Abstract
Background and Objectives: Titanium dioxide nanoparticles (TiO2-NP) are important materials used in commercial practice. Reportedly, TiO2-NP exposure during pregnancy can affect the development of the central nervous system in mouse offspring; however, the underlying mechanism remains unknown. In the present study, we investigated the impact of prenatal TiO2-NP exposure on global DNA methylation and mRNA expression patterns in the brains of neonatal mice.
Materials and Methods: Pregnant C57BL/6J mice were intratracheally administered a TiO2-NP suspension (100 μg/mouse) on gestational day 10.5, and brains were collected from male and female offspring at day 1 postpartum. After extraction of methylated DNA by immunoprecipitation, the DNA methylation profile was analyzed using a mouse CpG island microarray. Total RNA was obtained, and mRNA expression profiles were comprehensively assessed using microarray analysis.
Results: Among genes in the CpG island microarray, DNA methylation was increased in 614 and 2,924 genes and decreased in 6,220 and 6,477 genes in male and female offspring, respectively. Combined with mRNA microarray analysis, 88 and 89 genes were upregulated (≥1.5-fold) accompanied by demethylation of CpG islands, whereas 13 and 33 genes were downregulated (≤0.67-fold) accompanied by methylation of CpG islands in male and female offspring mice, respectively. Gene Set Enrichment Analysis (GSEA) revealed that these genes were enriched in gene ontology terms related to the regulation of transcription factors, cell proliferation, and organism development. Additionally, MeSH terms related to stem cells and morphogenesis were enriched.
Conclusion: Prenatal TiO2-NP exposure induced genome-wide alterations in DNA methylation and mRNA expression in the brains of male and female offspring. Based on GSEA findings, it can be speculated that prenatal TiO2-NP exposure causes adverse effects on brain functions by altering the DNA methylation state of the fetal brain, especially neural stem cells, resulting in the subsequent abnormal regulation of transcription factors that modulate development and differentiation.
Keywords: brain, DNA methylation, gene expression, titanium dioxide nanoparticle, prenatal exposure
Introduction
Nanomaterial research and development have gained momentum in recent years, with several new nanotechnology-based products now commercially available. Nanomaterials are characterized as objects measuring 1-100 nm in at least one plane of dimension. Notably, nanoparticulate substances impart new properties to the substance, for example, unique biokinetics and enhanced optical, catalytic, and biological activities, and are employed in several products, such as therapeutic agents, electronics, optics, and cosmetics (Oberdörster et al., 2005). Titanium dioxide nanoparticles (TiO2-NP) are important materials used in commercial paints for buildings (Shandilya et al., 2015), cosmetics (Kaida et al., 2004), and food additives (Weir et al., 2012). The increasing possibility of TiO2-NP exposure has evoked concerns regarding potential risks of TiO2-NP on human health (Borm et al., 2006). As possible exposure routes for TiO2-NP, inhalation, dermal and oral exposure are the most obvious (Borm et al., 2006). In particular, sunscreens and other cosmetics providing ultra violet protection may expose consumer lungs to TiO2-NP by inhalation because they can be available as sprayable products (Dréno et al., 2019). Pregnant women can be exposed by a similar route.
Previous studies have suggested that intranasal exposure to TiO2-NP (2.5 mg/kg body weight for 90 consecutive days) induces hippocampal neuroinflammation, with over-proliferation of all glial cells and impaired spatial recognition memory in adult mice (Ze et al., 2014). Induction of oxidative stress in the brain by TiO2-NP exposure (Shrivastava et al., 2014) would be related to these phenomena. On the other hand, following the subcutaneous administration of TiO2-NP to pregnant mice (500 μg/pregnant mouse), these nanoparticles were detected in the brains of offspring after birth (Takeda et al., 2009). The results have been suggested that TiO2-NP administered to pregnant mice can cross blood-placental barrier, and then affected the brain development. Male offspring mice exposed to TiO2-NP prenatally [subcutaneously injected; 100 μg/mouse on gestational day (GD) 6, 9, 12, and 15] reportedly present disruption of gene expression (Shimizu et al., 2009) and the dopaminergic system in the brain (Takahashi et al., 2010). Expression levels of genes associated with brain development were altered in 2 and 14 days after birth (Shimizu et al., 2009). Takahashi et al. (2010) showed that dopamine and its metabolites were increased in the prefrontal cortex and neostriatum in 6-week-old male pups. Furthermore, impaired learning and memory and decreased hippocampal cell proliferation were induced in offspring rats exposed to TiO2-NP (100 mg/kg body weight) by daily gavage administration from GD 2 to GD 21 (Mohammadipour et al., 2014). Moderate neurobehavioral alterations were observed in offspring mice following inhalation exposure of pregnant mice to a surface-coated TiO2-NP [1 h/day to 42 mg/m3 aerosolized powder (1.7 × 106 particle/cm3)] from GD 8 to GD 18 (Hougaard et al., 2010). As the result showed in this paper, offspring mice exposed to TiO2-NP prenatally tended to avoid the central zone of the open filed test, and female offspring exposed to TiO2-NP prenatally displayed enhanced prepulse inhibition (Hougaard et al., 2010). Thus, brain toxicity appeared after birth, although the mice were exposed to TiO2-NP during the prenatal period. It is necessary to elucidate underlying mechanisms through which nanoparticles affect human health to assess the safety of nanoparticles in association with advances in nanotechnology.
In the present study, we focused on the impact of TiO2-NP exposure on DNA methylation, a crucial mechanism for stable transcriptional silencing (Bird, 2002). Abnormal regulation of DNA methylation has been observed in mental and behavioral disorders (Robertson, 2005). DNA methylation patterns are initially constructed during the fetal stage and play an important role in development and differentiation (Bird, 2002). Genome-wide, oocyte DNA tends to be hypomethylated, whereas sperm DNA tends to be hypermethylated (Monk et al., 1987). After most methylation moieties present on the original parental chromosomes are removed from DNA at the morula stage, resulting in a predominantly unmethylated genome, a wave of de novo methylation follows, leading to an overall increase in genome methylation levels as newly implanted embryos develop and differentiate (Kafri et al., 1992). The deletion or inhibition of DNA methyltransferases induces abnormalities in embryonic development and organogenesis (Conerly and Grady, 2010). Most DNA methylation patterns are stably maintained following DNA replication and cell division, leading to normal embryonic development (Cheedipudi et al., 2014), and regulate postnatal neurodevelopment (Szulwach et al., 2011), neuronal synaptic functions (Feng et al., 2010), and astrocyte differentiation (Takizawa et al., 2001). We hypothesized that a change in reconstitution and maintenance of DNA methylation patterns could contribute to the effects of prenatal TiO2-NP exposure on brain development. Herein, we assessed the effects of prenatal TiO2-NP exposure on global DNA methylation patterns and gene expression profiles in the mouse brain.
Materials and Methods
Titanium Dioxide Nanoparticles
The TiO2-NP suspension (rutile: anatase = 20: 80; secondary particle diameter <150 nm; primary particle diameter of starting nanopowder, 21 nm; Cat. No. 700347-25 G, Sigma-Aldrich Co. St. Louis, MO, United States) was passed through a 0.45 μm filter (Millex®-HV; Cat. No. SLHU033RS; Merck Millipore Ltd., Burlington, MA, United States) to eliminate aggregated particles and then diluted to 0.25, 1, and 4 μg/μL with distilled water. The particles in the filtered suspension were characterized by transmission electron microscopy (TEM; JEM 1200EXII, JEOL Ltd., Akishima, Tokyo, Japan) on collodion-coated 200 Cu mesh (Cat. No. 6511; Nisshin EM, Tokyo, Japan). The size distribution of secondary TiO2-NP in the diluted suspension was determined by dynamic light scattering (DLS) measurements, using a Zetasizer Nano ZS (Malvern Instruments Ltd., Malvern, United Kingdom). For sham-treatment, a vehicle solution without any particles was prepared by centrifugation (18000 × g for 10 min at 4°C) of the TiO2-NP suspension, followed by passing through an Amicon® Ultra (3 K) device (Merck Millipore). In the DLS analysis, no particulate signal was detected in the vehicle solution.
Animals and Treatments
Pregnant C57BL/6J mice (10-week-old) at GD 6.5, 7.5, or 8.5 were purchased from Japan SLC, Inc. (Shizuoka, Japan). First, 11 pregnant mice were divided into two groups: sham exposure group: Sham (dams: n = 5); TiO2-NP (4 μg/μL) exposure group: TiO2-H (dams: n = 6) (Experiment 1). For dose-dependent analysis, a total of 19 pregnant mice were divided into TiO2-NP (0.25 μg/μL) exposure group: TiO2-L (dams: n = 3), TiO2-NP (1 μg/μL) exposure group: TiO2-M (Dams: n = 4), TiO2-H (Dams: n = 5), and Sham (Dams: n = 7) groups (Experiment 2). All animals were housed under controlled conditions (temperature: 22 ± 1°C, humidity: 50 ± 5%) with a 12-h light/12-h dark cycle and ad libitum access to food and water. On GD 10.5, 25 μL of the vehicle or TiO2-NP suspension was intratracheally administered to pregnant mice of Sham and TiO2-NP exposure groups using MicroSprayer (MicroSprayer/Syringe Assembly–MSA-250-M for Mouse, Penn-Century Inc., PA, United States). We selected GD 10.5 as the administration date because around this date is important time point for the differentiation of the mantle and marginal layers begins and eventually will form the gray matter and white matter after this date (Chen et al., 2017). Administration was performed under anesthetization with intraperitoneal injection of sodium pentobarbital according to manufacturer’s recommendation. The brains and tails of offspring mice were collected under anesthetized with isoflurane on postnatal day (PND) 1. All animal experiments were treated and handled in accordance with the Animal Research: Reporting In Vivo Experiments (ARRIVE) guidelines for the care and use of laboratory animals (Kilkenny et al., 2010), and with the approval of the Institutional Animal Care and Use Committee of Tokyo University of Science.
Genomic DNA and Total RNA Extraction From the Neonatal Brain
Whole-brain tissues were homogenized in 300 μL of phosphate-buffered saline (PBS) and divided into two tubes for extracting genomic DNA (200 μL) and total RNA (100 μL). Total RNA was extracted and purified using ISOGEN-LS (Nippon Gene Co., Tokyo, Japan) according to the manufacturer’s instructions. For DNA extraction, 150 μL of PBS and 350 μL of 2× extraction buffer [20 mM Tris-HCl (pH8.0), 0.2 M EDTA, 1% SDS, and 0.6 mg/ml Proteinase K] were added and incubated overnight at 55°C. After RNase A treatment, genomic DNA was extracted by phenol/chloroform extraction and ethanol precipitation, followed by purification using NucleoSpin® gDNA Clean-up (Macherey-Nagel GmbH & Co., KG, Germany), according to the manufacturer’s instructions.
Sex Determination
For each neonatal mouse from which brain samples were collected, genomic DNA was extracted from the tails and used for sex determination. For DNA extraction, 50 μL of TE buffer [10 mM Tris-HCl (pH 8.0), 1 mM EDTA] and 1.5 μL of proteinase K (10 mg/ml) was added to small pieces of tails and incubated overnight at 55°C. DNA was purified using NucleoSpin® gDNA Clean-up (Macherey-Nagel). Extraction of the genomic DNA was confirmed by PCR amplification of the genomic sequence of the glyceraldehyde-3-phosphate dehydrogenase (Gapdh) with specific primers (F: 5′- CACCCTGGCATTTTCTTCCA-3′, R: 5′- GACCCAGAGACCTGAATGCTG-3′). The sex of each offspring mouse was determined by PCR amplification of the genomic sequence of the sex-determining region Y (Sry) gene with specific primers (F: 5′-TGGGACTGGTGACAATTGTC-3′, R: 5′-GAGTACAGGTGTGCAGCTCT-3′). Genomic DNA obtained from male mouse was used for positive control for PCR amplification.
Methylated DNA Immunoprecipitation
Genomic DNA obtained from the brain of offspring was pooled for each of the following four groups: Sham male, Sham female, TiO2-H male, TiO2-H female. For Sham group, 2 offspring from 5 dams were pooled all together to have one sample. For TiO2-H group, 2 offspring from 6 dams were pooled all together to have one sample. The pooled DNA was then sonicated with Bioruptor (Cosmo Bio. Co., Ltd., Tokyo, Japan) to produce a random fragment, mainly ranging from 200 to 800 bp. The fragmented DNA (1.5 μg/100 μL) was denatured for 10 min at 95°C and incubated in a mixture of anti-5-methyl cytosine antibody (33D3) (ab10805; Abcam Inc., Cambridge, MA, United States) and Dynabeads® M-280 sheep anti-mouse IgG (Cat. No. 11201D; Life Technologies Inc., Gaithersburg, MD, United States) in 300 μL of immunoprecipitation (IP) buffer [20 mM Tris-HCl (pH8.0), 150 mM NaCl, 2 mM EDTA, 1% Triton X-100] overnight at 4°C. The beads combined with methylated DNA fragments were collected using a magnetic separator (MagnaRack™ Magnetic Separation Rack; Cat. No. CS15000; Thermo Fisher Scientific, Inc., Waltham, MA, United States), washed with IP buffer, and then incubated with elution buffer [25 mM Tris-HCl (pH8.0), 10 mM EDTA, 0.5% SDS, 0.25 mM DTT] for 15 min at 65°C. Finally, DNA fragments were removed from beads using a magnetic separator and purified by phenol/chloroform extraction and ethanol precipitation.
CpG Island Microarray for DNA Methylation Profiling
Input DNA and methylated DNA fragments (500 ng for each group) were labeled with Cy3 or Cy5 and purified using the SureTag DNA Labeling Kit (Agilent Technologies Inc., Santa Clara, CA, United States), respectively. The acquisition of CpG island microarray data using labeled DNA was performed with support from Agilent Technologies. Cy3-labeled input samples or Cy5-labeled methylated samples were mixed and then competitively hybridized to a Mouse CpG Island 2 × 105 K Microarray (Agilent Technologies), consisting of 88737 probes, using Oligo aCGH/ChIP-on-chip Hybridization Kit (Agilent Technologies), washed with Oligo aCGH/ChIP-on-chip Wash Buffer (Agilent Technologies), according to the manufacturer’s instructions, and scanned by SureScan G2600D (Agilent Technologies). Scanner output images were normalized and digitalized using Agilent Genomic Workbench (Agilent Technologies), and probes with a p-value less than 0.05 were extracted. Probes that presented higher values than the background, at least in either the Sham or TiO2 groups, were extracted. Relative DNA methylation was calculated by dividing the Cy5 signal by the Cy3 signal and then comparing Sham and TiO2-NP groups. Genes with CpG islands presenting ratios of relative DNA methylation ≥ 1.5-fold or ≤ 0.67-fold in the TiO2-NP group, compared with the Sham group, were determined as those demonstrating an increase or decrease in DNA methylation states, respectively. The number of flagged genes was identified using GenBank Accessions annotated on each probe in the microarray.
DNA Methylation Analysis of Specific Loci
Enrichment of methylated DNA was performed using a Methyl Collector Ultra kit (Active Motif Inc., Carlsbad, CA, United States) according to manufacturer’s instructions. Briefly, genomic DNA was digested with restriction enzyme Mse I (Nippon genetics, Co., Ltd., Tokyo, Japan). The digested DNA (0.5 μg) was incubated with a His-tagged recombinant MBD2b/MBD3L1 protein complex to capture methylated DNA. These protein–DNA complexes were captured with nickel-coated magnetic beads. Captured methylated DNA was eluted with elution buffer supplemented with proteinase K and subsequently purified using MinElute PCR clean-up kit (QIAGEN, Hilden, Germany). Obtained methylated DNA and input DNA were applied for quantitative real-time PCR to quantitate DNA methylation level of target genes. Relative methylation levels of target genes were calculated for each sample after normalization to input signals.
Microarray for mRNA Expression Profiling
Total RNA was equally pooled from each sample to obtain 45 μg for each of the four groups (Sham male, Sham female, TiO2-H male and TiO2-H female). For Sham group, 2 offspring from 5 dams were pooled all together to have one sample. For TiO2-H group, 2 offspring from 6 dams were pooled all together to have one sample. The pooled RNA was then purified using RNeasy Micro Kit (Qiagen). RNA integrity was evaluated using a Bioanalyzer 2100 (Agilent Technologies) and confirmed that the RNA integrity number (RIN) value is greater than 7. Each of the pooled RNA samples was labeled with Cy3 and hybridized to the SurePrint G3 Mouse GE 8 × 60 K Microarray (Agilent Technologies), consisting of 62976 spots, using the Gene Expression Hybridization Kit (Agilent Technologies) according to the protocol provided by Takara Bio, Inc. (Shiga, Japan). The microarray was then washed using Gene Expression Wash Pack (Agilent Technologies) and scanned using a DNA microarray scanner (G2565CA; Agilent Technologies). Scanner output images were normalized and digitalized using Feature Extraction software (Agilent Technologies) according to the Minimum Information About a Microarray Experiment (MIAME) guideline. The number of flagged genes (mRNAs) was identified using GenBank Accessions annotated on each probe in the microarray. The threshold value used to designate differentially expressed mRNAs was a fold change of ≥1.5 or ≤0.67.
Real-Time Reverse Transcription-Polymerase Chain Reaction
In brief, total RNA (1 μg) was employed as a template to generate the first-strand cDNA using M-MLV reverse transcriptase (Thermo Fisher Scientific), according to the manufacturer’s instructions. Quantitative RT-PCR was performed using ThunderbirdTM SYBR® qPCR Mix (Toyobo Co. Ltd., Osaka, Japan) and primers (Fasmac Co. Ltd., Kanagawa, Japan) for indicated genes (Supplementary Table S1) in Mx3000P (Stratagene, La Jolla, CA, United States). A dilution series of pooled cDNA samples was used for each primer pair to generate a relative standard curve. Relative expression levels of target genes were calculated for each sample after normalization to Gapdh.
Functional Analysis of Microarray Data by Gene Ontology
To better understand the biological implications of microarray results, functional analyses were performed using gene annotation by Gene Ontology (GO). All genes printed on the microarray were annotated with GO using an annotation file (ftp://ftp.ncbi.nih.gov/gene/DATA/gene2go.gz) provided by the National Center for Biotechnology Information (NCBI; Bethesda, MD). The annotations were updated in February 2021. Differentially expressed genes were classified by GO and pathways based on their functions. Enrichment factors for each GO were defined as (nf/n)/(Nf/N), where nf is the number of flagged genes within the category, Nf is the total number of genes within the same category, n is the number of flagged genes in the entire microarray, and N is the total number of genes in the microarray. Statistical analysis was performed with Fisher’s exact test, based on a hypergeometric distribution to calculate p values. The categories with enrichment factors ≥ 2, nf ≥ 5, and p < 0.05 were extracted.
Extraction of Cells, Biological Functions, or Brain Regions Susceptible to Prenatal TiO2-NP Exposure by Medical Subject Headings
All genes printed on the microarray were annotated with MeSH using the Gene-MeSH correspondence table provided by the National Center for Integrative Biotechnology Information (NCIBI). Annotations were updated in April 2018. Enrichment factors for each MeSH were defined as stated in Functional Analysis of Microarray Data by Gene Ontology. Statistical analysis was performed with Fisher’s exact test, based on a hypergeometric distribution to calculate p values. The categories with enrichment factors ≥ 2, nf ≥ 5, and p < 0.05 were extracted.
Statistical Analysis
Data values are presented as mean ± standard deviation (S.D). Data on litter size were analyzed using one-way analysis of variance (ANOVA), followed by post-hoc Tukey-Kramer test. The body weight of offspring mice and quantitative RT-PCR data were analyzed using two-way ANOVA with exposure and sex as factors, followed by post-hoc Tukey-Kramer test.
Results
Titanium Dioxide Nanoparticle Characterization
TiO2-NP suspension was characterized by TEM and DLS analyses. TEM analysis of the intratracheally administered suspension (4 μg/μL) revealed agglomerates ranging between 25–100 nm in diameter (Figure 1A). DLS analysis demonstrated small, agglomerated particles with a peak size of 78.6 nm (Figure 1B). The polydispersity index of 0.195 was less than 0.2, thus indicating a narrow size distribution (Tanvir et al., 2012). The observed 78.6 nm size corresponds well with the typical small agglomerate sizes of TiO2-NPs observed under TEM.
FIGURE 1.
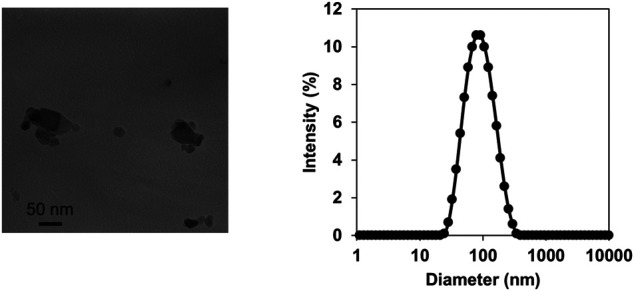
Characterization of titanium dioxide nanoparticles (TiO2-NPs). Transmission electron microscopy images of TiO2-NPs in the intratracheally administered suspension (A). Size distribution of TiO2-NP in the suspension determined by dynamic light scattering (B).
Litter Size, Sex Ratio, and Body Weight of Offspring
We assessed the effects of prenatal TiO2-NP (TiO2-L, -M, and -H) exposure on the number, sex ratio, and body weight of offspring. No significant differences in the number, sex ratio, and body weight of offspring were observed between the Sham and TiO2-NP groups (Table 1).
TABLE 1.
Number, sex ratio, and body weight of 1-day-old offspring.
| Group | Number of offspring per dam | Sex ratio (male %) a | Body weight of offspring (g) | ||
|---|---|---|---|---|---|
| Male | Female | ||||
| Experiment 1 | Sham (Dams: n = 5) | 7.00 ± 0.71 | 42.9 | 1.32 ± 0.17 | 1.23 ± 0.04 |
| TiO2-H (Dams: n = 6) | 7.67 ± 1.03 | 60.0 | 1.33 ± 0.08 | 1.32 ± 0.09 | |
| Experiment 2 | Sham (Dams: n = 7) | 7.14 ± 2.12 | 47.7 | 1.22 ± 0.10 | 1.22 ± 0.08 |
| TiO2-L (Dams: n = 3) | 6.00 ± 2.00 | 50.0 | 1.40 ± 0.12 | 1.23 ± 0.06 | |
| TiO2-M (Dams: n = 4) | 7.50 ± 1.29 | 57.7 | 1.25 ± 0.07 | 1.21 ± 0.11 | |
| TiO2-H (Dams: n = 5) | 6.80 ± 1.92 | 46.9 | 1.25 ± 0.14 | 1.22 ± 0.09 | |
Data are presented as mean ± SD.
Sex ratio (%) = male/(male + female) × 100.
DNA Methylation Profile in the Offspring Brain
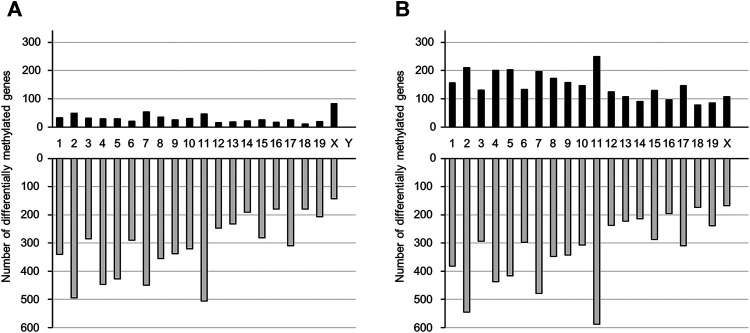
We performed genome-wide DNA methylation analysis using a Mouse CpG island 2 × 105 K Microarray, with 88738 probes designed for 15342 CpG islands. Probes were designed for multiple regions of each gene. In the TiO2-H group, the number of probes with differential methylation was 11266 (≥1.5-fold: 758, ≤0.67-fold: 10508) and 15223 (≥1.5-fold: 3,779, ≤0.67-fold: 11444) in male and female offspring brains, respectively (Supplementary Tables S2, S3). Overall, DNA methylation levels were increased in 614 and 2,924 genes and were decreased in 6,220 and 6,477 genes following TiO2-NP exposure in the male and female offspring, respectively (Supplementary Tables S4–S7). Prenatal TiO2-NP exposure altered the methylation state of genes in all chromosomes. (Figure 2).
FIGURE 2.
The number of genes that show altered DNA methylation states following prenatal TiO2-NP exposure. Number of genes presenting altered DNA methylation states between Sham and TiO2-H groups in the brains of 1-day-old offspring mice [(A): male, (B): female]. Black and gray bars indicate the number of genes with increased and decreased DNA methylation levels, respectively, in the TiO2-H groups when compared with the Sham group. The x-axis shows chromosome numbers.
mRNA Expression Profile in the Offspring Brain
We performed a comprehensive analysis of differentially expressed mRNAs following prenatal TiO2-NP exposure using SurePrint G3 Mouse GE 8 × 60 K Microarray (Design ID: 028005), containing 55,681 probes. The results revealed that 658 and 650 mRNAs were upregulated (≥1.5-fold) in male and female offspring brains of the TiO2-H group when compared with the Sham group (Supplementary Tables S8A, S9A); in contrast, prenatal TiO2-NP exposure downregulated 482 and 409 mRNAs (≤0.67-fold) in male and female offspring brains (Supplementary Tables S8B, S9B). Then, differentially expressed mRNAs that correlated with changes in the DNA methylation status were extracted. In the TiO2-H group, 88 and 89 genes were upregulated (≥1.5-fold) accompanied by demethylation of CpG islands (Figures 3A,B; Supplementary Tables S10A,B), while 13 and 33 genes were downregulated (≤0.67-fold) accompanied by methylation of CpG islands in male and female offspring mice, respectively (Figures 3A,B; Tables 2A, B). Unexpectedly, few of these extracted genes were common in male and female offspring (Supplementary Table S11).
FIGURE 3.

The number of differentially expressed genes with altered DNA methylation following TiO2-NP exposure.—Venn diagram analysis whether differentially expressed genes are accompanied by alteration of DNA methylation. Venn diagram showing the number of differentially expressed genes with altered DNA methylation following TiO2-NP (Sham vs. TiO2-H) in the brains of 1-day-old offspring mice [(A): male, (B): female]. The DNA methylation state and expression level of genes were identified by Mouse CpG Island Microarray and SurePrint G3 Mouse GE 8 × 60 K Microarray, respectively.
TABLE 2.
The genes of which expression levels were decreased with increased methylation of CpG island in the brain of male (A) and female (B) offspring.
| A | ||||
| GenBank accession | GeneSymbol | Target position of probe on CpG island microarray | mRNA | DNA methylation |
|---|---|---|---|---|
| Fold change | Fold change of relative methylation | |||
| NM_001161855 | 4933416C03Rik | chr10:115551150–115551194 | 0.165 | 2.153 |
| NM_008532 | Epcam | chr17:88039798–88039842 | 0.013 | 1.729 |
| NM_008192 | Gucy2e | chr11:69049315–69049359 | 0.105 | 1.541 |
| NR_027967 | Hhatl | chr9:121698137–121698187 | 0.600 | 2.288 |
| NM_213729 | Inca1 | chr11:70513486–70513530 | 0.196 | 1.704 |
| NM_027397 | Isl2 | chr9:55394278–55394322 | 0.045 | 1.522 |
| NM_001033250 | Lemd1 | chr1:134086976–134087020 | 0.188 | 1.840 |
| NM_015743 | Nr4a3 | chr4:48060289–48060333 | 0.056 | 2.393 |
| NM_008814 | Pdx1 | chr5:148081815–148081859 | 0.078 | 1.623 |
| NM_009027 | Rasgrf2 | chr13:92901976–92902020 | 0.263 | 3.225 |
| NM_001164704 | Renbp | chrX:71167531–71167575 | 0.199 | 3.621 |
| NM_001033415 | Shisa3 | chr5:67999755–67999799 | 0.306 | 1.502 |
| NM_173429 | Zfp775 | chr6:48569751–48569795 | 0.664 | 1.528 |
| NM_173429 | Zfp775 | chr6:48570111–48570155 | 0.664 | 1.549 |
| NM_173429 | Zfp775 | chr6:48570207–48570251 | 0.664 | 2.012 |
| NM_173429 | Zfp775 | chr6:48570342–48570386 | 0.664 | 2.204 |
| B | ||||
| GenBank accession | GeneSymbol | Target position of probe on CpG island microarray | mRNA | DNA methylation |
|---|---|---|---|---|
| Fold change | Fold change of relative methylation | |||
| NR_024323 | 4930426L09Rik | chr2:18919881–18919925 | 0.221 | 1.977 |
| NM_026185 | Abhd15 | chr11:77328807–77328851 | 0.058 | 1.661 |
| NM_009681 | Ap3s1 | chr18:46901926–46901970 | 0.541 | 2.070 |
| NM_001163614 | Ascl4 | chr10:85391455–85391499 | 0.513 | 1.767 |
| NM_001163614 | Ascl4 | chr10:85391524–85391568 | 0.513 | 1.602 |
| NM_016709 | Auh | chr13:53024981–53025025 | 0.247 | 1.692 |
| NM_009877 | Cdkn2a | chr4:88940707–88940751 | 0.084 | 1.630 |
| NM_007760 | Crat | chr2:30271133–30271177 | 0.144 | 2.214 |
| NR_028264 | Dleu2 | chr14:62301793–62301837 | 0.066 | 2.086 |
| NR_028264 | Dleu2 | chr14:62305865–62305909 | 0.066 | 1.558 |
| NR_028264 | Dleu2 | chr14:62300358–62300402 | 0.066 | 1.526 |
| NM_133763 | Dnttip1 | chr2:164571271–164571315 | 0.282 | 1.520 |
| NM_027828 | Fam110c | chr12:31758655–31758699 | 0.105 | 1.608 |
| NM_015821 | Fbxl8 | chr8:107788500–107788544 | 0.374 | 1.752 |
| NM_018789 | Foxo4 | chrX:98450245–98450289 | 0.134 | 1.854 |
| NM_010340 | Gpr50 | chrX:68917753–68917797 | 0.045 | 1.609 |
| NM_010468 | Hoxd3 | chr2:74578542-74578586 | 0.371 | 1.809 |
| NM_010468 | Hoxd3 | chr2:74550322–74550366 | 0.371 | 1.619 |
| NM_028708 | Jakmip3 | chr7:146132486–146132530 | 0.036 | 4.012 |
| NM_001033430 | Jhdm1d | chr6:39155596–39155640 | 0.122 | 2.359 |
| NM_001033430 | Jhdm1d | chr6:39157044–39157088 | 0.122 | 1.774 |
| NM_001037712 | Kcnh6 | chr11:105881863–105881907 | 0.153 | 1.967 |
| NM_030716 | Kcnip2 | chr19:45890581–45890625 | 0.500 | 1.823 |
| NM_152895 | Kdm5b | chr1:136457792–136457836 | 0.227 | 1.678 |
| NM_023061 | Mcam | chr9:43943262–43943306 | 0.444 | 2.490 |
| NM_001122667 | Mkl2 | chr16:13256418–13256462 | 0.100 | 1.806 |
| NM_181860 | Mkl2 | chr16:13257041–13257085 | 0.100 | 1.574 |
| NM_011037 | Pax2 | chr19:44830746–44830790 | 0.044 | 1.621 |
| NM_001162929 | Pom121l2 | chr13:22073387–22073431 | 0.142 | 1.679 |
| NM_001123362 | Prdm12 | chr2:31495757–31495801 | 0.420 | 1.676 |
| NM_001164570 | Rffl | chr11:82684465–82684509 | 0.474 | 1.706 |
| NM_017395 | Rfx5 | chr3:94758251–94758295 | 0.221 | 1.892 |
| NM_019732 | Runx3 | chr4:134708685–134708729 | 0.531 | 1.612 |
| NM_011432 | Snrpc | chr17:27977231–27977275 | 0.482 | 1.763 |
| NM_054103 | Stk33 | chr7:116582357–116582401 | 0.387 | 1.933 |
| NM_011517 | Sycp3 | chr10:87922666–87922710 | 0.427 | 1.532 |
| NM_009331 | Tcf7 | chr11:52096304–52096348 | 0.104 | 1.611 |
| NM_001085555 | Tcp11 | chr17:28217573–28217617 | 0.066 | 2.106 |
| NM_013687 | Tcp11 | chr17:28217206–28217250 | 0.066 | 1.703 |
| NR_001463 | Xist | chrX:100677336–100677380 | 0.050 | 1.721 |
| NR_001463 | Xist | chrX:100677473–100677517 | 0.050 | 1.547 |
The threshold value used to designate altered mRNA expression or DNA methylation was a fold change of ≥1.5 or ≤0.67.
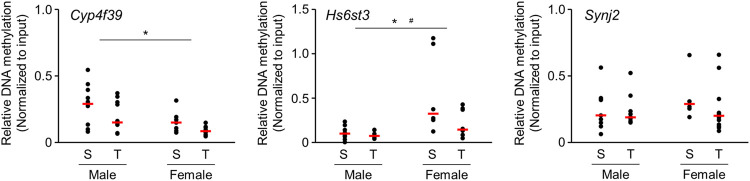
DNA Methylation Analysis of the Gene That Showed Altered DNA Methylation Accompanied by Changes in mRNA Expression
We further analyzed the alteration of DNA methylation using enrichment of methylated DNA fragment with methyl DNA binding proteins followed by quantitative PCR. We focused on the genes that showed altered DNA methylation accompanied by changes in mRNA expression. Experiments were performed on those genes for which PCR primers capable of specific amplification could be designed. DNA methylation state of Cyp4f39 and Hs6st3 genes were significantly decreased (Figure 4) in offspring brains. Significant difference of DNA methylation state was not detected in Synj2 gene, but it seems to decrease in female offspring brains.
FIGURE 4.
Scatter plots of brain DNA methylation levels of 1-day-old offspring mice from TiO2-H and Sham groups determined by Enrichment of Methylated DNA Fragments Followed by qPCR. Methylated DNA fragments were enriched from genomic DNA obtained from each sample {two offspring were randomly selected from each dam [Sham (S): n = 5, TiO2-H (T): n = 6]} using Methyl Collector Ultra Kit (Active Motif). Then, methylated DNA fragments were subjected to qPCR analysis. Two-way ANOVA with exposure and sex as factors, followed by post-hoc Tukey test. *p < 0.05: main effect of TiO2-NP exposure on relative DNA methylation; # p < 0.05: main effect of sex on relative DNA methylation (Sham vs. Exposure).
Quantitative Real-Time RT-PCR Analysis of mRNA Expression
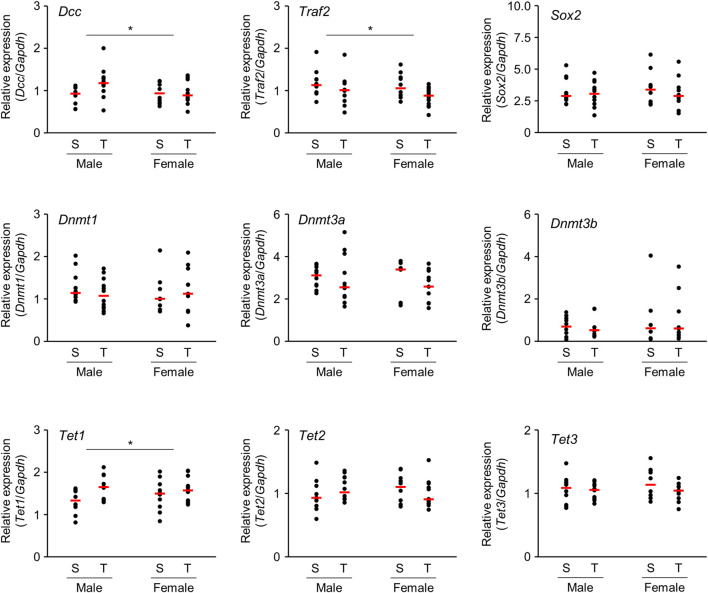
Microarray analysis revealed that several mRNAs were differentially expressed in the brains of offspring prenatally exposed to TiO2-NP. In particular, mRNAs with altered expression levels in both males and females are speculated to be closely related to adverse health effects induced following TiO2-NP exposure. We performed qRT-PCR analyses of Dcc and Traf2 mRNA, which demonstrate a high signal value on microarray analysis and are reportedly involved in nervous system development and inflammatory response, respectively. The tendency of expression change of these mRNAs determined by qRT-PCR was similar to those observed following the microarray analysis (Figure 5). Although we also examined gene expression levels of neural stem cell marker [SRY-box 2 (Sox2)] and Dnmts, no significant differences were detected (Figure 5). Further qRT-PCR analysis was performed for Ten-eleven translocation (TET) family genes associated with active DNA demethylation. The expression level of Tet1 mRNA was significantly upregulated following prenatal TiO2-NP exposure (Figure 5). No statistically significant effect of sex (male vs. female) was observed on the expression of analyzed genes, as well as no impact of interactions between TiO2-NP exposure and sex on analyzed genes was detected.
FIGURE 5.
Scatter plots of brain mRNA expression levels of 1-day-old offspring mice from TiO2-H and Sham groups determined by qRT-PCR. Total RNA obtained from each sample {two offspring were randomly selected from each dam [Sham (S): n = 5, TiO2-H (T): n = 6]} was transcribed to cDNA and subjected to qRT-PCR analysis. Two-way ANOVA with exposure and sex as factors, followed by post-hoc Tukey test. *p < 0.05: main effect of TiO2-NP exposure on expression (Sham vs. Exposure).
Functional Categorization of Potential Target Genes
We then aimed to obtain information regarding the biological function affected by altered DNA methylation in the brain of 1-day-old offspring mice following prenatal TiO2-H exposure. Accordingly, GSEA was performed for genes that showed altered DNA methylation accompanied by changes in mRNA expression. Genes upregulated accompanied by demethylation of CpG islands or those downregulated accompanied by methylation of CpG islands following TiO2-H exposure were functionally categorized with GO terms. The results revealed that 12 and 39 GO categories were enriched in male and female offspring, respectively (Tables 3A,B). Then, common GO terms between male and female offspring were extracted (Supplementary Table S12). The GO terms related to the regulation of transcription factors (“DNA-binding transcription factor activity,” “sequence-specific DNA binding,” and “regulation of transcription, DNA-templated”), cell proliferation (“negative regulation of cell population proliferation”), and organism development (“multicellular organism development”) were detected in genes that showed altered DNA methylation accompanied by differential mRNA expression in both male and female offspring.
TABLE 3.
GO terms enriched in the genes that showed differential expression accompanied by altered DNA methylation in male (A) and female (B) offspring in the TiO2-H group.
| A | |||
| ID | GO term | Enrichment factor | p-value |
|---|---|---|---|
| GO:0005923 | Bicellular tight junction | 8.99 | <0.001 |
| GO:0015629 | Actin cytoskeleton | 5.07 | 0.003 |
| GO:0003779 | Actin binding | 3.28 | 0.018 |
| GO:0008285 | Negative regulation of cell population proliferation | 3.07 | 0.024 |
| GO:0045893 | Positive regulation of transcription, DNA-templated | 2.71 | 0.014 |
| GO:0007275 | Multicellular organism development | 2.63 | 0.001 |
| GO:0003700 | DNA-binding transcription factor activity | 2.63 | 0.017 |
| GO:0043565 | Sequence-specific DNA binding | 2.50 | 0.033 |
| GO:0030054 | Cell junction | 2.42 | 0.012 |
| GO:0006355 | Regulation of transcription, DNA-templated | 2.34 | 0.010 |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 2.14 | 0.038 |
| GO:0045944 | Positive regulation of transcription by RNA polymerase II | 2.08 | 0.029 |
| B | |||
| ID | GO term | Enrichment factor | p-value |
|---|---|---|---|
| GO:0031175 | Neuron projection development | 6.05 | 0.001 |
| GO:0010468 | Regulation of gene expression | 5.20 | <0.001 |
| GO:0003700 | DNA-binding transcription factor activity | 5.01 | <0.001 |
| GO:0005667 | Transcription regulator complex | 4.94 | <0.001 |
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 4.86 | <0.001 |
| GO:1990837 | Sequence-specific double-stranded DNA binding | 4.71 | <0.001 |
| GO:0000977 | RNA polymerase II transcription regulatory region sequence-specific DNA binding | 4.67 | <0.001 |
| GO:0043565 | Sequence-specific DNA binding | 4.17 | <0.001 |
| GO:0031625 | Ubiquitin protein ligase binding | 3.97 | 0.004 |
| GO:0008134 | Transcription factor binding | 3.91 | 0.002 |
| GO:0045893 | Positive regulation of transcription, DNA-templated | 3.88 | <0.001 |
| GO:0007283 | Spermatogenesis | 3.79 | 0.001 |
| GO:0045892 | Negative regulation of transcription, DNA-templated | 3.70 | <0.001 |
| GO:0016604 | Nuclear body | 3.69 | 0.005 |
| GO:0045944 | Positive regulation of transcription by RNA polymerase II | 3.65 | <0.001 |
| GO:0008285 | Negative regulation of cell population proliferation | 3.58 | 0.003 |
| GO:0007275 | Multicellular organism development | 3.48 | <0.001 |
| GO:0000978 | RNA polymerase II cis-regulatory region sequence-specific DNA binding | 3.47 | <0.001 |
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 3.38 | <0.001 |
| GO:0006357 | Regulation of transcription by RNA polymerase II | 3.36 | <0.001 |
| GO:0006355 | Regulation of transcription, DNA-templated | 3.31 | <0.001 |
| GO:0001701 | In utero embryonic development | 3.18 | 0.021 |
| GO:0000122 | Negative regulation of transcription by RNA polymerase II | 2.97 | <0.001 |
| GO:0008270 | Zinc ion binding | 2.93 | 0.002 |
| GO:0019899 | Enzyme binding | 2.82 | 0.020 |
| GO:0043065 | Positive regulation of apoptotic process | 2.80 | 0.033 |
| GO:0003677 | DNA binding | 2.80 | 0.000 |
| GO:0030424 | Axon | 2.74 | 0.022 |
| GO:0003779 | Actin binding | 2.73 | 0.036 |
| GO:0010628 | Positive regulation of gene expression | 2.69 | 0.015 |
| GO:0016491 | Oxidoreductase activity | 2.68 | 0.010 |
| GO:0042803 | Protein homodimerization activity | 2.68 | 0.006 |
| GO:0030154 | Cell differentiation | 2.66 | 0.001 |
| GO:0019901 | Protein kinase binding | 2.62 | 0.018 |
| GO:0042995 | Cell projection | 2.53 | 0.001 |
| GO:0005694 | Chromosome | 2.43 | 0.037 |
| GO:0045202 | Synapse | 2.39 | 0.013 |
| GO:0030054 | Cell junction | 2.24 | 0.014 |
| GO:0031410 | Cytoplasmic vesicle | 2.21 | 0.021 |
The enrichment factor for each category was defined as described in Materials and Methods. Statistical analysis was performed using Fisher’s exact test with hypergeometric distribution and the level of statistical significance was set at p < 0.05.
We also attempted to identify cells, biological functions, or brain regions susceptible to prenatal TiO2-NP exposure from GSEA analysis using MeSH terms (Umezawa et al., 2012) (Tables 4A,B). Supplementary Table S13 presents MeSH terms enriched in genes showing altered DNA methylation accompanied by differential mRNA expression in male and female offspring. MeSH terms possibly related to the development and differentiation of cells from stem cells (“Stem Cells” and “Morphogenesis”) and transcription factors and gene expression (“Gene Expression Regulation, Developmental,” “Transcription Factors,” and “Homeodomain Proteins”) were detected in genes showing altered DNA methylation accompanied by differential mRNA expression, common in male and female offspring. We further analyzed whether MeSH terms related to brain regions were extracted by GSEA analysis because it has not been well investigated which brain region is susceptible to the effect of prenatal nanoparticle exposure in terms of DNA methylation and gene expression. But unfortunately, MeSH terms related to brain regions were not obtained by analysis.
TABLE 4.
MeSH terms enriched in the genes that showed differential expression accompanied by altered DNA methylation in male (A) and female (B) offspring in the TiO2-H group.
| A | |||
| ID | MeSH term | Enrichment factor | p-value |
|---|---|---|---|
| D060850 | LIM-Homeodomain Proteins | 8.73 | <0.001 |
| D019070 | Cell Lineage | 6.21 | <0.001 |
| D051792 | Basic Helix-Loop-Helix Transcription Factors | 4.98 | 0.001 |
| D015534 | Trans-Activators | 4.90 | 0.004 |
| D013234 | Stem Cells | 4.83 | 0.002 |
| D012097 | Repressor Proteins | 4.30 | 0.003 |
| D018398 | Homeodomain Proteins | 4.19 | <0.001 |
| D009024 | Morphogenesis | 3.32 | 0.018 |
| D018507 | Gene Expression Regulation, Developmental | 3.13 | <0.001 |
| D009419 | Nerve Tissue Proteins | 3.09 | 0.013 |
| D017403 | In Situ Hybridization | 3.07 | 0.005 |
| D004622 | Embryo, Mammalian | 3.01 | 0.003 |
| D014157 | Transcription Factors | 2.76 | 0.009 |
| B | |||
|---|---|---|---|
| ID | MeSH term | Enrichment factor | p-value |
| D014566 | Urogenital System | 15.60 | <0.001 |
| D029961 | Zebrafish Proteins | 13.00 | <0.001 |
| D014212 | Tretinoin | 11.10 | <0.001 |
| D001755 | Blastocyst | 11.00 | <0.001 |
| D010641 | Phenotype | 9.50 | <0.001 |
| D011518 | Proto-Oncogene Proteins | 8.10 | <0.001 |
| D011401 | Promoter Regions, Genetic | 8.00 | <0.001 |
| D004475 | Ectoderm | 7.50 | 0.001 |
| D051176 | beta Catenin | 7.20 | 0.001 |
| D005801 | Genes, Homeobox | 6.80 | <0.001 |
| D038081 | Organogenesis | 6.70 | <0.001 |
| D009432 | Neural Crest | 6.40 | 0.001 |
| D012249 | Rhombencephalon | 6.10 | 0.001 |
| D051153 | Wnt Proteins | 5.80 | <0.001 |
| D008648 | Mesoderm | 5.80 | <0.001 |
| D019521 | Body Patterning | 5.30 | <0.001 |
| D019070 | Cell Lineage | 5.20 | 0.001 |
| D004268 | DNA-Binding Proteins | 5.00 | <0.001 |
| D013234 | Stem Cells | 4.70 | 0.001 |
| D053823 | Hedgehog Proteins | 4.70 | 0.002 |
| D009024 | Morphogenesis | 4.40 | <0.001 |
| D014157 | Transcription Factors | 4.30 | <0.001 |
| D017403 | In Situ Hybridization | 4.20 | <0.001 |
| D009687 | Nuclear Proteins | 4.20 | 0.003 |
| D018507 | Gene Expression Regulation, Developmental | 3.90 | <0.001 |
| D004622 | Embryo, Mammalian | 3.90 | <0.001 |
| D018398 | Homeodomain Proteins | 3.90 | <0.001 |
| D015398 | Signal Transduction | 3.60 | 0.002 |
| D002454 | Cell Differentiation | 3.50 | 0.004 |
| D002874 | Chromosome Mapping | 3.00 | 0.010 |
| D009419 | Nerve Tissue Proteins | 2.60 | 0.030 |
The enrichment factor for each category was defined as described in Materials and Methods. Statistical analysis was performed using Fisher’s exact test with hypergeometric distribution and the level of statistical significance was set at p < 0.05.
Discussion
DNA methylation is a pivotal mechanism in epigenetic gene regulation. The construction of DNA methylation patterns during development is crucial for morphogenesis or the acquisition of biological functions (Leeb and Wutz, 2012). Several reports have indicated that prenatal exposure to environmental agents, such as bisphenol A, vinclozolin, tobacco smoke, diesel exhaust, and traffic particles, induces altered DNA methylation and gene expression (Anway et al., 2005; Baccarelli et al., 2009; Breton et al., 2009; Ma et al., 2013; Nahar et al., 2014; Tachibana et al., 2015; Nielsen et al., 2016). Although recent studies have revealed that nanoparticle exposure alters DNA methylation (Li et al., 2016; Pogribna et al., 2020), its impact on the DNA methylation pattern during the developmental stage, especially in the brain, remains poorly understood. In the present study, we analyzed the effects of prenatal TiO2-NP exposure on the DNA methylation status in brain samples of mouse offspring using a CpG island microarray designed to analyze CpG islands located around the transcription start site of genes. Alteration of DNA methylation attributable to gestational exposures is thought to occur before birth. Our previous study showed that prenatal diesel exhaust exposure caused abnormal DNA methylation in the brain of mouse offspring at PND1 (Tachibana et al., 2015). Hence, to capture the impacts of early life exposure to TiO2-NP, we selected PND1 as the time point of sample collection.
GSEA analysis of genes with altered DNA methylation accompanied by changes in mRNA expression revealed that these genes were enriched in GO terms related to the regulation of transcription factors, cell proliferation, and organism development. Furthermore, the results of categorization using MeSH terms indicated that these phenomena could be associated with the proliferation and differentiation of neural stem cells. Combined with these results and findings in previous reports, alterations in DNA methylation caused by prenatal TiO2-NP exposure might be involved in brain dysfunction, such as impairment of learning and memory or the dopaminergic system (Takahashi et al., 2010; Mohammadipour et al., 2014), by disrupting proliferation and differentiation of neural stem cells at the developmental stage. In particular, homeodomain proteins extracted by GSEA analysis using MeSH terms are known as transcription factors that play crucial roles in several developmental processes, including in the brain, by regulating the expression of other genes during development and differentiation (Prochiantz and Di Nardo, 2015). It can be speculated that abnormal development of the nervous system could be induced by disrupted expression of genes regulated by homeodomain protein. In the present study, gene expression level of Sox2 was not significantly affected by prenatal TiO2-NP exposure. Although it is just the result of the gene expression analysis, this result seems to demonstrate that the number of neural stem cells is still not significantly altered in PND1 although DNA methylation and expression of the genes related to cell proliferation and organism development were affected. Because the brain is still developing at this age, it will be necessary to investigate changes in the number of and distribution of neural stem cells in the brain occurred as the mice grow.
It is expected that genes that showed altered DNA methylation accompanied by changes in mRNA expression in male and female offspring are significantly involved in mediating the adverse effects of prenatal TiO2-NP exposure in the offspring. However, it remains unclear whether most of these genes, except for Slc1a2, are involved in brain function. Slc1a2, a glutamate transporter, is suggested to be involved in brain development, and its dysregulation has been associated with preterm brain injury (Pregnolato et al., 2019). Accordingly, increased Slc1a2 expression induced by TiO2-NP exposure might be associated with brain injury. Interestingly, although few genes are differentially methylated in common between male and female offspring, GSEA analysis extracted common GO terms and MeSH terms for DNA methylation alterations. Previous studies have reported that sex-dependent differentially methylated regions (S-DMRs) exist in the prefrontal cortex and liver tissues, and these differences are suggested to be constructed during developmental processes (Xu et al., 2014; Ito et al., 2015). These S-DMRs might be partially associated with differences in DNA methylation between male and female offspring. Further investigations are required to address these findings.
Furthermore, we observed that GO and MeSH terms related to morphological development (GO term: “cell junction” and “actin binding,” MeSH term: “Morphology”) were enriched in the GSEA analysis. A case-control study has revealed that global DNA hypomethylation is associated with neural tube defects (Chen et al., 2010). GO and MeSH terms extracted by GSEA analysis suggested that structural changes occurred in the cranial nerve system following prenatal TiO2-NP exposure. We searched for brain region that is susceptible to the effect of prenatal TiO2-NP exposure, in terms of DNA methylation and gene expression, using GSEA analysis. Since no brain region was extracted by GSEA analysis using MeSH terms, it was not possible to identify a region that is expected to be significantly more susceptible to TiO2-NP exposure at this developmental stage.
Our results showed that prenatal TiO2-NP exposure induced genome-wide alterations in DNA methylation in the offspring brain, with a higher number of demethylated than methylated genes detected. We examined gene expression levels of Dnmt1, Dnmt3a, and Dnmt3b, but no significant differences were observed in these genes. A recent study has revealed that TET family proteins actively induce demethylation of 5-methylcytosine (5-mC) (Wu and Zhang, 2010). TET family proteins catalyze the hydroxylation of 5-mC into 5-hydroxymethylcytosine (5-hmC), which can further oxidize 5-hmC to 5-formylcytosine (5-fC) and 5-carboxycytosine (5-caC) (Zhu et al., 2020). Then, 5-fC and 5-caC are rapidly excised by thymine DNA glycosylase in the active demethylation state, subsequently replaced by unmodified cytosines through base excision repair mechanisms (Ji et al., 2016). The generation of 5-hmC by TET proteins has been proposed to play a pivotal role in cell differentiation by controlling the methylation status of developmentally important enhancers (Schuermann et al., 2016). It can be speculated that Tet1 is involved in the decreased DNA methylation observed following prenatal TiO2-NP exposure in the present study, as qRT-PCR results revealed that, among Tet family genes, Tet1 mRNA expression was upregulated.
Notably, DNA methylation is important for neural development during the fetal period, and it has been suggested that dysregulated DNA methylation can be associated with psychosis (Fan et al., 2001; Mill et al., 2008). Abnormal DNA methylation has been reported in the promoter regions of several genes that control neurodevelopment and mitochondrial function in the brains of patients with schizophrenia, bipolar disorder, and Alzheimer’s disease (Mill et al., 2008; Mastroeni et al., 2009). Dcc was differentially expressed in offspring brain following prenatal TiO2-NP exposure. Reportedly, Dcc transcription is regulated by CpG island methylation (Sato et al., 2001; Carvalho et al., 2006); however, probes for analyzing this gene were not designed in the CpG island microarray used in the present study. Interestingly, Dcc is reported to be involved in neuronal differentiation and neurite outgrowth (Lawlor and Narayanan, 1992; Deiner and Sretavan, 1999; Li et al., 2002). Combined with these reports, DNA demethylation caused by prenatal TiO2-NP exposure could be associated with central nervous system function and mental disorders.
Numerous reports have revealed that exposure to nanoparticles, including TiO2-NP, induces inflammation (Borm et al., 2006; Grassian et al., 2007; Cui et al., 2011). As nanoparticle exposure in the mother can be transferred to the fetus (Takeda et al., 2009; Wick et al., 2010), it is speculated that inflammation within fetal tissues is associated with adverse health effects. In the present study, Traf2, which is related to proinflammatory signal transduction, showed a tendency toward decreased expression. As Traf2 reportedly affords protection against brain injury (Li et al., 2019), it can be postulated that a decrease in Traf2 may be associated with susceptibility to brain damage. In addition, inflammation that occurs in the mother can affect fetal biological functions. Maternal inflammation has been suggested to contribute to brain outgrowth, autism-associated behaviors, and schizophrenia (Venkatasubramanian and Debnath, 2013; Le Belle et al., 2014). Furthermore, recent studies have shown that chronic inflammation causes aberrant DNA methylation (Jangiam et al., 2015; Barnicle et al., 2017). Interestingly, prenatal inflammation alters the DNA methylation status in the brain and can be associated with neurodevelopmental disorders (Basil et al., 2014). Inflammation induced by prenatal TiO2-NP exposure in both the fetus and mother could be involved in altered DNA methylation in the offspring brain; however, further investigations are crucial to gain a comprehensive understanding of these implications.
In the present study, we indicated that prenatal intratracheal exposure to TiO2-NP induces abnormal DNA methylation status and mRNA expression profile in the brain of mouse offspring at PND1. However, the brain is still developing at this stage and DNA methylation pattern is dynamically modulated during development. Actually, Cisternas et al. indicated that global DNA methylation level in the preoptic area of the hypothalamus increase during development (Cisternas et al., 2020). It would be important to investigate how the abnormal methylation pattern cause by prenatal TiO2-NP exposure changes with development. We attempted to find the fundamental effects of TiO2-NP on the brain of newborns, in terms of DNA methylation and gene expression, by extracting common changes from the analysis of each male and female samples. On the other hand, sexual dimorphism is present in the brain development (Pilgrim and Reisert, 1992). Therefore, it can be speculated that sex difference in the effect of TiO2-NP exposure exists between male and female. It is necessary to examine whether there is a sex difference in the effects of prenatal exposure to TiO2-NP on the brain development. To address these issues, it would be required to examine the effects of TiO2-NP at multiple developmental stages in the future study. Furthermore, to know the risks of prenatal inhalation exposure to TiO2-NP correctly, it is required to investigate about not only hazard but also frequency or exposure levels of it. Unfortunately, it is not well known enough about frequency or exposure levels of TiO2-NP inhalation exposure at the time of writing. It is required to investigate these points to assess the risk of prenatal inhalation exposure to TiO2-NP.
Conclusion
Prenatal TiO2-NP exposure caused genome-wide alterations in DNA methylation and mRNA expression in the brains of both male and female offspring. These differential expression profiles may, in part, mediate the mechanism via which TiO2-NP toxicity affects brain development. GSEA analysis using GO terms suggested that these genes were associated with cell proliferation, organism development, and regulation of transcription factors. GSEA analysis using MeSH terms indicates that these phenomena are associated with the proliferation and differentiation of neural stem cells. It can be speculated that prenatal TiO2-NP exposure adversely impacts brain functions by altering the DNA methylation status of fetal brain tissues, especially neural stem cells, subsequently causing abnormal regulation of transcription factors that control development and differentiation, especially homeodomain protein. Future studies need to investigate molecular mechanisms underlying DNA methylation abnormalities that affect brain functions, especially from the perspective of neural stem cells.
Acknowledgments
We gratefully thank Rikio Niki and Rie Numazaki (Center for Environmental Health Science for the Next Generation, Research Institute for Science and Technology, Tokyo University of Science) for their skilled technical assistance. We also thank the graduate and undergraduate students at the Takeda laboratory for their help with the experiments.
Data Availability Statement
The datasets generated for this study can be found in the Gene Expression Omnibus (GSE178207) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE178207).
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Tokyo University of Science.
Author Contributions
KTc and SK conceived the study. KTc, AO, MU, and KTk wrote the manuscript. KTc, SK, and MU performed experiments and analyzed the data. AO and KTk supervised the project. All authors commented on the manuscript.
Funding
This work was supported in part by a JSPS KAKENHI Grant, Numbers 15K00558, and 19K12342 (KTc); a MEXT-Supported Program for the Strategic Research Foundation at Private Universities (KTk); and a Grant-in-Aid for the Health and Labour Science Research Grant, Research on the Risk of Chemical Substances, from the Ministry of Health, Labour and Welfare of Japan (KTk).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ftox.2021.705910/full#supplementary-material
References
- Anway M. D., Cupp A. S., Uzumcu M., Skinner M. K. (2005). Epigenetic Transgenerational Actions of Endocrine Disruptors and Male Fertility. Science 308, 1466–1469. 10.1126/science.1108190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A., Wright R. O., Bollati V., Tarantini L., Litonjua A. A., Suh H. H., et al. (2009). Rapid DNA Methylation Changes After Exposure to Traffic Particles. Am. J. Respir. Crit. Care Med. 179, 572–578. 10.1164/rccm.200807-1097oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnicle A., Seoighe C., Greally J. M., Golden A., Egan L. J. (2017). Inflammation-Associated DNA Methylation Patterns in Epithelium of Ulcerative Colitis. Epigenetics 12, 591–606. 10.1080/15592294.2017.1334023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basil P., Li Q., Dempster E. L., Mill J., Sham P.-C., Wong C. C. Y., et al. (2014). Prenatal Maternal Immune Activation Causes Epigenetic Differences in Adolescent Mouse Brain. Transl. Psychiatry 4, e434. 10.1038/tp.2014.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. (2002). DNA Methylation Patterns and Epigenetic Memory. Genes Dev. 16, 6–21. 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- Borm P. J., Robbins D., Haubold S., Kuhlbusch T., Fissan H., Donaldson K., et al. (2006). The Potential Risks of Nanomaterials: a Review Carried Out for ECETOC. Part. Fibre Toxicol. 3, 11. 10.1186/1743-8977-3-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton C. V., Byun H.-M., Wenten M., Pan F., Yang A., Gilliland F. D. (2009). Prenatal Tobacco Smoke Exposure Affects Global and Gene-Specific DNA Methylation. Am. J. Respir. Crit. Care Med. 180, 462–467. 10.1164/rccm.200901-0135oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho A. L., Chuang A., Jiang W.-W., Lee J., Begum S., Poeta L., et al. (2006). Deleted in Colorectal Cancer Is a Putative Conditional Tumor-Suppressor Gene Inactivated by Promoter Hypermethylation in Head and Neck Squamous Cell Carcinoma. Cancer Res. 66, 9401–9407. 10.1158/0008-5472.can-06-1073 [DOI] [PubMed] [Google Scholar]
- Cheedipudi S., Genolet O., Dobreva G. (2014). Epigenetic Inheritance of Cell Fates During Embryonic Development. Front. Genet. 5, 19. 10.3389/fgene.2014.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. S., Morrison J. P., Southwell M. F., Foley J. F., Bolon B., Elmore S. A. (2017). Histology Atlas of the Developing Prenatal and Postnatal Mouse Central Nervous System, With Emphasis on Prenatal Days E7.5 to E18.5. Toxicol. Pathol. 45, 705–744. 10.1177/0192623317728134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Guo J., Lei Y., Zou J., Lu X., Bao Y., et al. (2010). Global DNA Hypomethylation Is Associated With NTD-Affected Pregnancy: A Case-Control Study. Birth Defects Res. A: Clin. Mol. Teratology 88, 575–581. 10.1002/bdra.20670 [DOI] [PubMed] [Google Scholar]
- Cisternas C. D., Cortes L. R., Bruggeman E. C., Yao B., Forger N. G. (2020). Developmental Changes and Sex Differences in DNA Methylation and Demethylation in Hypothalamic Regions of the Mouse Brain. Epigenetics 15, 72–84. 10.1080/15592294.2019.1649528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conerly M., Grady W. M. (2010). Insights into the Role of DNA Methylation in Disease Through the Use of Mouse Models. Dis. Model. Mech. 3, 290–297. 10.1242/dmm.004812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Liu H., Zhou M., Duan Y., Li N., Gong X., et al. (2011). Signaling Pathway of Inflammatory Responses in the Mouse Liver Caused by TiO2 Nanoparticles. J. Biomed. Mater. Res. 96A, 221–229. 10.1002/jbm.a.32976 [DOI] [PubMed] [Google Scholar]
- Deiner M. S., Sretavan D. W. (1999). Altered Midline Axon Pathways and Ectopic Neurons in the Developing Hypothalamus of Netrin-1- and DCC-Deficient Mice. J. Neurosci. 19, 9900–9912. 10.1523/jneurosci.19-22-09900.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dréno B., Alexis A., Chuberre B., Marinovich M. (2019). Safety of Titanium Dioxide Nanoparticles in Cosmetics. J. Eur. Acad. Dermatol. Venereol. 33 Suppl 7 (Suppl. 7), 34–46. 10.1111/jdv.15943 [DOI] [PubMed] [Google Scholar]
- Fan G., Beard C., Chen R. Z., Csankovszki G., Sun Y., Siniaia M., et al. (2001). DNA Hypomethylation Perturbs the Function and Survival of CNS Neurons in Postnatal Animals. J. Neurosci. 21, 788–797. 10.1523/jneurosci.21-03-00788.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Zhou Y., Campbell S. L., Le T., Li E., Sweatt J. D., et al. (2010). Dnmt1 and Dnmt3a Maintain DNA Methylation and Regulate Synaptic Function in Adult Forebrain Neurons. Nat. Neurosci. 13, 423–430. 10.1038/nn.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassian V. H., O’Shaughnessy P. T., Adamcakova-Dodd A., Pettibone J. M., Thorne P. S. (2007). Inhalation Exposure Study of Titanium Dioxide Nanoparticles With a Primary Particle Size of 2 to 5 Nm. Environ. Health Perspect. 115, 397–402. 10.1289/ehp.9469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougaard K. S., Jackson P., Jensen K. A., Sloth J. J., Löschner K., Larsen E. H., et al. (2010). Effects of Prenatal Exposure to Surface-Coated Nanosized Titanium Dioxide (UV-Titan). A Study in Mice. Part. Fibre Toxicol. 7, 16. 10.1186/1743-8977-7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Hirabayashi K., Moriishi K., Matsui Y., Moriya K., Koike K., et al. (2015). Novel Sex-Dependent Differentially Methylated Regions Are Demethylated in Adult Male Mouse Livers. Biochem. Biophysical Res. Commun. 462, 332–338. 10.1016/j.bbrc.2015.04.137 [DOI] [PubMed] [Google Scholar]
- Jangiam W., Tungjai M., Rithidech K. N. (2015). Induction of Chronic Oxidative Stress, Chronic Inflammation and Aberrant Patterns of DNA Methylation in the Liver of Titanium-Exposed CBA/CaJ Mice. Int. J. Radiat. Biol. 91, 389–398. 10.3109/09553002.2015.1001882 [DOI] [PubMed] [Google Scholar]
- Ji H., Biagini Myers J. M., Brandt E. B., Brokamp C., Ryan P. H., Khurana Hershey G. K. (2016). Air Pollution, Epigenetics, and Asthma. Allergy Asthma Clin. Immunol. 12, 51. 10.1186/s13223-016-0159-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri T., Ariel M., Brandeis M., Shemer R., Urven L., Mccarrey J., et al. (1992). Developmental Pattern of Gene-Specific DNA Methylation in the Mouse Embryo and Germ Line. Genes Development 6, 705–714. 10.1101/gad.6.5.705 [DOI] [PubMed] [Google Scholar]
- Kaida T., Kobayashi K., Adachi M., Suzuki F. (2004). Optical Characteristics of Titanium Oxide Interference Film and the Film Laminated With Oxides and Their Applications for Cosmetics. J. Cosmet. Sci. 55, 219–220. [PubMed] [Google Scholar]
- Kilkenny C., Browne W., Cuthill I. C., Emerson M., Altman D. G. (2010). Animal Research: Reporting In Vivo Experiments: The ARRIVE Guidelines. Br. J. Pharmacol. 160, 1577–1579. 10.1111/j.1476-5381.2010.00872.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawlor K. G., Narayanan R. (1992). Persistent Expression of the Tumor Suppressor Gene DCC Is Essential for Neuronal Differentiation. Cell Growth Differ. 3, 609–616. [PubMed] [Google Scholar]
- Le Belle J. E., Sperry J., Ngo A., Ghochani Y., Laks D. R., López-Aranda M., et al. (2014). Maternal Inflammation Contributes to Brain Overgrowth and Autism-Associated Behaviors through Altered Redox Signaling in Stem and Progenitor Cells. Stem Cel. Rep. 3, 725–734. 10.1016/j.stemcr.2014.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M., Wutz A. (2012). Establishment of Epigenetic Patterns in Development. Chromosoma. 121, 251–262. 10.1007/s00412-012-0365-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Tian M., Cui L., Dwyer J., Fullwood N. J., Shen H., et al. (2016). Low-Dose Carbon-Based Nanoparticle-Induced Effects in A549 Lung Cells Determined by Biospectroscopy Are Associated With Increases in Genomic Methylation. Sci. Rep. 6, 20207. 10.1038/srep20207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang J., Zhang Y., Wang Z., Song Y., Wei S., et al. (2019). TRAF2 Protects Against Cerebral Ischemia-Induced Brain Injury by Suppressing Necroptosis. Cell Death Dis. 10, 328. 10.1038/s41419-019-1558-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Saint-Cyr-Proulx E., Aktories K., Lamarche-Vane N. (2002). Rac1 and Cdc42 but Not RhoA or Rho Kinase Activities Are Required for Neurite Outgrowth Induced by the Netrin-1 Receptor DCC (Deleted in Colorectal Cancer) in N1E-115 Neuroblastoma Cells. J. Biol. Chem. 277, 15207–15214. 10.1074/jbc.m109913200 [DOI] [PubMed] [Google Scholar]
- Ma Y., Xia W., Wang D. Q., Wan Y. J., Xu B., Chen X., et al. (2013). Hepatic DNA Methylation Modifications in Early Development of Rats Resulting From Perinatal BPA Exposure Contribute to Insulin Resistance in Adulthood. Diabetologia. 56, 2059–2067. 10.1007/s00125-013-2944-7 [DOI] [PubMed] [Google Scholar]
- Mastroeni D., Mckee A., Grover A., Rogers J., Coleman P. D. (2009). Epigenetic Differences in Cortical Neurons From a Pair of Monozygotic Twins Discordant for Alzheimer's Disease. PLoS One. 4, e6617. 10.1371/journal.pone.0006617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill J., Tang T., Kaminsky Z., Khare T., Yazdanpanah S., Bouchard L., et al. (2008). Epigenomic Profiling Reveals DNA-Methylation Changes Associated With Major Psychosis. Am. J. Hum. Genet. 82, 696–711. 10.1016/j.ajhg.2008.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadipour A., Fazel A., Haghir H., Motejaded F., Rafatpanah H., Zabihi H., et al. (2014). Maternal Exposure to Titanium Dioxide Nanoparticles During Pregnancy; Impaired Memory and Decreased Hippocampal Cell Proliferation in Rat Offspring. Environ. Toxicol. Pharmacol. 37, 617–625. 10.1016/j.etap.2014.01.014 [DOI] [PubMed] [Google Scholar]
- Monk M., Boubelik M., Lehnert S. (1987). Temporal and Regional Changes in DNA Methylation in the Embryonic, Extraembryonic and Germ Cell Lineages During Mouse Embryo Development. Development 99, 371–382. 10.1242/dev.99.3.371 [DOI] [PubMed] [Google Scholar]
- Nahar M. S., Kim J. H., Sartor M. A., Dolinoy D. C. (2014). Bisphenol A-Associated Alterations in the Expression and Epigenetic Regulation of Genes Encoding Xenobiotic Metabolizing Enzymes in Human Fetal Liver. Environ. Mol. Mutagen. 55, 184–195. 10.1002/em.21823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C. H., Larsen A., Nielsen A. L. (2016). DNA Methylation Alterations in Response to Prenatal Exposure of Maternal Cigarette Smoking: A Persistent Epigenetic Impact on Health From Maternal Lifestyle? Arch. Toxicol. 90, 231–245. 10.1007/s00204-014-1426-0 [DOI] [PubMed] [Google Scholar]
- Oberdörster G., Oberdörster E., Oberdörster J. (2005). Nanotoxicology: an Emerging Discipline Evolving From Studies of Ultrafine Particles. Environ. Health Perspect. 113, 823–839. 10.1289/ehp.7339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim C., Reisert I. (1992). Differences Between Male and Female Brains - Developmental Mechanisms and Implications. Horm. Metab. Res. 24, 353–359. 10.1055/s-2007-1003334 [DOI] [PubMed] [Google Scholar]
- Pogribna M., Koonce N. A., Mathew A., Word B., Patri A. K., Lyn-Cook B., et al. (2020). Effect of Titanium Dioxide Nanoparticles on DNA Methylation in Multiple Human Cell Lines. Nanotoxicology 14, 534–553. 10.1080/17435390.2020.1723730 [DOI] [PubMed] [Google Scholar]
- Pregnolato S., Chakkarapani E., Isles A. R., Luyt K. (2019). Glutamate Transport and Preterm Brain Injury. Front. Physiol. 10, 417. 10.3389/fphys.2019.00417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochiantz A., Di Nardo A. A. (2015). Homeoprotein Signaling in the Developing and Adult Nervous System. Neuron. 85, 911–925. 10.1016/j.neuron.2015.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K. D. (2005). DNA Methylation and Human Disease. Nat. Rev. Genet. 6, 597–610. 10.1038/nrg1655 [DOI] [PubMed] [Google Scholar]
- Sato K., Tamura G., Tsuchiya T., Endoh Y., Usuba O., Kimura W., et al. (2001). Frequent Loss of Expression Without Sequence Mutations of the DCC Gene in Primary Gastric Cancer. Br. J. Cancer 85, 199–203. 10.1054/bjoc.2001.1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuermann D., Weber A. R., Schär P. (2016). Active DNA Demethylation by DNA Repair: Facts and Uncertainties. DNA Repair 44, 92–102. 10.1016/j.dnarep.2016.05.013 [DOI] [PubMed] [Google Scholar]
- Shandilya N., Le Bihan O., Bressot C., Morgeneyer M. (2015). Emission of Titanium Dioxide Nanoparticles from Building Materials to the Environment by Wear and Weather. Environ. Sci. Technol. 49, 2163–2170. 10.1021/es504710p [DOI] [PubMed] [Google Scholar]
- Shimizu M., Tainaka H., Oba T., Mizuo K., Umezawa M., Takeda K. (2009). Maternal Exposure to Nanoparticulate Titanium Dioxide During the Prenatal Period Alters Gene Expression Related to Brain Development in the Mouse. Part. Fibre Toxicol. 6, 20. 10.1186/1743-8977-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava R., Raza S., Yadav A., Kushwaha P., Flora S. J. S. (2014). Effects of Sub-Acute Exposure to TiO2, ZnO and Al2O3nanoparticles on Oxidative Stress and Histological Changes in Mouse Liver and Brain. Drug Chem. Toxicol. 37, 336–347. 10.3109/01480545.2013.866134 [DOI] [PubMed] [Google Scholar]
- Szulwach K. E., Li X., Li Y., Song C.-X., Wu H., Dai Q., et al. (2011). 5-hmC-Mediated Epigenetic Dynamics During Postnatal Neurodevelopment and Aging. Nat. Neurosci. 14, 1607–1616. 10.1038/nn.2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana K., Takayanagi K., Akimoto A., Ueda K., Shinkai Y., Umezawa M., et al. (2015). Prenatal Diesel Exhaust Exposure Disrupts the DNA Methylation Profile in the Brain of Mouse Offspring. J. Toxicol. Sci. 40, 1–11. 10.2131/jts.40.1 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Mizuo K., Shinkai Y., Oshio S., Takeda K. (2010). Prenatal Exposure to Titanium Dioxide Nanoparticles Increases Dopamine Levels in the Prefrontal Cortex and Neostriatum of Mice. J. Toxicol. Sci. 35, 749–756. 10.2131/jts.35.749 [DOI] [PubMed] [Google Scholar]
- Takeda K., Suzuki K.-i., Ishihara A., Kubo-Irie M., Fujimoto R., Tabata M., et al. (2009). Nanoparticles Transferred From Pregnant Mice to Their Offspring Can Damage the Genital and Cranial Nerve Systems. J. Health Sci. 55, 95–102. 10.1248/jhs.55.95 [DOI] [Google Scholar]
- Takizawa T., Nakashima K., Namihira M., Ochiai W., Uemura A., Yanagisawa M., et al. (2001). DNA Methylation Is a Critical Cell-Intrinsic Determinant of Astrocyte Differentiation in the Fetal Brain. Developmental Cel. 1, 749–758. 10.1016/s1534-5807(01)00101-0 [DOI] [PubMed] [Google Scholar]
- Tanvir S., Oudet F., Pulvin S., Anderson W. A. (2012). Coenzyme Based Synthesis of Silver Nanocrystals. Enzyme Microb. Technology 51, 231–236. 10.1016/j.enzmictec.2012.07.002 [DOI] [PubMed] [Google Scholar]
- Umezawa M., Tainaka H., Kawashima N., Shimizu M., Takeda K. (2012). Effect of Fetal Exposure to Titanium Dioxide Nanoparticle on Brain Development − Brain Region Information. J. Toxicol. Sci. 37, 1247–1252. 10.2131/jts.37.1247 [DOI] [PubMed] [Google Scholar]
- Venkatasubramanian G., Debnath M. (2013). The TRIPS (Toll-like Receptors in Immuno-Inflammatory Pathogenesis) Hypothesis: a Novel Postulate to Understand Schizophrenia. Prog. Neuro-Psychopharmacology Biol. Psychiatry 44, 301–311. 10.1016/j.pnpbp.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Weir A., Westerhoff P., Fabricius L., Hristovski K., Von Goetz N. (2012). Titanium Dioxide Nanoparticles in Food and Personal Care Products. Environ. Sci. Technol. 46, 2242–2250. 10.1021/es204168d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick P., Malek A., Manser P., Meili D., Maeder-Althaus X., Diener L., et al. (2010). Barrier Capacity of Human Placenta for Nanosized Materials. Environ. Health Perspect. 118, 432–436. 10.1289/ehp.0901200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. C., Zhang Y. (2010). Active DNA Demethylation: Many Roads lead to Rome. Nat. Rev. Mol. Cel. Biol. 11, 607–620. 10.1038/nrm2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Wang F., Liu Y., Yu Y., Gelernter J., Zhang H. (2014). Sex-Biased Methylome and Transcriptome in Human Prefrontal Cortex. Hum. Mol. Genet. 23, 1260–1270. 10.1093/hmg/ddt516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze Y., Sheng L., Zhao X., Hong J., Ze X., Yu X., et al. (2014). TiO2 Nanoparticles Induced Hippocampal Neuroinflammation in Mice. PLoS One 9, e92230. 10.1371/journal.pone.0092230 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu T., Brown A. P., Ji H. (2020). The Emerging Role of Ten-Eleven Translocation 1 in Epigenetic Responses to Environmental Exposures. Epigenet. Insights. 13, 2516865720910155. 10.1177/2516865720910155 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for this study can be found in the Gene Expression Omnibus (GSE178207) (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=GSE178207).