Significance
Ion channels have evolved the ability to communicate with one another, either through protein–protein interactions, or indirectly via intermediate diffusible messenger molecules. In special cases, the channels are part of different membranes. In muscle tissue, the T-tubule membrane is in proximity to the sarcoplasmic reticulum, allowing communication between L-type calcium channels and ryanodine receptors. This process is critical for excitation–contraction coupling and requires auxiliary proteins like junctophilin (JPH). JPHs are targets for disease-associated mutations, most notably hypertrophic cardiomyopathy mutations in the JPH2 isoform. Here we provide high-resolution snapshots of JPH, both alone and in complex with a calcium channel peptide, and show how this interaction is targeted by cardiomyopathy mutations.
Keywords: muscle excitation–contraction coupling, calcium signaling, ion channels, calcium release, cardiomyopathy
Abstract
Junctophilins (JPH) are a class of proteins found at junctions between the plasma membrane and the endoplasmic or sarcoplasmic reticulum, allowing for communications between proteins embedded in different membranes. JPHs have been proposed to interact with lipids as well as several ion channels, allowing for specialized communication between them. The JPH3 isoform is the target for repeats that cause Huntington’s disease-like 2, whereas JPH2 is a hot spot for mutations linked to cardiomyopathy. Here we present crystal structures of two JPH isoforms, which resemble a twisted skeleton with ribs formed by membrane occupation recognition nexus repeats, and a backbone built by a long α-helix. We captured the structure of a complex between JPH2 and a C-terminal binding site in the L-type calcium channel (CaV1.1) and show that this interaction is required for clustering of these channels and for robust muscle excitation–contraction coupling. Over 80 sequence variants linked to cardiomyopathy are found in different structurally important regions of JPH2, most of which affect stabilizing interactions. A subset directly affects the interaction with the L-type calcium channel. In parallel, sequence variants in the L-type calcium channel, linked to cardiac arrhythmia, also affect critical interactions.
Junctions between the plasma membrane and the endoplasmic reticulum (ER) or sarcoplasmic reticulum (SR) are found in multiple cell types and allow for specialized communication between proteins embedded in these different membranes. Junctophilins (JPH) are key to enabling the formation of such junctions, by virtue of a C-terminal transmembrane helix, located in the ER or SR membrane, and an N-terminal domain containing MORN (membrane occupation recognition nexus) motifs, thought to interact with phospholipids in the plasma membrane (1, 2). As such, JPHs play critical roles in diverse signaling processes, often allowing functional or mechanical cross-talk between ion channels embedded in different membranes.
Four isoforms (JPH1–JPH4) are encoded in the human genome, with JPH1 primarily expressed in skeletal muscle, JPH2 in skeletal, cardiac, and smooth muscle, and JPH3/4 mostly found in the brain (3) and in sensory neurons (4). Additionally, JPHs have been found in multiple cell types, including pancreatic β cells (5) and T cells (6). In muscle tissue, JPHs allow for the communication between L-type voltage-gated calcium channels (CaV), located in the transverse-tubule (T-tubule) membrane, and ryanodine receptors (RyRs) in the SR membrane. In cardiac myocytes, a depolarization of the plasma membrane activates the CaV1.2 isoform, and the influx of Ca2+ then triggers opening of RyR2, in a process known as Ca2+-induced Ca2+ release (7). In skeletal muscle, direct mechanical coupling is thought to occur between the corresponding CaV1.1 and RyR1 isoforms, although any direct contacts between these two proteins remain to be elucidated (8, 9). In both scenarios, the coupling requires proximity between the T-tubule and SR membranes, for which JPHs are critical. JPHs have also been suggested to directly bind both CaVs and RyRs, as well as other ion channels including Ca2+-activated potassium channels (10–12) and KCNQ1 (13).
The importance of JPHs is underscored by the various disorders associated with them. JPH1 has been identified as a modifier of Charcot-Marie-Tooth disease (14). JPH2 is a hot spot for mutations linked to hypertrophic cardiomyopathy (HCM) (15–17), and repeats in JPH3 have been found to cause Huntington disease-like 2 (HDL-2) (18). In addition, cardiac stress results in activation of calpain, which cleaves JPH2 and drives heart failure progression (19–24). Cleaved fragments of JPH2 have also been shown to migrate to the nucleus, where, depending on the fragment type, they either attenuate (22) or promote (21) cellular remodeling. To date, no high-resolution structural information is available for any JPH isoform, hampering insights into its basic functions and disease mechanisms.
In this study, we describe high-resolution structures of two JPH isoforms, reveal how JPHs bind the C-terminal region of CaV1.1 via a groove formed by the MORN repeats, how this association is important for normal excitation–contraction coupling, and how disease-associated mutations may affect this process.
Results
Crystal Structures of JPH1 and JPH2.
All four JPH isoforms have been predicted to contain eight MORN repeats at the N terminus, an α-helical–rich domain (AHD), a divergent region, and a C-terminal transmembrane segment (Fig. 1A). As the divergent region contains low sequence complexity and is predicted to be intrinsically disordered (SI Appendix, Fig. S1), we set out to solve the crystal structure of the MORN and AHD region. MORN repeats 6 and 7 are separated by a long linker known as the “joining region.” It shows lower sequence conservation, low sequence complexity, and contains predicted disorder (SI Appendix, Fig. S1). Removal of most of this linker was required to obtain crystals for both JPH1 and JPH2. We used single-wavelength anomalous diffraction (SAD) phasing on selenomethionine-substituted JPH1. The corresponding structure was used as a molecular replacement model to solve the JPH2 structure.
Fig. 1.
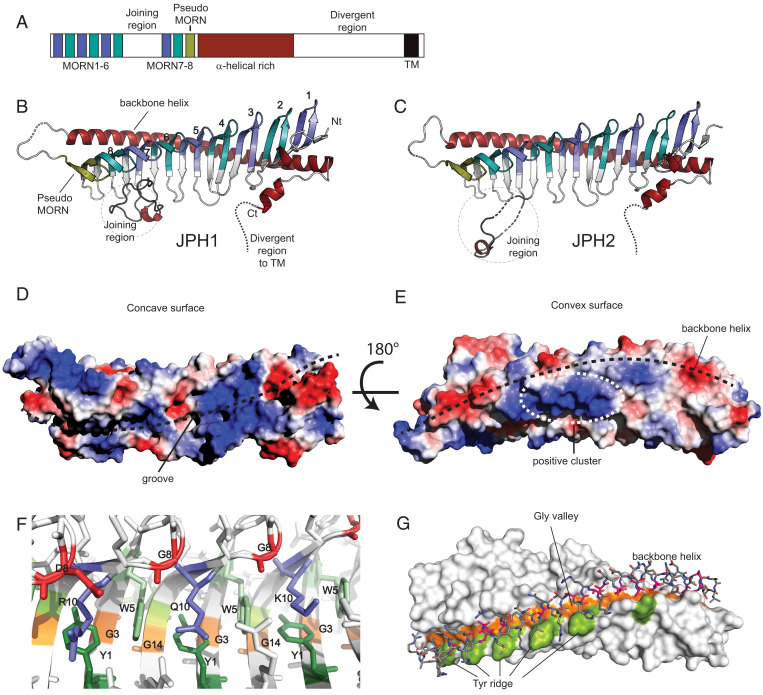
Crystal structure of JPH1 and JPH2 MORN-helical domain. (A) The primary sequence organization of JPH1-4 showing the 8 MORN repeats interrupted by a joining region, a pseudo-MORN repeat preceding a long α-helical–rich stretch, followed by a highly divergent region and ending in a short transmembrane helix. (B and C) The crystal structures of JPH1 and JPH2 (PDBs code 7RW4 and 7RXE) showing the MORN repeats, shortened joining region, and a long backbone helix. The conformation of the shortened joining region differs between the two isoforms. (D and E) The surface electrostatic potentials generated for JPH2 minus the shortened joining region, calculated in PyMOL, reveals a highly charged groove formed by the MORN repeats and an electropositive cluster at the center of the backbone helix. (F) A representation of the consensus MORN repeat-residues, including the Y1 and W5 consensus residues forming hydrophobic walls, accommodating R10, Q10, or K10 side chains. (G) The G3/G14 MORN residues forms a glycine valley, lined on one end by a tyrosine ridge formed by Y13 in some MORN repeats. The backbone helix fits in this valley using short alanine side chains.
Fig. 1 B and C show the overall structure of JPH1 (1.31 Å) and JPH2 (2.35 Å) (SI Appendix, Tables S1 and S2), which resemble the shape of a skeleton. The ribs are formed by a 19-stranded antiparallel β-sheet. The sheet is twisted, with an ∼90° angle between the first and last β-strand. The β-sheet forms a twisted cradle with a concave “inside” and a convex “outside.” The concave side forms a narrow groove poised to bind possible interaction partners (Fig. 1D). A 60-residue-long α-helix forms a “backbone” on the convex side of the β-sheet (Fig. 1E). This helix directly matches the overall twist of the sheet, thus contacting every β-hairpin at an equivalent position. The fold is completed with a C-terminal eight-residue helix that also contacts the convex outside. Together, these encode the previously annotated α-helical rich region of JPHs.
The MORN repeats directly map to the β-sheet. There are different definitions for MORN repeats, with either 14-residue or 23-residue lengths for a single MORN repeat. Within JPHs, there is high sequence conservation for the first 14 residues of each repeat, with a consensus sequence YxGxWxxGxxxGxG (SI Appendix, Fig. S1), although some of these positions can be altered. This conserved stretch encodes a β-hairpin that has a striking structural similarity in all eight repeats (SI Appendix, Fig. S1 B and C). The remaining stretches beyond these 14 residues are more varied in sequence and encode parts of the strands, along with a loop with varied conformation and length. Of the β-strands, 16 are thus encoded by the MORN repeats, with an additional flanking strand on the N-terminal end. On the C-terminal end of the β-sheet, an additional β-hairpin is present that resembles the MORN repeats, despite missing most consensus residues. This “pseudo-MORN” is observed for both JPH1 and JPH2.
A direct investigation of the conserved MORN repeat residues highlights their individual functions (Fig. 1F). The first Tyr of the repeat (Y1) forms an aromatic base platform that is typically flanked on both sides by Trp residues, corresponding to W5 from the same and the following repeat. In four of the repeats, an Arg residue (R10 in the MORN repeat), forms stacking and cation–π interactions with flanking aromatic residues (Fig. 1F). Among the three glycines within the motif, G8 is part of the β-turn.
An investigation of the interface between the backbone helix and the β-sheet shows a striking pattern of glycine residues within the β-sheet, encoded by G3 and G14 of the consensus MORN sequence. Within the second MORN repeat, the nonconserved G13 is also at the interface. Together, the systematic absence of side chains at the same positions in each β-hairpin results in a long continuous “glycine valley” in which the backbone helix can easily fit. Any other residue would cause steric hindrance, which explains the conservation of G3 and G14 within the MORN repeats. Within the backbone helix, there is also a selection for short residues at the interface, as most residues facing the glycine valley are alanines. Various additional interactions are made with residues on the ridge of the glycine valley. One of the two ridges is typically encoded by residue 2 or 15 in the MORN repeat. As this position shows no conservation, the interactions with the backbone are varied with either hydrophobic, ionic, or general hydrophilic interactions. The other ridge, however, is mostly made by residues 4 and 13 in the MORN repeat. Position 4 is not conserved, but position 13 is a Tyr or Phe in the last five repeats. This Tyr forms a regular interspersed hydrophobic platform that typically packs against hydrophobic residues in the backbone helix (Fig. 1G).
Thus, the conserved residues in the MORN repeats serve two structural purposes: to provide stabilizing interactions that drive the formation of the twisted β-sheet, and to allow interactions with the helical backbone.
JPHs have been proposed to form dimers (25), but the crystal structures did not show any stable dimeric interface. To confirm its behavior in solution, we performed size-exclusion chromatography multiangular light scattering (SEC-MALS), which shows that JPH, either with or without the joining region, are monomeric in solution (SI Appendix, Fig. S2A and Table S3). However, both the crystallization and SEC were performed under high ionic strength, which is required to prevent aggregation and precipitation. Thus, it is still possible that dimerization occurs under physiological conditions.
Comparison between JPH1 and JPH2.
The overall fold is very similar between both isoforms, but a large difference is seen in the loop connecting MORN repeats 6 and 7. This normally encodes a long, disordered stretch, most of which was removed to promote crystallization. In agreement with its flexibility, the remaining residues adopt different conformations in both isoforms. In JPH1, the remaining loop folds back and occludes part of the concave inside of the β-sheet, whereas in JPH2 it interacts with a neighboring molecule in the crystal lattice (SI Appendix, Fig. S3). In another JPH2 structure discussed in a later section, the loop is entirely invisible. Thus, the different conformations of the remaining stretch of the joining region are also suggestive of high flexibility.
Fig. 1 D and E and SI Appendix, Fig. S2 B and C show electrostatic surface potential maps for JPH2 and JPH1. In both cases, a distinct cluster of positive charge is located near the backbone helix, whereas the concave inside shows both negative and positive clusters near the N-terminal MORN repeats. The positive clusters may facilitate interactions with negatively charged phospholipids in the plasma membrane.
Interactions with L-Type Calcium Channels.
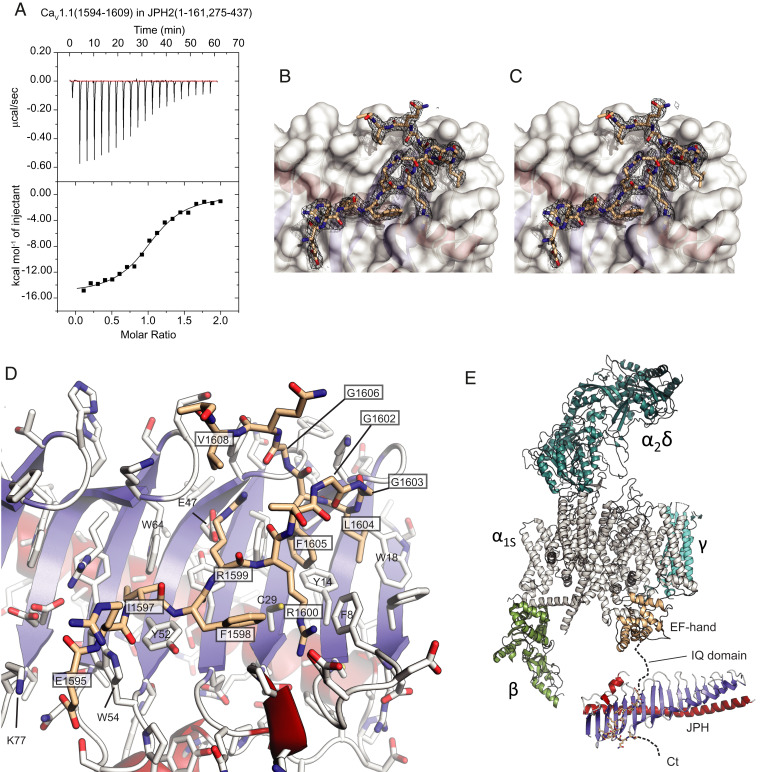
JPHs have been suggested to bind directly to various ion channels, including L-type CaVs (26–28) and RyRs (26). The interactions with the latter likely reside in the C-terminal disordered region of JPH (27, 29), but the N-terminal region has been proposed to interact with CaV directly, binding a site that is ∼50 residues downstream from the IQ domain in the channel’s cytoplasmic C terminus (27) (SI Appendix, Fig. S4). Using isothermal titration calorimetry (ITC) with the peptide from rabbit CaV1.1 (residues 1594 to 1609), we found that this site binds JPH1 and JPH2, both lacking the joining region, with an affinity of ∼1 to 2 μM and a 1:1 stoichiometry (Fig. 2A and SI Appendix, Fig. S4D and Table S4). Interestingly, when the joining region is included, the affinity reduces to ∼8 μM, suggesting that it can introduce some steric hindrance (SI Appendix, Fig. S4C and Table S4). We solved a crystal structure of JPH2 in complex with the corresponding peptide at 2.0-Å resolution. The structure shows unambiguous electron density for 15 of 16 residues of the peptide (Fig. 2 B and C).
Fig. 2.
Interactions between JPH2 and a CaV1.1 peptide. (A) Representative ITC titration of 300 μM CaV1.1 peptide into 30 μM JPH2 (1 to 161, 275 to 437) (Kd = 1.6 μM). (B) Final 2mFo–DFc density and (C) composite omit map for the CaV1.1 peptide shown as a black mesh, both contoured at σ = 1. The white surface corresponds to JPH2. (D) Close-up of the CaV1.1 peptide (beige) bound to JPH2. Boxed labels correspond to CaV1.1, with others corresponding to JPH2. Blue: β-strands; red: α-helices. Side chains are shown in stick representation with oxygens in red and nitrogens in blue. (E) Structure of the junctophilin complex in relationship to a cryoelectron microscopy structure of CaV1.1 (PDB ID code 5GJW). Different subunits are labeled. The α1S subunit is shown in gray, with the EF-hand domain in the C terminus in beige. The JPH binding site is 50 residues downstream of the IQ domain.
The peptide binds at the N-terminal end of the protein on the concave side of the β-sheet. Most interactions take place with residues on the first three MORN repeats and the N-terminal β-strand. The part of the peptide making contacts with JPH2 spans 14 residues, starting with Glu1595 and ending with Val1608.
The interaction is formed by hydrophobic, ionic, and H-bond interactions (Fig. 2D). The most prominent hydrophobic interactions are mediated by CaV1.1 residues Ile1597, Phe1598, Leu1604, and Phe1605. The latter residue is completely buried at the interface, forming part of a hydrophobic core that includes both CaV1.1 and JPH2 residues. A pair of arginine residues, encoded by CaV1.1 residues Arg1599 and Arg1600, is involved in ionic and H-bond interactions with JPH2. The CaV1.1 peptide also encodes two glycine residues (Gly1602 and Gly1603) that form the beginning of a sharp turn in the peptide. Both occupy regions of the Ramachandran plot that are not allowed for non-Gly residues, suggesting they are essential to allow the sharp twist of the CaV1.1 peptide. On the C-terminal side of this turn, the main chain of Gly1606 makes H-bond interactions with the Arg1599 side chain. In general, this binding profile matches a previous alanine scan using pull-down experiments (27). A sequence alignment with other L-type CaVs shows that these residues are conserved between CaV1.1 and CaV1.2, contain mild substitutions in CaV1.3, but are not conserved in CaV1.4 (SI Appendix, Fig. S4A). The motif is also absent from non–L-type CaVs. Fig. 2E shows the location of JPH in relation to CaV1.1, where it is on the same side as the Cavβ1 subunit.
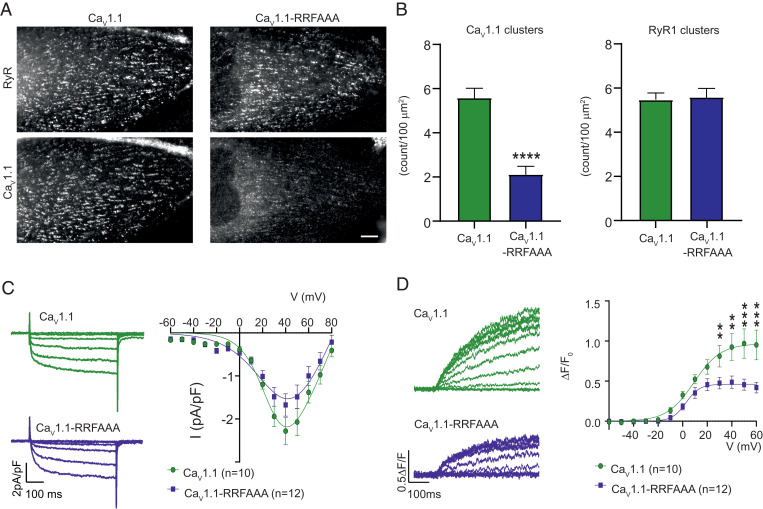
As JPHs are essential for muscle EC-coupling, we set out to determine the relative contribution of this interaction. We therefore created a mutant CaV1.1 that would no longer bind either JPH1 or JPH2 at this site. As individual point mutations may decrease affinity but not necessarily knock out the binding, we mutated three residues, involved in multiple interactions, to alanines: the Arg1599/Arg1600 pair, as well as Phe1605 (“RRFAAA” mutant). To confirm that this knocks out binding, we tested a peptide containing the triple mutation via ITC and could no longer detect any binding to JPH2 (SI Appendix, Fig. S4E). We then reconstituted dysgenic (CaV1.1-null) myotubes with either WT or RRFAAA CaV1.1 to investigate the importance of the interaction with JPHs for CaV1.1 targeting to the T-tubule/SR junctions. While the clustering of RyR1 appeared unperturbed in both conditions (Fig. 3 A and B), there was a 62% reduction in clustering of CaV1.1 when the interaction with JPHs was disrupted. Importantly, previous simultaneous knockdown of both JPH1 and JPH2 in myotubes had shown a much larger reduction in CaV1.1 cluster number (27), suggesting that there may be additional binding determinants between CaV1.1 and JPHs outside of this complex, or that JPHs organize other triadic proteins that help recruit CaV1.1.
Fig. 3.
The RRFAAA mutation results in reduced junctional localization and reduced EC coupling. (A) Double immunofluorescence labeling of CaV1.1 and RyR1 shows that the RRFAAA mutation results in reduced incorporation of CaV1.1 in the T-tubule/SR junctions of dysgenic myotubes. (Scale bar, 10 µm.) (B) CaV1.1 cluster count reveals a significant (n = 30, Student’s t-test ****P < 0.0001) decrease of CaV1.1-RRFAAA incorporation in clusters compared to CaV1.1, while the clustering of RyR1 is unchanged (n = 30, Student’s t-test P = 0.83). (C) Representative calcium currents (Vtest at −40, −20, 0, +20, +40 mV, Left) and average IV relationships (Right) in dysgenic myotubes show a modest but statistically not significant effect of the RRFAAA mutation on L-type currents [two-way repeated-measures ANOVA F(1, 14) = 1.706; P = 0.054]. (D) Representative whole-cell voltage-clamp measurements of calcium transients with Fluo-4 (Left) and average peak change in fluorescence normalized by baseline (ΔF/F) as a function of test potential (Right) show a significant reduction of EC coupling calcium release. [Two-way repeated measures ANOVA F(1, 12) = 3.831; P < 0.001. The asterisks in the figure refer to the post hoc analysis: at +30 mV **P = 0.004, at +40 mV **P = 0.001, at +50 mV and +60 mV ***P < 0.001.] Error bars correspond to SEMs in B–D. Details of the fits are indicated in Materials and Methods.
Knocking out the binding site also had a small, not statistically significant effect on the magnitude of the L-type Ca2+ currents (Fig. 3C). This reduction (22% at the peak) seems small in the context of the reduced clustering (62%), as channels that do not receive retrograde signals from RyRs would be expected to have much smaller currents. This suggest that many channels are still in clusters that are smaller and below the cutoff for the cluster size. Most importantly, the Ca2+ transients, reflective of RyR1 activity upon depolarization of the plasma membrane, significantly decreased by ∼44% (Fig. 3D). These results show that the interaction, as seen in the crystal structure, is required for normal EC-coupling, but that some degree of EC-coupling can still occur in its absence.
Palmitoylation Sites.
JPHs have been shown to associate with the plasma membrane, leading to the notion that they bind phospholipids directly. More specifically, phosphatidylserine (PS) and phosphatidylinositols (PIPs) have been suggested as ligands (2, 25). Although this classically has been assigned to the MORN repeats, the view of MORN repeats as bona fide binders of membranes has also been questioned (30, 31). We grew crystals of JPH1 in the presence of 500 μM 8:0 PI(4,5)P2 or 2 mM 6:0 PS, and collected diffraction data up to 2.0-Å and 1.9-Å resolution, respectively. However, no significant difference density could be identified for either ligand (SI Appendix, Fig. S5). This suggests that, if the MORN repeats bind PIP2 or PS lipids as previously suggested, they may do so with a very low affinity. In addition, it is possible that the high ionic strength used in the crystallization conditions may lower the affinity, especially if the binding is driven by ionic interactions with the phospholipid head groups.
An alternative model suggests that palmitoylation is required for stable association of JPHs with the plasma membrane (32). Four cysteines in JPH2 were shown to be palmitoylated, of which three reside in the structured region that was crystallized. Cys29 is conserved in all four JPH isoforms; Cys15 and 328 are conserved in JPH1-3. SI Appendix, Fig. S6 shows the location of these sites. Two of these (Cys15 and 328) are completely inaccessible to solvent and could thus only be palmitoylated upon local unfolding of the protein. It is therefore unlikely that these are major palmitoylation sites in native JPHs. Cys29, located in between MORN1 and MORN2, is accessible, but becomes fully buried when the CaV1.1 peptide is bound (Fig. 2D). This suggests that palmitoylation of this residue and binding to CaVs at this site are mutually exclusive.
Disease-Associated Mutations.
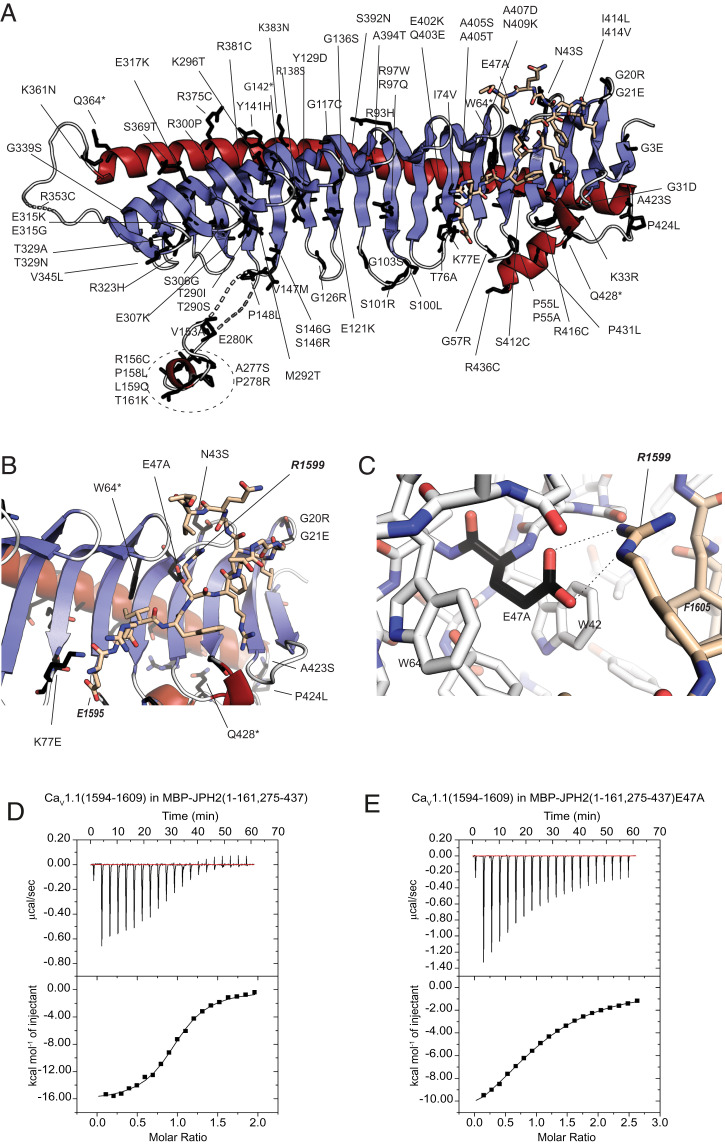
JPHs are targets for various disease-associated mutations. In particular, JPH2 is a hotspot for mutations linked to HCM (15, 16, 33–37). Up to 82 of these map directly to the crystallized region (Fig. 4A and SI Appendix, Table S6). Functional investigation has been carried out for only a small subset of these variants, but many of them are predicted to destabilize the folding of JPH2. For example, 16 variants affect the interface between the backbone helix and the β-sheet, either removing stabilizing interactions or creating steric hindrance (SI Appendix, Table S6). The backbone helix contains alanines that face the glycine valley, and several of these are substituted by residues that would create steric hindrance. In multiple cases, JPH2 glycines adopt backbone conformations not allowed for nonglycine residues, but are substituted, which would likely destabilize the fold. Six sequence variants directly affect consensus MORN residues, abolishing the stabilizing interactions.
Fig. 4.
Sequence variants in JPH2. (A) JPH2 sequence variants (labeled) are mapped to the crystallized JPH2 MORN-helical domain in complex with the CaV1.1 peptide. Affected residues are shown as black sticks. (B and C) Mutants in proximity to the peptide binding site, with residue E47 from JPH2 forming an ionic interaction with R1599 of the CaV1.1 peptide. (D) A representative ITC of 300 μM CaV1.1 peptide titrated into 35 μM MBP-tagged JPH2 (1 to 161, 275 to 437) (Kd = 1.8 μM). (E) A representative ITC of 750 μM CaV1.1 peptide titrated into 60 μM into the same MBP-tagged JPH2 construct carrying the E47A mutation (Kd = 26 μM).
Interestingly, several mutations map to the vicinity of the CaV peptide binding region (Fig. 4 B and C). JPH2 residue Lys77 forms ionic interactions with the N-terminal Glu1595 of the peptide (corresponding to CaV1.2 residue Glu1740). The K77E mutation has been linked to HCM and would disrupt this ionic interaction. JPH2 residue Glu47 is involved in ionic interactions with Arg1599 in CaV1.1 (corresponding to Arg1745 in CaV1.2) and the HCM-linked mutation E47A would abolish this. To investigate this mutation further, we prepared the E47A variant of JPH2 (residues 1 to 161, 275 to 437), but we found this mutant to readily precipitate at the concentrations required for ITC experiments. As the E47 side chain is also involved in packing interactions within JPH2 (SI Appendix, Table S6), loss of these may reduce the stability. However, ITC experiments could be performed for a fusion with maltose-binding protein (MBP), which show that the mutation results in an ∼15-fold decrease in affinity (Fig. 4 and SI Appendix, Table S4). This decrease may be a combined result of a loss of a salt bridge, together with structural perturbations of the binding site induced by the E47A mutation. Cardiomyopathy mutations may act through interfering with the JPH2-CaV1.2 interaction, but also with protein stability.
Interestingly, sequence variants have also been found in the CaV1.1 or CaV1.2 region that interacts with JPH (SI Appendix, Tables S7 and S8). Although none of these have been investigated functionally yet, several are predicted to affect the interface. In CaV1.2, three sequence variants have been found in patients with long QT syndrome, including the I1743T mutation which would affect substantial hydrophobic interactions with three aromatic residues (Y52, W54, and W64) in JPH2. Interestingly, the same ionic pair, targeted by the E47A mutation in JPH2, is also affected by sequence variants in the CaV channels: the CaV1.1 R1599W mutation was reported in a patient with susceptibility to malignant hyperthermia and hypokalemic periodic paralysis. Other sequence variants at the same site (R1599P, R1599Q) and the equivalent residue in CaV1.2 (R1745M) have been reported in the gnomAD database. All of these would abolish the ionic interactions with E47. Thus, the interaction between JPH2 and CaV1.2 is targeted by sequence variants on either side of the interface.
Discussion
We set out to provide structural insights into JPHs, proteins responsible for junctional membrane complex formation. This allows coupling between membrane proteins in the plasma membrane and the ER or SR. Junctional membrane complexes are present in nearly every excitable cell. The role of JPHs has been most extensively studied in the context of skeletal and cardiac myocytes, where they enable the communication between L-type CaVs and RyRs, a prerequisite for muscle excitation–contraction coupling.
Early on, JPHs were found to contain repeats coined MORN, because they were thought to directly bind membranes (1). MORN repeats have since been found in a plethora of proteins, but an unambiguous binding of such repeats to biological membranes remains to be reported. In contrast, several reports have questioned the role of the MORN repeats in membrane targeting. The plant type I/II phosphatidylinositol phosphate kinase contains eight MORN repeats, but its membrane targeting was found to rely on a separate catalytic domain instead (30). A thorough investigation of the Trypanosoma MORN1 protein found that it does not bind lipid vesicles (31). Although binding to lipids could be detected, this seemed to be mediated by binding to the hydrophobic lipid tails, inaccessible in lipid vesicles. Using cocrystallization between JPH1 and up 500 μM of PIP2 of 2 mM PS, we could not observe any density for a bound ligand. This suggests that any binding of PIP2 to this N-terminal region, if at all, is inherently very weak. Another report suggested instead that palmitoylation of JPHs is required for efficient membrane targeting (32), but two of the proposed palmitoylation sites are buried, and the remaining is at the interface with the CaV1.1 peptide. Thus, the exact role and mechanism of plasma membrane binding of JPHs remains enigmatic.
In contrast with this, the binding of JPHs to L-type CaVs appears more unambiguous. Previous reports had suggested that the C-terminal region of CaV1.1 and CaV1.2 is required for their targeting at junctional membranes (38, 39). Pull-down experiments showed that JPH1 and JPH2 could bind to a proximal C-terminal region, ∼50 residues downstream from the IQ domain in CaV1.1 (27). Our ITC experiments and crystal structure show that this interaction proceeds with a Kd of ∼1 to 2 μM and involves association of this site with the first three MORN repeats of JPH2. Critical residues in this motif are conserved among CaV1.1 and CaV1.2, though mild substitutions are present in CaV1.3. The motif is absent in CaV1.4 and non–L-type CaVs. The interface residues are strongly conserved in all four JPH isoforms, implying that all of them may associate in a similar fashion. Including the joining region reduces the affinity to ∼8 µM, suggesting that it introduces some mild steric hindrance. Although a low micromolar affinity may seem modest, this needs to be seen in the context of muscle dyads and triads, where the local concentration of these components may be much higher by virtue of many additional interactions with various components. Interestingly, the joining region in JPH2 has also been proposed to bind CaV1.2 directly (28), but our data suggest that this binding site would reside elsewhere in the channel.
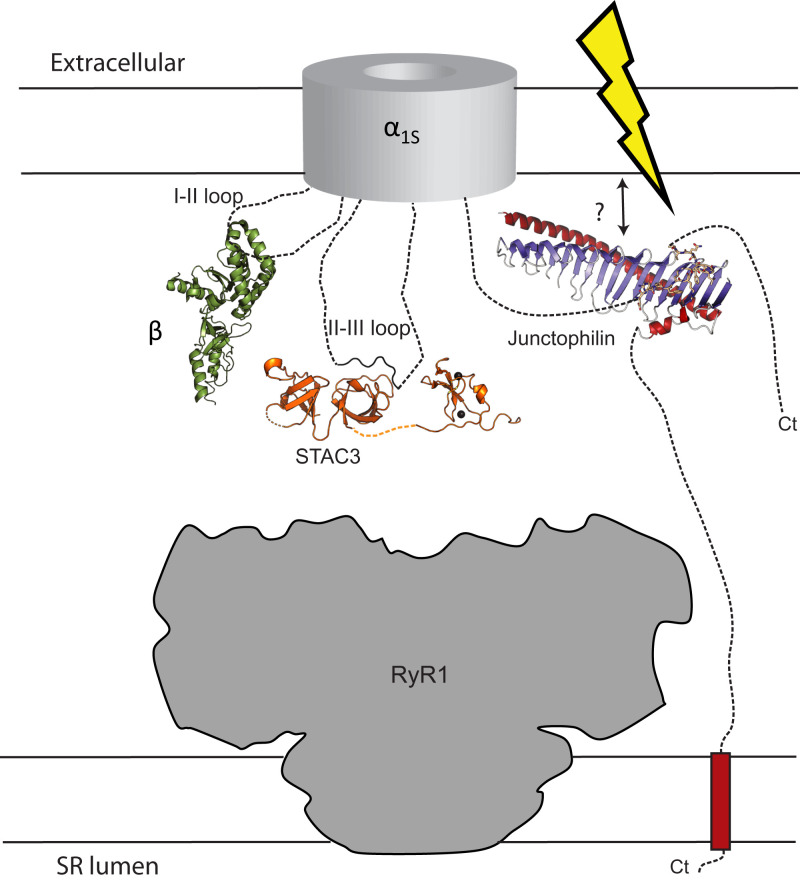
Rather than choosing individual point mutations, which may leave residual affinity, we introduced a triple-mutation in the CaV1.1 region that binds JPHs, removing side chains involved in extensive interactions. This is unlikely to affect the nearby IQ domain, as there are ∼50 residues in between. Using this triple mutant, we found that the interface is required for normal EC-coupling, as indicated by the reduced Ca2+-transients upon depolarization of the plasma membrane. However, still ∼56% remains, showing that the observed interface is not critical for EC-coupling. It is possible that an additional binding site for CaV1.1, formed by the joining region in JPH (28), compensates for this loss. In comparison, knocking out the interaction between STAC3 and the CaV1.1 II-III loop with a triple mutant in CaV1.1 or a single point mutation in STAC3 caused a much stronger reduction in EC-coupling (40–43). Fig. 5 shows a schematic overview of the five proteins deemed critical for EC-coupling (44). The strategic location of CaVβ1, STAC3, and JPH at the interface between Cav1.1 and RyR1 suggest that all three may cooperate in anchoring these two ion channels in skeletal muscle triads.
Fig. 5.
Essential components in skeletal muscle excitation-contraction coupling. Voltage-dependent conformational changes in the α1s subunit of CaV1.1 are transmitted mechanically to RyR1, located in the SR membrane. The CaVβ1 subunit, STAC3, and junctophilin are essential in this process, and all three are anchored to disordered regions of α1S (note: the CaVβ1 subunit is visible in cryoelectron microscopy structures of CaV1.1, but has poor local resolution, indicating its mobility). Possibly, all three may become immobilized upon interactions with RyR1 that remain to be identified.
Our JPH1 and JPH2 crystal structures also provide insights to pathogenic alternative splice variants of the homologous JPH3. Poly-CAG/CTG expansions in a variably spliced exon of the JPH3 gene cause HDL-2 (18, 45), with the disease phenotype either due to the mutant JPH3 transcripts causing cell toxicity (45) or lack of functional full length JPH3 in HDL-2 brains (18). These alternative splice variants only encode the first five MORN repeats of JPH3, followed by an alternative C-terminal sequence. Thus, lacking three MORNS and the α-helical region, such products are likely misfolded.
JPH2 forms a hot spot for sequence variants, most of which have been found in patients with HCM. Since JPH2 is also expressed in skeletal muscle, functional studies have also shown effects of several mutations in skeletal muscle (46, 47). Although only a limited number of them have been characterized functionally, our structures suggest that many may destabilize the folding of JPH2. A subset is found at the interface with the CaV target site and may act by reducing the affinity of this interaction.
In future experiments, it will be important to dissect how JPHs associate with RyRs and other ion channels.
Materials and Methods
Expression Constructs.
Clones for full-length human JPH1 and JPH2 were purchased from GenScript, corresponding to National Center for Biotechnology Information entry numbers NM_020647.4 and NM_020433.5. These were used as templates for PCR to produce clones used for ITC and X-ray crystallography. They were cloned into a modified pET28 vector (Novagen), containing a His6-tag, maltose-binding protein, and a cleavage site for the tobacco etch virus (TEV) protease (48). For the JPH1 (1 to 442) construct, we also prepared a version with a C-terminal strep tag, as this facilitated purification. SI Appendix, Table S4 outlines the exact constructs used. Constructs with the joining region removed were produced by overlapping extension PCR. Briefly, the cDNA sequences of JPH2 (residues 1 to 161) and JPH2 (residues 275 to 437) were amplified by PCR with overlapping primers in separate PCR reactions using full-length JPH2 as the template. The two separate PCR products were used as templates for a separate PCR using only the terminal primers, yielding the JPH2 construct (1 to 161, 275 to 437), used for crystallization. A similar strategy was used to produce JPH1 constructs with the joining region removed (1 to 161, 265 to 442) and (1 to 175, 265 to 442). The former allowed the formation of crystals, whereas the latter was used for ITC experiments due to its improved solubility. The E47A mutation in JPH2 (1 to 161, 275 to 442) was produced via the quikchange protocol (Agilent).
Protein Expression and Purification.
Proteins were expressed for 60 h in Escherichia coli Rosetta (DE3) pLacI (Novagen) grown in autoinduction medium (49) at 18 °C. Cells were lysed via sonication in 500 mM NaCl and 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes) at pH 7.4 (buffer A) supplemented with 1 mM PMSF, 25 µg/mL DNaseI, 25 µg/mL lysozyme, 10% (vol/vol) glycerol, and 40 mM CaCl2. Lysates were applied to HisTrap FF Crude columns (Cytiva), washed with 10 column volumes (CVs) of buffer A plus 20 mM imidazole, and eluted with buffer B (containing 500 mM NaCl, 250 mM imidazole and 20 mM Hepes pH 7.4). The elution was further purified by an amylose column (New England Biolabs), washed with 2 CVs of buffer A, and eluted with buffer A plus 10 mM maltose. Following cleavage of the fusion proteins by His-tagged TEV protease for 3 h at 4 °C, the proteins were applied to a Talon column (Clontech) in buffer A and eluted with buffer B. The collected flow-through fractions were diluted with 50 mM Hepes pH 7.0 to a final NaCl concentration of 250 mM, then applied to Resource S cation exchange column (Cytiva) and eluted with a gradient of 300 mM to 700 mM NaCl in 50 mM Hepes pH 7.0. The eluted protein was concentrated by using 10-kDa molelcular weight cutoff Amicon concentrators before loading onto a Superdex75 16/600 column (Cytiva) in buffer A supplemented with 2 mM Tris(2-carboxyethyl)phosphine (TCEP).
Crystallization and Data Collection.
The JPH1 construct (1 to 161, 265 to 442) was crystallized using sitting-drop or hanging-drop vapor diffusion at room temperature by mixing equal volumes of protein (3 mg/mL) and well solution, containing 0.5 M sodium acetate and 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) at pH 6.0. Crystals were soaked in a mixture of mother liquor and 25% (vol/vol) glycerol and flash-frozen in liquid N2. The JPH2 MORN-helical domain was crystallized by hanging-drop vapor diffusion at room temperature and by mixing equal volumes of protein (3 mg/mL) and well solution, which contained 5% (wt/vol) PEG 4000, 0.3 M sodium citrate, and 5% (vol/vol) isopropanol. Crystals were transferred to a cryosolution containing the mother liquor and 25% (vol/vol) glycerol and flash-frozen in liquid N2. The JPH2 construct (residues 1 to 161, 275 to 437) in complex with the synthetic minimal Oryctolagus cuniculus CaV1.1 peptide (obtained from LifeTein) was crystallized in a 1:50 ratio of JPH2:peptide by sitting-drop or hanging-drop vapor diffusion at room temperature by mixing an equal amount of protein and well solution, which contained 0.2 M K2SO4 and 12% (wt/vol) PEG 3350. The JPH2-peptide complex crystals were transferred to a drop containing mother liquor and 30% (vol/vol) glycerol before being flash-frozen in liquid N2. Diffraction data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline BL12-2 at wavelength 0.97946 Å and 80K and the Advanced Photon Source (APS) beamline 23-ID-D at wavelength 0.97937 Å and 80K. Datasets were processed by using HKL2000 (50) and xia2 (51) with SCALA (52).
Structural Determination and Refinement.
JPH1 construct (1 to 161, 265 to 442) was expressed in minimal media and supplemented with seleno-methionine and other amino acids, as described previously (53). SAD phasing was carried out via SHELXC/D/E (54); ARP/wARP (55) was used to build an initial model of the structure of JPH1 using high-resolution native datasets at 1.31 Å. The structure of JPH2 (residues 1 to 161, 275 to 437) was solved by molecular replacement via Phaser (56), using the JPH1 structure as the search model and refined at 2.35 Å. The structure of JPH2 in complex with the CaV1.1 peptide was solved by molecular replacement using the structure of JPH1 alone as the search model and refined at 2.03 Å. All models were completed with iterative cycles of manual model building in Coot (57) and refinement with Refmac5 in ccp4 (58) and Phenix (59). All structure files have been deposited in the Protein Data Bank (PDB) with accession codes 7RW4 (60) (JPH1), 7RXE (61) (JPH2), and 7RXQ (62) (JPH2 complex). For 7RW4, 98.11% and 1.89% of residues are in favored and allowed Ramachandran plot areas respectively. For 7RXE, 95.75% of residues are in favored areas and 4.28% are in allowed areas. For 7RXQ, 97.52% of residues are in favored and 2.48% are in allowed areas. No residues from the three structures are Ramachandran plot outliers.
Isothermal Titration Calorimetry.
All proteins were concentrated and dialyzed against 500 mM NaCl, 20 mM Hepes pH 7.4, and 2 mM TCEP at 4 °C. The synthetic CaV1.1 peptide was dissolved in the dialysis buffer. Protein concentrations were determined with the Edelhoch method (63). Titrations consisted of 20 injections of 2 µL with concentrations noted in the figure legends. Experiments were performed at 25 °C and using a stirring speed of 750 rpm on an ITC200 instrument (GE Healthcare). All data were processed by using Origin (v7.0), and isotherms were generated by following a point-by-point subtraction of a reference titration of ligand into buffer.
SEC-MALS.
Oligomeric states of various JPH1 and JPH2 were analyzed using SEC-MALS. A 100-μL sample of 1.5 mg/mL purified JPH were applied to a Superdex200 10/300 GL column (GE Healthcare) using a 1260 Infinity LC HPLC (Agilent Technologies). The mobile phase was 20 mM Hepes, 500 mM NaCl, 5 mM TCEP, pH 7.5 with a flow rate of 0.5 mL/min. Light-scattering data were collected using a miniDAWN TREOS MALS device (Wyatt Technologies) and the molecular masses were determined by applying a sphere model using the ASTRA6 program (Wyatt Technologies).
Dysgenic Myotube Culture and Transfection.
Cloning procedures for GFP-CaV1.1 were previously described (64). The triple RRFAAA mutation was introduced by PCR with overlapping extensions. Briefly, the cDNA of CaV1.1 (nucleotides 4483 to 5622) was amplified with overlapping mutagenesis primers in separate PCR reactions using GFP-CaV1.1 as template. The two PCR products were then used as templates for a final PCR with flanking primers to connect the nucleotide sequences. The obtained fragment was digested with BglII/AvrII and finally inserted in the corresponding sites of GFP-CaV1.1, yielding GFP-CaV1.1-RRFAAA. Myotubes of the dysgenic (mdg/mdg) cell line were cultured as previously described (65). At onset of myoblast fusion, 4 d after plating, cells plated on 35-mm dishes were transfected with 0.5 µg of CaV1.1 cDNA using FugeneHD (Promega).
Immunocytochemistry.
Paraformaldehyde-fixed cultures were double immunolabeled with the polyclonal rabbit anti-GFP antibody (serum, 1:10,000, A6455 Thermofisher Scientific) and the monoclonal mouse anti-RyR (34C, 1:500, MA3-925 Thermofisher Scientific) and with Alexa-488 and Alexa-594, respectively, as previously described (66). The 14-bit images were recorded with a cooled CCD camera (SPOT) and Metaview image processing software. Image composites were arranged in Adobe Photoshop CS6 and linear adjustments were performed to correct black level and contrast.
Clustering Quantification.
Clusters of CaV1.1 and RyR1 were quantified from acquired images by ImageJ software (NIH), as previously described (27). Briefly, the acquired images were converted to binary images using the intermodes threshold and particles larger than 0.198 µm2 were counted as clusters. The number of clusters of 30 myotubes from three separate experiments were counted. Graphs and statistical analysis (Student's t-test) were performed using the Graphpad software.
Electrophysiology and Fluorescent Calcium Measurements.
Calcium currents were measured at room temperature using whole-cell patch-clamp technique in voltage-clamp mode. The patch pipettes (borosilicate glass, Sutter Instruments) had resistance of 2 to 4 MΩ when filled with: 145 mM Cs-aspartate, 2 mM MgCl2, 10 mM Hepes, 0.1 mM Cs-EGTA, 2 mM Mg-ATP, and 0.2 mM Fluo-4, with pentapotassium salt to record calcium transients (pH 7.4 with CsOH). The extracellular bath solution contained: 10 mM CaCl2, 145 mM tetraethylammoniumchloride, 10 mM Hepes (pH 7.4 with CsOH). All recordings were performed with an HEKA amplifier (Harvard Bioscience). Voltage was stepped from the holding potential (−80 mV) to varying step potentials (from −60 mV to +80 mV) in steps of 10 mV for 500 ms. The current-voltage dependence was fitted according to: I = Gmax(V − Vrev)/(1 + exp(−(V − V1/2)/k)), where Gmax is the maximum conductance of CaV1.1, Vrev is the extrapolated reversal potential of the calcium current, V1/2 is the potential for half-maximal conductance, and k is the slope. The voltage dependence of Ca2+ transients were fitted according to: ΔF/F = (ΔF/F)max/(1 + exp(−(VF − V)/kF)) where (ΔF/F)max is the maximum fluorescence change, VF is the potential causing half of the maximal fluorescence change, and kF is the slope. SigmaPlot (v12.0; SPSS) was used for statistical analysis and curve fitting and Graphpad Prism (v9.1.2) was used to make the figures. All data are presented as mean ± SEM. Statistical comparisons of I − V and ΔF/F − V values at different voltages were obtained using a two-way repeated measures ANOVA. Statistical comparison of the fit parameters were obtained by using a Student's t-test.
Supplementary Material
Acknowledgments
We thank the staff at the Stanford Synchrotron Radiation Lightsource beamline BL12-2, and the Advanced Photon Source beamline 23-ID-D; Maren Becher, Nicole Kranebitten, and Enikō Török for excellent technical help; and Anson Chan for assistance with the in-house X-ray instrument, which was funded through the Canada Foundation for Innovation. F.V.P. acknowledges funding from the Canadian Institutes of Health Research (CIHR) (PJT159601). Z.F.Y. acknowledges the Frederick Banting and Charles Best Canada Graduate Scholarship-Master’s provided by the CIHR. M.C. acknowledges funding from the Austrian Science Fund (FWF P33776 and DOC30). W.E.T. is part of the CaVX PhD program co-funded by Fonds zur Förderung der Wissenschaftlichen Forschung (DOC30) and the Medical University Innsbruck.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
See online for related content such as Commentaries.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120416119/-/DCSupplemental.
Data Availability
The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 7RW4, 7RXQ, and 7RXE).
References
- 1.Takeshima H., Komazaki S., Nishi M., Iino M., Kangawa K., Junctophilins: A novel family of junctional membrane complex proteins. Mol. Cell 6, 11–22 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Bennett H. J., et al. , Human junctophilin-2 undergoes a structural rearrangement upon binding PtdIns(3,4,5)P3 and the S101R mutation identified in hypertrophic cardiomyopathy obviates this response. Biochem. J. 456, 205–217 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nishi M., Sakagami H., Komazaki S., Kondo H., Takeshima H., Coexpression of junctophilin type 3 and type 4 in brain. Brain Res. Mol. Brain Res. 118, 102–110 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Hogea A., et al. , Junctophilin-4 facilitates inflammatory signalling at plasma membrane-endoplasmic reticulum junctions in sensory neurons. J. Physiol. 599, 2103–2123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L., et al. , Junctophilin 3 expresses in pancreatic beta cells and is required for glucose-stimulated insulin secretion. Cell Death Dis. 7, e2275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woo J. S., et al. , Junctophilin-4, a component of the endoplasmic reticulum-plasma membrane junctions, regulates Ca2+ dynamics in T cells. Proc. Natl. Acad. Sci. U.S.A. 113, 2762–2767 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabiato A., Calcium-induced release of calcium from the cardiac sarcoplasmic reticulum. Am. J. Physiol. 245, C1–C14 (1983). [DOI] [PubMed] [Google Scholar]
- 8.Rufenach B., Van Petegem F., Structure and function of STAC proteins: Calcium channel modulators and critical components of muscle excitation-contraction coupling. J. Biol. Chem. 297, 100874 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flucher B. E., Campiglio M., STAC proteins: The missing link in skeletal muscle EC coupling and new regulators of calcium channel function. Biochim. Biophys. Acta Mol. Cell Res. 1866, 1101–1110 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Saeki T., Suzuki Y., Yamamura H., Takeshima H., Imaizumi Y., A junctophilin-caveolin interaction enables efficient coupling between ryanodine receptors and BKCa channels in the Ca2+ microdomain of vascular smooth muscle. J. Biol. Chem. 294, 13093–13105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan H. K., et al. , Functional interaction of junctophilin 2 with small- conductance Ca(2+)-activated potassium channel subtype 2(SK2) in mouse cardiac myocytes. Acta Physiol. (Oxf.) 222, e12986 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahu G., et al. , Junctophilin proteins tether a Cav1-RyR2-KCa3.1 tripartite complex to regulate neuronal excitability. Cell Rep. 28, 2427–2442.e6 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Jiang M., et al. , JPH-2 interacts with Cai-handling proteins and ion channels in dyads: Contribution to premature ventricular contraction-induced cardiomyopathy. Heart Rhythm 13, 743–752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pla-Martín D., et al. , Junctophilin-1 is a modifier gene of GDAP1-related Charcot-Marie-Tooth disease. Hum. Mol. Genet. 24, 213–229 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Landstrom A. P., et al. , Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J. Mol. Cell. Cardiol. 42, 1026–1035 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsushita Y., et al. , Mutation of junctophilin type 2 associated with hypertrophic cardiomyopathy. J. Hum. Genet. 52, 543–548 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Vanninen S. U. M., et al. , Heterozygous junctophilin-2 (JPH2) p.(Thr161Lys) is a monogenic cause for HCM with heart failure. PLoS One 13, e0203422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seixas A. I., et al. , Loss of junctophilin-3 contributes to Huntington disease-like 2 pathogenesis. Ann. Neurol. 71, 245–257 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Wang S., et al. , SERCA2a ameliorates cardiomyocyte T-tubule remodeling via the calpain/JPH2 pathway to improve cardiac function in myocardial ischemia/reperfusion mice. Sci. Rep. 11, 2037 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo A., et al. , Overexpression of junctophilin-2 does not enhance baseline function but attenuates heart failure development after cardiac stress. Proc. Natl. Acad. Sci. U.S.A. 111, 12240–12245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lahiri S. K., et al. , Nuclear localization of a novel calpain-2 mediated junctophilin-2 C-terminal cleavage peptide promotes cardiomyocyte remodeling. Basic Res. Cardiol. 115, 49 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo A., et al. , E-C coupling structural protein junctophilin-2 encodes a stress-adaptive transcription regulator. Science 362, eaan3303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy R. M., et al. , Ca2+-dependent proteolysis of junctophilin-1 and junctophilin-2 in skeletal and cardiac muscle. J. Physiol. 591, 719–729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C. Y., et al. , Calpain-dependent cleavage of junctophilin-2 and T-tubule remodeling in a mouse model of reversible heart failure. J. Am. Heart Assoc. 3, e000527 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi D., et al. , Molecular determinants of homo- and heteromeric interactions of Junctophilin-1 at triads in adult skeletal muscle fibers. Proc. Natl. Acad. Sci. U.S.A. 116, 15716–15724 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golini L., et al. , Junctophilin 1 and 2 proteins interact with the L-type Ca2+ channel dihydropyridine receptors (DHPRs) in skeletal muscle. J. Biol. Chem. 286, 43717–43725 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakada T., et al. , Physical interaction of junctophilin and the CaV1.1 C terminus is crucial for skeletal muscle contraction. Proc. Natl. Acad. Sci. U.S.A. 115, 4507–4512 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gross P., et al. , Interaction of the joining region in junctophilin-2 with the L-type Ca2+ channel is pivotal for cardiac dyad assembly and intracellular Ca2+ dynamics. Circ. Res. 128, 92–114 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perni S., Beam K., Neuronal junctophilins recruit specific CaV and RyR isoforms to ER-PM junctions and functionally alter CaV2.1 and CaV2.2. eLife 10, e64249 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mikami K., et al. , A dibasic amino acid pair conserved in the activation loop directs plasma membrane localization and is necessary for activity of plant type I/II phosphatidylinositol phosphate kinase. Plant Physiol. 153, 1004–1015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sajko S., et al. , Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats. PLoS One 15, e0242677 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang M., et al. , S-Palmitoylation of junctophilin-2 is critical for its role in tethering the sarcoplasmic reticulum to the plasma membrane. J. Biol. Chem. 294, 13487–13501 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beavers D. L., et al. , Mutation E169K in junctophilin-2 causes atrial fibrillation due to impaired RyR2 stabilization. J. Am. Coll. Cardiol. 62, 2010–2019 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seidelmann S. B., et al. , Application of whole exome sequencing in the clinical diagnosis and management of inherited cardiovascular diseases in adults. Circ. Cardiovasc. Genet. 10, e001573 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones E. G., et al. , Analysis of enriched rare variants in JPH2-encoded junctophilin-2 among Greater Middle Eastern individuals reveals a novel homozygous variant associated with neonatal dilated cardiomyopathy. Sci. Rep. 9, 9038 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quick A. P., et al. , Novel junctophilin-2 mutation A405S is associated with basal septal hypertrophy and diastolic dysfunction. JACC Basic Transl. Sci. 2, 56–67 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Bruijn S., et al. , A special case of hypertrophic cardiomyopathy with a differential diagnosis of isolated cardiac amyloidosis or junctophilin type 2 associated cardiomyopathy. Acta Clin. Belg. 76, 136–143 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Flucher B. E., Kasielke N., Grabner M., The triad targeting signal of the skeletal muscle calcium channel is localized in the COOH terminus of the alpha(1S) subunit. J. Cell Biol. 151, 467–478 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakada T., et al. , The proximal C-terminus of α(1C) subunits is necessary for junctional membrane targeting of cardiac L-type calcium channels. Biochem. J. 448, 221–231 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Wong King Yuen S. M., Campiglio M., Tung C. C., Flucher B. E., Van Petegem F., Structural insights into binding of STAC proteins to voltage-gated calcium channels. Proc. Natl. Acad. Sci. U.S.A. 114, E9520–E9528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rufenach B., et al. , Multiple sequence variants in STAC3 affect interactions with CaV1.1 and excitation-contraction coupling. Structure 28, 922–932.e5 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Horstick E. J., et al. , Stac3 is a component of the excitation-contraction coupling machinery and mutated in Native American myopathy. Nat. Commun. 4, 1952 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polster A., Nelson B. R., Olson E. N., Beam K. G., Stac3 has a direct role in skeletal muscle-type excitation-contraction coupling that is disrupted by a myopathy-causing mutation. Proc. Natl. Acad. Sci. U.S.A. 113, 10986–10991 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perni S., Lavorato M., Beam K. G., De novo reconstitution reveals the proteins required for skeletal muscle voltage-induced Ca2+ release. Proc. Natl. Acad. Sci. U.S.A. 114, 13822–13827 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rudnicki D. D., et al. , Huntington’s disease-like 2 is associated with CUG repeat-containing RNA foci. Ann. Neurol. 61, 272–282 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Woo J. S., et al. , S165F mutation of junctophilin 2 affects Ca2+ signalling in skeletal muscle. Biochem. J. 427, 125–134 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Woo J. S., et al. , Hypertrophy in skeletal myotubes induced by junctophilin-2 mutant, Y141H, involves an increase in store-operated Ca2+ entry via Orai1. J. Biol. Chem. 287, 14336–14348 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Petegem F., Clark K. A., Chatelain F. C., Minor D. L. Jr., Structure of a complex between a voltage-gated calcium channel beta-subunit and an alpha-subunit domain. Nature 429, 671–675 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Minor W., Cymborowski M., Otwinowski Z., Chruszcz M., HKL-3000: The integration of data reduction and structure solution--From diffraction images to an initial model in minutes. Acta Crystallogr. D Biol. Crystallogr. 62, 859–866 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Winter G., Lobley C. M., Prince S. M., Decision making in xia2. Acta Crystallogr. D Biol. Crystallogr. 69, 1260–1273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans P., Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J., Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993). [DOI] [PubMed] [Google Scholar]
- 54.Sheldrick G. M., Experimental phasing with SHELXC/D/E: Combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langer G., Cohen S. X., Lamzin V. S., Perrakis A., Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murshudov G. N., et al. , REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Z. F. Yang, P. Panwar, F. Van Petegem, Crystal structure of Junctophilin-1. Protein Data Bank (PDB). https://www.rcsb.org/structure/7RW4. Deposited 18 August 2021. [Google Scholar]
- 61.Z. F. Yang, P. Panwar, F. Van Petegem, Crystal structure of Junctophilin-2. Protein Data Bank (PDB). https://www.rcsb.org/structure/7RXE. Deposited 22 August 2021. [Google Scholar]
- 62.Z. F. Yang, P. Panwar, F. Van Petegem, Crystal structure of Junctophilin-1. Protein Data Bank (PDB). https://www.rcsb.org/structure/7RXQ. Deposited 23 August 2021. [Google Scholar]
- 63.Edelhoch H., Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry 6, 1948–1954 (1967). [DOI] [PubMed] [Google Scholar]
- 64.Grabner M., Dirksen R. T., Beam K. G., Tagging with green fluorescent protein reveals a distinct subcellular distribution of L-type and non-L-type Ca2+ channels expressed in dysgenic myotubes. Proc. Natl. Acad. Sci. U.S.A. 95, 1903–1908 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Powell J. A., Petherbridge L., Flucher B. E., Formation of triads without the dihydropyridine receptor alpha subunits in cell lines from dysgenic skeletal muscle. J. Cell Biol. 134, 375–387 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campiglio M., Di Biase V., Tuluc P., Flucher B. E., Stable incorporation versus dynamic exchange of β subunits in a native Ca2+ channel complex. J. Cell Sci. 126, 2092–2101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 7RW4, 7RXQ, and 7RXE).