Significance
While most small, regulatory RNAs are thought to be “noncoding,” a few have been found to also encode a small protein. Here we describe a 164-nucleotide RNA that encodes a 28-amino acid, amphipathic protein, which interacts with aerobic glycerol-3-phosphate dehydrogenase and increases dehydrogenase activity but also base pairs with two mRNAs to reduce expression. The coding and base-pairing sequences overlap, and the two regulatory functions compete.
Keywords: sRNA, catabolite repression, glycerol dehdrogenase, Hfq, ProQ
Abstract
Bacteria have evolved small RNAs (sRNAs) to regulate numerous biological processes and stress responses. While sRNAs generally are considered to be “noncoding,” a few have been found to also encode a small protein. Here we describe one such dual-function RNA that modulates carbon utilization in Escherichia coli. The 164-nucleotide RNA was previously shown to encode a 28-amino acid protein (denoted AzuC). We discovered the membrane-associated AzuC protein interacts with GlpD, the aerobic glycerol-3-phosphate dehydrogenase, and increases dehydrogenase activity. Overexpression of the RNA encoding AzuC results in a growth defect in glycerol and galactose medium. The defect in galactose medium was still observed for a stop codon mutant derivative, suggesting a second role for the RNA. Consistent with this observation, we found that cadA and galE are repressed by base pairing with the RNA (denoted AzuR). Interestingly, AzuC translation interferes with the observed repression of cadA and galE by the RNA and base pairing interferes with AzuC translation, demonstrating that the translation and base-pairing functions compete.
Bacteria are exposed to rapidly changing environmental conditions, such as variations in carbon availability and pH. To survive these fluctuating conditions, bacterial cells have fast, flexible, and energy-efficient mechanisms to regulate protein levels and activity at all levels. The cAMP receptor protein (CRP), a sequence-specific DNA binding protein, is a key regulator of transcription in response to changes in carbon source availability in Escherichia coli (1). Several small RNAs (sRNAs), which modulate the stability or translation of mRNAs through short base-pairing interactions and are major posttranscriptional regulators in bacteria (2), also impact carbon metabolism in E. coli. These include GlmZ, ChiX, SgrS, and the CRP-repressed sRNAs Spot 42 and CyaR (3, 4). Base-pairing sRNAs require RNA chaperones, such as Hfq and ProQ, for their stability and optimal pairing with their target mRNAs (5–7).
Base-pairing sRNAs generally are thought not to encode proteins and thus are often referred to as noncoding RNAs. However, a few sRNAs have been shown to be translated to produce small proteins and thus are denoted “dual-function RNAs” (8). Small proteins of fewer than 50 amino acids have long been overlooked due to many challenges related to their annotation and biochemical detection. Studies of a few have revealed that, by forming complexes with larger proteins, small proteins modulate diverse cellular functions, ranging from transporter and enzyme activities to regulatory networks and cell morphogenesis and division (9, 10).
To date, the only dual-function RNA characterized in E. coli is SgrS (11). The SgrS RNA was first found to protect cells against elevated levels of glucose phosphate by base pairing with and regulating the stability and translation of mRNAs encoding proteins involved in glucose transport and catabolism (12). The RNA was subsequently shown to encode a 43-amino acid protein, SgrT, which interacts with the glucose importer PtsG to block glucose transport and promote utilization of nonpreferred carbon sources to maintain growth during glucose-phosphate stress (11, 13). Thus, both the sRNA and its encoded small protein act together to repress glucose import to relieve glucose phosphate stress.
The 164-nt RNA initially denoted IS092 or IsrB (here denoted AzuCR) was identified in a bioinformatic search to find sRNA genes in E. coli (14), but not characterized. Later, this RNA was shown to encode a 28-amino acid sORF (15) (Fig. 1A). Synthesis of the small protein was documented by the detection of a sequential peptide affinity (SPA)-tagged derivative (15) and is supported by ribosome binding to the RNA (16) (SI Appendix, Fig. S1A). While the protein, denoted AzuC, is only conserved in a limited number of enteric bacteria (SI Appendix, Fig. S1B), expression of AzuC-SPA is highly regulated (17). The levels of the tagged small protein are elevated for growth in glucose compared to glycerol due to CRP-mediated repression in the absence of glucose. AzuC-SPA levels also are reduced under anaerobic conditions but induced upon exposure to low pH, high temperature, and hydrogen peroxide.
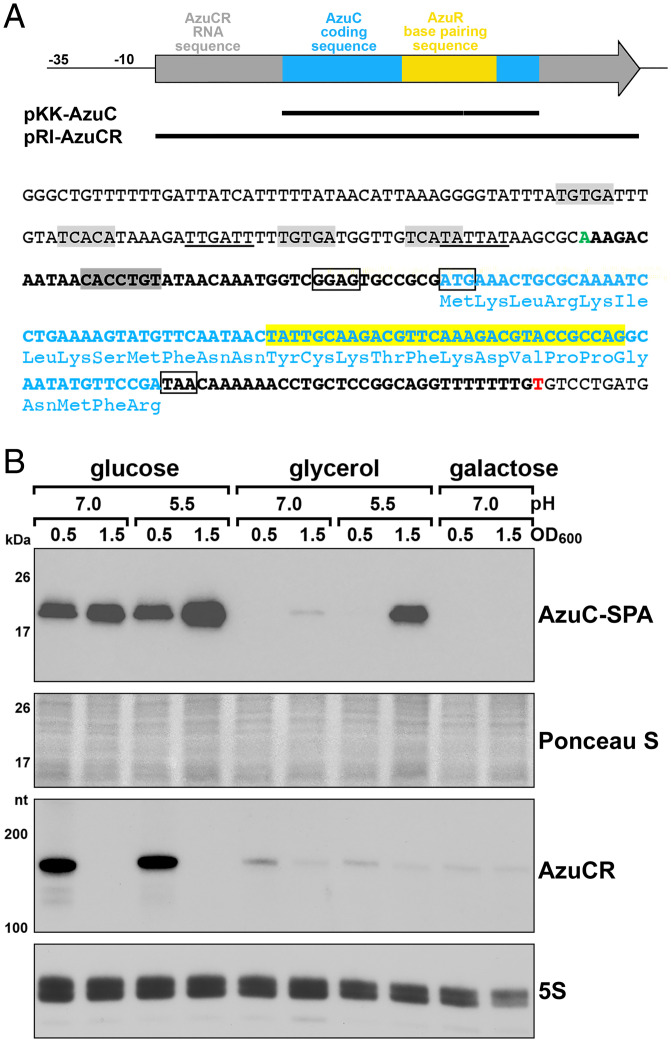
Fig. 1.
Discordance between AzuC protein and AzuCR RNA levels. (A) Diagram of the AzuCR RNA, overexpression constructs generated, and sequence of the promoter and coding region. Light blue boxes and font denote AzuC coding sequence, yellow box and highlighted text denote region of AzuR base pairing with target mRNAs, and gray arrow and bold black font denote the rest of the AzuCR RNA sequence. The start of the AzuCR transcript is indicated in green font (position 1988001 of the E. coli K-12 genome) and the 3′ end of the transcript is in red font. The ribosome binding site and the start and stop codons of the AzuC ORF are indicated by black boxes. Potential σ70-10 and -35 sequences are underlined, the predicted CRP binding sites are highlighted in light gray (17), and the region targeted by the FnrS sRNA is highlighted in dark gray. (B) Immunoblot blot analysis of AzuC-SPA levels (Top) in a azuC-SPA::kan strain (GSO351) and Northern blot analysis of AzuCR RNA levels (Bottom) in MG1655. Cultures were grown in M63 medium supplemented with glucose, glycerol, or galactose at pH 7.0 or glucose or glycerol at pH 5.5. Samples were taken at OD600 ∼0.5 and 1.5. α-FLAG antibody was used to detect the SPA tag. The membrane was stained with Ponceau S stain to control for loading. The AzuCR RNA and 5S RNA were detected by oligonucleotide probes specific to each of these transcripts.
Here we show that AzuC is associated with the membrane and binds GlpD, the aerobic glycerol-3-phosphate dehydrogenase enzyme required for glycerol catabolism. Dehydrogenase activity is increased upon AzuC overexpression. Additionally, we document that the transcript acts as a regulatory sRNA, denoted AzuR, repressing expression of cadA, a lysine decarboxylase involved in maintaining pH homeostasis, and galE, encoding UDP-glucose 4-epimerase, through direct base pairing. Thus, the AzuCR transcript has mRNA and sRNA activities impacting the utilization of different carbon sources. The protein coding and base-pairing sequences overlap, and we found that there is competition between the two activities. Intriguingly, while the transcript represses expression of target mRNAs as an sRNA, translation of AzuC itself is repressed by the FnrS sRNA.
Results
AzuC Protein and mRNA Levels Are Discordant under Some Growth Conditions.
Previous analysis showed that the levels of chromosomally encoded AzuC-SPA were higher in cells grown in minimal medium with glucose compared to glycerol, as well as in pH 5.5 compared to pH 7.5, and decreased under anerobic conditions (17). The decreased levels in minimal glycerol medium and part of the pH-induction were attributed to CRP-mediated repression of azuC mRNA transcription, given the elevated levels of AzuC-SPA observed in a Δcrp mutant (17). To further evaluate the conditions under which the levels of AzuC-SPA and the transcript (henceforth denoted AzuCR) are highest, strains were cultured in M63 media supplemented with glucose or glycerol at pH 7.0 or 5.5, or M63 galactose at pH 7.0 (the strain was unable to grow in M63 galactose at pH 5.5). Cells were collected in exponential (OD600 ∼0.5) and stationary (OD600 ∼1.5) phase (Fig. 1B). As observed previously, AzuC-SPA levels were significantly higher in glucose compared to glycerol and galactose. A notable exception was the high levels of AzuC-SPA observed in cells grown to stationary phase in glycerol pH 5.5. As expected for a CRP-repressed transcript, AzuCR RNA levels were low for all conditions except for cells growing exponentially in glucose. Though it is possible the SPA tag has an unnatural stabilizing effect on AzuC, the discordance between AzuC-SPA protein levels and AzuCR RNA levels in glycerol pH 5.5 raised the possibility that translation, RNA stability, and protein stability is regulated and that the protein and RNA may have different roles. However, before exploring the potential function of the RNA, we wanted to learn more about the function of the 28-amino acid protein.
AzuC Protein Is Localized to the Membrane.
Information about the subcellular localization of proteins can give clues about possible interacting partners and functions in the cell. Secondary structural predictions suggested that AzuC has the potential to fold into an amphipathic helix (Fig. 2A), indicating the protein might associate with the membrane. To test this, AzuC-SPA cells grown in M63 glucose to OD600 ∼1.0 were lysed, and cell extracts were homogenized and fractionated into soluble, inner membrane, and outer membrane fractions (18, 19). Consistent with the secondary structure prediction, immunoblot analysis showed that AzuC-SPA was enriched in the inner membrane fraction, while the OmpA control protein was enriched in the outer membrane fraction (Fig. 2B). Similar fractionation of untagged AzuC, expressed from a plasmid and detected by α-AzuC antiserum, also showed enrichment in the membrane fraction (SI Appendix, Fig. S2A). The localization of AzuC to the membrane was further confirmed by fluorescence microscopy imaging of chromosomally expressed AzuC C-terminally tagged with the green fluorescent protein (GFP) (Fig. 2C). While WT AzuC-GFP showed clear membrane localization, this was not observed for a mutant in which hydrophobic residues were replaced by charged residues (I6L7 to E6E7). Taken together, these data support the hypothesis that AzuC is associated with the membrane as an amphipathic protein.
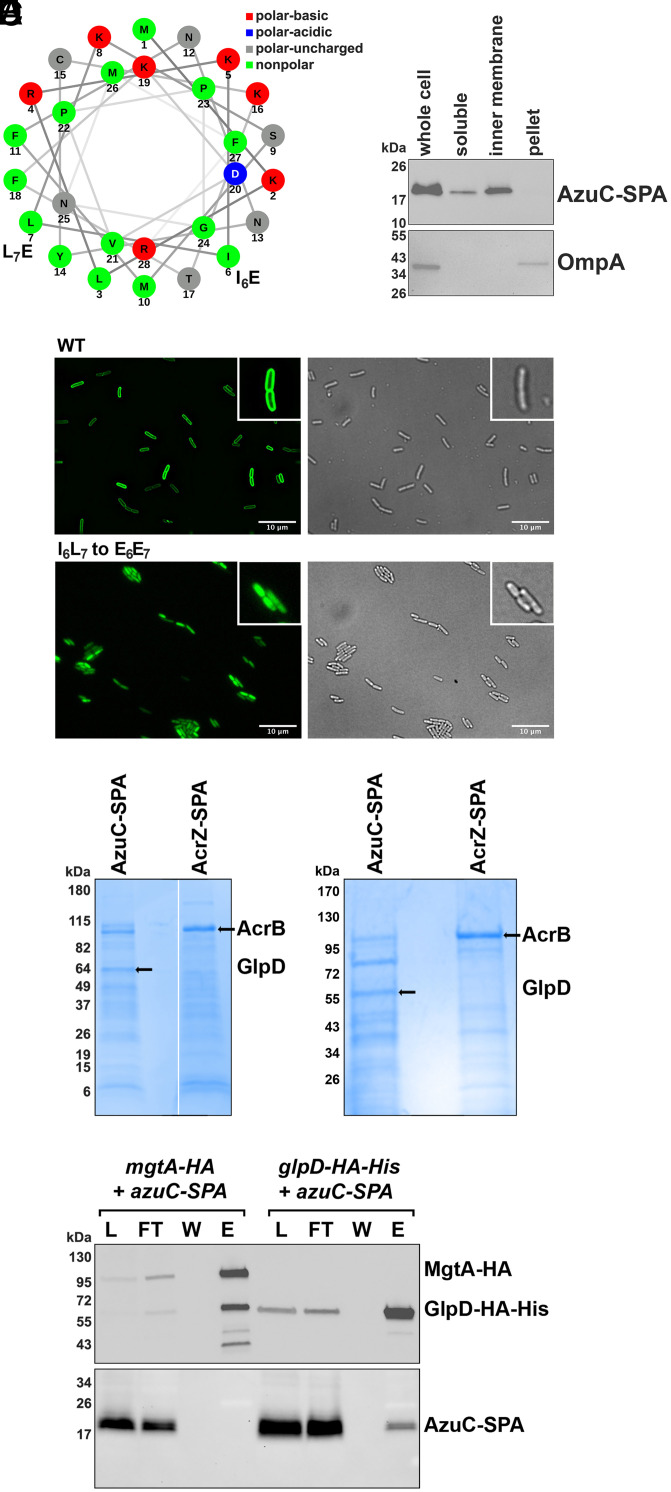
Fig. 2.
Membrane-associated AzuC protein copurifies with GlpD. (A) Helical wheel projection generated using NetWheels (37) showing amphipathic nature of AzuC. Mutations introduced in C are indicated. (B) Fractionation of AzuC-SPA strain. A culture expressing AzuC-SPA (GSO351) was grown in M63 glucose medium to OD600 ∼0.5, and cells were fractionated into a soluble, inner membrane, and pellet fractions, which were compared to the whole-cell lysate. The Upper panel shows AzuC-SPA as detected with α-FLAG antibody. The Lower panel shows the outer membrane OmpA control detected with α-OmpA antibody. (C) Microscopy of AzuC-GFP. AzuC-GFP (GSO1008) and AzuCI6L7 to E6E7-GFP (GSO1009) cells were grown in M63 glucose medium to OD600 ∼0.5 to observe membrane localization by fluorescent microscopy. Left panels are fluorescent images showing GFP labeled AzuC, and Right panels are the corresponding brightfield images. Insets provide a 2.3× enlargement of a few cells. (D) GlpD copurifies with AzuC-SPA. Cells expressing AzuC-SPA (GSO351) or AcrZ-SPA (GSO350) from the chromosome were grown in M63 glucose medium to OD600 ∼1.0 or in LB to OD600 ∼0.6, respectively. The cell lysates were split and passed over calmodulin beads. Eluants from each column were subjected to SDS/PAGE followed by Coomassie blue staining. The bands enriched in the eluant from the calmodulin beads and indicated by the arrows were excised from the gel, and proteins were identified by mass spectrometry. (E) AzuC-SPA copurifies with GlpD-HA-His6. Cells expressing either AzuC-SPA (GSO351) or GlpD-HA-His6 (GSO1011) from the chromosome were grown in M63 glucose or M63 glycerol media, respectively, to OD600 ∼1.0 and mixed in a 1:1 ratio. As a control, cells expressing MgtA-HA (GSO785) grown in N medium supplemented without added MgSO4 to OD600 ∼0.5, were mixed with the AzuC-SPA (GSO351) cells in the same ratio. The mixed cells were homogenized, cell lysates (L) were applied to α-HA beads and the flow-through (FT) samples were collected. The beads were washed (W), after which the bound proteins were eluted (E) and examined on immunoblots using either α-HA antibodies to detect MgtA-HA or GlpD-HA-His6 (Upper) or α-FLAG antibodies to detect AzuC-SPA (Lower).
AzuC Protein Copurifies with the Aerobic Glycerol 3-Phosphate Dehydrogenase.
To further investigate the role of AzuC in the cell, we carried out copurification assays to identify interacting proteins. Cells expressing chromosomally encoded AzuC-SPA or previously characterized AcrZ-SPA (20) were grown in M63 glucose medium. Cell lysates prepared from exponentially growing cells were applied to calmodulin beads, and the eluants from each column were separated by SDS/PAGE (Fig. 2D). Unique bands from each of the elutions were subjected to mass spectrometric analysis. AcrB, a known AcrZ interactor (20), was identified in the most prominent band in the AcrZ sample. For AzuC-SPA, the only protein identified in two independent experiments detected in the prominent band of ∼60 kDa, was the aerobic glycerol 3-phosphate dehydrogenase (GlpD). This enzyme catalyzes the oxidation of glycerol 3-phosphate and has been reported to be a peripheral membrane protein associated with the membrane through an amphipathic helix (21).
We tested the interaction between AzuC and GlpD by assessing reciprocal copurification of AzuC-SPA with GlpD-HA-His6. Cells with chromosomally encoded AzuC-SPA, grown to exponential phase in M63 glucose medium, were mixed with cells with chromosomally encoded GlpD-HA-His6 grown to exponential phase in M63 glycerol medium, a condition where GlpD is known to be expressed. The mixed cells were lysed and incubated with dodecyl β-D-maltoside (DDM) to facilitate mixing of the membrane fractions. The mixed lysate was then applied to α-HA magnetic beads, washed, and eluted (Fig. 2E). As controls, similar purifications were carried out by mixing the AzuC-SPA cells with cells lacking tagged proteins grown in M63 glycerol medium (SI Appendix, Fig. S2B) or cells expressing chromosomally encoded MgtA-HA (Fig. 2E). Consistent with the first purification, AzuC-SPA copurified with GlpD-HA-His6 and not with MgtA-HA. Similar copurification was observed when AzuC-SPA and GlpD-HA-His6 were expressed in the same cells grown in M63 glycerol pH 5.5 (SI Appendix, Fig. S2 C and D). All these observations support the conclusion that GlpD interacts with AzuC.
AzuC Overexpression Increases Dehydrogenase Activity and Cell Length.
As has been found for other small proteins (20, 22, 23), binding of AzuC to GlpD could potentially impact the stability, activity, and localization of the enzyme. To begin to distinguish among these possibilities, we first examined the effects of AzuC overexpression on the levels of chromosomally encoded GlpD-HA-His6. Cells were transformed with pKK-AzuC or pKK-AzuCL3STOP, in which the WT or mutant (harboring a stop codon mutation of the third codon) azuC ORF was cloned downstream of the heterologous Ptac promoter and ribosome binding site on the pKK177-3 (pKK) plasmid (Fig. 1A and SI Appendix, Fig. S3A). Transformed cells were grown in M63 glucose medium to OD600 ∼1.0 and then transitioned to glycerol pH 5.5 for 3 h given that chromosomally expressed AzuC-SPA levels are elevated under these conditions (Fig. 1B). The GlpD-HA-His6 protein levels were similar for all three strains grown under these conditions as well as in WT and ΔazuCR cells (SI Appendix, Fig. S3B).
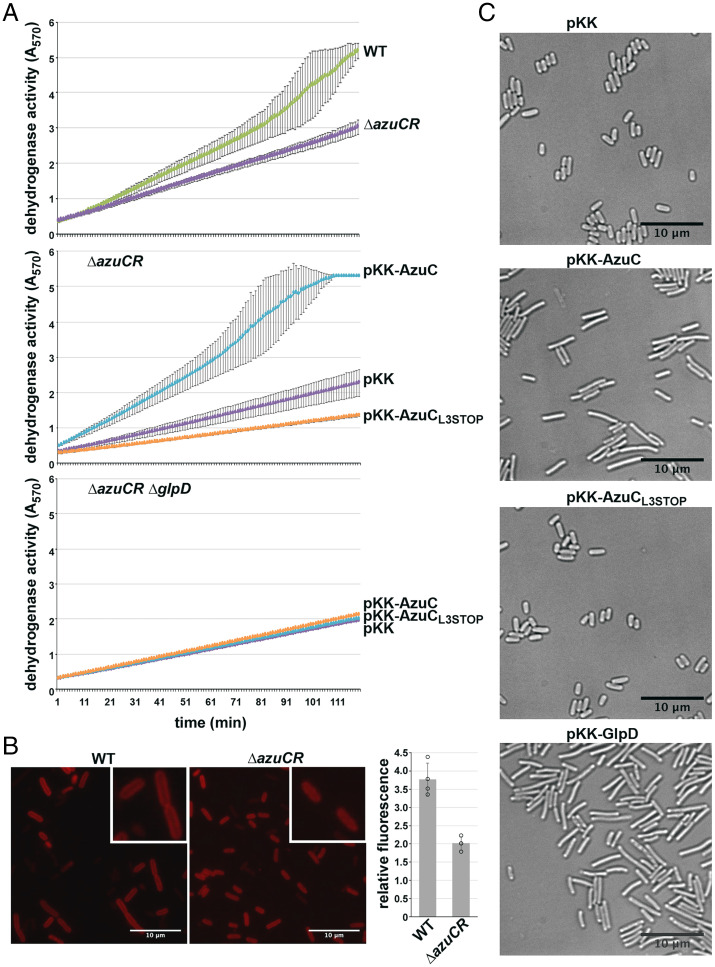
To test whether AzuC affects GlpD activity, we employed a dehydrogenase activity assay in which glycerol-3-phosphate oxidation to dihydroxyacetone phosphate (DHAP) is coupled to the reduction of yellow 2-(4,5-dimethyl-2-thiazolyl)-3,5-diphenyl-2H-tetrazolium bromide (MTT) to blue formazan, which is detected at OD570 (24) (Fig. 3A). Based on this assay, dehydrogenase activity was found to be almost twofold lower in the absence of AzuC when extracts were made from a WT or a ΔazuCR strain grown in M63 glucose medium and shifted to glycerol pH 5.5 for 3 h (Fig. 3 A, Top). In contrast, overexpression of WT AzuC, but not AzuCL3STOP, led to higher dehydrogenase activity in the extracts (Fig. 3 A, Middle). A ΔazuCR ΔglpD double mutant did not show the increase in activity upon AzuC overexpression, showing the increase in dehydrogenase activity was due to GlpD (Fig. 3 A, Bottom). These results indicate that AzuCR affects the levels of GlpD dehydrogenase activity. Interestingly, we observed a decrease in dehydrogenase activity with the pKK-AzuCL3STOP plasmid (Fig. 3 A, Middle). We hypothesize this may be due to regulatory activity of the RNA (see Fig. 5).
Fig. 3.
AzuC increases GlpD activity and membrane association. (A) Effect of AzuC overexpression on GlpD activity. WT or ΔazuCR::kan (GSO193) (Top), or ΔazuCR::kan transformed with pKK, pKK-AzuC, or pKK-AzuCL3STOP (Middle), or ΔazuCR ΔglpD::kan (GSO1015) transformed with the same plasmids (Bottom) were grown in M63 glucose medium to OD600 ∼1.0. Cells were then washed and resuspended in M63 glycerol medium, pH 5.5 for 3 h. MTT was added, and A570, reflecting the reduction of MTT to formazan, which is coupled to the oxidation of glycerol-3-phosphate to DHAP, was measured. Lines correspond to the average of three biological replicates, and error bars represent 1 SD. (B) Effect of ΔazuCR on GlpD-YFP membrane localization. WT (GSO1116) and ΔazuCR (GSO1117) cells expressing GlpD-YFP (38) grown in M63 glycerol pH 5.5 medium to OD600 ∼1.5 were examined by fluorescence microscopy. MG1655 cells grown in the same medium and mixed 1:1 with cells from either of the two GlpD-YFP strains prior to microscopy served as a control. Example images and quantitation of the membrane signal for 40 cells are shown (circles represent average of 10 cells for 4 different samples). Insets provide a 2.1× enlargement of a few cells. (C) Effect of AzuC overexpression on E. coli cell morphology. ΔazuCR::kan (GSO193) transformed with pKK, pKK-AzuC, pKK-AzuCL3STOP, or pKK-GlpD were grown in M63 glucose medium to OD600 ∼1.0. Cells were washed and resuspended in M63 glycerol medium, pH 5.5 for 3 h prior to microscopy. Example images are shown.
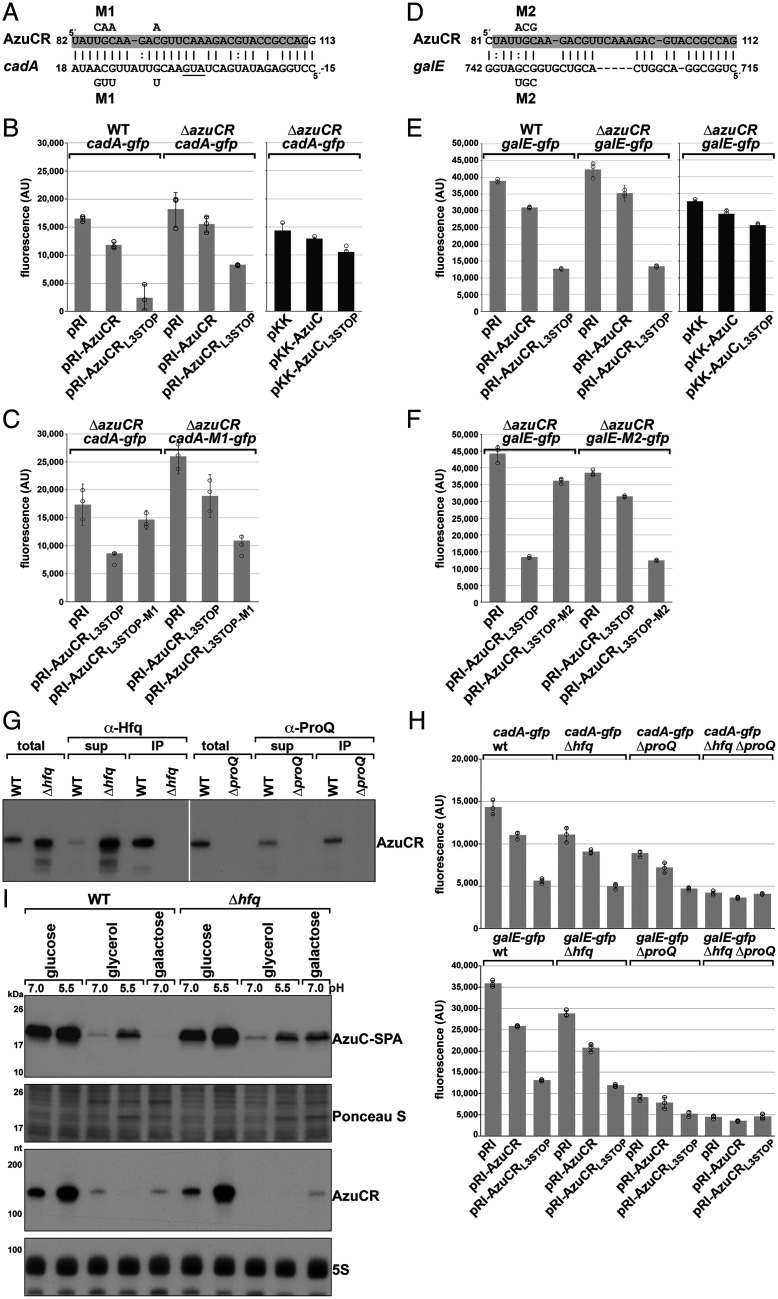
Fig. 5.
AzuCR represses cadA and galE expression. (A) AzuCR-cadA base pairing predicted by TargetRNA2 (27). AzuCR coordinates are relative to the +1 of the transcript while cadA coordinates are relative to the first nucleotide of the start codon. (B) Effect of AzuCR and AzuCRL3STOP overexpression on a cadA-gfp fusion in WT and ΔazuCR backgrounds. WT and ΔazuCR::kan (GSO193) cells were cotransformed with a reporter plasmid expressing a cadA-gfp translational fusion and either the empty pRI vector, AzuCR, or AzuCRL3STOP. (C) Test of AzuCR-cadA base pairing. ΔazuCR::kan (GSO193) cells were cotransformed with the WT cadA-gfp translational fusion reporter plasmid or a M1 derivative along with the empty pRI vector, AzuCRL3STOP, or mutant AzuCRL3STOP-M1. The mutations in the cadA-gfp translational fusion and AzuCRL3STOP are indicated (A). (D) AzuCR-galE base pairing predicted by IntaRNA (28). AzuCR coordinates are relative to the +1 of the transcript while galE coordinates are relative to the first nucleotide of the start codon. (E) Effect of AzuCR and AzuCRL3STOP overexpression on a galE-gfp fusion in WT and ΔazuCR backgrounds. WT and ΔazuCR::kan (GSO193) cells were cotransformed with a reporter plasmid expressing a galE-gfp translational fusion and either the empty pRI vector, AzuCR, or AzuCRL3STOP. (F) Test of AzuCR-galE base pairing. ΔazuCR::kan (GSO193) cells were cotransformed with the WT galE-gfp translational fusion reporter plasmid or a M2 derivative along with the empty pRI vector, AzuCRL3STOP, or mutant AzuCRL3STOP-M2. The mutations in the galE-gfp translational fusion and AzuCRL3STOP are indicated in D. For (B, C, E, and F), cells were grown in LB for 3 h before measuring the fluorescence corresponding to GFP expression. Bars correspond to the average of three biological replicates with circles corresponding to individual data points and error bars representing 1 SD. (G) AzuCR coimmunoprecipitation with Hfq and ProQ. Extracts from MG1655 (GSO982), Δhfq::cat-sacB (GSO954), and ΔproQ::kan (GSO956) cells grown in M63 glucose were incubated with α-Hfq or α-ProQ antiserum. Total and RNA chaperone-bound RNA was extracted and subjected to Northern analysis using an oligonucleotide probe specific for AzuCR. (H) Effect of Δhfq::kan (GSO955), ΔproQ::kan (GSO956) and Δhfq ΔproQ::kan (GSO959) double mutant on cadA-gfp and galE-gfp expression upon overexpression of AzuCR or AzuCRL3STOP. Bars correspond to the average of three biological replicates with circles corresponding to individual data points and error bars representing 1 SD. (I) Immunoblot blot analysis of AzuC-SPA levels (Top) and Northern blot analysis of AzuCR RNA levels (Bottom) in WT (GSO351) and Δhfq (GSO1007) cells grown with different carbon sources. The WT or Δhfq derivatives of the azuC-SPA::kan (GSO351) strain (Top) or unmarked (MG1655) strain (Bottom) were grown and analyzed as in Fig. 1B.
The dehydrogenase assay also was carried out with strains shifted to M63 glycerol pH 7.0, where we observed similar, although reduced, effects of ΔazuCR and AzuC overexpression on activity (SI Appendix, Fig. S3C). Finally, we assayed the mutant AzuC-I6L7 to E6E7 derivative, which is unable to localize to the membrane as well as the 26-amino acid, amphipathic helix SpoVM protein from Bacillus subtilis (23), but neither of these proteins increased dehydrogenase activity (SI Appendix, Fig. S3D).
Given that AzuC is an amphipathic helix and GlpD is a peripheral membrane protein (21) whose activity is reported to be increased by amphipaths (25), we hypothesized AzuC might function by increasing GlpD association with the inner membrane when cells are grown in M63 glycerol at low pH. Consistent with this hypothesis, we observed higher levels of GlpD, C-terminally tagged with yellow fluorescent protein (YFP), at the membrane in WT compared to ΔazuCR cells (Fig. 3B).
The substrate for GlpD, glycerol-3-phosphate, is a precursor for phospholipid biosynthesis. To test whether AzuC biases the flow of glycerol-3-phosphate toward glycerol metabolism rather than phospholipid biosynthesis, thereby impacting cell morphology, we conducted live-cell phase-contrast microscopy of cells carrying pKK, pKK-AzuC, or pKK-AzuCL3STOP (Fig. 3C). We found AzuC overexpressing cells, but not those carrying the vector or pKK-AzuCL3STOP, had an elongated morphology. The elongated morphology was similar to the morphology observed for cells upon GlpD overexpression as well as the morphology reported for cells lacking phosphatidylethanolamine (26).
AzuCR Overexpression Reduces Growth in Glycerol and Galactose.
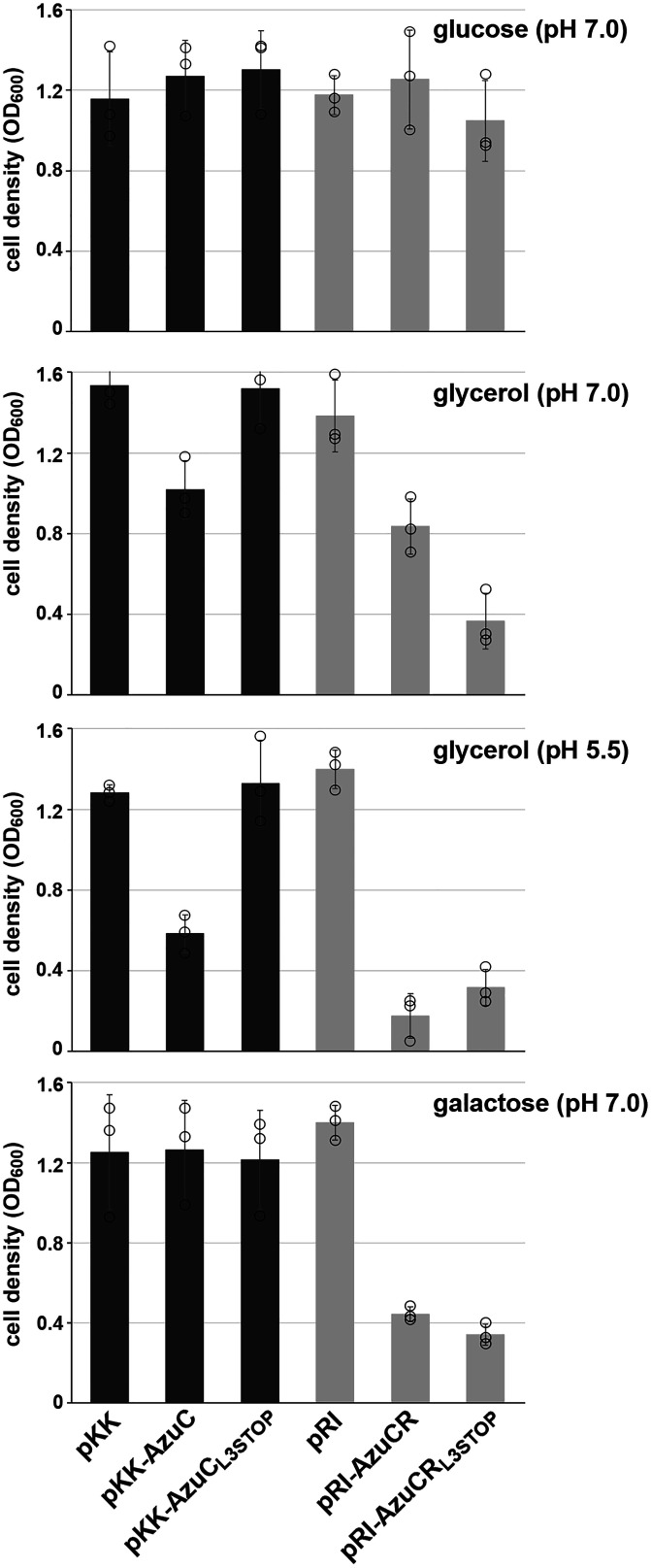
Given the AzuC effect on GlpD together with the different AzuC levels for cells grown in the presence of different carbon sources, we examined the consequences of AzuC overexpression for growth in glucose and glycerol at pH 7.0 and 5.5 and galactose at pH 7.0 (Fig. 4 and SI Appendix, Fig. S4A). For pKK-AzuC cells, but not the pKK vector control and pKK-AzuCL3STOP cells, we observed a significantly slower initial growth rate in M63 glycerol pH 5.5 and a partial defect in M63 glycerol pH 7.0, consistent with the larger effect of AzuC on GlpD activity in M63 glycerol pH 5.5 compared to pH 7.0. A similar phenotype was observed for overexpression of AzuC-SPA, indicating that the tagged derivative of AzuC is functional (SI Appendix, Fig. S4B). Growth in minimal medium with either glucose or galactose was not significantly changed by the pKK-AzuC plasmid.
Fig. 4.
AzuC and AzuCR overexpression lead to different growth phenotypes. OD600 of the ΔazuCR::kan strain (GSO193) transformed with pKK, pKK-AzuC, pKK-AzuCL3STOP, pRI, pRI-AzuCR, or pRI-AzuCRL3STOP in M63 medium with glucose pH 7.0, glycerol pH 7.0, glycerol pH 5.5, or galactose pH 7.0, was measured 16 h after dilution. Bars correspond to the average of three biological replicates with circles corresponding to individual data points and error bars representing 1 SD. Representative growth curves are shown in SI Appendix, Fig. S4A.
We also examined the effect of overexpressing the full-length AzuCR mRNA (pRI-AzuCR) without or with the L3STOP mutation (pRI-AzuCRL3STOP) (Fig. 1A and SI Appendix, Fig. S3A). Interestingly, we observed different effects on growth for these plasmids compared to the pKK derivatives expressing only the small protein (Fig. 4 and SI Appendix, Fig. S4A). While growth in M63 glucose pH 7.0 was not affected, pRI-AzuCR led to a growth defect in M63 glycerol pH 7.0 and even more so at pH 5.5, as well as in M63 galactose pH 7.0. Contrary to the relieving effect of the L3STOP mutation when only the azuC coding sequence was included, the L3STOP mutation in the full-length transcript still led to reduced growth and exacerbated the growth defect in M63 glycerol pH 7.0. These observations suggested the transcript could have an additional role as a regulatory RNA. AzuC denotes the small protein activity, AzuR denotes the sRNA activity, and AzuCR denotes the entire dual-function RNA.
AzuCR Functions as an sRNA to Repress cadA and galE.
Based on our findings that the AzuCR transcript could have an additional role as an sRNA, we investigated its potential as a base-pairing sRNA by searching for possible base pairing targets using the TargetRNA2 (27) and IntaRNA (28) prediction programs. Given the reduced growth in galactose and low pH due to AzuCR overexpression, we focused on potential target genes that might be important under these conditions. One predicted target with extensive potential base pairing was cadA, encoding lysine decarboxylase (Fig. 5A). Consistent with AzuR-mediated regulation of cadA, we observed decreased expression of a cadA-gfp fusion upon AzuCR overexpression and even more so for AzuCRL3STOP overexpression (Fig. 5B). Additionally, there were higher overall levels of cadA-gfp expression in the ΔazuCR strain compared to the WT strain, suggesting that chromosomally encoded AzuC contributes to the repression. Consistent with the base pairing predicted in Fig. 5A, the M1 mutations in AzuCRL3STOP reduced cadA-gfp repression, while regulation was restored when compensatory mutations were introduced in the cadA-gfp construct (Fig. 5C).
Another predicted target for base pairing with AzuR was galE (Fig. 5D), the first gene in the galETKM galactose operon. The AzuCRL3STOP derivative also repressed a galE-gfp fusion in both the WT and ΔazuCR backgrounds, with partial repression by WT AzuCR (Fig. 5E). M2 mutations in AzuCRL3STOP or in galE-gfp alone reduced AzuCR-mediated galE-gfp repression, while repression was restored when both compensatory mutations were present (Fig. 5F) documenting direct base pairing between AzuCR and galE.
Since the base-pairing region is encompassed within the azuC coding sequence, we also examined the effects of the pKK-AzuC and pKK-AzuCL3STOP derivatives, which only carry the ORF. Both had some repressive effects (Fig. 5 B and E) but less than the pRI derivatives, possibly due to the absence of a sequence element or lower levels of the transcripts (SI Appendix, Fig. S3A).
AzuCR RNA Association with Hfq Is Not Required for cadA and galE Repression.
Most base-pairing sRNAs in E. coli require the RNA chaperones Hfq or ProQ for their stability and optimal pairing with their target mRNAs (5–7). To determine if the AzuCR RNA binds either of these two chaperones, we carried out coimmunoprecipitation experiments with antibodies to Hfq and ProQ and examined the levels of the AzuCR RNA in the Δhfq and ΔproQ mutant backgrounds (Fig. 5G). The RNA coimmunoprecipitates with both proteins, consistent with binding to both chaperones. Interestingly, unlike most sRNAs, AzuCR levels increased in the Δhfq background. In contrast, the levels were significantly decreased in the ΔproQ background.
To test whether AzuCR function as a regulatory sRNA is mediated by Hfq or ProQ, we examined repression of the cadA-gfp and galE-gfp fusions in Δhfq and ΔproQ single mutants, as well as in a Δhfq ΔproQ double-mutant background (Fig. 5H). In the Δhfq background, GFP activity levels overall were somewhat lower, but we still observed clear cadA-gfp and galE-gfp repression by AzuCR and AzuCRL3STOP overexpression. The effects of ΔproQ were more significant with less repression, particularly of the galE-gfp fusion. Regulation of both fusions was eliminated in the Δhfq ΔproQ double mutant. These observations indicate that although both Hfq and ProQ bind to AzuCR, Hfq is less critical for the repression of cadA and galE, though some of the requirements for an RNA chaperone may be suppressed by overexpression of AzuCR.
AzuC Protein Levels Are Repressed by the FnrS sRNA.
Given the increased AzuCR RNA levels in the Δhfq strain, as well as the lack of an Hfq requirement for AzuCR repression of the cadA-gfp and galE-gfp fusions, we wondered whether azuCR binds to Hfq as an mRNA targeted by other sRNAs. This hypothesis is supported by the observed discordance between AzuC-SPA protein and AzuCR RNA levels for cells grown with different carbon sources (Fig. 1C). Additionally, AzuC-SPA protein levels were elevated in the Δhfq strain for cells grown in M63 galactose in the absence of increased AzuCR RNA levels (Fig. 5I). This observation is consistent with potential posttranscriptional repression of AzuC synthesis by Hfq and base-pairing sRNAs. As in Fig. 1C, we noted relatively high AzuC-SPA protein levels and low AzuCR RNA levels for cells grown in M63 glycerol pH 5.5, but this was not affected by Hfq.
We previously found that AzuC levels are higher under aerobic compared to anaerobic conditions (17). Interactions between FnrS, an sRNA induced under anaerobic conditions, and AzuCR were detected in genome-wide assays of RNA–RNA interactions on the Hfq chaperone (29, 30), and we predicted base pairing between the 5′ end of FnrS and a sequence 25 to 31 nt upstream of the azuC ORF (SI Appendix, Fig. S5A). Consistent with FnrS-mediated repression of AzuC synthesis, we observed higher AzuC-SPA levels in a ΔfnrS strain (SI Appendix, Fig. S5B) and lower AzuC-SPA levels upon overexpression of WT FnrS and previously generated FnrS-I and FnrS-II mutants (31), but not FnrS-III, for which base pairing is predicted to be disrupted (SI Appendix, Fig. S5C). We similarly observed that WT FnrS, FnrS-I, and FnrS-II, but not FnrS-III, repressed an azuC-lacZ translational fusion expressed from the heterologous PBAD promoter (SI Appendix, Fig. S5D). Repression was restored for the FnrS-III mutant but not WT FnrS, FnrS-I, and FnrS-II by compensatory mutations in the azuC-lacZ-III mutant fusion, demonstrating direct base pairing between FnrS and AzuCR.
GlpD is down-regulated during anaerobic growth when a second glycerol dehydrogenase, GlpABC, is induced. We thus found it interesting that interactions between FnrS and the glpD mRNA were also observed in the RNA interactome data (29, 30), and there was predicted pairing between the 5′ end of FnrS and glpD (SI Appendix, Fig. S5E). While the effects were subtle, we consistently observed lower levels of the GlpD-HA-His6 protein upon overexpression of WT FnrS, FnrS-I and FnrS-II, but not FnrS-III (SI Appendix, Fig. S5F). Collectively, these results suggest that AzuCR binds Hfq as an mRNA and indicate that the 5′ end of FnrS base pairs with the AzuCR and glpD mRNAs to repress synthesis of the two encoded proteins.
AzuC Translation and AzuR Base-Pairing Activity Interfere.
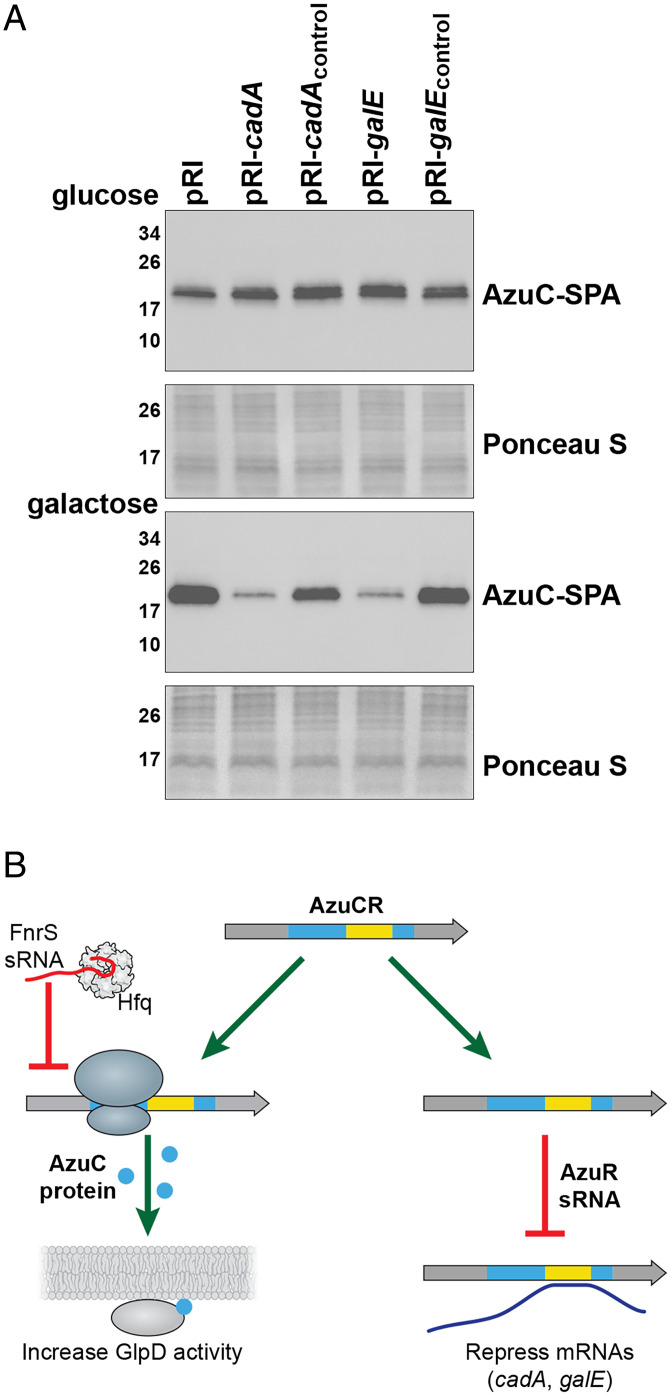
The region of base pairing between AzuR and cadA or galE overlaps the azuC coding sequence (40 to 126 nt relative to the transcription start), raising the question of whether the mRNA and base-pairing activities of the AzuCR RNA interfere with each other. The model that translation interferes with base pairing is supported by the observations that AzuCRL3STOP was more effective at repressing the cadA-gfp and galE-gfp fusions than AzuCR (Fig. 5 B and E). To determine if base-pairing activity reciprocally interferes with translation, we examined the levels of chromosomally encoded AzuC-SPA upon overexpression of the base-pairing regions of cadA and galE or control regions of these genes not predicted to base pair with AzuCR. Interestingly, no repression was observed for cells grown in M63 glucose. In contrast, the base-pairing fragments, but not the control fragments, led to decreased AzuC-SPA levels for cells grown in M63 with galactose (Fig. 6A). These observations suggest that base pairing can interfere with translation, particularly when AzuC protein levels are low as is the case for cells grown in M63 galactose.
Fig. 6.
AzuC and AzuR activities compete. (A) Effect of cadA, cadAcontrol, galE, and galEcontrol on AzuC-SPA levels in cells (GSO351) transformed with the respective overexpression plasmid and grown in M63 medium supplemented with glucose or galactose. Samples were taken at OD600 ∼0.5 and α-FLAG antibody was used to detect the SPA tag. The membranes stained with Ponceau S stain serves as a loading control. (B) Model for the different functions of the AzuCR RNA. For growth in M63 glycerol, pH 5.5, the RNA can be translated to give the 28-amino acid amphipathic AzuC protein, which increases the activity of GlpD glycerol-3-phosphate dehydrogenase. Under anaerobic conditions, translation of AzuC, as well as GlpD, is blocked by the FnrS sRNA. The RNA also can act as the AzuR base-pairing sRNA to repress synthesis of CadA and GalE.
Discussion
Successful adaptation to varying environmental conditions requires regulation that can rapidly lead to changes in metabolism. Along with transcription factors, sRNAs and small proteins are emerging as important regulators. While base-pairing sRNAs generally are not thought to be translated, a few have been reported to encode small proteins. Even fewer of these dual-function RNAs have been characterized. Here we report that the 164-nt RNA previously reported to encode a 28-amino acid small protein (AzuC) (17) also functions as a regulatory RNA (AzuR). We show the AzuC protein binds GlpD, an enzyme at the junction of respiration, glycolysis, and phospholipid biosynthesis, and increases dehydrogenase activity (Fig. 3), while the RNA base pairs with and represses expression from the cadA and galETKM mRNAs (Fig. 5).
AzuCR Is a Unique Dual-Function RNA.
There are several features of AzuCR that are unique compared to other dual-function RNAs. First, while the regions involved in base pairing and protein coding are separate for the well-characterized E. coli SgrS-SgrT and Staphylococcus aureus RNAIII RNAs (8), as well as the Vibrio cholerae VcdRP RNA (32), the region of AzuCR involved in base pairing with the cadA and galE targets overlaps the azuC coding sequence (Fig. 1A). Another notable feature of this dual-function RNA is that while the gene is not broadly conserved, the levels of the RNA and protein are highly regulated in response to multiple environmental conditions (17). The regulation in response to glucose availability was shown to be at the transcriptional level via CRP derepression (17). In our present study, we show that AzuC repression under anaerobic conditions is mediated by the Hfq-dependent sRNA FnrS, which base pairs near the AzuC ribosome binding site (SI Appendix, Fig. S5). We think FnrS regulation of AzuCR is a unique example of an sRNA regulating the translation of a dual-function RNA. The observations that there is discordance between RNA and protein levels under some conditions (Fig. 1B) and that AzuC protein levels are elevated in the Δhfq strain for cells grown in M63 galactose medium (Fig. 5I) and are higher in Δhfq compared to ΔfnrS mutant cells (SI Appendix, Fig. S5) suggests that additional Hfq-dependent sRNAs repress AzuC synthesis.
Consistent with the discordant expression of the AzuCR RNA compared to the AzuC protein, we found that the small protein and base-pairing activities modulate overlapping but also distinct pathways; AzuC plays a role in glycerol metabolism (Figs. 3 and 4) and AzuCR impacts galactose and glycerol metabolism (Figs. 4 and 5). The regulation of different pathways by the two activities of AzuCR (Fig. 6B) contrasts with SgrST RNA, where the SgrT protein and the base-pairing SgrS RNA both down-regulate PtsG glucose transporter activity.
AzuC Stimulation of Dehydrogenase Activity.
While the functions of only a few small proteins have been described, most are inhibitory (9). Thus, AzuC is unusual in that it increases GlpD-dependent dehydrogenase activity. GlpD exists in both soluble and membrane-bound forms and is only fully active when associated with the cytoplasmic membrane through lipid–enzyme interactions or when reconstituted with phospholipids in vitro (25, 33, 34). AzuC is an amphipathic protein localized to the cytoplasmic membrane (Fig. 2). Given that we see increased association of GlpD-YFP with the membrane in azuCR+ compared to ΔazuCR cells (Fig. 3B), we suggest that AzuC increases dehydrogenase activity by promoting GlpD binding to the cytoplasmic membrane. The 29-amino acid V. cholerae VcdP also was found to increase citrate synthase activity, but in this case the small protein counteracts the inhibitory effects of NADH (32).
The physiological role of AzuC, particularly at pH 5.5, is an interesting question. We suggest AzuC binds GlpD under acidic conditions to modulate the levels of glycerol-3-phosphate, which can undergo two acylation steps to form phosphatidic acid, a precursor for the phospholipids phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and cardiolipin (CL). Bacterial adaptation to environmental stress can be accompanied by changes in the lipopolysaccharide structure, the phospholipid composition, and the protein content of the inner and outer membranes (26). These changes in turn impact cell division, energy metabolism, and osmoregulation, as well as resistance to cationic antimicrobial peptides (CAMPs). In support of the hypothesis that the AzuC-dependent increase in dehydrogenase activity affects membrane composition, we observed that AzuC- or GlpD-overexpressing cells grown in M63 glycerol pH 5.5 medium showed increased cell length (Fig. 3C) and reduced growth (Fig. 4), phenotypes that have also been observed in cells lacking the phospholipids PE and PG/CL (26).
AzuR Repression of the cadA and the galETKM mRNAs.
We found that as a base-pairing RNA, AzuCR represses expression of CadA (Fig. 5 A–C), which is induced under acidic growth conditions and confers resistance to weak organic acids produced during carbohydrate fermentation under anaerobiosis and phosphate starvation (35). This regulation could partially explain the growth defect for cells growing in glycerol pH 5.5 observed upon AzuCR overexpression both with and without a stop codon (Fig. 4). We also identified galETKM mRNA as another direct target (Fig. 5 D–F). Consistent with this regulation, we saw a drastic growth defect with galactose as the sole carbon source upon overexpression of AzuCR and AzuCRL3STOP (Fig. 4). While AzuCR base pairing near the ribosome binding site of the cadA mRNA likely blocks ribosome binding, the base pairing with the galETKM mRNA is internal to the galE coding sequence. We suggest that for this mRNA, base pairing may lead to changes in mRNA stability or alternatively the Rho-dependent transcription termination reported for the galETKM mRNA (36). Since we also observed an RNA-dependent growth phenotype for cells grown in glycerol pH 7.0, we think AzuCR might target other genes, particularly genes related to glycerol metabolism.
Competition between Two AzuCR Activities.
Several of our experiments indicate there is competition between the mRNA and base-pairing activities of AzuCR. We observed that a stop codon blocking translation improves base-pairing activity (Fig. 5 B and E) and overexpression of fragments of the base-pairing targets cadA and galE inhibits AzuC translation (Fig. 6A). The conflict between base pairing and translation raises intriguing questions about what activity predominates under different growth conditions, whether the RNA can transition from one function to the other, what factors determine which activity predominates, and how the two activities evolved. We suggest that there are several scenarios for how AzuCR could act. There may be conditions where AzuCR acts solely as a riboregulator and other conditions where AzuCR is solely an mRNA. There also may be conditions where there are two populations of AzuCR, some transcripts acting as an sRNA and others being translated, where the function of the RNA is determined stochastically depending on whether the ribosome, Hfq, or ProQ bind first. Additionally, AzuCR could first act as an mRNA and subsequently act as a riboregulator.
The factors that regulate the distribution of AzuCR between these regulatory roles are not fully understood but clearly depend on the levels of the AzuCR RNA, sRNAs that repress AzuC translation, the levels of the mRNA targets of AzuCR, the levels of the Hfq and ProQ chaperones, and likely other factors. The observed FnrS-dependent repression of AzuC synthesis is at least partially dependent on Hfq, while the stability of the RNA appears to depend on ProQ. The finding that AzuCR may only be a base-pairing RNA under specific conditions raises caveats for global approaches, such as RNA interaction by ligation and sequencing (30) or RNA sequencing after pulse overexpression for identifying sRNA targets and function. If these experiments are carried out under conditions where translation predominates, the effects of the base-pairing activity may not be detected. These questions about AzuCR likely are relevant for other dual-function RNAs and are important directions for future research.
Materials and Methods
Bacterial Strains and Plasmids.
Bacterial strains, plasmids, and oligonucleotides used in this study are listed in Dataset S1. Details about strain and plasmid construction are provided in SI Appendix.
Bacterial Growth.
Bacterial growth was monitored in LB-rich or M63 minimal medium supplemented with 0.001% vitamin B1 and glucose or galactose at 0.2% or glycerol at 0.4% (with pH adjusted to 5.5 for some samples), and appropriate antibiotics as described in SI Appendix.
Immunoblot and Northern Analysis.
Specific tagged proteins were detected by immunoblot analysis and specific RNAs were detected by northern analysis as described in detail in SI Appendix. For some experiments, protein was isolated from cell fractions and RNA was coimmunoprecipitated with RNA chaperones, as described in SI Appendix.
Microscopy.
The bright field and fluorescence microscopy were carried out as described in SI Appendix.
Protein Purification.
Tagged proteins were purified based on their tags as described in detail in SI Appendix.
Dehydrogenase, β-Galactosidase, and GFP-Reporter Assays.
Dehydrogenase, β-galactosidase, and GFP-reporter assays were carried out as described in detail in SI Appendix.
Supplementary Material
Acknowledgments
We thank P. Backlund for carrying out mass spectrometry in the Eunice Kennedy Shriver National Institute of Child Health and Human Development core; A. Buskirk for the AzuCR browser image; M. Hemm, N. Soltanzad, L. Stamper, and A. Romero for help with initial experiments; and members of the G.S. group for comments on the manuscript. This research was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
Reviewers: P.B., Pennsylvania State University; and C.V., University of Illinois at Urbana Champaign.
Competing interest statement: M.R. and C.V. are coauthors on a review published in 2018.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117930119/-/DCSupplemental.
Data Availability
All study data are included in the main text and supporting information.
References
- 1.Soberón-Chávez G., Alcaraz L. D., Morales E., Ponce-Soto G. Y., Servín-González L., The transcriptional regulators of the CRP family regulate different essential bacterial functions and can be inherited vertically and horizontally. Front. Microbiol. 8, 959 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner E. G. H., Romby P., Small RNAs in bacteria and archaea: Who they are, what they do, and how they do it. Adv. Genet. 90, 133–208 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Papenfort K., Vogel J., Small RNA functions in carbon metabolism and virulence of enteric pathogens. Front. Cell. Infect. Microbiol. 4, 91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durica-Mitic S., Göpel Y., Görke B., Carbohydrate utilization in bacteria: Making the most out of sugars with the help of small regulatory RNAs. Microbiol. Spectr. 6, 229–248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holmqvist E., Vogel J., RNA-binding proteins in bacteria. Nat. Rev. Microbiol. 16, 601–615 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Updegrove T. B., Zhang A., Storz G., Hfq: The flexible RNA matchmaker. Curr. Opin. Microbiol. 30, 133–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olejniczak M., Storz G., ProQ/FinO-domain proteins: Another ubiquitous family of RNA matchmakers? Mol. Microbiol. 104, 905–915 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raina M., King A., Bianco C., Vanderpool C. K., Dual-function RNAs. Microbiol. Spectr. 6, 10.1128/microbiolspec.RWR-0032-2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Storz G., Wolf Y. I., Ramamurthi K. S., Small proteins can no longer be ignored. Annu. Rev. Biochem. 83, 753–777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemm M. R., Weaver J., Storz G., Escherichia coli small proteome. Ecosal Plus 9, 10.1128/ecosalplus.ESP-0031-2019 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadler C. S., Vanderpool C. K., A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. U.S.A. 104, 20454–20459 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanderpool C. K., Gottesman S., Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54, 1076–1089 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Lloyd C. R., Park S. Y., Fei J., Vanderpool C. K., The small protein SgrT controls transport activity of the glucose-specific phosphotransferase system. J. Bacteriol. 199, e00869-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S., et al. , A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome. Biosystems 65, 157–177 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Hemm M. R., Paul B. J., Schneider T. D., Storz G., Rudd K. E., Small membrane proteins found by comparative genomics and ribosome binding site models. Mol. Microbiol. 70, 1487–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weaver J., Mohammad F., Buskirk A. R., Storz G., Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio 10, e02819-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemm M. R., et al. , Small stress response proteins in Escherichia coli: Proteins missed by classical proteomic studies. J. Bacteriol. 192, 46–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontaine F., Fuchs R. T., Storz G., Membrane localization of small proteins in Escherichia coli. J. Biol. Chem. 286, 32464–32474 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhoads D. B., Tai P. C., Davis B. D., Energy-requiring translocation of the OmpA protein and alkaline phosphatase of Escherichia coli into inner membrane vesicles. J. Bacteriol. 159, 63–70 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hobbs E. C., Yin X., Paul B. J., Astarita J. L., Storz G., Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc. Natl. Acad. Sci. U.S.A. 109, 16696–16701 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walz A. C., Demel R. A., de Kruijff B., Mutzel R., Aerobic sn-glycerol-3-phosphate dehydrogenase from Escherichia coli binds to the cytoplasmic membrane through an amphipathic alpha-helix. Biochem. J. 365, 471–479 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H., et al. , Increasing intracellular magnesium levels with the 31-amino acid MgtS protein. Proc. Natl. Acad. Sci. U.S.A. 114, 5689–5694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramamurthi K. S., Lecuyer S., Stone H. A., Losick R., Geometric cue for protein localization in a bacterium. Science 323, 1354–1357 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wegener M., Vogtmann K., Huber M., Laass S., Soppa J., The glpD gene is a novel reporter gene for E. coli that is superior to established reporter genes like lacZ and gusA. J. Microbiol. Methods 131, 181–187 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Robinson J. J., Weiner J. H., The effect of amphipaths on the flavin-linked aerobic glycerol-3-phosphate dehydrogenase from Escherichia coli. Can. J. Biochem. 58, 1172–1178 (1980). [DOI] [PubMed] [Google Scholar]
- 26.Rowlett V. W., et al. , Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J. Bacteriol. 199, e00849-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kery M. B., Feldman M., Livny J., Tjaden B., TargetRNA2: Identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 42, W124-9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann M., Wright P. R., Backofen R., IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 45 (W1), W435–W439 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melamed S., Adams P. P., Zhang A., Zhang H., Storz G., RNA-RNA interactomes of ProQ and Hfq reveal overlapping and competing roles. Mol. Cell 77, 411–425.e7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melamed S., et al. , Global mapping of small RNA-target interactions in bacteria. Mol. Cell 63, 884–897 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durand S., Storz G., Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol. Microbiol. 75, 1215–1231 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkat K., et al. , A dual-function RNA balances carbon uptake and central metabolism in Vibrio cholerae. EMBO J. 40, e108542 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh J. I., Chinte U., Du S., Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism. Proc. Natl. Acad. Sci. U.S.A. 105, 3280–3285 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schryvers A., Lohmeier E., Weiner J. H., Chemical and functional properties of the native and reconstituted forms of the membrane-bound, aerobic glycerol-3-phosphate dehydrogenase of Escherichia coli. J. Biol. Chem. 253, 783–788 (1978). [PubMed] [Google Scholar]
- 35.Kanjee U., Houry W. A., Mechanisms of acid resistance in Escherichia coli. Annu. Rev. Microbiol. 67, 65–81 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Wang X., et al. , Expression of each cistron in the gal operon can be regulated by transcription termination and generation of a galk-specific mRNA, mK2. J. Bacteriol. 196, 2598–2606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mól A. R., Castro M. S., Fontes W., NetWheels: A web application to create high quality peptide helical wheel and net projections. BioRxiv [Preprint] (2019). 10.1101/416347. Accessed 5 October 2019. [DOI]
- 38.Taniguchi Y., et al. , Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and supporting information.