Abstract
Increasing lines of evidence indicate that chloroplast‐related genes are involved in plant–virus interactions. However, the involvement of photosynthesis‐related genes in plant immunity is largely unexplored. Analysis of RNA‐Seq data from the soybean cultivar L29, which carries the Rsv3 resistance gene, showed that several chloroplast‐related genes were strongly induced in response to infection with an avirulent strain of soybean mosaic virus (SMV), G5H, but were weakly induced in response to a virulent strain, G7H. For further analysis, we selected the PSaC gene from the photosystem I and the ATP‐synthase α‐subunit (ATPsyn‐α) gene whose encoded protein is part of the ATP‐synthase complex. Overexpression of either gene within the G7H genome reduced virus levels in the susceptible cultivar Lee74 (rsv3‐null). This result was confirmed by transiently expressing both genes in Nicotiana benthamiana followed by G7H infection. Both proteins localized in the chloroplast envelope as well as in the nucleus and cytoplasm. Because the chloroplast is the initial biosynthesis site of defence‐related hormones, we determined whether hormone‐related genes are involved in the ATPsyn‐α‐ and PSaC‐mediated defence. Interestingly, genes involved in the biosynthesis of several hormones were up‐regulated in plants infected with SMV‐G7H expressing ATPsyn‐α. However, only jasmonic and salicylic acid biosynthesis genes were up‐regulated following infection with the SMV‐G7H expressing PSaC. Both chimeras induced the expression of several antiviral RNA silencing genes, which indicate that such resistance may be partially achieved through the RNA silencing pathway. These findings highlight the role of photosynthesis‐related genes in regulating resistance to viruses.
Keywords: ATPsyn‐α, photosynthesis, plant hormones, plant–virus interactions, PSaC, RNA silencing, soybean, soybean mosaic virus
Two photosynthesis‐related genes, PS aC and ATPsyn‐α, contribute to resistance against soybean mosaic virus via the RNA silencing pathway.
1. INTRODUCTION
Symptoms of viral infection in plants usually include a change in the green pigmentation such as mottling, mosaic, chlorosis, and yellowing. Most of these symptoms indicate changes in photosynthetic activity in the infected plants (Liu et al., 2020; Scholthof et al., 2011). It has long been known that viral infection leads to reduced photosynthesis and major changes in chloroplast ultrastructure (Bhattacharyya & Chakraborty, 2018; Lehto et al., 2003). The roles of chloroplasts in virus replication, virus movement, and plant defence have only recently been investigated (Azim & Burch‐Smith, 2020; Bhattacharyya & Chakraborty, 2018; Ganusova et al., 2020; Zhao et al., 2016).
Photosynthesis includes two major stages: a light‐dependent stage and a light‐independent stage. In the light‐dependent stage, photosystem I (PSI), cytochrome, photosystem II (PSII), and ATPase synthesis sequentially contribute to the production of NADPH and then ATP, which are used in the light‐independent stage to produce sugar through the Calvin cycle (Moejes et al., 2017; Nevo et al., 2012; Yu et al., 2020). Virus interference with chloroplasts in general, and with photosynthesis in particular, can occur on different levels. Because the chloroplast is the site for the biosynthesis of several defence‐related hormones and helps control plasmodesmata (PD) permeability, some viruses reduce host defences by targeting the chloroplast with specific viral proteins (Alazem & Lin, 2015, 2020; Ganusova et al., 2020). The P25 protein of potato virus X (PVX), for example, interferes with the function of ferredoxin 1 (FD1), an important protein involved in electron transfer between PSII and PSI, resulting in reduced levels of the defence‐related hormones abscisic acid (ABA) and salicylic acid (SA) (Yang et al., 2020). This reduction decreases callose accumulation at PD, and consequently increases PD permeability and PVX spread in the host plant (Yang et al., 2020). Because the chloroplast is also the site for the replication of several RNA viruses, viral effectors are expected to recruit specific chloroplast proteins into their viral replication complex (Budziszewska & Obrepalska‐Steplowska, 2018; Cheng et al., 2013; Ganusova et al., 2020; Zhang et al., 2017). Bamboo mosaic virus (BaMV), for example, recruits the chloroplast phosphoglycerate kinase (chl‐PGK) protein, that is, the viral RNA genome binds to chl‐PGK and transports it to the chloroplast (Cheng et al., 2013). Once in the chloroplast, BaMV recruits further chloroplast proteins into the viral replication complex to complete the infection cycle (Huang et al., 2017). In another example, infection with rice stripe virus (RSV) dramatically changes the proteome profiles of the Nicotiana benthamiana protoplast and chloroplast, resulting in a significant decrease in the number of nuclear‐encoded chloroplast‐localized proteins; the decrease is caused by RSV interference with three host factors (K4CSN4, K4CR23, and K4BXN9) that are involved in protein delivery to the chloroplast (Zhao et al., 2019).
It follows that viral interference with the functions of chloroplast proteins explains why photosynthesis is reduced in susceptible plants (i.e., in compatible interactions). In contrast, some resistant plants show increased expression of photosynthesis‐related genes. For example, expression of photosynthesis‐related genes in soybean cultivar L29 (which carries the resistance [R]‐gene Rsv3) was increased in response to infection by the avirulent G5H strain but not in response to the virulent G7H strain of soybean mosaic (SMV) (Alazem et al., 2018).
Soybean mosaic virus is a member of the genus Potyvirus and has a single‐stranded, positive‐sense RNA genome that encodes 11 viral proteins and is about 10 kb in length (Hajimorad et al., 2018; Liu et al., 2016). SMV has many strains distributed worldwide and, depending on the phenotypic responses of various soybean cultivars, these strains have been classified into seven distinct strains in the United States (G1 to G7) and into 21 strains in China (SC1 to SC21) (Hajimorad et al., 2018). Genetic resistance to SMV is mainly achieved through different strain‐specific NLR‐type R‐genes such as the Rsv and the Rsc groups (Widyasari et al., 2020). There are several other non‐NLR host factors that have been found to be critical for resistance, either because they are key components in the signalling pathway downstream of the R‐gene or because they are part of a plant system that degrades viral RNA or protein (i.e., antiviral RNA silencing and double‐stranded RNA ribonuclease) (Ishibashi et al., 2019; Liu et al., 2014; Widyasari et al., 2020).
Here, we investigated the roles of two photosynthesis‐related proteins, PSaC and ATPsyn‐α, in the resistance to SMV in soybean cultivar L29, which is resistant to G5H but not to G7H. Both proteins were strongly up‐regulated in cultivar L29 in response to G5H, whereas the response to G7H infection was rather weak. However, their roles in resistance to SMV have not been investigated. Constitutive expression of PSaC, a member of PSI, and ATPsyn‐α, a component in the ATPase synthase complex, increased resistance to SMV‐G7H infection in Lee74 (a susceptible rsv‐null soybean cultivar) and in N. benthamiana plants. Genes involved in the antiviral RNA pathways were up‐regulated in the plants transiently expressing PSaC or ATPsyn‐α, which may account for the resistance phenotype induced by both genes.
2. RESULTS
2.1. Chloroplast‐related genes are induced in the resistant cultivar L29 in response to SMV‐G5H infection
The soybean cultivar L29 carries the R‐gene Rsv3, which confers resistance against the SMV avirulent strain G5H but is ineffective against the virulent strain G7H (Seo, Lee, & Kim, 2009). We previously obtained RNA‐Seq data from L29 plants infected with strains G5H and G7H (Alazem et al., 2018). The data showed that, in the incompatible interaction (resistance against G5H), a large number of differentially regulated genes were photosynthesis‐related (Alazem et al., 2018). To examine this list more closely, we searched for the top up‐regulated genes (fold change >1) that were induced only in response to G5H infection at any time point (Figure 1a). Most of these genes have different functions related to photosynthesis/chloroplasts (Table 1). While the expression of most of these genes was induced in response to G5H, the expression of several was temporarily and slightly increased in response to G7H at 8 h postinfection (hpi) but then decreased at 24 and 54 hpi (Figure 1a). This suggests a possible relationship between their suppression and G7H virulence. We selected two genes, Glyma.18G155300.1 and Glyma.12G232000.1, which were strongly down‐regulated in response to G7H but up‐regulated in response to G5H (Figure 1a), for further analysis. In the soybean DB (Soybase) assembly 4 v. 1, Glyma.18G155300.1 and Glyma.12G232000.1 were reported to encode the PSaC subunit of the PSI subunit (PSaC) and the ATP‐synthase α‐subunit (ATPsyn‐α), respectively (Table 1) (Brown et al., 2021; Grant et al., 2010).
FIGURE 1.
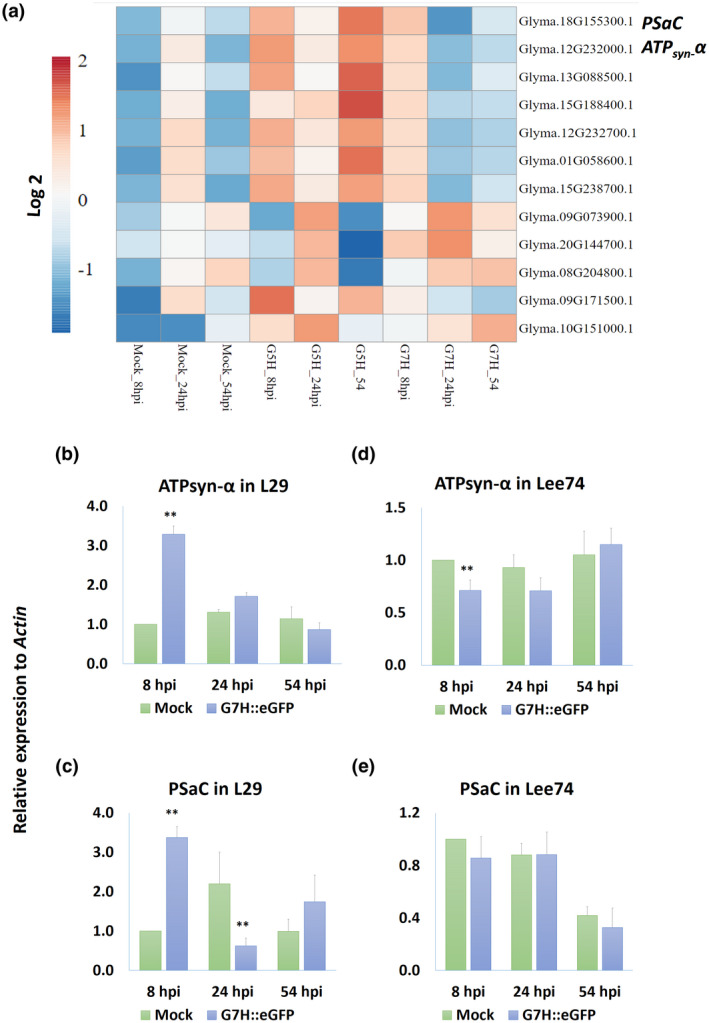
Expression of photosynthesis‐related genes in response to soybean mosaic virus (SMV) infection. (a) Heat‐map of photosynthesis‐related genes regulated by infection with the avirulent strain G5H or the virulent strain G7H of SMV. Expression of ATPsyn‐α (b) and PSaC (c) in L29 plants (which carry the Rsv3 resistance gene) at 8, 24, and 54 h postinfection (hpi) by G7H::eGFP. Expression of ATPsyn‐α (d) and PSaC (e) in Lee74 plants (rsv‐null) at 8, 24, and 54 hpi by G7H::eGFP. Actin11 was used as the internal control. In (b–e), values are means + SD of three biological replicates. Values were compared to that of the corresponding mock‐treated plants (the bar on the left) with one‐sided Student's t tests; * and ** indicate a significant difference at p < 0.05 and p < 0.01, respectively
TABLE 1.
Functional analysis and gene ontology of the photosynthesis‐related genes regulated by SMV infection
| Gene ID | Annotation | Predicted localization based on Arabidopsis ortholog | Database ID | Pathway | Arabidopsis relative ortholog | BLAST results against Arabidopsis ortholog |
|---|---|---|---|---|---|---|
| Glyma.18G155300.1 | PSaC subunit of photosystem I subunit | Chloroplast, nucleus | PF12838 | Photosystem I/4Fe‐4S dicluster domain | ATCG01060.1 | Identities = 223/243 (88%), gaps = 0/246 (0%) |
| Glyma.12G232000.1 | ATP synthase subunit α (ATPsyn‐α) | Chloroplast, cytosol, mitocondrion, plasma membrane | K02887 | Ribosome | ATCG00120.1 | Identities = 1345/1514 (89%), gaps = 0/1514 (0%) |
| Glyma.13G088500.1 | 4Fe‐4S binding domain/photosystem I | Chloroplast, nucleus | PF00037 | Photosystem I | ATCG01060.1 | Identities = 132/143 (92%), gaps = 0/143 (0%) |
| Glyma.15G188400.1 | Photosystem II reaction centre N protein (psbN) | Chloroplast, nucleus | PF02468 | Photosystem II reaction centre protein N | ATCG00700.1 | Identities = 121/132 (92%), gaps = 0/132 (0%) |
| Glyma.12G232700.1 | Photosystem II cytochrome b559 subunit α (psbE) | Chloroplast | K02711 | Photosystem II PsbJ protein (psbJ) | ATCG00580.1 | Identities = 220/238 (92%), gaps = 0/238 (0%) |
| Glyma.01G058600.1 | Cytochrome b6 (petB) | Chloroplast | K02704 | Photosystem II CP47 chlorophyll apoprotein | ATCG00720.1 | Identities = 305/337 (91%), gaps = 0/337 (0%) |
| Glyma.15G238700.1 | Photosystem II cytochrome b559 subunit α (psbE) | Chloroplast | K02707 | Photosystem II cytochrome b559 subunit α | ATCG00580.1 | Identities = 228/252 (90%), gaps = 0/252 (0%) |
| Glyma.09G073900.1 | Photosystem II subunit X | Chloroplast | PF06596 | Photosystem II | AT2G06520.1 | Identities = 152/226 (67%), gaps = 9/226 (4%) |
| Glyma.20G144700.1 | Photosystem I subunit D‐2 | Chloroplast, cytosol | PF02531 | Photosystem I | AT4G02770.1 | Identities = 365/466 (78%), gaps = 3/466 (1%) |
| Glyma.08G204800.1 | Photosystem I subunit H2 | Chloroplast, nucleus | PF03244 | Photosystem I | AT1G52230.1 | Identities = 327/438 (75%), gaps = 3/438 (1%) |
| Glyma.09G171500.1 | ATP synthase subunit α/defence response to bacteria | Chloroplast, cytosol, mitocondrion, plasma membrane | PF02874 | Chloroplast ATP synthase complex | ATCG00120.1 | Identities = 312/357 (87%), gaps = 0/357 (0%) |
| Glyma.10G151000.1 | Large subunit ribosomal protein L22e | Nucleus | K02891 | Ribosome/coronavirus disease—COVID‐19 | AT1G56220.1 | Identities = 218/288 (76%), gaps = 17/288 (6%) |
Gene annotation information was obtained from the Soybase database (DB) assembly 4 v. 1. The Phytozome soybean DB was used when the Soybase DB did not provide information regarding gene annotation. The predicted pathways of the genes were obtained from the following DBs: (a) Pfam v. 33.1 (http://pfam.xfam.org/), (b) Tair DB (https://www.arabidopsis.org/), and (c) KEGG Pathway (https://www.genome.jp/kegg/pathway.html).
To confirm the RNA‐Seq data, we used reverse transcription quantitative PCR (RT‐qPCR) to measure the expression of both genes in L29 plants infected with G7H. Expression of GmATPsyn‐α significantly increased at 8 hpi but then declined at 24 and 54 hpi to levels comparable to that in mock treatments (Figure 1b). GmPSaC increased only at 8 hpi, then decreased to a level lower than that of the mock treatment at 24 hpi (Figure 1c). We then analysed the expression of these genes in Lee74 plants, a susceptible rsv‐null soybean cultivar. Interestingly, the expression pattern of both genes did not differ with G7H infection compared with mock treatment at any time point, except for a slight decrease of GmATPsyn‐α at 8 hpi (Figure 1d,e). This suggests that although the interaction is compatible with L29, the Rsv3 gene might be involved in the early induction of both genes in L29 plants but that G7H was able to suppress the responses as the infection progressed.
Sequence analysis revealed that GmPSaC is a small protein composed of 81 amino acid residues and has two copies of the ferredoxin‐like 4Fe‐4S binding site in the specific Fer4‐7 domain located between amino acids 10 and 61 (Figure 2a). PSaC is an essential member of PSI (iron‐sulphur protein PSaC) in the chloroplast and functions in the fast electron transfer to ferrodoxin through the Fer4‐7 domain (Fischer et al., 1998; Kubota‐Kawai et al., 2018). The other protein, GmATPsyn‐α, encodes the ATPase α subunit of 510 amino acids from the ATP synthase α/β family with three domains, including the β‐barrel domain positioned between amino acids 29 and 93, the nucleotide‐binding domain positioned between amino acids 150 and 365, and the C‐terminal domain positioned between amino acids 372 and 496 (Figure 2b). The enzyme complexes catalyse the conversion of ADP to ATP using proton motive force, confer redox regulatory properties, and are located in the thylakoid membrane of the chloroplast (Table 1) (Hahn et al., 2018; Hisabori et al., 2013).
FIGURE 2.
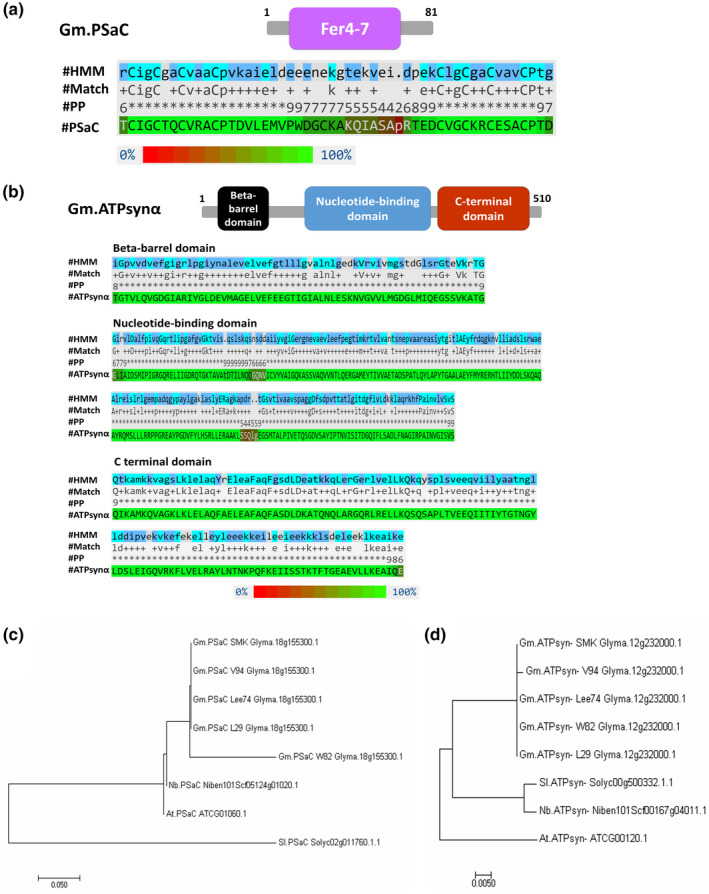
Domains and phylogenetic analyses of GmPSaC and GmATPsyn‐α. (a) Conserved domain in GmPSaC. Protein sequence of GmPSaC BLASTed against the Pfam database showed that GmPSaC belongs to the Fer4‐7 superfamily. Alignment result between the hidden Markov model (HMM) and GmPSaC (positions 10–61, E‐value 1.55e−07). (b) Conserved domains in GmATPsyn‐α. Protein sequence of GmATPsyn‐α BLASTed against the Pfam database showed that GmATPsyn‐α belongs to the ATP synthase α/β family and has three domains. Alignment between HMM and GmATPsyn‐α showed that the protein has three domains: the 1‐β‐barrel domain (positions 29–93, E‐value 3.66e−16), the 2‐nucleotide‐binding domain (positions 150–365, E‐value 5.21e−114), and the 3‐C terminal domain (positions 372–496, E‐value 3.34e−59). #HMM is the hidden Markov model, and #Match indicates the match between the query sequence and the HMM. #PP indicates the posterior probability (or degree of confidence) in each individual aligned residue. The coloured PSaC or ATPsyn‐α sequences indicate the posterior probability according to the scale from 0% to 100% at the bottom. Analysis was carried out in the Pfam database v. 33.1. (c, d) Phylogenetic analysis (nucleic acid sequences) of GmPSaC (c) and GmATPsyn‐α (d) with homologs from five soybean cultivars: William 82 (W82), Lee74, Somyoungkong (SMK), V94, and L29, as well as orthologs from Arabidopsis thaliana, Nicotiana benthamiana, and Solanum lycopersicum. The phylogeny was generated using the neighbour‐joining method with MEGA 7 software. Numbers represent relative phylogenetic distance
Analysis of amino acid sequences from five soybean cultivars with different resistance backgrounds (L29, Rsv3; William 82 (W82), rsv‐null; Lee74, rsv‐null; Somyoungkong (SMK), rsv‐null; and V94, Rsv4) showed that the sequence for GmATPsyn‐α is identical in all five cultivars (Figure S1a). However, the sequence of GmPSaC in W82 differed in six amino acids relative to the other cultivars (Figure S1b). Phylogenetic analysis clustered GmPSaC close to orthologs from Arabidopsis thaliana and N. benthamiana, and only the ortholog from Solanum lycopersicum was genetically distant from the others (Figure 2c). Analysis revealed much closer relatedness for most orthologs except for At.GmATPsyn‐α, which clustered far from the others (Figure 2d). Hereafter, the genes Glyma.18G155300.1 and Glyma.12G232000.1 will be referred to as GmPSaC and GmATPsyn‐α, respectively.
2.2. GmPSaC and GmATPsyn‐α genes are induced in cultivars with different resistance backgrounds
The finding that GmPSaC and GmATPsyn‐α are temporarily induced in L29, which is immune to G5H via the Rsv3 gene but is susceptible to G7H, prompted us to determine the expression of both genes in other cultivars with different resistance backgrounds. For this, three rsv‐null cultivars (Lee74, W82, and SMK), one Rsv4 cultivar (V94), and one Rsv3 cultivar (L29) were assessed for their susceptibility to G7H. Infection by G7H (which expresses green fluorescent protein, GFP) induced visual symptoms in the systemically infected leaves (SL) of all cultivars except V94 at 10 days postinoculation (dpi) (Figure 3a). Confirming this, a protein blot revealed that GFP from G7H was undetectable in cultivar V94 but accumulated to different levels in the other cultivars, with Lee74 being the most susceptible to infection (Figure 3b). Reactive oxygen species (ROS), which is a sign of activated antiviral defence (Calil & Fontes, 2017; Wu et al., 2017), was not detected in any of the tested cultivars regardless of the resistance levels exhibited in response to G7H infection (Figure 3c). The expression level of GmPSaC and GmATPsyn‐α was then measured in the inoculated leaves (IL) of the five infected cultivars at 5 dpi. Interestingly, only the resistant cultivar V94 showed a significant increase in the expression of both genes; the other cultivars did not exhibit significant changes in the expression except for a c.50% increase in ATPsyn‐α in SMK plants, which accumulated less G7H than the other susceptible cultivars (Figure 3d,e). These findings indicate that tolerance/resistance to G7H infection might be related to the function of both genes in soybean plants, and that the presence of an anti‐SMV R‐gene may enhance their regulation in response to SMV infection.
FIGURE 3.
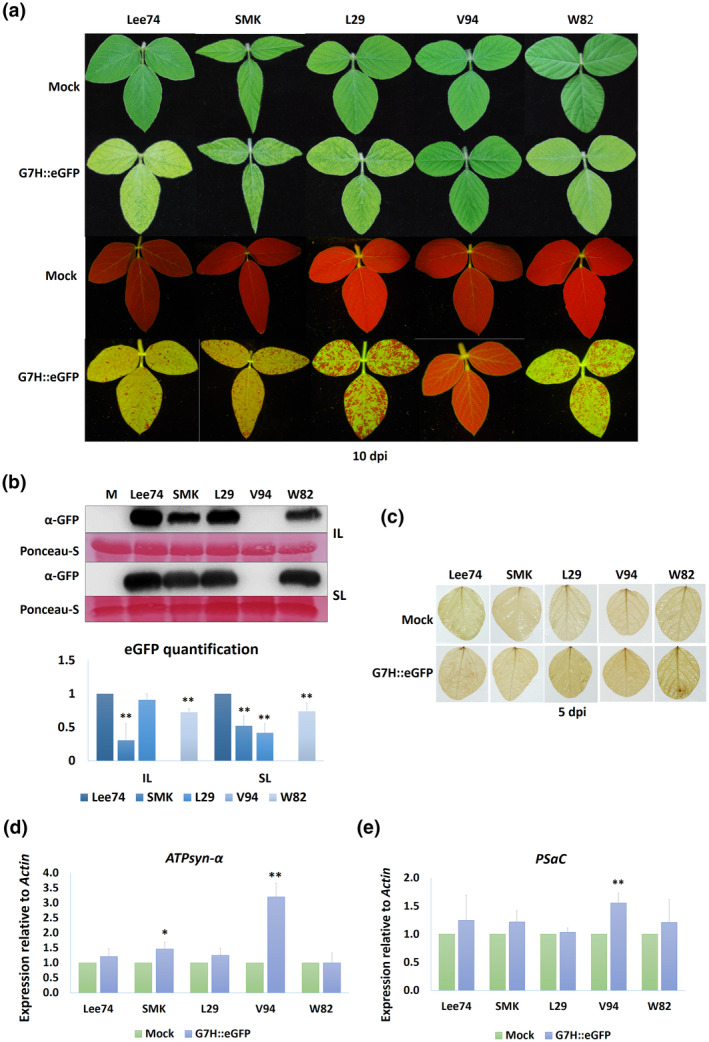
Soybean susceptibility to infection by SMV‐G7H. (a) Visual symptoms on the following five soybean cultivars infected with pSMV‐G7H::eGFP: Lee74, Somyoungking (SMK), L29, V94, and William 82 (W82). (b) Western protein blot for green fluorescent protein (GFP) levels (upper panel) in soybean cultivars infected with pSMV‐G7H::eGFP and their quantified levels (lower panel). Inoculated leaves (IL) were assayed at 5 days postinoculation (dpi) and systemically infected leaves (SL) were assayed at 10 dpi. M is mock from uninfected Lee74 plants, which were used as a negative control. Ponceau S staining of RuBisCO was used on the loading control. The blot is a representative of three biological replicates with similar results. (c) Accumulation of reactive oxygen species (ROS) in soybean cultivars as indicated by 3,3′‐diaminobenzidine staining at 5 dpi of pSMV‐G7H::eGFP. (d, e) Relative expression levels of ATPsyn‐α (d) and PSaC (e) in the five soybean cultivars infected with pSMV‐G7H::eGFP at 5 dpi. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in the legend of Figure 1; * and ** indicate a significant difference at p < 0.05 and p < 0.01, respectively
2.3. Overexpression of ATPsyn‐α and PSaC induces resistance against G7H in the susceptible cultivar Lee74
To determine the effect of ATPsyn‐α and PSaC on resistance to G7H, the coding sequence (CDS) of each gene was cloned from L29 plants into the G7H genome to create pSMV‐G7H::eGFP::ATPsyn‐α and pSMV‐G7H::eGFP::PSaC constructs (Figure 4a). As a member of the Potyvirus genus, SMV uses the host's cellular translation machinery to translate its RNA into one single polyprotein, which undergoes self‐cleavage to produce 11 different viral proteins (Hajimorad et al., 2018). We previously took advantage of this characteristic by inserting Green fluorescent protein (GFP) as a reporter gene within the SMV‐infectious clone pSMV‐G7H::eGFP (Seo et al., 2014). Here, we inserted both genes downstream of the GFP within the G7H genome (Figure 4a). The rsv‐null cultivar Lee74 was rub‐inoculated at the unifoliate stage with plasmids of both constructs (the seedlings were about 12 days old) and the accumulation level was measured in IL and SL at 7 and 14 dpi, respectively. While Lee74 developed strong GFP fluorescence in the SL following infection with pSMV‐G7H::eGFP, GFP fluorescence was weak in the case of pSMV‐G7H::eGFP::PSaC and undetectable in the case of pSMV‐G7H::eGFP::ATPsyn‐α (Figure 4b). A western protein blot confirmed this observation, that is, GFP protein accumulation was lower for pSMV‐G7H::eGFP::PSaC than for pSMV‐G7H::eGFP, and was undetectable for pSMV‐G7H::eGFP::ATPsyn‐α in both IL and SL (Figure 4c). Expression levels of eGFP RNA were also confirmed by RT‐qPCR for both constructs, that is, expression was significantly lower in the chimeras than in the pSMV‐G7H::eGFP control, and was lowest in pSMV‐G7H::eGFP::ATPsyn‐α (Figure 4d). These findings indicate that both genes contribute to resistance against G7H in soybean plants, although to different degrees.
FIGURE 4.
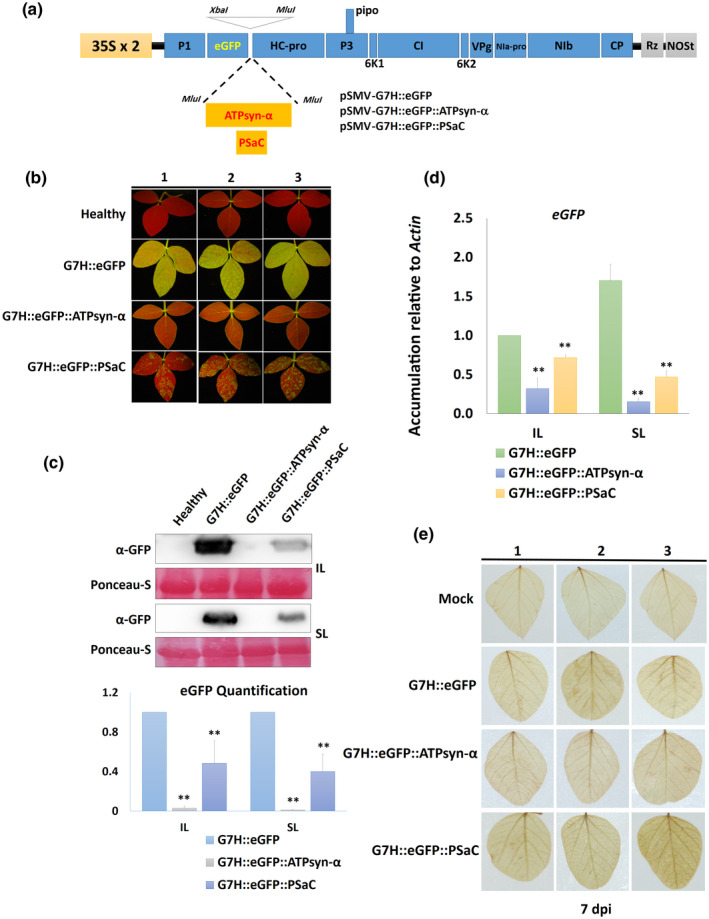
Effect of overexpressing ATPsyn‐α and PSaC on resistance against G7H in the susceptible cultivar Lee74. (a) Schematic drawing of pSMV‐G7H::eGFP construct with the insertion site for ATPsyn‐α and PSaC downstream of the GFP coding sequence; the expression of the construct is driven by two copies of the CaMV 35S promoter (35S × 2) and is terminated by an NOS terminator (NOSt). Rz is a cis‐cleaving ribozyme sequence. (b) Green fluorescent protein (GFP) visual levels in the systemically infected leaves (SL) of Lee74 plants. The first unifoliate leaves of 12‐day‐old seedlings were infected with pSMV‐G7H::eGFP, pSMV‐G7H::eGFP::ATPsyn‐α, or pSMV‐G7H::eGFP::PSaC. Fourteen days later, the SL from three plants (1, 2, and 3) were photographed under UV light. (c) Western protein blot for GFP levels (upper panel) in the inoculated leaves (IL) and the SL, and their quantified levels (lower panel). Ponceau S staining of RuBisCO was used on the loading control. (d) Relative expression levels of GFP RNA in IL and SL of Lee74 infected with pSMV‐G7H::eGFP constructs. Actin11 was used as the internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in the legend of Figure 1; ** indicates a significant difference at p < 0.01. (e) Detection of reactive oxygen species in Lee74 as indicated by 3,3′‐dimainobenzidine staining at 7 days postinoculation (dpi)
The effect of ATPsyn‐α and PSaC was also assayed on resistance to the avirulent strain G5H. Lee74 plants were infected with G5H::eGFP, G5H::eGFP::ATPsyn‐α or G5H::eGFP::PSaC infectious clones (Figure S2a). Plants developed strong GFP patches following the infection with G5H::eGFP. However, GFP fluorescence was less in G5H::ATPsyn‐α or G5H::PSaC constructs than in the G5H::eGFP control (Figure S2b), and the western protein blot showed very low accumulation of eGFP in plants infected with G5H::eGFP::ATPsyn‐α or G5H::eGFP::PSaC compared to those infected with G5H::eGFP (Figure S2c). This result indicates that both genes induce a common defence mechanism against SMV virulent and avirulent strains.
To confirm that both inserts translated into proteins, we first checked for the presence of both genes in the pSMV‐G7H::eGFP genome from RNA extracted from the soybean SL using primers targeting the flanking regions of the insert site. Indeed, both inserts were detected in the pSMV‐G7H::eGFP genome (Figure S3a), and sequencing of PCR products showed that both insets remained intact throughout the replication and movement of pSMV‐G7H::eGFP (Figure S3b,c). Next, an HA‐tag was fused to each insert to generate G7H::eGFP::PSaC::HA and G7H::eGFP::ATPsyn‐α::HA clones. A western protein blot showed that both genes were translated into proteins and that they were not lost or missed in the translation of the SMV polyprotein in the SL (Figure S3d). In addition, the expression of these genes in pSMV‐G7H::eGFP might trigger their silencing in plants. To examine this, RT‐qPCR with primers annealing to the 3′ untranslated regions of both genes showed that endogenous transcripts of both genes were not affected by the constitutive expression via pSMV‐G7H::eGFP (Figure S4a,b). To determine whether this resistance is connected to ROS, 3,3′‐diaminobenzidine staining on the IL 7 dpi showed no ROS in response to G7H::eGFP or the constructs expressing either gene (Figure 4e). This indicated that ROS may not be part of the resistance induced by PSaC or ATPsyn‐α.
2.4. Knockdown of ATPsyn‐α and PSaC increased Lee74 susceptibility against G7H infection
To confirm the role of both genes in resistance against G7H, virus‐induced gene silencing was employed using the silencing vector bean pod mottle virus (BPMV). Knocking down either gene significantly reduced its expression by c.60% compared with the empty vector of BPMV (BPMV‐EV) (Figure 5a). No visual symptoms were developed on the knocked‐down plants other than the typical BPMV mottling symptoms observed at 12 dpi (Figure 5b). Lee74 plants were then infected with G7H::eGFP, which developed a strong GFP signal in the SL at 10 dpi in ATPsyn‐α‐silenced plants, but was of similar intensity to that of PSaC‐silenced plants (Figure 5c). RT‐qPCR and western blot for eGFP confirmed that G7H::eGFP accumulated more in the ATPsyn‐α knocked‐down plants, and that G7H accumulation level was similar between BPMV‐EV and BPMV‐PSaC plants (Figure 5d,e). These data indicated that silencing ATPsyn‐α has a strong influence on plant susceptibility to G7H infection, unlike that of PSaC, which was similar to the control BPMV‐EV treatment.
FIGURE 5.
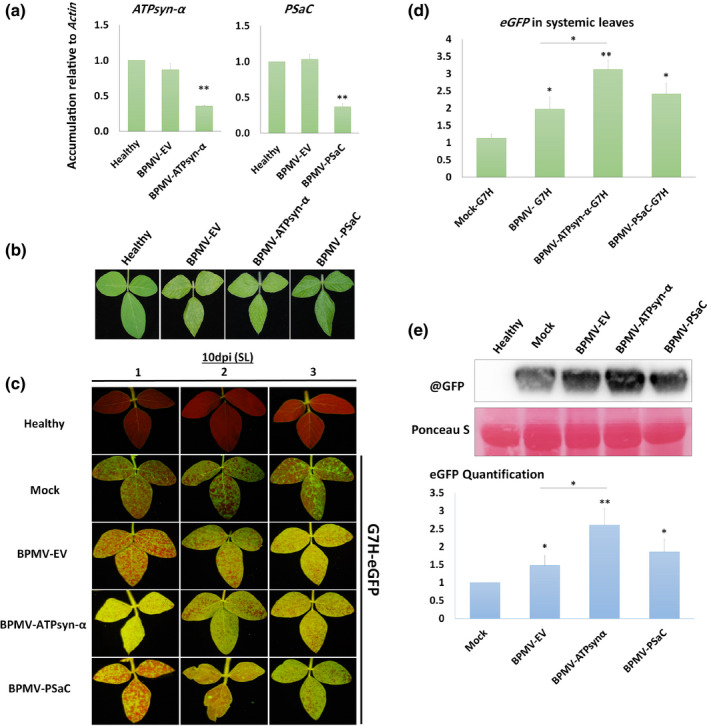
Effect of silencing GmATPsyn‐α and GmPSaC on soybean susceptibility to SMV‐G7H infection. Lee74 plants were silenced in GmATPsyn‐α and GmPSaC using BPMV silencing vector. (a) Relative expression levels of ATPsyn‐α (left) and PSaC (right) in the upper systemic leaves of Lee74 plants 14 days after BPMV infection in the empty vector (BPMV‐EV), ATPsyn‐α‐silenced (BPMV‐ATPsyn‐α), and PSaC‐silenced plants (BPMV‐PSaC). Healthy plants were used as negative control. Actin was used as internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in the legend of Figure 1; ** indicates a significant difference at p < 0.01. (b) Mottling symptoms developed in silenced Lee74 plants compared to BPMV‐EV control or healthy plants. (c) Green fluorescent protein (GFP) fluorescence from the upper systemic leaves of silenced plants infected with pSMV‐G7H::eGFP at 10 days postinoculation (dpi). Mock plants were treated with phosphate buffer as a control for BPMV infection. (d) Relative expression levels of eGFP in the Lee74 systemically infected leaves with pSMV‐G7H::eGFP at 10 dpi. Healthy plants were used as negative control. Actin was used as internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in the legend of Figure 1; significant difference at *p < 0.05, **p < 0.01. (e) Protein blot of GFP levels (upper panel) in the Lee74 systemically infected leaves with pSMV‐G7H::e GFP at 10 dpi, and their quantified levels (lower panel). Ponceau S staining of RuBisCO was used on the loading control. The blot is a representative of three biological replicates with similar results
To determine whether the silencing process may affect off‐target transcripts, a BLAST search using both genes was made in the Soybase database in a search for paralogs. Only ATPasesyn‐α had two close paralogs: Glyma.16G115300.1 (which encodes a chloroplast ATP synthase subunit α) and Glyma.05G092300.1 (which encodes a mitochondrial ATP synthase subunit α). However, the designated fragment for silencing shares low similarity with the two paralogs (Figure S5). Expression levels of either gene were not affected by the silencing of ATPsyn‐α (Figure S6a,b), which indicates that silencing probably did not affect off‐target transcripts.
2.5. Localization of ATPsyn‐α and PSaC in N. benthamiana and their effects on N. benthamiana resistance against SMV‐G7H
To investigate the localization of ATPsyn‐α and PSaC, we expressed both genes in the binary vector pBin61‐HA‐mCherry (Alazem et al., 2020). We used the chloroplast‐localized protein from Arabidopsis, EMB1303, fused with eGFP as a marker protein (Huang et al., 2009). AtEMB1303 localized in the chloroplast membrane, and the GFP signal was also detected in the extended stromules (Figure 6a and Figure S7). Both PSaC and ATPsyn‐α localized in the chloroplast envelope, the nucleus, and the cytoplasm (Figure 6b,c). We next examined the effect of the transient expression of both genes on G7H accumulation in N. benthamiana. Although N. benthamiana is not a preferred host for SMV, the virus can accumulate to detectable levels in this host. Interestingly, both soybean genes reduced the accumulation of SMV‐G7H in N. benthamiana plants, indicating that the resistance mechanism regulated by these genes could be similar in the two hosts and independent of the Rsv3‐mediated resistance (Figure 6d).
FIGURE 6.
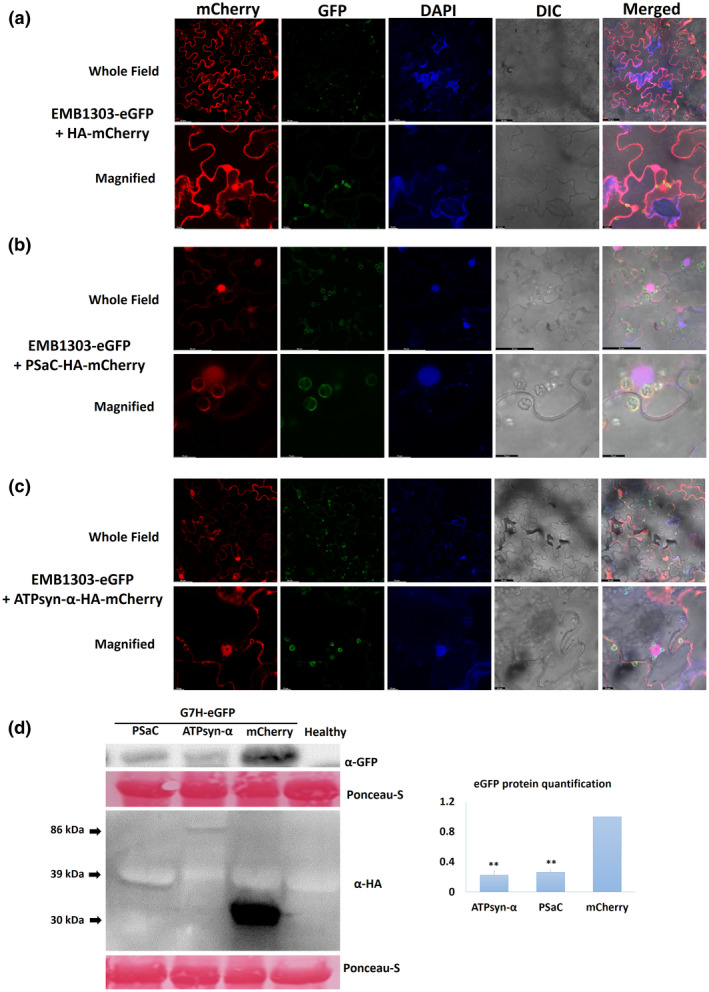
Localization and effects of GmATPsyn‐α and GmPSaC on resistance to SMV‐G7H in Nicotiana benthamiana leaves. (a) Localization of the chloroplast‐marker protein AtEMB1303‐eGFP with pBin‐3HA‐mCherry as a control. (b) Co‐localization of EMB1303‐eGFP and PSaC‐HA‐mCherry. (c) Co‐localization of EMB1303‐eGFP and ATPsyn‐α‐3HA‐mCherry. N. benthamiana plants were agroinfiltrated with pBin‐eGFP‐AtEMB1303 (chloroplast‐marker protein) with pBin‐3HA‐mCherry constructs carrying GmATPsyn‐α or GmP S aC, and pPZP‐2b, which carries the CMV suppressor of RNA‐silencing protein gene (2b) to enhance the transient expression. DAPI was used to stain nuclei. Leaves were photographed 3 days after agroinfiltration. Scale bars measure 50 µm for the whole field and 10 µm for the magnified field. (d) Effect of transient overexpression of PSaC and ATPsyn‐α on resistance to SMV‐G7H in N. benthamiana plants. The same agrobacterial cultures used for the localization test were used without pPZP2b for the SMV‐G7H::eGFP infection. One day after agroinfiltration, N. benthamiana leaves were sap‐infected with SMV‐G7H::eGFP prepared from infected soybean plants. Samples were collected at 5 days postinoculation, and western protein blots were hybridized with anti‐GFP to detect eGFP from SMV‐G7H, and anti‐HA to detect GmPSaC (39 kDa), GmATPsyn‐α (80 kDa), and the empty vector 3HA‐mCherry (30 kDa). eGFP levels were quantified using ImageJ (right panel). Ponceau S staining of RuBisCO was used as the internal control, and the blots are representatives of three biological replicates. ** indicates a significant difference at p < 0.01
2.6. Involvement of defence‐related hormones in ATPsyn‐α‐ and PSaC‐mediated resistance
The chloroplast plays a critical role in plant immunity because it is a major site for the production of several plant hormones such as SA, ABA, jasmonic acid (JA), and ethylene (ET) (Alazem & Lin, 2015; Bhattacharyya & Chakraborty, 2018; Zhao et al., 2016). To investigate whether ATPsyn‐α and PSaC have any effect on the signalling pathway of defence‐related hormones, the expression levels of key genes in the signalling pathways of SA, ABA, JA, and ET were measured in Lee74 plants infected with pSMV‐G7H::eGFP, pSMV‐G7H::eGFP::PSaC, and pSMV‐G7H::eGFP::ATPsyn‐α. SMV‐G7H::eGFP infection of Lee74 plants decreased the expression of ICS1 in the SA pathway and of ABA1 in the ABA pathway (Figure 7a,e). However, the following genes belonging to different pathways were increased in response to G7H infection: PAD4 in the SA pathway (Figure 7b), JAR1 in the JA pathway (Figure 7c), ABA2 in the ABA pathway (Figure 7f), and DREB1A‐1 and DREB1A‐2 in the ET pathway (Figure 8g,h). This indicated that SMV‐G7H infection disrupts the hormone balance in the infected plant by inducing several antagonistic hormone signalling pathways.
FIGURE 7.
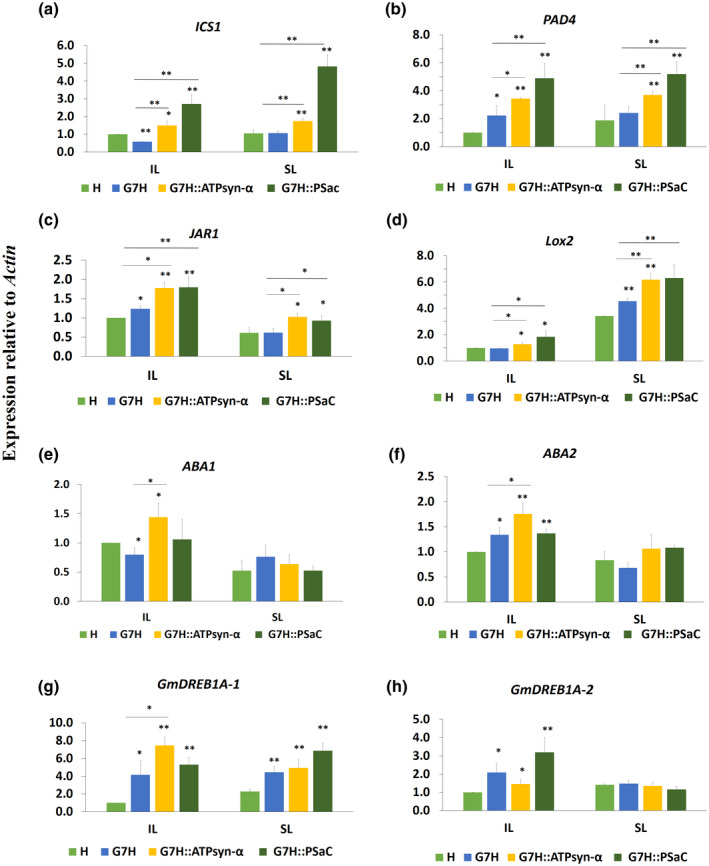
Expression levels of key genes of defence‐related hormones in Lee74 plants in response to SMV‐G7H expressing ATPsyn‐α and PSaC genes. Relative expression levels in Lee74 plants of salicylic acid‐related genes ICS1 (a) and PAD4 (b); jasmonic acid‐related genes JAR1 (c) and Lox2 (d), abscisic acid biosynthesis genes ABA1 (e) and ABA2 (f), and ethylene‐related genes Gm DREB1A‐1 (g) and Gm DREB1A‐2 (h). The unifoliate leaves of Lee74 plants were inoculated with pSMV‐G7H::eGFP expressing ATPsyn‐α or PSaC genes (pSMV‐G7H::eGFP::ATPsyn‐α or pSMV‐G7H::eGFP::PSaC, respectively); the inoculated leaves (IL) and systemically infected leaves (SL) were collected at 7 and 14 days postinoculation, respectively. Actin11 was used as the internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in Figure 1; * and ** indicate a significant difference at p < 0.05 and p < 0.01, respectively. An additional t test was carried out to compare expressions in the pSMV‐G7H::eGFP::ATPsyn‐α and pSMV‐G7H::eGFP::PSaC treatments to that in pSMV‐G7H::eGFP
FIGURE 8.
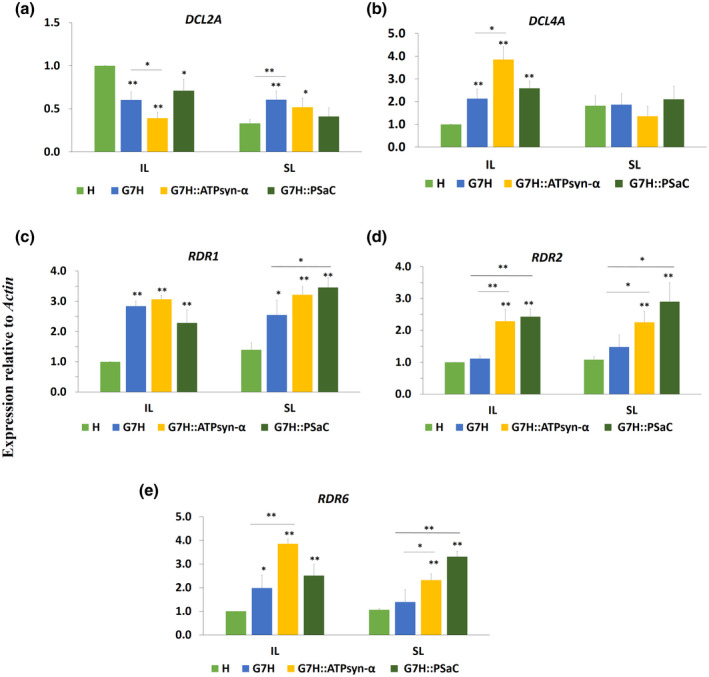
Expression levels of RNA silencing genes in Lee74 plants in response to SMV‐G7H expressing ATPsyn‐α and PSaC genes. Relative expression of Dicer‐like (DCL) 2a (a) and DCL4a (b), and of RNA‐dependent RNA polymerase (RDR) 1a (c), RDR2a (d), and RDR6a (e) in Lee74 plants. Actin11 was used as the internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in Figure 1; * and ** indicate a significant difference at p < 0.05 and p < 0.01, respectively. An additional t test was carried out to compare expression in the pSMV‐G7H::eGFP::ATPsyn‐α and pSMV‐G7H::eGFP::PSaC treatment with that in the pSMV‐G7H::e GFP treatment
The expression levels of ICS1 and PAD4 in the SA biosynthesis pathway were significantly higher in the SL of plants infected with pSMV‐G7H::eGFP::PSaC and pSMV‐G7H::eGFP::ATPsyn‐α than in plants infected with pSMV‐G7H::eGFP. Such an increase was only recorded for PAD4 in the SL of plants infected with both constructs (Figure 7a,b). Similarly, the expression levels of the JA‐related genes JAR1 and Lox2 were significantly higher in both IL and SL of plants infected with both constructs than in plants infected with pSMV‐G7H::eGFP (Figure 7c,d). However, only the IL of pSMV‐G7H::eGFP::ATPsyn‐α infected plants exhibited increased levels of ABA1 and ABA2 from the ABA biosynthesis pathway (Figure 7e,f). Compared to its expression in response to pSMV‐G7H::eGFP infection, expression of the ET‐related transcription factor (TF) GmDREB1A‐1 increased only in response to infection by SMV‐G7H::eGFP::ATPsyn‐α in the IL (Figure 7g). The other ET TF GmDREB1A‐2 was not affected by infection of either constructs compared to SMV‐G7H::eGFP infection (Figure 7h). These data indicate that ATPsyn‐α has a strong effect on the expression of SA‐, JA‐, and ABA‐related genes, although they function antagonistically under abiotic stress conditions, and that PSaC increased the expression of the SA‐ and JA‐related genes.
2.7. Antiviral RNA silencing genes are regulated in ATPsyn‐α‐ and PSaC‐mediated resistance
Because SA and ABA affect the expression of RNA silencing genes (Alazem & Lin, 2020) the expression levels of the key genes in this pathway were measured in response to infection by pSMV‐G7H::eGFP, pSMV‐G7H::eGFP::PSaC, or pSMV‐G7H::eGFP::ATPsyn‐α. The expression levels of the Dicer‐like (DCL) genes DCL2a and DCL4a, and of the RNA‐dependent RNA polymerase (RDR) genes RDR1a, RDR2a, and RDR6a were up‐regulated in response to infection with either construct (Figure 8). Compared to infection of Lee74 plants with pSMV‐G7H::eGFP, infection with pSMV‐G7H:e:GFP::ATPsyn‐α significantly increased the expression of DCL4a, RDR2a, and RDR2a in the IL (Figure 8b,d,e), and this effect was evident only for RDR2a and RDR6a in the SL (Figure 8d). In contrast, DCL2a and RDR1a were down‐regulated or unchanged, respectively, in response to pSMV‐G7H::eGFP::ATPsyn‐α local infection (Figure 8a,c). The effect of pSMV‐G7H::eGFP::PSaC infection was weaker than that of pSMV‐G7H::eGFP::ATPsyn‐α infection with only RDR2a induced locally and systemically (Figure 8d), and RDR6a induced systemically (Figure 8e). We next determined if this effect on the RNA silencing genes was similar to that in N. benthamiana plants infected with G7H::eGFP expressing either gene. NbDCL2, NbDCL4, NbRDR2, and NbRDR6 were significantly increased response to virus infection (Figure S8). However, the expression was significantly higher for plants infected with G7H::eGFP::ATPsyn‐α or G7H::eGFP::PSaC than those infected with G7H::eGFP for NbDCL4, NbRDR2, and NbRDR6 genes (Figure S8b–d). These data indicate that the defence mechanisms affected by both genes are similar between soybean and N. benthamiana plants. Collectively, the antiviral RNA silencing genes may partially contribute to the ATPsyn‐α‐ and PSaC‐mediated resistance in soybean plants, and the influence of ATPsyn‐α on RNA silencing genes is greater than that of PSaC.
3. DISCUSSION
An increasing body of evidence connects plant virus replication and movement with the chloroplast. The effects of chloroplast genes on viruses are diverse and vary among virus groups. While some viruses recruit specific chloroplast proteins to their replication or movement complex, others reduce the expression of specific chloroplast genes to facilitate their replication and spread (Cheng et al., 2013; Ganusova et al., 2020; Jiang et al., 2020; Zhao et al., 2016, 2019). The current study provides evidence of positive roles of two photosynthesis‐related genes, GmPSaC and GmATPsyn‐α, in inducing resistance against SMV infection in the susceptible soybean cultivar Lee74. Previous studies reported a similar role for other ATPsyn subunits in resistance to other viruses. For example, infection with tobacco mosaic virus (TMV) reduced the expression levels of the ATPsyn‐γ subunit, and when ATPsyn‐γ was silenced in N. benthamiana plants, TMV accumulation and pathogenicity were greatly enhanced, indicating that ATPsyn‐γ is involved in limiting the intracellular trafficking of TMV as well as in inducing defence signalling pathways (Bhat et al., 2013). Interestingly, an opposite effect was found for ATP‐syn‐γ in response to infection with PVX or tomato bushy stunt virus, that is, their spread was decreased in ATP‐syn‐γ‐silenced plants (Bhat et al., 2013). In another example, infection with potato virus Y reduced the photosynthesis rate through the HC‐Pro protein in Nicotiana tabacum plants; HC‐Pro interacted with the ATPsyn‐β subunit but did not affect the enzymatic activity of ATP synthase, leading to a reduced ATP synthase content in HC‐Pro‐transgenic plants (Tu et al., 2015). In other words, we cannot generalize about the effects of ATPsyn subunits on host plant resistance to viruses; the influence on resistance can vary depending on the virus group.
ATPsyn‐α and ‐β form the hydrophilic head (cF1) powered by the membrane‐embedded‐cF0 rotary motor in the ATP synthase complex. ATPsyn‐α guides protons to and from the c‐ring protonation site (Hahn et al., 2018). In general, ATP synthase is redox‐regulated and controlled by the chloroplast thioredoxin system, which is connected with photosynthesis (Hisabori et al., 2013). Regulation of redox controls the accumulation of ROS and nitrogen species, both of which are important for resistance against several pathogens (Bentham et al., 2020; Frederickson Matika & Loake, 2014). Given the absence of necrotic lesions in soybean expressing PSaC or ATPsyn‐α, however, it is unlikely that ROS is involved in ATPsyn‐α‐ or PSaC‐mediated‐defence against SMV‐G7H.
PSaC encodes a subunit in the PSI complex and functions in electron transfer and ferrodoxin docking on the stromal side of PSI (Rantala et al., 2020). Although studies on the role of PSaC in plant–virus interactions are lacking, a previous report indicated a positive role for another member of the PSI complex, PSaK, in resistance against plum pox virus (PPV) (Jimenez et al., 2006). Infection with PPV decreased PSaK expression in N. benthamiana, and when PSaK was knocked down, PPV accumulation was enhanced. In addition, the cylindrical inclusion protein of PPV interacted with PSaK and possibly interfered with its function (Jimenez et al., 2006). Our data showed that, in response to SMV‐G7H infection, expression of PSaC and ATPsyn‐α increased in resistant soybean plants but did not decrease in susceptible plants (Figure 3d,e). That their overexpression reduced SMV‐G7H accumulation (Figure 4b,d) suggests that both genes partially contributed to resistance against SMV. In line with this finding, silencing ATPsyn‐α, but not PSaC, increased soybean susceptibility to SMV‐G7H infection (Figure 5). This confirms the role of ATPsyn‐α in resistance, but also suggests functional redundancy for genes might interrelate with PSaC, which could be members of the PSI.
The resistance conferred by ATPsyn‐α is stronger than that conferred by PSaC in both N. benthamiana and soybean plants (Figure 4b,d). This could be attributed to the simultaneous induction of several genes in the defence signalling pathways of SA, JA, and ABA in response to pSMV‐G7H::eGFP::ATPsyn‐α, but for pSMV‐G7H::eGFP::PSaC the response was limited to SA and JA (Figure 7). Pathways of all of these hormones are involved in soybean resistance to SMV (Alazem et al., 2018, 2019; Zhang et al., 2012). In fact, the connection between defence hormones and the antiviral RNA silencing pathway is well established (Alazem et al., 2019; Alazem & Lin, 2015, 2020). We previously showed that SA and ABA enhance the expression of the antiviral RNA silencing genes in soybean and A. thaliana, and that the enhanced expression confers partial resistance against SMV, BaMV, and PVX (Alazem et al., 2017, 2019). Our current findings show that ATPsyn‐α induced the expression of more genes (DCL4a, RDR2a, and RDR6a) in the antiviral RNA silencing pathway than PSaC, which only induced the expression of RDR2a and only in the IL (Figure 8). It is therefore likely that the stronger resistance triggered by ATPsyn‐α than PSaC is due to the greater influence of ATPsyn‐α on the antiviral RNA‐silencing genes.
Because trafficking through PD is strongly regulated by light and the circadian clock (Brunkard & Zambryski, 2019; Ganusova et al., 2020), it is highly probable that chloroplast‐related genes can adversely affect viruses in two ways, that is, the gene products may hinder cell‐to‐cell trafficking through PD and may also induce defence‐related hormone signalling pathways. Our results provide evidence that induction of these photosynthesis genes induces hormone signalling pathways that eventually trigger antiviral RNA silencing pathways that partially contribute to local and systemic resistance to SMV (Figure 8). Whether SMV trafficking through PD is affected by photosynthesis genes requires further investigation. The effect of enhanced photosynthesis on plant resistance to viruses is incompletely understood and also warrants additional research.
We expected to detect ATPsyn‐α and PSaC inside the chloroplast, but, surprisingly, we found that they were localized in the chloroplast envelope. In addition, both proteins were localized in the cytoplasm and the nucleus (Figure 6b,c). We did not detect any degradation of either protein by western blot (Figure 6d), which indicates that both proteins can be distributed to the cytoplasm and the nucleus for further functions that remain to be examined.
In conclusion, strong photosynthesis can increase resistance against viruses. Additional research is needed to clarify how chloroplasts in general, and photosynthesis in particular, enhance resistance against plant viruses.
4. EXPERIMENTAL PROCEDURES
4.1. Construction of the SMV vector expressing ATPsyn‐α and PSaC genes
The CDS of ATPsyn‐α and PSaC genes were amplified and cloned from several soybean cultivars and were then cloned into a TA vector (pGEM‐T Easy; Promega). The clones were confirmed by sequencing with gene‐specific primers (Table S1). The CDS of both genes from L29 plants were then cloned into the pSMV‐G7H::eGFP infectious clone to generate pSMV‐G7H::eGFP::ATPsyn‐α and pSMV‐G7H::eGFP::PSaC as previously described (Seo, Lee, Choi, et al., 2009).
4.2. Plant materials, growth conditions, and virus infections
The following five soybean cultivars were used in this study: Lee74, L29, V94, Somyoungkong (SMK), and William 82 (W82). Soybean and N. benthamiana plants were grown in growth chambers at 25°C with 70% relative humidity and a 16/8 h photoperiod. To prepare infectious sap, the first unifoliate leaf from Lee74 plants was inoculated with 10 µg per leaf of the infectious clones pSMV‐G7H::eGFP, pSMV‐G7H::eGFP::PSaC, and pSMV‐G7H::eGFP::ATPsyn‐α as previously described (Seo et al., 2009). About 15 dpi, a pool of SL from three plants was mixed and divided into 0.1‐g portions as a source of virus inoculum. After each 0.1‐g portion was ground into powder in liquid nitrogen, it was mixed with 1 ml of phosphate buffer. The mixture was placed on ice for 10 min and was then centrifuged for 10 min at 4°C and 13,580 × g. A 50‐µl volume of the supernatant was rub‐inoculated onto each leaflet of the trifoliate leaf on each soybean plant, or on two leaves on each N. benthamiana plant. Samples were collected from three plants (a total of nine leaves for soybean and six leaves for N. benthamiana) at 5 and 10 dpi for further analyses.
To investigate the effects of PSaC and ATPsyn‐α on the accumulation of SMV‐G7H::eGFP in Lee74 plants, plasmids of the infectious clones pSMV‐G7H::eGFP, pSMV‐G7H::eGFP::PSaC, and pSMV‐G7H::eGFP::ATPsyn‐α were directly rub‐inoculated on Lee74 plants using 10 µg of plasmid per leaf. The ILs and SLs were collected at 7 and 14 dpi, respectively, for RNA and protein extraction.
4.3. Silencing ATPsyn‐α and PSaC in soybean plants
The BPMV silencing vector was used to silence ATPsyn‐α and PSaC genes in Lee74 plants. In brief, fragments of 173 bp from PSaC CDS, and 347 bp from ATPsyn‐α CDS were cloned in the antisense direction in the multiple cloning site of RNA2 of BPMV, as described previously (Zhang et al., 2010). Ten micrograms of BPMV plasmids (RNA1 and RNA2) were rub‐inoculated onto the first unifoliate leaves of Lee74 plants, and the silencing efficiency was tested at 14 dpi in the second trifoliate leaf. The same leaf was sap‐inoculated with G7H::eGFP as described in section 4.2. Samples were collected from the SL 10 days after G7H::eGFP infection for further analyses.
4.4. RNA extraction and RT‐qPCR
Total RNA was extracted using TRIzol (Sigma) following the manufacturer's instructions. A 1‐μg quantity of total RNA was used for cDNA synthesis using the GoScript kit (Promega). RT‐qPCR was carried out with SYBR Green (Promega) to measure the relative expression of target genes using the ΔΔC t method. Actin11 was used as an internal control, and the primers used in this study are listed in Table S1. One‐sided Student's t tests (p < 0.05) were used to determine whether the expression level of each gene in each line was up‐regulated or down‐regulated relative to the mock‐treated plants.
4.5. Statistical analysis
RT‐qPCR was carried out in three biological replicates, and each biological replicate was repeated in three technical replicates. In Figures 1 and 3–8, values were compared to that of the mock‐treated, uninfected plants (the bar on the left) with one‐sided Student's t tests; * and ** indicate a significant difference at p < 0.05 and p < 0.01, respectively. Error bars in the charts are means of standard deviation of three biological replicates.
4.6. Western protein blot
Total protein was extracted from 0.1 g of tissue collected from a pool of IL and SL from three plants, as described previously (Alazem et al., 2018). Constructs expressing GFP were detected by western blot using polyclonal anti‐GFP antibody, and those expressing HA were detected using monoclonal anti‐HA antibody (Sigma); Ponceau S staining was used on the loading control.
4.7. Phylogenetic analysis
Amino acid sequences of ATPsyn‐α and PSaC for Glycine max, N. benthamiana, S. lycopersicum, and A. thaliana were obtained from the soybean database (DB) (Soybase), the Sol Genomics Network, and the Tair DB (Brown et al., 2021; Fernandez‐Pozo et al., 2015). The phylogenetic trees were generated using MEGA 7.0 software and by applying the neighbour‐joining method (Kumar et al., 2016).
4.8. Gene description, function, and pathways
Information about gene annotations and functions was obtained from the Soybase DB assembly 4, v. 1 (https://www.soybase.org/). The Phytozome soybean DB was used when the Soybase DB did not have gene annotation information. Both DBs predicted the pathways of the genes from the following DBs: Pfam v. 33.1, release 2019/08 (http://pfam.xfam.org/) (El‐Gebali et al., 2019), Tair DB (https://www.arabidopsis.org/), and KEGG Pathway (https://www.genome.jp/kegg/pathway.html).
4.9. Subcellular localization of ATPsyn‐α and PSaC proteins
ATPsyn‐α and PSaC were cloned into the binary vector pBin61‐3HA‐mCherry (Alazem et al., 2020). Agrobacterium infiltration was carried out on N. benthamiana plants using Agrobacterium tumefaciens C58C1 at OD600 = 0.5, with the aid of 2b, the viral suppressor of RNA silencing (pPZP‐2b), to enhance the expression of both genes. Infection with pSMV‐G7H::eGFP was carried out 1 day after agroinfiltration using 50 µl of infectious sap extract/leaf. Samples were collected at 3 dpi for confocal microscopy, and at 5 dpi for protein and RNA analysis. The chloroplast marker protein gene At EMB1301 was cloned into pBin‐eGFP and used as a marker for the localization of ATPsyn‐α and PSaC proteins.
4.10. Visualization of GFP expression and localization of the target proteins in plants
GFP fluorescence of the IL and SL was examined with UV light and with a digital camera (D700; Nikon) with a green filter. A Leica confocal microscope was used to determine the subcellular localization of AtEMB1303, ATPsyn‐α, and PSaC with a 40× lens (HC PL APO CS2 40×/1.10 WATER), and the detectors HyD (421–467 nm) and PMT (654–711 nm), with bidirectional scanning at a speed of 400 Hz and a resolution of 2048 × 2048. Leica application suite X package was used to analyse images.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
M.A. and K.‐H.K. conceived the ideas and designed the experiments, J.B. and M.A. carried out the experiments, analysed the data, and wrote the manuscript. M.A. and K.‐H.K. revised and edited the manuscript and acquired the funding. All authors have read and agreed to the published version of the manuscript.
Supporting information
FIGURE S1. Sequence alignment of ATPsyn‐α (a) and PSaC (b) proteins from five soybean cultivars: William 82 (W82), L29, Lee74, Somyongkong (SMK), and V94
FIGURE S2. Effect of over‐expressing ATPsyn‐α and PSaC on resistance against SMV‐G5H in the susceptible cultivar Lee74. (a) Schematic drawing of G5H::eGFP construct with the insertion site for ATPsyn‐α and PSaC downstream of the eGFP gene. (b) GFP visual levels in the systemically infected leaves of Lee74 plants. The first unifoliate leaves of 12‐day‐old seedlings were infected with G5H::eGFP, G5H::eGFP::ATPsyn‐α, or G5H::eGFP::PSaC. Fourteen days later, the systemically infected leaves from three plants (1, 2, and 3) were photographed under UV light. (c) Western blot for GFP in the systemically infected leaves. Ponceau S staining was used on the loading control
FIGURE S3. Stability of ATPsyn‐α and PSaC inserts in pSMV::G7H::eGFP vector. (a) Detection of ATPsyn‐α and PSaC inserts in the pSMV‐G7H::eGFP by RT‐PCR. RNA was extracted from the systemically infected Lee74 leaves with pSMV‐G7H::eGFP::ATPsny‐α or pSMV:‐G7H::eGFP::PSaC. The primers anneal to the flanking regions of the insert site. Each lane represents a reverse transcription PCR from one biological replicate. The PCR product of each chimera was purified, sequenced, and then aligned with the sequences of ATPsyn‐α (b) and PSaC (c) proteins identified in Figure 2 from L29 soybean plants. (d) Western blots for the HA‐tagged ATPsyn‐α‐HA and PSaC‐HA expressed in the pSMV‐G7H::eGFP::HA vector from the systemically infected leaves of Lee74 plants
FIGURE S4. Relative expression levels of endogenous ATPsyn‐α and PSaC in Lee74 plants infected with pSMV‐G7H::eGFP chimeras. (a) Expression levels of endogenous ATPsyn‐α in leaves inoculated with pSMV‐G7H::eGFP or pSMV‐G7H::eGFP::ATPsyn‐α. (b) Expression levels of endogenous PSaC in leaves inoculated with pSMV‐G7H::eGFP or pSMV‐G7H::eGFP::PSaC. Primers were designed to target the 3′ untranslated regions (UTRs) of either transcripts so their expression can be distinguished from that expressed within the pSMV‐G7H::eGFP infectious clone. Actin11 was used as internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in the legend of Figure 1
FIGURE S5. Multiple sequence alignment of ATPsyn‐α and its paralogs from chloroplast (Glyma.16G115300.1) and mitochondria (Glyma.05G092300.1). The sequence highlighted in red is the fragment used to initiate the virus‐induced gene silencing of ATPsyn‐α in Lee74 plants
FIGURE S6. Expression levels of ATPsyn‐α paralogs from (a) chloroplast “Glyma.16G115300.1” and (b) mitochondria “Glyma.05G092300.1” in Lee74 plants silenced with either gene. Actin11 was used as internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in the legend of Figure 1
FIGURE S7. Localization of the chloroplast marker protein AtEMB1303 in Nicotiana benthamiana leaves. Chloroplast autofluorescence is pseudocoloured in red. DAPI was used to stain the nuclei. Bars measure 50 µm
FIGURE S8. Expression levels of RNA silencing genes in Nicotiana benthamiana in response to infection with chimeras of pSMV‐G7H::eGFP. Relative expression levels of Dicer‐like (DCL) 2a (a) DCL4a (b), RNA‐dependent RNA polymerase (RDR) 2a (c), and RDR6a (d) in N. benthamiana plants infected with pSMV‐G7H::eGFP, pSMV‐G7H::eGFP::ATPsyn‐α, or pSMV‐G7H::eGFP::PSaC. Actin was used as internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in Figure 1; * and ** indicate a significant difference at p < 0.05 and p < 0.01, respectively. An additional t test was carried out to compare expression in the pSMV‐G7H::eGFP::ATPsyn‐α and pSMV‐G7H::eGFP::PSaC treatment with that in the pSMV‐G7H::eGFP treatment
TABLE S1. Primers used in this study
ACKNOWLEDGEMENTS
The authors thank Kristin Widyasari (Seoul National University) for her technical assistance. This research was supported in part by grants from the Korea Research Fellowship program (KRF grant no. 2017H1D3A1A01054585), funded by the Ministry of Science and ICT through the National Research Foundation of Korea, and from the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (120080‐05‐1‐HD030), funded by the Ministry of Agriculture, Food and Rural Affairs, Republic of Korea. J.B. was supported by a research fellowship from the Brain Korea 21 Four Program.
Bwalya, J. , Alazem, M. & Kim, K.‐H. (2022) Photosynthesis‐related genes induce resistance against soybean mosaic virus: Evidence for involvement of the RNA silencing pathway. Molecular Plant Pathology, 23, 543–560. 10.1111/mpp.13177
Contributor Information
Mazen Alazem, Email: m.alazem@gmail.com.
Kook‐Hyung Kim, Email: kookkim@snu.ac.kr.
DATA AVAILABILITY STATEMENT
Data are available on request from the authors.
REFERENCES
- Alazem, M. , He, M.H. , Chang, C.H. , Cheng, N. & Lin, N.S. (2020) Disrupting the homeostasis of high mobility group protein promotes the systemic movement of Bamboo mosaic virus. Frontiers in Plant Science, 11, 597665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem, M. , He, M.H. , Moffett, P. & Lin, N.S. (2017) Abscisic acid induces resistance against Bamboo mosaic virus through argonaute2 and 3. Plant Physiology, 174, 339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem, M. , Kim, K.H. & Lin, N.S. (2019) Effects of abscisic acid and salicylic acid on gene expression in the antiviral RNA silencing pathway in Arabidopsis. International Journal of Molecular Sciences, 20, 2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem, M. & Lin, N.S. (2015) Roles of plant hormones in the regulation of host–virus interactions. Molecular Plant Pathology, 16, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem, M. & Lin, N.S. (2020) Interplay between ABA signaling and RNA silencing in plant viral resistance. Current Opinion in Virology, 42, 1–7. [DOI] [PubMed] [Google Scholar]
- Alazem, M. , Tseng, K.C. , Chang, W.C. , Seo, J.K. & Kim, K.H. (2018) Elements involved in the Rsv3‐mediated extreme resistance against an avirulent strain of soybean mosaic virus. Viruses, 10, 581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azim, M.F. & Burch‐Smith, T.M. (2020) Organelles‐nucleus‐plasmodesmata signaling (ONPS): an update on its roles in plant physiology, metabolism and stress responses. Current Opinion in Plant Biology, 58, 48–59. [DOI] [PubMed] [Google Scholar]
- Bentham, A.R. , De la Concepcion, J.C. , Mukhi, N. , Zdrzałek, R. , Draeger, M. , Gorenkin, D. et al. (2020) A molecular roadmap to the plant immune system. Journal of Biological Chemistry, 295, 14916–14935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, S. , Folimonova, S.Y. , Cole, A.B. , Ballard, K.D. , Lei, Z. , Watson, B.S. et al. (2013) Influence of host chloroplast proteins on tobacco mosaic virus accumulation and intercellular movement. Plant Physiology, 161, 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, D. & Chakraborty, S. (2018) Chloroplast: the Trojan horse in plant–virus interaction. Molecular Plant Pathology, 19, 504–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A.V. , Conners, S.I. , Huang, W. , Wilkey, A.P. , Grant, D. , Weeks, N.T. et al. (2021) A new decade and new data at soybase, the USDA‐ARS soybean genetics and genomics database. Nucleic Acids Research, 49, D1496–D1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkard, J.O. & Zambryski, P. (2019) Plant cell‐cell transport via plasmodesmata is regulated by light and the circadian clock. Plant Physiology, 181, 1459–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budziszewska, M. & Obrepalska‐Steplowska, A. (2018) The role of the chloroplast in the replication of positive‐sense single‐stranded plant RNA viruses. Frontiers in Plant Science, 9, 1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calil, I.P. & Fontes, E.P.B. (2017) Plant immunity against viruses: antiviral immune receptors in focus. Annals of Botany, 119, 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, S.F. , Huang, Y.P. , Chen, L.H. , Hsu, Y.H. & Tsai, C.H. (2013) Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiology, 163, 1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Gebali, S. , Mistry, J. , Bateman, A. , Eddy, S.R. , Luciani, A. , Potter, S.C. et al. (2019) The Pfam protein families database in 2019. Nucleic Acids Research, 47, D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Pozo, N. , Menda, N. , Edwards, J.D. , Saha, S. , Tecle, I.Y. , Strickler, S.R. et al. (2015) The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Research, 43, D1036–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, N. , Hippler, M. , Setif, P. , Jacquot, J.P. & Rochaix, J.D. (1998) The PSAC subunit of photosystem I provides an essential lysine residue for fast electron transfer to ferredoxin. EMBO Journal, 17, 849–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederickson Matika, D.E. & Loake, G.J. (2014) Redox regulation in plant immune function. Antioxidants and Redox Signaling, 21, 1373–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganusova, E.E. , Reagan, B.C. , Fernandez, J.C. , Azim, M.F. , Sankoh, A.F. , Freeman, K.M. et al. (2020) Chloroplast‐to‐nucleus retrograde signalling controls intercellular trafficking via plasmodesmata formation. Philosophical Transactions of the Royal Society B: Biological Sciences, 375, 20190408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, D. , Nelson, R.T. , Cannon, S.B. & Shoemaker, R.C. (2010) Soybase, the USDA‐ARS soybean genetics and genomics database. Nucleic Acids Research, 38, D843–D846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn, A. , Vonck, J. , Mills, D.J. , Meier, T. & Kuhlbrandt, W. (2018) Structure, mechanism, and regulation of the chloroplast ATP synthase. Science, 360, 620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajimorad, M.R. , Domier, L.L. , Tolin, S.A. , Whitham, S.A. & Saghai Maroof, M.A. (2018) Soybean mosaic virus: a successful potyvirus with a wide distribution but restricted natural host range. Molecular Plant Pathology, 19, 1563–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisabori, T. , Sunamura, E. , Kim, Y. & Konno, H. (2013) The chloroplast ATP synthase features the characteristic redox regulation machinery. Antioxidants and Redox Signaling, 19, 1846–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X.Z. , Zhang, X.Y. & Yang, S.H. (2009) A novel chloroplast‐localized protein EMB1303 is required for chloroplast development in Arabidopsis . Cell Research, 19, 1225. [DOI] [PubMed] [Google Scholar]
- Huang, Y.P. , Chen, I.H. & Tsai, C.H. (2017) Host factors in the infection cycle of Bamboo mosaic virus. Frontiers in Microbiology, 8, 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi, K. , Saruta, M. , Shimizu, T. , Shu, M. , Anai, T. , Komatsu, K. et al. (2019) Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nature Communications, 10, 4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, Z. , Zhang, K. , Li, Z. , Li, Z. , Yang, M. , Jin, X. et al. (2020) The Barley stripe mosaic virus gammab protein promotes viral cell‐to‐cell movement by enhancing ATPase‐mediated assembly of ribonucleoprotein movement complexes. PLoS Pathogens, 16, e1008709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez, I. , Lopez, L. , Alamillo, J.M. , Valli, A. & Garcia, J.A. (2006) Identification of a plum pox virus CI‐interacting protein from chloroplast that has a negative effect in virus infection. Molecular Plant‐Microbe Interactions, 19, 350–358. [DOI] [PubMed] [Google Scholar]
- Kubota‐Kawai, H. , Mutoh, R. , Shinmura, K. , Sétif, P. , Nowaczyk, M.M. , Rögner, M. et al. (2018) X‐ray structure of an asymmetrical trimeric ferredoxin‐photosystem I complex. Nature Plants, 4, 218–224. [DOI] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. & Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto, K. , Tikkanen, M. , Hiriart, J.B. , Paakkarinen, V. & Aro, E.M. (2003) Depletion of the photosystem II core complex in mature tobacco leaves infected by the Flavum strain of Tobacco mosaic virus . Molecular Plant‐Microbe Interactions, 16, 1135–1144. [DOI] [PubMed] [Google Scholar]
- Liu, J.‐Z. , Braun, E. , Qiu, W.‐L. , Shi, Y.‐F. , Marcelino‐Guimarães, F.C. , Navarre, D. et al. (2014) Positive and negative roles for soybean MPK6 in regulating defense responses. Molecular Plant‐Microbe Interactions, 27, 824–834. [DOI] [PubMed] [Google Scholar]
- Liu, J.Z. , Fang, Y. & Pang, H. (2016) The current status of the soybean–soybean mosaic virus (SMV) pathosystem. Frontiers in Microbiology, 7, 1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Liu, Y. , Spetz, C. , Li, L. & Wang, X. (2020) Comparative transcriptome analysis in Triticum aestivum infecting wheat dwarf virus reveals the effects of viral infection on phytohormone and photosynthesis metabolism pathways. Phytopathology Research, 2, 3. [Google Scholar]
- Moejes, F.W. , Matuszyńska, A. , Adhikari, K. , Bassi, R. , Cariti, F. , Cogne, G. et al. (2017) A systems‐wide understanding of photosynthetic acclimation in algae and higher plants. Journal of Experimental Botany, 68, 2667–2681. [DOI] [PubMed] [Google Scholar]
- Nevo, R. , Charuvi, D. , Tsabari, O. & Reich, Z. (2012) Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. The Plant Journal, 70, 157–176. [DOI] [PubMed] [Google Scholar]
- Rantala, M. , Rantala, S. & Aro, E.M. (2020) Composition, phosphorylation and dynamic organization of photosynthetic protein complexes in plant thylakoid membrane. Photochemical and Photobiological Sciences, 19, 604–619. [DOI] [PubMed] [Google Scholar]
- Scholthof, K.‐B. , Adkins, S. , Czosnek, H. , Palukaitis, P. , Jacquot, E. , Hohn, T. et al. (2011) Top 10 plant viruses in molecular plant pathology. Molecular Plant Pathology, 12, 938–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.K. , Kwon, S.J. , Cho, W.K. , Choi, H.S. & Kim, K.H. (2014) Type 2c protein phosphatase is a key regulator of antiviral extreme resistance limiting virus spread. Scientific Reports, 4, 5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, J.K. , Lee, H.G. , Choi, H.S. , Lee, S.H. & Kim, K.H. (2009) Infectious in vivo transcripts from a full‐length clone of soybean mosaic virus strain G5H. Plant Pathology Journal, 25, 54–61. [Google Scholar]
- Seo, J.K. , Lee, S.H. & Kim, K.H. (2009) Strain‐specific cylindrical inclusion protein of soybean mosaic virus elicits extreme resistance and a lethal systemic hypersensitive response in two resistant soybean cultivars. Molecular Plant‐Microbe Interactions, 22, 1151–1159. [DOI] [PubMed] [Google Scholar]
- Tu, Y. , Jin, Y. , Ma, D. , Li, H. , Zhang, Z. , Dong, J. et al. (2015) Interaction between PVY Hc‐Pro and the NTCF1β‐subunit reduces the amount of chloroplast ATP synthase in virus‐infected tobacco. Scientific Reports, 5, 15605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widyasari, K. , Alazem, M. & Kim, K.H. (2020) Soybean resistance to soybean mosaic virus. Plants, 9, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Yang, R. , Yang, Z. , Yao, S. , Zhao, S. , Wang, Y. et al. (2017) ROS accumulation and antiviral defence control by microRNA28 in rice. Nature Plants, 3, 16203. [DOI] [PubMed] [Google Scholar]
- Yang, X. , Lu, Y. , Wang, F. , Chen, Y. , Tian, Y. , Jiang, L. et al. (2020) Involvement of the chloroplast gene ferredoxin 1 in multiple responses of Nicotiana benthamiana to potato virus X infection. Journal of Experimental Botany, 71, 2142–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, A. , Xie, Y. , Pan, X. , Zhang, H. , Cao, P. , Su, X. et al. (2020) Photosynthetic phosphoribulokinase structures: enzymatic mechanisms and the redox regulation of the Calvin‐Benson‐Bassham cycle. The Plant Cell, 32, 1556–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Bradshaw, J.D. , Whitham, S.A. & Hill, J.H. (2010) The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant Physiology, 153, 52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Grosic, S. , Whitham, S.A. & Hill, J.H. (2012) The requirement of multiple defense genes in soybean Rsv1‐mediated extreme resistance to soybean mosaic virus. Molecular Plant‐Microbe Interactions, 25, 1307–1313. [DOI] [PubMed] [Google Scholar]
- Zhang, K. , Zhang, Y. , Yang, M. , Liu, S. , Li, Z. , Wang, X. et al. (2017) The Barley stripe mosaic virus gammab protein promotes chloroplast‐targeted replication by enhancing unwinding of RNA duplexes. PLoS Pathogens, 13, e1006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Xu, J. , Chen, B. , Cui, W. , Zhou, Z. , Song, X. et al. (2019) Characterization of proteins involved in chloroplast targeting disturbed by rice stripe virus by novel protoplast(‐)chloroplast proteomics. International Journal of Molecular Sciences, 20, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J. , Zhang, X. , Hong, Y. & Liu, Y. (2016) Chloroplast in plant–virus interaction. Frontiers in Microbiology, 7, 1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1. Sequence alignment of ATPsyn‐α (a) and PSaC (b) proteins from five soybean cultivars: William 82 (W82), L29, Lee74, Somyongkong (SMK), and V94
FIGURE S2. Effect of over‐expressing ATPsyn‐α and PSaC on resistance against SMV‐G5H in the susceptible cultivar Lee74. (a) Schematic drawing of G5H::eGFP construct with the insertion site for ATPsyn‐α and PSaC downstream of the eGFP gene. (b) GFP visual levels in the systemically infected leaves of Lee74 plants. The first unifoliate leaves of 12‐day‐old seedlings were infected with G5H::eGFP, G5H::eGFP::ATPsyn‐α, or G5H::eGFP::PSaC. Fourteen days later, the systemically infected leaves from three plants (1, 2, and 3) were photographed under UV light. (c) Western blot for GFP in the systemically infected leaves. Ponceau S staining was used on the loading control
FIGURE S3. Stability of ATPsyn‐α and PSaC inserts in pSMV::G7H::eGFP vector. (a) Detection of ATPsyn‐α and PSaC inserts in the pSMV‐G7H::eGFP by RT‐PCR. RNA was extracted from the systemically infected Lee74 leaves with pSMV‐G7H::eGFP::ATPsny‐α or pSMV:‐G7H::eGFP::PSaC. The primers anneal to the flanking regions of the insert site. Each lane represents a reverse transcription PCR from one biological replicate. The PCR product of each chimera was purified, sequenced, and then aligned with the sequences of ATPsyn‐α (b) and PSaC (c) proteins identified in Figure 2 from L29 soybean plants. (d) Western blots for the HA‐tagged ATPsyn‐α‐HA and PSaC‐HA expressed in the pSMV‐G7H::eGFP::HA vector from the systemically infected leaves of Lee74 plants
FIGURE S4. Relative expression levels of endogenous ATPsyn‐α and PSaC in Lee74 plants infected with pSMV‐G7H::eGFP chimeras. (a) Expression levels of endogenous ATPsyn‐α in leaves inoculated with pSMV‐G7H::eGFP or pSMV‐G7H::eGFP::ATPsyn‐α. (b) Expression levels of endogenous PSaC in leaves inoculated with pSMV‐G7H::eGFP or pSMV‐G7H::eGFP::PSaC. Primers were designed to target the 3′ untranslated regions (UTRs) of either transcripts so their expression can be distinguished from that expressed within the pSMV‐G7H::eGFP infectious clone. Actin11 was used as internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in the legend of Figure 1
FIGURE S5. Multiple sequence alignment of ATPsyn‐α and its paralogs from chloroplast (Glyma.16G115300.1) and mitochondria (Glyma.05G092300.1). The sequence highlighted in red is the fragment used to initiate the virus‐induced gene silencing of ATPsyn‐α in Lee74 plants
FIGURE S6. Expression levels of ATPsyn‐α paralogs from (a) chloroplast “Glyma.16G115300.1” and (b) mitochondria “Glyma.05G092300.1” in Lee74 plants silenced with either gene. Actin11 was used as internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in the legend of Figure 1
FIGURE S7. Localization of the chloroplast marker protein AtEMB1303 in Nicotiana benthamiana leaves. Chloroplast autofluorescence is pseudocoloured in red. DAPI was used to stain the nuclei. Bars measure 50 µm
FIGURE S8. Expression levels of RNA silencing genes in Nicotiana benthamiana in response to infection with chimeras of pSMV‐G7H::eGFP. Relative expression levels of Dicer‐like (DCL) 2a (a) DCL4a (b), RNA‐dependent RNA polymerase (RDR) 2a (c), and RDR6a (d) in N. benthamiana plants infected with pSMV‐G7H::eGFP, pSMV‐G7H::eGFP::ATPsyn‐α, or pSMV‐G7H::eGFP::PSaC. Actin was used as internal control. Values are means + SD of three biological replicates. Statistical analysis was carried out as described in Figure 1; * and ** indicate a significant difference at p < 0.05 and p < 0.01, respectively. An additional t test was carried out to compare expression in the pSMV‐G7H::eGFP::ATPsyn‐α and pSMV‐G7H::eGFP::PSaC treatment with that in the pSMV‐G7H::eGFP treatment
TABLE S1. Primers used in this study
Data Availability Statement
Data are available on request from the authors.


