Artificial intelligence is ushering in a revolution in structural biology. How far will it go?
Twenty-eight years ago, computational biologists led by John Moult then at the University of Maryland, Baltimore, launched an ambitious, large-scale experiment designed to answer one of the most challenging open questions in biology: How can researchers determine the structure of any protein?
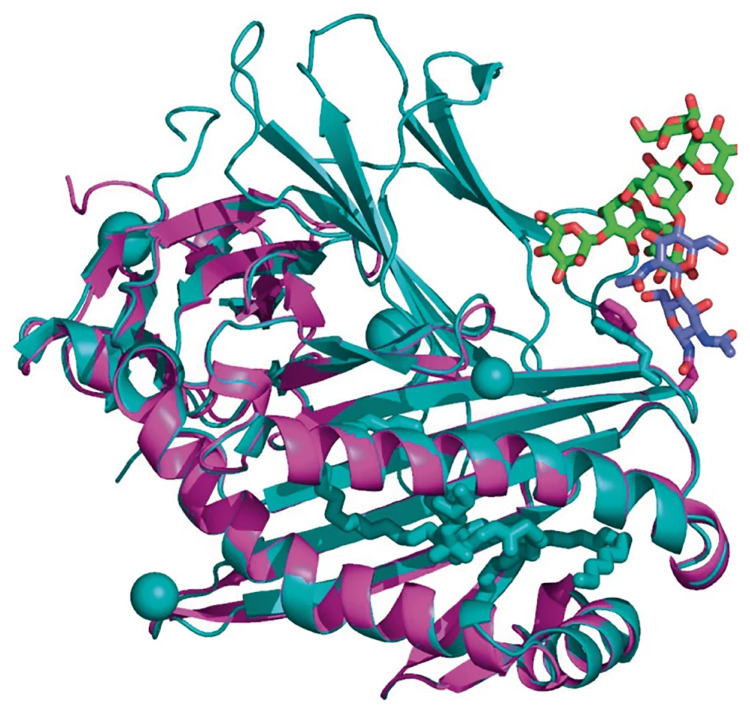
AlphaFold uses AI to predict the shapes of proteins; structural biologists are using the program to deepen our understanding of the big molecules. This image shows AlphaFold's predicted structure (in magenta) of a glycoprotein found on the surface of a T cell. Researchers used other data to complete the structure (in cyan). Image credit: Reprinted with permission from Springer Nature: ref. 10.
Structure is key to understanding biological function because a the shape of a protein directly determines what it does. But proteins are complex molecules comprising linked chains of hundreds or even thousands of amino acids, wound and folded into tortuous coils and pleats that bond and twist into myriad configurations. If that weren’t enough, proteins can also change over time, further bedeviling researchers’ efforts to define the structure of a protein in any given context. “It’s such a complicated problem with so many parameters, so many ways to go wrong,” says structural biologist Andriy Kryshtafovych at the University of California, Davis, who since 2000 has been a co-organizer of Moult’s experiment. “We couldn’t believe that it could be solved.”
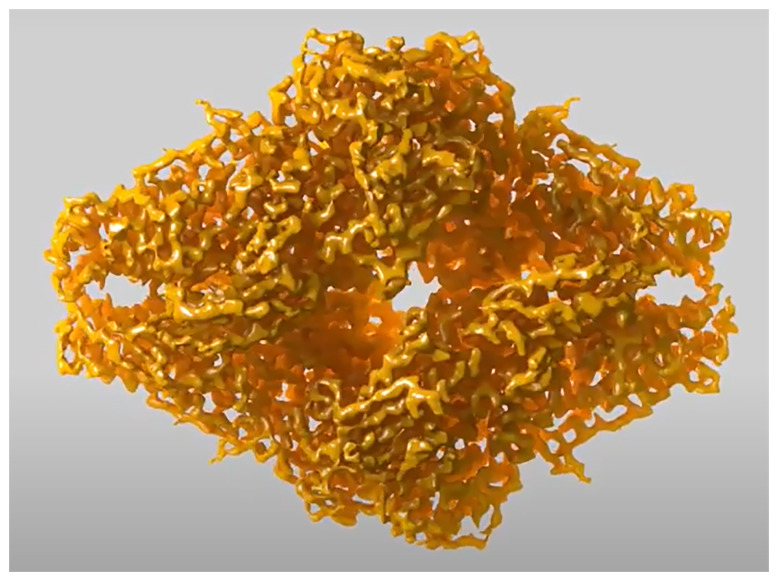
The revolution in structural biology isn’t attributable to AI alone; the algorithms have to train on big datasets of high-resolution crystal structures generated by technologies such as nuclear magnetic resonance spectroscopy or cryogenic electron microscopy (cryo-EM), which produced the above image of a protein complex called β-galactosidase. Image credit: Veronica Falconieri and Sriram Subramaniam (National Cancer Institute, Bethesda, MD).
In the 1970s, biologists ran experiments suggesting that the structure of a protein could be predicted from its amino acid sequence alone. And over the decades, protein structures have been laboriously determined, one by one, by studying the molecules using experimental tools like X-ray crystallography. But with no general how-to manual, researchers in fields ranging from biophysics to chemistry to biological evolution have sought strategies for exploiting the sequence–structure connection. Progress has often been incremental: for example, small assemblages of certain amino acids give rise to predictable shapes that can serve as templates for protein sub-structures. If a protein were a 1,000-piece LEGO house, these templates might provide general plans with details on how many rooms the house contains and the arrangement of doors.
In 1994, when Moult and his colleagues launched their computation-driven experiment, called CASP (for Critical Assessment of protein Structure Prediction), researchers had just begun to develop computer programs to tackle the question. Its most recent iteration, however, has largely upended Kryshtafovych’s belief that the problem couldn’t be solved—thanks in large part to a growing wave of artificial intelligence (AI) approaches that use deep learning algorithms to map out the structure of proteins. In 2020, the frontrunner method in CASP used AI to predict protein structures with an average accuracy approaching 90%, putting it on par with the most sophisticated experimental techniques. Most importantly, it showed that AI could do in a matter of minutes what used to take years, if not decades.
“This advance is just phenomenal,” says Frances Arnold, a chemical engineer at the California Institute of Technology in Pasadena, who in 2018 was awarded a Nobel Prize for her work using directed evolution to create enzymes. “It will revolutionize structural biology.” However, she notes, structure is just one piece in the much larger puzzle of understanding how these large molecules function.
Bolstered by the emerging trend among researchers to make code and data repositories open access and freely available, AI-driven acceleration of structure discovery is spreading to uses far beyond the exploration of basic protein structures. Many researchers are now harnessing AI to guide related applications, such as designing pharmaceuticals, predicting how proteins will interact, and mapping the structure of other biomolecules like RNA. These efforts, in the future, could both answer big questions about the micromachinery of life and enable a more precise approach to health care and disease treatment.
Finding the Problem
For most of its history, the field of structural biology has been driven by advances in crystallography and other imaging technologies that help researchers determine the shape and structure of large, essential biomolecules, such as proteins and RNA. But in the past, resolving a single structure could take years of imaging, computation, and analysis. Determining a structure requires finding clever ways to stabilize and still proteins long enough to get a non-blurry image.
The quest to know those protein structures dates back nearly 200 years. Proteins were first identified in the 19th century by European chemists who recognized a distinct class of macromolecules in substances like egg whites and wheat gluten. The word “protein” initially appeared as a description of these molecules in an 1838 letter from Swedish chemist Jöns Jacob Berzelius to Dutch chemist Gerard Johann Mulder (1). It wasn’t until the 20th century, however, that researchers began to make headway on figuring out how proteins were put together.
In a landmark series of articles published in PNAS in early 1951, Linus Pauling, Robert Corey, and Herman Branson described a predicted structure of proteins. Their approach was straightforward: They reasoned that if they knew the basic ingredients and had information about how those ingredients interacted at the atomic level, they could predict the molecular architecture (2). They were the first to recognize—a full decade before crystallographers would image the proteins—that amino acids could bend and bond into the alpha helix and the beta sheet, two telltale structures in the backbone of almost every protein. Experiments conducted in the 1960s confirmed their predictions, and their work remained unsurpassed in accuracy for more than four decades.
The 2000s brought a surge of interest in using computer algorithms to improve understanding of protein shapes, but progress was slow, as documented by CASP results, says Kryshtafovych. By the early 2010s, researchers were experimenting with more sophisticated computational methods and AI tools like artificial neural networks. These algorithms, inspired by and named for the wiring of the brain, use large training datasets to develop abstract rules that connect inputs to known outputs. This is how AI systems can identify objects in photos—or, in the case of structural biology, put amino acids together to build a protein.
Early efforts, however, were laborious and largely ineffective, says Kryshtafovych. It wasn’t until the recent explosion in accuracy in the CASP experiments that AI began to deliver by exploiting deep learning architectures. That’s largely because structural biology is simultaneously delicate and complicated, says Amir Farimani at Carnegie Mellon University in Pittsburgh, PA, who has been using deep learning to design synthetic antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The number of possible combinations of amino acids in peptides or proteins can lead to mathematical questions that invoke thousands of dimensions—an ideal task for deep learning, which excels at recognizing patterns and applying them to new cases. A protein made of 10 amino acids entails 1020 different possible combinations.
“The design space is just enormous,” says computational biologist Arvind Ramanathan at Argonne National Laboratory in Lemont, IL. “The AI revolution allows us to peer inside something that is not visible to us from the outset,” he says.
Hints of a Revolution
CASP, run like a contest to spur innovation, pulled deep learning from the fringes of structural biology into the leading edge of research. Here’s how it works: Over the course of a few months, competing research groups develop models to predict the structure of a few dozen target proteins selected by CASP organizers. Entrants receive only the amino acid sequences of each target. The process is double-blind, so the organizers don’t know the protein shapes beforehand, and the targets are proteins that have been recently solved but not yet submitted to the Protein Data Bank, a database of known large molecule structures. At the end of the contest, the organizers rank the entrants by the accuracy of their models in a few categories.
Since Moult and his colleagues launched CASP, the experiment has been held every two years. Until 2016, entrants rarely achieved higher than 20% accuracy. Then AI entered the scene. That year, during CASP12, computational biologist David Jones from the University College London, UK, more than doubled previous accuracy levels using a model powered by deep learning algorithms. In the wake of his work, neural networks took the CASP community—and the broader field of structural biology—by storm.
By CASP13, in 2018, most groups were using deep learning to predict protein structures, pushing accuracy levels up to about 60%. Top marks that year went to AlphaFold, a model designed by researchers at DeepMind, a London-based AI company owned by Alphabet Inc., which also owns Google. (Frances Arnold sits on the board of Alphabet, which is the parent company of AlphaFold.) At CASP14, AlphaFold achieved scores above 90% on many of the target proteins. Other AI-driven entrants in the contest reached accuracies above 70%, which just two years earlier would have been unimaginable.
With AlphaFold, “if you give us the sequence, I’ll give you the structure,” says Farimani. He notes that the revolution isn’t attributable to AI alone; the algorithms have to train on big datasets of high-resolution crystal structures generated by sophisticated technologies such as nuclear magnetic resonance (NMR) spectroscopy or cryogenic electron microscopy (cryo-EM). “They go hand in hand,” he says, noting that AI models are nearly as good at predicting structure as those advanced experimental methods. “Now, the problem is basically solved,” Kryshtafovych adds.
Putting AI to Work
Since the debut of AlphaFold, a growing chorus of groups around the world have been rolling out new AI-based applications and advances that continue to push structural biology forward. In July 2021, DeepMind, in collaboration with the European Bioinformatics Institute, publicly released the structures of hundreds of thousands of proteins, including not only those from CASP but all of the roughly 20,000 known human proteins, as well as the entire proteomes of other scientifically important organisms such as mice and fruit flies. (For comparison, approximately 100,000 protein structures were known in 2016.)
Other big breakthroughs followed. That same July, a group at the University of Washington in Seattle unveiled RoseTTAFold, a program that uses neural networks to predict protein structures based on scant genomic information (3). In August, computer scientists at Stanford University, CA, debuted a machine-learning approach that predicts the structure of RNA using very little training data (4, 5). Prediction of RNA structure has been challenging owing to a lack of experimental data for training, says Stanford computer scientist Ron Dror, who led the machine learning work, but a new deep learning method addresses this challenge. “Structure prediction is especially valuable for types of molecules for which it’s hard to determine structures experimentally,” like RNA, he says.
The AI revolution allows us to peer inside something that is not visible to us from the outset.
—Arvind Ramanathan
The ultimate goal of these advances, says Ramanathan, is a machine learning tool “that will help biologists do their experiments even better.” At Argonne, he uses deep learning to study protein interactions, such as how the SARS-CoV-2 spike protein interacts with host cells, as well as exceptionally complicated molecules called “intrinsically disordered proteins.” These complex proteins don’t reliably fold into predictable three-dimensional shapes and are associated with many diseases, including cancer, diabetes, and neurodegenerative disorders. Ramanathan’s goal is to use deep learning models to predict some of these irregular protein structures, then verify them using diverse experimental techniques including crystallography and electron microscopy.
Recent predictions of complex protein interactions can shed light on the mechanistic machinery underlying important biological processes. In October 2021, DeepMind researchers used an AlphaFold model to predict complexes made up of multiple proteins (6). In November 2021, an international group combined the strengths of AlphaFold and RoseTTAFold to evaluate interactions among 8.3 million pairs of proteins and predict large protein assemblies, involved in important biological functions, in the yeast Saccharomyces cerevisiae (7). Advances like these have also pushed the field further into protein design.
Researchers are also increasingly deploying deep learning to predict structures that may bind target molecules, for uses such as personalized cancer treatments or synthetic antibodies able to neutralize SARS-CoV-2 (8, 9). Farimani’s work at Carnegie Mellon shows one way that AI can help unearth new treatments for COVID-19. He and his collaborators first amassed data on the amino acid sequences of antibodies—proteins that can fight off an invader in the body—against viruses including HIV, dengue, SARS, influenza, and Ebola. They then trained a few candidate machine learning models to identify antibodies able to neutralize the target virus. Finally, they fed data about SARS-CoV-2 into the most accurate of the models to identify sequences for antibodies with the best chance of inhibiting the virus.
What’s remarkable about AlphaFold, says Farimani, is that it can make accurate predictions based on the amino acid sequence alone. “You don’t need the mathematics or the understanding of the physics of the molecule,” he says. “It’s really an amazing tool.” And because it’s open-access, he even has students in his classes use it for assignments.
Black Box Warnings
AI-driven advances in structural biology are not only pushing forward the basic science but also exposing new questions and opportunities. “I believe it’s still early days,” says Dror. “The recent results are really exciting and impressive, and at the same time there’s a great deal left to be done.”
For example, proteins in biological systems change structure continuously. They wiggle and deform, changing shape and moving with the system. Current methods—both computational and experimental—yield average protein structures. “The average structure isn’t the only thing that’s important,” Dror says. “It’d be wonderful to go beyond that, to predict whole sets of structures and determine which ones will be adopted in a cell and under which conditions.” AI models will likely play a role in answering that challenge as well, predicts Ramanathan.
But structures tell only part of the story of how life’s machines work: Researchers will still need to connect those structures with function of the molecules. “You can get the structure of an enzyme and still have no clue about how it works,” Arnold says. Deep learning will likely help biologists crack open that mystery as well, she says, but it’s not going to happen right away. “We will at some point have sufficient data and modeling requirements to do the same for function,” she says. “But it’s an orders-of-magnitude more complex problem and requires different kinds of data. Structure is just one part of that data.”
There are also challenges associated with neural nets themselves; namely, that researchers don’t know exactly how the algorithms make such accurate predictions. “We don’t know what the neural net learns,” Kryshtafovych says. (See News Feature: What are the limits of deep learning?*) The algorithm generates its own abstract rules based on the training data and not on natural laws, which means its reasoning is likely impossible to decode, even if one could crack open the program and peer inside. Taking the model apart won’t reveal the rules that the neural net invented. “There is no mathematical function, no analytical explanation of how this is able to make this complex connection,” says Farimani.
Whatever the reasoning, it’s likely that the neural net doesn’t use the natural, mechanical rules that guide protein shapes in biological systems. “Why does nature pick this pathway and not that pathway?” Kryshtafovych asks. “These recent successes don’t help us know that.”
In the future, researchers see a role for deep learning not only in understanding a protein’s shape but also how it interacts within a living system. “Let’s say I want to build a protein and I have some idea of the shape, but I want to insert it into an organism and make it functional,” says Ramanathan. Deep learning models may predict not only the sequence of amino acids that would produce the needed shape, but also how they’ll behave—and interact with other molecules in their biological neighborhood—once they’re in place.
Knowing the structure of proteins is a step toward answering even bigger questions about how big molecules interact, evolve, and drive life itself, says Arnold. “We need a lot more to revolutionize our full understanding of biology. It’s more of a game than just solving a problem,” she says, “but it’s a fantastic game.”
Footnotes
References
- 1.Hartley H., Origin of the word ‘protein.’ Nature 168, 244 (1951). [DOI] [PubMed] [Google Scholar]
- 2.Eisenberg D., The discovery of the α-helix and β-sheet, the principal structural features of proteins. Proc. Natl. Acad. Sci. U.S.A. 100, 11207–11210 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek M., et al. , Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Townshend R. J. L., et al. , Geometric deep learning of RNA structure. Science 373, 1047–1051 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eismann S., et al. , Hierarchical, rotation-equivariant neural networks to select structural models of protein complexes. Proteins 89, 493–501 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Evans R., et al. , Protein complex prediction with AlphaFold-Multimer. bioRxiv [Preprint] (2021). https://www.biorxiv.org/content/10.1101/2021.10.04.463034v1.full.pdf (Accessed 22 February 2022).
- 7.Humphreys I. R., et al. , Computed structures of core eukaryotic protein complexes. Science 374, eabm4805 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elemento O., Leslie C., Lundin J., Tourassi G., Artificial intelligence in cancer research, diagnosis and therapy. Nat. Rev. Cancer 21, 747–752 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Magar R., Yadav P., Barati Farimani A., Potential neutralizing antibodies discovered for novel corona virus using machine learning. Sci. Rep. 11, 5261 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bagdonas H., Fogarty C. A., Fadda E., Agirre J., The case for post-predictional modifications in the AlphaFold Protein Structure Database. Nat. Struct. Mol. Biol. 28, 869–870 (2021). [DOI] [PubMed] [Google Scholar]