Significance
While increasing evidence associates the disruption of circadian rhythms with pathologic conditions, including obesity, type 2 diabetes, and nonalcoholic fatty liver diseases (NAFLD), the involved mechanisms are still poorly described. Here, we show that, in both humans and mice, the pathogenesis of NAFLD is associated with the disruption of the circadian clock combined with perturbations of the growth hormone and sex hormone pathways. However, while this condition protects mice from the development of fibrosis and insulin resistance, it correlates with increased fibrosis in humans. This suggests that the perturbation of the circadian clock and its associated disruption of the growth hormone and sex hormone pathways are critical for the pathogenesis of metabolic and liver diseases.
Keywords: circadian clock, nonalcoholic fatty liver disease, insulin resistance, growth hormone, estrogen
Abstract
Obesity and liver diseases are associated with the disruption of the circadian clock that orchestrates mammalian physiology to optimize nutrient metabolism and storage. Here, we show that the activity of the circadian clock regulator Brain and Muscle Aryl hydrocarbon receptor nuclear translocator-like 1 (BMAL1) is perturbed during liver fibrosis in humans. To understand the impact of BMAL1 perturbation in obesity and liver diseases, we assessed the impact of a high fat diet or leptin deficiency on Bmal1 knockout mice. While Bmal1 knockout mice were prone to obesity, they were protected against insulin resistance, hepatic steatosis, inflammation, and fibrosis. In addition, to direct the transcriptional regulation of metabolic programs by BMAL1, we show that the disruption of the growth hormone and sex hormone pathways plays a critical role in this protection. Similar endocrine perturbations correlate with the development of liver fibrosis in humans but were absent in hepatocyte-specific Bmal1 knockout mice. This suggests that systemic endocrine perturbation associated with the global disruption of BMAL1 activity is critical for the pathogenesis of metabolic and liver diseases.
Nonalcoholic fatty liver disease (NAFLD) refers to a spectrum of liver diseases ranging from noninflammatory isolated steatosis to nonalcoholic steatohepatitis (NASH), which can progress to cirrhosis and hepatocellular carcinoma (HCC). NAFLD is a severe health burden, affecting around 25% of the world’s population and up to 90% of people with morbid obesity, and is associated with cardiometabolic risk factors and a sedentary lifestyle. Increasing evidence directly links obesity and NAFLD with disrupted circadian rhythms caused by a Western lifestyle. This phenomenon is termed “chronodisruption,” and people with a perturbed circadian clock reportedly exhibit a higher risk of developing obesity, NAFLD, and HCC (1–3). However, the underlying mechanisms remain poorly understood.
The circadian clock is an endogenous timing system that orchestrates most aspects of physiology and behavior, including nutrient uptake, storage, and metabolism. In this context, the circadian clock likely contributes to the evolutionary acquired genetic program that optimizes nutrient storage during periods of resource limitation (4). The mammalian circadian clock is hierarchically organized. Daily synchronization of the central clock localized in the suprachiasmatic nucleus of the hypothalamus is coordinated by light acting via the retina–hypothalamus tract. This, in turn, orchestrates clocks localized in peripheral tissues through systemic signals such as metabolic cues provided by food intake (5). In virtually all mammalian cells, rhythmicity is generated by a molecular machinery consisting of interconnected transcriptional and translational feedback loops. In mammals, the transcription factor protein Brain and Muscle Aryl hydrocarbon receptor nuclear translocator-like 1 (BMAL1, also named ARNTL) plays a critical role through the transcriptional regulation of the repressor complex composed of the CRYPTOCHROMEs and PERIODs proteins. A second loop involving the nuclear receptors of the ROR and REV-ERB families regulates Bmal1 expression (6).
Because of the central role of the circadian clock in the regulation of mammalian physiology, most animal models that are deficient in circadian clock genes exhibit metabolic phenotypes (7). For example, Bmal1-deficient mice have several metabolic defects, including high serum triglycerides (TAG) (8–10) and low circulating insulin associated with high insulin sensitivity (9, 11, 12). Associated with these metabolic defects, they present signs of early aging that start around 18 wk (13). However, the impact of Bmal1 deletion on obesity remains controversial; some studies reported higher (11, 14) or lower (8, 10, 13, 15) body weight as well as higher (8, 11, 14) or lower (10) body fat. To date, information about clock gene expression in human disease is still limited. In addition, the impact of global Bmal1 deletion on the pathogenesis of liver diseases remains unknown.
Here, we investigated correlations between obesity and liver diseases and the expression of clock genes in humans, as well as the impact of perturbing the circadian clock on obesity and liver diseases in mouse models. Surprisingly, while the expression of BMAL1-regulated circadian clock genes decreased with the evolution of liver fibrosis, we found that Bmal1 deficiency protects against steatohepatitis, inflammation, and fibrosis, explaining the high insulin sensitivity of Bmal1 knockout (KO) mice. These phenotypes are likely explained by endocrine adaptations associated with circadian clock depletion during mouse development and during the progression of liver disease in humans.
Results
Regulation of BMAL1 Activity in Obesity and Liver Diseases in Humans.
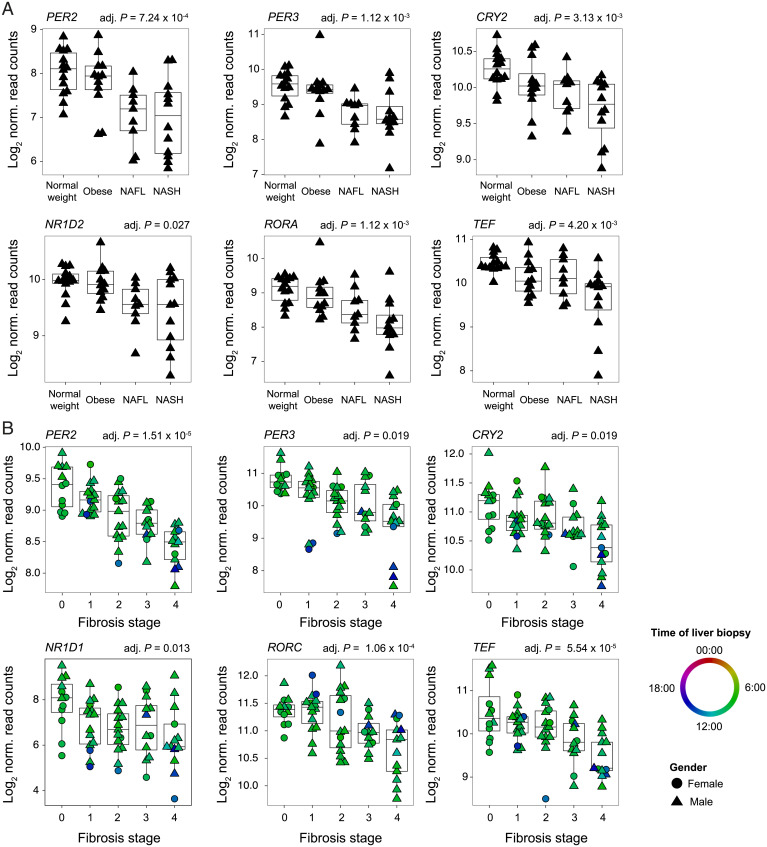
To study the impact of obesity and NAFLD pathogenesis on the expression of circadian clock genes, we analyzed RNA sequencing (RNA-seq) data from human liver biopsies collected from healthy volunteers as well as from obese volunteers and from patients with a spectrum of NAFLD, including simple steatosis (nonalcoholic fatty liver [NAFL]), and NASH (16). As shown in Fig. 1A, most of the genes encoding the core clock repressors (PER2, PER3, and CRY2), the ROR/REV-ERB loop (NR1D2 and RORA), and clock-regulated PARbZip transcription factors (TEF) showed reduced expression, mainly in patients with NAFL and NASH (Dataset S1). The genes encoding the core clock activators BMAL1, as well as its heterodimerization partners CLOCK and NPAS2, appeared marginally affected (SI Appendix, Fig. S1A). This decreased expression correlated with the level of fibrosis and inflammation as indicated by increased expression of related marker genes (SI Appendix, Fig. S1B). Because no information about the time of tissue collection was available for this dataset, we next performed a similar analysis in a cohort with recorded time of liver biopsy (tissues collected between 8:20 AM and 3:10 PM). Liver gene expression was analyzed in liver tissue from patients with various liver disease etiologies (chronic Hepatitis C virus infection, NASH, and alcohol-related liver disease) at different stages of liver fibrosis (17). While the expression of the genes encoding the circadian clock activators did not significantly change (SI Appendix, Fig. S1C), the expression of the genes encoding the circadian clock repressors (PER2, PER3, and CRY2), the ROR/REV-ERB loop (NR1D1 and RORC), and PARbZip transcription factors was significantly decreased, correlating with the increased expression of genes linked to fibrosis and inflammation (Fig. 1B, SI Appendix, Fig. S1D, and Dataset S1).
Fig. 1.
The regulation of BMAL1 activity in obesity and liver disease in humans. (A) The expression of circadian clock genes in a male human liver of normal weight, obese, simple steatosis (NAFL), or NASH. n ≥ 9 subjects per condition. (B) The expression of circadian clock genes in the liver of humans at different stages of fibrosis. Circles and triangles represent female and male individuals, respectively. n ≥ 12 subjects per stage. The color code represents the time of liver biopsy. P values were determined with a generalized linear model (A and B; for details, see Materials and Methods).
BMAL1, as a heterodimer with CLOCK and NPAS2, is the key regulator of circadian clock genes (6). To characterize a potential modification of BMAL1 activity that can explain these changes in gene expression, we predicted the activity of BMAL1 based on RNA-seq data from patients with liver fibrosis. This analysis confirmed a decreased activity of BMAL1 with the level of fibrosis (SI Appendix, Fig. S1E). These results are consistent with the hypothesis that liver disease–associated inflammation can interfere with the DNA-binding activity of BMAL1 and the organization of the circadian clock (18, 19) and suggest a link between BMAL1 activity and the progression of liver diseases.
Bmal1 KO Mice Fed a High Fat Diet Exhibit an Early Obese Phenotype but no Sign of Prediabetes.
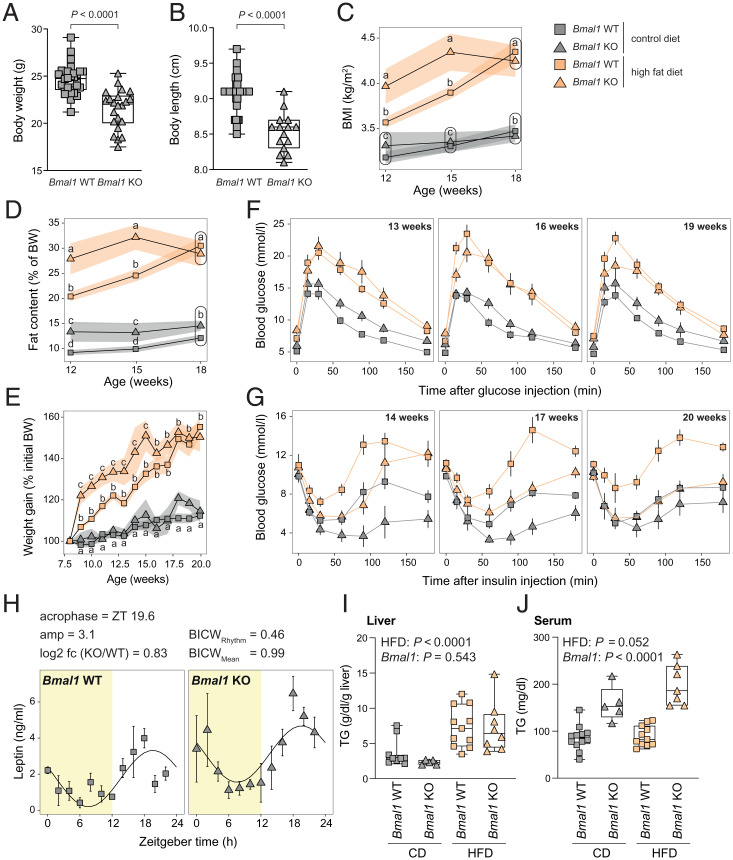
To explore the mechanistic aspects of this link, we studied the impact of Bmal1 deletion on the development of obesity and liver disease in mice. As reported previously (8, 10, 13, 15), at 8 wk of age, Bmal1 KO mice weighed less than their wild-type (WT) littermates (Fig. 2A). However, their lower body weight was accompanied by an unreported lower body length (Fig. 2B), leading to similar body mass indexes (BMI) between WT and KO (Fig. 2C). Low body weight and length likely reflect the poor growth of Bmal1 KO mice caused by a perturbed growth hormone (GH) secretion (20, 21) rather than a lean phenotype. The higher body fat content of Bmal1 KO mice detected throughout the experiment confirmed that Bmal1 KO mice are not leaner than their WT littermates (Fig. 2D). To study the kinetics of the metabolic impact of a high fat diet (HFD) on circadian clock–depleted animals, 8-wk-old WT and Bmal1 KO mice were placed on a control diet (CD) or an HFD for a total duration of 12 wk. Several metabolic tests were performed during the whole experiment to evaluate the appearance of metabolic disorders (SI Appendix, Fig. S2A). WT mice on an HFD expectedly had a continuous increase in body weight, BMI, and fat content (Fig. 2 C–E). While Bmal1 KO mice gained weight more rapidly than their WT littermates, both animal groups exhibited similar weight gain at the end of the experiment (Fig. 2E). Similar observations were made for BMI and fat content measured by NMR (Fig. 2 C and D). This observation suggested that while Bmal1 KO mice present signs of early obesity under HFD, they can rapidly adapt to this feeding regimen.
Fig. 2.
Bmal1 KO mice under HFD exhibit an early obese phenotype but no sign of prediabetes. (A and B) Body weight (A) and size (B) of Bmal1 of WT and KO mice before the treatment with HFD or CD. n = 22 to 32 mice (A) and 15 to 37 mice (B) per genotype. (C) BMI of indicated genotype and feeding regimen throughout the experiment. The size and weight were measured at ZT3. n = 6 to 19 mice per genotype and week. (D) Relative fat content measured by EchoMRI throughout the experiment. n = 6 to 19 mice per genotype and week. (E) The relative weight gain of Bmal1 WT and KO mice fed under HFD or CD. The body weight of the mice was measured at ZT3. n = 4 to 17 mice per condition and week. (F) Glucose tolerance tests performed on 13-, 16-, and 19-wk-old mice at ZT3 after 15 h of fasting. n = 6 to 17 mice per condition and week. (G) Insulin tolerance tests performed on 14-, 17-, and 20-wk-old mice at ZT3. n = 4 to 14 mice per condition and week. (H) The temporal serum leptin concentration in Bmal1 WT and KO under CD. n = 3 mice per time point and genotype. (I and J) Liver (I) and serum (J) triglycerides concentration at ZT12 in Bmal1 WT and KO mice fed with CD or HFD. n = 5 to 12 mice per condition. ZT is defined as follows: ZT0, lights on; ZT12, lights off. Statistical tests included unpaired Student’s t test (A and B), dryR (H), linear models (C–E; see details in Materials and Methods), and two-way ANOVA (I and J). Means with different letters are significantly different (P < 0.05; C–E).
While WT mice expectedly exhibited an increase in glucose levels after 6 h of refeeding, Bmal1 KO mice did not (SI Appendix, Fig. S2B). Under HFD, Bmal1 KO mice initially exhibited increased fasting glucose levels (SI Appendix, Fig. S2C). However, this phenotype was lost at the experimental endpoint. As observed previously, Bmal1 KO mice fed on either diet failed to increase glucose levels after 6 h of refeeding to the levels observed in WT mice (SI Appendix, Fig. S2C). This result suggests that glucose metabolism is impaired in Bmal1 KO mice. Therefore, we examined glucose tolerance, finding that Bmal1 KO mice on CD showed mild glucose intolerance (Fig. 2F and SI Appendix, Fig. S2D). Under HFD, both WT and Bmal1 KO mice exhibited similar glucose intolerance throughout the experiment. This detected glucose intolerance in Bmal1 KO mice is probably a consequence of blunted rhythmic glycogen synthesis, resulting in an overall 49% decreased level (SI Appendix, Fig. S2E) and is likely caused by a reduced expression of Glycogen Synthase 2 (Gys2) (22, 23).
As expected, WT mice on an HFD exhibited insulin resistance and a prediabetic state at the end of the experiment (Fig. 2G and SI Appendix, Fig. S2F). As previously reported (9, 11), Bmal1 KO mice under CD exhibited high insulin sensitivity (Fig. 2G and SI Appendix, Fig. S2F). Astonishingly, this high insulin sensitivity in Bmal1 KO mice further increased over time under HFD. Indeed, at the end of the experiment, Bmal1 KO on HFD mice showed the same insulin sensitivity as WT mice under CD (Fig. 2G, SI Appendix, Fig. S2F). As previously described (12), Bmal1 KO mice had very low insulin levels that did not increase under HFD, contrary to the expected increase seen in WT mice (SI Appendix, Fig. S2G). Altogether, we observed that, despite glucose intolerance, glucose levels in Bmal1 KO mice were maintained in a normal range after 12 wk of HFD due to their high and increasing insulin sensitivity and capacity to adapt to the HFD. We speculated that this effect may be mediated by leptin, which is able to maintain glucose levels even when insulin levels are low (24). This is a consequence of increased insulin sensitivity through effects on hepatic lipid and glucose metabolism (25–27). As previously described (8), circulating leptin levels are increased in Bmal1 KO mice (Fig. 2H), confirming this hypothesis and drawing our attention to lipid metabolism. Both WT and KO animals showed enlarged liver and epididymal white adipose tissue (eWAT) under HFD; however, this increased weight was lower in Bmal1 KO mice (SI Appendix, Fig. S2H). At the histological level, animals of both genotypes exhibited features of hepatic steatosis (SI Appendix, Fig. S2I) that appeared to be reduced in Bmal1 KO under HFD despite identical liver TAG content (Fig. 2I). On the other hand, Bmal1 KO mice showed increased circulating TAG levels under both CD and HFD (Fig. 2J).
Altogether, these data indicate that Bmal1 KO mice are prone to obesity but can adapt to this regimen and present limited signs of prediabetes or liver disease after 12 wk of HFD. Bmal1 KO animals also exhibited increased circulating TAG but decreased eWAT, suggesting a potential defect in the import and/or storage of TAG. However, we cannot exclude that the premature aging and the arthropathy developed by Bmal1 KO mice around 20 wk of age may interfere with this phenotype (13, 28). In addition, it remains unclear to what extent this phenotype can be directly linked to the deletion of Bmal1 or the compensatory effect of leptin in the condition of low insulin levels in Bmal1 KO.
Consequences of Bmal1 Deletion in the Absence of Leptin.
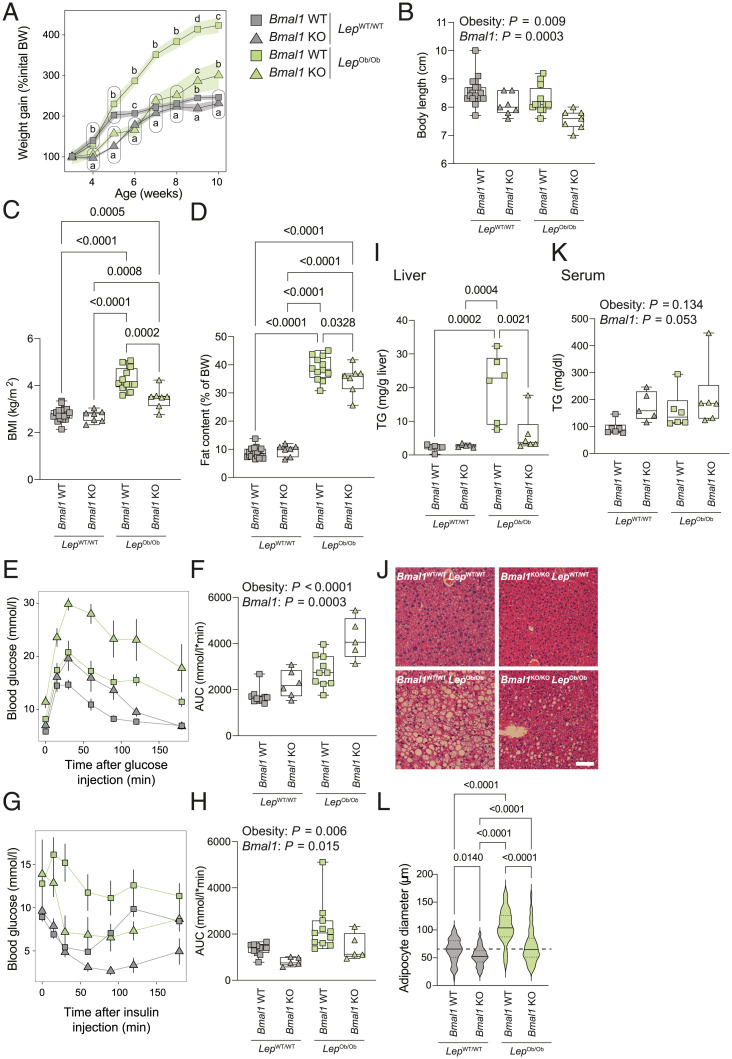
To decipher the importance of leptin in the phenotype of Bmal1 KO mice under HFD, we crossed Bmal1 KO mice with mice harboring the Ob allele, a spontaneous loss-of-function mutation of the Leptin gene (29). A first striking observation was that, while animals were born with the expected Mendelian ratio (SI Appendix, Fig. S3A), around 47% of the Bmal1KO/KO–LepOb/Ob (KO–Ob) died at 3 to 6 wk of age, suggesting a strong deleterious effect of the mutation of the Leptin gene in Bmal1 KO mice (SI Appendix, Fig. S3B). The animals that survived this stage were weighed weekly and subjected to several metabolic tests to study the impact of the deletion of Bmal1 on the obese phenotype of Ob/Ob mice (SI Appendix, Fig. S3C). All the experiments were performed in animals younger than 13 wk to limit the potential effect of premature aging observed in Bmal1 KO mice. While Bmal1WT/WT–LepOb/Ob (WT–Ob) expectedly gained weight rapidly, KO–Ob mice showed a similar weight gain compared to Bmal1WT/WT–LepWT/WT (WT–WT) and Bmal1KO/KO–LepWT/WT (KO–WT) mice (Fig. 3A). However, KO–Ob mice additionally showed a severe growth deficiency and a decreased body length (Fig. 3B), probably as a result of the cumulative effects of decreased insulin and GH caused by both the deletion of Bmal1 and the mutation of the Leptin gene (30–32). Therefore, both WT–Ob and KO–Ob mice exhibited an obese phenotype, though the BMI and fat content were slightly reduced in KO–Ob mice (Fig. 3 C and D). As observed when fed with HFD, the deletion of Bmal1 increased fasting blood glucose whether in association or not with the Ob mutation (SI Appendix, Fig. S3D). While WT–Ob and KO–WT were expectedly glucose intolerant, KO–Ob were severely glucose intolerant (Fig. 3 E and F and SI Appendix, Fig. S3E), confirming that the high leptin level in Bmal1 KO mice has a positive impact on their glucose tolerance in low insulin conditions. However, while WT–Ob mice were insulin resistant, Bmal1 deletion conferred increased insulin sensitivity even in obese KO–Ob mice (Fig. 3 G and H).
Fig. 3.
The consequences of Bmal1 deletion in the absence of leptin. (A) The evolution of the body weight of Bmal1KO/KO–LepOb/Ob (KO–Ob), Bmal1KO/KO–LepWT/WT (KO–WT), Bmal1WT/WT–LepOb/Ob (WT–Ob), and Bmal1WT/WT–LepWT/WT (WT–WT) mice. The body weight was measured at ZT3. n = 3 to 22 mice per genotype and week. (B–D) Size (B), BMI (C), and fat content (D) of 6-wk-old mice from the four genotypes of interest. The size, weight, and body fat measured by EchoMRI were measured At ZT3. n = 7 to 19 mice per genotype. (E and F) Glucose tolerance tests performed on 11-wk-old mice from the four genotypes at ZT3 after a 15-h fasting. Data represent glucose concentrations over time (E) and area under the curve (F). n = 5 to 10 mice per genotype. (G and H) Insulin tolerance tests performed on 12-wk-old mice from the four genotypes at ZT3. Data represent glucose concentrations over time (G) and area under the curve (H). n = 5 to 11 mice per genotype. (I) Liver triglycerides concentration measured at ZT12. n = 5 to 6 mice per genotype. (J) Representative liver sections stained with hematoxylin and eosin from each genotype of mice at ZT12. (Scale bar, 50 μm.) (K) Serum triglycerides concentration measured at ZT12. n = 5 to 6 mice per genotype. (L) The distribution of adipocytes size in eWAT from the different groups of mice at ZT12. The diameter (µm) of the adipocytes was determined from eWAT slices stained with hematoxylin and eosin (SI Appendix, Fig. S3H). n = 45 to 174 adipocytes per condition in representative mice of the cohort. Statistical tests included linear models (A; see details in Materials and Methods), and two-way ANOVA (B, F, H, and K) followed by a Holm–Šídák multiple comparison test (C, D, I, K, and L). Means with different letters are significantly different (P < 0.05; A).
While WT–Ob exhibited an enlarged liver and eWAT compared to WT–WT mice, only the eWAT was enlarged in KO–Ob and to a lesser extent than observed in WT–Ob mice (SI Appendix, Fig. S3F). The reduced liver weight of KO–Ob mice compared to WT–Ob mice correlated with lower liver TAG concentrations (Fig. 3I) and reduced steatosis (Fig. 3J and SI Appendix, Fig. S3G). However, KO–Ob showed higher circulating TAG levels like KO–WT mice (Fig. 3K), a characteristic of deficient lipid storage. This assumption was confirmed by the normal-sized adipocytes of KO–Ob mice compared to the enlarged adipocytes of WT–Ob mice (Fig. 3L and SI Appendix, Fig. S3H).
BMAL1 Controls Lipid Metabolism and Storage.
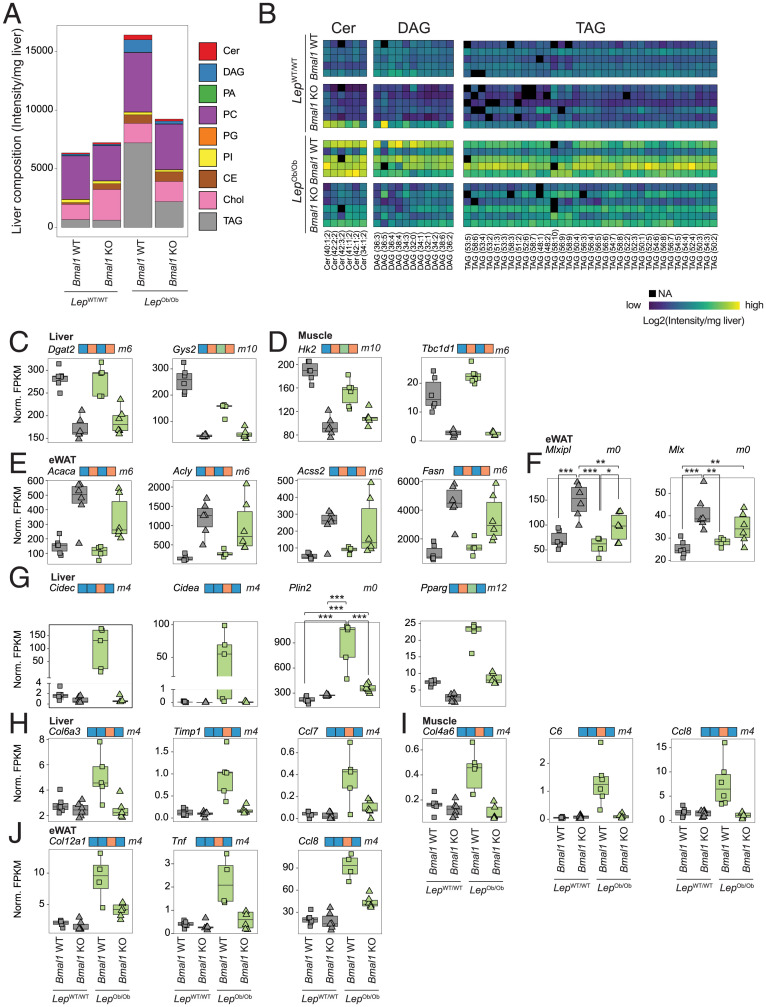
Abnormal lipid storage during obesity is associated with insulin resistance (33). Thus, we further studied the impact of Bmal1 deletion on lipid accumulation in the liver. To this end, we compared the lipidomic profiles of the livers from the four genotypes. We performed a differential analysis and grouped detected lipid species into statistical models to predict differential mean levels between the four genetic backgrounds (SI Appendix, Fig. S4A and Dataset S2). Unsurprisingly, liver lipid composition was strongly altered in WT–Ob mice at both quantitative and qualitative levels (Fig. 4 A and B and SI Appendix, Fig. S4 B–D). However, KO–Ob mice showed lower levels of glycerol esters (TAG and diacylglycerols [DAG]) compared to WT–Ob mice but also decreased Ceramides (Cer) and most phospholipids. Because DAG and Cer accumulations play a key role in hepatic inflammation and insulin resistance (34), their low levels in of KO–Ob mice livers likely contribute to the insulin sensitivity of these animals.
Fig. 4.
BMAL1 controls lipid metabolism and storage. (A) Global liver lipids composition analyzed by lipidomics in each group of mice liver at ZT12. Values represent average of n = 5 to 6 mice per genotype. TAG: triacylglycerides, Chol: cholesterol, CE: cholesteryl esters, PI: phosphatidylinositols, PG: phosphatidylglycerols, PC: phosphatidylcholines, PA: phosphatidic acid, DAG: diacylglycerides, Cer: ceramides. (B) A heatmap of the abundance of the lipid species associated with insulin resistance (Cer, DAG, and TAG) in the livers of individual mice. (C–E) The expression of genes encoding enzymes involved in glucose and lipid metabolism measured by RNA-seq at ZT12 in liver (C), muscle (D), and eWAT (E). n = 5 to 6 mice per genotype. (F) The expression of the carbohydrate-responsive element-binding protein and its heterodimerization partner measured by RNA-seq at ZT 12 in eWAT. N = 4 to 6 mice per genotype. (G) The expression of genes encoding proteins involved in lipid storage measured by RNA-seq in the liver at ZT12. N = 4 to 6 mice per genotype. (H–J) The expression of markers of inflammation and fibrosis measured by RNA-seq at ZT12 in liver (H), muscle (I), and eWAT (J). n = 4 to 6 mice per genotype. For statistical analysis, generalized linear models were employed (C–I; see details in Materials and Methods). Differences in expression level between the genotypes are defined in SI Appendix, Fig. S4A and are indicated on top of the graph. Genes that follow an ambiguous model are marked m0, and P values for each significant contrast are provided. ***P < 0.001, **P < 0.01, and *P < 0.05.
To further characterize the impact of Bmal1 deletion on lipid metabolism, we performed RNA-seq on liver, eWAT, and skeletal muscle (quadriceps) samples harvested at Zeitgeber time (ZT)12 in WT–WT, KO–WT, WT–Ob, and KO–Ob mice. We performed a similar differential gene expression analysis, grouping genes in the same models as described for lipids (SI Appendix, Fig. S4A and Dataset S3). This analysis revealed that the genes differentially expressed between WT and Bmal1 KO irrespective of their leptin status (WT and Ob background) (models 6 and 10) are expectedly involved in the regulation of the circadian clock. However, many genes of these models are also associated with metabolism and the storage of carbohydrate and lipids across all three tissues (SI Appendix, Fig. S5 and Dataset S4). This includes the decreased expression of Dgat2 and Gys2 in the liver (Fig. 4C) and Hk2 and Tbc1d1 in the muscle (Fig. 4D) of Bmal1 KO mice. More surprisingly, we found that Bmal1 deficiency resulted in an increased expression of most enzymes involved in de novo lipogenesis in eWAT, including Acaca, Acly, Acss2, Acsl1, Fasn, Elovl6, and Scd1 (Fig. 4E and SI Appendix, Fig. S6A). eWAT de novo lipogenesis is mainly regulated by the carbohydrate-responsive element binding protein (encoded by the Mlxipl gene), and its activation and expression are associated with increased insulin sensitivity (35, 36). Accordingly, the expression of Mlxipl and its dimerization partner Mlx are both increased in the eWAT of KO–WT and KO–Ob mice (Fig. 4F), likely also contributing to their increased insulin sensitivity.
Genes that are differentially expressed specifically in WT–Ob mice (models 4 and 14) are enriched in genes involved in inflammation, cytoskeleton and extracellular structure organizations, and lipid transport, suggesting that the decreased steatohepatitis in KO–Ob animals is associated with decreased inflammation and fibrosis (SI Appendix, Fig. S5 and Dataset S4). Accordingly, many liver PPAR-γ target genes involved in lipid droplet formation were increased only in WT–Ob mice. This is the case, for example, for cell-death–inducing DNA Fragmentation Factor subunit alpha (DFFA)–like effector C (Cidec or Fsp27) and Cidea (37), Perilipin 2 (Plin2) (38), and Pparg itself (Fig. 4G). A similar observation is apparent in livers from Bmal1 KO mice fed an HFD (SI Appendix, Fig. S6B), supporting the decreased steatosis in Bmal1 KO mice under HFD (SI Appendix, Fig. S2I). Interestingly, the rhythmic activity of the PPAR-γ pathway has been shown to be reprogrammed by HFD and obesity (39). This result provides evidence that Bmal1 is required for this reprogramming and the development of steatohepatitis by HFD via the transcriptional activation of Cidea and Cidec by PPAR-γ (37, 40). Thus, this confirms that the deletion of Bmal1 inhibits the formation of lipid droplets and lipid storage.
The decreased lipid accumulation in Bmal1 KO mice likely explains the decreased inflammation and fibrosis observed in these animals. Indeed, the high expression of markers of inflammation and fibrosis was observed only in WT–Ob mice: Col6a3, Timp1, or Ccl7 in the liver; Col4a6, C6, or Ccl8 in muscle; and Col12a1, Tnf, or Ccl8 in eWAT (Fig. 4 H–J). This inflammation is correlated with an increased infiltration of immune cells in WT–Ob mice as depicted by the expression of the macrophage markers Clec4e in the liver or Adgre1 (F4/80) in the eWAT and muscle (SI Appendix, Fig. S6C). Inflammation and fibrosis are important drivers of insulin resistance (41); hence, the observed global decrease in inflammation in all three tissues likely participates in the high insulin sensitivity of KO–Ob mice.
Gene Expression Analysis Revealed the Impact of Bmal1 Deletion on the Feminization of Gene Expression.
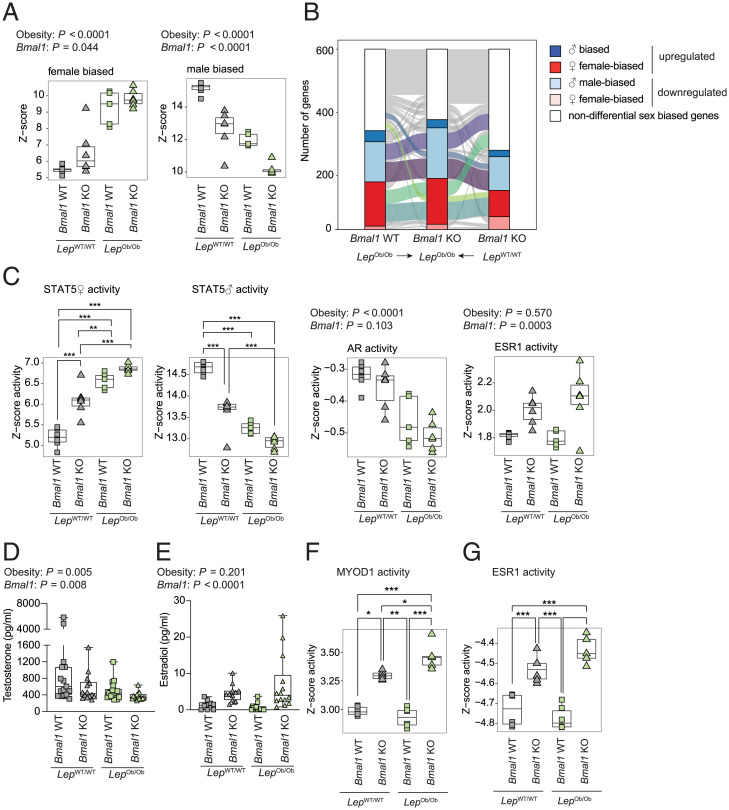
While direct regulation of transcription by BMAL1 likely plays a role in glucose and lipid metabolism, this effect unlikely explains the impact on fibrosis and inflammation. We have previously shown that Bmal1 deletion and obesity cause a feminization of liver gene expression (20), a condition known to protect against insulin resistance and NAFLD in rodents (42–45). Therefore, we studied sex-dimorphic gene expression in the liver transcriptome of the four genotypes (Dataset S3). A comparison of the differentially expressed sex-biased genes showed that KO–WT, WT–Ob, and KO–Ob mice exhibited a feminization of liver gene expression compared to WT–WT mice (Fig. 5A and SI Appendix, Fig. S7A). Obesity and the Bmal1 deletion contribution to feminization appear only partially redundant, as only 32% of the up-regulated female-biased and 39% of down-regulated male-biased genes were common among the three mouse models (Fig. 5B). Strikingly, the specific contribution of obesity and Bmal1 deletion to feminization is cumulative. Indeed, KO–Ob mice showed stronger feminization of liver gene expression compared to WT–Ob and KO–WT mice, particularly for the male-biased genes (Fig. 5 A and B and SI Appendix, Fig. S7A). To better evaluate the extent of the feminization of differential gene expression in KO–Ob mice, we compared differential sex-biased gene expression to WT female mice. Strikingly, among the differentially expressed genes in male KO–Ob mice, the reduction of male-biased gene expression was similar to WT female mice, while the expression of female-biased genes was very close (SI Appendix, Fig. S7B). This shows that the fraction of sex-biased genes that have an altered gene expression in the liver of KO–Ob mice strongly shifts its expression pattern toward a female liver. This change likely contributes to the improved inflammation and fibrosis observed in male KO–Ob mice.
Fig. 5.
Gene expression analysis revealed the impact of Bmal1 deletion on feminization of gene expression. (A) The differential expression of female- and male-biased genes in the liver of mice from the four genotypes at ZT12. n = 5 to 6 mice per genotype. (B) The clustered expression of the genes with similar sex-biased genes in the different genotypes at ZT12. (C) The predicted activity of transcription factors known to be involved in sex-biased gene expression in the liver. Activity is computed based on female (♀) and male (♂) STAT5, AR, and ESR1 target genes. n = 5 to 6 mice per genotype. (D and E) Concentrations of testosterone (D) and estradiol (E) in the serum of the mice of indicated genotypes at ZT12. n = 14 to 18 mice per genotype (D) and n = 10 to 16 mice per genotype (E). (F and G) The predicted activity of MYOD1 (F) and ESR1 (G) in skeletal muscle. n = 4 to 6 mice per genotype. P values were determined by a two-way ANOVA (A, C–G) followed by a Holm–Šídák multiple comparison test if interaction term was significant (P < 0.05). ***P < 0.001, **P < 0.01, and *P < 0.05.
To gain more insights into the pathways involved, we used published chromatin immunoprecipitation sequencing data to analyze the respective contribution of the two pathways involved in liver sex dimorphism: the GH pathway [male and female STAT5-binding sites (46)] and the sex hormones pathway [Androgen Receptor (AR) (47) and Estrogen Receptor (ESR1) (48)] (Fig. 5C). The sex-specific activity of the STAT5 pathway confirmed a cumulative impact of the Leptin mutation and Bmal1 deletion on the feminization of the GH pathway activity (Fig. 5C). This is further supported by the observed effects on animal growth (Fig. 3B), as growth changes reflect the predicted GH activity. A similar result was found with the AR motif analysis that predicted a decreased AR activity that further contributes to the observed feminization. Conversely, the increased activity of ESR1 appears to be only a consequence of the deletion of Bmal1 (Fig. 5C). These findings are further corroborated by the cognate changes in serum testosterone and 17β-estradiol (E2) (Fig. 5 D and E). Altogether, the differential activity of these pathways reflects the feminization of Bmal1 KO mice, contributing to their high insulin sensitivity, as already described in obese mouse models and humans (49–52).
Another striking observation in skeletal muscle RNA-seq data consisted of the shifts in muscle fibers composition in the mice from the four genotypes. This change is characterized by the differential expression of the type-specific myosin heavy chains 7 (Myh7, Type I), 2 (Myh2, Type IIa), 1 (Myh1, Type IIX), and 4 (Myh4, Type IIb) and slow type-specific Troponin I or T1 (Tnni1 and Tnnt1) and Myoglobin (Mb) (SI Appendix, Fig. S7C). This confirmed the previously described decrease in slow oxidative type I and IIa fibers in Bmal1-deficient mice that has been linked to decreased insulin-associated glucose uptake (53). Interestingly, obesity induced a similar shift in muscle fiber type (54, 55) known to correlate with inflammation (56, 57) and insulin resistance (58). Strikingly, KO–Ob mice showed a shift in muscle fibers composition, with a strong increase in slow fibers markers (Tnni1, Tnnt1, and Mb) and type IIx glycolytic fibers (SI Appendix, Fig. S7C). This increase in slow type fibers could likely contribute to the increased insulin sensitivity of KO–Ob mice. This shift is accompanied by the expression of the markers for muscle regeneration Myogenin (Myog) and embryonic Myh3 (SI Appendix, Fig. S7D) and a higher activity of MYOD1 (Fig. 5F). A similar increase in predicted ESR1 activity (Fig. 5G) in KO mice suggests a direct role for E2 in the observed muscle regeneration. Indeed, E2 is known to promote muscle regeneration and to reduce inflammation and fibrosis (59, 60). The high E2 concentration in Bmal1 KO mice likely also participates in the decreased fibrosis and inflammation in skeletal muscle, thus contributing to their high insulin sensitivity.
Feminization of Gene Expression Increases during Development of Liver Fibrosis in Humans.
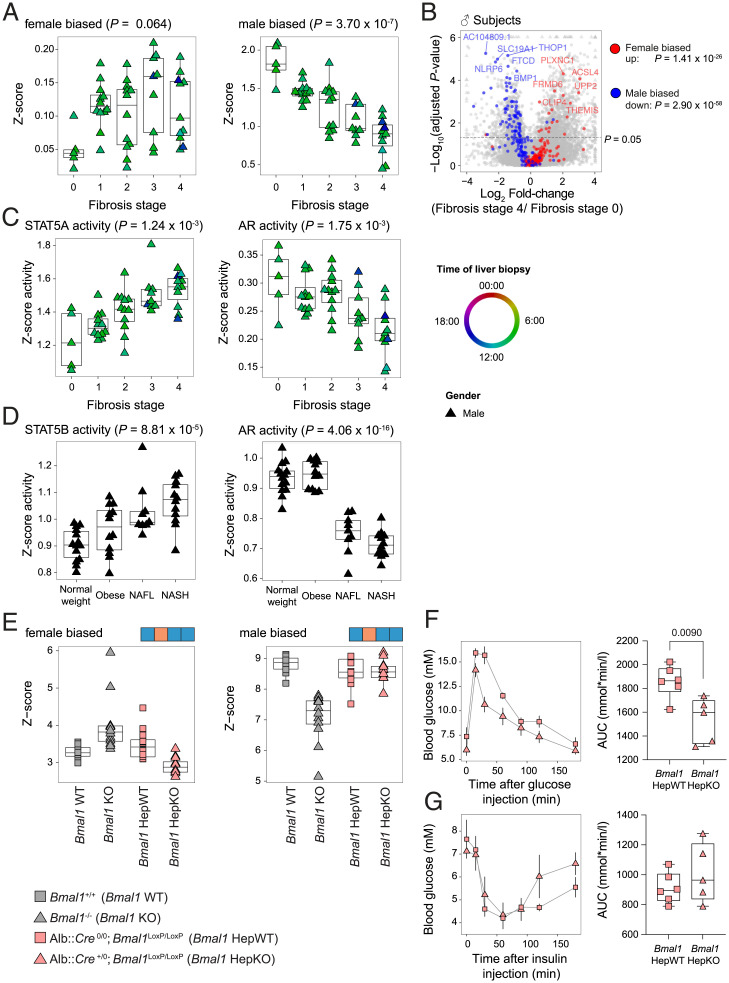
Our mouse model experiments pointed to a protective role for feminized liver gene expression associated with perturbed sex hormone concentrations against liver fibrosis. To test whether this is also the case in humans, we decided to next analyze sex-specific liver gene expression in male patients with liver fibrosis for investigating its potential perturbation by the disease. Therefore, we studied samples from patients with fibrosis stage 0 (5 males, 7 females) from the second cohort (17) to define the human liver sex-specific transcriptome (Dataset S1). Interestingly, despite the diverse etiologies of liver fibrosis in this cohort, we observed a clear alteration of the sex-biased gene expression during liver fibrosis that correlated with the stage of the disease. This is particularly pronounced for male-biased genes (Fig. 6 A and B). This feminization correlated with a decreased predicted activity of AR and an increased activity of STAT5, the transcription factor that conveys GH signaling (Fig. 6C). Because the healthy controls of the first cohort were only males (16), we were not able to identify sex-biased gene expression in this dataset. Nevertheless, a motif activity analysis confirmed the decreased predicted activity for AR and the increased predicted activity for STAT5 (Fig. 6D). Altogether, these results confirmed that simultaneous perturbations of the activity of the circadian clock regulator BMAL1 (Fig. 1 and SI Appendix, Fig. S1E) and the GH pathway, as well as the feminization of the liver associated with a decreased AR activity, are hallmarks of the progression of liver fibrosis and diseases.
Fig. 6.
The feminization of gene expression increases during development of NAFL and NASH in humans but not in Bmal1 HepKO mice. (A) The differential expression of female- and male-biased genes in the livers of humans at different stages of fibrosis. n ≥ 12 subjects per stage. (B) A volcano plot representing the differential expression of the sex-biased genes in the livers of individuals with fibrosis stage 0 or 4. Colors indicate male-biased (blue), female-biased (red), and unbiased (gray) expressed genes. n ≥ 12 subjects per stage. (C) The predicted activity of STAT5 and AR in the livers of individuals at different stage of fibrosis. (D) The predicted activity of STAT5 and AR in the livers of individuals with normal weight, obesity, simple steatosis (NAFL), or NASH. n ≥ 9 subjects per condition. (E) The differential expression of female- and male-biased genes in the livers of Bmal1 KO and Bmal1 HepKO mice (n = 12 per genotype). (F) Glucose tolerance tests performed on 9-wk-old Bmal1 HepKO and hepatocyte-specific WT (HepWT) littermates at ZT3 after a 15-h fasting. Data represent glucose concentrations over time (Left) and area under the curve (Right). n = 5 to 6 mice per genotype. (G and H) Insulin tolerance tests performed at ZT3 on 10-wk-old Bmal1 HepKO and HepWT littermates. Data represent glucose concentrations over time (Left) and area under the curve (Right). n = 5 to 6 mice per genotype. Statistical tests included linear models (A, C, and D; for details, see Materials and Methods), mean-rank gene set test (B), dryR (E), and two-tailed unpaired Student’s t test (F and G).
Finally, we wanted to investigate the possibility that the disruption of the circadian clock in hepatocytes initiates these detected endocrine perturbations. Therefore, we generated RNA-seq data from hepatocyte-specific Bmal1 KO mice (Bmal1 HepKO) and performed a similar analysis of differential expression of sex-biased genes in total Bmal1 KO and Bmal1 HepKO mice. In contrast to total Bmal1 KO mice, Bmal1 HepKO mice did not show any change of expression of sex-biased genes (Fig. 6E and Dataset S5). We rather observed a slight but not significant decrease in the expression of female-biased genes. An analysis of biological pathways confirmed that the genes exclusively different between total Bmal1 KO and Bmal1 HepKO livers (model 9) were enriched in sex biased functions like lipid, steroids, and drug metabolisms (Dataset S6). This observation confirms that the difference in gene expression between total and hepatocyte-specific Bmal1 KO is mainly explained by the absence of endocrine perturbation in Bmal1 HepKO. The absence of feminization was associated with no difference in insulin sensitivity but a slightly improved glucose tolerance as already described (11) (Fig. 6 F and G). This result suggests that the feminization of liver gene expression in humans with liver fibrosis or in Bmal1 KO mice is mainly caused by a systemic metabolic dysfunction than a specific effect of the hepatocyte circadian clock.
Discussion
While perturbations of the circadian clock have been associated with several human diseases, the molecular mechanisms involved are still poorly described. Here, we provided evidence in humans that BMAL1 activity is perturbed during liver fibrosis and NAFLD. Counterintuitively, our studies revealed that the depletion of Bmal1 protects against insulin resistance, steatohepatitis, inflammation, and fibrosis. This result suggests that while a high amplitude circadian clock is key to protect against the development of numerous diseases and pathologies, a fully functional BMAL1 can also have detrimental effects. In support of this concept, Bmal1 deficiency in the intestine protects against HFD obesity (61), while its deletion in macrophages improves defense against bacterial infection (62). Moreover, Bmal1 deletion in myeloid cells attenuates the development of atherosclerosis (63). Thus, our understanding of the potential consequences of perturbations of the circadian clock on physiology remains very limited.
Animal models have been used to decipher the role of the circadian clock in metabolic diseases, but many aspects are still controversial. While Bmal1 KO mice showed the same BMI as their WT littermates, we here describe that they gain weight more rapidly under HFD. However, obese Bmal1 KO mice had glucose concentrations in the normal range despite low insulin levels. This is likely due to their increased insulin sensitivity and higher leptin levels. In the absence of leptin, Bmal1 KO mice became severely glucose intolerant despite preserved high insulin sensitivity, confirming the protective impact of leptin in the Bmal1 KO mice. Nevertheless, more than half of these animals died at the time of weaning when they started to eat solid food (3 to 6 wk of age). The cause of death at weaning age is not clear, but a similar mortality phenotype is observed in numerous animal models of defective lipid metabolism and storage (64–68) and models of deficiency in the GH pathway (69, 70). Interestingly, most of these models are protected from HFD-induced obesity. Collectively, these results suggest that the circadian clock optimizes nutrient storage in peripheral tissues during the feeding phase and orchestrates their utilization during the fasting phase (71). Accordingly, Bmal1 KO mice present a deficient storage of glucose and lipids. Consequently, Bmal1 KO mice can adapt their physiology to an HFD regimen (10, 72), although they cannot properly adapt and survive under a 30% caloric restriction (73). Therefore, while the optimization of metabolism by the circadian clock provides an advantage under conditions of limited resources, it could contribute to the obesity associated with irregular feeding of a poor-quality diet as observed in mice and humans (74, 75).
While largely underestimated, endocrine perturbations caused by the deletion of Bmal1 seem to play a pivotal role in the phenotype of these KO mice. Here, we show that the changes in leptin, GH, and sex hormones are integral parts of the Bmal1 KO mice phenotype and contribute to their high insulin sensitivity as well as reduced fibrosis and inflammation. These systemic endocrine perturbations likely explain why Bmal1 KO mice can adapt to obesity and are protected from steatohepatitis and fibrosis. In contrast, hepatocyte-specific Bmal1 KO mice do not present a similar feminization of gene expression (Fig. 6E) but rather exhibit exacerbated steatohepatitis that ultimately leads to HCC (76–78). The increased E2 levels in male total Bmal1 KO mice likely play a role in this reduced inflammation and fibrosis. While elevated E2 levels have never been reported before for male Bmal1 KO mice, the described impairment in sex hormone synthesis and reproductive functions observed in Bmal1 KO mice likely play a causative role (79, 80). This impaired sex hormone synthesis could also be a consequence of the perturbed GH secretion observed in the KO mice (81, 82).
In mammals, the GH and sex hormone pathways play a critical role in establishing the sexual dimorphism of the liver and thus have a strong impact on liver physiology (83). Consequently, the prevalence of liver disease is also sex biased, and the incidence of NAFLD among premenopausal women is very low (84). This can be explained by the critical role of E2 in preventing insulin resistance, inflammation, and fibrosis (85). Interestingly, perturbations of GH and sex hormone pathways are frequently observed in patients with NAFL, NASH, and HCC, along with other pathologies associated with the disruption of circadian rhythms (86). Surprisingly, while this condition protects mice from NAFL and NASH, it is associated with poor outcomes in humans (87–90). Moreover, chronic liver diseases are correlated with an altered GH pathway (91, 92), and GH deficiency is associated with NAFLD (89, 93). This suggests that GH deficiency could have a protective role in the early stages of the disease but could be detrimental at later stages. GH deficiency is also associated with hypogonadism, characterized by low testosterone and high E2 (87, 94–96). While a lower activity of AR appears to protect from the development of HCC at the early stages (88, 97, 98), higher AR activity has a protective action at later stages (99). Therefore, early deficiency of the GH and AR pathways could protect from the development of NASH and continue to decrease during the course of the disease in an attempt to counteract its progression. At later stages, and as observed at the time of diagnosis, this leads to a more severe deficiency that correlates with a more advanced disease. This observation could also be true for the observed perturbed BMAL1 activity in NAFL and NASH (Fig. 1 and SI Appendix, Fig. S1E). In support of this hypothesis, the inactivation of the BMAL1 heterodimerization partner NPAS2 also protects against liver fibrosis (100), while the inactivation of the BMAL1 inhibitors PER2 and CRY1/CRY2 exacerbates fibrosis and inflammation (101, 102).
The secretion of GH and testosterone, both of which follow a diurnal rhythm with highest levels during sleep, are altered by sleep deprivation (103, 104). On the other hand, patients with liver disease and cirrhosis show perturbed sleep and daily rhythm of body temperature (105, 106). Decreased testosterone and GH levels as well as sleep problems caused by NAFLD are improved after liver transplantation, suggesting that the liver could play a role in these endocrine and circadian perturbations (107–109). However, inactivating Bmal1 only in hepatocytes has no impact on the circadian rhythm of the animals (11) and is not sufficient to drive the endocrine perturbations observed in animals with a global deletion of Bmal1 (Fig. 6E). Therefore, while the endocrine and circadian perturbations observed in patients with NAFLD certainly contribute to their complex metabolic disruption, the cause of these perturbations remains poorly understood.
Taken together, our comprehensive phenotyping of Bmal1 KO mice demonstrates a central role for the circadian clock in lipid and glucose metabolism. Furthermore, these results provide evidence that systemic endocrine perturbations caused by the disruption of BMAL1 activity play a role in the development of inflammation and liver fibrosis in both humans and mice. While some findings are associative in nature, these results support the idea that endocrine perturbations play a pivotal role in conveying the impact of circadian clock disruption on mammalian physiology.
Materials and Methods
Details about the described procedures and data analysis are available in SI Appendix.
Animal Experiments.
All animal studies were conducted in accordance with the regional committee for ethics in the regulations of the veterinary office of the Canton of Vaud, Switzerland, and the University of Queensland Animal Ethics Committee. The 8-wk-old male Bmal1 WT and KO mice (110) were fed with HFD or CD (no. D12492 and no. D12450J from Research Diet, respectively) for 12 wk to study the diet-induced obesity. Obese Bmal1 KO mice and control littermates were generated by crossing heterozygous mice for Bmal1 KO and the Ob mutated allele of Leptin, respectively (Bmal1KO/WT–LepOb/WT). The 6-wk-old male mice harboring the selected genotypes of interest were used for the experiments. The hepatocytes-specific deletion of Bmal1 was created by crossing Bmal1 floxed mice (111) with mice expressing the Cre recombinase under the control of the Albumin promoter (Alb–Cre) (112). In all experiments, unless noted otherwise, the mice were maintained under standard animal housing conditions, with free access to the food and water and in 12-h light/12-h dark cycles. Additional descriptions of the phenotyping procedures and metabolites measurements are provided in SI Appendix.
Lipid Extraction and Analysis.
Lipidomic analysis were performed as described previously (113, 114).
RNA Extractions and Analysis.
Liver RNAs were extracted as previously described (115). Muscle and epididymal white adipose tissue were disrupted using the tissue lyser II (Qiagen) and RNA extracted using the miRNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. Liver RNA were analyzed by real-time qRT-PCR as previously described (115). Sequencing libraries were prepared using the TruSeq Stranded RNA Sample Prep Kit v2 (Illumina) with the Ribo-Zero Gold depletion set.
Statistical Analysis.
Statistical analyses were performed with Prism 6.0 (GraphPad Software, Inc.) and R (116). Unless otherwise stated, statistical significance was determined for the comparison of two groups by Student’s t test for data following normal distribution. For analyses following a 2 × 2 factorial design, a two-way ANOVA was performed. If the interaction term was significant (P ≤ 0.05), we performed a Holm–Sidak multiple comparisons test. Differential rhythmicity analyses were performed using dryR (117). Longitudinal data that have been acquired over several weeks were analyzed using a linear model, and pairwise comparisons were computed using the CRAN package emmeans for each time point. Multiple testing was corrected with a Benjamini–Krieger–Yekutieli two-stage procedure across all contrasts (118). Except for box and violin plots, data are represented as mean ± SEM throughout the manuscript.
Supplementary Material
Acknowledgments
We thank Anabela Dacosta and Frédéric Preitner from the phenotyping facility of the University of Lausanne and José-Luis Sanchez-Garcia and Frédéric Raymond from Nestlé Research for helping in mouse and RNA-seq experiments, respectively. We would like to acknowledge the Mouse Pathology Facility of the University of Lausanne and the École Polytechnique Fédérale de Lausanne Histology Core Facility for performing histology. This work was supported by the European Research Council (through individual Starting Grant ERC-2010-StG-260988 to F.G.) and the Swiss NSF (P2LAP3_164906 to C.J.). C.J. and N.H.U. are supported by the Deutsche Forschungsgemeinschaft Trans-Regio TRR205 adrenal gland and CRC1064 chromatindynamics as well as the Deutsches Zentrum für Diabetesforschung. B.D.W. is supported by a University of Queensland Early Career Researcher Grant (UQECR2058233) and M.J.S. by a National Health and Medical Research Council of Australia Investigator Grant (APP1194406). F.G. receives support from the Institute for Molecular Bioscience, The University of Queensland.
Footnotes
Competing interest statement: C.J., B.D.W., E.M., F.A., C.G., A.C., D.M.-R., M.M., and F.G. are or were employees of Société des Produits Nestlé SA.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2200083119/-/DCSupplemental.
Data Availability
Raw files and technical details about the RNA-seq data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (119) and are accessible through GEO Series accession number GSE150102 (Bmal1-KO–Ob/Ob) (120) and GSE190221 (Bmal1 HepKO) (121). Previously published data were used for this work (122, 123). All other study data are included in the article and/or supporting information.
References
- 1.Mukherji A., Bailey S. M., Staels B., Baumert T. F., The circadian clock and liver function in health and disease. J. Hepatol. 71, 200–211 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.VoPham T., et al. , Circadian misalignment and hepatocellular carcinoma incidence in the United States. Cancer Epidemiol. Biomarkers Prev. 27, 719–727 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cedernaes J., Ramsey K. M., Bass J., “The role of circadian biology in health and disease” in Harrison’s Principles of Internal Medicine, 20E, Jameson J. L., et al., Eds. (McGraw-Hill Education, New York, NY, 2018). [Google Scholar]
- 4.Speakman J. R., Evolutionary perspectives on the obesity epidemic: Adaptive, maladaptive, and neutral viewpoints. Annu. Rev. Nutr. 33, 289–317 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Hastings M. H., Maywood E. S., Brancaccio M., Generation of circadian rhythms in the suprachiasmatic nucleus. Nat. Rev. Neurosci. 19, 453–469 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Patke A., Young M. W., Axelrod S., Molecular mechanisms and physiological importance of circadian rhythms. Nat. Rev. Mol. Cell Biol. 21, 67–84 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Mayeuf-Louchart A., Zecchin M., Staels B., Duez H., Circadian control of metabolism and pathological consequences of clock perturbations. Biochimie 143, 42–50 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Kennaway D. J., Varcoe T. J., Voultsios A., Boden M. J., Global loss of bmal1 expression alters adipose tissue hormones, gene expression and glucose metabolism. PLoS One 8, e65255 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudic R. D., et al. , BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2, e377 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shimba S., et al. , Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One 6, e25231 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamia K. A., Storch K.-F., Weitz C. J., Physiological significance of a peripheral tissue circadian clock. Proc. Natl. Acad. Sci. U.S.A. 105, 15172–15177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcheva B., et al. , Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature 466, 627–631 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondratov R. V., Kondratova A. A., Gorbacheva V. Y., Vykhovanets O. V., Antoch M. P., Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 20, 1868–1873 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo B., et al. , The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 26, 3453–3463 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang G., et al. , Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci. Transl. Med. 8, 324ra16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suppli M. P., et al. , Hepatic transcriptome signatures in patients with varying degrees of nonalcoholic fatty liver disease compared with healthy normal-weight individuals. Am. J. Physiol. Gastrointest. Liver Physiol. 316, G462–G472 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Ramnath D., et al. , Hepatic expression profiling identifies steatosis-independent and steatosis-driven advanced fibrosis genes. JCI Insight 3, e120274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong H.-K., et al. , Requirement for NF-κB in maintenance of molecular and behavioral circadian rhythms in mice. Genes Dev. 32, 1367–1379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maury E., Navez B., Brichard S. M., Circadian clock dysfunction in human omental fat links obesity to metabolic inflammation. Nat. Commun. 12, 2388 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weger B. D., et al. , The mouse microbiome is required for sex-specific diurnal rhythms of gene expression and metabolism. Cell Metab. 29, 362–382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schoeller E. L., Tonsfeldt K. J., Sinkovich M., Shi R., Mellon P. L., Growth hormone pulses and liver gene expression are differentially regulated by the circadian clock gene Bmal1. Endocrinology 162, bqab023 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doi R., Oishi K., Ishida N., CLOCK regulates circadian rhythms of hepatic glycogen synthesis through transcriptional activation of Gys2. J. Biol. Chem. 285, 22114–22121 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irimia J. M., et al. , Impaired glucose tolerance and predisposition to the fasted state in liver glycogen synthase knock-out mice. J. Biol. Chem. 285, 12851–12861 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chinookoswong N., Wang J. L., Shi Z. Q., Leptin restores euglycemia and normalizes glucose turnover in insulin-deficient diabetes in the rat. Diabetes 48, 1487–1492 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Shimomura I., Hammer R. E., Ikemoto S., Brown M. S., Goldstein J. L., Leptin reverses insulin resistance and diabetes mellitus in mice with congenital lipodystrophy. Nature 401, 73–76 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Kamohara S., Burcelin R., Halaas J. L., Friedman J. M., Charron M. J., Acute stimulation of glucose metabolism in mice by leptin treatment. Nature 389, 374–377 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Barzilai N., et al. , Leptin selectively decreases visceral adiposity and enhances insulin action. J. Clin. Invest. 100, 3105–3110 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bunger M. K., et al. , Progressive arthropathy in mice with a targeted disruption of the Mop3/Bmal-1 locus. Genesis 41, 122–132 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Lindström P., The physiology of obese-hyperglycemic mice [ob/ob mice]. Scientific World J. 7, 666–685 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raisingani M., Preneet B., Kohn B., Yakar S., Skeletal growth and bone mineral acquisition in type 1 diabetic children; abnormalities of the GH/IGF-1 axis. Growth Horm. IGF Res. 34, 13–21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Upadhyay J., Farr O. M., Mantzoros C. S., The role of leptin in regulating bone metabolism. Metabolism 64, 105–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luque R. M., Huang Z. H., Shah B., Mazzone T., Kineman R. D., Effects of leptin replacement on hypothalamic-pituitary growth hormone axis function and circulating ghrelin levels in ob/ob mice. Am. J. Physiol. Endocrinol. Metab. 292, E891–E899 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Borén J., Taskinen M. R., Olofsson S. O., Levin M., Ectopic lipid storage and insulin resistance: A harmful relationship. J. Intern. Med. 274, 25–40 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Petersen M. C., Shulman G. I., Roles of diacylglycerols and ceramides in hepatic insulin resistance. Trends Pharmacol. Sci. 38, 649–665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herman M. A., et al. , A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 484, 333–338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song Z., Xiaoli A. M., Yang F., Regulation and metabolic significance of de novo lipogenesis in adipose tissues. Nutrients 10, 1383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsusue K., et al. , Hepatic steatosis in leptin-deficient mice is promoted by the PPARgamma target gene Fsp27. Cell Metab. 7, 302–311 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bildirici I., et al. , The lipid droplet-associated protein adipophilin is expressed in human trophoblasts and is regulated by peroxisomal proliferator-activated receptor-γ/retinoid X receptor. J. Clin. Endocrinol. Metab. 88, 6056–6062 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Eckel-Mahan K. L., et al. , Reprogramming of the circadian clock by nutritional challenge. Cell 155, 1464–1478 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu W., et al. , Differential roles of cell death-inducing DNA fragmentation factor-α-like effector (CIDE) proteins in promoting lipid droplet fusion and growth in subpopulations of hepatocytes. J. Biol. Chem. 291, 4282–4293 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olefsky J. M., Glass C. K., Macrophages, inflammation, and insulin resistance. Annu. Rev. Physiol. 72, 219–246 (2010). [DOI] [PubMed] [Google Scholar]
- 42.Kamada Y., et al. , Estrogen deficiency worsens steatohepatitis in mice fed high-fat and high-cholesterol diet. Am. J. Physiol. Gastrointest. Liver Physiol. 301, G1031–G1043 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Riant E., et al. , Estrogens protect against high-fat diet-induced insulin resistance and glucose intolerance in mice. Endocrinology 150, 2109–2117 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Zhu L., et al. , Estrogen treatment after ovariectomy protects against fatty liver and may improve pathway-selective insulin resistance. Diabetes 62, 424–434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hewitt K. N., Pratis K., Jones M. E. E., Simpson E. R., Estrogen replacement reverses the hepatic steatosis phenotype in the male aromatase knockout mouse. Endocrinology 145, 1842–1848 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Laz E. V., Waxman D. J., Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell. Biol. 32, 880–896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang F.-P., et al. , Lack of androgen receptor SUMOylation results in male infertility due to epididymal dysfunction. Nat. Commun. 10, 777 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gertz J., et al. , Distinct properties of cell-type-specific and shared transcription factor binding sites. Mol. Cell 52, 25–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dakin R. S., Walker B. R., Seckl J. R., Hadoke P. W. F., Drake A. J., Estrogens protect male mice from obesity complications and influence glucocorticoid metabolism. Int. J. Obes. 39, 1539–1547 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dominici F. P., Hauck S., Argentino D. P., Bartke A., Turyn D., Increased insulin sensitivity and upregulation of insulin receptor, insulin receptor substrate (IRS)-1 and IRS-2 in liver of Ames dwarf mice. J. Endocrinol. 173, 81–94 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Coschigano K. T., et al. , Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased life span. Endocrinology 144, 3799–3810 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Guevara-Aguirre J., et al. , Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci. Transl. Med. 3, 70ra13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dyar K. A., et al. , Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 3, 29–41 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mastrocola R., et al. , Accumulation of advanced glycation end-products and activation of the SCAP/SREBP lipogenetic pathway occur in diet-induced obese mouse skeletal muscle. PLoS One 10, e0119587 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kaneko S., et al. , Changes in triacylglycerol-accumulated fiber type, fiber type composition, and biogenesis in the mitochondria of the soleus muscle in obese rats. Anat. Rec. (Hoboken) 294, 1904–1912 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Pellegrinelli V., et al. , Human adipocytes induce inflammation and atrophy in muscle cells during obesity. Diabetes 64, 3121–3134 (2015). [DOI] [PubMed] [Google Scholar]
- 57.Warfel J. D., et al. , Mitochondrial fat oxidation is essential for lipid-induced inflammation in skeletal muscle in mice. Sci. Rep. 6, 37941 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stuart C. A., et al. , Slow-twitch fiber proportion in skeletal muscle correlates with insulin responsiveness. J. Clin. Endocrinol. Metab. 98, 2027–2036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kitajima Y., Ono Y., Estrogens maintain skeletal muscle and satellite cell functions. J. Endocrinol. 229, 267–275 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Sugiura T., et al. , Estrogen administration attenuates immobilization-induced skeletal muscle atrophy in male rats. J. Physiol. Sci. 56, 393–399 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Yu F., et al. , Deficiency of intestinal Bmal1 prevents obesity induced by high-fat feeding. Nat. Commun. 12, 5323 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kitchen G. B., et al. , The clock gene Bmal1 inhibits macrophage motility, phagocytosis, and impairs defense against pneumonia. Proc. Natl. Acad. Sci. U.S.A. 117, 1543–1551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang G., et al. , Bmal1 deletion in myeloid cells attenuates atherosclerotic lesion development and restrains abdominal aortic aneurysm formation in hyperlipidemic mice. Arterioscler. Thromb. Vasc. Biol. 40, 1523–1532 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aksentijević D., et al. , Cardiac dysfunction and peri-weaning mortality in malonyl-coenzyme A decarboxylase (MCD) knockout mice as a consequence of restricting substrate plasticity. J. Mol. Cell. Cardiol. 75, 76–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fan Y.-Y., et al. , Characterization of an arachidonic acid-deficient (Fads1 knockout) mouse model. J. Lipid Res. 53, 1287–1295 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fischer J., et al. , Inactivation of the Fto gene protects from obesity. Nature 458, 894–898 (2009). [DOI] [PubMed] [Google Scholar]
- 67.Moitra J., et al. , Life without white fat: A transgenic mouse. Genes Dev. 12, 3168–3181 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shan T., Xiong Y., Kuang S., Deletion of Lkb1 in adult mice results in body weight reduction and lethality. Sci. Rep. 6, 36561 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu J.-L., LeRoith D., Insulin-like growth factor I is essential for postnatal growth in response to growth hormone. Endocrinology 140, 5178–5184 (1999). [DOI] [PubMed] [Google Scholar]
- 70.Zhu X., et al. , Disruption of PC1/3 expression in mice causes dwarfism and multiple neuroendocrine peptide processing defects. Proc. Natl. Acad. Sci. U.S.A. 99, 10293–10298 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yeung J., et al. , Transcription factor activity rhythms and tissue-specific chromatin interactions explain circadian gene expression across organs. Genome Res. 28, 182–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hemmeryckx B., Himmelreich U., Hoylaerts M. F., Lijnen H. R., Impact of clock gene Bmal1 deficiency on nutritionally induced obesity in mice. Obesity (Silver Spring) 19, 659–661 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Patel S. A., Chaudhari A., Gupta R., Velingkaar N., Kondratov R. V., Circadian clocks govern calorie restriction-mediated life span extension through BMAL1- and IGF-1-dependent mechanisms. FASEB J. 30, 1634–1642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gill S., Panda S., A smartphone app reveals erratic diurnal eating patterns in humans that can be modulated for health benefits. Cell Metab. 22, 789–798 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hatori M., et al. , Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kettner N. M., et al. , Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 30, 909–924 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang D., et al. , The hepatic BMAL1/AKT/lipogenesis axis protects against alcoholic liver disease in mice via promoting PPARα pathway. Hepatology 68, 883–896 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacobi D., et al. , Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab. 22, 709–720 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alvarez J. D., et al. , The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice. J. Biol. Rhythms 23, 26–36 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Boden M. J., Varcoe T. J., Voultsios A., Kennaway D. J., Reproductive biology of female Bmal1 null mice. Reproduction 139, 1077–1090 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Chandrashekar V., Bartke A., Coschigano K. T., Kopchick J. J., Pituitary and testicular function in growth hormone receptor gene knockout mice. Endocrinology 140, 1082–1088 (1999). [DOI] [PubMed] [Google Scholar]
- 82.Keene D. E., et al. , Puberty is delayed in male growth hormone receptor gene-disrupted mice. J. Androl. 23, 661–668 (2002). [PubMed] [Google Scholar]
- 83.Lefebvre P., Staels B., Hepatic sexual dimorphism - implications for non-alcoholic fatty liver disease. Nat. Rev. Endocrinol. 17, 662–670 (2021). [DOI] [PubMed] [Google Scholar]
- 84.Lonardo A., Suzuki A.; Focus on Clinical Aspects and Implications for Practice and Translational Research, Sexual dimorphism of NAFLD in adults. J. Clin. Med. 9, 1278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Della Torre S., Non-alcoholic fatty liver disease as a canonical example of metabolic inflammatory-based liver disease showing a sex-specific prevalence: Relevance of estrogen signaling. Front. Endocrinol. (Lausanne) 11, 572490 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weger B. D., Rawashdeh O., Gachon F., At the intersection of microbiota and circadian clock: Are sexual dimorphism and growth hormones the missing link to pathology?: Circadian clock and microbiota: Potential effect on growth hormone and sexual development. BioEssays 41, e1900059 (2019). [DOI] [PubMed] [Google Scholar]
- 87.Grossmann M., et al. , Low testosterone levels as an independent predictor of mortality in men with chronic liver disease. Clin. Endocrinol. (Oxf.) 77, 323–328 (2012). [DOI] [PubMed] [Google Scholar]
- 88.Tanaka K., Sakai H., Hashizume M., Hirohata T., Serum testosterone:estradiol ratio and the development of hepatocellular carcinoma among male cirrhotic patients. Cancer Res. 60, 5106–5110 (2000). [PubMed] [Google Scholar]
- 89.Ichikawa T., et al. , Non-alcoholic steatohepatitis and hepatic steatosis in patients with adult onset growth hormone deficiency. Gut 52, 914 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagasue N., Ogawa Y., Yukaya H., Ohta N., Ito A., Serum levels of estrogens and testosterone in cirrhotic men with and without hepatocellular carcinoma. Gastroenterology 88, 768–772 (1985). [DOI] [PubMed] [Google Scholar]
- 91.Cuneo R. C., et al. , Altered endogenous growth hormone secretory kinetics and diurnal GH-binding protein profiles in adults with chronic liver disease. Clin. Endocrinol. (Oxf.) 43, 265–275 (1995). [DOI] [PubMed] [Google Scholar]
- 92.Assy N., et al. , Growth hormone-stimulated IGF-1 generation in cirrhosis reflects hepatocellular dysfunction. J. Hepatol. 49, 34–42 (2008). [DOI] [PubMed] [Google Scholar]
- 93.Takahashi Y., et al. , Growth hormone reverses nonalcoholic steatohepatitis in a patient with adult growth hormone deficiency. Gastroenterology 132, 938–943 (2007). [DOI] [PubMed] [Google Scholar]
- 94.Green G. R., Mechanism of hypogonadism in cirrhotic males. Gut 18, 843–853 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaymakoğlu S., et al. , Hypogonadism is not related to the etiology of liver cirrhosis. J. Gastroenterol. 30, 745–750 (1995). [DOI] [PubMed] [Google Scholar]
- 96.Castilla-Cortazar I., et al. , Insulin-like growth factor-I reverts testicular atrophy in rats with advanced cirrhosis. Hepatology 31, 592–600 (2000). [DOI] [PubMed] [Google Scholar]
- 97.Li H., Li Y., Lu J.-W., Huo X., Gong Z., Liver-specific androgen receptor knockout attenuates early liver tumor development in zebrafish. Sci. Rep. 9, 10645 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma W. L., et al. , Androgen receptor is a new potential therapeutic target for the treatment of hepatocellular carcinoma. Gastroenterology 135, 947–955 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ma W.-L., et al. , Hepatic androgen receptor suppresses hepatocellular carcinoma metastasis through modulation of cell migration and anoikis. Hepatology 56, 176–185 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang T., et al. , NPAS2 contributes to liver fibrosis by direct transcriptional activation of Hes1 in hepatic stellate cells. Mol. Ther. Nucleic Acids 18, 1009–1022 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen P., et al. , Loss of clock gene mPer2 promotes liver fibrosis induced by carbon tetrachloride. Hepatol. Res. 40, 1117–1127 (2010). [DOI] [PubMed] [Google Scholar]
- 102.Mteyrek A., et al. , Critical cholangiocarcinogenesis control by cryptochrome clock genes. Int. J. Cancer 140, 2473–2483 (2017). [DOI] [PubMed] [Google Scholar]
- 103.Brandenberger G., Gronfier C., Chapotot F., Simon C., Piquard F., Effect of sleep deprivation on overall 24 h growth-hormone secretion. Lancet 356, 1408 (2000). [DOI] [PubMed] [Google Scholar]
- 104.Luboshitzky R., Zabari Z., Shen-Orr Z., Herer P., Lavie P., Disruption of the nocturnal testosterone rhythm by sleep fragmentation in normal men. J. Clin. Endocrinol. Metab. 86, 1134–1139 (2001). [DOI] [PubMed] [Google Scholar]
- 105.Bruyneel M., Sersté T., Sleep disturbances in patients with liver cirrhosis: Prevalence, impact, and management challenges. Nat. Sci. Sleep 10, 369–375 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garrido M., et al. , Abnormalities in the 24-hour rhythm of skin temperature in cirrhosis: Sleep-wake and general clinical implications. Liver Int. 37, 1833–1842 (2017). [DOI] [PubMed] [Google Scholar]
- 107.Bassanello M., et al. , Growth hormone/insulin-like growth factor 1 axis recovery after liver transplantation: A preliminary prospective study. Liver Transpl. 10, 692–698 (2004). [DOI] [PubMed] [Google Scholar]
- 108.Bhat M., et al. , Prevalence and predictors of sleep disturbance among liver diseases in long-term transplant survivors. Can. J. Gastroenterol. Hepatol. 29, 440–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Madersbacher S., Grünberger T., Maier U., Andrological status before and after liver transplantation. J. Urol. 151, 1251–1254 (1994). [DOI] [PubMed] [Google Scholar]
- 110.Jouffe C., et al. , The circadian clock coordinates ribosome biogenesis. PLoS Biol. 11, e1001455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Storch K.-F., et al. , Intrinsic circadian clock of the mammalian retina: Importance for retinal processing of visual information. Cell 130, 730–741 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Postic C., et al. , Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic β cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274, 305–315 (1999). [DOI] [PubMed] [Google Scholar]
- 113.Chakrabarti A., et al. , Transcriptomics-driven lipidomics (TDL) identifies the microbiome-regulated targets of ileal lipid metabolism. NPJ Syst. Biol. Appl. 3, 33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chakrabarti A., et al. , Impact of multi-micronutrient supplementation on lipidemia of children and adolescents. Clin. Nutr. 39, 2211–2219 (2020). [DOI] [PubMed] [Google Scholar]
- 115.Gachon F., Olela F. F., Schaad O., Descombes P., Schibler U., The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 4, 25–36 (2006). [DOI] [PubMed] [Google Scholar]
- 116.Team R. C., R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2018). [Google Scholar]
- 117.Weger B. D., et al. , Systematic analysis of differential rhythmic liver gene expression mediated by the circadian clock and feeding rhythms. Proc. Natl. Acad. Sci. U.S.A. 118, e2015803118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Benjamini Y., Krieger A. M., Yekutieli D., Adaptive linear step-up procedures that control the false discovery rate. Biometrika 93, 491–507 (2006). [Google Scholar]
- 119.Edgar R., Domrachev M., Lash A. E., Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.C. Jouffe et al., Effect of leptin deficiency in Bmal1 knockout mice across different tissues. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE150102. Deposited 8 May 2020. [Google Scholar]
- 121.C. Jouffe et al., Diurnal profiling of mouse liver of hepatocyte specific BMAL1 KO mice (Bmal1 HepKO). Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE190221. Deposited 6 December 2021. [Google Scholar]
- 122.K. Rigbolt, J. C. Nielsen, Hepatic transcriptome signatures in patients with varying degrees of non-alcoholic fatty liver disease compared to healthy normal-weight individuals. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126848. Accessed 26 March 2019. [DOI] [PubMed] [Google Scholar]
- 123.D. Ramnath et al., RNA-seq of hepatic biopsies taken from patients with chronic liver disease presenting with different aetiologies (HCV, FLD) and fibrosis stages. European Nucleotide Archive. https://www.ebi.ac.uk/ena/browser/view/PRJEB27201. Accessed 28 July 2018. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw files and technical details about the RNA-seq data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (119) and are accessible through GEO Series accession number GSE150102 (Bmal1-KO–Ob/Ob) (120) and GSE190221 (Bmal1 HepKO) (121). Previously published data were used for this work (122, 123). All other study data are included in the article and/or supporting information.