Significance
ΔNp63 is a master regulator of skin homeostasis since it finely controls keratinocyte differentiation and proliferation. Here, we provide cellular and molecular evidence demonstrating the functional role of a ΔNp63 interactor, the R-loop–resolving enzyme Senataxin (SETX), in fine-tuning keratinocyte differentiation. We found that SETX physically binds the p63 DNA–binding motif present in two early epidermal differentiation genes, Keratin 1 (KRT1) and ZNF750, facilitating R-loop removal over their 3′ ends and thus allowing efficient transcriptional termination and gene expression. These molecular events translate into the inability of SETX-depleted keratinocytes to undergo the correct epidermal differentiation program. Remarkably, SETX is dysregulated in cutaneous squamous cell carcinoma, suggesting its potential involvement in the pathogenesis of skin disorders.
Keywords: Senataxin, p63, skin differentiation
Abstract
ΔNp63, a master regulator of epithelial biology, is involved in regulating epithelial stem cell function, maintaining the integrity of stratified epithelial cells, and committing epidermal cells to the differentiation program. To this end, ΔNp63 exploits several direct mechanisms. Here, we elucidated a mechanism whereby ΔNp63 efficiently sustains the expression of epidermal differentiation genes. We show that ΔNp63 interacts with Senataxin (SETX), an RNA/DNA helicase able to resolve the R-loop intermediates over the GC-rich termination sites of coding genes. Notably, we found that SETX and ΔNp63 coregulate a subset of genes involved in the early step of the keratinocyte differentiation program. At the molecular level, SETX physically binds the p63 DNA–binding motifs present in two early epidermal differentiation genes, Keratin 1 (KRT1) and ZNF750, facilitating R-loop removal over their 3′ ends and thus promoting efficient transcriptional termination and gene expression. Remarkably, SETX loss affects the activation of the proper epidermal differentiation program in vitro and impacts epidermal layer stratification in organotypic human skin. Furthermore, we found that SETX is mutated or downmodulated in squamous cell carcinoma (SCC), and SETX gene mutation is a negative prognostic factor for cutaneous SCC patient survival. Collectively, our results unveil SETX as a molecular player of skin homeostasis potentially involved in hyperproliferative skin disorders.
The development and maintenance of the functional epidermal barrier is the result of a highly dynamic process that coordinates the proliferation, migration, and terminal differentiation of keratinocytes. This differentiation program is finely orchestrated by the action of transcription factors, cell signaling pathways, and epigenetic regulators that coordinate the stage-dependent expression of early differentiation proteins (e.g., keratin 1 and keratin 10) and late cornified envelope proteins (e.g., filaggrin and loricrin) in distinct epidermal layers (1, 2).
A master regulator of keratinocyte differentiation and skin homeostasis is the transcription factor p63 (3). The TP63 gene is expressed as multiple isoforms, among which the N-terminal truncated isoforms (hereinafter referred to as ΔNp63) are the most abundant isoforms expressed in the basal layer of stratified epithelia, including the epidermis (3–6). The critical role of ΔNp63 in maintaining the integrity of stratified epithelial cells was formally demonstrated by an analysis of a mouse model devoted to ΔNp63 isoforms. Similar to mice lacking all p63 isoforms, ΔNp63 knockout mice showed defects of the skin, oral epithelium, mammary glands, limb, and craniofacial region (7, 8). In humans, heterozygous mutations in the TP63 gene cause several developmental disorders, which partially resemble the developmental defects observed in ΔNp63-null mice (9, 10).
At the molecular level, classical and genome-wide approaches have allowed the identification and characterization of the ΔNp63-dependent transcriptome, which includes transcriptional targets involved in sustaining stem cell function, regulating cell adhesion, and, paradoxically, activating the differentiation program (11–19). The current view is that ΔNp63 activity is fundamental to maintain the proliferative potential of basal epithelial cells and, at the same time, initiate the early step of the differentiation program. Accordingly, ΔNp63 activity has been involved in the transcriptional regulation of several genes involved in triggering the terminal differentiation, such as the transcription factor ZNF750 (20).
To directly regulate its numerous target genes, ΔNp63 recruits epigenetic modulators and chromatin remodeling factors (21–23). For instance, ΔNp63 associates with DNMT3a to maintain high levels of DNA hydroxymethylation at the center of enhancers of epidermal genes in a Tet2-dependent manner (24). ΔNp63 also acts as a transcriptional repressor by recruiting the histone deacetylases HDAC1 and HDAC2 (25) or interacting with the epigenetic modifier ACTL6a (26).
Here, we elucidated a mechanism exploited by ΔNp63 to efficiently sustain the expression of epidermal differentiation genes. We show that ΔNp63 is able to interact with Senataxin (SETX), an RNA/DNA helicase able to resolve R-loop intermediates over the GC-rich termination sites of RNA polymerase II and III genes (27, 28). Notably, we found that SETX and ΔNp63 coregulate a subset of genes involved in skin development and keratinocyte differentiation and that depletion of SETX markedly affects the activation of the epidermal differentiation program in vitro and impacts epidermal layer stratification in organotypic human skin. At the molecular level, we found that SETX physically binds the p63 DNA–binding motif present in two epidermal differentiation genes, Keratin 1 (KRT1) and ZNF750, facilitating R-loop removal over their 3′ ends and thus allowing efficient transcriptional termination and gene expression. Collectively, our results unveil a molecular mechanism by which ΔNp63 finely tunes keratinocyte differentiation and skin homeostasis.
Results
The RNA/DNA Helicase SETX Is a ΔNp63 Interactor.
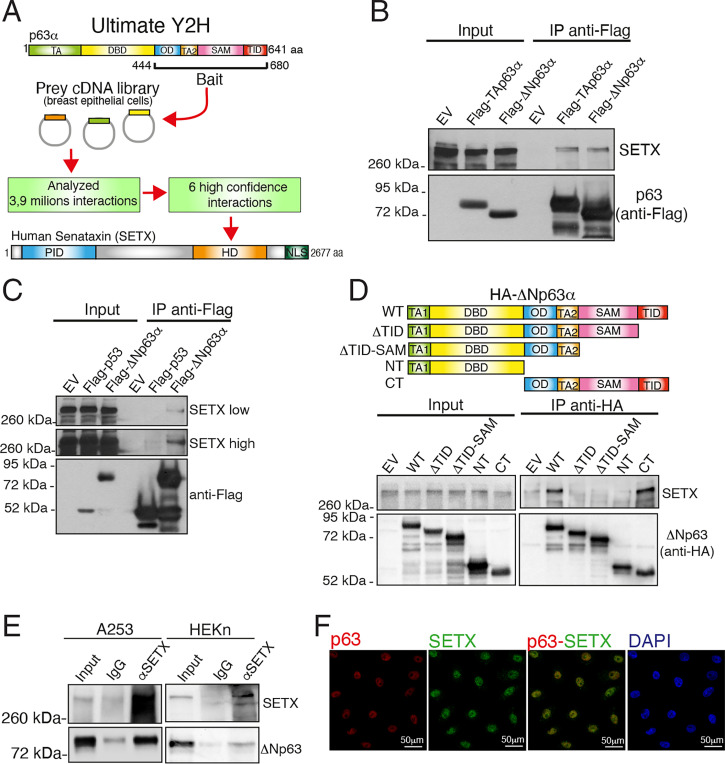
Compared to p53, p63 and p73 alpha isoforms are characterized by a unique C terminus containing the sterile alpha motif (SAM) domain and the trans inhibitory (TI) domain (29). To gain insight into the functional relevance of these p63 C-terminal domains, we performed a yeast two-hybrid screen using the C-terminal fragment of p63α (amino acids 444 to 680), which contains the oligomerization, SAM, and TI domains, as bait (Fig. 1A). As prey, we utilized the Human Breast Tumor Epithelial cDNA library based on the pivotal role of p63 in regulating breast epithelial homeostasis (8, 30, 31). We analyzed ∼3.9 million hits and found several high-confidence p63 interactors. Known p63 interactors such as PIAS1 and RACK1 were selected by this screen (Dataset S1). Among the high-confidence p63 interactors, we focused our attention on SETX, an RNA/DNA helicase implicated in transcriptional regulation and the DNA damage response through the resolution of R-loop structures (27, 32). We first confirmed the interaction of p63 isoforms with SETX at semiendogenous levels. As shown in Fig. 1B, endogenous SETX was able to bind to both ectopically expressed TAp63 and ΔNp63 alpha isoforms. Remarkably, SETX does not bind to p53 (Fig. 1C). To gain insight into the specificity of the SETX-p63 interaction, we mapped the p63 domains responsible for SETX binding. To achieve this aim, we generated HA-tagged ΔNp63α deletion mutants and performed coimmunoprecipitation experiments. As shown in Fig. 1D, wild-type ΔNp63α and the CT mutant, lacking the TA and the DBD domains, efficiently bound endogenous SETX. On the contrary, neither the carboxyl terminus–deleted NT mutant nor the TID and the TID/SAM domain–deleted mutants (respectively named ΔTID and ΔTID-ΔSAM) were able to interact with SETX. These data indicate that the C-terminal portion of p63 is necessary for the SETX-ΔNp63 interaction, thus validating the Y2H results. To confirm this interaction at the endogenous level, we analyzed the SETX-ΔNp63α interaction in human epidermal keratinocytes, neonatal (HEKn) and in two epithelial cancer cell lines, A253 (squamous cell carcinoma) and MCF-7 (breast cancer), which express predominantly the ΔNp63α isoform (33, 34). In all three cell types, we confirmed the formation of a complex between SETX and ΔNp63 at endogenous levels (Fig. 1E and SI Appendix, Fig. S1A). We also performed colocalization studies in immortalized keratinocytes Ker-CT (Fig. 1F). For this approach, we firstly validated the anti-SETX antibody for immunofluorescence studies. As shown in SI Appendix, Fig. S1B and Fig. 1F, the anti-SETX antibody is specific and recognizes a nuclear protein which colocalizes with endogenous ΔNp63.
Fig. 1.
SETX is a p63 interactor. (A) Schematic workflow recapitulating the Ultimate Yeast two-hybrid screening approach utilized to identify p63 interactors. The p63α isoform contains the following protein domains: the transactivation domain (TA), DNA-binding domain (DBD), oligomerization domain (OD), transactivation domain 2 (TA2), sterile alpha motif (SAM), and transactivation inhibitory domain (TID). The following protein domains of SETX are also indicated: the protein interaction domain (PID), helicase domain (HD), and nuclear localization signals (NLS). (B) H1299 cells were transfected with the indicated FLAG-tagged p63 constructs or the empty vector (EV). Exogenous proteins were immunoprecipitated (IP) from cell extracts with anti-FLAG antibody, and immunocomplexes were analyzed with the indicated antibodies. (C) H1299 cells were transfected with the indicated FLAG-tagged constructs or the EV. Exogenous proteins were subjected to IP with an anti-FLAG antibody, and immunocomplexes were analyzed with anti-Flag and anti-SETX antibodies. (D) H1299 cells were transfected with EV, HA-tagged ΔNp63 (wild type, WT), or the HA-tagged deletion mutants (ΔTID, ΔTID-ΔSAM, NT, CT) as shown in the schematic model (Upper). Cell extracts were subjected to IP with an anti-HA antibody followed by immunoblotting with antibodies to the indicated protein. (E) Whole-cell extracts from A253 tumor cell line and human primary keratinocytes HEKn cells were subjected to IP with an anti-SETX antibody followed by immunoblotting with the indicated antibodies. (F) Immunofluorescence staining of SETX (green) and p63 (red) in immortalized keratinocytes (Ker-CT). DAPI (blue) was used to visualize nuclei.
Taken together, these results identify the RNA/DNA helicase SETX as a ΔNp63 interactor.
ΔNp63 and SETX Coregulate a Subset of Genes Involved in the Early Step of the Keratinocyte Differentiation Program.
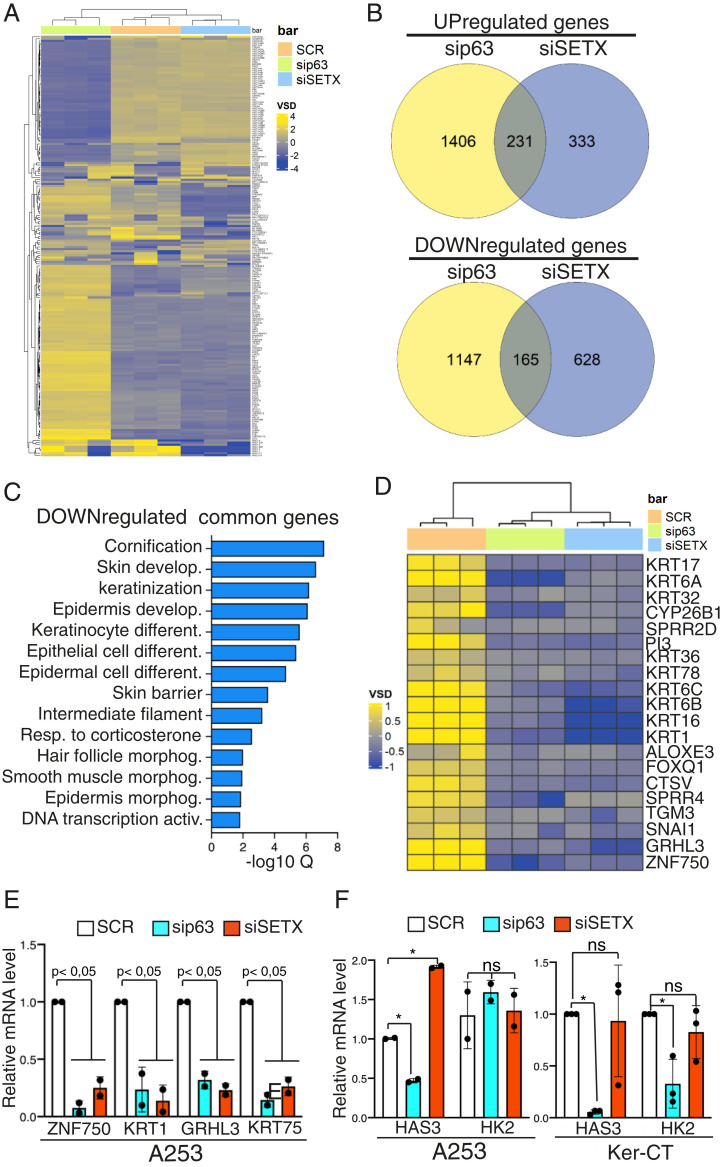
As SETX is an RNA/DNA helicase able to resolve R-loop structures, we investigated whether the association of ΔNp63 with SETX might trigger changes in R-loop formation. To achieve this aim, we silenced ΔNp63 or SETX in normal human keratinocytes and then measured R-loop signals by dot blot and immunofluorescence utilizing the S9.6 antibody that specifically recognizes RNA/DNA hybrids (35, 36). As shown in SI Appendix, Fig. S2, depletion of SETX or ΔNp63 led to a modest, not statistically significant, change in R-loop formation, suggesting that the global R-loop signature is not affected by dysregulation of SETX or ΔNp63 levels, at least in our experimental conditions. Since ΔNp63 controls the transcription of a subset of genes and R-loop forms cotranscriptionally (37), we then tested the hypothesis that the functional relevance of the ΔNp63-SETX interaction might be related to the transcriptional regulation of specific genes. To address this issue, we performed RNA sequencing (RNA-seq) analysis to compare the transcriptome of the epithelial cell line A253 treated with a control small interfering RNA (siRNA) (scramble [SCR]), siSETX, or sip63 oligos (SI Appendix, Fig. S3). SETX and ΔNp63 silencing affected ∼1,350 and 2,900 genes, respectively, with similar proportions between down- and up-regulated genes (Fig. 2B). To test whether ΔNp63 and SETX might influence the transcription of common pathways, we performed Gene Ontology (GO) analysis of those genes coderegulated in SETX- and ΔNp63-depleted cells. While we did not find any relevant enrichment of pathways regulated by coup-regulated genes, the GO analysis of common down-regulated genes revealed statistically relevant enrichment of genes related to cornification, skin development, epidermis development, and keratinization (Fig. 2 C and D). To validate these results, we performed RT-qPCR analysis of RNA extracted from SETX- or ΔNp63-depleted cells, and we confirmed that depletion of SETX or ΔNp63 affects the expression of genes involved in epithelial morphogenesis, such as ZNF750, GRHL3, keratin 1 (KRT1), and keratin 75 (KRT75) (Fig. 2E). Remarkably, SETX loss does not affect the mRNA levels of two well-established ΔNp63 target genes (HAS3 and HK2) (Fig. 2F), indicating that SETX specifically regulates a subset of epidermal differentiation genes. Importantly, the RNA-seq analysis performed in immortalized human keratinocytes upon depletion of SETX or ΔNp63 also confirmed the coregulation of a subset of genes involved in epithelial morphogenesis and epidermal differentiation (SI Appendix, Fig. S4 A–D).
Fig. 2.
SETX and ΔNp63 control a common transcriptional signature involved in epidermal differentiation. (A) Heatmap showing the unsupervised hierarchical clustering of the most variables (Deseq2 VSD) genes (n = 200) in the SCR (orange), sip63 (green), and siSETX (blue) transfected cells. Color scheme: violet (highest), yellow (lowest) VSD score. (B) Venn diagram showing the shared down-regulated genes (Top) and up-regulated genes (Bottom) in sip63 (yellow) and siSTEX (violet) transfected cells. (C) Bar plot showing the top GO terms for Biological Process, Molecular Function, and Cell Compartment ordered by false discovery rate (FDR). (D) Heatmap showing the unsupervised hierarchical clustering of genes associated to the top 5 GO terms in SCR (orange), sip63 (green), and siSETX (blue) transfected cells. Color scheme: violet (highest), yellow (lowest) VSD score. (E) A253 cells were transfected with siRNA targeting p63 (sip63), SETX (siSETX), or a nonrelevant mRNA (scramble, SCR), and mRNA levels of the indicated genes were quantified by RT-qPCR. Data are shown as the mean ± SD of two biological replicates (n = 2). P value was calculated by Student’s t test. (F) A253 and immortalized keratinocytes (Ker-CT) were transfected as in E, and mRNA levels of the indicated genes were quantified by RT-qPCR. Data are shown as the mean ± SD of two biological replicates (n = 2) for A253 and as the mean ± SD of three biological replicates (n = 3) for Ker-CT. P value was calculated by Student’s t test. *P < 0.05, ns = not significant.
Overall, these data indicate that SETX and ΔNp63 control a subset of genes involved in the early step of the keratinocyte differentiation program.
SETX Is Required for Efficient Transcription Termination of KRT1 and ZNF750 Genes.
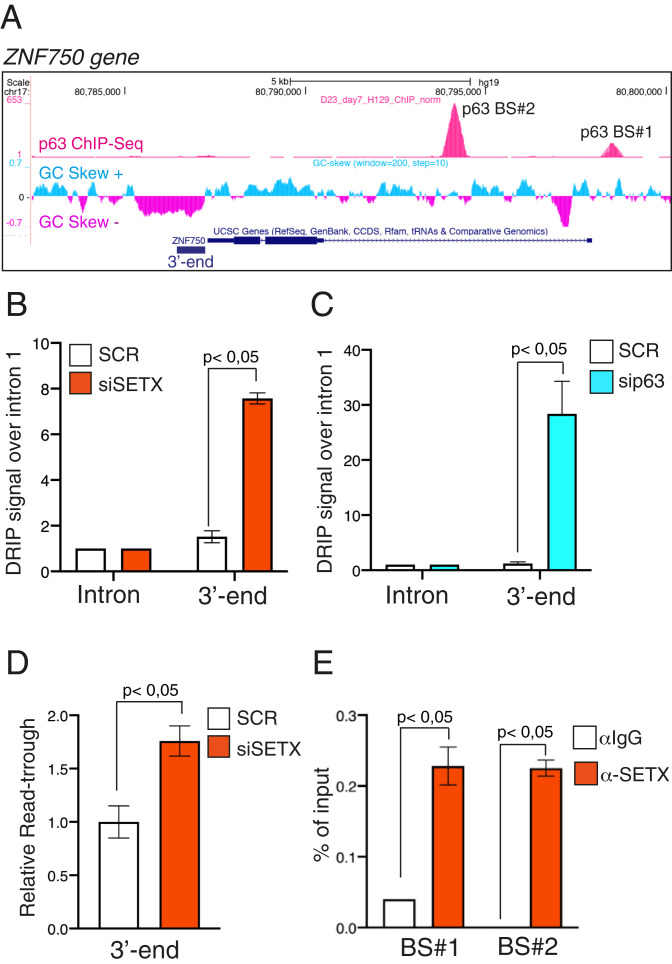
The transcriptional signature coregulated by SETX and ΔNp63 includes epidermal differentiation genes that have been shown to be bona fide transcriptional targets of ΔNp63. Since SETX promotes gene expression mainly by regulating transcription termination, we reasoned that the ΔNp63/SETX complex might facilitate the resolution of termination-associated R-loops of epidermal genes. To prove this hypothesis, we analyzed the ability of SETX to promote transcription termination over the epidermal differentiation gene ZNF750 since 1) it is an important player in skin homeostasis, and its expression is down-regulated by ΔNp63 or SETX silencing; 2) it is a direct target of ΔNp63 (20); and 3) its genomic locus is characterized by an extensive GC skew sequence located downstream of the poly(A) signal, which might favor the formation of R-loops and, as a consequence, impact termination efficiency (Fig. 3A). We first assessed whether SETX depletion led to an increase in R-loop signals at the 3′ ends of the ZNF750 gene. To achieve this aim, we employed the DNA–RNA immunoprecipitation (DRIP) assay, which relies on the immunoprecipitation of noncross-linked purified RNA/DNA hybrids, using the S9.6 antibody (35). We found that in immortalized keratinocytes and in A253 cells, SETX depletion increases R-loop signals that were specifically enriched in the termination region of the endogenous ZNF750 gene (Fig. 3B and SI Appendix, Fig. S5A). Importantly, p63 depletion exerts a similar increase of the DRIP signal (Fig. 3C), suggesting that the SETX-ΔNp63 complex is functionally important to resolve the R-loops structure in the termination region of the ZNF750 gene. To test the specificity of the signal detected by the S9.6 antibody, we performed RNase H1 nuclease treatment prior to the immunoprecipitation step. As demonstrated in SI Appendix, Fig. S5B, treatment of the samples with RNase H1 abolished the R-loop signals. RNase H1 also decreased R-loop signals formed over the transcribed regions of three R-loop–positive loci (RPL13A, CALM3, and TFTP), strengthening the reliability of our DRIP-based experimental approach (SI Appendix, Fig. S5C). To formally prove that SETX depletion resulted in transcription termination defects, we analyzed the production of readthrough transcripts at the 3′ ends. Using this approach, we detected readthrough transcripts in the absence of SETX at the ZNF750 3′ terminal region (Fig. 3D). To functionally link these events to the ability of SETX to bind ΔNp63, we tested whether SETX is able to occupy the p63 DNA–binding sites located in the proximal promoter and in the first intron of the ZNF750 locus (Fig. 3A) (12, 20). To achieve this aim, we performed a chromatin immunoprecipitation (ChIP) assay using the ChIP grade anti-SETX antibody, and we found that SETX physically binds the two p63 DNA–binding motifs in the ZNF750 gene (Fig. 3E and SI Appendix, Fig. S5D), suggesting a looped gene architecture that might facilitate functional cross-talk between the initiation and termination processes as described for several genes (38). Remarkably, SETX function seems to be specific for epidermal differentiation genes since its depletion does not affect neither R-loop signals nor the production of readthrough transcripts at the 3′ ends of another ΔNp63 transcriptional target gene, the hyaluronic acid synthase HAS3 (SI Appendix, Fig. S6 A and B).
Fig. 3.
SETX regulates R-loop formation on the 3′ end of the p63 target gene ZNF750. (A) Schematic representation of ZNF750 genetic locus (modified from Genome Browser, https://genome.ucsc.edu) showing from top to bottom p63 ChIP-seq peaks in human keratinocytes (12), GC skew signal, and gene structure with exons (boxes) and introns (lines). (B) Quantification by qPCR of DRIP signal in differentiated keratinocytes treated with SCR or SETX siRNAs. Values at 3′ ends were normalized to ZNF750 intron 1 signal. Bars represent the mean of three replicates (n = 3, PCR runs) ± SD and are representative of three independent experiments (n = 3 biological replicates). P value was calculated by Student’s t test. (C) Quantification by qPCR of DRIP signal in differentiated keratinocytes treated with SCR or p63 siRNAs. Values at 3′ ends were normalized to ZNF750 intron 1 signal. Bars represent the mean of three replicates (n = 3, PCR runs) ± SD and are representative of two independent experiments (n = 2 biological replicates). P value was calculated by Student’s t test. (D) RT-qPCR analysis of readthrough transcripts at the ZNF750 3′ region in SCR or siSETX differentiated keratinocytes. Values are normalized to ZNF750 intron 1 and showed as fold enrichment of siSETX over SCR. Bars represent the mean of three replicates (n = 3, PCR runs) ± SD and are representative of three independent experiments (n = 3 biological replicates). P value was calculated by Student’s t test. (E) Quantification by qPCR of ChIP analysis of endogenous SETX occupancy at p63-binding sites (BS#1 and BS#2) in differentiated human keratinocytes. Bars represent the mean of three replicates (n = 3, PCR runs) ± SD and are representative of three independent experiments (n = 3 biological replicates). P value was calculated by Student’s t test.
To provide additional proof of the ability of SETX to specifically modulate the termination of epidermal genes, we analyzed the impact of SETX anti–R-loop activity in regulating the early differentiation gene KRT1, which, unlike the ZNF750 gene, has a p63 DNA–binding motif located downstream of the poly(A) signal (SI Appendix, Fig. S7A). We found that SETX silencing increased RNase H1-sensitive R-loop signals downstream of poly(A) (SI Appendix, Fig. S7 B and C) and led to the production of readthrough transcripts at the 3′ ends of the KRT1 gene (SI Appendix, Fig. S7D). Furthermore, SETX was able to physically bind the genomic locus overlapping the p63-binding site downstream of the poly(A) signal (SI Appendix, Fig. S7E). Taken together, these results demonstrate that SETX promotes the efficient transcriptional termination of epidermal genes, thus sustaining their expression.
SETX Regulates Skin Differentiation.
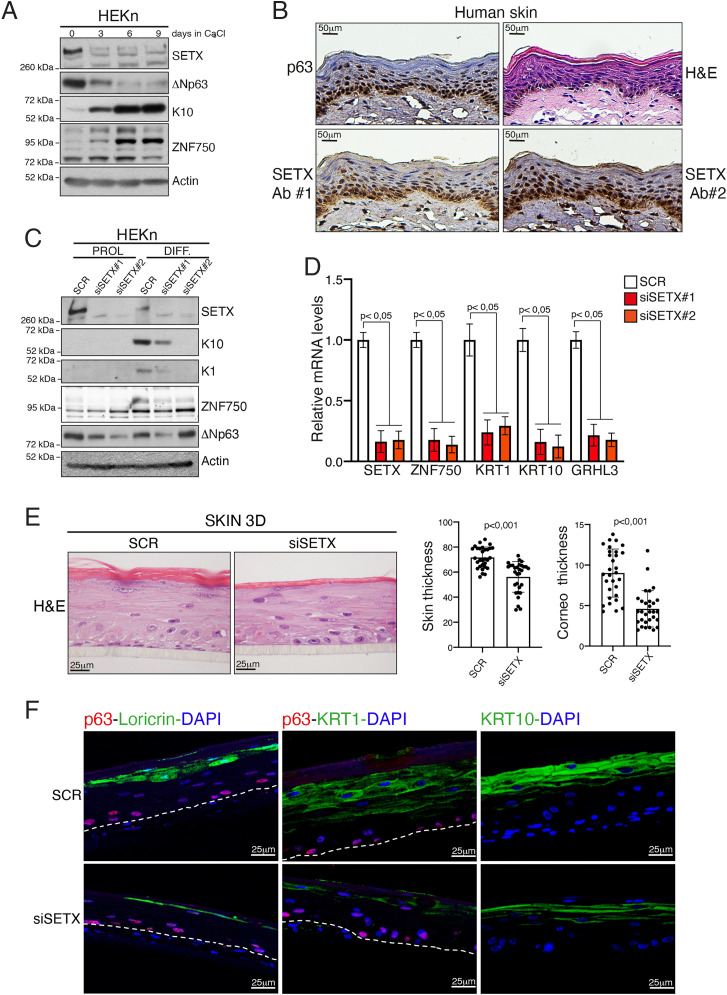
To assess the physiological relevance of the SETX-mediated control of the transcriptional termination of epithelial genes, we tested the role of SETX in skin differentiation since both KRT1 and ZNF750 genes are important players of the epidermal differentiation program. We utilized the well-established model of HEKn that are able to undergo calcium-induced differentiation in vitro. We first analyzed the expression of SETX upon the onset of keratinocyte differentiation. Similar to that of ΔNp63, the expression of the SETX protein decreased upon calcium treatment, concomitant with the up-regulation of differentiation markers such as keratin 10 (KRT10) and ZNF750 (Fig. 4A). SETX down-regulation is likely due to posttranscriptional events, as its mRNA levels do not change upon induction of keratinocyte differentiation (SI Appendix, Fig. S8A). To assess the in vivo localization of SETX, we performed an immunohistochemistry (IHC) and immunofluorescence analysis in human skin sample. As shown in Fig. 4B and SI Appendix, Fig. S8B, SETX localization overlaps with that of ΔNp63, being its expression mainly restricted to the basal compartment and reduced in the upper layer of human epidermis.
Fig. 4.
SETX depletion impairs keratinocyte differentiation. (A) Immunoblotting analysis of cell lysates extracted from HEKn at different time points (0, 3, 6, and 9 d) upon CaCl2 treatment. (B) Representative images of hematoxylin/eosin (H&E) staining or IHC analysis of p63 and SETX expression in normal skin samples. SETX antibody #1 (SETX Ab#1) is from Novus Biologicals (NBP1-94712), and SETX antibody #2 (SETX Ab#2) is a kind gift from Domenico Delia (University of Milan, Italy). (C) HEKn cells transfected with nonrelevant siRNA (SCR) or two different siRNAs targeting SETX mRNA (siSETX#1 or siSETX#2) were treated with CaCl2 to induce differentiation. Protein lysates from proliferating (PROL) or differentiating keratinocytes (DIFF, 4 d of differentiation) were analyzed by Western blotting using antibodies to the indicated proteins. (D) mRNA extracted from differentiated keratinocytes was used for RT-qPCR quantification of different epithelial differentiation markers. Bars represent the mean of three replicates (n = 3, PCR runs) ± SD and are representative of two independent experiments (n = 2 biological replicates). (E) H&E staining of SETX-depleted (siSETX) organotypic human epidermis compared to scramble control organotypic epidermis (SCR). Histograms on Right show quantification of whole skin or stratum corneum thickness in SCR (n = 30 measurement) and siSETX (n = 30 measurement) organotypic epidermis. (F) Immunofluorescence staining of the differentiation markers loricrin, KRT1, and KRT10 (green) in SCR and siSETX organotypic skin cultures. p63 staining (red) and DAPI (blue) were used to visualize the basal layer and nuclei, respectively. Dotted lines underline the keratinocyte–fibroblast border.
Since ΔNp63 and SETX are similarly modulated during the onset of keratinocyte differentiation and skin stratification and ΔNp63 critically regulates the skin differentiation program, we analyzed the effect of SETX silencing on the differentiation capabilities of normal human keratinocytes. As shown in Fig. 4 C and D, SETX depletion impairs the keratinocyte differentiation program as revealed by the lack of calcium-mediated up-regulation of several differentiation markers at both the mRNA and protein levels.
To validate these results in a more physiological model of epidermal development, we generated an organotypic human epidermal model that recapitulates the structure and the gene expression profile of the human epidermis. Although normally stratified, the SETX‐deficient epidermis appeared less thick than the wild-type epidermis, with a marked decrease in stratum corneum thickness (Fig. 4E). In parallel, we observed a marked decrease of the expression of key differentiation genes, including ZNF750 (Fig. 4F and SI Appendix, Fig. S8C). Notably, SETX depletion affects the expression of both late (loricrin) and early differentiation markers (KRT10 and KRT1). Taken together, these results uncover a role of the DNA/RNA helicase SETX in modulating keratinocyte differentiation.
SETX Is Mutated or Downmodulated in Squamous Cell Carcinoma, and SETX Gene Mutation Is a Negative Prognostic Factor for Cutaneous SCC Patient Survival.
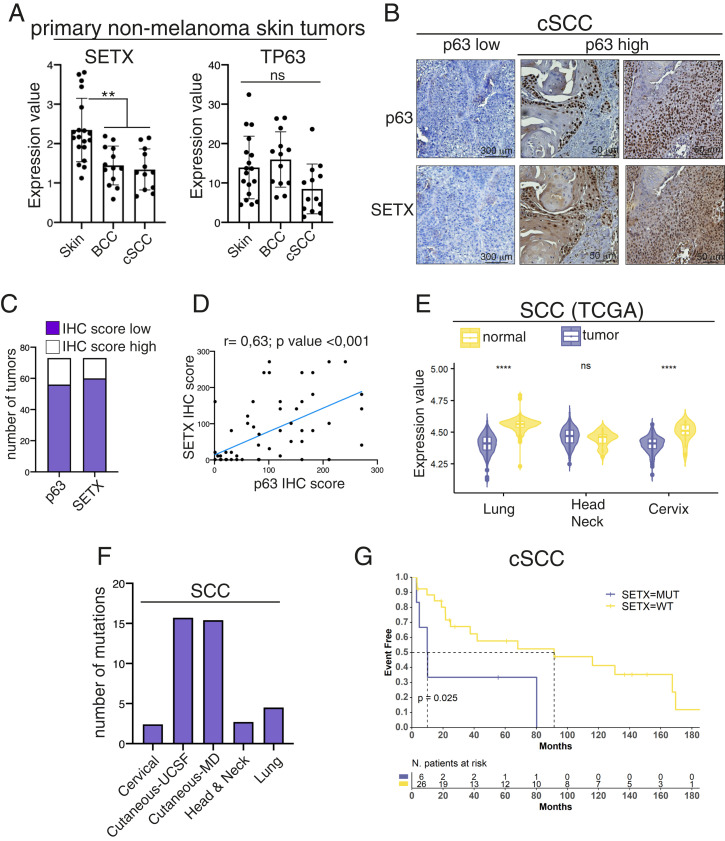
The results described so far clearly indicate that the SETX-ΔNp63 circuit controls keratinocyte differentiation. Since several skin pathologies are associated with alteration of the keratinocyte differentiation program, we decided to analyze SETX expression in two hyperproliferative skin disorders: psoriasis and squamous carcinogenesis. Regarding psoriasis disease, we analyzed SETX expression in the publicly available GSE13355 dataset, which comprises microarray data obtained from normal and psoriatic lesions. As shown in SI Appendix, Fig. S9, SETX expression is downmodulated in psoriatic lesions respect to normal skin. Regarding the squamous carcinogenesis, we firstly analyzed SETX mRNA levels in primary basal cell carcinoma and cutaneous squamous cell carcinoma (cSCC). Similarly to what we observed in psoriatic lesions, SETX expression is down-regulated in these primary tumors (Fig. 5A). To validate these data in cSCC tissues, we analyzed SETX protein levels by IHC in the TMA comprising 73 cSCC samples. We observed that the majority of cSCC samples express low levels of SETX, and its expression is confined mainly in those basal-derived cells expressing p63 (Fig. 5 B and C). Accordingly, we found a positive correlation between p63 and SETX protein levels in cSCC (Fig. 5D).
Fig. 5.
SETX is mutated or downmodulated in SCC, and SETX gene mutation is a negative prognostic factor for cutaneous SCC patient survival. (A) Total RNA extracted from normal skin (n = 18), basal cell carcinoma (BCC, n = 13), and cutaneous SCC (n = 12) samples was utilized for RT-qPCR quantification of SETX and p63 mRNA. **P < 0.01, ns = not significant. (B) Representative images of IHC analysis of SETX and p63 expression in skin SCC. (C) IHC score for p63 and SETX staining was calculated for each tumor sample. Distribution among high- and low-expressing tumors is shown for SETX and p63. (D) Correlation plot of SETX and p63 IHC scores in 73 human cSCC. Pearson’s correlation coefficient (r) and the P value of the correlation study are indicated. (E) Violin plot illustrating SETX expression values in SCCs and in the respective normal tissues. Asterisks indicate statistically significant differences (****P < 0.05), ns = not significant. (F) Bar plot showing the number of SETX mutations in the indicated types of SCCs. (G) Kaplan–Meier survival curves of Event Free related cSCC patients harboring wild-type (WT) or mutated SETX allele.
To extend this analysis to other SCC arising from different anatomical sites, we performed a bioinformatic analysis on The Cancer Genome Atlas datasets and found that SETX expression is significantly decreased in cervical and lung squamous cell carcinoma (SCC) (Fig. 5E). In addition to mRNA and protein dysregulation, we also found that the SETX gene is mutated in lung (4.5%), head and neck (2.7%), cervical (2.4%), and cSCC (15%) (Fig. 5F), and cSCC patients harboring SETX mutation showed a decrease of the overall survival compared to those with wild-type SETX (Fig. 5G).
To test whether SETX dysregulation might impact some cancer-related phenotypes, we analyzed the effect of SETX downmodulation on tumor proliferation and migration using as cellular models the oral SCC cell line A253 and the cutaneous SCC cell line A431. These cell lines express ΔNp63 and SETX and are derived from tumors in which SETX is down-regulated or mutated (SI Appendix, Fig. S10A). We did not find any changes of the proliferative and migratory capabilities upon SETX depletion (SI Appendix, Fig. S10 B–F), suggesting that SETX does not controls these processes, at least in our experimental models.
All together, these data suggest that SETX downmodulation is a cancer-related event occurring in some SCC, raising the possibility that SETX might function as a tumor suppressor gene during squamous carcinogenesis.
Discussion
R-loops are three-stranded structures formed by RNA:DNA hybrids and displaced single-stranded DNA that are physiological intermediates in both DNA and RNA synthesis processes (39). The turnover of R-loops is finely regulated since their unscheduled formation can interfere with important physiological processes such as gene expression, DNA replication, and DNA repair and ultimately lead to pathological events such as neurodegeneration. SETX is an R-loop–resolving enzyme involved mainly in the removal of transcriptional R-loops over the GC-rich termination sites of coding genes, thus allowing efficient transcription termination (27, 28).
In this manuscript, we characterized SETX as an important regulator of the epidermal differentiation program. We found that SETX is able to control the transcription termination of genes involved in the onset of keratinocyte differentiation such as KRT1 and ZNF750. The depletion of SETX increases the R-loop signals over the 3′ ends of these genes, causing transcriptional readthrough and diminished gene expression. Importantly, these molecular events translate into the inability of SETX-depleted keratinocytes to undergo a correct epidermal differentiation program as revealed by defects in the expression of early epidermal differentiation markers in primary human keratinocytes and organotypic human skin.
These data represent an example of the involvement of SETX in the epidermal differentiation program. The physiological relevance of SETX in skin homeostasis is also supported by its ability to functionally interact with ΔNp63, a transcription factor critically involved in keratinocyte proliferation and differentiation. We showed that SETX physically associates with ΔNp63 at endogenous levels and that this complex might be important to recruit SETX over epidermal genes. Indeed, we found that SETX is able to occupy the p63 DNA–binding motif located in the KRT1 and ZNF750 genes, suggesting that ΔNp63 might facilitate the recruitment of SETX over epidermal differentiation genes, thus promoting efficient transcription termination. Remarkably, there are other examples of SETX-interacting factors that might dictate the gene specificity function of SETX. For instance, the DNA repair enzyme BRCA1 mediates the recruitment of SETX to R-loops that normally form over a subset of transcription termination regions (40). The disruption of the BRCA1-SETX complex led to transcription termination defects, R-loop persistence, and DNA damage at those regions. In addition, SETX is able to interact with several RNA transcription and processing factors (41). Collectively, these observations and our findings support a fascinating model in which SETX might regulate transcription termination at specific loci through the recruitment of factors that dictate gene specificity. This consideration might also explain the ability of SETX to modulate the expression of a specific subset of genes in a cell-specific manner. Initial studies indeed reported that human fibroblasts with defects in SETX function are characterized by deregulation of several stress response genes such as SOD1 (42, 43). More recently, SETX depletion has been reported to impact the expression of autophagy-related genes, which results in autophagic flux defects.
The ability of SETX to regulate the expression of specific genes might also explain the absence of a global effect of SETX depletion on R-loop signals. Indeed, we did not find any global changes in R-loop formation in SETX-deficient keratinocytes; rather, we found a trend, although not statistically significant, toward R-loop loss. This result appears to contradict studies that associated the loss of SETX with R-loop gains. Although this discrepancy might be explained by the low sensitivity of our experimental approaches or the redundancy of enzymes with anti–R-loop activities, we believe that the ability of SETX to resolve R-loops is restricted to specific loci and that the interaction of SETX with ΔNp63 might ensure this gene specificity in the cellular context of epidermal tissue.
In addition to physically interacting with each other, SETX and ΔNp63 are also similarly regulated during the onset of keratinocyte differentiation. We showed that the SETX and ΔNp63 proteins are down-regulated concomitantly with the induction of the keratinocyte differentiation program, likely via a post-transcriptional mechanism. Although this result seems to be in contrast with the positive role of SETX in modulating the expression of early differentiation genes, it is worth noting that the same scenario has been described for ΔNp63, which is able to paradoxically regulate the proliferation potential of basal epithelial cells and control commitment to epidermal differentiation. The differentiation-mediated downmodulation of SETX is also quite interesting considering our poor understanding of the mechanisms regulating SETX expression and turnover. Uncovering the circuits controlling SETX expression might be relevant for the pathogenesis of human disorders whereby SETX is mutated, such as ataxia with oculomotor apraxia type 2 (AOA2), amyotrophic later sclerosis type 4 (ALS4) (44), and SCC (our data). We indeed found that SETX is either down-regulated or mutated in a subset of SCC. Although we did not find any changes of the proliferative and migratory capabilities of SCC cells upon SETX depletion, we found that cutaneous SCC patients harboring SETX mutations have a worse prognosis with respect to patients with the wild-type allele, suggesting a potential tumor suppressor role of SETX in SCC. This hypothesis is also strengthened by the finding that some neuro-related SETX mutations have been found in human tumors. In detail, two ataxia with AOA2-related SETX mutations, R1363X and P413L, have been reported in colon adenocarcinoma and cervical SCC, respectively (cBIOPortal for Cancer Genomics; http://www.cbioportal.org/public-portal). Additionally, a study involving 32 ALS4 patients has revealed that eight patients have benign or malignant neoplasia of the colon (colonic polyps or colon adenocarcinoma) (45). Markedly, one patient with colon adenocarcinoma is a 35-y-old male, a relatively early onset given that the colon adenocarcinoma frequency is just 0.4% in men younger than 49 y. These observations raise the possibility that the SETX mutation might be an early and causative tumor event, playing a role during the early onset of tumor initiation. Future investigations will be necessary to test this intriguing hypothesis.
In conclusion, our data revealed the involvement of the R-loop–resolving activity of SETX in the regulation of the expression of early keratinocyte differentiation genes and suggests its potential involvement in the pathogenesis of skin disorders.
Materials and Methods
Cell Lines, Transfection, and Plasmids.
A253 (submaxillary salivary gland carcinoma) cells were cultured in McCoy’s medium (Gibco, Thermo Fisher); H1299 (non-small cell lung carcinoma) cells were cultured in Roswell Park Memorial Institute 1640 medium (Gibco, Thermo Fisher). MCF-7 (mammary gland adenocarcinoma) and A431 (skin squamous cell carcinoma) cells were cultured in Dulbecco’s Modified Eagle Medium (Lonza). FaDu (Pharynx squamous cell carcinoma) cells were cultured in Eagle's Minimum Essential Medium (Corning). All media were supplemented with 10% vol/vol fetal bovine serum, 100 μg/mL penicillin, and 100 μg/mL streptomycin (Gibco, Thermo Fisher). Cells were cultured at 37 °C with 5% CO2. Tumoral cell lines were purchased from American Type Culture Collection (ATCC) and routinely tested for mycoplasma contaminations. HEKn (Cascade Biologics, Thermo Fisher) and Ker-CT (HEKn immortalized with hTERT; ATCC) were grown in EpiLife medium supplemented with Human Keratinocyte Growth Supplement (Thermo Fisher). Differentiation was induced by adding 1.2 mM CaCl2 to culture medium. siRNA oligos were transfected in A253, HEKn, and Ker-CT using Lipofectamine RNAiMAX (Thermo Fisher) according to the manufacturer’s instructions. HEKn and Ker-CT were transfected twice, 48 h apart, before inducing differentiation. Smart pool siRNA oligos directed against p63 and SETX (siSETX #1) mRNAs and nonrelevant siRNA were purchased by Dharmacon (ON-TARGETplus Human SETX siRNA, L-021420–00-0010, Thermo Scientific). siSETX #2 oligos were purchased by Qiagen (Hs_SETX_5 FlexiTube siRNA, SI04300828). H1299 cells were transfected to express Flag-tagged constructs with Lipofectamine 2000 reagent (Thermo Fisher) according to the manufacturer’s instructions. HA- and FLAG-tagged p63 constructs were previously described (46).
Antibodies.
We utilized the following antibodies: mouse monoclonal anti-FLAG (clone M2, Sigma-Aldrich), rabbit polyclonal anti-FLAG (Sigma-Aldrich), rabbit polyclonal anti-Senataxin (Novus Biologicals, NBP1-94712), rabbit polyclonal anti-Senataxin (kind gift from Domenico Delia, University of Milan, Milan, Italy), mouse monoclonal anti-HA (BioLegend), rabbit monoclonal anti-p63-α (clone D2K8X; Cell Signaling Technology), mouse monoclonal anti-DNA–RNA hybrids clone S9.6 (Millipore), mouse monoclonal anti-vinculin (Sigma), rabbit polyclonal anti‐KRT1 (BioLegend, PRB‐149P), rabbit polyclonal anti‐KRT10 (Covance PRB159P), rabbit polyclonal anti ZNF750 (Sigma-Aldrich, HPA023012), rabbit polyclonal anti-Loricrin (Covance PRB145P), and rabbit polyclonal anti-Adenosine (Biovision).
Supplementary Material
Acknowledgments
We thank Prof. Domenico Delia for providing us the rabbit anti-Senataxin antibody and Dr. Maurizio Fanciulli for RNA-seq analysis. This work has been supported by the Associazione Italiana per la Ricerca contro il Cancro (AIRC) to A.P. (Investigator Grant IG#24678) and grant PRIN2017XCXAFZ to A.P. This work has been also partially supported by the LazioInnova Progetto Gruppo di Ricerca 2020 grant A0375-2020-36585 to G.M. and A.P., the AIRC grant to G.M. (IG#20473), and the Italian Ministry of Health, Istituto Dermopatico dell'Immacolata (IDI-IRCCS) grant (RC, CRI-NMSC, grant RF-2019-123698888) to E.C.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2104718119/-/DCSupplemental.
Data Availability
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE193205) (47).
References
- 1.Candi E., Schmidt R., Melino G., The cornified envelope: A model of cell death in the skin. Nat. Rev. Mol. Cell Biol. 6, 328–340 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Botchkarev V. A., The molecular revolution in cutaneous biology: Chromosomal territories, higher-order chromatin remodeling, and the control of gene expression in keratinocytes. J. Invest. Dermatol. 137, e93–e99 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soares E., Zhou H., Master regulatory role of p63 in epidermal development and disease. Cell. Mol. Life Sci. 75, 1179–1190 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Candi E., et al. , p63 in epithelial development. Cell. Mol. Life Sci. 65, 3126–3133 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Medawar A., et al. , DeltaNp63 is essential for epidermal commitment of embryonic stem cells. PLoS One 3, e3441 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shalom-Feuerstein R., et al. , ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 18, 887–896 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang A., et al. , p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 398, 714–718 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Romano R. A., et al. , ΔNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development 139, 772–782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Celli J., et al. , Heterozygous germline mutations in the p53 homolog p63 are the cause of EEC syndrome. Cell 99, 143–153 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Rinne T., Brunner H. G., van Bokhoven H., p63-associated disorders. Cell Cycle 6, 262–268 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Kouwenhoven E. N., van Bokhoven H., Zhou H., Gene regulatory mechanisms orchestrated by p63 in epithelial development and related disorders. Biochim. Biophys. Acta 1849, 590–600 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Kouwenhoven E. N., et al. , Transcription factor p63 bookmarks and regulates dynamic enhancers during epidermal differentiation. EMBO Rep. 16, 863–878 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melino G., Memmi E. M., Pelicci P. G., Bernassola F., Maintaining epithelial stemness with p63. Sci. Signal. 8, re9 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Smirnov A., et al. , ZNF185 is a p63 target gene critical for epidermal differentiation and squamous cell carcinoma development. Oncogene 38, 1625–1638 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truong A. B., Kretz M., Ridky T. W., Kimmel R., Khavari P. A., p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 20, 3185–3197 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ihrie R. A., et al. , Perp is a p63-regulated gene essential for epithelial integrity. Cell 120, 843–856 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Carroll D. K., et al. , p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 8, 551–561 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Gatti V., et al. , p63 at the crossroads between stemness and metastasis in breast cancer. Int. J. Mol. Sci. 20, 2683 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatti V., Fierro C., Annicchiarico-Petruzzelli M., Melino G., Peschiaroli A., ΔNp63 in squamous cell carcinoma: Defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol. Oncol. 13, 981–1001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen G. L., et al. , ZNF750 is a p63 target gene that induces KLF4 to drive terminal epidermal differentiation. Dev. Cell 22, 669–677 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fessing M. Y., et al. , p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J. Cell Biol. 194, 825–839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mardaryev A. N., et al. , p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development 141, 101–111 (2014). Correction in: Development 141, 3437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mardaryev A. N., et al. , Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J. Cell Biol. 212, 77–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rinaldi L., et al. , Dnmt3a and Dnmt3b associate with enhancers to regulate human epidermal stem cell homeostasis. Cell Stem Cell 19, 491–501 (2016). [DOI] [PubMed] [Google Scholar]
- 25.LeBoeuf M., et al. , Hdac1 and Hdac2 act redundantly to control p63 and p53 functions in epidermal progenitor cells. Dev. Cell 19, 807–818 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saladi S. V., et al. , ACTL6A is co-amplified with p63 in squamous cell carcinoma to drive YAP activation, regenerative proliferation, and poor prognosis. Cancer Cell 31, 35–49 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skourti-Stathaki K., Proudfoot N. J., Gromak N., Human Senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol. Cell 42, 794–805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivosecchi J., et al. , Senataxin homologue Sen1 is required for efficient termination of RNA polymerase III transcription. EMBO J. 38, e101955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chillemi G., et al. , Structural evolution and dynamics of the p53 proteins. Cold Spring Harb. Perspect. Med. 7, a028308 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Memmi E. M., et al. , p63 sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc. Natl. Acad. Sci. U.S.A. 112, 3499–3504 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senoo M., Pinto F., Crum C. P., McKeon F., p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell 129, 523–536 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Cohen S., et al. , Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 9, 533 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Compagnone M., et al. , ΔNp63-mediated regulation of hyaluronic acid metabolism and signaling supports HNSCC tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 114, 13254–13259 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gatti V., et al. , ΔNp63 regulates the expression of hyaluronic acid-related genes in breast cancer cells. Oncogenesis 7, 65 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boguslawski S. J., et al. , Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89, 123–130 (1986). [DOI] [PubMed] [Google Scholar]
- 36.Phillips D. D., et al. , The sub-nanomolar binding of DNA-RNA hybrids by the single-chain Fv fragment of antibody S9.6. J. Mol. Recognit. 26, 376–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richard P., et al. , SETX (Senataxin), the helicase mutated in AOA2 and ALS4, functions in autophagy regulation. Autophagy 17, 1889–1906 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan-Wong S. M., French J. D., Proudfoot N. J., Brown M. A., Dynamic interactions between the promoter and terminator regions of the mammalian BRCA1 gene. Proc. Natl. Acad. Sci. U.S.A 105, 5160–5165 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brambati A., Zardoni L., Nardini E., Pellicioli A., Liberi G., The dark side of RNA:DNA hybrids. Mutat. Res. Rev. Mutat. Res. 784, 108300 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Hatchi E., et al. , BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol. Cell 57, 636–647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yüce Ö., West S. C., Senataxin, defective in the neurodegenerative disorder ataxia with oculomotor apraxia 2, lies at the interface of transcription and the DNA damage response. Mol. Cell. Biol. 33, 406–417 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suraweera A., et al. , Senataxin, defective in ataxia oculomotor apraxia type 2, is involved in the defense against oxidative DNA damage. J. Cell Biol. 177, 969–979 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sariki S. K., et al. , Sen1, the homolog of human Senataxin, is critical for cell survival through regulation of redox homeostasis, mitochondrial function, and the TOR pathway in Saccharomyces cerevisiae. FEBS J. 283, 4056–4083 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Bennett C. L., La Spada A. R., Senataxin, a novel helicase at the interface of RNA transcriptome regulation and neurobiology: From normal function to pathological roles in motor neuron disease and cerebellar degeneration. Adv. Neurobiol. 20, 265–281 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Grunseich C., et al. , Clinical and molecular aspects of Senataxin mutations in amyotrophic lateral sclerosis 4. Ann. Neurol. 87, 547–555 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Regina C., et al. , Setdb1, a novel interactor of ΔNp63, is involved in breast tumorigenesis. Oncotarget 7, 28836–28848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A. Peschiaroli et al., ΔNp63-Senataxin circuit controls keratinocyte differentiation by promoting the transcriptional termination of epidermal genes. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE193205. Deposited 7 January 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE193205) (47).