDF). Integration of data across the AOP-KB (AOP-Wiki), AOP-DB, and expanding research frameworks through WikiPathways and the EU funded OpenRiskNet, creates additional ontological linkages and improves capabilities for computational analyses. These tools are useful to AOP users trying to retrieve information for AOP development, as well as those trying to understand and characterize existing AOPs.
As part of OpenRiskNet, a 3 years project supported by the European Commission within Horizon2020 EINFRA-22-2016 Programme, the US EPA AOP-DB was selected as an Implementation Challenge winner. The Implementation Challenge was created to select external tools for use in risk assessment to be prioritized for integration in the OpenRiskNet e-Infrastructure (https://openrisknet.org/) and foster collaborative interaction between project partners. In contribution to this effort, US EPA and Maastricht University project partners have completed the semantic mapping of several AOP-DB data tables into RDF, which is a standard model for data interchange (W3C, 2014). The application of RDF defines relationships between data objects using triplestores that include three positional statements (subject, predicate and object). The mapping of AOP-DB data to the RDF data model stores relevant AOP information in a computer-readable format, and contributes to the identification, disambiguation, and meaningful linkage of AOP data with other data structures, following FAIR (findable, accessible, interoperable, and reusable) principles (Wilkinson et al., 2019a; Wilkinson et al., 2019b).
Materials and Methods
We selected seven AOP-DB data tables for semantic integration, specifically the Gene Interaction, Biological Pathway, Toxcast Assay, Taxonomy, Chemical-Gene, Gene Info, and Key Event tables. In developing the AOP-DB RDF, we implemented the most recent version of the SQL AOP-DB (Mortensen, 2020) to map each table of interest into RDF triples. Each table was filtered using the R version 3.6 and Rstudio version 1.2.83 (R Core Team, 2020) to include only records involving a molecular initiating event (MIE) or key event (KE) that maps to a molecular identifier (e.g., gene, protein, cytokine). Code was developed to implement each record as input, modify and filter the AOP-DB table data, and output each modified record to an RDF triple. Additionally, subjects were created for Ensembl and UniProt identifiers. Ontology terms were referenced using BioPortal (Whetzel et al., 2011) in order to find the most appropriate ontology terms for each entity, in line with the AOP-Wiki RDF (Martens et al., 2021a) for optimal interoperability between the two resources. Terms were selected with the most accurate description from ontologies that are relevant to the context of the field. For the development of the AOP-DB RDF, several ontologies and consistent vocabularies have been included. Furthermore, publicly available datasets included in the AOP-DB for RDF mapping are described in detail in Mortensen et al. (2021). Table 1 provides an overview of the included ontologies and database links, including their prefix in the RDF and their corresponding Internationalized Resource Identifier (IRI).
TABLE 1.
Overview of ontologies, consistent vocabularies and databases included in the AOP-DB RDF.
Testing the AOP-DB RDF
Using a Jupyter notebook (Jupyterlab version 3.2.5, Python version 3.8.5), the AOP-DB SPARQL endpoint has been tested by executing SPARQL queries, using the SPARQLWrapper Python library (version 1.8.5). SPARQL queries were used to extract statistics of the data, and a federated SPARQL query was constructed to explore the integrative capabilities of the AOP-DB RDF. The Jupyter notebook, SPARQL queries for extracting data counts, and instructions for setting up the AOP-DB SPARQL endpoint are available on https://github.com/BiGCAT-UM/AOP-DB-RDF.
Results
The AOP-DB Semantic Mapping
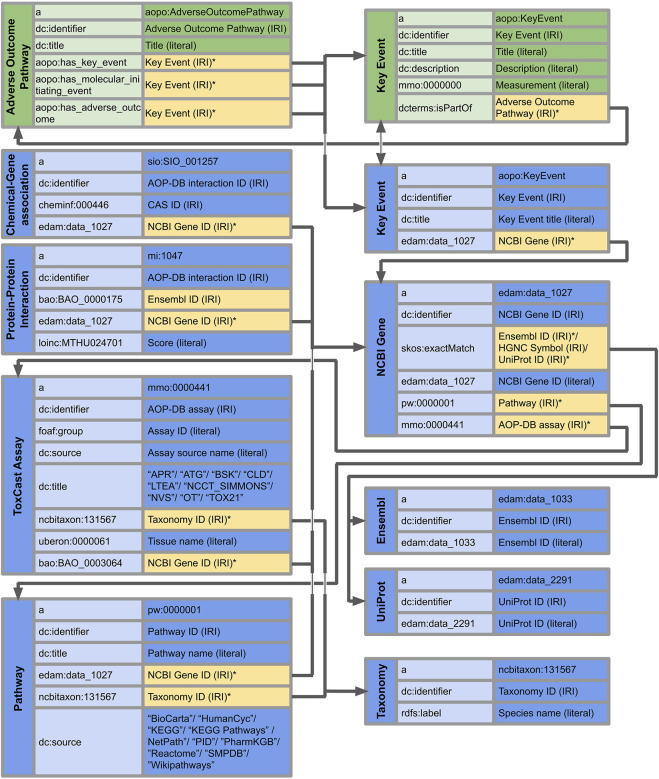
The AOP-DB RDF schema developed according to the methods described above resulted in the primary and secondary table structure, as illustrated in Figure 2. The AOP-DB extends AOP-Wiki RDF with the inclusion of gene/protein, chemical, ToxCast, and biological pathway and taxonomy information. In total, the RDF contains 157 kEs, 376 NCBI genes linked to KEs, 93,449 Chemical-Gene Interactions (3,982 unique chemicals and 122 unique genes), 763,446 Protein-Protein Interactions, 1,143 ToxCast Assays 110,833 Biological Pathways from 10 sources, and 22 taxonomies. Also, the NCBI Gene IDs were matched to 299 Ensembl IDs and 1,026 UniProt IDs. The AOP-DB RDF data tables associate the gene and protein information of AOP genes to chemical, pathway, and assay information organized within the AOP-DB (Mortensen, 2020; Mortensen, 2021).
FIGURE 2.
AOP-DB Semantic Mapping using illustrating the predicates and objects of the nine core subject types in the AOP-DB RDF (in blue). Vertical columns show subjects, and the middle and right columns indicate predicates and objects, respectively. Where applicable, the type of entry is indicated (literal or IRI). Yellow objects with an asterisk (*) indicate the connection between their subjects and the subjects of other tables. The interaction with the AOP-Wiki RDF is highlighted at the Key Events and Adverse Outcome Pathways (in green). Forward slashes indicate the inclusion of multiple objects as part of the subject-predicate-object triple.
The Key Event subjects are linked to NCBI Genes through the ‘data_1,027’ term of the EDAM ontology, which in turn is linked to pathways and assays with respectively the terms ‘pw:0000001’ from the Pathway Ontology and ‘mmo:0000441’ from the Measurement Method Ontology. Furthermore, matching identifiers were linked with ‘skos:exactMatch’, providing IRIs of Ensembl IDs, HGNC Symbols, and UniProt IDs. On the other hand, Chemical-Gene interactions, Protein-protein interactions, ToxCast assays, and Pathways have links to NCBI Gene subjects through the term ‘data_1,027’ from the EDAM ontology. Finally, taxonomy is referenced by ToxCast assay and pathway subjects through the term ‘ncbitaxon:131,567’ indicating cellular organism.
The AOP-DB SPARQL Endpoint
The AOP-DB RDF can be explored through the AOP-DB SPARQL (https://aopdb.rdf.bigcat-bioinformatics.org/sparql). It allows custom SPARQL queries to return output tables in a variety of formats, where it is possible to directly combine different resources with federated SPARQL queries.
AOP-DB RDF Use Case Example
SPARQL queries can be used to query the RDF in order to answer biological and toxicological questions, such as which molecular targets (e.g. genes/proteins), chemical stressors, key events, or in vitro assays are relevant for adverse outcomes of interest. The use case examples provided herein (Supplementary 1) illustrate the utility of the AOP-DB RDF content, as well as the power of integrating these data with other diverse, external databases using federated queries. Our first use case implements the AOP-DB RDF to identify AOP-relevant molecular targets that have associated ToxCast assay targets, which has previously not been possible. The automated linkage of ToxCast assays and KEs in AOP-Wiki can serve as a prioritization tool by exploring the activation of KEs by the many chemicals that have been investigated in ToxCast. The second use case shows the integration of the AOP-DB RDF with other databases that provide access to their data through SPARQL endpoints. A single SPARQL query can be executed to extract AOP IDs, KE IDs, KE titles and protein names from the AOP-Wiki RDF, extract protein descriptions from the Protein Ontology, and the names and descriptions of pathways in WikiPathways, all based on the NCBI Gene IDs captured in the AOP-DB. Through the integration of these diverse data sources, we can effectively explore the data and build automated computational workflows to address questions of toxicological concern.
Conclusion
A central goal of computational toxicology is to predict and explain how the human body responds after exposure to specific xenobiotics or other chemicals in silico. This effort has been hampered by several major limiting factors, including fragmented and poorly structured data, and insufficient access to computational resources and expertise. The AOP-DB RDF and SPARQL endpoint created and discussed herein allow improved access to rigorously structured AOP data and other associated data of toxicological interest. This work improves computational organization and efficiency, through improved data integration, for toxicological and related datasets, and contributes to continued progress in computational toxicology, chemical screening and the improvement of human health risk assessment.
The AOP-DB RDF will be improved with regular data updates and continued data integration with relevant datasets. Future work includes semantic integration of AOP-DB disease-gene data, tissue-specific gene interaction networks, AOP functional single nucleotide polymorphism (SNP) and population SNP frequency information and chemical-specific datasets.
Acknowledgments
The authors gratefully acknowledge Weston Slaughter for his contribution to the queries presented in the supplemental materials, and Dr. Antony Williams for his critical comments of the earlier version of this manuscript. The training of data curators, and necessary travel were all accomplished through the identification of the EPA AOP-DB resource by Douglas Connect GmbH and with the generous contributions to this project by the EU funded OpenRiskNet.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material and are also made available at: https://github.com/BiGCAT-UM/AOP-DB-RDF. Further inquiries can be directed to the corresponding author.
Author Contributions
CTE, HM—Developed EPA AOP-DB and submitted project and data to OpenRiskNet MM—Determined project feasibility, Trained TL and facilitated project JS—modified AOP-DB RDF code TL—wrote initial RDF code for AOP-DB tables CTE Supported project, ELW—Trained TL and facilitated project TE—Supported AOP-DB RDF Research through EU funded OpenRiskNet Project.
EPA Disclaimer
This manuscript has been reviewed by the Center for Public Health and Environmental Assessment, United States Environmental Protection Agency and approved for publication. Approval does not signify that the contents necessarily reflect the views and policies of the Agency nor does mention of trade names or commercial products constitute endorsement or recommendation for use. The authors declare no conflict of interest.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AOP, Adverse Outcome Pathway; AOP-DB, Adverse Outcome Pathway Database; AOP-KB, Adverse Outcome Pathway Knowledgebase; AOP-Wiki, The Collaborative Adverse Outcome Pathway Wiki; FAIR, Findable, Accessible, Interoperable, Reusable; ID, Identifier; IRI, Internationalized Resource Identifier (IRI); KE, Key Event; MIE, Molecular Initiating Event; NCBI; National Center for Biotechnology Information; NRC, National Research Council; NAMs, New Approach Methodologies, OECD, Organisation for Economic Co-operation and Development; RDF, Research Description Framework; SPARQL, SPARQL Protocol and RDF Query Language; SQL, Standard Query Language; ToxCas, tEPA’s Toxicity ForecasterUS EPA, United States Environmental Protection Agency; W3C, World Wide Web Consortium.
References
- Abeyruwan S., Vempati U. D., Küçük-McGinty H., Visser U., Koleti A., Mir A., et al. (2014). Evolving BioAssay Ontology (BAO): Modularization, Integration and Applications. J. Biomed. Semant 5 (Suppl. 1 Proceedings of the Bio-Ontologies Spec Interest G), S5. 10.1186/2041-1480-5-S1-S5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenreider O. (2004). The Unified Medical Language System (UMLS): Integrating Biomedical Terminology. Nucleic Acids Res. 32 (Database issue), 267D–270D. 10.1093/nar/gkh061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braschi B., Denny P., Gray K., Jones T., Seal R., Tweedie S., et al. (2019). Genenames.org: the HGNC and VGNC Resources in 2019. Nucleic Acids Res. 47 (D1), D786–D792. 10.1093/nar/gky930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoon L. (2017). Aop-Ontology. San Francisco, CA: GitHub inc. [Google Scholar]
- Dumontier M., Baker C. J., Baran J., Callahan A., Chepelev L., Cruz-Toledo J., et al. (2014). The Semanticscience Integrated Ontology (SIO) for Biomedical Research and Knowledge Discovery. J. Biomed. Sem 5 (1), 14. 10.1186/2041-1480-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings J., Chepelev L., Willighagen E., Adams N., Steinbeck C., Dumontier M. (2011). The Chemical Information Ontology: Provenance and Disambiguation for Chemical Data on the Biological Semantic Web. PLoS One 6 (10), e25513. 10.1371/journal.pone.0025513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ison J., Kalas M., Jonassen I., Bolser D., Uludag M., McWilliam H., et al. (2013). EDAM: an Ontology of Bioinformatics Operations, Types of Data and Identifiers, Topics and Formats. Bioinformatics 29 (10), 1325–1332. 10.1093/bioinformatics/btt113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ives C., Campia I., Wang R.-L., Wittwehr C., Edwards S. (2017). Creating a Structured Adverse Outcome Pathway Knowledgebase via Ontology-Based Annotations. Appl. Vitro Toxicol. 3 (4), 298–311. 10.1089/aivt.2017.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., et al. (2020). The Reactome Pathway Knowledgebase. Nucleic Acids Res. 48 (D1), D498–D503. 10.1093/nar/gkz1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewison T., Su Y., Disfany F. M., Liang Y., Knox C., Maciejewski A., et al. (2014). SMPDB 2.0: Big Improvements to the Small Molecule Pathway Database. Nucl. Acids Res. 42 (Database issue), D478–D484. 10.1093/nar/gkt1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy K., Mohan S., Raju R., Keerthikumar S., Kumar G. S. S., Venugopal A. K., et al. (2010). NetPath: a Public Resource of Curated Signal Transduction Pathways. Genome Biol. 11 (1), R3. 10.1186/gb-2010-11-1-r3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M. (2021). KEGG: Integrating Viruses and Cellular Organisms. Nucleic Acids Res. 49 (D1), D545–D551. 10.1093/nar/gkaa970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp P. D., Billington R., Caspi R., Fulcher C. A., Latendresse M., Kothari A., et al. (2019). The BioCyc Collection of Microbial Genomes and Metabolic Pathways. Brief Bioinform 20 (4), 1085–1093. 10.1093/bib/bbx085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens M., Ammar A., Riutta A., Waagmeester A., Slenter D. N., Hanspers K., et al. (2021b). WikiPathways: Connecting Communities. Nucleic Acids Res. 49 (D1), D613–D621. 10.1093/nar/gkaa1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens M., Evelo E., Willighagen E. (2021a). Providing Adverse Outcome Pathways from the AOP-Wiki in Semantic Web Format to Increase Usability and Accessibility of the Content. ChemRxiv. Cambridge: Cambridge Open Engage. 10.26434/chemrxiv.13524191.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens M., Verbruggen T., Nymark P., Grafström R., Burgoon L. D., Aladjov H., et al. (2018). Introducing WikiPathways as a Data-Source to Support Adverse Outcome Pathways for Regulatory Risk Assessment of Chemicals and Nanomaterials. Front. Genet. 9, 661. 10.3389/fgene.2018.00661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald C. J., Huff S. M., Suico J. G., Hill G., Leavelle D., Aller R., et al. (2003). LOINC, a Universal Standard for Identifying Laboratory Observations: a 5-year Update. Clin. Chem. 49 (4), 624–633. 10.1373/49.4.624 [DOI] [PubMed] [Google Scholar]
- Millán P. P. (2020). Molecular Interactions Controlled Vocabulary. San Francisco, CA: GitHub inc. [Google Scholar]
- Mortensen H. M. (2021). The EPA Adverse Outcome Pathway Database Version 2.0 (AOP-DB_v2). London United Kingdom: Nature. [Google Scholar]
- Mortensen H. M., Chamberlin J., Joubert B., Angrish M., Sipes N., Lee J. S., et al. (2018). Leveraging Human Genetic and Adverse Outcome Pathway (AOP) Data to Inform Susceptibility in Human Health Risk Assessment. Mamm. Genome 29 (1-2), 190–204. 10.1007/s00335-018-9738-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen H. M. L. T. A., Martens M., Evelo C. T., Willighagen E. L. (2020). Enhancing the EPA Adverse Outcome Pathway Database (AOP-DB): Recent Updates and Sematic Integration The Toxicologist 174 (1) [Google Scholar]
- Mortensen H. M., Senn J., Levey T., Langley P., Williams A. J. (2021). The 2021 Update of the EPA's Adverse Outcome Pathway Database. Sci. Data 8 (1), 169. 10.1038/s41597-021-00962-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall C. J., Torniai C., Gkoutos G. V., Lewis S. E., Haendel M. A. (2012). Uberon, an Integrative Multi-Species Anatomy Ontology. Genome Biol. 13 (1), R5. 10.1186/gb-2012-13-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Research Council (NRC) (2007). Toxicity Testing in the 21st Century. A Vision and a Strategy. Committee on Toxicity Testing and Assessment of Environmental Agents; [Google Scholar]
- National Research Council (NRC) (2009). Science and Decisions. Advancing Risk Assessment [PubMed] [Google Scholar]
- National Research Council (NRC) (2010). Toxicity-Pathway-Based Risk Assessment. Preparing for Paradigm Change. Washington, DC: A Symposium Summary; [PubMed] [Google Scholar]
- National Research Council (NRC) (2017). Using 21st Century Science to Improve Risk-Related Evaluations. 10.17226/24635 [DOI] [PubMed] [Google Scholar]
- Petri V., Jayaraman P., Tutaj M., Hayman G., Smith J. R., De Pons J., et al. (2014). The Pathway Ontology - Updates and Applications. J. Biomed. Sem 5 (1), 7. 10.1186/2041-1480-5-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman M. E., Edwards S. W., Ives C., Mortensen H. M. (2018). AOP-DB: A Database Resource for the Exploration of Adverse Outcome Pathways through Integrated Association Networks. Toxicol. Appl. Pharmacol. 343, 71–83. 10.1016/j.taap.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2020). R: A language and environment for statistical computing. [Google Scholar]
- Schaefer C. F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., et al. (2009). PID: the Pathway Interaction Database. Nucleic Acids Res. 37 (Database issue), D674–D679. 10.1093/nar/gkn653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. R., Park C. A., Nigam R., Laulederkind S. J., Hayman G. T., Wang S.-J., et al. (2013). The Clinical Measurement, Measurement Method and Experimental Condition Ontologies: Expansion, Improvements and New Applications. J. Biomed. Semant 4 (1), 26. 10.1186/2041-1480-4-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R. S., Bahadori T., Buckley T. J., Cowden J., Deisenroth C., Dionisio K. L., et al. (2019). The Next Generation Blueprint of Computational Toxicology at the U.S. Environmental Protection Agency. Toxicol. Sci. 169 (2), 317–332. 10.1093/toxsci/kfz058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium (2019). UniProt: a Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 47 (D1), D506–D515. 10.1093/nar/gky1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- W3C (2014). RDF Schema 1.1. Retrieved from https://www.w3.org/TR/2014/REC-rdf-schema-20140225/ . (Accessed October 2021).
- Wheeler A. R. (2019). Memorandum: Directive to Prioritize Efforts to Reduce Animal Testing. Retrieved from https://www.epa.gov/sites/production/files/2019-09/documents/image2019-09-09-231249.pdf . [Google Scholar]
- Whetzel P. L., Noy N. F., Shah N. H., Alexander P. R., Nyulas C., Tudorache T., et al. (2011). BioPortal: Enhanced Functionality via New Web Services from the National Center for Biomedical Ontology to Access and Use Ontologies in Software Applications. Nucleic Acids Res. 39 (Web Server issue), W541–W545. 10.1093/nar/gkr469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirl‐Carrillo M., Huddart R., Gong L., Sangkuhl K., Thorn C. F., Whaley R., et al. (2021). An Evidence‐Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 110 (3), 563–572. 10.1002/cpt.2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M. D., Dumontier M., Jan Aalbersberg I., Appleton G., Axton M., Baak A., et al. (2019a). Addendum: The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 6 (1), 6. 10.1038/s41597-019-0009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M. D., Dumontier M., Sansone S.-A., Bonino da Silva Santos L. O., Prieto M., Batista D., et al. (2019b). Evaluating FAIR Maturity through a Scalable, Automated, Community-Governed Framework. Sci. Data 6 (1), 174. 10.1038/s41597-019-0184-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A. J., Grulke C. M., Edwards J., McEachran A. D., Mansouri K., Baker N. C., et al. (2017). The CompTox Chemistry Dashboard: a Community Data Resource for Environmental Chemistry. J. Cheminform 9 (1), 61. 10.1186/s13321-017-0247-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A. D., Achuthan P., Akanni W., Allen J., Allen J., Alvarez-Jarreta J., et al. (2020). Ensembl 2020. Nucleic Acids Res. 48 (D1), D682–D688. 10.1093/nar/gkz966 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material and are also made available at: https://github.com/BiGCAT-UM/AOP-DB-RDF. Further inquiries can be directed to the corresponding author.