Significance
Our work focuses on the critical longstanding question of the nontranscriptional role of p53 in tumor suppression. We demonstrate here that poly(ADP-ribose) polymerase (PARP)–dependent modification of p53 enables rapid recruitment of p53 to damage sites, where it in turn directs early repair pathway selection. Specifically, p53-mediated recruitment of 53BP1 at early time points promotes nonhomologous end joining over the more error-prone microhomology end-joining. Similarly, p53 directs nucleotide excision repair by mediating DDB1 recruitment. This property of p53 also correlates with tumor suppression in vivo. Our study provides mechanistic insight into how certain transcriptionally deficient p53 mutants may retain tumor-suppressive functions through regulating the DNA damage response.
Keywords: p53, tumor suppression, DNA repair, laser microirradiation, recruitment kinetics
Abstract
p53 is primarily known as a downstream transcriptional effector in the DNA damage-response cascade. We report that endogenous p53 rapidly accumulates at DNA damage sites within 2 s of UVA microirradiation. The kinetics of p53 recruitment mimics those of known DNA damage-response proteins, such as Ku70 and poly(ADP-ribose) polymerase (PARP), and precedes recruitment of Nbs1, 53BP1, and DDB1. Mutations in the DNA-binding and C-terminal domains significantly suppress this rapid recruitment. The C-terminal domain of p53 contains key residues for PARP interaction that are required for rapid recruitment of p53 to DNA damage sites, as is PARP-dependent modification. The presence of p53 at damage sites influences the recruitment kinetics of 53BP1 and DDB1 and directs the choice of nonhomologous end joining repair (NHEJ) and nucleotide excision repair. Mutations that suppressed rapid recruitment of p53 promoted error-prone alternative end-joining (alt-NHEJ) and inhibited nucleotide excision repair. Our finding that p53 is a critical early responder to DNA damage stands in contrast with its extensively studied role as a downstream transcriptional regulator in DNA damage repair. We highlight an unrecognized role of p53 in directing DNA repair dynamics and integrity and suggest a parallel mode of p53 tumor suppression apart from its function as a transcription factor.
The tumor suppressor p53 is best known for its capacity as a master transcriptional regulator (1). The expansive network of p53-regulated genes accounts for p53 functions in cell cycle arrest, apoptosis, metabolism, and DNA repair. While this has reinforced p53 transcriptional activity as a critical aspect of tumor suppression, significant transcription-independent functions have been uncovered (2–4). Recent work has identified p53 as a component of the replication machinery or replisome with a direct role in safeguarding against replicative stress (5–7). Through protein interactions, p53 influences the restart and processing of stalled forks and suppresses mutagenic pathways, such as single-strand annealing and microhomology-mediated end joining (MMEJ) (8). Similarly, transcription-independent functions of p53 in influencing cellular repair outcomes are evidenced from the protein complexes p53 interacts with (9–11). This stands apart from p53 transcriptional regulation of components of nucleotide excision repair (NER), base excision repair (BER), and mismatch repair (MMR) (12). These findings indicate that p53 might play a previously unknown role in sensing and mitigating cellular stress and DNA damage that undermine genome stability. That this might occur much earlier than and separate from p53’s role as a major transcriptional effector in the DNA damage response (DDR) signaling cascade is substantiated by findings that effects of p53 deficiency on genome stability and tumor suppression cannot be entirely explained by systematic perturbation of downstream DDR and repair transcriptional targets of p53 (13, 14).
This prompted us to reexamine the kinetics and structural aspects of p53 response to DNA damage using localized laser microirradiation techniques. We found that p53 has an unexpected role in rapid detection of DNA lesions. In our study, we examined a range of domain-specific and clinically relevant p53 mutants to determine the structural requirements for rapid recruitment of p53 to sites of DNA damage. We show that both the DNA-binding domain (DBD) and C-terminal domain (CTD) are critical for this function. Furthermore, poly(ADP-ribose) polymerase (PARP)–dependent modification of the CTD is a prerequisite for p53 recruitment. Rapid recruitment of p53 to sites of DNA damage is unique, evolutionarily distinct, and not shared by other related transcription factors, nor its structural homologs, such as p63 and p73. Importantly, the rapid accumulation of p53 did not correlate with presence of transcriptional activity. We also demonstrate the impact of this property of p53 on directing recruitment of major repair factors, such as 53BP1 and DDB1, and subsequent repair choice with important implications for tumor suppression.
Results
Wild-Type p53 Rapidly Accumulates at Localized Sites of DNA Damage.
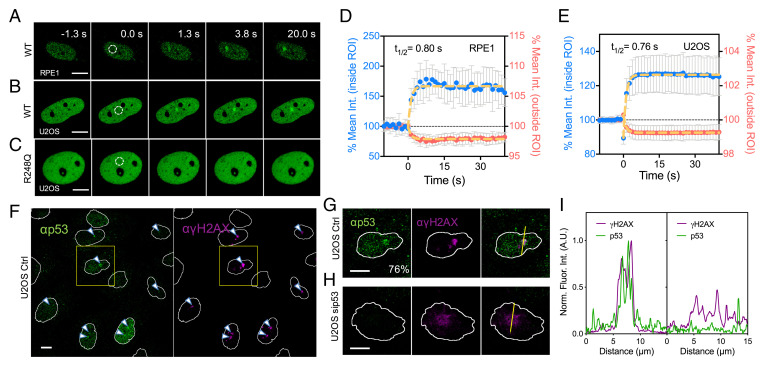
To investigate the potential of p53 to function as a sensor of DNA damage, we sensitized cells with Hoechst 33342, microirradiated them with a 355-nm laser and monitored the kinetics of p53 recruitment. This method ensured localized damage of a specific region of the nucleus, allowing for direct measurement of the dynamics, of p53 recruitment. This differs from induction of global DNA damage with methods such as ionizing radiation (IR) and chemical treatments. When RPE1 nuclei, in which p53 was endogenously tagged with a mNeonGreen (mNG) reporter (SI Appendix, Fig. S1A), were irradiated, p53 was recruited to the specific site of damage within 2 s (Fig. 1A, Movie S1). A measure of intranuclear redistribution of p53 within and outside the site of irradiation revealed an exponential time constant of around 0.8 s (Fig. 1D). The rate of rapid accumulation could not be decoupled from that of bleach recovery, hence suggesting that rapid p53 recruitment was diffusion-limited. We also verified that mNG-tagged p53 was functionally similar to untagged wild-type (WT) p53 in various assays. Successful biallelic insertion of the mNG tag was demonstrated by PCR and Western blot (SI Appendix, Fig. S1B). Western blots comparing lysates from WT and p53-mNG RPE1 cells treated with the Plk4 inhibitor, centrinone, or the MDM2 inhibitor, R7112, showed that both compounds induced increased levels of p53-WT and p53-mNG in a similar fashion (SI Appendix, Fig. S1 C and D). Further examination revealed that levels of p53-mNG in RPE1 were elevated in the presence of the MPS1 inhibitor, NMS-P7145, or the DNA damaging agent, doxorubicin (Dox) (SI Appendix, Fig. S1 D and E). Similarly, we observed rapid recruitment of enhanced green fluorescent protein (EGFP)–tagged WT p53 in U2OS (Fig. 1B, Movie S2). The halftimes of p53 accumulation in both cases were similar (Fig. 1 D and E). Hence, rapid accumulation of p53 was a property of the protein and not an artifact of the presence of fluorescent tags.
Fig. 1.
Rapid accumulation of WT p53, but not the R248Q mutant, at sites of DNA damage upon laser microirradiation. (A) Representative montage showing the rapid recruitment of endogenous WT p53 with a CRISPR-KI mNG tag in RPE1 cells. (B) Similar montage showing that overexpressed WT p53-EGFP also accumulated at DNA damage sites in U2OS cells. (C) The rapid accumulation is however not detected with the p53 DNA-binding mutant, R248Q. White dotted circle indicates irradiated area. (D and E) Quantification of fluorescence intensity within ROIs, whose diameter is set at 2.5 μm: (D) endogenous WT p53-mNG in RPE1 cells (mean ± SD, n = 19) and (E) WT p53-EGFP in U2OS cells (mean ± SD, n = 30). Signal intensity measurements outside the ROI indicate that p53 protein is being redistributed rapidly following irradiation. Yellow dotted lines indicate single exponential fitting. (F) Global and (G) zoomed-in views of immunostaining of endogenous WT p53 and γH2AX in U2OS cells 5 min after irradiation. (H) The staining of p53 at DNA damage sites is not detectable in U2OS treated with siRNA against p53 and the staining of γH2AX appears more diffuse. Yellow lines indicate the region used for intensity profile analysis. (I) Corresponding line intensity profile of the yellow lines demonstrating the colocalization of p53 and γH2AX at early time points after laser irradiation (Left, G) or the lack thereof (Right, H). (Scale bars, 10 μm.)
We further verified this recruitment property of endogenous p53 using immunofluorescence staining. U2OS cells were fixed within 5 min following laser microirradiation. Discernible p53 foci that colocalized with γH2AX foci were identified in 76% of the cells (Fig. 1F). Higher-magnification micrographs and the line profiles measuring fluorescence intensity across damage sites showed clear colocalization between p53 and γH2AX (Fig. 1 G and I). In contrast, p53-specific fluorescence was undetectable following p53 depletion by RNAi, and γH2AX staining at the damage site was more diffuse (Fig. 1 H and I). Further characterization demonstrated that the magnitude of p53 accumulation was dependent on the laser power, and hence degree of DNA damage. The magnitude of p53 accumulation plateaued when the 355-nm laser power reached 3 nW at the back aperture of the objective (SI Appendix, Fig. S2 C and D). Irreversible bleaching and nuclear breakdown were observed with higher laser powers of 5 nW and 10 nW measured at the back aperture of the focal plane, respectively. As such, the laser power employed throughout the study was set at 1 nW to avoid extensive and irreparable DNA damage. Under such conditions, cells survived for at least 12 h following laser microirradiation and were able to resolve DNA lesions.
Importantly, we ruled out any photoconversion artifacts arising from exposure of Hoechst 33342-sensitized cells to a 355-nm UVA laser. In control experiments using untransfected U2OS cells that were sensitized with Hoechst 33342, microirradiation with the 355-nm laser at optimal settings (15) did not result in discernable photoconversion at locally ablated sites (SI Appendix, Fig. S2E). Imaging of untransfected, Hoechst 33342-sensitized U2OS cells at 405 nm naturally resulted in a global photoconversion and green fluorescent signals (SI Appendix, Fig. S2 E, Lower). Local photoconversion remained undetectable (SI Appendix, Fig. S2F) in the absence of global photoconversion (when the 405-nm laser was switched off). Combined with the observation that the p53 mutant, R248Q, did not accumulate at the damage site (Fig. 1C, Movie S3), we validated that our system was free of photoconversion artifacts, as discussed elsewhere (16).
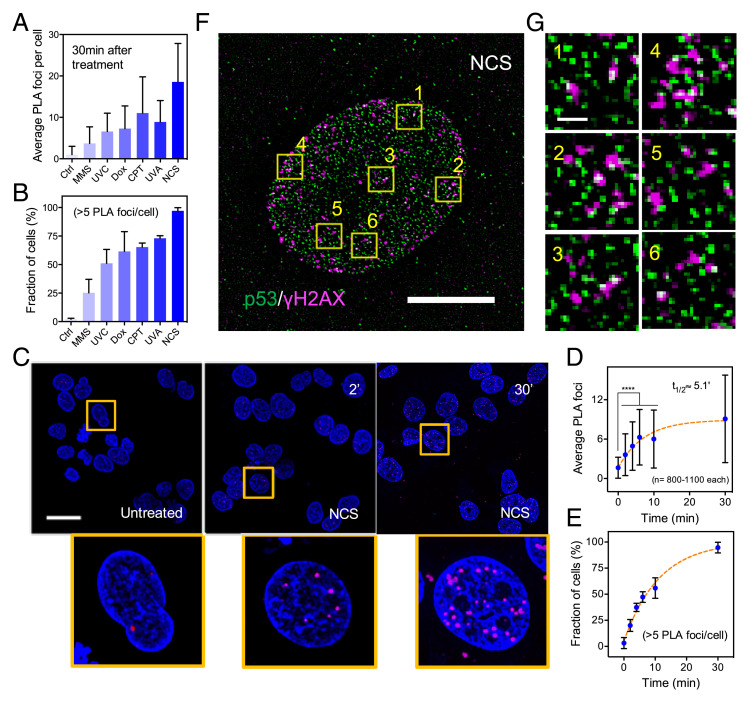
UVA microirradiation of Hoechst 33342-sensitized cells generally results in a broad spectrum of DNA lesions due to oxidative damage (17, 18). We employed a variety of DNA damaging agents, such as methyl methanesulfonate (MMS), camptothecin (CPT), Dox, and global UVA and UVC, all of which are known to induce a variety of lesions, including double-strand breaks (DSBs), via different mechanisms, such as DNA cross-linking and disruption of the replication machinery (19–21). Using proximity ligation assays (PLA), we observed an increased number of p53-γH2AX PLA foci at 30 min posttreatment under the various conditions (Fig. 2 A and B). These findings strongly support the role of p53 in rapid detection of DNA lesions. To better characterize the dynamics of complex formation between p53 and γH2AX, we employed neocarzinostatin (NCS), a potent radiomimetic drug that rapidly generates DSBs and is quickly hydrolyzed in aqueous media. We found p53-γH2AX PLA foci as early as 2 min post-NCS treatment of U2OS cells (Fig. 2C). Temporal analysis showed that the number of PLA foci plateaued at about 30 min post-NCS treatment, with a half-time around 5 min (Fig. 2 D and E). Colocalization between p53 and γH2AX foci following NCS treatment was further examined using an instant structural illuminated microscopy spinning disk confocal. Interaction between p53 and γH2AX was evident from magnified micrographs (Fig. 2 F and G). Our observations were also consistent with reports describing preferential accumulation of phosphorylated p53 (Ser15) and γH2AX at DSBs and sites of DNA base damage following IR and MMS treatment, respectively (22, 23), as well as the general ability of p53 to bind nonspecifically to a wide variety of DNA damage and repair intermediates (24, 25).
Fig. 2.
Complex formation between p53 and γH2AX at sites of DNA damage following different forms of chemical- or UV-induced DNA damage. (A) Average p53-γH2AX PLA counts (mean ± SD) and (B) Fraction of U2OS cells with more than 5 PLA foci at 30 min following different treatments (mean ± SD). Conditions of various treatments are listed below. Ctrl: untreated; UVC: 2 min; UVA: 8 min; CPT: 4 μM; MMS: 0.01%; Dox: 4 μM; NCS: 0.5 μg/mL. (C) Fluorescent micrographs from a PLA showing complex formation between p53 and γH2AX at 2 and 30 min following NCS treatment at 0.5 ng/mL. Zoomed-in views of areas marked by the yellow boxes are below. (Scale bar, 30 μm.) (D) Average p53-γH2AX PLA foci counts increased rapidly over time and plateau within 30 min following NCS treatment (mean ± SD, ****P < 0.001). (E) Fraction of cells with more than five PLA foci per nuclei also increased significantly over time (mean ± SD). (F) Superresolution confocal fluorescent micrographs of cells stained with p53 and γH2AX at 30 min after NCS treatment. (Scale bar, 10 μm.) (G) Zoomed-in views of areas marked by the yellow boxes. (Scale bar, 1 μm.)
Rapid Recruitment of p53 Requires Both the DBD and CTD and Is Not Correlated with Transcriptional Activity.
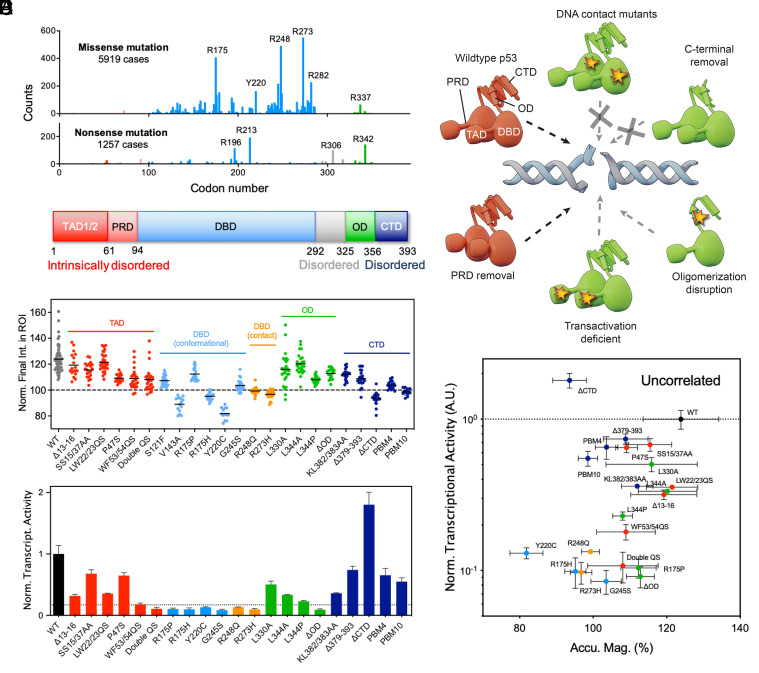
TP53 is one of the most frequently mutated genes in cancer, with mutants exhibiting diverse functional consequences, which include loss- and gain-of-function (26). Mutations in the DBD of p53 constitute “hotspots” and are commonly identified in human cancers (Fig. 3A). To investigate the structural requirements of p53 recruitment, we generated various p53 domain-specific mutants (Fig. 3B and SI Appendix, Table S1) and tested their recruitment to sites of DNA damage. Most p53 DBD hotspot mutants, including conformational (i.e., R175H, V143A, G245S) and DNA-binding contact mutants (i.e., R273H, R248Q, except R175P), were defective in recruitment to damage sites (Fig. 3C and SI Appendix, Fig. S5). Interestingly, R175P, which was more proficient in recruitment to DNA damage sites compared with other DBD mutants, has been reported to be capable of tumor suppression in vivo despite lack of transcriptional capability (8). We also found that deletion of the CTD (ΔCTD) abolished the early recruitment of p53 to sites of damage (Fig. 3C). Partial loss of the CTD (Δ379-393) also impaired p53 recruitment. It appeared that both the DBD and CTD were critical for p53’s function as a rapid sensor of and responder to DNA damage.
Fig. 3.
Structural requirements for p53 rapid accumulation that is not correlated with transcriptional activity. (A) Frequency analysis of missense and nonsense mutations of TP53 in human cancer based on data available on the COSMIC (Catalogue of Somatic Mutations in Cancer) database. Common “hotspot” areas for mutations are R175, R248, and R273. (B) Diagram of human WT p53 with various domains indicated. (C) Magnitude of accumulation of WT and various mutant p53 as measured by fluorescence intensity within ROI at the end of the time course (40 s after irradiation) in U2OS cells. Each dot represents a measurement from a different cell (n = 15 to 54, N = 3). (D) Summary of different p53 domain-specific mutants and their ability to be rapidly recruited to damage sites. Dotted black arrow represents rapid recruitment comparable to that of WT p53; dotted gray arrow represents partial recruitment at a lower magnitude; dotted gray arrow with a cross represents no recruitment. (E) Transcriptional activity of various p53 mutants were determined using a β-galactosidase reporter assay in ARN8 cells. Dotted line is an arbitrary threshold that separates transcriptionally active from inactive mutants. The expression levels of p53 were relatively normalized through cell sorting of high-intensity EGFP+ cells prior to measurement (SI Appendix, Fig. S2G). (F) The accumulation magnitudes of WT and mutant p53 were not correlated with their transcriptional activity (slope = 0.0050 and R2 = 0.0011 in a simple linear regression model).
Examining the N terminus of p53, we found that mutations in the second, but not the first, transactivation domain (TAD) disrupted p53 recruitment (Fig. 3C). In reported mouse models, the first TAD has been shown to be required for the transcriptional activation of the majority of p53 responsive genes; however, the second TAD is sufficient for tumor suppression and only loss of both TADs inactivated tumor suppression completely (27). In our study, the double TAD (double QS) mutant was also impaired in its recruitment. A functionally important mutant, P47S, which is associated with increased cancer risk (28), was also partially defective in recruitment to damage sites. P47S has been reportedly linked to defective replication fork restart (8). Thus, mutations that disrupted early recruitment of p53 to DNA damage sites also appeared to strongly correlate with loss of tumor suppression. P53 has often been reported to be altered by posttranslational modifications (PTMs) following DNA damage. Multisite phosphorylation of serine residues at the N terminus of p53 is carried out by major kinases, such as ATM and ATR, which contribute to induction of p53-mediated transcriptional regulation (29, 30). The detection of phosphorylated p53 in damage-related foci hours after an insult is generally accepted as an indicator of downstream DDR signaling processes (31, 32). We found that a p53 mutant that cannot be phosphorylated on both Ser15 and Ser37 (the major target residues) was still recruited to sites of irradiation (Fig. 3C). This indicated that rapid recruitment of p53 was, at least, less dependent on PTMs, which instead, are known to be a predominant feature of modulating p53’s downstream transcription-related functions in the DDR cascade.
P53 functions optimally as a transcription factor by binding target promoters as a tetramer. L344A, a mutant that abolishes the formation of tetramers but forms dimers, was recruited rapidly to damage sites. In contrast, L344P, a mutant which abolishes both tetramer and dimer formation, was defective in recruitment (Fig. 3C). Surprisingly, deletion of the oligomerization domain (ΔOD) was dispensable for rapid recruitment of p53, although the magnitude of recruitment was lower than that of L344A. This is likely due to the ability of p53 to also dimerize via its DBD and the inherent stability of p53 as a dimer over a monomer (33). Our analysis also revealed cell-type–dependent differences in the degree of WT and mutant p53 recruitment to damage sites. Compared to U2OS cells, which contained WT p53, we found a dramatic increase in damage-dependent recruitment of exogenous WT p53 in Saos2 cells, which are p53-null (34), and a slight reduction in three other cancer cell lines that expressed either mutant or WT p53 (SI Appendix, Fig. S5 B and C). However, the DBD and ΔCTD mutants consistently failed to be recruited to damage sites in U2OS and Saos2 cell lines (SI Appendix, Fig. S5A). The effects of domain-specific mutations and deletions on p53 protein recruitment are summarized in Fig. 3D.
We next investigated the relationship between magnitude of p53 recruitment and transcriptional capability of various mutants using ARN8 cells stably transfected with a p53-responsive β-galactosidase reporter construct (35) (Fig. 3E). ARN8 cells were transiently transfected with EGFP-tagged WT or mutant p53 and expression levels relatively normalized by sorting for high-intensity EGFP+ cells. Mutations in the second TAD, DBD, and loss of the OD resulted in loss of transcriptional activity in our assay. Meanwhile, the ΔCTD mutant demonstrated a significant increase in transcriptional activity, consistent with previous findings (36). These findings demonstrate that the recruitment magnitude of mutant p53 did not correlate with transcriptional capability as measured by our reporter assay.
The Polybasic Stretch in the p53 CTD Mediates p53 PARylation, Which Is Critical for p53 Rapid Recruitment.
We further investigated how perturbation of the CTD of p53 can impede protein accumulation. The CTD is the main site for interaction with PARP (37–40) and studies have demonstrated that PARP binds to p53 through the polybasic stretch of amino acids located at the CTD, before PAR-modification of p53 at multiple residues across the protein (40). We investigated PARylation-deficient p53 mutants where either 4 (PBM4) or 10 (PBM10) critical basic amino acids in the CTD were substituted with alanine (SI Appendix, Fig. S3 A, C, and D) in live cells with laser microirradiation. We showed that the EGFP fusion of p53-WT was PARylated shortly after 2-min UVA treatment and that this was suppressed in the presence of PARP inhibition. No PARylation was detected with the PBM4 and PBM10 mutants after UVA treatment (SI Appendix, Fig. S3B). Given that we have identified the CTD and DBD as necessary components required for rapid recruitment of p53, we also tested the binding ability of the individual DBD and CTD domains and found that they were not capable of being recruited to damage sites (SI Appendix, Fig. S3E). A fusion protein comprised of just the DBD and CTD also failed to be recruited (SI Appendix, Fig. S3F). Thus, we conclude that p53’s rapid response to DNA damage likely required these elements to act in cis and depended on appropriate protein folding and structure.
Next, we sought to determine the enzymatic requirements for p53 recruitment. Pretreatment with PARP inhibitors significantly blocked the early recruitment of p53 to damage sites in Saos2 cells, which lacked endogenous p53 (SI Appendix, Fig. S4 A–D, Movie S4). Furthermore, the presence of PARP inhibitor did not suppress rapid recruitment of PARP itself. This indicated that the rapid recruitment of p53 required the enzymatic activity of PARP and suggested that PARP and p53 could not interact with each other if one or both proteins were not PARylated. We also measured the time-dependency of PARP and p53 recruitment on a longer time scale. Interestingly, PARP dissociated from sites of irradiation over time, while p53 accumulation persisted (SI Appendix, Fig. S4E). PARP inhibition delayed recruitment of p53 by about an hour but did not suppress the eventual recruitment of p53 (SI Appendix, Fig. S4F). This indicated that the recruitment of p53 at later time points likely involved different mechanisms.
We then investigated other transcription factors known to be modified by PARP, such as NF-κB and CEBPβ. The results revealed different and delayed recruitment dynamics for these proteins in comparison with p53 (SI Appendix, Fig. S4H), suggesting that the rapid recruitment of p53 was not a general feature of proteins subject to PARylation. Additionally, we also investigated whether p53-binding protein 1 (53BP1) was required for or promoted the recruitment of p53. 53BP1 contains a BRCA1 C-terminal (BRCT) domain that is involved in facilitating DNA binding, protein interactions, and importantly, PAR binding (41). Our results revealed that 53BP1 was not recruited within the same time window where p53 was initially recruited (SI Appendix, Fig. S4I), but was only recruited at a much later time (approximately 10 min following laser irradiation) (Fig. 4D). Concordance of our findings between PARylation mutants (PBM4, PBM10) and PARP inhibition provide genetic evidence that rapid recruitment of p53 selectively required both p53–PARP interaction through the CTD and subsequent PARylation of p53.
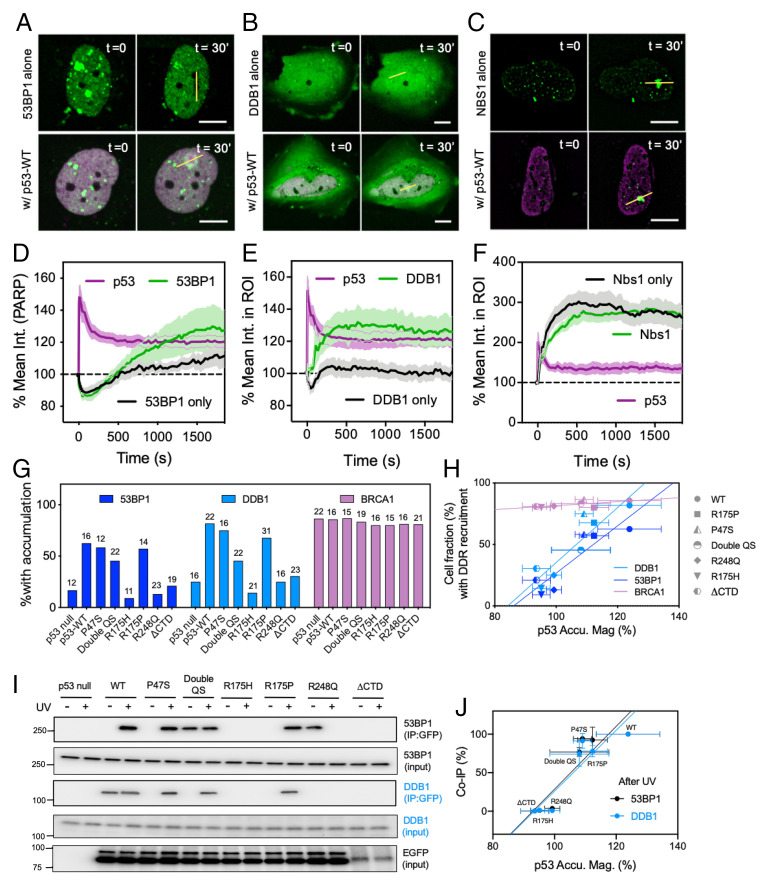
Fig. 4.
Rapid accumulation of p53 promotes recruitment of DDB1 and 53BP1 to the sites of damage through direct interaction. (A–C) Representative micrographs of (A) 53BP1-EGFP, (B) DDB1-EGFP, and (C) Nbs1-EGFP with and without coexpression of p53-mCherry. (Dotted circles indicate damage sites. Scale bars, 10 μm.) (D–F) Average recruitment time courses of (D) 53BP1 alone (n = 12) vs. 53BP1 and p53-WT coexpression (n = 16), (E) DDB1 alone (n = 16) vs. DDB1 and p53-WT coexpression (n = 22), and (F) NBS1 alone (n = 11) vs. Nbs1 and p53 coexpression (n = 11) following laser microirradiation in Saos2 cells (mean ± SEM). (G) The percentage of Saos2 cells demonstrating recruitment of 53BP1, DDB1, and BRCA1 within 30 min following UVA microirradiation in the absence or presence of p53-WT or selected p53 mutants. Numbers on top of each bar represent sample numbers. (H) Cell fractions where 53BP1 and DDB1 were recruited to damage sites correlated with accumulation magnitudes of the given WT or mutant p53. No correlation was observed in the case of BRCA1 (mean ± SD). (I) IP of WT p53 and select mutants in Saos2 cells prior to and immediately following UVA irradiation (<3 min). The experiment was performed with stable Saos2 cell lines with doxycycline-inducible expression of EGFP fusion of p53-WT or mutants. (J) Corresponding quantification of the blot demonstrated a positive correlation between normalized co-IP% of 53BP1/DDB1 with various p53 mutants following UVA treatment and the ability of these mutants to be recruited to sites of damage following laser microirradiation (mean ± SD).
p53 Family Members p63 and p73 Are Not Recruited to Damage Sites.
To determine whether the rapid recruitment of p53 was a shared feature among orthologs in the p53 family, we compared the damage response of p53 with that of p63, p73, and their splicing isoforms. Strikingly, these orthologs failed to be recruited to sites of damage (SI Appendix, Fig. S4G). It indicated that this property of p53 was not shared by related transcription factors in response to similar DNA damaging agents. We next compared the recruitment kinetics of p53 to other known sensors of DNA damage using our laser microirradiation system to determine the relative temporal sequence of these events. We found that the kinetics of p53 recruitment mirrored that of Ku70 (a major component of the nonhomologous end-joining [NHEJ] pathway) and was faster than that of another known early sensor, NBS1, which is part of the MRN complex (SI Appendix, Fig. S2 A and B). The measured halftimes of Ku70 and Nbs1 recruitment in our system were consistent with prior studies (42, 43). Together, these results placed p53 among other bona fide early sensors of DNA damage.
Rapid Recruitment of p53 Is an Evolved Property.
Genome sequencing has revealed remarkable conservation of the p53 family of genes across primitive metazoans and the oldest group of jawless vertebrates, as well as some of the most primordial cartilaginous fish amounting to over 500 million y of species evolution. To investigate whether rapid recruitment of p53 to damage sites was a conserved property or one that evolved in higher vertebrates, we selected the following species: a primitive lineage of metazoans, Trichoplax adhaerens; an ancient extant lineage of jawless fish, the Japanese lamprey Lethenteron japonicum; the first cartilaginous fish, the elephant shark Callorhinchus milii; and the rodent Mus musculus. The overall domain structure of p53 was relatively conserved during evolution although sequence homology diverged significantly. We analyzed protein sequence similarity using an alignment tool Clustal Omega (SI Appendix, Fig. S8A). The values in parentheses in SI Appendix, Fig. S8A indicate the percentage similarity of each species-specific p53 with human p53. The basic p53 domains are displayed next to the phylogram and color-coded. The values above each domain represent the domain size as measured by the number of amino acids. The degree of similarity of each species-specific domain compared to the human equivalent was represented by shaded patterning (see “Alignment score” in SI Appendix, Fig. S8A). While the core structure of the DBD and OD were relatively conserved, the CTDs differed significantly between species.
Strikingly, the magnitude of p53 rapid accumulation followed a clear trend that correlated with the sequence homology of the species-specific p53 to human p53, with mouse p53 demonstrating a greater order of magnitude followed by elephant shark, lamprey, and finally, trichoplax p53 (SI Appendix, Fig. S8B). Elephant shark, lamprey, and trichoplax p53 were not recruited to sites of laser microirradiation when exogenously expressed in U2OS cells, while mouse p53 demonstrated a clear enrichment albeit with different kinetics from that of human p53 (SI Appendix, Fig. S8 B and C). To determine whether the presence of the human CTD might rescue recruitment capability, we generated respective chimeric p53 constructs. The human CTD improved the magnitude of mouse p53 accumulation at sites of laser microirradiation but did not alter the magnitudes for elephant shark, lamprey, and trichoplax p53 (SI Appendix, Fig. S8D). Additional structural studies would be necessary to identify key elements unique to human p53 that contribute to its rapid recruitment to sites of DNA damage.
p53 Rapid Accumulation Influences the Recruitment of Downstream Effectors.
We next investigated the effect of rapid p53 binding to damage sites on the recruitment kinetics of key repair factors. The rapid recruitment of coexpressed WT p53 to damage sites promoted recruitment of 53BP1, which is implicated in NHEJ repair, from approximately 4 min following microirradiation, and accumulation increased over time. In contrast, the recruitment of 53BP1 occurred at a later time and was of lower magnitude in the absence of WT p53 (Fig. 4 A and D, Movie S5). Previous studies have also reported that 53BP1 is recruited to UVA laser microirradiated stripes at later times and in the presence of p53 (44, 45), corroborating our findings. The effect of p53 on recruitment of 53BP1 was significant in comparison to other core NHEJ factors, such as Ku70 and XRCC4, which we found to be present at sites of microirradiation regardless of p53 (SI Appendix, Table S2). We also measured DDB1 accumulation since it is involved in the NER repair mechanism as part of the cellular response to UV-induced damage and is responsible for recruiting factors involved in NER (46). The presence of p53 influenced accumulation of DDB1, which did not accumulate at irradiated sites in the absence of p53 (Fig. 4 B and E, Movie S6). In the presence of p53, DDB1 was recruited approximately 1 min following irradiation with sustained accumulation over time. Meanwhile, we found that other NER factors—such as XPA, XPB, and XPC—were largely present at sites of microirradiation regardless of p53 (SI Appendix, Table S2).
We then compared recruitment of 53BP1 and DDB1 in cells expressing different p53 mutants. Mutants such as P47S and R175P retained the ability to corecruit 53BP1 and DDB1 (Fig. 4 G and H). Next, we performed immunoprecipitation (IP) on different mutant-expressing Saos2 cells before and immediately following UVA treatment. We observed pull down of 53BP1 and DDB1 in the case of WT, P47S, and R175P expression, consistent with our observations of their recruitment to damage sites (Fig. 4 I and J). Our data demonstrated that the kinetics of p53 recruitment was faster than that of either 53BP1 or DDB1, implying that p53 involvement in processes at the damage sites facilitated binding of 53BP1 and DDB1. Furthermore, reintroduction of p53 in a p53-null line like Saos2 did not significantly alter levels of 53BP1 and DDB1 (Fig. 4I). Thus, both 53BP1 and DDB1 accumulated at damage sites well after the rapid binding of p53 because of p53-dependent effects at those sites. Homologous recombination (HR)–mediated repair is another mechanism in response to UV damage and DSB formation. We observed that the presence of p53 at damage sites did not significantly alter the recruitment dynamics of NBS1 (Fig. 4 C and F, Movie S7), Rad51, or BRCA1 (SI Appendix, Table S2).
p53 Rapid Accumulation Influences Choice of the Downstream Repair Pathway and Tumor Growth In Vivo.
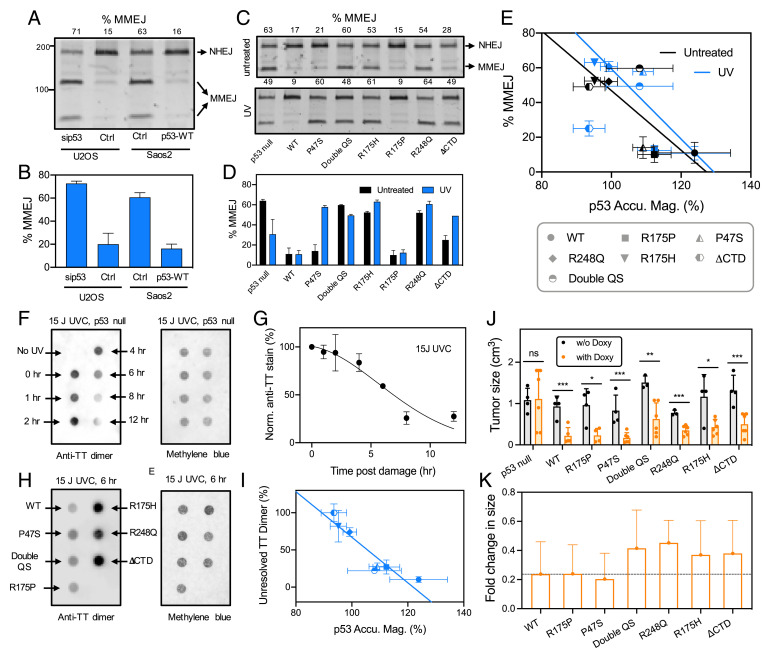
To understand if the early recruitment of p53 influenced the choice of end-joining repair pathways, we conducted plasmid-based recircularization assays to determine the relative efficiencies of direct end-joining and MMEJ. The latter is also known as alt-NHEJ and is highly error-prone and results in deletion of DNA (47). U2OS cells (containing endogenous p53), U2OS cells treated with small-interfering RNA (siRNA) against p53, Saos2 cells (p53-null), and doxycycline-inducible Saos2 cells stably expressing WT p53 were transfected with a linearized plasmid (pDvG94) (48, 49) and end-joining products were analyzed. The absence of p53 in siRNA-treated U2OS or Saos2 cells was associated with increased use of MMEJ over direct end-joining while levels of direct end-joining were increased in the presence of WT p53 (Fig. 5 A and B). Here we defined %MMEJ as the fraction of MMEJ products over the sum of MMEJ and NHEJ products. Thus, the rapid recruitment of p53 to damage sites favored the NHEJ repair pathway.
Fig. 5.
Recruitment-competent p53 mutants bias DSB repair toward NHEJ over MMEJ and facilitate TT-dimer resolution. (A) Electrophoresis of PCR products from an end-joining assay using U2OS cells and Saos2 cell lines in the presence and absence of WT p53. The lower two bands were PCR products of plasmids repaired via the MMEJ pathway resulting in creation of a BstXI digestion site. The top most band was the PCR product of plasmids repaired via the NHEJ pathway that did not result in a BstXI digestion site. The numbers on top indicate percentages of repaired plasmids that were repaired via the MMEJ pathway (%MMEJ). (B) Graphical summary of A (n = 3 replicates, mean ± SD). (C) End-joining assay in control Saos2 cells (p53 null) and Saos2 cells stably expressing WT and mutant p53 in the absence and presence of UVA treatment. (D) Graphical summary of C (n = 3 replicates, mean ± SD). (E) Mutants that showed lower levels of %MMEJ correlated with higher recruitment magnitudes of p53 mutants at damage sites, and vice versa (mean ± SD). (F) DNA dot blot assay using genomic DNA extracted from Saos2 cells at different time points post-UVC treatment (Left). A duplicate blot stained with methylene blue served as a loading control (Right). (G) Normalized levels of TT dimers showed a monotonic decrease over time suggesting typical NER repair dynamics in control Saos2 cells. (H) The dot plot was probed for the unresolved TT dimers at 6 h post-UVC treatment in Saos2 cells (p53-null) and Saos2 cells expressing WT and selected p53 mutants (Left). A duplicate blot stained with methylene blue served as a loading control (Right). (I) The normalized level of unresolved TT dimers inversely correlated with the accumulation magnitude of p53 mutants. E and I share the same symbol legend. (J) Tumor sizes of Saos2 cells with and without doxycycline-induced expression of p53 WT or selected p53 mutants in immunocompromised NSG mice. Each dot represents an independent technical repeat (n ≥ 2 mice, mean± SD, *P < 0.05; **P < 0.01; ***P < 0.005). (K) Fold-change in tumor volume with and without doxycycline induction. Error bars represent SD calculated following error propagation principles.
Next, we sought to determine the efficiency of direct end-joining and MMEJ under different mutant p53 conditions. Doxycycline-inducible Saos2 cells stably expressing WT and mutant p53 were transfected with linearized pDvG94. In the presence of WT p53 and mutants, such as R175P, which had demonstrated the ability to be recruited to damage sites, we observed reduced levels of MMEJ in the absence of UV treatment (Fig. 5 C–E). Meanwhile, mutants that were defective in recruitment to damage sites displayed higher levels of MMEJ. Surprisingly, in the case of P47S, which was defective in recruitment to damage sites (Fig. 3C), levels of MMEJ in the absence of UV treatment were similar to that of WT p53 and R175P. Following UV irradiation, the recruitment of WT p53 and R175P effectively suppressed the use of MMEJ (Fig. 5 C–E). MMEJ levels increased significantly in the case of R175H-, R248Q-, ΔCTD-, and P47S-expressing cells. The increase in MMEJ observed in P47S-expressing cells only post-UV treatment was likely due to downstream mechanisms involving other players in the MMEJ pathway. Likewise, given p53-mediated recruitment of DDB1, we sought to determine NER-mediated resolution of thymine-thymine cyclobutane pyrimidine dimers (TT-CPD dimer) in a DNA dot blot assay using an anti-TT dimer antibody (50). In p53-null Saos2 cells, we observed a decrease in TT dimers over time (Fig. 5 F and G). We then compared TT-dimer resolution at 6 h post-UVC in various p53 mutant-expressing cells and found that these lesions were more efficiently resolved in conditions where p53 was capable of rapid accumulation at damage sites and mediated DDB1 recruitment (Fig. 5 H and I). Together, these findings further reinforce the hypothesis that rapid recruitment of p53 to damage sites favored choice of the NHEJ and NER pathways in response to UV treatment by influencing recruitment of key repair factors, such as 53BP1 and DDB1 (Fig. 6).
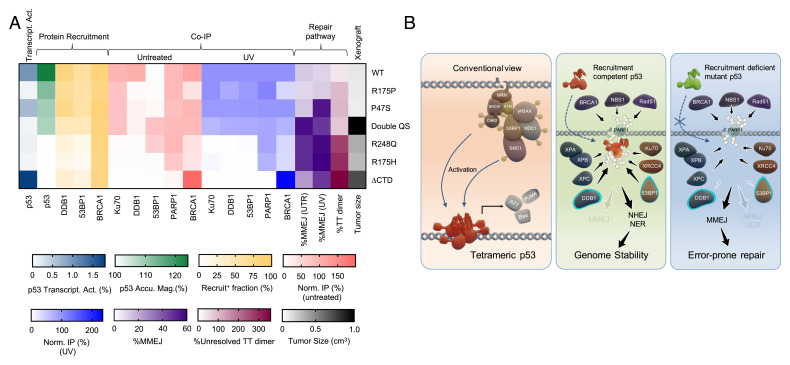
Fig. 6.
(A) Summary heatmap demonstrating that p53 recruitment, but not its transcriptional activity, strongly correlated with the DSB repair pathway choice, efficiency of TT-dimer removal, and tumor suppression in vivo. (B) Proposed model for a nontranscriptional role of p53 in rapid DNA damage sensing, downstream factor recruitment and repair pathway selection.
To assess the functional significance of differential DDRs and repair mechanisms mediated by the various p53 mutants on tumorigenesis, we transplanted Saos2 cells expressing the p53 mutants into immunocompromised NSG (NOD-scid IL2Rγnull) mice. Strikingly, we observed that Saos2 cells stably expressing WT p53 and the mutants P47S or R175P formed smaller tumors after 1 mo (Fig. 5J). These tumors were undifferentiated and of high grade (SI Appendix, Fig. S9A). By contrast, at the same time point, p53-null Saos2 cells and cells expressing double QS, R175H, R248Q, or ΔCTD mutants formed larger cystic tumors (Fig. 5J). These tumors were also undifferentiated and of high grade, but with regions of pronounced nuclear atypia and adipocyte infiltration (SI Appendix, Fig. S9B). Based on tumor size ratio between doxycycline-treated and nontreated controls, there was an increased reduction in volume of recruitment-competent WT and mutant p53-expressing tumors (Fig. 5K). Collectively, these findings demonstrate that there is indeed a better correlation between p53 recruitment and 53BP1 and DDB1 recruitment, choice of repair pathway, and tumor suppression in vivo as compared to the correlation between transcriptional capability and these aspects (SI Appendix, Fig. S7). While both rapid recruitment and transcriptional activity of p53 likely play active roles in tumor suppression, these two functions may, in part, be independent of each other.
Given that we have identified PARP-dependent modification of p53 as important for its rapid recruitment to damage sites, we next investigated the effect of PARP inhibition or expression of PARylation-deficient mutants, PBM4 and PBM10, on choice of repair pathway. Strikingly, PARP inhibition reduced 53BP1 recruitment (SI Appendix, Fig. S6 A and C). PARP inhibition also significantly decreased the rate and magnitude of DDB1 recruitment (SI Appendix, Fig. S6B), although the overall percentage of cells that exhibited DDB1 recruitment was unaltered (SI Appendix, Fig. S6C). Likewise, expression of the PARylation-deficient mutants, PBM4 and PBM10, led to a decrease in the percentage of cells that exhibited 53BP1 or DDB1 recruitment (SI Appendix, Fig. S6D). Next, we investigated the effect of PARP inhibition and PARylation-deficient mutants on the balance between NHEJ and MMEJ using plasmid-based recircularization assays. Cells were briefly exposed to UVA to activate the DDR and induce p53 PARylation. %MMEJ was significantly increased in U2OS (p53 WT) treated with PARP inhibitor, while there was no change in %MMEJ in p53-null Saos2 cells (SI Appendix, Fig. S6 E and G). Expression of PBM4 or PBM10 mutants in Saos2 cells did not alter %MMEJ as compared to expression of WT p53, which suppressed MMEJ in favor of NHEJ (SI Appendix, Fig. S6 F and H). We also investigated the tumorigenic potential of Saos2 cells stably expressing PBM4 and PBM10 in xenograft models. Consistent with our previous findings of recruitment-deficient mutant tumors, PBM4- and PBM10-expressing tumors were significantly larger in size compared to WT tumors (SI Appendix, Fig. S6J). These observations are consistent with our in vitro findings above and support a role for PARP-dependent modification of p53 on recruitment of downstream repair factors and choice of repair pathway.
Discussion
We report the striking rapid recruitment of p53 to laser-induced sites of DNA damage. The intranuclear redistribution of p53 occurred with a half-time of 0.8 s upon laser microirradiation. Both endogenous and transiently expressed p53 accumulated at damage sites at very early time points in a variety of different cell lines and, furthermore, p53 accumulation was precipitated by a variety of DNA damaging agents. This rapid binding depended upon PARylation of p53 and closely mirrored the recruitment of PARP and Ku70 to damage sites. Rapid binding of p53 to DNA damage sites is a unique property of human p53. Analysis of various domain-specific p53 mutants revealed that rapid accumulation of p53 did not correlate with presence of transcriptional activity. We also report that rapid accumulation of p53 influenced recruitment of 53BP1 and DDB1 and subsequent choice of end-joining repair pathway and NER, respectively. We put forward the following model (Fig. 6B): upon induction of DNA damage, WT p53 (and recruitment-competent mutants) rapidly accumulate at these sites. Subsequent recruitment of 53BP1 preferentially activates NHEJ repair over the more error-prone MMEJ with implications for tumor suppression.
Structural Contributions of the DBD and CTD to p53 Recruitment to Damage Sites.
From our analysis of various domain-specific p53 mutants, we found that mutations within the DBD or CTD are able to completely abolish the rapid recruitment of p53 to damage sites. Meanwhile, mutations in other domains affected the magnitude of recruitment to varying degrees. Both the DBD and CTD of p53 are essential for interaction with DNA. Interaction of the p53 DBD with damaged DNA likely involves elements that recognize some particular feature of DNA structures, as well as damage-related response factors. On the other hand, the CTD facilitates PARP interaction and p53 PARylation and has a strong affinity for sequence nonspecific DNA and DNA damage lesions (25, 51). The CTD facilitates p53 “sliding” along DNA in a sequence-nonspecific manner, not only as part of the search for p53 target gene promoters (52–54) but as part of its checkpoint function given its ability to recognize a variety of DNA lesions, such as mismatched bases, bulges, hairpins, loops, single-strand DNA, and single-strand DNA–double-strand DNA transition intermediates (51, 55, 56). It is known that DNA repair enzymes use such a sliding mechanism to locate DNA lesions (57). In line with our findings, binding of the CTD to DNA potentially enhances rapid p53 recruitment to DNA damage lesions. Furthermore, as a result of the ability of the CTD to recognize and bind unusual DNA structures, p53 has been implicated in recruitment of other proteins to such sites (25, 51, 58). Notably, the common hotspot cancer mutations that affect the specific DNA binding function of p53 abolished early binding to damage sites. Our findings that other domain-specific mutations do impact this property of p53 are likely due to critical intramolecular interactions that can alter the configuration of the DBD or CTD. Specifically, interactions between the N-terminal domain, which is comprised of the TADs and the DBD, modulate the DNA-binding affinity and specificity of p53 (56). Our findings that a DBD–CTD fusion protein alone is incapable of being recruited reinforces the notion that proper protein conformation and interactions between the different domains of p53 are likely critical in fine-tuning the ability of the protein to be rapidly recruited to damage sites, as well as the complexities of p53 structure and function.
We have also demonstrated that PARP-dependent modification of the CTD is necessary for the rapid recruitment of p53. The PARylation-deficient PBM4 and PBM10 mutants have been shown to impair PARP-1 binding capability, which significantly decreases PARP1-mediated PARylation of p53 (40). While PARP can affect a variety of enzymatic substrates, the functional consequence of both PARP inhibition and the PARylation-deficient mutants on p53 recruitment and choice of repair pathway provide strong genetic evidence for the significance of PARP-dependent modification of p53 on p53 functions in DNA repair and maintenance of genomic stability.
Implications of Rapid p53 Binding to Damaged DNA.
Previous studies have emphasized the transcriptional functions of p53 in response to DNA damage but the rapid recruitment of p53 to sites of DNA damage that we demonstrate here implicates p53 in other aspects of DNA damage repair. Our laser microirradiation technique allowed for analysis of the real-time dynamics of p53 protein kinetics at damaged DNA in a localized area within the nucleus. This method also allowed us to investigate the sequential order of recruitment of other repair factors that were both dependent and independent of the presence of p53 at damage sites. Prior studies have not investigated the kinetics of p53 accumulation immediately following a localized, isolated DNA damaging insult. Instead, studies have often focused on detection of phosphorylated p53 in damage foci at late timepoints (30 min or more postdamage) (22, 23, 31, 59). Furthermore, those studies have also relied on IR and chemical-based methods that induce higher, global levels of damage across a cell, which might obscure the rapid binding of p53 to damage sites. We have optimized our laser conditions to avoid extensive and irreparable damage. The rapid recruitment of p53 in our system might hence be more reflective of how p53 typically responds to routine DNA damage that arises from endogenous sources of cellular stress, such as those characteristic of replication and transcription processes. Given that a cell possesses multiple, fine-tuned mechanisms for dealing with varying degrees and types of DNA damaging lesions, a given factor, such as p53, might likely exhibit very different accumulation dynamics under different conditions. At later time points following DNA damage, p53 may primarily be functioning as a transcription factor that is activated by extensive DDR-associated PTMs. In contrast, the rapid binding of p53 to damage sites offers insight into a different role of p53 in guiding DNA repair mechanisms, as such cellular decisions, are often made early on following DNA damage.
The rapid binding of p53 to damage sites raises the possibility that there are specific DNA lesions and early processing events detected by p53 as part of DDR signaling. This has potential implications for the subsequent choice of repair mechanisms, repair efficiency, and ultimately, cell fate. For example, Caenorhabditis elegans p53 (cep-1) and zebrafish p53 facilitate removal of UV lesions and DNA repair (60, 61), suggesting potential conservation of such a critical property of p53. To this end, we have demonstrated that rapid accumulation of p53 directly promoted recruitment of 53BP1 and DDB1, impacting choice of end-joining repair mechanisms and the cellular ability to resolve UV-induced damage. Furthermore, we observed differences in tumor latency in vivo among various mutant p53 that correlated well with whether the given mutant was capable of rapid recruitment to damage sites or not. In this regard, the ability of transcription factors to function in a transcription-independent manner and modulate DDR signaling at DNA lesions is not unprecedented. ATF2, E2F1, Sp1, and the NR4A family of nuclear receptors have been shown to recruit chromatin remodelers to damage sites and, more importantly, regulate NER (of which transcription-coupled repair is an associated subpathway) (62). Interestingly, E2F1 accumulates at sites of UV laser microirradiation in a manner independent of its DBD (63). Like p53, E2F1 is modified by ATM and ATR, and E2F1-deficiency impairs recruitment of NER factors XPA and XPC.
p53 has been shown to have an effect on enhancing end-joining using in vitro I-SceI–based assays, although its effect on precise versus error-prone end-joining is less conclusive (64). Here, we demonstrate that the rapid presence of WT p53 (and select mutants) at damage sites influenced the recruitment of 53BP1, and that this resulted in selection of direct end-joining over the more error-prone MMEJ. With increasing evidence for MMEJ not simply being an alternative “back-up” repair mechanism, but rather one that can occur despite intact HR and NHEJ mechanisms (65), p53’s active suppression of this pathway following damage insults is important in preventing mutagenic repair. Following UV irradiation, p53 has also been shown to interact with NER components, such as XPC and XPB, and this correlates with their increased recruitment to photoproducts and DNA lesions (66). p53 has also been implicated in global relaxation of chromatin in order to promote lesion detection and facilitate NER (67). We show here that p53-dependent recruitment of DDB1 is critical in determining NER efficiency.
Recruitment magnitude of various forms of mutant p53 studied correlated better with tumor suppression, as observed in our in vivo xenograft models as compared to presence of transcriptional capability (Fig. 6A and SI Appendix, Fig. S7 G–J). While we cannot exclude the potential contribution of transcriptional activity of certain mutant p53 to repair choice and overall tumor suppression, our findings indicate that rapid recruitment of p53 to damage sites and p53 transcriptional capability may, in part, be independent of one another. The role of p53 in tumor suppression has long been attributed to its transcriptional regulation of cell cycle and cell death mechanisms. More recently, its transcriptional regulation of metabolic processes and DNA repair proteins, as well as nontranscriptional functions in replication stability and genome stability, have broadened p53’s repertoire of tumor-suppressive mechanisms. Our findings of rapid binding of p53 to sites of DNA damage and the ability of p53 to influence subsequent formation of repair complexes support other hypotheses of p53 function in DDR and repair that are tumor suppressive and has important implications for genomic stability and integrity.
Methods
Tissue Culture.
U2OS and Saos2 (human osteosarcoma), SW-13 cells (human primary small cell carcinoma in adrenal gland/cortex), BxPC3 (pancreatic adenocarcinoma), and SK-BR-3 (mammary adenocarcinoma) were obtained from the American Type Culture Collection (ATCC, catalog no. HTB-96TM, HTB-85, CCL-105TM, CRL-1687TM, and HTB-30, respectively). All cell lines except BxPC-3 and SK-BR-3 were maintained in high glucose (4.5 g L−1) Dulbecco’s modified Eagle’s medium (Thermo Fisher Scientific). BxPC-3 cells were maintained in RPMI medium (Thermo Fisher Scientific) and SK-BR-3 cells were maintained in McCoy medium (Thermo Fisher Scientific), respectively. All media were supplemented with 10% fetal bovine serum (FBS, Thermo Fisher Scientific) without antibiotics for cell culture at 37 °C in 5% CO2. All cell lines were free from mycoplasma contamination after testing with the MycoAlert PLUS mycoplasma detection kit (Lonza) (LT07-701).
Generation of p53-mNG RPE1 Cells Using CRISPR.
A 19-bp guide RNA (gRNA) (GAGAATGTCAGTCTGAGTC) targeting the junction of the last exon and 3′ UTR of the endogenous human p53 locus was used for Cas9-mediated cutting. To introduce the mNG tag, we generated a plasmid with a cassette consisting of mNG followed by a P2A sequence and a Tn5 neomycin-resistance gene. This cassette was flanked by 1-kb sequences around the p53 cut site, to facilitate homology-directed repair (SI Appendix, Fig. S1A). The Cas9-p53-gRNA and mNG-P2A-NeoR plasmids were simultaneously transfected into cells using Lipofectamine 3000, following the manufacturer’s protocols. Cells that had undergone the correct cutting and repair events were selected on the basis of antibiotic resistance by applying 0.4 mg/mL G418 for 14 d. Single-cell clones were derived from the selected population and validated by PCR of the endogenous p53 locus to confirm correct biallelic incorporation of the mNG cassette. Subsequently, expression of p53-mNG protein was confirmed by Western blot and examination by fluorescent microscopy.
Plasmid Constructs and DNA Manipulations.
Plasmids encoding p53-WT-EGFP were a gift from Tyler Jacks (Addgene #12091; MIT, Boston, MA). All mutant constructs used in this study were generated from this construct either by a QuikChange II XL kit (Stratagene) or a Q5 Site-directed mutagenesis kit (New England Biolabs) as per the manufacturer’s instructions. EGFP-FLAG-Ku70 was a gift from Steven Jackson (Addgene #46957; University of Cambridge, Cambridge, UK). 53BP1-EGFP fusions were subcloned from pcDNA5-FRT/TO-eGFP-53BP1 (gift from Daniel Durocher, Addgene #60813; Lunenfeld-Tanenbaum Research Institute, Toronto, ON, Canada). TurboGFP-fusions of NBS1 were obtained from OriGene and the turboGFP tag at its C-terminal was replaced with an EGFP tag by conventional molecular biology techniques. PARP1-chromobody (RFP-tag) plasmid was obtained from Chromotek (#xcr). All constructs were amplified with a Plasmid Midi Kit (Qiagen). All constructs tested in this study, except p53-mNG, were transiently expressed in selected cell lines using either Neon electroporation transfection system (Invitrogen) or Lipofectamine 2000 (Thermo Fisher Scientific).
Localized DNA Damage by Laser Microirradiation.
Localized DNA damage was induced by a UV picosecond laser. The majority of the work, except those presented in Fig. 4, was performed with a Nikon A1R confocal with a pulsed UVA laser (PowerChip, Teem Photonics: 355 nm; 1-kHz repetition rate; 0.3-ns pulse width) through a 60× oil objective (NA 1.4, Nikon). Detailed procedure was described elsewhere (15). The power of UV laser used in this study ranged 0.2 to 3 nW measured from the back-scattering light of a neutral density filter wheel in the optical path, which corresponds to 1 to 15 nW measured at the back aperture of the objective. For convenience, the laser power reported in this study referred to the intensity of the scattering light measured from the optical path, which was set at 0.2 nW unless indicated otherwise. In short, cells expressing constructs of interest were sensitized with 10 μg mL−1 Hoechst 33342 (Sigma-Aldrich) for 10 min. Localized DNA damage was introduced by exposing regions of interest (ROIs) within nuclei to a 355-nm (UVA) laser for 500 ms at 0.2-nW laser power. The rapid dynamics of p53 recruitment was captured at 1.25 s per frame with a Galvano scanner. The long-term observations (up to several hours) were taken at a rate of 30 s or 5 min per frame. For immunofluorescence studies, cells were seeded in grid-pattered glass-bottom dishes. Fifteen to 20 cells were selected from each dish with coordinates recorded. UV laser beam were introduced to different cells at a 15- to 20-s interval. The cells were fixed within 5 min following laser microirradiation. To investigate the late response of p53, cells were fixed 12 h after damage instead. The fixed samples were sequentially stained with primary antibodies against antiphospho-histone H2A.X Ser139 (Abcam, ab2893) and p53 (clone DO-1; Cell Signaling Technology #18032), followed by costaining of secondary antibodies including Alexa Fluor-488 goat anti-mouse IgG (A11001, Invitrogen) and Alexa Fluor-647 donkey anti-rabbit IgG (A31573, Invitrogen). The cells were then imaged again using recorded coordinates with fixed parameters for all samples from the same experiment.
Due to laboratory relocation, a second laser microirradiation system was constructed for this study. Most major experiments were repeated using a different UVA laser (UGA-42 Caliburn, Rapp OptoElectronics: 355 nm; 1-kHz repetition rate; 4.2-ns pulse width) and an ORCA Fusion sCMOS camera (Hamamatsu) installed on an Olympus SpinSR-10 Yokogawa spinning disk confocal. The 355-nm laser beam was introduced to the sample through an UIS2 XLine Plan Apochromat (UPLAPO) 100× oil objective (NA 1.5, Olympus) with an ND2 filter. The UV laser exposure time was set at 250 ms. Image acquisition was taken in two stages. The first set of time-lapse images was taken at 2.5-s intervals for the first 1 min following laser microirradiation to capture the rapid recruitment of p53. The rest of the movie was taken at 30-s intervals for 30 min to monitor DDR protein recruitment.
Global UVA Damage.
Global DNA damage induced by UV was achieved by sensitizing the cells with Hoechst 33342 (Sigma-Aldrich) at 10 μg mL−1 for 10 min and was kept in the incubator for an hour prior to being exposed to UV. The culture medium was replaced with phosphate-buffered saline (PBS) and the lid removed during UV irradiation. After being UV-irradiated for a stipulated time, PBS was replaced with regular culture medium and cells were placed back to the incubator for 30 min before fixation followed by staining with antibodies for monitoring damage resolution or persistence. UVA irradiation at 13.3 W m−2 was performed using a hand-carry UVP 3UV UV lamp (Thermo Scientific) for 8 min with a 4.5-cm spacer. The power was measured using a HS116K silicon photodiode (BaseLine Chromtech).
Chemical Inhibitors.
Plk4 inhibitor, centrinone (125 nM), and MDM2 inhibitor, R7112 (350 nM to 1 μM), were synthesized by Sundia Meditech, Shanghai, China and used as previously reported (68). The following chemical inhibitors were purchased from commercial sources, with their working concentrations and incubation time indicated in parentheses: CPT (Fisher Scientific, 4 μM for 30 min), MMS (Fisher Scientific, 0.01% for 30 min), Dox (Fisher Scientific, 4 μM for 30 min), and NCS (Sigma-Aldrich, 0.25 to 0.5 μg/mL for 30 min).
Quantification of Recruitment Kinetics.
Image analysis of DDR protein recruitment was performed using a homemade FIJI macro. The images were first stabilized using either the Image Stabilizer or Manual Drift Correction plugin to minimize translational movement of the nuclei. Following image stabilization, a 2.5-μm diameter ROI was chosen for analysis while the full width at half maximum of the UVA laser at 1 nW was 0.85 μm, as measured by the bleaching profile of a fluorescently labeled agarose gel. The mean fluorescence intensity of the whole nucleus being ablated was used for bleach correction. The data were then processed using a double normalization method following a background subtraction. The data were averaged from experiments with triplicates with about 20 to 40 cells per condition.
Proximity Ligation Assay.
The early recruitment of p53 and its interaction with γH2AX was probed by Duolink In Situ Orange Starter Kit Mouse/Rabbit (Millipore-Sigma) following the manufacturer’s instructions. In this assay, the fixed samples were probed with primary antibodies against phospho-histone H2A.X Ser139 (20E3) (Cell Signaling Technology, #9718) and p53 (DO-1) (Millipore-Sigma, #MABE327). The image acquisition was performed on an Olympus Spin-SR10 spinning disk confocal. Images were collected using z-stacks in combination with a tile scan. Maximum intensity z-projection were used for quantifying average PLA foci count per cell. The quantification was performed using a homemade FIJI macro.
Statistical Analysis.
Statistical analysis was carried out using GraphPad Prism v9.0.1 (GraphPad Software). Two-tailed t test with Welch’s correction, which does not assume equal SD, was performed throughout this study.
P53 Reporter Assay.
ARN8 cells were derived from the melanoma cell line A375 and stably express the p53 reporter RGCΔFos-LacZ, as described previously (69, 70). Cells were transiently transfected with EGFP-tagged WT and mutant p53 constructs and high-intensity EGFP+ cells were FACS-sorted and replated. β-Galactosidase activity was measured using the FluoReporter LacZ/Galactosidase Quantitation kit (Invitrogen). Measurements were carried out on a Safire II multiplate reader (Tecan).
Xenografts.
Animal experimentation were performed in accordance to the A*STAR Biological Resource Center guidelines. Live Saos2 WT and p53 mutant cells were resuspended in 50% Matrigel (Corning Basement Membrane Matrix) and 50% cell culture media. Three million cells were injected per site, subcutaneously, into NSG mice (InVivos). For the doxycycline-treatment group, mice water supply was supplemented with 2 mg/mL doxycycline (Sigma, D9891). Mice were killed and tumors were harvested before individual tumor exceeded 1.5 cm3 in volume.
Histopathology.
Tumors were fixed in 4% PFA, paraffin-embedded, sectioned, and stained with H&E (A*STAR Advanced Molecular Pathology Lab). Representative images were taken on a Leica DM LB2 brightfield microscope with an attached Leica DMC 4500 digital camera.
Endogenous p53 IP.
U2OS cells (untreated and UVA) were lysed with EBC buffer 1 (50 mM Tris⋅HCl, pH 7.5, 100 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, 1 mM DTT) containing NaV (New England Biolabs), NEM (Sigma), and cOmplete Mini protease inhibitor mixture (Sigma) followed by EBC buffer 2 (50 mM Tris⋅HCl, pH 7.5, 300 mM NaCl, 5 mM CaCl2) containing NaV (New England Biolabs), NEM (Sigma), and cOmplete Mini protease inhibitor mixture (Sigma). Lysates were incubated with 3 μg of anti-p53 (DO1, homemade) preconjugated to Protein G Dynabeads (Invitrogen) overnight at 4 °C.
End-Joining Assay.
End-joining assay using the construct pDVG94 was performed as previously described (49). Saos2 cells stably expressing WT and mutant p53 were transfected with EcoRV-AfeI double-digested pDVG94 4 h after UVA irradiation. Cells were harvested for plasmid extraction 24 h later.
DNA Dot Blot Assay.
Prior to UVC irradiation, cell culture medium was replaced with PBS. Cells were kept in PBS and the lid were removed prior to being exposed to UV. Untransfected Saos2 cells (p53-null) were irradiated with UVC at 15 J using an UVC 500 Crosslinker (Amersham Biosciences) and harvested at designated time points. Saos2 cells transfected with various selected p53 mutants were similarly irradiated and harvested at 6 h postdamage. Genomic DNA were extracted using PureLink Genomic DNA Mini Kit (Invitrogen). An equal amount of DNA from each sample (250 ng) was diluted in 50 µL 2× saline sodium citrate buffer and topped up with water to a final volume of 100 µL. The samples were then loaded on a prewet Hybond+ membrane (GE) using a Hybri-Dot Manifold (Life Technologies). Duplicates were created for each blot. One of the blots was probed with an antithymine dimer antibody (#MC-062, Kamiya Biomed). The other blot was stained with methylene blue and served as a loading control. For methylene blue stain, the blot was rinsed with 5% acetic acid solution then immersed in 1% methylene blue-containing 5% acetic acid solution for 2 min. Images of the blots were developed by a ChemiDoc MP (Bio-Rad).
Supplementary Material
Acknowledgments
We thank Prof. Yusuke Toyama for his expertise in laser ablation microscopy; various members of the M.P.S. and p53/DITL laboratories and Dr. Jung-Hoon Yoon for feedback and suggestions; Stephen Kunkel for his assistance in proximity ligation assay experiments; Dr. Jia Min Loo and Xiaoqian Zhang for their help in xenograft studies; and Dr. Diego Pitta de Araujo for his assistance in graphic design. D.P.L. and T.L.F.H. were supported by A*STAR core funding. M.P.S. and Y.-H.W. were supported by the Mechanobiology Institute at the National University of Singapore, and, more recently, by a Cancer Prevention and Research Institute of Texas grant at the University of Texas Medical Branch. W.L.T. and M.Y.L. were supported by funding from the National Medical Research Council, Singapore (OFIRG17may-061, OFIRG19nov-0106, CTGIIT18may-0012, NMRC/OFLCG/002-2018), the National Research Foundation, Singapore (NRF-NRFF2015-04, NRF-CRP22-2019-0003, NRF-CRP23-2019-0004), and the Singapore Ministry of Education under its Research Centers of Excellence initiative.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2113233119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Laptenko O., Prives C., Transcriptional regulation by p53: One protein, many possibilities. Cell Death Differ. 13, 951–961 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Kastenhuber E. R., Lowe S. W., Putting p53 in context. Cell 170, 1062–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Joerger A. C., Fersht A. R., The p53 pathway: Origins, inactivation in cancer, and emerging therapeutic approaches. Annu. Rev. Biochem. 85, 375–404 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Vousden K. H., Prives C., Blinded by the light: The growing complexity of p53. Cell 137, 413–431 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Gottifredi V., Wiesmüller L., The tip of an iceberg: Replication-associated functions of the tumor suppressor p53. Cancers (Basel) 10, 250 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klusmann I., et al. , p53 Activity results in DNA replication fork processivity. Cell Rep. 17, 1845–1857 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Yeo C. Q. X., et al. , p53 maintains genomic stability by preventing interference between transcription and replication. Cell Rep. 15, 132–146 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Roy S., et al. , p53 orchestrates DNA replication restart homeostasis by suppressing mutagenic RAD52 and POLθ pathways. eLife 7, e31723 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ho T., Tan B. X., Lane D., How the other half lives: What p53 does when it is not being a transcription factor. Int. J. Mol. Sci. 21, 13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams A. B., Schumacher B., p53 in the DNA-damage-repair process. Cold Spring Harb. Perspect. Med. 6, a026070 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sengupta S., Harris C. C., p53: Traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol. 6, 44–55 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Janic A., et al. , DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat. Med. 24, 947–953 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Chen X., Ko L. J., Jayaraman L., Prives C., p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 10, 2438–2451 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Chen J., The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb. Perspect. Med. 6, a026104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y. H., et al. , DNA damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat. Commun. 8, 2118 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurst V., Gasser S. M., The study of protein recruitment to laser-induced DNA lesions can be distorted by photoconversion of the DNA binding dye Hoechst. F1000 Res. 8, 104 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinant C., et al. , Activation of multiple DNA repair pathways by sub-nuclear damage induction methods. J. Cell Sci. 120, 2731–2740 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Mouret S., et al. , Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc. Natl. Acad. Sci. U.S.A. 103, 13765–13770 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akopiants K., et al. , Tracking the processing of damaged DNA double-strand break ends by ligation-mediated PCR: Increased persistence of 3′-phosphoglycolate termini in SCAN1 cells. Nucleic Acids Res. 42, 3125–3137 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob S., Miquel C., Sarasin A., Praz F., Effects of camptothecin on double-strand break repair by non-homologous end-joining in DNA mismatch repair-deficient human colorectal cancer cell lines. Nucleic Acids Res. 33, 106–113 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swift L. P., Rephaeli A., Nudelman A., Phillips D. R., Cutts S. M., Doxorubicin-DNA adducts induce a non-topoisomerase II-mediated form of cell death. Cancer Res. 66, 4863–4871 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Al Rashid S. T., et al. , Evidence for the direct binding of phosphorylated p53 to sites of DNA breaks in vivo. Cancer Res. 65, 10810–10821 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Fayzullina S., Martin L. J., DNA damage response and DNA repair in skeletal myocytes from a mouse model of spinal muscular atrophy. J. Neuropathol. Exp. Neurol. 75, 889–902 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed M., et al. , The C-terminal domain of p53 recognizes DNA damaged by ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 92, 9455–9459 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S., Elenbaas B., Levine A., Griffith J., p53 and its 14 kDa C-terminal domain recognize primary DNA damage in the form of insertion/deletion mismatches. Cell 81, 1013–1020 (1995). [DOI] [PubMed] [Google Scholar]
- 26.Freed-Pastor W. A., Prives C., Mutant p53: One name, many proteins. Genes Dev. 26, 1268–1286 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raj N., Attardi L. D., The transactivation domains of the p53 protein. Cold Spring Harb. Perspect. Med. 7, a026047 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy M. E., et al. , A functionally significant SNP in TP53 and breast cancer risk in African-American women. NPJ Breast Cancer 3, 5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meek D. W., Tumour suppression by p53: A role for the DNA damage response? Nat. Rev. Cancer 9, 714–723 (2009). [DOI] [PubMed] [Google Scholar]
- 30.Meek D. W., Anderson C. W., Posttranslational modification of p53: Cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 1, a000950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bekker-Jensen S., et al. , Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173, 195–206 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loughery J., Cox M., Smith L. M., Meek D. W., Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Res. 42, 7666–7680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho W. C., Fitzgerald M. X., Marmorstein R., Structure of the p53 core domain dimer bound to DNA. J. Biol. Chem. 281, 20494–20502 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Leroy B., et al. , Analysis of TP53 mutation status in human cancer cell lines: A reassessment. Hum. Mutat. 35, 756–765 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karlsson G. B., et al. , Activation of p53 by scaffold-stabilised expression of Mdm2-binding peptides: Visualisation of reporter gene induction at the single-cell level. Br. J. Cancer 91, 1488–1494 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sauer M., et al. , C-terminal diversity within the p53 family accounts for differences in DNA binding and transcriptional activity. Nucleic Acids Res. 36, 1900–1912 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumari S. R., Mendoza-Alvarez H., Alvarez-Gonzalez R., Functional interactions of p53 with poly(ADP-ribose) polymerase (PARP) during apoptosis following DNA damage: Covalent poly(ADP-ribosyl)ation of p53 by exogenous PARP and noncovalent binding of p53 to the M(r) 85,000 proteolytic fragment. Cancer Res. 58, 5075–5078 (1998). [PubMed] [Google Scholar]
- 38.Wesierska-Gadek J., Wojciechowski J., Schmid G., Central and carboxy-terminal regions of human p53 protein are essential for interaction and complex formation with PARP-1. J. Cell. Biochem. 89, 220–232 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Vaziri H., et al. , ATM-dependent telomere loss in aging human diploid fibroblasts and DNA damage lead to the post-translational activation of p53 protein involving poly(ADP-ribose) polymerase. EMBO J. 16, 6018–6033 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischbach A., et al. , The C-terminal domain of p53 orchestrates the interplay between non-covalent and covalent poly(ADP-ribosyl)ation of p53 by PARP1. Nucleic Acids Res. 46, 804–822 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leung C. C., Glover J. N., BRCT domains: Easy as one, two, three. Cell Cycle 10, 2461–2470 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haince J. F., et al. , PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283, 1197–1208 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Yang G., et al. , Super-resolution imaging identifies PARP1 and the Ku complex acting as DNA double-strand break sensors. Nucleic Acids Res. 46, 3446–3457 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bekker-Jensen S., Lukas C., Melander F., Bartek J., Lukas J., Dynamic assembly and sustained retention of 53BP1 at the sites of DNA damage are controlled by Mdc1/NFBD1. J. Cell Biol. 170, 201–211 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aleksandrov R., et al. , Protein dynamics in complex DNA lesions. Mol. Cell 69, 1046–1061.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Li J., et al. , DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res. 66, 8590–8597 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Seol J. H., Shim E. Y., Lee S. E., Microhomology-mediated end joining: Good, bad and ugly. Mutat. Res. 809, 81–87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weterings E., et al. , The Ku80 carboxy terminus stimulates joining and artemis-mediated processing of DNA ends. Mol. Cell. Biol. 29, 1134–1142 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verkaik N. S., et al. , Different types of V(D)J recombination and end-joining defects in DNA double-strand break repair mutant mammalian cells. Eur. J. Immunol. 32, 701–709 (2002). [DOI] [PubMed] [Google Scholar]
- 50.Xiang Y., et al. , RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y., Kulesz-Martin M., p53 protein at the hub of cellular DNA damage response pathways through sequence-specific and non-sequence-specific DNA binding. Carcinogenesis 22, 851–860 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Liu Y., Kulesz-Martin M. F., Sliding into home: Facilitated p53 search for targets by the basic DNA binding domain. Cell Death Differ. 13, 881–884 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Terakawa T., Kenzaki H., Takada S., p53 searches on DNA by rotation-uncoupled sliding at C-terminal tails and restricted hopping of core domains. J. Am. Chem. Soc. 134, 14555–14562 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Kamagata K., Itoh Y., Subekti D. R. G., How p53 molecules solve the target DNA search problem: A review. Int. J. Mol. Sci. 21, 1031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tafvizi A., Huang F., Fersht A. R., Mirny L. A., van Oijen A. M., A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. U.S.A. 108, 563–568 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He F., et al. , Interaction between p53 N terminus and core domain regulates specific and nonspecific DNA binding. Proc. Natl. Acad. Sci. U.S.A. 116, 8859–8868 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blainey P. C., van Oijen A. M., Banerjee A., Verdine G. L., Xie X. S., A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. U.S.A. 103, 5752–5757 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Q. E., et al. , Tumor suppressor p53 dependent recruitment of nucleotide excision repair factors XPC and TFIIH to DNA damage. DNA Repair (Amst.) 2, 483–499 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Kodama M., et al. , Requirement of ATM for rapid p53 phosphorylation at Ser46 without Ser/Thr-Gln sequences. Mol. Cell. Biol. 30, 1620–1633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hoffman S., Martin D., Meléndez A., Bargonetti J., C. elegans CEP-1/p53 and BEC-1 are involved in DNA repair. PLoS One 9, e88828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zeng Z., Richardson J., Verduzco D., Mitchell D. L., Patton E. E., Zebrafish have a competent p53-dependent nucleotide excision repair pathway to resolve ultraviolet B-induced DNA damage in the skin. Zebrafish 6, 405–415 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adam S., Polo S. E., Blurring the line between the DNA damage response and transcription: The importance of chromatin dynamics. Exp. Cell Res. 329, 148–153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malewicz M., Perlmann T., Function of transcription factors at DNA lesions in DNA repair. Exp. Cell Res. 329, 94–100 (2014). [DOI] [PubMed] [Google Scholar]
- 64.Menon V., Povirk L., Involvement of p53 in the repair of DNA double strand breaks: Multifaceted roles of p53 in homologous recombination repair (HRR) and non-homologous end joining (NHEJ). Subcell. Biochem. 85, 321–336 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang H., Xu X., Microhomology-mediated end joining: New players join the team. Cell Biosci. 7, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang X. W., et al. , p53 modulation of TFIIH-associated nucleotide excision repair activity. Nat. Genet. 10, 188–195 (1995). [DOI] [PubMed] [Google Scholar]
- 67.Rubbi C. P., Milner J., p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. EMBO J. 22, 975–986 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wong Y. L., et al. , Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155–1160 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hupp T. R., Sparks A., Lane D. P., Small peptides activate the latent sequence-specific DNA binding function of p53. Cell 83, 237–245 (1995). [DOI] [PubMed] [Google Scholar]
- 70.Böttger A., et al. , Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr. Biol. 7, 860–869 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.