Significance
Understanding and treating neurological disorders are global priorities. Some of these diseases are engendered by mutations that cause defects in the cellular synthesis of transfer RNAs (tRNAs), which function as adapter molecules that translate messenger RNAs into proteins. During tRNA biogenesis, ribonuclease P catalyzes removal of the transcribed sequence upstream of the mature tRNA. Here, we focus on a cytoplasmic tRNAArgUCU that is expressed specifically in neurons and, when harboring a particular point mutation, contributes to neurodegeneration in mice. Our results suggest that this mutation favors stable alternative structures that are not cleaved by mouse ribonuclease P and motivate a paradigm that may help to understand the molecular basis for disease-associated mutations in other tRNAs.
Keywords: tRNA processing, neurodegeneration, conformational toggling, tRNA-Arg-TCT-4-1
Abstract
n-Tr20 is a neuron-specific, cytoplasmic transfer RNAArgUCU (tRNAArgUCU) isodecoder that affects seizure susceptibility, neuronal excitability, and translation signaling in mice. In addition, the C50U substitution in n-Tr20 (n-Tr20C50U), which is found in the widely used C57BL/6J (B6J) inbred mouse line, contributes to neurodegeneration by epistatic interactions with mutations in Gtpbp1 or Gtpbp2, two factors that rescue ribosomal stalling. The brains of B6J mice have high levels of immature and low levels of mature n-Tr20C50U, implicating defective tRNA biogenesis as the basis for neuronal dysfunction, a hypothesis tested in this study. We demonstrate that partially purified mouse brain ribonuclease P, the endonuclease responsible for 5′ maturation of tRNAs, exhibits at 1 mM magnesium a 20-fold lower apparent cleavage rate for processing the n-Tr20C50U precursor than the wild-type precursor. Thermal denaturation studies unexpectedly revealed that substituting the native C50-G64 base pair with the U50-G64 wobble pair in n-Tr20C50U increased the Tm by as much as 8 °C, with the magnitude of the change dependent on [Mg2+]. Moreover, results from thermal denaturation, native gel electrophoresis, 1H nuclear magnetic resonance spectroscopy, and kinetic studies collectively support the presence of stable alternative folds for n-Tr20C50U that may also be sampled transiently and in low abundance by the wild-type tRNA. These findings suggest that mutation-driven restructuring could foster nonnative folds and that conformational toggling of tRNAs poses an intrinsic risk for dysfunction when point mutations selectively stabilize nonnative states. This model may be applicable for understanding the molecular basis of other diseases that are associated with tRNA mutations.
Transfer RNAs (tRNAs) are the most highly expressed, small noncoding RNAs in all domains of life (1, 2). While many diseases arise from mutations in processing/modifying enzymes that act on multiple tRNAs (3–5), mutations in single tRNA genes in higher eukaryotes have largely been overlooked as drivers of disease due to the multiplicity of genes encoding tRNAs with the same anticodon (isodecoders) and the expectation that these RNA polymerase III–transcribed genes are ubiquitously expressed. However, there is growing evidence that the expression of individual tRNAs can vary between tissues and with cellular state, stress, and disease (6–11). In this regard, n-Tr20 (Fig. 1A) and its human ortholog (tRNA-Arg-TCT-4-1), members of the cytoplasmic tRNA-Arg-TCT family (five genes in mice and six in humans), we found to be tissue-specifically expressed. n-Tr20 is specifically expressed in neurons where it is the most prominently expressed member of this tRNA family (7, 12, 13). Moreover, a mutant form of n-Tr20 contributes to neurodegeneration (7, 12, 14).
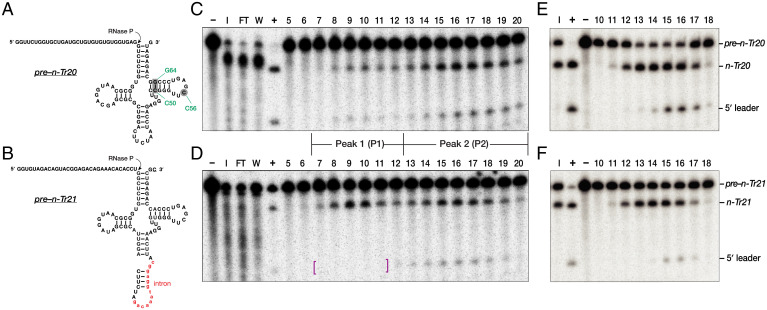
Fig. 1.
Purification of mouse brain RNase P. (A, C, and E) Pre–n-Tr20 and (B, D, and F) pre–n-Tr21 were used to assay individual purification fractions for RNase P activity. While both pre-tRNAs contain a 32-nt 5′ leader, pre–n-Tr21 also has a 13-nt intron (red lowercase) and a 1-nt 3′ trailer (underlined). The arrows indicate the expected RNase P cleavage site. In A, the three positions that are mutated in different n-Tr20 variants used in this study are shown in green. (C and D) RNase P assays of the elution fractions from the DEAE column. In D, magenta-colored brackets are used to highlight degradation of the cleaved 5′ leader of pre–n-Tr21 in fractions 7 to 11. The two reproducible peaks of activity are indicated: P1 (fractions 7 to 12) and P2 (fractions 13 to 20). (E and F) Further purification of P1 using a 10 to 35% (vol/vol) glycerol gradient. Fractions 14 to 16 were pooled to yield P1F (the final P1 preparation). Lanes are as follows: I, input; FT, flow through; W, wash; –, negative control containing substrate but no enzyme; +, positive control with substrate and in vitro–reconstituted E. coli RNase P (49).
The C50U mutation in n-Tr20 (n-Tr20C50U) found in the C57BL/6J (B6J) inbred mouse strain leads to an accumulation of immature n-Tr20C50U with a 5′ leader and a 3′ trailer, which are normally removed during tRNA maturation in preparation for aminoacylation. Interestingly, this mutation has also been found in a human Arg-TCT tRNA (15). Accumulation of this immature tRNA was accompanied by a corresponding decrease in the level of mature aminoacylated tRNA and, ultimately, ribosome stalling at AGA codons (7). Homozygosity for this mutation resulted in neurodegeneration when associated with the loss of guanosine-5'-triphosphate (GTP) binding protein 1 (GTPBP1) or GTPBP2, which relieves ribosome stalling (7, 14). Both B6J.Gtpbp1−/− and B6J-Gtpbp2−/− mice exhibit pronounced truncal ataxia starting at 4 wks of age, concomitant with apoptosis of neurons in the inner granule layer of the cerebellum, as well as degeneration of hippocampal, cortical, and retinal neurons. This neurological phenotype results from a breach in the GTPBP1/2-mediated translational surveillance mechanism; ribosome profiling revealed that n-Tr20C50U–induced stalling at AGA codons increased in the absence of these proteins (7, 14). Furthermore, loss of n-Tr20 expression (caused by either the C50U mutation or knocking out n-Tr20) is sufficient to confer resistance to electrically and pharmacologically induced seizures (12). n-Tr20C50U homozygosity also modified the effect of an epilepsy-associated mutation in the gamma-aminobutyric acidA receptor γ2-subunit as evidenced by a decreased frequency of spike-wave discharges (12). Decreased levels of n-Tr20 represses translation and activates the integrated stress response via general control nonderepressible 2, a protein kinase that is activated by ribosomal stalling (12).
The above studies, which unmasked the disease potential of tRNA genes, motivated further biochemical and structural characterization of n-Tr20. Here, we sought to dissect the molecular basis for defective processing of n-Tr20C50U that has a U50-G64 base pair (bp) instead of a C50-G64 bp in the T stem (7). Since most cytoplasmic precursor tRNAs (pre-tRNAs) are processed in the 5′-before-3′ order in eukaryotes (16, 17) and the brains of B6J mice accumulated a 105-nucleotide (nt) immature n-Tr20C50U with a 5′ leader and a 3′ trailer instead of the 77-nt mature n-Tr20C50U (7), we hypothesized that the C50U mutation impairs cleavage by ribonuclease (RNase) P, the endonuclease that removes the 5′ leader of pre-tRNAs (18–22). Although the composition [one catalytic RNA and ≤10 protein cofactors,] and cryogenic electron microscopy structures [that reveal subunit arrangement and substrate–recognition determinants (23, 24)] of eukaryotic nuclear RNase P are known, it is difficult to explain why this mutation that is distal to the RNase P cleavage site would adversely affect 5′ processing. Moreover, U50 is not uncommon in tRNAs (25) and is even present in two other Arg-TCT isodecoders, n-Tr21 (tRNA-Arg-TCT-3-1) (Fig. 1B) and n-Tr25 (tRNA-Arg-TCT-2-1). Our thermal denaturation, native gel electrophoresis, nuclear magnetic resonance (NMR) spectroscopy, and kinetic studies of wild-type (WT) and mutant n-Tr20 led to the surprising finding that n-Tr20C50U adopts stable nonnative structures. These results form the basis for a conformational toggling model that posits how intrinsic vulnerabilities associated with tRNA folding are amplified by point mutations.
Results
Purification of RNase P from Mouse Cortex.
To determine whether the C50U mutation affects 5′ processing of pre–n-Tr20C50U, we partially purified RNase P from the brain cortices of B6J mice. The cortical lysate was loaded on a diethylaminoethyl (DEAE) (anion-exchange) column, and the eluted fractions were assayed for RNase P activity with two in vitro–transcribed WT pre-tRNAArgUCU isodecoders: pre–n-Tr20 and pre–n-Tr21, both with a 5′ leader but only the latter had a single-nt 3′ trailer (Fig. 1 A and B). We consistently observed two reproducible and separable peaks (P) of RNase P activity. Because these two biochemically distinct forms could potentially process pre–n-Tr20 and –n-Tr20C50U differently, active fractions from each peak were pooled separately as P1 and P2 (Fig. 1 C and D).
Both P1 and P2 activities were unstable and steadily lost activity within a week of purification, likely due to degradation of the RNase P RNA by coeluting nucleases (Fig. 1D). Therefore, P1 was further purified using a 10 to 35% (vol/vol) glycerol gradient (Fig. 1 E and F) and then concentrated sevenfold using a centrifugal filter. While P2 lost all activity when subjected to further purification, its activity could be prolonged when concentrated 10-fold. The final preparations, P1F and P2F, had a longer shelf-life and lasted about a month. Importantly, both activities catalyzed site-specific cleavage of n-Tr20 (SI Appendix, Fig. S1).
The 3′ Trailer Does Not Interfere with 5′ Processing of Pre–n-Tr20 by Mouse Brain RNase P.
The presence of a 3′ trailer in pre-tRNAs has no effect on cleavage by eukaryotic nuclear RNase P (16) (an exception is in ref. 26). However, it was unclear whether this attribute holds for n-Tr20C50U because B6J brains accumulated an immature n-Tr20C50U with both a 5′ leader and a 3′ trailer (7). Therefore, we assayed P1F and P2F for their ability to cleave in vitro–transcribed pre–n-Tr20 + 3′ containing a 5′ leader and a 3′ trailer (SI Appendix, Fig. S2). P1F and P2F cleaved the 5′ leader of both pre–n-Tr20 + 3′ and pre–n-Tr20C50U+3′, albeit the latter was much poorer, revealing that it is the C50U mutation, not the trailer, that hampers RNase P cleavage. In addition to this 5′ maturation activity, we observed that the 3′ trailer was also cleaved in our assays due to a coeluting 3′ processing activity (likely RNase Z) in both P1F and P2F (SI Appendix, Fig. S2). Notably, the pattern of product formation indicated that 3′ processing took place only with the RNase P–processed intermediate, consistent with previous reports that most eukaryotic pre-tRNAs are cleaved by RNase P before RNase Z (17).
Mg2+-Dependent Cleavage Rates of Mouse Brain RNase P Vary with Substrate Identity.
Site-specific coordination of Mg2+ in binding pockets (particularly the elbow) of tRNAs is critical for their overall three-dimensional architecture which, in turn, is important for function (e.g., aminoacylation) (27–29). We hypothesized that the C50U mutation may weaken the T stem and consequently, tertiary contacts in n-Tr20C50U. Therefore, we tested P1F and P2F for their ability to cleave pre–n-Tr21, –n-Tr20, and –n-Tr20C50U (all with only a 5′ leader) in 0.5, 1, 1.5, or 5 mM MgCl2. P1F and P2F displayed slightly different MgCl2 optima during cleavage of pre–n-Tr21 and –n-Tr20, with maximal activity in near-physiological [MgCl2] (Fig. 2A). P2F also appeared to be less sensitive than P1F to [MgCl2] changes between 0.5 and 1.5 mM. Interestingly, pre–n-Tr20C50U was cleaved poorly by both P1F and P2F regardless of the [MgCl2] tested, with minimal rescue even in 5 mM MgCl2 (Fig. 2A). While their differential elution from DEAE is suggestive of differences in the compositional makeup of P1F and P2F, both cleaved pre–n-Tr20C50U with an apparent cleavage rate that is 20-fold slower than the WT in 1 mM MgCl2, the optimal concentration for processing pre–n-Tr20 (Fig. 2). Despite this weak cleavage, processing fidelity was maintained with the mutant pre-tRNA (SI Appendix, Fig. S1).
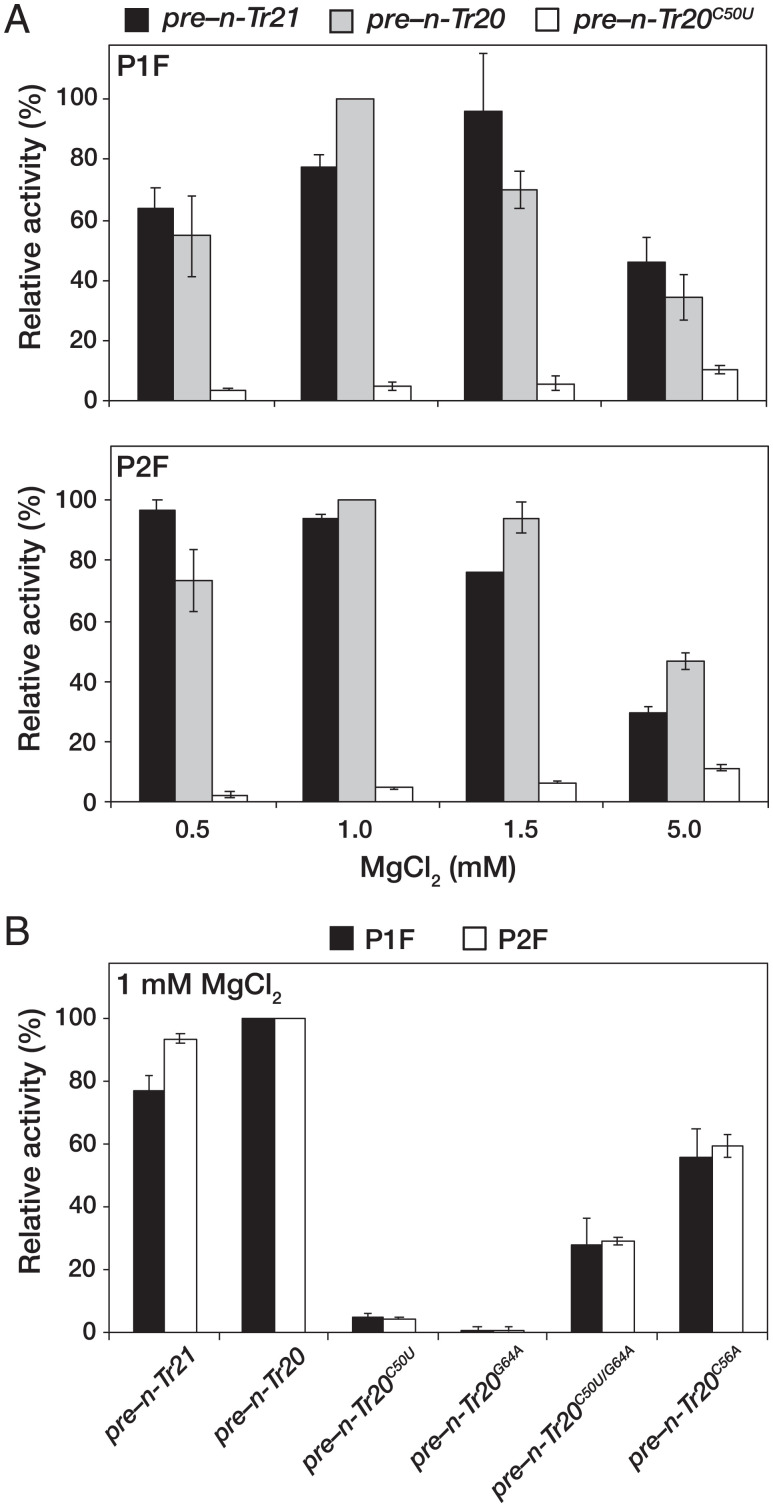
Fig. 2.
Comparison of the apparent cleavage rates of different pre-tRNAs by mouse brain RNase P. (A) Relative Mg2+-dependent cleavage activities for processing pre–n-Tr21, pre–n-Tr20, and pre–n-Tr20C50U by the two peaks of RNase P activity, P1F (Upper) and P2F (Lower). (B) Relative cleavage activities for processing pre–n-Tr21, pre–n-Tr20, and different mutant derivatives of pre–n-Tr20 in 1 mM MgCl2. All values are normalized to the activity observed with pre–n-Tr20. Mean and standard deviation values were calculated from three independent initial velocity measurements.
To investigate whether tissue-specific factors in the mouse brain RNase P preparations somehow dampen the enzyme’s ability to process specific mutant pre-tRNAs, we partially purified RNase P from the livers of 4-wk-old B6J mice and conducted a cleavage assay with pre–n-Tr20 and –n-Tr20C50U at 1 and 5 mM MgCl2. Again, we observed a profound decrease in pre–n-Tr20C50U processing (SI Appendix, Fig. S3), suggesting that this cleavage defect arises from structural perturbations in pre–n-Tr20C50U rather than tissue-specific variations in RNase P.
The N50-N64–bp Identity in n-Tr20 Influences the Rate of RNase P Cleavage.
While the C50U mutation does not eliminate base pairing (C50-G64 is replaced by U50-G64), poor cleavage of pre–n-Tr20C50U by both P1F and P2F prompted us to examine the importance of the identity of N50 and N50-N64 bp itself. Therefore, we constructed two mutant derivatives and examined their cleavage by P1F and P2F in 1 mM MgCl2. Pre–n-Tr20G64A (with a C50-A64 mismatch) was processed at only 1% of the apparent cleavage rate observed for n-Tr20 (Fig. 2B), suggesting that the N50-N64 bp is important for promoting a pre-tRNA structure recognized by RNase P. Surprisingly, despite the isosteric replacement of C50-G64 with U50-A64, the apparent cleavage rate for pre–n-Tr20C50U/G64A was only ∼30% of that determined for pre–n-Tr20 (Fig. 2B). These data indicate a preference for a C50-G64 bp in n-Tr20 to promote an optimal structure for recognition and cleavage by RNase P. Although the identity of N50 may not be an absolute determinant for mouse RNase P activity (as supported by the robust cleavage of pre–n-Tr21, which has a U50-A64 bp) (Fig. 1B), it is conceivable that substitution of C50 with U50 in n-Tr20 alters its conformational ensemble (see below).
n-Tr20C50U Is More Stable than WT n-Tr20.
We used thermal denaturation experiments to investigate the stability of in vitro–transcribed mature n-Tr20 and its mutant derivatives and to gain insights into whether the observed differences in the apparent RNase P cleavage rates of these substrates reflect their respective distribution between “cleavable” and “noncleavable” conformers (28, 30). Monitoring hyperchromicity as a function of increasing temperature traces the transition from a folded to an unfolded state, and the resulting Tm values can provide insights into the relative stabilities of different RNAs (28). For tRNAs, the nuanced low-temperature transition (∼40 °C) is typically attributed to unraveling of the elbow (tertiary contacts) (30), while a sharper higher-temperature transition corresponds to cooperative melting of the secondary structure (28, 31). Thermal denaturation experiments were conducted in 0, 1, or 5 mM MgCl2 to assess whether potential nonnative structural realignments in tRNA mutants might be reversed by increasing [MgCl2]. Normalized absorbance at 260 nm was plotted vs. temperature (20 to 95 °C) (Fig. 3A), and the corresponding first-derivative plots (dA/dT vs. temperature) (Fig. 3A) were generated. The Tm values for each tRNA were obtained from the maxima of the first-derivative plots.
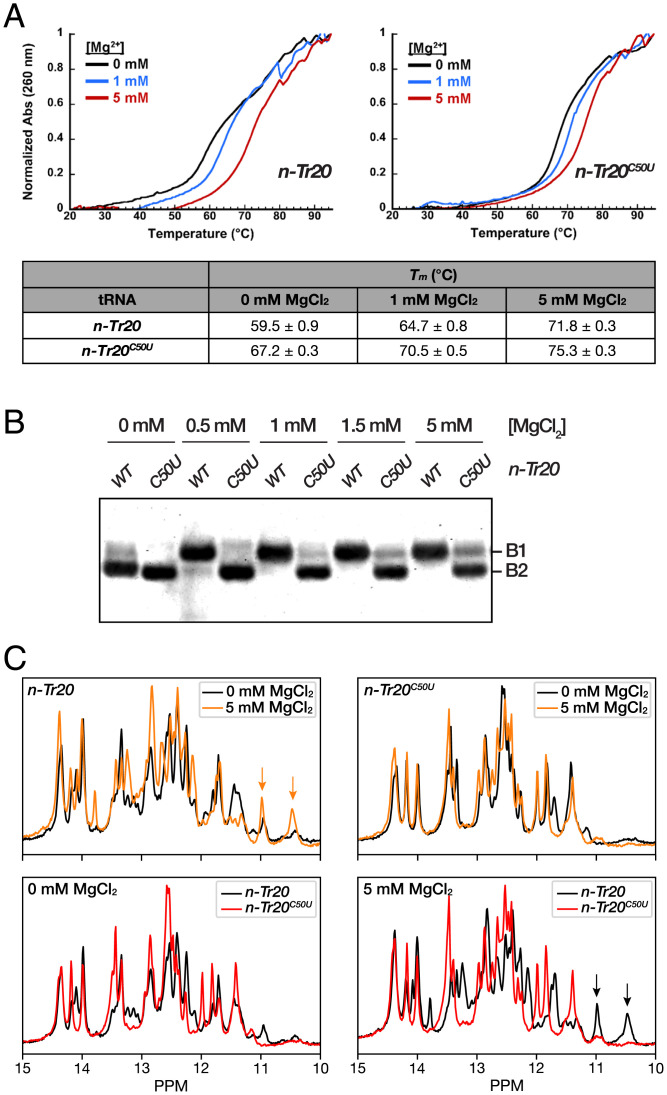
Fig. 3.
Structural characterization of mature n-Tr20 and n-Tr20C50U. (A) Thermal denaturation experiments reveal increased structural stability conferred by the C50U mutation. (Upper) Normalized absorbance at 260 nm is plotted as a function of increasing temperature in 0, 1, or 5 mM MgCl2. Mean and SD values were calculated from three independent thermal melts. (B) Native PAGE analyses reveal Mg2+-dependent tRNA conformational changes. The two tRNAs migrated as two bands, labeled B1 and B2, with B1 likely constituting the native fold. (C) The imino proton region of 1H NMR spectra of n-Tr20 and n-Tr20C50U exhibits two distinct conformers, with n-Tr20 showing more Mg2+-induced alterations than n-Tr20C50U. (Upper) Overlaid spectra for each tRNA in 0 vs. 5 mM MgCl2. (Lower) Comparison of the two tRNAs in 0 vs. 5 mM MgCl2. Orange and black arrows indicate the two upfield peaks in n-Tr20 that form in the presence of MgCl2 and that likely indicate tertiary contacts (SI Appendix, Figs. S6 and S7).
Based on previous studies that examined disease-associated mutant tRNAs with single-nt substitutions (32, 33), the C50U mutation was expected to decrease the Tm of n-Tr20. However, we were surprised to find that n-Tr20C50U exhibited a Tm that was ∼8 °C higher than the WT in the absence of MgCl2 (Fig. 3A). Increasing [MgCl2] from 0 to 5 mM raised the Tm by 12 °C for n-Tr20 and 8 °C for n-Tr20C50U (Fig. 3A). While the Tm observed with n-Tr20C50U remained higher than n-Tr20 in all three MgCl2 concentrations, the rift narrowed with increasing [MgCl2] (Fig. 3A). Remarkably, the compensatory base substitution mutant n-Tr20C50U/G64A also displayed increased stability, with similar Tm values as n-Tr20C50U in 0, 1, and 5 mM MgCl2 (SI Appendix, Fig. S4). The C50-A64 mismatch mutant n-Tr20G64A had Tm values between those of n-Tr20 and n-Tr20C50U/n-Tr20C50U/G64A, suggesting that n-Tr20G64A adopts a secondary structure that is distinct from the other tRNAs.
Conformational Sampling by n-Tr20 and Its Mutants.
Our unexpected finding of the increased structural stability of native n-Tr20C50U prompted us to examine the conformational attributes of all in vitro–transcribed variants of mature n-Tr20 using native polyacrylamide gel electrophoresis (PAGE), which allows for the visualization and quantitation of multiple conformations with distinct electrophoretic mobilities (34). In 0 mM MgCl2, we detected two species for n-Tr20: a slower-migrating band (B1) that is much fainter than a faster-migrating band (B2) (Fig. 3B). Consistent with the expectation that n-Tr20 attains a native-like fold at higher [MgCl2], the proportion of n-Tr20 found in B1 increased with increasing [MgCl2], while B2 disappeared completely by 1 mM MgCl2. Thus, B1 likely reflects the native conformation. n-Tr20C50U migrated as a single band (B2) in 0 mM MgCl2. The slight increase in the amount of B1 from 0.5 to 5 mM MgCl2 suggests that only a small fraction of n-Tr20C50U attains a native-like fold even in 5 mM MgCl2 (Fig. 3B), in sharp contrast to the trend observed with n-Tr20.
The other n-Tr20 mutants also showed interesting conformational differences. Like n-Tr20C50U, n-Tr20C50U/G64A and n-Tr20G64A migrated only as B2 in 0 mM MgCl2 (SI Appendix, Fig. S5). More than half of n-Tr20C50U/G64A shifted to B1 in 1 mM MgCl2, with a further shift in 5 mM MgCl2. While MgCl2 also induced a slower migration for n-Tr20G64A, the shift seemed incremental and was more discernible in 5 mM MgCl2, where n-Tr20G64A existed in two bands that appeared to migrate between B1 and B2. As observed in the thermal denaturation curves, n-Tr20G64A behaved differently from n-Tr20, n-Tr20C50U, and n-Tr20C50U/G64A. Interestingly, the native PAGE migration profiles for all n-Tr20 mutants reflected roughly their cleavability by RNase P (Fig. 2B and SI Appendix, Fig. S5).
To fingerprint the putative alternative secondary structure of n-Tr20C50U predicted by thermal denaturation and native PAGE analyses, we performed one-dimensional (1D) 1H solution-state NMR experiments on all the n-Tr20 variants described above. The imino protons of G-H1 and U-H3 are excellent reporters of the secondary structure as they depend on the nature of base pairing (35). Experiments were performed at varying MgCl2 concentrations (0, 0.5, 1, 2, 5, or 10 mM) to assess RNA folding (Fig. 3C and SI Appendix, Fig. S6). The 1D 1H spectra of n-Tr20 showed many overall changes with increasing [MgCl2], with some already apparent even between 0 and 0.5 mM MgCl2, most notably the stabilization of two resonances at ∼11 and ∼10.5 ppm (Fig. 3C and SI Appendix, Fig. S6). Although unassigned, these distinct upfield peaks are consistent with Mg2+-stabilized tertiary contacts between the T and D loops in the tRNA elbow (36). In contrast, the spectra of n-Tr20C50U changed very little with increasing [MgCl2] as high as 10 mM (SI Appendix, Fig. S6), which is consistent with results from native PAGE analyses (Fig. 3B). The absence of the upfield peaks at ∼11 and ∼10.5 ppm suggested the lack of T loop–D loop interactions. Further differences throughout the spectra of n-Tr20C50U relative to n-Tr20 (Fig. 3C) suggest broad secondary structural changes that involve multiple G-C bps, at least one A-U bp, and the possible formation of one or two new G-U wobble pairs.
Next, we obtained 1D 1H NMR spectra for n-Tr20G64A and n-Tr20C50U/G64A in 1 mM Mg2+. Like n-Tr20C50U, both n-Tr20G64A and n-Tr20C50U/G64A lack the upfield peaks at ∼11 and ∼10.5 ppm. Consistent with the observations made with thermal melt and native PAGE analyses, the imino spectrum of n-Tr20G64A shows a unique pattern that is unlike that of n-Tr20 or n-Tr20C50U (SI Appendix, Fig. S7). In contrast, the spectrum of n-Tr20C50U/G64A appeared to result from an ensemble resembling partly that of n-Tr20 and n-Tr20C50U and suggested that the U50-A64 bp does not stabilize the structure to the same extent as the native G50-C64 bp (SI Appendix, Fig. S7).
Our hypothesis that C50U destabilizes tertiary contacts prompted us to construct the n-Tr20C56A mutant, expecting that it would provide a spectral signature indicative of a destabilized elbow. C56, the third position in the penta-nt T loop, forms a canonical bp with G19 in the D loop (29). However, we found that the imino spectrum of n-Tr20C56A largely resembled that of n-Tr20 including the two upfield peaks ∼11 and ∼10.5 ppm (SI Appendix, Fig. S7), suggestive of similar structures, an inference further supported by their concordant Tm values and similar behavior in native PAGE (Fig. 3A and SI Appendix, Figs. S4 and S5). Thus, the C56A mutation alone may be inadequate to completely disrupt the tertiary brace that is also buttressed by the intercalation of D-loop G18 between T-loop G57 and A58 to create a contiguous stack of 5 nt (37). Nevertheless, since pre–n-Tr20C56A is cleaved by RNase P approximately twofold slower than n-Tr20 (Fig. 3B), tertiary interactions must be weakened in n-Tr20C56A, and conformational switching is likely, as supported by native PAGE where its B2 band was detectable even in 5 mM MgCl2 (SI Appendix, Fig. S5).
A Model Depicting Structural Toggling of n-Tr20 and Its Mutant Derivatives.
While we expected the C50U mutation to be destabilizing, it actually increased the Tm of n-Tr20C50U by ∼4 to 8 °C relative to that of n-Tr20 (Fig. 3A). A more compact n-Tr20C50U structure would also be consistent with the faster mobility observed in native PAGE (Fig. 3B). In addition to an RNAFold prediction (http://gtrnadb.ucsc.edu/genomes/eukaryota/Mmusc39/genes/tRNA-Arg-TCT-4-1.html), we also considered an alternative fold for n-Tr20C50U (nonnative 2) (Fig. 4A). Both nonnative folds have more bp and disrupted tertiary contacts compared with the canonical structure predicted for n-Tr20. While additional data are needed to confirm the assignment of individual resonances in the NMR spectra and to obtain a high-resolution tertiary structure, the spectral “fingerprints” for n-Tr20 mutants (Fig. 3C and SI Appendix, Figs. S6 and S7) support the presence of nonnative secondary structures that lack the tertiary contacts required to form the tRNA elbow in n-Tr20C50U and other variants. There is a clear correlation between this conformational switching and the apparent rate of cleavage for each tRNA by mouse RNase P (Fig. 4B), despite some minor discrepancies likely due to the differences in the conditions used for RNase P assays, thermal melts, native PAGE, and NMR.
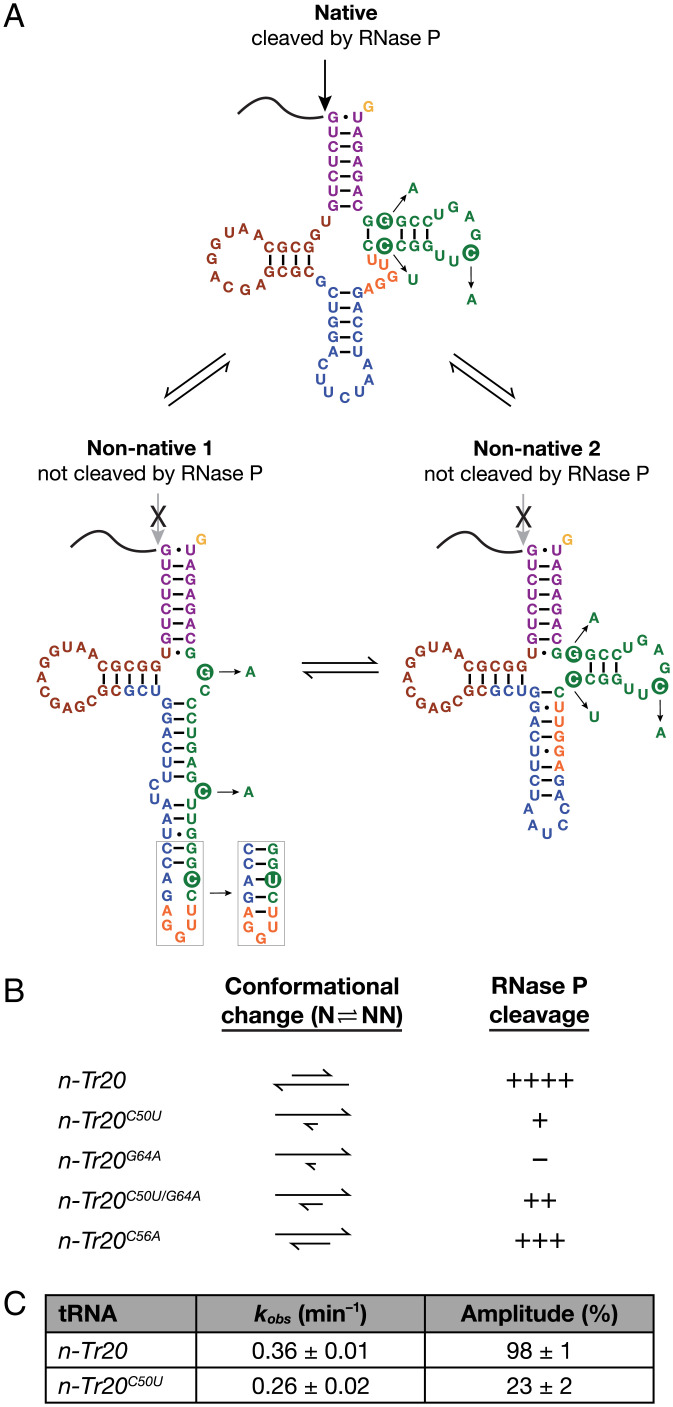
Fig. 4.
Postulated nonnative conformers of n-Tr20 and its mutant derivatives. (A) n-Tr20 may toggle between the native (favored in the presence of Mg2+) and several nonnative conformations, including the two shown here. Both nonnative configurations likely lack the native tRNA elbow, a key recognition determinant necessary for RNase P cleavage (22–24). (B) The propensity of mouse RNase P to cleave each n-Tr20 variant (data are from Fig. 2B) likely reflects the proportion of tRNA that adopts the native conformation. Cleavage efficiency: ++++, fully competent; +/++/+++, partially competent to varying extents; –, defective. N, native structure; NN, nonnative structure. (C) Summary of data from single-turnover kinetic studies conducted with in vitro–reconstituted E. coli RNase P in 5 mM Mg2+ and at pH 5.5. Mean and standard deviation values were determined from three independent measurements.
As an independent test of the conformational toggling idea, we performed single-turnover kinetic studies where [enzyme] ≫ [substrate]. If n-Tr20 mostly adopts a conformation that is cleavable by RNase P while n-Tr20C50U samples two or more conformations and only one of which coincides with the predominantly native conformation assumed by n-Tr20, then the kobs values for processing n-Tr20 and n-Tr20C50U should be similar under single-turnover conditions; moreover, the maximal product formation should be lower for the mutant tRNA and proportional to the fraction of n-Tr20C50U that adopts the RNase P–cleavable conformation. Because we sought to test a substrate structural attribute and needed a firm knowledge of the enzyme concentration for the single-turnover kinetic studies, we used recombinant bacterial RNase P. In 5 mM Mg(OAc)2 and at pH 5.5 [to dampen the cleavage rate (38, 39)], 1 μM in vitro–reconstituted Escherichia coli RNase P cleaved ∼1 fM n-Tr20 or n-Tr20C50U with similar kobs values of 0.36 ± 0.01 or 0.26 ± 0.02 min−1, respectively, and different amplitudes of 98 ± 1 or 23 ± 2, respectively (Fig. 4C and SI Appendix, Fig. S8). Our finding that only one-fourth of n-Tr20C50U is cleaved in 5 mM Mg2+ by E. coli RNase P is consistent with two findings: native PAGE analysis that showed predominantly nonnative states for n-Tr20C50U in 5 mM Mg2+ (Fig. 3B) and a three- to fourfold greater apparent cleavage rate for n-Tr20 than n-Tr20C50U in 5 mM Mg2+ by mouse RNase P (Fig. 2A).
Discussion
Our biochemical and biophysical studies shed light on why C50U, which changes a C-G bp in n-Tr20 to a U-G wobble bp, leads to profound neuronal dysfunction. We demonstrate here that poor processing of n-Tr20C50U by RNase P accounts for the low levels of mature and high levels of immature n-Tr20 in the brains of B6J mice (Fig. 2). Together, our findings from thermal denaturation, native PAGE, NMR, and kinetic studies point to nonnative structure(s) associated with the C50U mutation as the basis for weak tRNA cleavage by RNase P (Fig. 4).
Why does the C50U mutation cause dramatic structural perturbations in n-Tr20? Instead of a largely intact native structure with replacement of a C50-G64 with a U50-G64 bp, we consider the possibility of a highly favorable realignment to a nonnative state. Like many RNAs, conformational sampling of the functional native fold as well as several inactive nonnative structures is an intrinsic, sequence context–dependent property of n-Tr20 and other tRNAs (40). Such conformational toggling, however, poses an inherent risk for dysfunction, particularly when a mutation selectively stabilizes the nonnative state and skews the equilibrium to favor the inactive conformer. While n-Tr20 may sample a few nonnative folds (two of which are shown in Fig. 4A), Mg2+-stabilized tertiary contacts as well as binding to RNase P nudge it toward the native state. In contrast, C50U likely promotes nonnative states with long helices, as exemplified in the nonnative 1 structure (Fig. 4A); here, U50 forms a nonnative A42-U50 bp and promotes G43-C49 and A44-U48 bp, thereby stabilizing a long, nonnative helix. Such secondary structure stability, however, comes at the cost of the canonical tRNA L-shaped tertiary structure. The cryogenic electron microscopy structures of yeast and human nuclear RNase P bound to tRNA (22–24) highlight the enzymes’ use of a shallow pocket to bind the acceptor-T arm and of π−π interactions with the G19-C56 bp of the pre-tRNA elbow to guide cleavage. The absence of this essential recognition determinant in the two predicted nonnative folds for n-Tr20C50U (Fig. 4A) likely accounts for its poor cleavage by RNase P. Even for the fraction that is processed, the nonnative states adopted by the mature tRNA will continue to engender adverse functional consequences (7).
While the conformational ensemble sampled by n-Tr20 appears to be an intrinsic vulnerability, as evidenced by the C50U mutation, it may serve a biological role. n-Tr20 could act as a precursor for generating small RNA(s), as reported in the human brain (13), and the alternate nonnative folds allow for apportioning the tRNA to this pathway instead of translation. tRNA-derived fragments play various cellular roles, including stress responses, RNA silencing, translational control, and apoptosis (1, 2, 13). In the case of a tRNAArg-like n-Tr20, it can also act as an arginine donor for protein arginylation even after fragmentation (41). Moreover, pre-tRNA leaders and trailers have been found to accumulate as well (42). In this regard, the proximal 30-nt 5′ leader of n-Tr20 is GU rich and is conserved completely in mammals and up to 75% in birds and reptiles (revealed by the basic local alignment search tool at National Center for Biotechnology Information). When cleaved by RNase P, this leader can potentially bind CA-rich sequences in the introns of messenger RNAs to influence alternative splicing (38) or in 3′ untranslated regions and long noncoding RNAs to affect the stability of targets RNAs (39). Thus, the equilibrium between the native and alternative folds of n-Tr20 may regulate cleavage by RNase P to modulate the amount of free 5′ leader available for binding to other RNAs.
Other questions warrant future study. First, what causes the buildup of n-Tr20 precursor in the postnatal mouse brain, and why does more accumulate in the cerebellum than in the cortex or hippocampus (7)? Is the native–nonnative equilibrium dependent on spatiotemporal gene expression and cellular cofactors? Second, while our studies were performed with in vitro transcripts, it would be informative to examine how other cellular events, such as cotranscriptional RNA folding and modification (8, 43, 44), affect the conformational sampling of tRNAs.
The severity of a tRNA mutation likely depends on the fractional population of mutant tRNA in its native vs. nonnative states, which is predicated on the innate propensity of the WT tRNA to embrace alternative conformations (40). While mutations that alter the stability of tRNA structures, either through modest local rearrangements or dimerization, have been documented (32, 33, 45–47), our concept of a mutation-driven large-scale restructuring that fosters alternative folds already infrequently sampled by the WT tRNA merits consideration both to better understand diseases arising from tRNA mutations [e.g., cancer, MELAS (3, 4)] and to design small molecules that steer tRNAs away from nonnative folds.
Materials and Methods
See SI Appendix for details.
Preparation of n-Tr20 Variants and n-Tr21.
Sequences of pre–n-Tr20+3′ and –n-Tr21 were PCR amplified from C57BL/6NJ genomic DNA to include a 32-nt 5′ leader and the entire 3′ trailer upstream of the polyT. These amplicons were cloned downstream of the T7 RNA polymerase promoter in pBT7 (48). PCR was also used to generate other precursor and mature n-Tr20 variants using pre–n-Tr20+3′ as template. PCR with gene-specific reverse primers was used to generate templates from all these clones for runoff in vitro transcription with T7 RNA polymerase. The tRNA substrates were labeled 1) uniformly with [α-32P]-GTP for multiple-turnover RNase P assays and 2) at the 5′ end with [γ-32P]-ATP for RNase P cleavage-site mapping and for single-turnover RNase P assays.
Partial Purification of RNase P from Mouse Cortices/Livers.
Cortices (∼1.8 g) from 4-wk-old B6J mice were lysed in extraction buffer (EB; 20 mM Tris⋅HCl, pH 8, 5 mM MgCl2, 14 mM β-mercaptoethanol, 0.2 mM PMSF) supplemented with 100 mM NaCl and loaded on a HiTrap DEAE-FF column (Cytiva). After elution with a linear 100–700 mM NaCl gradient, the 20 fractions were assayed for RNase P activity. The two reproducible activity peaks, P1 and P2, were pooled. P1 was further purified using a 10 to 35% (vol/vol) glycerol gradient and then concentrated approximately sevenfold using Ultra-4 centrifugal filters (3,000 nominal molecular weight limit; Amicon) to yield P1F (P1 final). Because further purification (including a glycerol gradient) led to loss of activity, P2 was concentrated ∼10-fold to yield P2F. Liver RNase P was purified using the same scheme up to the DEAE-FF elution step.
Mouse (Multiple-Turnover) and E. coli (Single-Turnover) RNase P Assays.
All assays were performed at 37 °C. Purified mouse RNase P fractions were assayed in EB with 50 nM pre-tRNA and a trace amount (∼1 nM) of uniformly radiolabeled pre-tRNA. For other multiple-turnover assays, the above reactions were supplemented with 140 mM KCl. For the single-turnover assays in which only 5′-radiolabeled (∼1-fM) pre-tRNA was used, cleavage by reconstituted recombinant E. coli RNase P (49) was performed at pH 5.5 where the rate is slower (38, 39). After electrophoresis, gels were scanned using a Typhoon phosphorimager (GE Healthcare), and band intensities were determined using ImageQuant (GE Healthcare). In multiple-turnover assays, linear curve fits (product vs. time) were obtained using Microsoft Excel to calculate the initial velocities. In single-turnover assays, the percentage of product formed at time t (Pt) was fit to Pt = P∞(1 − e−kt) using Kaleidagraph (Synergy Software).
Thermal Denaturation of Mature n-Tr20 and Its Mutant Derivatives.
Mature tRNAs (500 nM) in 20 mM sodium cacodylate (pH 7); 150 mM NaCl; and 0, 1, or 5 mM MgCl2 were analyzed in parallel using a Cary 3500 UV-visible double-beam, eight-cell Peltier spectrophotometer (Agilent). Abs260 (A) was monitored as the temperature (T) was increased from 20 to 95 °C at a rate of 0.5 °C/min, and Tm values were calculated from first-derivative plots (dA/dT vs. T).
Native PAGE.
Mature tRNA (5 pmol) was refolded in water (95 °C for 3 min, 37 °C for 10 min) before adding 20 mM Hepes (pH 7.5), 20 mM NaCl, and either water or MgCl2 at the specified concentration. After a 30-min incubation at 37 °C, 11.5% (vol/vol) glycerol and trace amounts of bromophenol blue and xylene cyanol (0.01%) were added. The samples were electrophoresed at 180 V and 4 °C in a 5% (wt/vol) polyacrylamide (19:1 acrylamide/bis-acrylamide) gel made in 89 mM Tris, 89 mM boric acid, and 2 mM MgCl2, which were also the components in the running buffer. Gels were stained with SYBR Gold (Invitrogen) in TBE buffer for 5 min and scanned in a Typhoon phosphorimager (GE Healthcare) using the Cy2 imaging mode.
NMR Experiments.
Diafiltration was used to buffer exchange mature n-Tr20 variants into 15 mM sodium phosphate, pH 6.8; 25 mM NaCl; 10% (vol/vol) D2O; and 0, 0.5, 1, 2, 5, or 10 mM MgCl2. Mg2+-free samples contained 0.1 mM EDTA in addition. Imino 1H NMR spectra were acquired at 25 °C using ∼0.4 mM tRNA and a 14.1-T Bruker Avance III spectrometer equipped with an HCPN (hydrogen/carbon/phosphorus/nitrogen) cryogenic probe. All data were processed and analyzed using NMRpipe (50).
Supplementary Material
Acknowledgments
We thank Prof. Anita Hopper (The Ohio State University [OSU]) for feedback on the manuscript, one of the reviewers (Prof. William Scott) for valuable input, and Dr. Besik Kankia (OSU) for generously sharing the Cary 3500 spectrophotometer used for the thermal denaturation experiments in this study. We are grateful for funding from NIH Grants NS096600 (to S.L.A. and V.G.), GM-120582 (to V.G.), and GM-132899 (to H.M.A.-H.), United States–Israel Binational Science Foundation Grant 2015157 (to V.G.), and the Behrman Research Fund (to V.G.). Research reported in this publication was also supported by Office of Director, NIH Award S10OD023582. S.L.A. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Reviewers: S.A., Yale University; and W.S., University of California, Santa Cruz.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119529119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Avcilar-Kucukgoze I., Kashina A., Hijacking tRNAs from translation: Regulatory functions of tRNAs in mammalian cell physiology. Front. Mol. Biosci. 7, 610617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su Z., Wilson B., Kumar P., Dutta A., Noncanonical roles of tRNAs: tRNA fragments and beyond. Annu. Rev. Genet. 54, 47–69 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abbott J. A., Francklyn C. S., Robey-Bond S. M., Transfer RNA and human disease. Front. Genet. 5, 158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirchner S., Ignatova Z., Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 16, 98–112 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Schaffer A. E., Pinkard O., Coller J. M., tRNA metabolism and neurodevelopmental disorders. Annu. Rev. Genomics Hum. Genet. 20, 359–387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodarzi H., et al. , Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 165, 1416–1427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishimura R., et al. , RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orellana E. A., et al. , METTL1-mediated m7G modification of Arg-TCT tRNA drives oncogenic transformation. Mol. Cell 81, 3323–3338.e14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavon-Eternod M., et al. , tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 37, 7268–7280 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gingold H., et al. , A dual program for translation regulation in cellular proliferation and differentiation. Cell 158, 1281–1292 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Kutter C., et al. , Pol III binding in six mammals shows conservation among amino acid isotypes despite divergence among tRNA genes. Nat. Genet. 43, 948–955 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapur M., et al. , Expression of the neuronal tRNA n-Tr20 regulates synaptic transmission and seizure susceptibility. Neuron 108, 193–208.e9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torres A. G., Reina O., Stephan-Otto Attolini C., Ribas de Pouplana L., Differential expression of human tRNA genes drives the abundance of tRNA-derived fragments. Proc. Natl. Acad. Sci. U.S.A. 116, 8451–8456 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terrey M., et al. , GTPBP1 resolves paused ribosomes to maintain neuronal homeostasis. eLife 9, e62731 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lant J. T., Berg M. D., Heinemann I. U., Brandl C. J., O’Donoghue P., Pathways to disease from natural variations in human cytoplasmic tRNAs. J. Biol. Chem. 294, 5294–5308 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levinger L., et al. , Sequence and structure requirements for Drosophila tRNA 5′- and 3′-end processing. J. Biol. Chem. 270, 18903–18909 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Maraia R. J., Lamichhane T. N., 3′ processing of eukaryotic precursor tRNAs. Wiley Interdiscip. Rev. RNA 2, 362–375 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altman S., A view of RNase P. Mol. Biosyst. 3, 604–607 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Esakova O., Krasilnikov A. S., Of proteins and RNA: The RNase P/MRP family. RNA 16, 1725–1747 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarrous N., Roles of RNase P and its subunits. Trends Genet. 33, 594–603 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Lai L. B., Vioque A., Kirsebom L. A., Gopalan V., Unexpected diversity of RNase P, an ancient tRNA processing enzyme: Challenges and prospects. FEBS Lett. 584, 287–296 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phan H. D., Lai L. B., Zahurancik W. J., Gopalan V., The many faces of RNA-based RNase P, an RNA-world relic. Trends Biochem. Sci. 46, 976–991 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan P., et al. , Structural insight into precursor tRNA processing by yeast ribonuclease P. Science 362, eaat6678 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Wu J., et al. , Cryo-EM Structure of the human ribonuclease P holoenzyme. Cell 175, 1393–1404.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Marck C., Grosjean H., tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8, 1189–1232 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kufel J., Tollervey D., 3′-processing of yeast tRNATrp precedes 5′-processing. RNA 9, 202–208 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindahl T., Adams A., Fresco J. R., Renaturation of transfer ribonucleic acids through site binding of magnesium. Proc. Natl. Acad. Sci. U.S.A. 55, 941–948 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strulson C. A., Boyer J. A., Whitman E. E., Bevilacqua P. C., Molecular crowders and cosolutes promote folding cooperativity of RNA under physiological ionic conditions. RNA 20, 331–347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi H., Moore P. B., The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: A classic structure revisited. RNA 6, 1091–1105 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serebrov V., Vassilenko K., Kholod N., Gross H. J., Kisselev L., Mg2+ binding and structural stability of mature and in vitro synthesized unmodified Escherichia coli tRNAPhe. Nucleic Acids Res. 26, 2723–2728 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brion P., Westhof E., Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 26, 113–137 (1997). [DOI] [PubMed] [Google Scholar]
- 32.Hao R., Yao Y.-N., Zheng Y.-G., Xu M.-G., Wang E.-D., Reduction of mitochondrial tRNALeu(UUR) aminoacylation by some MELAS-associated mutations. FEBS Lett. 578, 135–139 (2004). [DOI] [PubMed] [Google Scholar]
- 33.Wang Y., et al. , A natural non-Watson-Crick base pair in human mitochondrial tRNAThr causes structural and functional susceptibility to local mutations. Nucleic Acids Res. 46, 4662–4676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodson S. A., Koculi E., Analysis of RNA folding by native polyacrylamide gel electrophoresis. Methods Enzymol. 469, 189–208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marušič M., Schlagnitweit J., Petzold K., RNA dynamics by NMR spectroscopy. ChemBioChem 20, 2685–2710 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yue D., Kintanar A., Horowitz J., Nucleoside modifications stabilize Mg2+ binding in Escherichia coli tRNA(Val): An imino proton NMR investigation. Biochemistry 33, 8905–8911 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Ferré-D’Amaré A. R., The tRNA elbow in structure, eecognition and evolution. Life (Basel) 6, 3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsieh J., Fierke C. A., Conformational change in the Bacillus subtilis RNase P holoenzyme--pre-tRNA complex enhances substrate affinity and limits cleavage rate. RNA 15, 1565–1577 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun L., Campbell F. E., Zahler N. H., Harris M. E., Evidence that substrate-specific effects of C5 protein lead to uniformity in binding and catalysis by RNase P. EMBO J. 25, 3998–4007 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madore E., Florentz C., Giegé R., Lapointe J., Magnesium-dependent alternative foldings of active and inactive Escherichia coli tRNA(Glu) revealed by chemical probing. Nucleic Acids Res. 27, 3583–3588 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Avcilar-Kucukgoze I., et al. , tRNA(Arg)-derived fragments can serve as arginine donors for protein arginylation. Cell Chem. Biol. 27, 839–849.e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olvedy M., et al. , A comprehensive repertoire of tRNA-derived fragments in prostate cancer. Oncotarget 7, 24766–24777 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jarrous N., Mani D., Ramanathan A., Coordination of transcription and processing of tRNA. FEBS J., 10.1111/febs.15904 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Yu A. M., et al. , Computationally reconstructing cotranscriptional RNA folding from experimental data reveals rearrangement of non-native folding intermediates. Mol. Cell 81, 870–883.e10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kelley S. O., Steinberg S. V., Schimmel P., Fragile T-stem in disease-associated human mitochondrial tRNA sensitizes structure to local and distant mutations. J. Biol. Chem. 276, 10607–10611 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Wittenhagen L. M., Kelley S. O., Dimerization of a pathogenic human mitochondrial tRNA. Nat. Struct. Biol. 9, 586–590 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Jones C. N., Jones C. I., Graham W. D., Agris P. F., Spremulli L. L., A disease-causing point mutation in human mitochondrial tRNAMet results in tRNA misfolding leading to defects in translational initiation and elongation. J. Biol. Chem. 283, 34445–34456 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsai H. Y., Lai L. B., Gopalan V., A modified pBluescript-based vector for facile cloning and transcription of RNAs. Anal. Biochem. 303, 214–217 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Gopalan V., Baxevanis A. D., Landsman D., Altman S., Analysis of the functional role of conserved residues in the protein subunit of ribonuclease P from Escherichia coli. J. Mol. Biol. 267, 818–829 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Delaglio F., Grzesiek S., Vuister G., Zhu G., Pfeifer J., Bax A., NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, (1995). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.