Abstract
Salt stress is one of the most important abiotic stresses as it persists throughout the plant life cycle. The productivity of crops is prominently affected by soil salinization due to faulty agricultural practices, increasing human activities, and natural processes. Approximately 10% of the total land area (950 Mha) and 50% of the total irrigated area (230 Mha) in the world are under salt stress. As a consequence, an annual loss of 12 billion US$ is estimated because of reduction in agriculture production inflicted by salt stress. The severity of salt stress will increase in the upcoming years with the increasing world population, and hence the forced use of poor-quality soil and irrigation water. Unfortunately, majority of the vegetable crops, such as bean, carrot, celery, eggplant, lettuce, muskmelon, okra, pea, pepper, potato, spinach, and tomato, have very low salinity threshold (ECt, which ranged from 1 to 2.5 dS m–1 in saturated soil). These crops used almost every part of the world and lakes’ novel salt tolerance gene within their gene pool. Salt stress severely affects the yield and quality of these crops. To resolve this issue, novel genes governing salt tolerance under extreme salt stress were identified and transferred to the vegetable crops. The vegetable improvement for salt tolerance will require not only the yield influencing trait but also target those characters or traits that directly influence the salt stress to the crop developmental stage. Genetic engineering and grafting is the potential tool which can improve salt tolerance in vegetable crop regardless of species barriers. In the present review, an updated detail of the various physio-biochemical and molecular aspects involved in salt stress have been explored.
Keywords: oxidative stress, physio-biochemical responses, antioxidant, transgenic crops, gene regulation, yield loss
Introduction
Nearly, three thousand species of plants are being utilized for the food by human; anyhow, presently, the total global population mainly depends mostly upon 20 species of crops for its major calorie needs from which 50% is contributed by eight cereal crop species (Krishna et al., 2019). The insufficient availability of vegetables is mainly due to increasing population, abiotic (drought, salt, heat, water logging, etc.) and biotic (virus, viroids, bacteria, fungi, nematodes, and insects) stresses, which potentially reduce the production and quality of the vegetable crops (Ingram, 2011; Prasanna et al., 2015; Karkute et al., 2019; Krishna et al., 2021a,b; Singh et al., 2021; Soumia et al., 2021). The key challenge of modern agriculture is to fulfill the nutritional and food security of the global growing population. Among abiotic stresses, salt stress is the second most destructive stress as it persists throughout the crop life cycle. Salinity stress is one of the most important environmental constraints that limits the economic productivity of vegetable crops (Hong et al., 2021). Salinity in soil is influenced by a regular fluctuation in climatic conditions, irrigation of crops with low quality water, excessive use of ground water, and massive introduction of irrigation associated intensive farming (Tiwari et al., 2010). Further, prolonged water stress conditions in soil could result in increased salinity in soil profile due to lack of leaching rain, increased bore water salinity, and evaporation from irrigation dams. Vegetable crops are more prone to climatic changes compared with other horticultural crops (Giordano et al., 2021), and particularly, salinity stress influences the growth and development throughout their ontogeny. Salinity-induced oxidative stressed in vegetables could affect the qualitative and quantitative value of vegetables as this oxidative stress could lead to a plethora of biochemical and physiological changes in plants (Kashyap et al., 2020, 2021). The most common of them include membrane damage, leakage of substances causing water imbalance and plasmolysis, disturbance in ROS detoxification system, changes in nutrient flux and dynamics, and photosynthetic attributes. These changes ultimately affect the physiological activities like respiration, photosynthesis, transpiration, hormonal regulation, water use efficiency, germination, production of antioxidants, and plasma membrane permeability (Chourasia et al., 2021). The most common approach adopted by plants during such extreme conditions is transcriptional reprogramming of stress responsive genes (Aamir et al., 2017; Tolosa and Zhang, 2020), although the conventional breeding approaches have helped a lot in developing stress-tolerant breeds of vegetables. However, we do not still have developed optimum solutions to prevent the economic losses of vegetables from salt stress, particularly, in intensively irrigated areas (Machado and Serralheiro, 2017). Transgenic technology for salt stress tolerance has been reported as one of the most crucial tool in developing the stress-tolerant vegetable crops (Kumar et al., 2017). For example, to avoid salt tolerance in plants, genes encoding for proteins like Na+ “exclusion” (PM-ATPases with SOS1 antiporter, and HKT1 transporter), vacuolar compartmentalization of Na+ V-H+-ATPase and V-H+-PPase with NHX antiporter, and also other genes encoding proteins such as aquaporins and dehydrins that are involved in mitigation of water stress during salinity have been transferred and/or overexpressed in tomato or Arabidopsis through transgenic technology (Kotula et al., 2020). Since tomato is one of the most important vegetable crop and experimental model for molecular biology studies, most of the research done so far with respect to abiotic and biotic stresses have been done in tomato (Meena et al., 2016, 2018; Zehra et al., 2017a,b). For example, overexpression of LeNHX2 and SlSOS2 proteins resulted in salinity tolerance in tomato transgenic lines (Maach et al., 2021). The stress-responsive genes expressed during salinity stress and their fine-tuning could be an eminent tool for developing stress-resistant varieties.
On an average every year approximately 12 billion USD are lost worldwide due to the salinity stress which greatly affects the agriculture production (Zahedi et al., 2019). Almost 10% of the world’s entire land area (950 Mha), 20% of the world’s cultivated land (300 Mha), and approximately 50% of the total irrigated land (230 Mha) are consequently distressed with extreme salinity (Abiala et al., 2018).
Salt Stress Responses in Plants
Important physiological and biochemical processes in plants are adversely affected by salinity in various ways through an intense concentration of salts and unavoidably leading to a gradual reduction in plant growth. High salt concentration in rhizosphere of plant cell causes osmotic effect, which remains as a chief contributor to growth reduction during the preliminary stages of a plant life cycle. Amendment in K+/Na+ ratio arises when ions reach the plant cell through saline water, leading to augmented Na+ and Cl– ion, inflicting extensive damage of numerous physiological processes like protein metabolism and enzyme activities (Tester and Davenport, 2003). The interactions between salts and essential mineral nutrients may consequently result in significant nutrient deficiencies and disproportion. Ionic imbalances may also result in decreased uptake of various significant minerals like potassium, manganese, and calcium to the plants. However, in response to ionic and nutrient imbalances, salt-tolerant plants have uniquely developed the capability of accumulation and compartmentalization of Na+ and Cl– in their matured leaves, but sensitive species at absurdly high salinity stage cannot manage to compartmentalize the ions or Na+ transport, leading to the ionic or osmotic effect. Considerable reduction in plant height has been documented under different abiotic stresses. Due to salinity, plants are exposed to serious water deficit conditions that reduces the leaf growth and leaf areas in several species such as wheat (Sacks et al., 1997), poplar (Wullschleger et al., 2005), and cowpea (Manivannan et al., 2007). One example of the physiological changes in response to salt is shedding of the older leaves of plants (Shao et al., 2008). The upsurge in root to shoot ratio due to salinity conditions was found to be associated with the ABA content of plants (Sharp and LeNoble, 2002). Plant productivity under salinity is strongly correlated with biomass distribution.
Mechanism of Salinity Tolerance
Salinity tolerance is related to a list of morphological, biochemical, molecular, and physiological traits that govern the plant growth and productivity (Alexieva et al., 2001). Morphological and physiological adaptation toward tolerance to the salt-induced osmotic stress is also facilitated by reducing water loss from cuticle and stomata and maximized uptake of water by root to maintain the osmotic adjustment (Rai et al., 2021). Tolerance and adaptation to salt stress are governed by a cascade of molecular networks, which trigger response processes like production of stress proteins, upregulation of antioxidants, and accumulation of compatible solutes (Nahakpam and Shah, 2011) to provide homeostatic reestablishment of cells and to repair and protect the damaged membranes and proteins. On the basis of responses to salinity, plants are categorized as either halophytes or glycophytes (Flowers and Flowers, 2005). Under high saline conditions, glycophytes are unable to survive, whereas halophytes can easily grow and reproduce. Tissue tolerance and salt avoidance are two main approaches implemented by plants to overcome the salt stress. Plants also execute the compartmentalization of ions in the plant tissues. To regulate their osmotic pressure, plants continuously generate water-soluble and low molecular-weight compatible solutes like sugars, glycinebetaine, and proline and the metabolic processes of plants are not disturbed. Plants also produce many enzymatic and non-enzymatic antioxidants to minimize the adverse effect of salinity. In the procedure of plant tissue tolerance, ions compartmentalization occurs in the vacuole, resulting in sustained salt concentration in cytosol, and thus the cytoplasm of the plant cell can be protected from water stress and ion toxicity (Chinnusamy et al., 2005).
To cope with salt stress many strategies have been evolved and developed to secure the vegetable yield under salt stress like transgenic development, regulation of transcription factors (TFs), and grafting. In this review, we have presented updated information on biotechnological interventions in vegetable crops for salt stress.
Transgenic Vegetables for Salt Stress Tolerance
Vegetables are the cheapest source of minerals, vitamins, antioxidative phytochemicals, and consumed all over the world in raw, semicooked, cooked, and/or in processed forms. Salt stress does not affect vegetable yield but it also affects the nutritional quality of the vegetables. Due to lack of novel salt tolerance in the gene in many vegetable crops gene pools, the transgene has been transferred from the non-parent sources like bacteria, fungi, plant, and animals. For developing transgenic vegetable crops, firstly gene is being identified, characterized, then transferred in the desired vegetables for salt stress. The transgene is being induced under salt stress and the upregulation or downregulation of transgenes initiates a cascade of stress regulatory phenomenon, which ultimately results in salt tolerance (Figure 1). Among vegetable crops, Solanaceous crops like potato, tomato, capsicum, and chili constitute major group of vegetables consumed all over the world, and out of these, potato is the most important and ranks third in the world in terms of economic importance and a key agriculture crop for food and nutritional security. Potato is cultivated globally and very sensitive to salt stress, and more than 60% crop loss is caused abiotic stresses including salt (Upadhyaya et al., 2011; Xu et al., 2014; Shafi et al., 2017). To improve potato yield and quality under salt stress condition, many transgenic potato plants have been developed using different genes with different modes of action (Shafi et al., 2017; Wang et al., 2019; Ali et al., 2020). Many osmoprotectant genes like P5CS, mtlD, and AtBADH have been transferred to potato, which significantly improves the salt tolerance under salt stress (Karthikeyan et al., 2011; Rahnama et al., 2011; Zhang et al., 2011). Like potato, tomato is the second most important vegetable fruit crop that belongs to the Solanaceae family and it is the highest processed crop in the world. Tomato is a rich source of proteins, minerals, carbohydrates, and vitamins, especially vitamin C. Tomato also contains many phytochemicals like carotenes and lycopenes which have anticancer properties and other health benefits (Krishna et al., 2021a,b; Rai et al., 2021). In tomato far salt stress tolerance, osmoprotectants genes like BADH-1, ToOsmotin, Ectoine (ectA, ectB, and ectC), and coda gene have been transformed in tomato, which reduces the impact of salt stress by encoding osmoprotectant solutes (Moghaieb et al., 2000, 2011; Goel et al., 2010; Wei et al., 2017). To maintain the cellular acidity under salt stress many Na+/H+ antiporter genes also have been transformed like NHX1, TaNHX2, and LeNHX4 which regulate the Na+/H+ to maintain cellular homeostasis (Zhang and Blumwald, 2001; Yarra et al., 2012; García-Abellan et al., 2014). Transgenes like cAPX, MdSOS2L1, AnnSp2, LeNHX2 and SlSOS2, At FeSOD, and BcZAT12 have been also transformed and works with different modes of action, details of the gene transformed in tomato and their mode of action are summarized in Table 1. Like potato and tomato, other important Solanaceous crops like brinjal and chili face the salt stress; in these crops also transgenics have been developed for salt stress, and details of the transgenic crop in Solanaceae family are given in Table 1. In vine crops group like cucumber, cucurbits, and bottle gourds are also a very popular in vegetable crops and play an important role in food and nutritional security, salt significantly reduced the yield and quality of vine crops also (Park et al., 2014; Kim et al., 2015; Sun et al., 2018; Li et al., 2020). In water melon HAL1 transferred which encodes for 32 kDa water soluble proteins which protects from salt induced osmotic stress (Bordas et al., 1997). Park et al. (2014), Han et al. (2015), and Kim et al. (2015) transformed bottle gourds with AVP1 which encodes vacuolar H+-pyrophosphatase, which regulate the proton pump and ultimately maintains the cellular acidity to avoid salt stress (Table 2). Cole crops like cabbage, cauliflower, mustard green, rape seed, and Chinese cabbage constitute a major group of leafy vegetable, which are considered as cheapest and richest sources of mineral, vitamins, and oils and they play an important role in nutrition and food security (Wang et al., 2010; Kim et al., 2016; Ahmed et al., 2017; Luo et al., 2017). The cole crops are very sensitive to the salt stress, and different genes like CodA, PgNHX1, OsNASI, BnSIP1-1, APX, SOD, and LEA4-1 have been transferred to sustain salt stress (Park et al., 2005; Wang et al., 2010; Kong et al., 2011). Details of the gene transferred and their mechanism of action in cole crops is given in Table 3.
FIGURE 1.
Mechanisms of transgene action in transgenic plants; downstream signaling process and transcription controls that stimulates stress-responsive mechanisms to reestablish cellular homeostasis and damage repair.
TABLE 1.
Transgene used for development of salt stress tolerance, their function and mechanism of action.
| S. N. | Genes | Function | Mechanism of action | References |
| Potato (Solanum tuberosum) | ||||
| 1 | P5CS | Encodes for pyrroline-5-carboxylate synthetase (P5CS) | P5CS gene expression enhances proline content in the cells, proline is a potential osmolyte and tolerate to salt stress | Karthikeyan et al., 2011 |
| 2 | StCYS1 | Encodes for, Cysteine protease inhibitors (CPI) | Cysteine protease inhibitors (CPI) is a cystatin protein superfamily and facilities biological activities by cysteine protease inhibition | Liu et al., 2020 |
| 3 | Glycinebetaine | Glycinebetaine (GB) synthesizing enzymes | Glycinebetaine GB is a osmolytes and potent compatible compound, its accumulation does not hamper plants normal activities and help in salt tolerance | Ahmad et al., 2014 |
| 4 | StDREB1 | Transcription factors | Regulate differential gene expressions in the different signaling pathways due to their different DNA-binding specificity | Bouaziz et al., 2013 |
| 5 | AcBADH | Encodes for betaine aldehyde dehydrogenase | Betaine aldehyde dehydrogenase converts betaine aldehyde to glycine betaine which predominantly accumulate in the leaves and stems in dicot and monocot and enhance salt tolerance | Ali et al., 2020 |
| 6 | AtNHX1 | Na+/H+ antiporters | AtNHX1 gene improves the absorption and transportation of the Na+ of the host plant species and enhances salt stress tolerance | Wang et al., 2013 |
| 7 | mtlD | Encodes for mannitol 1-phosphate dehydrogenase | Mannitol accumulation increases in plants in response to osmotic stresses like salt | Rahnama et al., 2011 |
| 8 | AtHKT1 | Facilitates high-affinity potassium transporter | HKTs actively involve at the plasma membrane level, HKT transporters exclude Na+ from the leaves while increasing K+ transportation to resist salt stress | Wang et al., 2019 |
| 9 | PaSOD and RaAPX | Encodes for superoxide dismutase (SOD) and ascorbate peroxide (APX) enzymes | SOD and APX enzyme system converts superoxide radical to hydrogen peroxide (H2O2), followed by conversion of H2O2 to water and oxygen, respectively | Shafi et al., 2017 |
| 10 | Rat GLOase | Over-expressing L-gulono-c-lactone oxidase | Enhanced ascorbic acid accumulation have been reported to have salt/osmotic stress | Upadhyaya et al., 2010 |
| 11 | GalUR | L-gulono-1,4-lactone conversion to AsA | D galacturonic acid reductase (GalUR over-expression enhances AsA production enhances salt tolerance) | Upadhyaya et al., 2009 |
| 12 | GalUR | Encodes for D galacturonic acid reductase | Overexpression of GalUR, an ascorbate pathway enzyme enhances its ascorbic acid content (L-AsA) and enhances salt tolerance | |
| 13 | StNAC2 | Regulates NAC transcription factors | NAC proteins are plant-specific TFs and to play important roles in abiotic biotic stresses | Xu et al., 2014 |
| 14 | AtBADH | Encodes for betaine aldehyde dehydrogenase | Converts betaine aldehyde to glycine betaine, the elevated glycine betaine level enhances cellular buffering capacity and stress tolerance | Zhang et al., 2011 |
| Tomato (Solanum lycopersicum) | ||||
| 1 | BADH-1 | Over expression of betaine aldehyde dehydrogenase | Betaine aldehyde dehydrogenase catalyzes conversion of betaine aldehyde into glycine betaine which improves abiotic stresses tolerance | Moghaieb et al., 2000 |
| 2 | NHX1 | Over expression the NHX1 antiporter | Over expressed NHX1 vacuolar Na+/H+ antiporter helps in maintaining cellular integrity and improve salt stress tolerance | Zhang and Blumwald, 2001 |
| 3 | cAPX | Over expression of APX | Enhanced activity of ascorbate peroxidase activity reduces cellular damage by scavenging the superoxides under salt stress | Wang et al., 2005 |
| 4 | CaKR1 | Over expression of LeSOD2, LeAPX2, and LeAPX3 | High transcript level of antioxidative enzyme machinery scavenge the ROS under abiotic stresses | Seong et al., 2007 |
| 5 | ToOsmotin | Osmotic adjustment | Over expression leads accumulation or compartmentalization of solutes and also protect proteins denaturation under salt stress | Goel et al., 2010 |
| 6 | Ectoine (ectA, ectB and ectC) | Compatible solute | Enhance peroxidase activity and decrease MDA contents by ectoine accumulation | Moghaieb et al., 2011 |
| 7 | AtSlSOS2 (AtSlSOS2) | Homeostasis of Na+ and K+ | Upregulation of the plasma membrane Na+/H+ (SlSOS1) and endosomal-vacuolar K+, Na+/H+ (LeNHX2 and LeNHX4) antiporters, responsible for Na+ extrusion out of the root, active loading of Na+ into the xylem, and Na+ and K+ compartmentalization | Huertas et al., 2012 |
| 8 | TaNHX2 | Na+/H+ antiporter | Na+/H+ antiporters are involved in intracellular ion (Na+), pH regulation and K+ homeostasis in plants under salt stress | Yarra et al., 2012 |
| 9 | HAL5 | Maintaining Na+/K+ homeostasis | Maintenance of Na+ and K+ transporters like SlHKT1;2 and SlHAK5 improve homeostasis | García-Abellan et al., 2014 |
| 10 | MdSOS2L1 | Codes for MdSOS2L1 protein kinase | MdSOS2L1 protein kinase physically interacts with MdCBL1, MdCBL4, and MdCBL10 proteins to increase tolerance against salt | Hu et al., 2016 |
| 11 | coda | Encode for glycine betaine | Glycine betaine enhanced NaCl-induced expression of genes encoding the K+ transporter, Na+/H+ antiporter, and H+-ATPase | Wei et al., 2017 |
| AnnSp2 | Encodes annexins proteins | AnnSp2 alleviated ABA sensitivity in tomato in the germination and seedling stages under salt stress | Ijaz et al., 2017 | |
| 12 | SlCMO | Choline monooxygenase (CMO) | Is a key enzyme involved in the synthesis of glycine betaine, which is a osmoprotectant that plays an important role in plant salt tolerance | Li Q. L. et al., 2018 |
| 13 | LeNHX2 and SlSOS2 | Homeostasis of Na+ and K+ and Na+/H+ antiporter | Involves Na+ and/or K+ intracellular accumulation mediated by NHX transporters | Baghour et al., 2019 |
| 14 | SlMYB102 | Decrease the transcripts of ABA-dependent genes | Suppress the expression of PP2Cs or protein phosphatases of PP2Cs to help plants adapt to higher salt concentrations | Zhang et al., 2020 |
| 15 | At FeSOD | Encodes for super oxide dismutase enzyme | The main function of these enzymes is the enzymatic conversion of such a highly toxic molecule for cells as superoxide into hydrogen peroxide (H2O2) | Bogoutdinova et al., 2020 |
| 16 | LeNHX4 | K+, Na+/H+ antiporter | An important mechanism to overcome salt stress is the exclusion of Na+ from the cytoplasm, by the operation of Na+/H+ antiporters at the plasma membrane or tonoplast. Plant NHX antiporters play a key role in NaCl tolerance by the extrusion of Na+ out of cytosol | Maach et al., 2020 |
| 17 | COMT1 | Promote the synthesis of melatonin | SlCOMT1 overexpression could maintain the balance of Na+/K+ and decrease ion damage by activating salt overly sensitive (SOS) pathway under salt treatment | Sun et al., 2020 |
| 18 | BcZAT12 | Encodes for C2H2 type zinc finger protein | The C2H2 type zinc finger protein is known to confer tolerance to dehydration, heat stress, salt and/or cold stresses | Rai et al., 2021 |
| Brinjal (Solanum melongena) | ||||
| 1 | Yeast HAL1 | Encodes a water-soluble protein | HAL1 and HAL3, which were involved in the regulation of K+ and Na+ transport, respectively, and considerably enhanced salt tolerance in egg plants | Kumar et al., 2014 |
| 2 | TaNHX2 | Vacuolar Na+/H+ antiporter | Na+/H+ antiporters are involved in intracellular ion (Na+), pH regulation, and K+ homeostasis in plants | Yarra and Kirti, 2019 |
| 3 | adc | Biosynthetic of polyamine by arginine decarboxylase | Accumulation of higher polyamine in cells works as a osmoprotectants | Prabhavathi and Rajam, 2007 |
| 4 | mtlD | Mannitol-1-phosphate dehydrogenase | The accumulation of mannitol in the cytoplasm and increased tolerance to salt stress | Prabhavathi et al., 2002 |
| Chili pepper (Capsicum annuum L.) | ||||
| 1 | TaNHX2 | Vacuolar Na+/H+ antiporter | Na+/H+ antiporters are involved in intracellular ion (Na+), pH regulation, and K+ homeostasis in plants | Bulle et al., 2016 |
| 2 | PDH45 | Encodes for Pea DNA Helicase 45 | DNA and RNA helicases have proved their translational efficacy in multiple crops by improving tolerance to salinity and drought stress. DNA and RNA helicases, also known as molecular motors, are involved in myriad cellular processes of protein turnover and protection | Shivakumara et al., 2017 |
| 3 | Osmotin | Encodes for Osmotin is a stress-responsive protein | Osmotin is a stress-responsive protein adapted to salinity and desiccation and accumulates in saltadapted cells. Osmotin is an abundant cationic 26-kDa protein that belongs to the family of PR-5 type proteins. Osmotin provides osmotolerance to plants probably by facilitating the compartmentation of solutes | Subramanyam et al., 2011 |
TABLE 2.
Transgenic crops developed in vine crops for salt stress tolerance.
| S. N. | Genes | Function | Mechanism of action | References |
| Cucumber (Cucumis sativus L.) | ||||
| 1 | LOS5 | Encodes a molybdenum cofactor (MoCo) sulfurase | Molybdenum cofactor (MoCo) sulfurase catalyzes the last step of ABA biosynthesis in plants | Liu Z. et al., 2013 |
| 2 | HAL1 | HAL1 encodes a water soluble protein (32 kDa) | Water soluble protein (32 kDa) that may modulate monovalent ion channels, by affecting the set point of intracellular potassium determined by the feedback regulation of the uptake system | Bordas et al., 1997 |
| 3 | CsbHLH041 | Encodes Basic helix-loop-helix (bHLH) transcription factors | The bHLH genes are involved in processes such as metabolic regulation, plant growth and development, and response to environmental signals | Li et al., 2020 |
| CmHKT1;1 | Encodes a Na+ preferential transporter | (HKT1) encodes a Na+ preferential transporter that principally controls root-to-shoot Na+ delivery via the withdrawal of Na+ from the xylem sap | Sun et al., 2018 | |
| 4 | Bottle gourds | |||
| 5 | AVP1 | Encodes vacuolar H+-pyrophosphatase | A vacuolar H+-pyrophosphatase encoded by the AVP1 gene is one of the proton pumps in Arabidopsis and generates an H+ electrochemical gradient across the tonoplast | Kim et al., 2015 |
| 6 | AVP1 | Encodes vacuolar H+-pyrophosphatase | A vacuolar H+-pyrophosphatase encoded by the AVP1 gene is one of the proton pumps in Arabidopsis and generates an H+ electrochemical gradient across the tonoplast | Park et al., 2014 |
| AVP1 | Encodes vacuolar H+-pyrophosphatase | A vacuolar H+-pyrophosphatase encoded by the AVP1 gene is one of the proton pumps in Arabidopsis and generates an H+ electrochemical gradient across the tonoplast | Han et al., 2015 | |
| Watermelon [Citrullus lanatus (Thunb.) Matsum. & Nakai] | ||||
| HAL1 | A vacuolar Na+/H+ antiport | Water soluble protein (32 kDa) that may modulate monovalent ion channels, by affecting the set point of intracellular potassium determined by the feedback regulation of the uptake system | Ellul et al., 2003 | |
TABLE 3.
Transgenic crops developed in cole crops for salt stress tolerance.
| S. N. | Genes | Function | References |
| Mustard green (Brassica juncea) | |||
| 1 | CodA | Choline oxidase | Prasad et al., 2000 |
| 2 | PgNHX1 | Vacuolar Na+/H+ antiporter | Rajagopal et al., 2007 |
| 3 | AtLEA4-1 | LEA4 protein | Saha et al., 2016 |
| 4 | Lectin | Induced fungal resistance | Kumar et al., 2015 |
| 5 | Gly I | Detoxification of methylglyoxal | Rajwanshi et al., 2016 |
| 6 | Gly II | Detoxification of methylglyoxal | Saxena et al., 2011 |
| AnnBj2 | Upregulated expression of ABA-dependent (RAB18) and ABA independent (DREB2B) genes | Ahmed et al., 2017 | |
| Rape seed (Brassica napus) | |||
| CodA | Choline oxidase | Huang et al., 2000 | |
| AtNHX1 | Vacuolar Na+/H+ antiporter | Zhang et al., 2001 | |
| YHem1 | Accelerate endogenous 5-ALA metabolism | Sun et al., 2015 | |
| LEA4-1 | LEA4 protein | Dalal et al., 2009 | |
| OsNASI | 11 proteins upregulated including dehydrogenase, GST, POD and Rubisco | Kong et al., 2011 | |
| DREB | Expression of many stress-inducible genes | Qamarunnisa et al., 2015 | |
| BnSIP1-1 | Regulates BnABI5, BnNAC485 or other stress-related genes | Luo et al., 2017 | |
| PR10 | Pathogenesis related | Srivastava et al., 2004 | |
| Chinese cabbage (Brassica campestris L. spp. Chinensis) | |||
| CodA | Choline oxidase | Wang et al., 2010 | |
| Cabbage (Brassica campestris) | |||
| LEA4-1 | LEA protein | Park et al., 2005 | |
| Cauliflower (B. oleracea var. botrytis) | |||
| APX, SOD | Antioxidants | ||
| Napa cabbage (Brassica rapa ssp. Pekinensis) | |||
| BrGI | Reduced expression of GI, enhanced salt tolerance | Kim et al., 2016 | |
Transcriptional Regulation of Salinity Stress Signaling in Vegetables
Recently, it was demonstrated that during salt stress, the expression level of multiple TFs increases much more compared with their basal trends, which reflects their crucial role in regulating the function mechanism and stress-dynamics of stress-tolerance (Franzoni et al., 2019, 2020). TFs are key regulators that play important roles in various stress responses (Debbarma et al., 2019). In fact, TFs are the key players that actually bind with the cis acting elements to regulate the spatial and temporal expression of specific genes, or genes regulating the functional activities of signal transduction and/or other genes regulating the transcriptional efficiency of stress-responsive genes under environmental stresses (Liu et al., 2014). Therefore, transcriptomic characterization for identification of stress-responsive TFs in plants or vegetable crops could be useful as a prominent tool or may provide a genetic resource for transgenic technology to improve the stress-responsive traits in different crops (Hong et al., 2021). The TFs belonging to WRKY, NAC, bZIP, MYB, and AP2/ERF play a crucial role in modifying and fine-tuning of the different stress-responsive genes involved in stress avoidance (Golldack et al., 2014). We have provided a comparative pie chart showing the functional annotation of two different transcriptional factors WRKY and NAC having WRKY and NAC domain. Based on functional annotation and gene ontology structured around three ontological terms, biological processes, molecular function, and cellular component, we reported and confirmed the function of WRKY and NAC TFs as to bind with DNA and also playing an important role in metabolism, stress response, and nucleic acid binding transcriptional factor activity (2) (Figure 2). During the last few years, many TFs have been deployed for transgenic overexpression of different TFs to mitigate various abiotic stresses (Tran et al., 2010). For example, the overexpression of moso bamboo WRKY (Phyllostachys edulis) in Arabidopsis uncovered the importance of PeWRKY83 in imparting salinity tolerance in transgenic Arabidopsis (Wu et al., 2017). SlAREB1, a bZIP transcriptional activator that belongs to ABA-responsive element binding protein (AREB)/ABA-responsive element binding factor (ABF) subfamily overexpression in tomato lines, reported enhanced salt and drought tolerance (Orellana et al., 2010). Furthermore, with the help of CRISPR/Cas9 genome editing technology it has now become possible to edit specific transcriptional factors that could be directly or indirectly fine-tune the expression and regulation of stress-responsive genes against salinity tolerance in plants (Debbarma et al., 2019). For example, CRISPR/Cas9 mediated genome editing of SlMAPK3 gene in tomato affected the expression level of other drought stress-responsive genes, particularly, SlDREB, SlLOX, and SlGST in tomato. The downregulation of these genes indirectly affected the salinity response and provided tolerance to salinity. Moreover, genetic engineering, gene silencing, CRISPR/Cas9 mediated-genome editing, transgenic overexpression, gene complementation and genetic transformation, mutant analysis studies done so far for engineering better salt-tolerance strategies in various vegetable and horticultural crops have of course identified novel signaling pathways, interconnected networks, transcriptional activators in mitigating salinity as well as other environmental stresses. Recently, CRISPR/Cas9 technology has provided a novel platform for precise editing of alleles that could assist in providing stress tolerance in plants. Further, the latest advancement in CRISPR-Cas system has sparked the genome editing revolution in plant genetics and breeding. We have discussed the role of some of the important TFs that regulate the stress-tolerance mechanism in plants.
FIGURE 2.
Functional annotation of WRKY and NAC proteins. These functional annotation were retrieved from Cello2GO server and were structured around gene ontologies like biological process, molecular function, and cellular component along with their localization probability. It is to be noted that both WRKY and NAC proteins have diversified function with more specifically targeted to DNA binding or Nucleic acid binding transcriptional factor activity.
WRKY Gene Family in Salt Tolerance in Vegetables
WRKY gene family is one of the most important transcriptional regulators that regulates stress tolerance mechanism in plants. Many studies done till so far have highlighted the functional role of WRKY gene signaling against various abiotic and biotic stress response in plants (Aamir et al., 2017, 2018, 2019; Hichri et al., 2017; Bai et al., 2018; Li et al., 2020). It has been well documented that WRKYs gene-mediated plant defense is controlled by both crossregulation and autoregulation, and extensive signaling involving multiple protein partners like histone acetylases, MAP kinases, MAP kinases kinases (MAPKK), calmodulin, 14-3-3 proteins, and other associated WRKYs partners in a complex network and dynamic web with built in redundancy to fine tune the transcriptional reprogramming, genetic-expression, and stress-tolerance (Rushton et al., 2010). WRKYs prominent role in both abiotic as well as biotic stress tolerance is well-documented (Rushton et al., 2010; Phukan et al., 2016; Bai et al., 2018). WRKYs role in salinity tolerance against various horticultural crops and other plants is well reported (Table 4). For example, Kashyap et al. (2020) reported the relevance of tomato WRKY1, WRKY3, and WRKY72 in mitigating salt stress in wild tomato Solanum chilense as the expression of these WRKYs was more prominent and increased the expression in wild genotype compared with domestic and cultivated genotype DVRT1. Villano et al. (2020) characterized the list of putative WRKYs involved in various abiotic and biotic stresses in two wild relatives of potato Solanum commersonii and Solanum chacoense, and revealed the ScWRKY23 as multiple stress regulator WRKY in wild potato (Villano et al., 2020). Likewise, Hichri et al. (2016) also reported the expression of tomato WRKY3 in alleviating salt stress (Hichri et al., 2016). Transcriptomic characterization and function validation through qRT-PCR analysis unraveled the expression profiling and importance of sweet potato WRKYs after treatment with 150 mM salt stress (Qin et al., 2020). In one work, Yue et al. (2019) provided the genome-wide identification and characterization of 92 WRKY genes in Chenopodium quinoa, and reported the importance of 25 WRKYs in both development and stress tolerance. Similarly, genome-wide identification and characterization of WRKYs in Cucumis sativus (cucumber) unraveled the importance of CsWRKY9, CsWRKY18, CsWRKY48, and CsWRKY57, in both heat and salt stress tolerance (Chen et al., 2020). In another study, based on Illumina RNA-seq transcriptomic studies, Tang et al. (2014) reported the tissue-specific and differential expression profiles of the Brassica rapa ssp. pekinensis (Chinese cabbage) and further validated their role in different abiotic and biotic stresses (Tang et al., 2014). Karanja et al. (2017a) reported 126 WRKYs in Raphanus sativus out of which 35 WRKYs had differential expression in various abiotic stresses. Further, the relevance of WRKY3 in salt tolerance could be better understood as CRISPR/Cas9-mediated WRKY3 and WRKY4 mutagenesis in Arabidopsis, decreasing both MeJA stress as well as decreased salt tolerance in Arabidopsis (Li et al., 2020).
TABLE 4.
Transcriptional regulation and their mode of action in vegetable crops for salt stress tolerance.
| S. No. | Transcription Factor/Gene/Protein | Vegetable | Functional aspects | References |
| 1. | JUNGBRUNNEN1 (JUB1), a NAC transcription factor | Solanum lycopersicum (tomato) | Overexpression of JUNGBRUNNEN1 (JUB1) increases salinity tolerance in tomato | Alshareef et al., 2019 |
| 2. | BZR/BES transcription factor | S. lycopersicum (tomato) | BRASSINAZOLE RESISTANT 1 (BZR1) and BRI1-EMS-SUPPRESSOR 1 (BES1) homologs in have potential role in salt tolerance. SlBZR1D played positive role in salt stress tolerance | Jia et al., 2021 |
| 3. | Wild tomato WRKY1, WRKY3, and WRKY72 | S. chilense (wild tomato) | Salinity stress tolerance | Kashyap et al., 2020 |
| 4. | Transgenic overexpression of tomato ERF84 in Arabidopsis | Arabidopsis thaliana | Salt and Drought | Li Q. L. et al., 2018 |
| 5. | Transgenic overexpression of tomato ERF1 | S. lycopersicum | Salt Stress | Lu et al., 2011 |
| 6. | Microarray analysis for salt-tolerant genes in wild tomato uncovered putative 5ERFs in alleviating salt stress | S. pimpinellifolium | Salt stress tolerance | Yang et al., 2018 |
| 7. | ScbZIP and SlbZIP | S. chilense (wild tomato) and S. lycopersicum | Salinity stress tolerance | Zhu et al., 2018; Kashyap et al., 2020 |
| 8. | Tomato SRN1 (Solanum lycopersicum stress-related NAC1) plasma membrane-localized protein with transactivation activity in yeast | S. lycopersicum | Positively regulates defense response against biotic stress but negatively regulates abiotic stress response | Liu et al., 2014 |
| 9. | Tomato NAC35 | S. lycopersicum | Induced by drought stress, salt stress, bacterial pathogen, and signaling molecules | Wang et al., 2016 |
| 10. | Tomato SlAREB1, a bZIP transcription factor, member of the ABA-responsive element binding protein (AREB)/ABA-responsive element binding factor (ABF) subfamily | S. lycopersicum | Salt stress and Drought stress tolerance | Orellana et al., 2010 |
| 11. | Tomato NAC4 and NAC35 | S. lycopersicum | Salt, Drought tolerance Biotic stress | Zhu et al., 2014; Wang et al., 2016 |
| 12. | ZFP179, a salt responsive gene encoding a Cys2/His2 zinc finger protein | Oryza sativa | Overexpression of ZFP179 provided salt tolerance | Sun et al., 2010 |
| 13. | BnaABF2, a bZIP transcription factor | Brassica napus | Salt tolerance in Transgenic Arabidopsis | Zhao et al., 2016 |
| 14. | Chili NAC46 | Capsicum annum | Salt tolerance in transgenic Arabidopsis | Ma et al., 2021 |
| 15. | Tomato DREB2 | S. lycopersicum and A. thaliana | Salt tolerance | Hichri et al., 2016 |
| 16. | Tomato ERF84, ERF5 | S. lycopersicum | Positive regulation for Salt and drought tolerance; negative regulation for biotic stress | Pan et al., 2012; Li Q. L. et al., 2018 |
| 17. | Chenopodium WRKY | Chenopodium quinoa | Stress tolerance and development | Yue et al., 2019 |
| 18. | CsWRKY9, CsWRKY18, CsWRKY48 and CsWRKY57 | Cucumis sativus | Heat and salt stress tolerance | Ling et al., 2011; Chen et al., 2020 |
| 19. | Radish WRKY | Raphanus sativus | Abiotic Stress tolerance | Karanja et al., 2017a |
| 20. | Carrot WRKY20 | Daucus carota | DcWRKY20 made interaction with DcMAPK1 and DcMAPK4 Abiotic and biotic stress tolerance | Li et al., 2016 |
| 21. | Tomato NAC1; NAC3 | S. lycopersicum | Salt stress tolerance; NAC3 suppressed by salt stress | Yang et al., 2011; Han et al., 2012, 2014 |
| 22. | Illumina RNA-seq transcriptomic studies of root, stem and leaves in Chinese cabbage | Brassica rapa ssp. pekinensis | Abiotic and biotic stress tolerance | Tang et al., 2014 |
| 23. | Carrot WRKYs in hormonal regulation and mechanical injuries | Daucus carota | Hormone and mechanical injuries | Nan and Gao, 2019 |
| 24. | Transcriptomic studies of sweet potato under salt stress | Ipomoea batatas | Salt stress tolerance | Qin et al., 2020 |
| 25. | Genome-wide identification and characterization of tomato WRKYs under drought, salt and biotic stress | S. lycopersicum | Drought, Salt, and Biotic stress | Huang et al., 2012 |
| 26. | Genome-wide identification and characterization of WRKYs in wild potato | S. commersonii and S. chacoense | ScWRKY045 as multiple stress-responsive regulator | Villano et al., 2020 |
| 27. | Identification of biotic-stress responsive WRKY from Brassica oleracea var. italica | B. oleracea var. italica | Increased expression of BoWRKy6 against biotic stress | Jiang et al., 2016 |
| 28. | Genome-wide characterization of potato WRKYs and expression analysis of potato 22 WRKYs under different stresses | S. tuberosum | Increased upregulation of StWRKY01 and StWRKY39 under different abiotic stresses. StWRKY58 had highest expression profile under drought and salt stress | Zhang et al., 2017 |
| 29. | Genome-wide identification and characterization of WRKYs in brinjal and Turkey berry | S. melongena L. S. torvum Sw.) | Biotic stress response | Yang et al., 2015 |
| 30 | Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L | S. lycopersicum | Negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance | Li et al., 2014 |
| 31 | Radish NAC145 | Raphanus sativus | Salt, heat and drought stresses | Karanja et al., 2017b |
| 32 | Melon NAC14 | Cucumis melo | Overexpression of CmNAC14 increased the sensitivity of transgenic Arabidopsis lines to salt stress | Wei et al., 2016 |
| 33. | Potato NAC proteins StNAC072 and StNAC101; StNAC2 | S. tuberosum | StNAC072 and StNAC101 are orthologs of known stress-responsive Arabidopsis RESPONSIVE TO DEHYDRATION 26 (RD26) involved in abiotic stress tolerance; Overexpression of StNAC2 in transgenic potato increased salt tolerance | Singh et al., 2013; Xu et al., 2014 |
| 34. | Watermelon WRKY ClWRKYs | Citrullus lanatus | Growth, Development, Biotic and Abiotic stress response | Yang et al., 2018 |
| 35. | Wild turnip WRKY (BsWRKYs) | Brassica rapa | Biotic and abiotic stress response | Kayum et al., 2015 |
| 36. | BjABR1, an AP2/ERF superfamily gene, from tuber mustard | Brassica juncea var. tumida Tsen et Lee | Abscisic acid and abiotic stress responses | Xiang et al., 2018 |
| 37. | Arabidopsis NAC2 | A. thaliana | Stress response and lateral root development | He et al., 2005 |
| 38. | Comparative transcriptome and proteome analysis of salt-tolerant and salt-sensitive genotypes of sweet potato and expression profiling of IbNAC07 | Ipomoea batatas | Salinity stress tolerance | Meng et al., 2020 |
| 39. | Genome wide characterization of WRKY genes in summer squash | Cucurbita pepo | Water and salt stress tolerance | Bankaji et al., 2019 |
| 40. | Genome-Wide Identification of AP2/ERF transcription Factors in cauliflower | Brassica oleracea L. var. botrytis | Salt and drought stress tolerance | Li et al., 2017 |
| 41 | Genome wide characterization of NAC family in celery and further transcriptomic characterization under salt stress AgNAC47 and AgNAC63 were key player | Apium graveolens | Heat, salinity, cold stress | Duan et al., 2020 |
| 42 | Genome-wide characterization of homeobox-leucine zipper gene family in tomato (Solanum lycopersicum) | S. lycopersicum | Functional analysis of SlHDZ34 (III sub-family member)under salinity stress revealed salt stress tolerance | Hong et al., 2021 |
| 43 | SlMYB02, a R2R3-type MYB transcription factor | S. lycopersicum | Salt tolerance | Zhang et al., 2020 |
| 44. | CabZIP25 | Capsicum annum | Salt tolerance | Gai et al., 2021 |
| 45. | Sweet potato bZIP IbbZIP1; IbABF4 | A. thaliana | Transgenic overexpression of IbbZIP1 in Arabidopsis provided salt tolerance; IbABF4 imparted multiple stress tolerance | Wang et al., 2019 |
| 46. | Tomato bZIP transcription factor SlAREB | S. lycopersicum | Salt tolerance | Hsieh et al., 2010 |
| 47. | SlbZIP38 tomato bZIP transcription factor | S. lycopersicum | Negative regulator of drought and Salt Stress Tolerance | Pan et al., 2017 |
| 48. | SlbHLH22 a Basic Helix-Loop-Helix (bHLH) transcription factor in tomato | S. lycopersicum | Transgenic over expression imparted high tolerance to both salinity and drought | Waseem et al., 2019 |
| 49. | AtMYB20 Arabidopsis R2R3-MYB transcription factor | A. thaliana | Negatively regulated type 2C serine/threonine protein phosphatases to positively regulate salt tolerance | Xu et al., 2014 |
| 50. | SlMYB102, R2R3-type MYB gene | S. lycopersicum | Transgenic overexpression provided salt tolerance | Zhang et al., 2020 |
Ethylene Response Factors
The APETALA2/ethylene responsive factor (AP2/ERF) family of transcription factor is one of the prominent groups of transcriptional activator/regulator during various abiotic and biotic stress responses in plants. ERF group has been further classified or subdivided into the dehydration-responsive element-binding proteins (DREBs). The AP2/ERF families in different plants have been further subdivided based on the presence of double AP2 domain, single AP2 domain, and/or single AP2 domain along with presence of a B3-DNA binding domain (Nakano et al., 2006). The interaction of ERF proteins with DRE/CRT motif and cis-acting elements is generally associated with stress-responsive genes and plays a crucial role in mitigation of various environmental stresses. For example, transgenic overexpression of ERF1-V (Haynaldia villosa) in wheat provided salt tolerance (Xing et al., 2017). Similarly, a ERF gene from wheat (TaERF3) overexpression had significant results against salt stress compared with control counterparts in wheat (Rong et al., 2014). In one work, Yang et al. (2018) performed the microarray analysis on salt-tolerant genes in wild tomato lines Solanum pimpinellifolium PI365967’ under the effect of salt treatment and reported the increased expression of five ERF genes (SpERF). Sequence analysis and transcriptomic characterization of these five SpERFs uncovered the crucial seven amino acid residues that were involved in binding with GCC box in the promoter of ethylene responsive genes and were shown to be conserved in all the reported ERFs (Yang et al., 2018). In recent years, ERF TFs have been investigated in depth in various horticultural as well as vegetable crops to enhance the breeding program as well as crop improvement, with respect to various environmental stresses in plants (Table 4). For example, transgenic overexpression of tomato ERF84 (SlERF84) in Arabidopsis provided resistance against salt and drought. Further, tomato ERFTF (Sl-ERF.B.3) expression was found to be decreased/downregulated under salinity stress and drought conditions, whereas it has been found to be upregulated/increased following the exposure to cold, flood, and heat response.
NAC Transcription Family
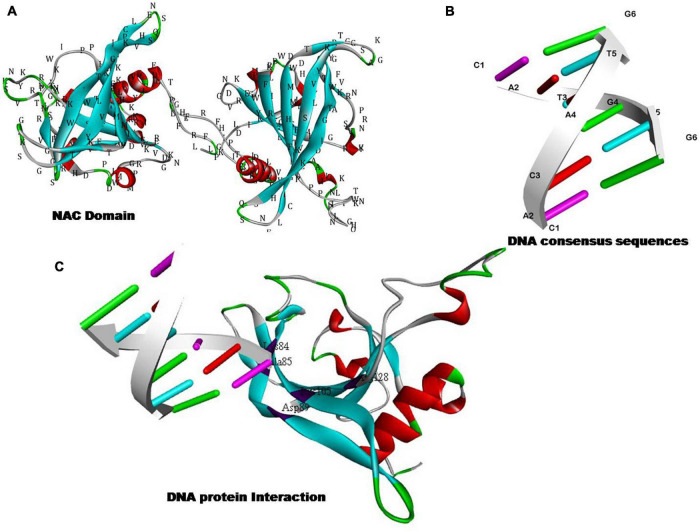
NAC (NAM, ATAF, and CUC) TFs have been considered an important group of transcriptional activators that play an important role in developmental programming as well as to encounter challenges against various environmental constraints (Tran et al., 2010; Puranik et al., 2012). The DNA binding property of NAC TFs lie at their N-terminal end (Figure 3). The expression of NAC proteins is highly dependent on the promoter region as each and every NAC gene is characterized by the presence of at least one unique cis-element type in their promoter (Li et al., 2016). NAC TFs role in various abiotic and biotic stress responses is well-documented (Table 4). For example, salt-tolerance in tomato is well regulated by NAC1 transcription factor as the enhanced expression of SlNAC1 in root, flower, seeds, and green fruits following the salt stress are well known (Yang et al., 2011). Transcriptomic characterization unraveled the importance of 10 NAC genes in tomato against abiotic stresses (Song et al., 2015). In this context, Liu et al. (2014) reported the relevance of tomato NAC transcription factor SlSRN1 in mediating the positive defense response against biotic stresses while regulating negatively to abiotic stress signaling (Liu et al., 2014). Yang et al. (2011) reported the expression profiling of tomato NAC1 (SlNAC1), an ATAF subfamily transcription factor in different tissues (root, leaves, seeds, and fruit) under salt stress (Yang et al., 2011). Likewise, potato NAC genes StNAC072 and StNAC101 that have been reported as orthologs of known stress-responsive Arabidopsis responsive to dehydration 26 (RD26) were found to play a crucial role in mitigating abiotic stress response (Singh et al., 2013). Wei et al. (2016) performed the genome-wide characterization of NAC transcription factor family in melon (Cucumis melo L.) and evaluated their expression profile during salt stress. Further, transgenic overexpression of CmNAC14 in Arabidopsis resulted in increased salt-tolerance (Wei et al., 2016). Karanja et al. (2017b) reported the tissue-specific expression profiling of radish NAC TFs and reported the positive regulation of RsNAC023 and RsNAC080 toward all types to abiotic stresses. Further, RsNAC145 had much more active expression profile under salt, heat, and drought stresses when compared with other genes that were expressed under different abiotic stresses (Karanja et al., 2017b). Li et al. (2016) investigated and characterized the list of putative NAC TFs in water melon (Citruluslanatus) across the genome and also checked the expression profile and potential function of several NAC TFs in different stresses. Overall, transgene overexpression of IbNAC7 in Arabidopsis provided salt tolerance. Recently, Duan et al. (2020) provided the genome-wide characterization of NAC gene family in leafy vegetable Apium graveolens and studied the characterized WRKYs for their stress-tolerance attribute. It was found that a total 111 NAC member were present based on genomic studies. Further, transcriptomic characterization under various abiotic stresses uncovered the AgNAC63 (ortholog of ANAC072/RD26 role in mitigating salt, cold, and heat stresses). However, the study reported tissue-specific higher expression profiles AgNAC63 and AgNAC47 in leaves under the different treatments (Duan et al., 2020).
FIGURE 3.
Hypothetical models showing the DNA binding and/or nucleic acid binding transcription factor activity of NAC proteins. (A) The structure of NAC domain. (B) Structure of NAC binding DNA consensus sequences. (C) DNA–protein interaction of NAC proteins with relevant DNA sequences to fine tune the gene regulatory aspects.
Basic Leucine Zipper
The basic leucine (Leu) zipper (bZIP) family also includes one of the most important group of transcriptional activator against abiotic and biotic stress response (Pan et al., 2017). bZIP family also plays an essential role in growth and development of plants (Sornaraj et al., 2016). The bZIP name was based on the presence of the bZIP domain. The bZIP domain is characterized by some specific structural features that is located on an alpha-helix. It has been reported that the first 18 amino acid residues constitute the basic group followed by an invariant N-x7-R/K-x9 motifs for nuclear localization and sequence-specific DNA binding. However, for the second part, the Leu zipper region composed of several heptad repeats of Leu amino acid or other bulky amino acids, such as isoleucine, valine, phenyl nine, tryptophan, or methionine, positioned exactly nine amino acids toward the C-terminus, creating an amphipathic helix (Jakoby et al., 2002; Nijhawan et al., 2008). It has been reported that in bZIP transcriptional proteins, apart from the bZIP domain, some other transcriptional active domains are found and play an essential role in bZIP functioning. The most common site that function as transcriptional activator include phosphorylation site [R/KxxS/T (Furihata et al., 2006; Liao et al., 2008)] and a string of glutamine rich motif. The basic part of the Leu zipper interact with ACGT core region of the B-DNA sequences, particularly, at A-box (TACGTA), G-box (CACGTG), and C-box (GACGTC) (Izawa et al., 1993; Foster et al., 1994). In fact, during the DNA–protein interaction of bZIP proteins with DNA motifs, the half N-terminal of the bZIP domain interacts with DNA major grove region, whereas the other C-terminal end of the Leu zipper constitutes dimer formation for a coiled superimposed structure, defined as zipper superimposed coiled structure, the so-called zipper (Landschulz et al., 1988; Ellenberger et al., 1992). bZIP proteins also play an essential role in imparting tolerance against several abiotic as well as biotic stresses (Table 4). For example, Gai et al. (2021) characterize the list of different bZIP TFs in pepper and reported the relevance of CabZIP25 in imparting salt tolerance as overexpression done in transgenic Arabidopsis. Wang et al. (2018) identified the list of 54 and 50 bZIP proteins from whole-genome sequences of Vigna radiata and Vigna angularis, respectively.
Grafting Strategies in Vegetable Crops for Salt Stress Tolerance
Improving the productivity of vegetable crops is a challenge under salt-affected soil or water. Hence, increasing salt tolerance in vegetable crops will have a greater impact in nutritional and economic security, particularly of (semi) arid regions, where salinity in soil and water are widespread (Singh et al., 2020). Traditional breeding programs have been attempted to improve salt tolerance in crop plants (Borsani et al., 2003), but the commercial success is limited due to the trait’s complexity. Currently, major efforts are being directed toward genetic transformation in plants to increase their tolerance, and despite the trait’s complexity, the transfer of a single gene or a few genes has resulted in claims of improved salt tolerance, such as the expression of genes involved in the control of Na+ transport (Gaxiola et al., 2001). But, the genetically complicated mechanisms of abiotic stress tolerance, as well as the possibility for adverse side effects make this a challenging task (Flowers, 2004). However, unless a full proof practical and faster breeding tool comes in vogue, a well-proven fast and eco-friendly technique “vegetable grafting” can be deployed to increase tolerance to stresses in vegetables. Vegetable grafting, in fact, has emerged as an efficient tool to sustainably increase vigor and yield of commercial cultivars under challenged growth environment by mechanically attaching with resistant root genotypes.
Grafting Alleviates Salt Stress
Salinity disturbs dry mass partitioning between vegetative and reproductive organs, whereas grafted plants exhibited less alteration (Parthasarathi et al., 2021). In grafting, some rootstocks may have better performance than the others, though their response may change depending on level of salt concentration in the growth medium (Singh et al., 2020; Bayoumi et al., 2021). Numerous reports have demonstrated the ameliorative response of grafting to salinity stress in cucurbitaceous crops (e.g., melon, watermelon, and cucumber) involving the Cucurbita interspecific hybrid rootstocks (Goreta et al., 2008; Rouphael et al., 2012). The agronomic performance of pepper cv. “Adige” under natural salinity condition was clearly evident with 75% higher yield and with 31% lesser fruit damage (blossom end rot) when it was grafted onto a salt-tolerant accession “A 25” as rootstock in comparison with non-grafted control plants (Penella et al., 2016). Eggplant (“SuqiQie”) grafting onto the rootstock of wild eggplant (Solanum torvum cv. “Torvum vigor”) provided salinity tolerance by minimizing the yield reduction under saline stress (Wei et al., 2009). In contrary, Chen et al. (2003) found that scion genotypes had a significant impact on the growth of grafted tomato plants, regardless of the salinity of the growing environment, but rootstock had no impact.
Mechanism of Salt Tolerance in Grafted Plants
Grafting is a reciprocal integrative process; the salt tolerance of grafted plants is influenced by both scion and rootstock (Etehadnia et al., 2008). The positive response of grafting can be attributed to more vigorous and robust root systems, greater efficiency of roots for water and nutrient uptake with efficiency to exclude salt-ions, higher photosynthesis, and better oxidative defense system, hormonal regulations, and osmotic adjustment of the grafted plants as compared with the non-grafted plants (Amaro et al., 2014; Rouphael et al., 2017; Singh et al., 2020).
Root Characteristics
Root, besides providing physical support and anchor to the plants, plays a crucial role in water and ion uptake and their supply to aerial part that help regulate various plant processes (Kumar et al., 2017). However, the alteration in root characteristics is expected to occur since roots being the foremost plant organ exposed to saline growth medium (Singh et al., 2020), consequently the subsequent effects on water and mineral uptake (Rouphael et al., 2017). The numerous reports claim that grafting onto genetically strong root system can effectively mitigate the effect of salinity on the performance of salt-sensitive scion cultivars (Colla et al., 2013); hence it prompts the emphasis of selecting the vigorous root stock for increasing salt tolerance (Colla et al., 2013; Singh et al., 2020). Salinity depressed shoot and root parameters, but grafted plants of tomato onto potato rootstocks were able to avoid the changes in their growths with balanced partitioning between vegetative and reproductive dry masses (Parthasarathi et al., 2021). Furthermore, the tolerance ability of grafting provided by the rootstocks is often associated with the root morphological characteristics to exclude Na+ and/or Cl– under saline medium.
Regulation of Salt and Mineral Ions
Grafted plants tend to restrain Na+ and Cl– ions in their root tissues, preventing them from being translocated to the shoots and leaves in high concentrations. The diverse agronomic responses of grafted plants to salinity in numerous studies were resulted by the differential abilities of root genotypes to regulate the uptake and/or translocation of ions of the salts, and of nutrients, due to their competitive interactions (Rouphael et al., 2017). The ability of rootstocks to minimize toxicity of Na+ and/or Cl– by exclusion and/or reduction of Cl– absorption by the roots, as well as the replacement or substitution of K+ by total Na+ in the foliage has been related to the enhancement of salt tolerance by grafting (Martinez-Rodriguez et al., 2008). In pepper, salt-tolerant wild pepper rootstocks “ECU-973” (Capsicum chinense) and “BOL-58” (Capsicum baccatum var. pendulum) provided salinity tolerance in pepper (“Adige”) plants by controlling Na+ and Cl– ions accumulation in shoots (Penella et al., 2015). In spite of maintaining better control over Na+ and Cl– accumulation in their shoots, grafted plants were able to maintain higher ratio of K+/Na+ in grafted cucumber on pumpkin rootstock (Usanmaz and Abak, 2019), and higher K+ and Ca++ in grafted eggplant (Talhouni et al., 2019). Salinity tolerance in a salt-sensitive cucumber (“Jinchun No. 2”) was enhanced by grafting onto a salt-tolerant pumpkin rootstock (“Chaojiquanwang”); this tolerance mechanism shows the better ability of pumpkin rootstock to exclude Na+, and thus lesser amount of Na+ ions (−69%) reaches the cucumber shoots (Huang et al., 2013).
Physio-Biochemical Alterations
The tolerance response of grafting on vigorous rootstock with efficiency to control Na+ ion accumulation in shoots has been associated with the efficiency of rootstocks to modulate water uptake by roots and losses by transpiration. Grafting onto some rootstock was useful to maintain better leaf water status than the others. Grafting tomato (“Ikram”) on potato rootstock (“Charlotte”) was found promising to increase salinity tolerance of 5.0 dS/m in grafted tomato with enhanced water productivity (+56.8%) (Parthasarathi et al., 2021). Grafting onto certain rootstocks was able to mitigate salt induced photoinhibition of photosynthesis and consequently growth of grafted plants (Liu Z. et al., 2013). As a coping mechanism of oxidative damage, plants activate enzymatic (i.e., ascorbate peroxidase, catalase, superoxide dismutase, monodehydroascorbate reductase, dehydro ascorbate reductase, and glutathione reductase) as well as non-enzymatic (i.e., reduced glutathione, reduced ascorbate, carotenoids, and tocopherols) antioxidant systems (Rouphael et al., 2017; Singh et al., 2020). Grafting studies demonstrated that some of the graft combinations have a better ability to mitigate salinity stress by regulating antioxidative defense system than the others. Salt-stressed eggplants experienced oxidative stress (higher malondialdehyde, MDA), whereas grafted eggplants were capable of mitigating ROS induced by oxidative stress as a result of increased level of antioxidant enzymes (SOD, CAT, and APX) (Talhouni et al., 2019). Polyamines increase under salinity stress and hence increases plants tolerance; the increased level of polyamines (free, soluble, and conjugated polyamines) provided better tolerance to salinity (i.e., excess calcium nitrate) in grafted seedlings than the non-grafted tomatoes (Wei et al., 2009). Stegemann and Bock (2009) reported that plant grafting can result in the exchange of genetic information via either large DNA pieces or entire plastid genomes. However, gene transfer is restricted to the contact zone between scion and rootstock. Thus, the use of rootstock cannot change the sensitivity of scion itself to salt stress. Working with model plant Arabidopsis, Shi et al. (2002) reported that SOS1 (salt excessively sensitive) gene is expected to play a role in the loading of Na+ into the xylem tracheids from xylem parenchyma cells. Further reports suggest that in Arabidopsis, high affinity K+ transporters (HKTs) were involved in the removal of Na+ from the xylem (Rus et al., 2001, 2006; Sunarpi et al., 2005; Davenport et al., 2007), and hence the leaves were safe from Na+ ion toxicity. Furthermore, it was recently discovered that expressing the Na+ transporter HKT1;1 in the mature root stele of Arabidopsis thaliana utilizing an enhancer trap expression system reduced Na+ build up in the shoot by 37–64%, and hence increased salinity tolerance (Møller et al., 2009). Using grafting experiments, it was discovered that HKT1;1 expressed in the root rather than the shoot regulates Na+ accumulation in Arabidopsis shoots (Rus et al., 2006), implying that the SOS1 analogous gene and HKTs are likely involved in Na+ transport in the pumpkin rootstock, allowing it to limit Na+ transport from the root to the shoot.
Grafting onto some rootstocks has shown to also increase scion’s tolerance to salinity by modulating the hormonal balance namely of ABA, cytokinins, and polyamines (Rouphael et al., 2017). The reduced transpiration with maintained leaf water relations by elevated level of shoot ABA concentration under salt stress have been reported (Singh et al., 2020). The enhanced salinity tolerance in tomato was related to increased level of ABA content in scion shoots, irrespective of the rootstocks raised under saline condition (Chen et al., 2003). Likewise, increased root-to-shoot cytokine transport by rootstock that overexpressed cytokinin biosynthesis genes (e.g., isopentenyl transferase) was associated with the increased salinity tolerance in tomato presented by maintained stomatal conductance and photosystem II efficiency accompanied with lesser accumulation of toxic ions, consequently producing higher shoot and fruit growths (Ghanem et al., 2011).
Conclusion
Abiotic stresses like salt stress which persists throughout plant’s whole life cycle negatively affects plant yield and nutritional quality. To ensure vegetable production under salt stress many transgenes have been transferred to the vegetables. Transforming vegetables are the one of the most reliable technique to cope salt stress as most of the vegetable gene pool lack novel gene for salt stress in its gene pool. To manage salt stress at molecular level, the native TFs in the vegetable crops are also being regulated to sustain yield and quality under salt stress. Grafting has shown potential to alleviate salinity stress (water or soil) on the selected vigorous and tolerant rootstocks. Certain wild accessions which possess resistance to salinity but are difficult to introgress these traits into commercial cultivars through traditional breeding tools, can be utilized as rootstock to increase grafted scion’s efficiency or tolerance to salinity. Plant biologists around the world are grappling with the dilemma of exponential population growth and rising food demand. Abiotic stress, such as salt, is a major threat to agricultural productivity and has been linked to worsening food security trends since the beginning. Soil salinity and degradation of soil quality are linked to lower agricultural yields. The production of salinity-tolerant crops is the only way to ensure global food. The actual yield produced by saline soils is more than half of what was originally predicted for normal soils. Organic matter and biodiversity are quite low in these soils.
Author Contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aamir M., Kashyap S. P., Zehra A., Dubey M. K., Singh V. K., Ansari W. A., et al. (2019). Trichoderma erinaceum bio-priming modulates the WRKYs defense programming in tomato against the Fusarium oxysporum f. sp. lycopersici (Fol) challenged condition. Front. Plant Sci. 10:911. 10.3389/fpls.2019.00911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aamir M., Singh V. K., Dubey M. K., Kashyap S. P., Zehra A., Upadhyay R. S., et al. (2018). Structural and functional dissection of differentially expressed tomato WRKY transcripts in host defense response against the vascular wilt pathogen (Fusarium oxysporum f. sp. lycopersici). PLoS One 13:e0193922. 10.1371/journal.pone.0193922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aamir M., Singh V. K., Meena M., Upadhyay R. S., Gupta V. K., Singh S. (2017). Structural and functional insights into WRKY3 and WRKY4 transcription factors to unravel the WRKY–DNA (W-Box) complex interaction in tomato (Solanum lycopersicum L.). A computational approach. Front. Plant Sci. 8:819. 10.3389/fpls.2017.00819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abiala M. A., Abdelrahman M., Burritt D. J., Tran L. S. P. (2018). Salt stress tolerance mechanisms and potential applications of legumes for sustainable reclamation of salt-degraded soils. Land. Degrad. Dev. 29 3812–3822. 10.1002/ldr.3095 [DOI] [Google Scholar]
- Ahmed I., Yadav D., Shukla P., Vineeth T. V., Sharma P. C., Kirti P. B. (2017). Constitutive expression of Brassica junceaannexin, AnnBj2 confers salt tolerance and glucose and ABA insensitivity in mustard transgenic plants. Plant Sci. 265 12–28. 10.1016/j.plantsci.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Ahmad R., Hussain J., Jamil M., Kim M. D., Kwak S. S., Shah M. M., et al. (2014). Glycinebetaine synthesizing transgenic potato plants exhibit enhanced tolerance to salt and cold stresses. Pak. J. Bot. 46 1987–1993. [Google Scholar]
- Alexieva V., Sergiev I., Mapelli S., Karanov E. (2001). The effect of drought and ultraviolet radiation on growth and stress markers in pea and wheat. Plant. Cell Environ. 24 1337–1344. 10.1046/j.1365-3040.2001.00778.x [DOI] [Google Scholar]
- Ali A., Ali Q., Iqbal M. S., Nasir I. A., Wang X. (2020). Salt tolerance of potato genetically engineered with the atriplex canescens BADH gene. Biol. Plant. 64 271–279. 10.32615/bp.2019.080 [DOI] [Google Scholar]
- Alshareef N. O., Wang J. Y., Ali S., Al-Babili S., Tester M., Schmöckel S. M. (2019). Overexpression of the NAC transcription factor JUNGBRUNNEN1 (JUB1) increases salinity tolerance in tomato. Plant. Phys. Bioch. 140 113–121. 10.1016/j.plaphy.2019.04.038 [DOI] [PubMed] [Google Scholar]
- Amaro A. C. E., Macedo A. C., Ramos A. R. P., Goto R., Ono E. O., Rodrigues J. D. (2014). The use of grafting to improve the net photosynthesis of cucumber. Theor. Exp. Plant Physiol. 26 241–249. 10.1007/s40626-014-0023-1 [DOI] [Google Scholar]
- Baghour M., Gálvez F. J., Sánchez M. E., Aranda M. N., Venema K., Rodríguez-Rosales M. P. (2019). Overexpression of LeNHX2 and SlSOS2 increases salt tolerance and fruit production in double transgenic tomato plants. Plant Phys. Biochem. 135 77–86. 10.1016/j.plaphy.2018.11.028 [DOI] [PubMed] [Google Scholar]
- Bai Y., Sunarti S., Kissoudis C., Visser R. G., Van Der Linden C. G. (2018). The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front. Plant Sci. 9:801. 10.3389/fpls.2018.00801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaji I., Sleimi N., Vives-Peris V., Gómez-Cadenas A., Pérez-Clemente R. M. (2019). Identification and expression of the Cucurbita WRKY transcription factors in response to water deficit and salt stress. Sci. Hort. 256:108562. 10.1016/j.scienta.2019.108562 [DOI] [Google Scholar]
- Bayoumi Y., Abd-Alkarim E., El-Ramady H., El-Aidy F., Hamed E.-S., Taha N., et al. (2021). Grafting improves fruit yield of cucumber plants grown under combined heat and soil salinity stresses. Horticulturae 7:61. 10.3390/horticulturae7030061 [DOI] [Google Scholar]
- Bogoutdinova L. R., Lazareva E. M., Chaban I. A., Kononenko N. V., Dilovarova T., Khaliluev M. R., et al. (2020). Salt stress-induced structural changes are mitigated in transgenic tomato plants over-expressing superoxide dismutase. Biology 9:297. 10.3390/biology9090297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordas M., Montesinos C., Dabauza M., Salvador A., Roig L. A., Serrano R., et al. (1997). Transfer of the yeast salt tolerance gene HAL1 to Cucumis melo L. cultivars and in vitro evaluation of salt tolerance. Transgenic Res. 6 41–50. 10.1023/A:1018453032336 [DOI] [PubMed] [Google Scholar]
- Borsani O., Valpuesta V., Botella M. A. (2003). Developing salt-tolerant plants in a new century: a molecular biology approach. Plant Cell Tiss. Org. Cult. 73 101–115. 10.1023/A:1022849200433 [DOI] [Google Scholar]
- Bouaziz D., Pirrello J., Charfeddine M., Hammami A., Jbir R., Dhieb A., et al. (2013). Overexpression of StDREB1 transcription factor increases tolerance to salt in transgenic potato plants. Mol. Biotec. 54 803–817. 10.1007/s12033-012-9628-2 [DOI] [PubMed] [Google Scholar]
- Bulle M., Yarra R., Abbagani S. (2016). Enhanced salinity stress tolerance in transgenic chilli pepper (Capsicum annuum L.) plants overexpressing the wheat antiporter (TaNHX2) gene. Mol. Breed. 36:36. 10.1007/s11032-016-0451-5 [DOI] [Google Scholar]
- Chen C., Chen X., Han J., Lu W., Ren Z. (2020). Genome-wide analysis of the WRKY gene family in the cucumber genome and transcriptome-wide identification of WRKY transcription factors that respond to biotic and abiotic stresses. BMC Plant Biol. 20:443. 10.1186/s12870-020-02625-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Fu X., Lips S. H., Sagi M. (2003). Control of plant growth resides in the shoot, and not in the root, in reciprocal grafts of flacca and wild-type tomato (Lysopersicon esculentum), in the presence and absence of salinity stress. Plant Soil 256 205–215. 10.1023/A:1026279719242 [DOI] [Google Scholar]
- Chinnusamy V., Jagendorf A., Zhu J. K. (2005). Understanding and improving salt tolerance in plants. Crop Sci. 45 437–448. 10.2135/cropsci2005.0437 34798789 [DOI] [Google Scholar]
- Chourasia K. N., Lal M. K., Tiwari R. K., Dev D., Kardile H. B., Patil V. U., et al. (2021). Salinity stress in potato: understanding physiological, biochemical and molecular responses. Life 11:545. 10.3390/life11060545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colla G., Rouphael Y., Jawad R., Kumar P., Rea E., Cardarelli M. (2013). The effectiveness of grafting to improve NaCl and CaCl2 tolerance in cucumber. Sci. Hortic. 164 380–391. 10.1016/j.scienta.2013.09.023 [DOI] [Google Scholar]
- Dalal M., Tayal D., Chinnusamy V., Bansal K. C. (2009). Abiotic stress and ABA-inducible Group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J. Biotechnol. 139 137–145. 10.1016/j.jbiotec.2008.09.014 [DOI] [PubMed] [Google Scholar]
- Davenport R. J., Munoz-Mayor A., Jha D., Essah P. A., Rus A., Tester M. (2007). The Na+ transporter AtHKT1;1 controls retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 30 497–507. 10.1111/j.1365-3040.2007.01637.x [DOI] [PubMed] [Google Scholar]
- Debbarma J., Sarki Y. N., Saikia B., Boruah H. P. D., Singha D. L., Chikkaputtaiah C. (2019). Ethylene response factor (ERF) family proteins in abiotic stresses and CRISPR–Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: a review. Mol. Biotech. 61 153–172. 10.1007/s12033-018-0144-x [DOI] [PubMed] [Google Scholar]
- Duan A. Q., Yang X. L., Feng K., Liu J. X., Xu Z. S., Xiong A. S. (2020). Genome-wide analysis of NAC transcription factors and their response to abiotic stress in celery (Apium graveolens L.). Comp. Biochem. 84:107186. 10.1016/j.compbiolchem.2019.107186 [DOI] [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. (1992). The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell 71 1223–1237. 10.1016/S0092-8674(05)80070-4 [DOI] [PubMed] [Google Scholar]
- Ellul P., Rios G., Atares A., Roig L. A., Serrano R., Moreno V. (2003). The expression of the Saccharomyces cerevisiae HAL1 gene increases salt tolerance in transgenic watermelon [Citrullus lanatus (Thunb.) Matsun.&Nakai.]. Theor. Appl. Genet. 107 462–469. 10.1007/s00122-003-1267-3 [DOI] [PubMed] [Google Scholar]
- Etehadnia M., Waterer D., Jong H. D., Tanino K. K. (2008). Scion and rootstock effects on ABA-mediated plant growth regulators and salt tolerance of acclimated and unacclimated potato genotypes. J. Plant Growth Regul. 27 125–140. 10.1007/s00344-008-9039-6 [DOI] [Google Scholar]
- Flowers T. J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55 307–319. 10.1093/jxb/erh003 [DOI] [PubMed] [Google Scholar]
- Flowers T. J., Flowers S. A. (2005). Why does salinity pose such a difficult problem for plant breeders? Agric. Water Manag. 78 15–24. 10.1016/j.agwat.2005.04.015 [DOI] [Google Scholar]
- Foster R., Izawa T., Chua N. H. (1994). Plant bZIP proteins gather at ACGT elements. FASEB J. 8 192–200. 10.1096/fasebj.8.2.8119490 [DOI] [PubMed] [Google Scholar]
- Franzoni G., Cocetta G., Trivellini A., Ferrante A. (2020). Transcriptional regulation in rocket leaves as affected by salinity. Plants 9:20. 10.3390/plants9010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzoni G., Trivellini A., Bulgari R., Cocetta G., Ferrante A. (2019). “Bioactive molecules as regulatory signals in plant responses to abiotic stresses,” in Plant Signaling Molecules, eds Khan M. I., Reddy P. S., Ferrante A., Khan N. A. (Sawston: Woodhead Publishing; ), 169–182. 10.1016/B978-0-12-816451-8.00010-1 [DOI] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103 1988–1993. 10.1073/pnas.0505667103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai Y., Li L., Liu B., Ma H., Chen Y., Zheng F., et al. (2021). Critical roles of bZIP transcription factors in fungal development, conidiation and stress response in Alternaria alternata. Microbiol. Res. 256:126915. 10.1016/j.micres.2021.126915 [DOI] [PubMed] [Google Scholar]
- García-Abellan J. O., Egea I., Pineda B., Sanchez-Bel P., Belver A., Garcia-Sogo B., et al. (2014). Heterologous expression of the yeast HAL5 gene in tomato enhances salt tolerance by reducing shoot Na+ accumulation in the long term. Phys. Plant 152 700–713. 10.1111/ppl.12217 [DOI] [PubMed] [Google Scholar]
- Gaxiola R. A., Li J. S., Undurraga S., Dang L. M., Allen G. J., Alper S. L., et al. (2001). Drought- and salt-tolerant plants result from overexpression of the AVP1 H+- pump. Proc. Natl. Acad. Sci. U.S.A. 98 11444–11449. 10.1073/pnas.191389398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanem M. E., Albacete A., Smigocki A. C., Frébort I., Pospíšilová H., Martínez-Andújar C., et al. (2011). Root-synthesized cytokinins improve shoot growth and fruit yield in salinized tomato (Solanum lycopersicum L.) lants. J. Exp. Bot. 62 125–140. 10.1093/jxb/erq266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M., Petropoulos S. A., Rouphael Y. (2021). Response and defence mechanisms of vegetable crops against drought, heat and salinity stress. Agriculture 11:463. 10.3390/agriculture11050463 [DOI] [Google Scholar]
- Goel D., Singh A. K., Yadav V., Babbar S. B., Bansal K. C. (2010). Overexpression of osmotin gene confers tolerance to salt and drought stresses in transgenic tomato (Solanum lycopersicum L.). Protoplasma 245 133–141. 10.1007/s00709-010-0158-0 [DOI] [PubMed] [Google Scholar]
- Golldack D., Li C., Mohan H., Probst N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151. 10.3389/fpls.2014.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goreta S., Bucevic-Popovic V., Selak G. V., Pavela-Vrancic M., Perica S. (2008). Vegetative growth, superoxide dismutase activity and ion concentration of salt-stressed watermelon as influenced by rootstock. J. Agric. Sci. 146 695–704. 10.1017/S0021859608007855 [DOI] [Google Scholar]
- Han J. S., Park K. I., Jeon S. M., Park S., Naing A. H., Kim C. K. (2015). Assessments of salt tolerance in a bottle gourd line expressing the Arabidopsis H+-pyrophosphatase AVP 1 gene and in a watermelon plant grafted onto a transgenic bottle gourd rootstock. Plant Breed. 134 233–238. 10.1111/pbr.12253 [DOI] [Google Scholar]
- Han Q., Zhang J., Li H., Luo Z., Ziaf K., Ouyang B., et al. (2012). Identification and expression pattern of one stress-responsive NAC gene from Solanum lycopersicum. Mol. Biol. Rep. 39 1713–1720. 10.1007/s11033-011-0911-2 [DOI] [PubMed] [Google Scholar]
- Han Q. Q., Song Y. Z., Zhang J. Y., Liu L. F. (2014). Studies on the role of the SlNAC3 gene in regulating seed development in tomato (Solanum lycopersicum). J. Hort. Sci. Biot. 89 423–429. 10.1080/14620316.2014.11513101 [DOI] [Google Scholar]
- He X. J., Mu R. L., Cao W. H., Zhang Z. G., Zhang J. S., Chen S. Y. (2005). AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44 903–916. 10.1111/j.1365-313X.2005.02575.x [DOI] [PubMed] [Google Scholar]
- Hichri I., Muhovski Y., Clippe A., Žižková E., Dobrev P. I., Motyka V., et al. (2016). SlDREB2, a tomato dehydration-responsive element-binding 2 transcription factor, mediates salt stress tolerance in tomato and A rabidopsis. Plant. Cell Environ. 39 62–79. 10.1111/pce.12591 [DOI] [PubMed] [Google Scholar]
- Hichri I., Muhovski Y., Žižková E., Dobrev P. I., Gharbi E., Franco-Zorrilla J. M., et al. (2017). The Solanum lycopersicum WRKY3 transcription factor SlWRKY3 is involved in salt stress tolerance in tomato. Front. Plant Sci. 8:1343. 10.3389/fpls.2017.01343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y., Liu Y., Zhang Y., Jia L., Yang X., Zhang X., et al. (2021). Genome-wide characterization of homeobox-leucine zipper gene family in tomato (Solanum lycopersicum) and functional analysis of SlHDZ34 (III sub-family member) under salinity stress. Environ. Exper Bot. 192:104652. 10.1016/j.envexpbot.2021.104652 [DOI] [Google Scholar]
- Hsieh T. H., Li C. W., Su R. C., Cheng C. P., Tsai Y. C., Chan M. T. (2010). A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 231 1459–1473. 10.1007/s00425-010-1147-4 [DOI] [PubMed] [Google Scholar]
- Huang S., Gao Y., Liu J., Peng X., Niu X., Fei Z., et al. (2012). Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Gen. 287 495–513. 10.1007/s00438-012-0696-6 [DOI] [PubMed] [Google Scholar]
- Huang Y., Bie Z., Liu P., Niu M., Zhen A., Liu Z., et al. (2013). Reciprocal grafting between cucumber and pumpkin demonstrates the roles of the rootstock in the determination of cucumber salt tolerance and sodium accumulation. Sci. Hortic. 149 47–54. 10.1016/j.scienta.2012.04.018 [DOI] [Google Scholar]
- Hu D. G., Ma Q. J., Sun C. H., Sun M. H., You C. X., Hao Y. J. (2016). Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol. Plant. 156 201–214. 10.1111/ppl.12354 [DOI] [PubMed] [Google Scholar]
- Huang J., Hirji R., Adam L., Rozwadowski K. L., Hammerlindl J. K., Keller W. A., et al. (2000). Genetic engineering of glycinebetaine production toward enhancing stress tolerance in plants: metabolic limitations. Plant Physiol. 122 747–756. 10.1104/pp.122.3.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas R., Olias R., Eljakaoui Z., Gálvez F. J., Li J., De Morales P. A., et al. (2012). Overexpression of SlSOS2 (SlCIPK24) confers salt tolerance to transgenic tomato. Plant Cell Environ. 35 1467–1482. 10.1111/j.1365-3040.2012.02504.x [DOI] [PubMed] [Google Scholar]
- Ijaz R., Ejaz J., Gao S., Liu T., Imtiaz M., Ye Z., et al. (2017). Overexpression of annexin gene AnnSp2, enhances drought and salt tolerance through modulation of ABA synthesis and scavenging ROS in tomato. Sci. Rep. 7:12087. 10.1038/s41598-017-11168-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram J. (2011). A food systems approach to researching food security and its interactions with global environmental change. Food Sec. 3 417–431. 10.1007/s12571-011-0149-9 [DOI] [Google Scholar]
- Izawa T., Foster R., Chua N. H. (1993). Plant bZIP protein DNA binding specificity. J. Mol. Biol. 230 1131–1144. 10.1006/jmbi.1993.1230 [DOI] [PubMed] [Google Scholar]
- Jakoby M., Weisshaar B., Droge-Laser W., Vicente-Carbajosa J., Tiedemann J., Kroj T., et al. (2002). BZIP transcription factors in Arabidopsis. Trends Plant Sci. 7 106–111. 10.1016/S1360-1385(01)02223-3 [DOI] [PubMed] [Google Scholar]
- Jia C., Zhao S., Bao T., Zhao P., Peng K., Guo Q., et al. (2021). Tomato BZR/BES transcription factor SlBZR1 positively regulates BR signaling and salt stress tolerance in tomato and Arabidopsis. Plant Sci. 302:110719. 10.1016/j.plantsci.2020.110719 [DOI] [PubMed] [Google Scholar]
- Jiang M., Liu Q. E., Liu Z. N., Li J. Z., He C. M. (2016). Over-expression of a WRKY transcription factor gene BoWRKY6 enhances resistance to downy mildew in transgenic broccoli plants. Aust. Plant Path. 45 327–334. 10.1007/s13313-016-0416-5 [DOI] [Google Scholar]
- Karanja B. K., Fan L., Xu L., Wang Y., Zhu X., Tang M., et al. (2017a). Genome-wide characterization of the WRKY gene family in radish (Raphanus sativus L.) reveals its critical functions under different abiotic stresses. Plant Cell Rep. 36 1757–1773. 10.1007/s00299-017-2190-4 [DOI] [PubMed] [Google Scholar]
- Karanja B. K., Xu L., Wang Y., Muleke E. M. M., Jabir B. M., Xie Y., et al. (2017b). Genome-wide characterization and expression profiling of NAC transcription factor genes under abiotic stresses in radish (Raphanus sativus L.). Peer. J. 5:e4172. 10.7717/peerj.4172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkute S. G., Krishna R., Ansari W. A., Singh B., Singh P. M., Singh M., et al. (2019). Heterologous expression of the AtDREB1A gene in tomato confers tolerance to chilling stress. Biol. Plant 63 268–277. 10.32615/bp.2019.031 [DOI] [Google Scholar]
- Karthikeyan A., Pandian S. K., Ramesh M. (2011). Transgenic indica rice cv. ADT 43 expressing a Δ 1-pyrroline-5-carboxylate synthetase (P5CS) gene from Vigna aconitifolia demonstrates salt tolerance. Plant Cell Tis. Org. Cult. 107 383–395. 10.1007/s11240-011-9989-4 [DOI] [Google Scholar]
- Kashyap S. P., Kumari N., Mishra P., Moharana D. P., Aamir M. (2021). Tapping the potential of Solanum lycopersicum L. pertaining to salinity tolerance: perspectives and challenges. Genet. Res. Crop Evol. 68 2207–2233. 10.1007/s10722-021-01174-9 [DOI] [Google Scholar]
- Kashyap S. P., Kumari N., Mishra P., Moharana D. P., Aamir M., Singh B., et al. (2020). Transcriptional regulation-mediating ROS homeostasis and physio-biochemical changes in wild tomato (Solanum chilense) and cultivated tomato (Solanum lycopersicum) under high salinity. Saud. J. Biol. Sci. 27 1999–2009. 10.1016/j.sjbs.2020.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayum M. A., Jung H. J., Park J. I., Ahmed N. U., Saha G., Yang T. J., et al. (2015). Identification and expression analysis of WRKY family genes under biotic and abiotic stresses in Brassica rapa. Mol. Genet. Genomics 290 79–95. 10.1007/s00438-014-0898-1 [DOI] [PubMed] [Google Scholar]
- Kim B. O., Han J. S., Park K. I., Jeon S. M., Kim C. K. (2015). Absence of AVP1 transcripts in wild type watermelon scions grafted onto transgenic bottle gourdrootstocks. J. Plant. Biotec. 42 13–18. 10.5010/JPB.2015.42.1.13 [DOI] [Google Scholar]
- Kim J. A., Jung H. E., Hong J. K., Hermand V., McClung C. R., Lee Y. H., et al. (2016). Reduction of GIGANTEA expression in transgenic Brassica rapa enhances salt tolerance. Plant Cell Rep. 35 1943–1954. 10.1007/s00299-016-2008-9 [DOI] [PubMed] [Google Scholar]
- Kong F., Mao S., Du K., Wu M., Zhou X., Chu C., et al. (2011). Comparative proteomics analysis of OsNAS1 transgenic Brassica napus under salt stress. Chin. Sci. Bull. 56 2343–2350. 10.1007/s11434-011-4585-x [DOI] [Google Scholar]
- Kotula L., Garcia Caparros P., Zörb C., Colmer T. D., Flowers T. J. (2020). Improving crop salt tolerance using transgenic approaches: an update and physiological analysis. Plant Cell Environ. 43 2932–2956. 10.1111/pce.13865 [DOI] [PubMed] [Google Scholar]
- Krishna R., Ansari W. A., Jaiswal D. K., Singh A. K., Prasad R., Verma J. P., et al. (2021a). Overexpression of AtDREB1 and BcZAT12 genes confers drought tolerance by reducing oxidative stress in double transgenic tomato (Solanum lycopersicum L.). Plant Cell Rep. 40 2173–2190. 10.1007/s00299-021-02725-1 [DOI] [PubMed] [Google Scholar]
- Krishna R., Ansari W. A., Jaiswal D. K., Singh A. K., Verma J. P., Singh M. (2021b). Co-overexpression of AtDREB1A and BcZAT12 increases drought tolerance and fruit production in double transgenic tomato (Solanum lycopersicum) plants. Environ. Exp. Bot. 184:104396. 10.1016/j.envexpbot.2021.104396 [DOI] [PubMed] [Google Scholar]
- Krishna R., Karkute S. G., Ansari W. A., Jaiswal D. K., Verma J. P., Singh M. (2019). Transgenic tomatoes for abiotic stress tolerance: status and way ahead. 3 Biotech 9:143. 10.1007/s13205-019-1665-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J., Singh S., Singh M., Sri vastava P. K., Mishra R. K., Singh V. P., et al. (2017). Transcriptional regulation of salinity stress in plants: a short review. Plant Gene 11 160–169. 10.1016/j.plgene.2017.04.001 [DOI] [Google Scholar]
- Kumar P., Lucini L., Rouphael Y., Cardarelli M., Kalunke R. M., Colla G. (2015). Insight into the role of grafting and arbuscular mycorrhiza on cadmium stress tolerance in tomato. Front. Plant Sci. 6:477. 10.3389/fpls.2015.00477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. K., Sivanesan I., Murugesan K., Jeong B. R., Hwang S. J. (2014). Enhancing salt tolerance in eggplant by introduction of foreign halotolerance gene, HAL1 isolated from yeast. Hort. Environ. Biotech. 55 222–229. 10.1007/s13580-014-0141-3 [DOI] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. (1988). The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240 1759–1764. 10.1126/science.3289117 [DOI] [PubMed] [Google Scholar]
- Li H., Wang Y., Wu M., Li L., Li C., Han Z., et al. (2017). Genome-wide identification of AP2/ERF transcription factors in cauliflower and expression profiling of the ERF family under salt and drought stresses. Front. Plant Sci. 8:946. 10.3389/fpls.2017.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang T., Han J., Ren Z. (2020). Genome-wide identification and characterization of cucumber bHLH family genes and the functional characterization of CsbHLH041 in NaCl and ABA tolerance in Arabidopsis and cucumber. BMC Plant Biol. 20:272. 10.1186/s12870-020-02440-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. Y., Xu Z. S., Tian C., Huang Y., Wang F., Xiong A. S. (2016). Genomic identification of WRKY transcription factors in carrot (Daucus carota) and analysis of evolution and homologous groups for plants. Sci. Rep. 6:23101. 10.1038/srep23101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. L., Lai L. D., Yu X. D., Wang J. Y. (2018). The SlCMO gene driven by its own promoter enhances salt tolerance of transgenic tomato without affecting growth and yield. Plant. Mol. Biol. Rep. 36 596–604. 10.1007/s11105-018-1104-0 [DOI] [Google Scholar]
- Li X., Huang L., Zhang Y., Ouyang Z., Hong Y., Zhang H., et al. (2014). Tomato SR/CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 14:286. 10.1186/s12870-014-0286-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Zou H. F., Wei W., Hao Y. J., Tian A. G., Huang J., et al. (2008). Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228 225–240. 10.1007/s00425-008-0731-3 [DOI] [PubMed] [Google Scholar]
- Ling J., Jiang W., Zhang Y., Yu H., Mao Z., Gu X., et al. (2011). Genome-wide analysis of WRKY gene family in Cucumis sativus. BMC Genomics 12:471. 10.1186/1471-2164-12-471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Ouyang Z., Zhang Y., Li X., Hong Y., Huang L., et al. (2014). Tomato NAC transcription factor SlSRN1 positively regulates defense response against biotic stress but negatively regulates abiotic stress response. PLoS One 9:e102067. 10.1371/journal.pone.0102067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M. M., Li Y. L., Li G. C., Dong T. T., Liu S. Y., Pei L. I. U., et al. (2020). Overexpression of StCYS1 gene enhances tolerance to salt stress in the transgenic potato (Solanum tuberosum L.) plant. J. Integ. Agric. 19 2239–2246. 10.1016/S2095-3119(20)63262-2 [DOI] [Google Scholar]
- Liu Z., Bie Z., Huang Y. (2013). Rootstocks improve cucumber photosynthesis through nitrogen metabolism regulation under salt stress. Acta Physiol. Plant 35 2259–2267. 10.1007/s11738-013-1262-5 [DOI] [Google Scholar]
- Lu C. W., Shao Y., Li L., Chen A. J., Xu W. Q., Wu K. J., et al. (2011). Overexpression of SlERF1 tomato gene encoding an ERF-type transcription activator enhances salt tolerance. Russ. J. Plant Phys. 58 118–125. 10.1134/S1021443711010092 [DOI] [Google Scholar]
- Luo J., Tang S., Mei F., Peng X., Li J., Li X., et al. (2017). BnSIP1-1, a trihelix family gene, mediates abiotic stress tolerance and ABA signaling in Brassica napus. Front. Plant Sci. 8:44. 10.3389/fpls.2017.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Wang L. Y., Dai J. X., Wang Y., Lin D. (2021). The NAC-type transcription factor CaNAC46 regulates the salt and drought tolerance of transgenic Arabidopsis thaliana. BMC Plant Biol. 21:11. 10.1186/s12870-020-02764-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maach M., Baghour M., Akodad M., Gálvez F. J., Sánchez M. E., Aranda M. N., et al. (2020). Overexpression of LeNHX4 improved yield, fruit quality and salt tolerance in tomato plants (Solanum lycopersicum L.). Mol. Biol. Rep. 47 4145–4153. 10.1007/s11033-020-05499-z [DOI] [PubMed] [Google Scholar]
- Maach M., Rodríguez-Rosales M. P., Venema K., Akodad M., Moumen A., Skalli A., et al. (2021). Improved yield, fruit quality, and salt resistance in tomato co-overexpressing LeNHX2 and SlSOS2 genes. Phys. Mol. Biol. Plant 27 703–712. 10.1007/s12298-021-00974-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado R. M. A., Serralheiro R. P. (2017). Soil salinity: effect on vegetable crop growth. Management practices to prevent and mitigate soil salinization. Horticulturae 3:30. 10.3390/horticulturae3020030 [DOI] [Google Scholar]
- Manivannan P., Jaleel C. A., Kishorekumar A., Sankar B., Somasundaram R., Sridharan R., et al. (2007). Changes in antioxidant metabolism of Vignaunguiculata (L.) Walp. bypropiconazole under water deficit stress. Colloids Surf. B 57 69–74. 10.1016/j.colsurfb.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Martinez-Rodriguez M. M., Estan M. T., Moyano E., Garcia A., Bellan J. O., Flores F. B., et al. (2008). The effectiveness of grafting to improve salt tolerance in tomato when an excluder genotype is used as scion. Environ. Exp. Bot. 63 392–401. 10.1016/j.envexpbot.2007.12.007 [DOI] [Google Scholar]
- Meena M., Aamir M., Kumar V., Swapnil P., Upadhyay R. S. (2018). Evaluation of morpho-physiological growth parameters of tomato in response to Cd induced toxicity and characterization of metal sensitive NRAMP3 transporter protein. Environ. Exp. Bot. 148 144–167. 10.1016/j.envexpbot.2018.01.007 [DOI] [Google Scholar]
- Meena M., Zehra A., Dubey M. K., Aamir M., Gupta V. K., Upadhyay R. S. (2016). Comparative evaluation of biochemical changes in tomato (Lycopersicon esculentum Mill.) infected by Alternaria alternata and its toxic metabolites (TeA, AOH, and AME). Front. Plant Sci. 7:1408. 10.3389/fpls.2016.01408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Liu S., Dong T., Xu T., Ma D., Pan S., et al. (2020). Comparative transcriptome and proteome analysis of salt-tolerant and salt-sensitive sweet potato and overexpression of IbNAC7 confers salt tolerance in Arabidopsis. Front. Plant Sci. 11:572540. 10.3389/fpls.2020.572540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaieb R. E., Nakamura A., Saneoka H., Fujita K. (2011). Evaluation of salt tolerance in ectoine-transgenic tomato plants (Lycopersicon esculentum) in terms of photosynthesis, osmotic adjustment, and carbon partitioning. GM Crops. 2 58–65. 10.4161/gmcr.2.1.15831 [DOI] [PubMed] [Google Scholar]
- Moghaieb R. E. A., Tanaka N., Saneoka H., Hussein H. A., Yousef S. S., Ewada M. A. F., et al. (2000). Expression of betaine aldehyde dehydrogenase gene in transgenic tomato hairy roots leads to the accumulation of glycine betaine and contributes to the maintenance of the osmotic potential under salt stress. Soil Sci. Plant Nutr. 46:3. 10.1080/00380768.2000.10409153 [DOI] [Google Scholar]
- Møller I. S., Gilliham M., Jha D., Mayo G. M., Roy S. J., Coates J. C., et al. (2009). Shoot Na+ exclusion and increased salinity tolerance engineered by cell type–specific alteration of Na+ transport in Arabidopsis. Plant Cell 21 2163–2178. 10.1105/tpc.108.064568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahakpam S., Shah K. (2011). Expression of key antioxidant enzymes under combined effect of heat and cadmium toxicity in growing rice seedlings. PlantGro Reg. 63 23–35. 10.1007/s10725-010-9508-3 [DOI] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Phys. 140 411–432. 10.1104/pp.105.073783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan H., Gao L. Z. (2019). Genome-wide analysis of WRKY genes and their response to hormone and mechanic stresses in carrot. Front. Gent. 10:363. 10.3389/fgene.2019.00363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhawan A., Jain M., Tyagi A. K., Khurana J. P. (2008). Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Phys. 146 333–350. 10.1104/pp.107.112821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana S., Yañez M., Espinoza A., Verdugo I., González E., Ruiz-Lara S., et al. (2010). The transcription factor SlAREB1 confers drought, salt stress tolerance and regulates biotic and abiotic stress-related genes in tomato. Plant Cell Environ. 33 2191–2208. 10.1111/j.1365-3040.2010.02220.x [DOI] [PubMed] [Google Scholar]
- Park B. J., Liu Z., Kanno A., Kameya T. (2005). Genetic improvement of Chinese cabbage for salt and drought tolerance by constitutive expression of a B. napus LEA gene. Plant Sci. 169 553–558. 10.1016/j.plantsci.2005.05.008 [DOI] [Google Scholar]
- Pan Y., Hu X., Li C., Xu X., Su C., Li J., et al. (2017). SlbZIP38, a tomato bZIP family gene downregulated by abscisic acid, is a negative regulator of drought and salt stress tolerance. Genes 8:402. 10.3390/genes8120402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Seymour G. B., Lu C., Hu Z., Chen X., Chen G. (2012). An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rept. 31 349–360. 10.1007/s00299-011-1170-3 [DOI] [PubMed] [Google Scholar]
- Park M., Han J., Ahn Y., Kim J., Lee H., Jang Y., et al. (2014). Ectopic expression of Arabidopsis H+-pyrophosphatase AVP1 enhances drought resistance in bottle gourd (Lagenaria siceraria Standl.). Plant Cell Tiss. Org. Cult. 118 383–389. 10.1007/s11240-014-0490-8 [DOI] [Google Scholar]
- Parthasarathi T., Ephrath J. E., Lazarovitch N. (2021). Grafting of tomato (Solanum lycopersicum L.) onto potato to improve salinity tolerance. Sci. Hortic. 282:110050. 10.1016/j.scienta.2021.110050 [DOI] [Google Scholar]
- Penella C., Nebauer S. G., Quiñones A., Bautista A. S., López-Galarza S., Calatayud A. (2015). Some rootstocks improve pepper tolerance to mild salinity through ionic regulation. Plant Sci. 230 12–22. 10.1016/j.plantsci.2014.10.007 [DOI] [PubMed] [Google Scholar]
- Penella P., Landi M., Guidi L., Nebauer S. G., Pellegrini E., Bautista A. S., et al. (2016). Salt-tolerant rootstock increases yield of pepper under salinity through maintenance of photosynthetic performance and sinks strength. J. Plant Physiol. 193 1–11. 10.1016/j.jplph.2016.02.007 [DOI] [PubMed] [Google Scholar]
- Phukan U. J., Jeena G. S., Shukla R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7:760. 10.3389/fpls.2016.00760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhavathi V., Yadav J. S., Kumar P. A., Rajam M. V. (2002). Abiotic stress tolerance in transgenic eggplant (Solanum melongena L.) by introduction of bacterial mannitol phosphodehydrogenase gene. Mol. Breed. 9 137–147. 10.1023/A:1026765026493 [DOI] [Google Scholar]
- Prabhavathi V. R., Rajam M. V. (2007). Polyamine accumulation in transgenic eggplant enhances tolerance to multiple abiotic stresses and fungal resistance. Plant Biot. 24 273–282. 10.5511/plantbiotechnology.24.273 [DOI] [Google Scholar]
- Prasad K. V. S. K., Sharmila P., Kumar P. A., Saradhi P. P. (2000). Transformation of Brassica juncea (L.) Czern with bacterial codA gene enhances its tolerance to salt stress. Mol. Breed. 6 489–499. 10.1023/A:1026542109965 [DOI] [Google Scholar]
- Prasanna H. C., Sinha D. P., Rai G. K., Krishna R., Kashyap S. P., Singh N. K., et al. (2015). Pyramiding Ty-2 and Ty-3 genes for resistance to monopartite and bipartite tomato leaf curl viruses of India. Plant Path. 64 256–264. 10.1111/ppa.12267 [DOI] [Google Scholar]
- Puranik S., Sahu P. P., Srivastava P. S., Prasad M. (2012). NAC proteins: regulation and role in stress tolerance. Trend. Plant Sci. 17 369–381. 10.1016/j.tplants.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Qamarunnisa S., Jamil I., Raza S., Azhar A., Naqvi S. M. (2015). Genetic improvement of canola against abiotic stress through incorporation of DREB gene. Asian J. Agric. Biol. 3 77–104. [Google Scholar]
- Qin Z., Hou F., Li A., Dong S., Wang Q., Zhang L. (2020). Transcriptome-wide identification of WRKY transcription factor and their expression profiles under salt stress in sweetpotato (Ipomoea batatas L.). Plant Biot. Rep. 14 599–611. 10.1007/s11816-020-00635-4 [DOI] [Google Scholar]
- Rahnama H., Vakilian H., Fahimi H., Ghareyazie B. (2011). Enhanced salt stress tolerance in transgenic potato plants (Solanum tuberosum L.) expressing a bacterial mtlD gene. Acta Phys. Plant 33 1521–1532. 10.1007/s11738-010-0690-8 [DOI] [Google Scholar]
- Rai A. C., Rai A., Shah K., Singh M. (2021). Engineered BcZAT12 gene mitigates salt stress in tomato seedlings. Phys. Mol. Biol. Plant 27 535–541. 10.1007/s12298-021-00948-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal D., Agarwal P., Tyagi W., Singla-Pareek S. L., Reddy M. K., Sopory S. K. (2007). Pennisetum glaucum Na+/H+ antiporter confers high level of salinity tolerance in transgenic Brassica juncea. Mol. Breed. 19 137–151. 10.1007/s11032-006-9052-z [DOI] [Google Scholar]
- Rajwanshi R., Kumar D., Yusuf M. A., DebRoy S., Sarin N. B. (2016). Stress-inducible overexpression of glyoxalase I is preferable to its constitutive overexpression for abiotic stress tolerance in transgenic Brassica juncea. Mol. Breed. 36:76. 10.1007/s11032-016-0495-6 [DOI] [Google Scholar]
- Rong W., Qi L., Wang A., Ye X., Du L., Liang H., et al. (2014). The ERF transcription factor Ta ERF 3 promotes tolerance to salt and drought stresses in wheat. Plant Biotech. J. 12 468–479. 10.1111/pbi.12153 [DOI] [PubMed] [Google Scholar]
- Rouphael Y., Cardarelli M., Rea E., Colla G. (2012). Improving melon and cucumber photosynthetic activity, mineral composition, and growth performance under salinity stress by grafting onto Cucurbita hybrid rootstocks. Photosynthetica 50 180–188. 10.1007/s11099-012-0002-1 [DOI] [Google Scholar]
- Rouphael Y., Venema J. H., Edelstein M., Savvas D., Colla G., Ntatsi G., et al. (2017). “Grafting as a tool for tolerance of abiotic stress,” in Vegetable Grafting: Principles and Practices, eds Colla G., Perez-Alfocea F., Schwarz D. (Wallingford: CAB International; ), 171–215. 10.1079/9781780648972.0171 [DOI] [Google Scholar]
- Rus A., Baxter I., Muthukumar B., Gustin J., Lahner B., Yakubova E., et al. (2006). Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2:e210. 10.1371/journal.pgen.0020210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A. M., Estan M. T., Gisbert C., Garcia-Sogo B., Serrano R., Caro M., et al. (2001). Expressing the yeast HAL1 gene in tomato increases fruit yield and enhances K+/Na+ selectivity under salt stress. Plant Cell Environ. 24 875–880. 10.1046/j.1365-3040.2001.00719.x [DOI] [Google Scholar]
- Rushton P. J., Somssich I. E., Ringler P., Shen Q. J. (2010). WRKY transcription factors. Trend. Plant Sci. 15 247–258. 10.1016/j.tplants.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Saha B., Mishra S., Awasthi J. P., Sahoo L., Panda S. K. (2016). Enhanced drought and salinity tolerance in transgenic mustard [Brassica juncea (L.) Czern&Coss.] overexpressing Arabidopsis group 4 late embryogenesis abundant gene (AtLEA4-1). Environ. Exp. Bot. 128 99–111. 10.1016/j.envexpbot.2016.04.010 [DOI] [Google Scholar]
- Sacks M. M., Silk W. K., Burman P. (1997). Effect of water stress on cortical cell division rates within the apical meristem of primary roots of maize. Plant Phys. 114 519–527. 10.1104/pp.114.2.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena M., Deb Roy S., Singla-Pareek S. L., Kumar Sopory S., Bhalla-Sarin N. (2011). Overexpression of the glyoxalase II gene leads to enhanced salinity tolerance in Brassica juncea. Open Plant Sci. J. 5 23–28. 10.2174/1874294701105010023 [DOI] [Google Scholar]
- Seong E. S., Cho H. S., Choi D., Joung Y. H., Lim C. K., Hur J. H., et al. (2007). Tomato plants overexpressing CaKR1 enhanced toleranceto salt and oxidative stress. Biochem. Biophys. Res. Commun. 363 983–988. 10.1016/j.bbrc.2007.09.104 [DOI] [PubMed] [Google Scholar]
- Shafi A., Pal A. K., Sharma V., Kalia S., Kumar S., Ahuja P. S., et al. (2017). Transgenic potato plants overexpressing SOD and APX exhibit enhanced lignification and starch biosynthesis with improved salt stress tolerance. Plant Mol. Biol. Rep. 35 504–518. 10.1007/s11105-017-1041-3 [DOI] [Google Scholar]
- Shao H. B., Chu L. Y., Jaleel C. A., Zhao C. X. (2008). Water-deficit stress-induced anatomical changes in higher plants. Compt. Rendus Biol. 331 215–225. 10.1016/j.crvi.2008.01.002 [DOI] [PubMed] [Google Scholar]
- Sharp R. E., LeNoble M. E. (2002). ABA, ethylene and the control of shoot and root growth under water stress. J. Exp. Bot. 53 33–37. 10.1093/jexbot/53.366.33 [DOI] [PubMed] [Google Scholar]
- Shi H. Z., Quintero F. J., Pardo J. M., Zhu J. K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transportin plants. Plant Cell 14 465–477. 10.1105/tpc.010371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumara T. N., Sreevathsa R., Dash P. K., Sheshshayee M. S., Papolu P. K., Rao U., et al. (2017). Overexpression of Pea DNA Helicase 45 (PDH45) imparts tolerance to multiple abiotic stresses in chili (Capsicum annuum L.). Sci. Rep. 7:2760. 10.1038/s41598-017-02589-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. K., Sharma V., Pal A. K., Acharya V., Ahuja P. S. (2013). Genome-wide organization and expression profiling of the NAC transcription factor family in potato (Solanum tuberosum L.). DNA Res. 20 403–423. 10.1093/dnares/dst019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A. K., Yadav B. K., Krishna R., Kumar R. V., Mishra G. P., Karkute S. G., et al. (2021). Bhendi yellow vein mosaic virus and bhendi yellow vein mosaic betasatellite cause enation leaf curl disease and alter host phytochemical contents in okra. Plant Dis. 105 2595–2600. 10.1094/PDIS-12-20-2655-RE [DOI] [PubMed] [Google Scholar]
- Singh H., Kumar P., Kumar A., Kyriacou M. C., Colla G., Rouphael Y. (2020). Grafting tomato as a tool to improve salt tolerance. Agronomy 10:263. 10.3390/agronomy10020263 [DOI] [Google Scholar]
- Song F., Su H., Cheng X., Zhu L., Wang L. (2015). Characterization and expression analysis of TNAC genes of tomato. Bull. Bot. Res. 35 898–903. [Google Scholar]
- Sornaraj P., Luang S., Lopato S., Hrmova M. (2016). Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta (BBA)-Gen. Subj. 1860 46–56. 10.1016/j.bbagen.2015.10.014 [DOI] [PubMed] [Google Scholar]
- Soumia P. S., Pandi G. G. P., Krishna R., Ansari W. A., Jaiswal D. K., Verma J. P., et al. (2021). “Whitefly-transmitted plant viruses and their management,” in Emerging Trends in Plant Pathology. eds Singh K. P., Jahagirdar S., Sarma B. K. (Singapore: Springer; ), 175–195. 10.1007/978-981-15-6275-4_8 [DOI] [Google Scholar]
- Srivastava S., Fristensky B., Kav N. N. (2004). Constitutive expression of a PR10 protein enhances the germination of Brassica napus under saline conditions. Plant Cell Physiol. 45 1320–1324. 10.1093/pcp/pch137 [DOI] [PubMed] [Google Scholar]
- Stegemann S., Bock R. (2009). Exchange of genetic material between cells in plant tissue grafts. Science 324 649–651. 10.1126/science.1170397 [DOI] [PubMed] [Google Scholar]
- Subramanyam K., Sailaja K. V., Subramanyam K., Rao D. M., Lakshmidevi K. (2011). Ectopic expression of an osmotin gene leads to enhanced salt tolerance in transgenic chilli pepper (Capsicum annum L.). Plant Cell Tiss. Org. Cult. 105 181–192. 10.1007/s11240-010-9850-1 [DOI] [Google Scholar]
- Sun J., Cao H., Cheng J., He X., Sohail H., Niu M., et al. (2018). Pumpkin CmHKT1; 1 controls shoot Na+ accumulation via limiting Na+ transport from rootstock to scion in grafted cucumber. Inter. J. Mol. Sci. 19:2648. 10.3390/ijms19092648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Wen D., Yang W., Meng Q., Shi Q., Gong B. (2020). Overexpression of caffeic acid O-methyltransferase 1 (COMT1) increases melatonin level and salt stress tolerance in tomato plant. J. Plant Growth Regulat. 39 1221–1235. 10.1007/s00344-019-10058-3 [DOI] [Google Scholar]
- Sun S. J., Guo S. Q., Yang X., Bao Y. M., Tang H. J., Sun H., et al. (2010). Functional analysis of a novel Cys2/His2 type zinc finger protein involved in salt tolerance in rice. J. Exp. Bot. 61 2807–2818. 10.1093/jxb/erq120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. E., Feng X. X., Li C., Zhang Z. P., Wang L. J. (2015). Study on salt tolerance with YHem1 transgenic canola (Brassica napus). Physiol. Plant. 154 223–242. 10.1111/ppl.12282 [DOI] [PubMed] [Google Scholar]
- Sunarpi, Horie T., Motoda J., Kubo M., Yang H., Yoda K., Horie R., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 44 928–938. 10.1111/j.1365-313X.2005.02595.x [DOI] [PubMed] [Google Scholar]
- Talhouni M., Sonmez K., Kiran S., Beyaz R., Yildiz M., Kusvuran S., et al. (2019). Comparison of salinity effects on grafted and non-grafted eggplants in terms of ion accumulation, MDA content and antioxidative enzyme activities. Adv. Hortic. Sci. 33 87–95. [Google Scholar]
- Tang J., Wang F., Hou X. L., Wang Z., Huang Z. N. (2014). Genome-wide fractionation and identification of WRKY transcription factors in Chinese cabbage (Brassicarapa ssp. pekinensis) reveals collinearity and their expression patterns under abiotic and biotic stresses. Plant Mol. Biol. Rep. 32 781–795. 10.1007/s11105-013-0672-2 [DOI] [Google Scholar]
- Tester M., Davenport R. (2003). Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 91 503–527. 10.1093/aob/mcg058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari J. K., Munshi A. D., Kumar R., Pandey R. N., Arora A., Bhat J. S., et al. (2010). Effect of salt stress on cucumber: Na+–K+ ratio, osmolyte concentration, phenols and chlorophyll content. Acta Phys. Plant. 32 103–114. 10.1007/s11738-009-0385-1 [DOI] [Google Scholar]
- Tolosa L. N., Zhang Z. (2020). The role of major transcription factors in Solanaceous food crops under different stress conditions: current and future perspectives. Plants 9:56. 10.3390/plants9010056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L. S. P., Nishiyama R., Yamaguchi-Shinozaki K., Shinozaki K. (2010). Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1 32–39. 10.4161/gmcr.1.1.10569 [DOI] [PubMed] [Google Scholar]
- Upadhyaya C. P., Akula N., Young K. E., Chun S. C., Kim D. H., Park S. W. (2010). Enhanced ascorbic acid accumulation in transgenic potato confers tolerance to various abiotic stresses. Biotec. Lett. 32 321–330. 10.1007/s10529-009-0140-0 [DOI] [PubMed] [Google Scholar]
- Upadhyaya C. P., Venkatesh J., Gururani M. A., Asnin L., Sharma K., Ajappala H., et al. (2011). Transgenic potato overproducing L-ascorbic acid resisted an increase in methylglyoxal under salinity stress via maintaining higher reduced glutathione level and glyoxalase enzyme activity. Biotec. Lett. 33 2297–2307. 10.1007/s10529-011-0684-7 [DOI] [PubMed] [Google Scholar]
- Upadhyaya C. P., Young K. E., Akula N., Soon Kim H., Heung J. J., Oh O. M., et al. (2009). Over-expression of strawberry D-galacturonic acid reductase in potato leads to accumulation of vitamin C with enhanced abiotic stress tolerance. Plant. Sci. 177 659–667. 10.1016/j.plantsci.2009.08.004 [DOI] [Google Scholar]
- Usanmaz S., Abak K. (2019). Plant growth and yield of cucumber plants grafted on different commercial and local rootstocks grown under salinity stress. Saudi J. Biol. Sci. 26 1134–1139. 10.1016/j.sjbs.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villano C., Esposito S., D’Amelia V., Garramone R., Alioto D., Zoina A., et al. (2020). WRKY genes family study reveals tissue-specific and stress-responsive TFs in wild potato species. Sci. Rep. 10:7196. 10.1038/s41598-020-63823-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Zhang S., Ma X., Wang Y., Kong F., Meng Q. (2016). A stress-associated NAC transcription factor (SlNAC35) from tomato plays a positive role in biotic and abiotic stresses. Physiol. Plant. 158 45–64. 10.1111/ppl.12444 [DOI] [PubMed] [Google Scholar]
- Wang Q. B., Xu W., Xue Q. Z., Su W. A. (2010). Transgenic Brassica chinensis plants expressing a bacterial codA gene exhibit enhanced tolerance to extreme temperature and high salinity. J. Zhejiang Univ. Sci. B 11 851–861. 10.1631/jzus.B1000137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhang J., Wang D., Zhang J., Cui Y., Liu Y., et al. (2013). Assessment of salt tolerance in transgenic potato carrying AtNHX1 gene. Crop Sci. 53 2643–2651. 10.2135/cropsci2013.03.0179 34798789 [DOI] [Google Scholar]
- Wang W., Qiu X., Yang Y., Kim H. S., Jia X., Yu H., et al. (2019). Sweetpotato bZIP transcription factor IbABF4 confers tolerance to multiple abiotic stresses. Front. Plant Sci. 10:630. 10.3389/fpls.2019.00630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wisniewski M., Meilan R., Cui M., Webb R., Fuchigami L. (2005). Overexpression of cytosolic ascorbate peroxidase in tomato confers tolerance to chilling and salt stress. J. Am. Soc. Hortic. Sci. 130 167–173. 10.21273/JASHS.130.2.167 [DOI] [Google Scholar]
- Wang Y., Zhang Y., Zhou R., Dossa K., Yu J., Li D., et al. (2018). Identification and characterization of the bZIP transcription factor family and its expression in response to abiotic stresses in sesame. PLoS One 13:e0200850. 10.1371/journal.pone.0200850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waseem M., Rong X., Li Z. (2019). Dissecting the role of a basic helix-loop-helix transcription factor, SlbHLH22, under salt and drought stresses in transgenic Solanum lycopersicum L. Front. Plant Sci. 10:734. 10.3389/fpls.2019.00734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D., Zhang W., Wang C., Meng Q., Li G., Chen T. H., et al. (2017). Genetic engineering of the biosynthesis of glycinebetaine leads to alleviate salt-induced potassium efflux and enhances salt tolerance in tomato plants. Plant Sci. 257 74–83. 10.1016/j.plantsci.2017.01.012 [DOI] [PubMed] [Google Scholar]
- Wei G., Yang L., Zhu Y., Chen C. (2009). Changes in oxidative damage, antioxidant enzymes activity and polyamine contents in the leaves of grafted and non-grafted eggplant seedlings under stress by excess of calcium nitrate. Sci. Hortic. 120 443–451. 10.1016/j.scienta.2008.12.009 [DOI] [Google Scholar]
- Wei S., Gao L., Zhang Y., Zhang F., Yang X., Huang D. (2016). Genome-wide investigation of the NAC transcription factor family in melon (Cucumis melo L.) and their expression analysis under salt stress. Plant. Cell Rep. 35 1827–1839. 10.1007/s00299-016-1997-8 [DOI] [PubMed] [Google Scholar]
- Wu M., Liu H., Han G., Cai R., Pan F., Xiang Y. (2017). A moso bamboo WRKY gene PeWRKY83 confers salinity tolerance in transgenic Arabidopsis plants. Sci. Rept. 7:11721. 10.1038/s41598-017-10795-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S. D., Yin T. M., DiFazio S. P., Tschaplinski T. J., Gunter L. E., Davis M. F., et al. (2005). Phenotypic variation in growth and biomass distribution for two advanced-generation pedigrees of hybrid poplar. Can. J. For. Res. 35 1779–1789. 10.1139/x05-101 [DOI] [Google Scholar]
- Xiang L., Liu C., Luo J., He L., Deng Y., Yuan J., et al. (2018). A tuber mustard AP2/ERF transcription factor gene, BjABR1, functioning in abscisic acid and abiotic stress responses, and evolutionary trajectory of the ABR1 homologous genes in Brassica species. PeerJ 6:e6071. 10.7717/peerj.6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Di Z., Yang W., Liu J., Li M., Wang X., et al. (2017). Overexpression of ERF1-V from Haynaldia villosa can enhance the resistance of wheat to powdery mildew and increase the tolerance to salt and drought stresses. Front. Plant Sci. 8:1948. 10.3389/fpls.2017.01948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q., He Q., Li S., Tian Z. (2014). Molecular characterization of StNAC2 in potato and its overexpression confers drought and salt tolerance. Acta Phys. Plant 36 1841–1851. 10.1007/s11738-014-1558-0 [DOI] [Google Scholar]
- Yang R., Liu J., Lin Z., Sun W., Wu Z., Hu H., et al. (2018). ERF transcription factors involved in salt response in tomato. Plant Growth Regulat. 84 573–582. 10.1007/s10725-017-0362-4 [DOI] [Google Scholar]
- Yang S. D., Seo P. J., Yoon H. K., Park C. M. (2011). The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23 2155–2168. 10.1105/tpc.111.084913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Deng C., Zhang Y., Cheng Y., Huo Q., Xue L. (2015). The WRKY transcription factor genes in eggplant (Solanum melongena L.) and Turkey Berry (Solanum torvum Sw.). Inter. J. Mol. Sci. 16 7608–7626. 10.3390/ijms16047608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarra R., He S. J., Abbagani S., Ma B., Bulle M., Zhang W. K. (2012). Overexpression of a wheat Na+/H+ antiporter gene (TaNHX2) enhances tolerance to salt stress in transgenic tomato plants (Solanum lycopersicum L.). Plant Cell Tiss. Org. Cult. 111 49–57. 10.1007/s11240-012-0169-y [DOI] [Google Scholar]
- Yarra R., Kirti P. B. (2019). Expressing class I wheat NHX (TaNHX2) gene in eggplant (Solanum melongena L.) improves plant performance under saline condition. Funct. Integ. Gen. 19 541–554. 10.1007/s10142-019-00656-5 [DOI] [PubMed] [Google Scholar]
- Yue H., Chang X., Zhi Y., Wang L., Xing G., Song W., et al. (2019). Evolution and identification of the WRKY gene family in quinoa (Chenopodium quinoa). Genes 10:131. 10.3390/genes10020131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahedi S. M., Abdelrahman M., Hosseini M. S., Hoveizeh N. F., Tran L. S. P. (2019). Alleviation of the effect of salinity on growth and yield of strawberry by foliar spray of selenium-nanoparticles. Environ. Poll. 253 246–258. 10.1016/j.envpol.2019.04.078 [DOI] [PubMed] [Google Scholar]
- Zehra A., Meena M., Dubey M. K., Aamir M., Upadhyay R. S. (2017a). Activation of defense response in tomato against Fusarium wilt disease triggered by Trichoderma harzianum supplemented with exogenous chemical inducers (SA and MeJA). Braz. J. Bot. 40 651–664. 10.1007/s40415-017-0382-3 [DOI] [Google Scholar]
- Zehra A., Meena M., Dubey M. K., Aamir M., Upadhyay R. S. (2017b). Synergistic effects of plant defense elicitors and Trichoderma harzianum on enhanced induction of antioxidant defense system in tomato against Fusarium wilt disease. Bot. Stud. 58 1–14. 10.1186/s40529-017-0198-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Wang D., Yang C., Kong N., Shi Z., Zhao P., et al. (2017). Genome-wide identification of the potato WRKY transcription factor family. PLoS One 12:e0181573. 10.1371/journal.pone.0181573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. X., Blumwald E. (2001). Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 19 765–768. 10.1038/90824 [DOI] [PubMed] [Google Scholar]
- Zhang H. X., Hodson J. N., Williams J. P., Blumwald E. (2001). Engineering salt-tolerant Brassica plants: characterization of yield and seed oil quality in transgenic plants with increased vacuolar sodium accumulation. Proc. Natl. Acad. Sci. U.S.A. 98 12832–12836. 10.1073/pnas.231476498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Si H. J., Wen G., Du H. H., Liu B. L., Wang D. (2011). Enhanced drought and salinity tolerance in transgenic potato plants with a BADH gene from spinach. Plant Biotech. Rep. 5 71–77. 10.1007/s11816-010-0160-1 [DOI] [Google Scholar]
- Zhang X., Chen L., Shi Q., Ren Z. (2020). SlMYB102, an R2R3-type MYB gene, confers salt tolerance in transgenic tomato. Plant Sci. 291:110356. 10.1016/j.plantsci.2019.110356 [DOI] [PubMed] [Google Scholar]
- Zhao B. Y., Hu Y. F., Li J. J., Yao X. (2016). BnaABF2, a bZIP transcription factor from rapeseed (Brassica napus L.), enhances drought and salt tolerance in transgenic Arabidopsis. Bot. Stud. 57 1–12. 10.1186/s40529-016-0127-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M., Chen G., Zhang J., Zhang Y., Xie Q., Zhao Z., et al. (2014). The abiotic stress-responsive NAC-type transcription factor SlNAC4 regulates salt and drought tolerance and stress-related genes in tomato (Solanum lycopersicum). Plant Cell Rep. 33 1851–1863. 10.1007/s00299-014-1662-z [DOI] [PubMed] [Google Scholar]
- Zhu M., Meng X., Cai J., Li G., Dong T., Li Z. (2018). Basic leucine zipper transcription factor SlbZIP1 mediates salt and drought stress tolerance in tomato. BMC Plant Biol. 18:83. 10.1186/s12870-018-1299-0 [DOI] [PMC free article] [PubMed] [Google Scholar]