Abstract
Reef-building corals maintain an intracellular photosymbiotic association with dinoflagellate algae. As the algae are hosted inside the symbiosome, all metabolic exchanges must take place across the symbiosome membrane. Using functional studies in Xenopus oocytes, immunolocalization, and confocal Airyscan microscopy, we established that Acropora yongei Rh (ayRhp1) facilitates transmembrane NH3 and CO2 diffusion and that it is present in the symbiosome membrane. Furthermore, ayRhp1 abundance in the symbiosome membrane was highest around midday and lowest around midnight. We conclude that ayRhp1 mediates a symbiosomal NH4+-trapping mechanism that promotes nitrogen delivery to algae during the day—necessary to sustain photosynthesis—and restricts nitrogen delivery at night—to keep algae under nitrogen limitation. The role of ayRhp1-facilitated CO2 diffusion is less clear, but it may have implications for metabolic dysregulation between symbiotic partners and bleaching. This previously unknown mechanism expands our understanding of symbioses at the immediate animal-microbe interface, the symbiosome.
Corals have a molecular mechanism to regulate ammonium and carbon dioxide delivery to their symbiotic algae.
INTRODUCTION
Photosymbiotic associations between invertebrates and microalgae are widespread in aquatic environments. Perhaps the most well known of these partnerships is that of reef-building corals (phylum: Cnidaria) and dinoflagellate symbiotic algae (family: Symbiodiniaceae), which is key to the evolutionary success of coral reef ecosystems (1). In an otherwise oligotrophic environment, the cnidarian host satisfies the majority of its energetic needs using photosynthates derived from its symbiotic algae (2). The host cells are believed to exercise considerable control over the metabolism of their symbionts, which favors both the production and release of algal photosynthates. This control is possible because of an architectural arrangement whereby coral gastrodermal cells host the algal symbionts intracellularly within an arrested phagosome known as the symbiosome [reviewed in (3)]. Because the symbiosome isolates the alga from the cytosol of the host cell, the symbiosome membrane necessarily mediates all metabolic exchanges between the symbiotic partners. In addition, the symbiosome membrane may serve as an interface for the coral to manipulate the alga’s microenvironment. For example, the coral symbiosome is markedly acidic (pH ~4) because of active H+ pumping by V-type H+-ATPases (VHAs) located in the symbiosome membrane (4). The acidic nature of the symbiosome drives CO2 accumulation as part of a carbon concentrating mechanism (CCM) that helps overcome the low affinity of algal Rubisco for CO2, thereby promoting algal photosynthesis (4). This H+ gradient has been proposed to additionally energize the movement of other essential nutrients and metabolites into or out of the symbiosome including nitrogen, phosphorus, and sugars (3, 4). However, no additional molecular players or regulatory mechanisms have been definitely identified to date.
The vast majority of the symbiotic algae’s nitrogen demand is supplied by protein catabolism by their animal host, which produces waste as ammonia gas (NH3) and ammonium ion (NH4+), which exist in a pH-dependent equilibrium [collectively referred to as “total ammonia” (Tamm)] (2, 5). Rather than excreting its nitrogenous waste into the environment like most other aquatic animals (6), the coral symbiosis recycles a substantial portion of Tamm via the glutamine synthase/glutamate dehydrogenase/glutamine oxoglutarate aminotransferase pathways (GS/GDH/GOGAT) (5, 7, 8). In addition, corals are able to take up NH4+ from seawater and transport it to their algal symbionts (9), and isolated algal symbionts take up and use NH4+ (10). Moreover, coral host cells are known to regulate Tamm delivery to their symbionts, and as a result, the algae accumulate significantly more nitrogen in the light than in the dark (9, 11). The diel regulation of Tamm delivery by corals allows for host control over the carbon and nitrogen metabolisms of symbionts (12) and, by extension, the growth rate and biomass of the symbiont population to prevent symbiont overgrowth that would disrupt the symbiosis (13). Transcriptomic analyses on whole coral colonies have identified candidate transporters proposed to mediate Tamm delivery to symbionts (14), but a lack of localization studies precludes a definite assessment of their involvement in symbiosis. Overall, the mechanisms that mediate and regulate nitrogen transport to symbionts across the symbiosome membrane remain unknown.
NH3 and NH4+ exist in pH-dependent equilibrium with pKa ~9.25, and thus >96% of Tamm is found as NH4+ both in seawater (pH ~8) and in coral host cells (pH ~7.4) (15). However, the much lower pH in the symbiosome space has three critical and interlinked implications: first, a virtually nil NH3 partial pressure (pNH3) in the symbiosome space that should drive NH3 gas diffusion from the host cytoplasm; second, the immediate “trapping” of NH3 as NH4+ in the symbiosome space, which can be taken up by the alga, thus maintaining the inwardly directed NH3 diffusion gradient; and lastly, an unfavorable electrochemical gradient for NH4+ transport into the symbiosome.
However, despite being a gas, NH3 has limited permeability through lipidic membranes because of its strong dipole moment that makes it a polar molecule [reviewed in (16)]. In some plant-bacteria symbioses, NH3 transport across the symbiosome membrane is facilitated by nodulin-intrinsic proteins (17, 18); however, this protein family is exclusive to plants. In addition, NH3 diffusion across biological membranes can be significantly enhanced by Rhesus (Rh) channels, a family of evolutionary conserved proteins present in eubacterial, invertebrate, and vertebrate lineages (19–21). On the basis of the observed up-regulation of an Rh-like mRNA transcript upon establishment of symbiosis in anemones (22–24), Rh channels have been suggested to play important roles in cnidarian-algae symbioses. However, the Rh-like mRNA was expressed in many soft coral cell subtypes (25), and therefore, the coded protein probably plays multiple physiological roles. In addition, Rh channels are typically present in the cell outer plasma membrane [reviewed in (16, 26)], and few studies have localized Rh-like proteins to intracellular compartments or organelles (27, 28). Last, the various Rh protein isoforms have different substrate specificity: some may transport both NH3 and NH4+ (16), some act as dual NH3 and CO2 gas channels (19, 29, 30), and others do not facilitate Tamm/CO2 transport across membranes at all and have structural functions instead (31, 32). However, detailed functional studies about transport properties by “primitive” Rh proteins from invertebrate animals (termed “Rhp”) are very scarce. As a result, assessing the physiological role of the coral Rh-like coded protein and its potential involvement in delivering Tamm to algal endosymbionts requires elucidating its actual function as well as its cellular and subcellular localizations. Furthermore, if coral Rh facilitated CO2 diffusion and was present on the symbiosome membrane, it would provide a pathway for CO2 backflow from the symbiosome into the coral gastrodermal cells and affect interactions between nitrogen transport and the CCM.
Given that NH3 diffusion through biological membranes is limited, we hypothesized that corals use Rh-like proteins to deliver NH3 to their algae across the symbiosome membrane, which would subsequently get trapped as NH4+ in the acidic symbiosome. To investigate this possibility, we cloned an Rh-like gene from the coral Acropora yongei (ayRhp1) and determined its phylogenetic relationship to other Tamm-transporting proteins. Then, we heterologously expressed ayRhp1 protein in Xenopus oocytes and measured Tamm transport under a range of pHs to determine whether it transports NH3, NH4+, or both. In additional oocyte experiments, we determined whether ayRhp1 facilitates CO2 diffusion. Using custom-made antibodies and immunocytochemistry, we established the localization of ayRhp1 protein in the various cell subtypes throughout the coral colony, and, using confocal Airyscan microscopy, we investigated whether ayRhp1 was specifically located in the symbiosome membrane. Last, we quantified the subcellular localization of ayRhp1 within algae-containing coral gastrodermal cells throughout a diel cycle to explore a potential mechanism whereby coral host cells could regulate Tamm delivery to their algal symbionts.
RESULTS AND DISCUSSION
Rhp1 genes are widespread in corals
The cloned ayRhp1 cDNA open reading frame contains 1440 base pairs encoding a protein with a predicted molecular weight of 51.8 kDa. BLAST searches in genomic and transcriptomic databases revealed predicted ayRhp1 homologs in multiple coral species from both the robust and complex clades, which diverged from each other 300 million to 400 million years ago (33). These coral Rh proteins clustered together with Rhp1 genes from invertebrate animals (fig. S1).
The protein features of ayRhp1 are similar to those of well-studied Rh channels from mammals (fig. S2). It has 12 transmembrane helices and an N-linked glycosylation site (N61), which differentiate all animal Rh50 channels capable of Tamm transport (Rhag-cg, Rhp1-2) (16) from the Rh30 proteins involved in structural functions (27). Crystallography and simulation studies have identified several key amino acid residues that are required for NH3 transport across mammalian RhCG (20, 21): a phenylalanine gate (F130 and F235) and a cytosolic shunt (L193, T325, L328, I334, N341, and N342), which recruit NH4+ at the external and internal vestibules, respectively, twin histidines that deprotonate NH4+ to NH3 (H185 and H344), two highly conserved aspartic acid residues that help shuttle the H+ back to the original compartment (D177 and D336), and a hydrophobic transmembrane channel that selectively conducts NH3 but not NH4+ (32, 34). An alignment of ayRhp1 with RhCG reveals that the phenylalanine gate (F147 and F251), the twin histidines (H202 and H364), and analogous aspartate residues (D195 and D356) are all conserved in ayRhp1, while the cytosolic shunt and hydrophobic channel-lining residues are highly conserved (~83 and ~70%, respectively). In addition, ayRhp1 contains the nine residues that form the putative cytoplasmic CO2 binding pocket of mammalian RhCG [which facilitates CO2 transport (19)]: L91, F94, D234, A237, M238, M299, V300, Q303, and N304. Six of these residues are also conserved in the Rh protein from the bacterium Nitrosomonas europaea, where the CO2 binding pocket was definitely identified using x-ray crystallography (35). In summary, the overall high conservation of these key structures suggests that ayRhp1 can facilitate both NH3 and CO2 transport; this was experimentally tested through functional studies.
ayRhp1 facilitates NH3 and CO2 diffusion
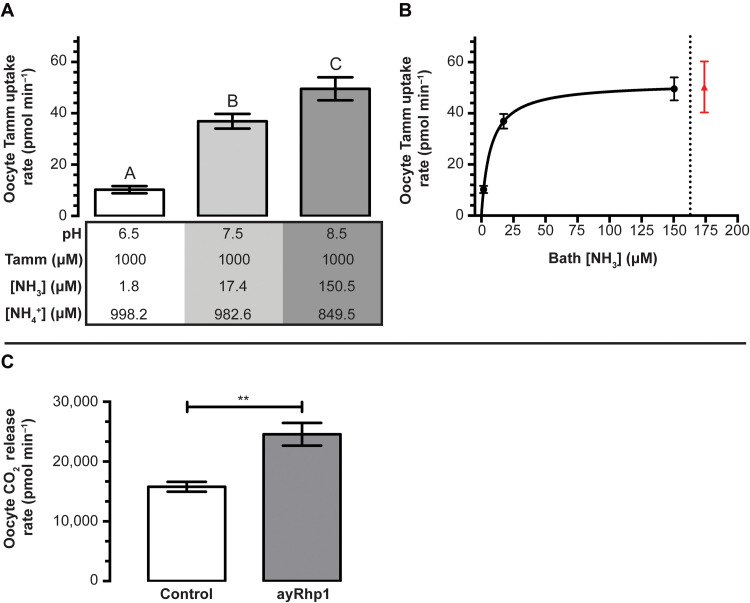
ayRhp1 was functionally characterized by measuring Tamm uptake rates in Xenopus oocytes injected with ayRhp1 cRNA. To avoid potential artifacts resulting from using radiolabeled [14C]-methylammonium as a Tamm analog (16), we used a hypochlorite-salicylate-nitroprusside–based colorimetric assay to directly measure Tamm accumulation in oocytes and estimate Tamm uptake rate. The bath solutions contained 1 mM Tamm at pH 6.5, 7.5, or 8.5, resulting in 10-fold pKa-dependent [NH3] increases for every pH unit (1.8, 17.4, and 150.5 μM, respectively). ayRhp1-expressing oocytes had consistently higher Tamm uptake rates than those of controls in all conditions tested (P < 0.001; fig. S3). In addition, Tamm uptake rate in ayRhp1 oocytes significantly increased from 10.2 ± 1.4 pmol Tamm min−1 at pH 6.5, to 36.9 ± 2.8 pmol Tamm min−1 at pH 7.5, and to 49.6 pmol Tamm min−1 at pH 8.5 (Fig. 1A), an apparent Jmax and Km of 51.93 ± 1.45 pmol Tamm min−1 and 7.14 ± 0.91 μmol Tamm liter−1, respectively (Fig. 1B). These results indicate that ayRhp1 transports NH3 following the partial pressure difference. In addition, Tamm uptake rate of oocytes incubated in a solution with 10 mM Tamm at pH 7.5 was 50.3 ± 10.0 pmol Tamm min−1 (i.e., indistinguishable from the rate in the 1 mM Tamm pH 8.5 solution) (Fig. 1B). These two solutions have similar [NH3] (174.0 versus 150.5 μM), but the former has >10-fold greater [NH4+] than the latter (9826.0 μM versus 849.5 μM). Together, these results established that ayRhp1 can facilitate NH3 diffusion following pH-dependent partial pressure gradients and that Tamm transport is not directly dependent on the [NH4+] difference.
Fig. 1. Functional characterization of total ammonia (Tamm) and CO2 transport by Acropora yongei Rhesus protein (ayRhp1).
(A) Effect of [NH3] on Tamm uptake rate in Xenopus oocytes expressing ayRhp1. Control Tamm uptake rates have been subtracted. Data show means ± SEM of six to eight oocytes; the letters denote significant differences [one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test; pH 6.5 versus pH 7.5, P < 0.0001; pH 7.5 versus pH 8.5, P = 0.0222; pH 6.5 versus pH 8.5, P < 0.0001]. (B) Michaelis-Menten Tamm uptake kinetics calculated from the data shown in (A) (black dots to the left of the dotted line). Apparent Jmax = 51.93 ± 1.45 pmol Tamm min−1 and Km = 7.14 ± 0.91 μmol Tamm liter−1. The red triangle indicates Tamm uptake rate obtained in a solution with 10 mM Tamm at pH 7.5 (175 μM NH3 and 9.825 mM NH4+) (i.e., similar [NH3] to the previous data point, but ~10-fold higher [NH4+]). (C) Functional characterization of CO2 transport by ayRhp1. Xenopus oocytes expressing ayRhp1 (ayRhp1) display a higher rate of CO2 release than control oocytes after equal CO2 preloading. Data show means ± SEM of n = 8, 25 oocytes per n; ** denotes significant differences (Welch’s t test; P = 0.0019).
Next, we preloaded control and ayRhp1 cRNA-injected oocytes with 5% CO2 and measured CO2 release into normocapnic media using a custom-built CO2 analyzer (36). These experiments revealed that ayRhp1-expressing oocytes released CO2 at a rate ~50% faster than control oocytes (P = 0.0019; Fig. 1C). As wild-type Xenopus oocytes lack notable HCO3− efflux (37), this demonstrates that ayRhp1 can facilitate the diffusion of CO2 in addition to NH3.
ayRhp1 protein is present in multiple coral cell types
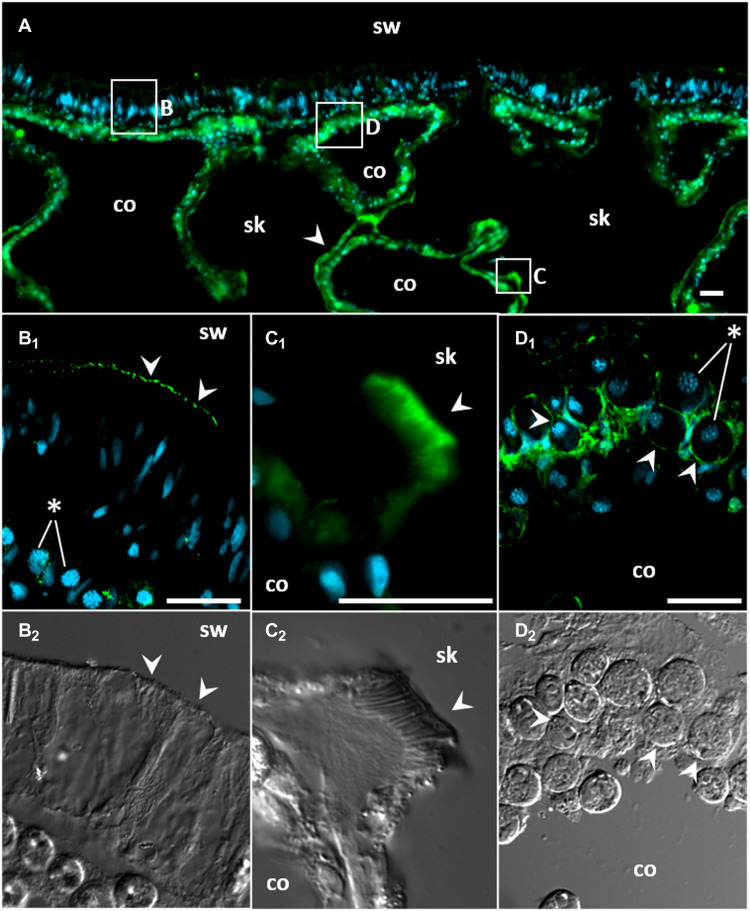
Immunofluorescence microscopy using custom-made–specific antibodies revealed high ayRhp1 protein expression throughout A. yongei coral tissue sections (Fig. 2A). In the epidermis, ayRhp1 was present in the apical membrane of columnar cells along the seawater-coral interface (Figs. 2B1 and 3A). Although corals recycle most of their nitrogen waste through their algal symbionts, they also excrete some Tamm to the environment (2, 7). Thus, we hypothesize that ayRhp1 in epidermal cells aids in nitrogenous waste excretion as previously described in gills and skin from fish and aquatic invertebrates (38, 39). Moreover, Tamm excretion may be facilitated by stirring of the boundary layer by ciliary beating, akin to mussels and polychaetes (38, 40).
Fig. 2. Immunolocalization of Acropora yongei Rhesus protein (ayRhp1).
(A) Overview of A. yongei tissues; the boxes indicate regions of interest shown at higher magnification below, and the white arrowhead indicates ayRhp1-labeled calcifying cells. (B1) Apical membrane of columnar cells in the oral epidermis. (C1) Desmocyte with intense signal in its apical region. (D1) Alga-containing gastrodermal cells. (B2, C2, and D2) Corresponding bright-field differential interference contrast images; the white arrowheads mark corresponding locations in (B), (C), and (D). Coral and algal nuclei are shown in blue, and ayRhp1 immunofluorescence is shown in green. Several algal nuclei are marked with asterisks in (B1) and (D1) for clarity. This coral was sampled at midday. sw, seawater; co, coelenteron; sk, skeleton. Scale bars, 20 μm.
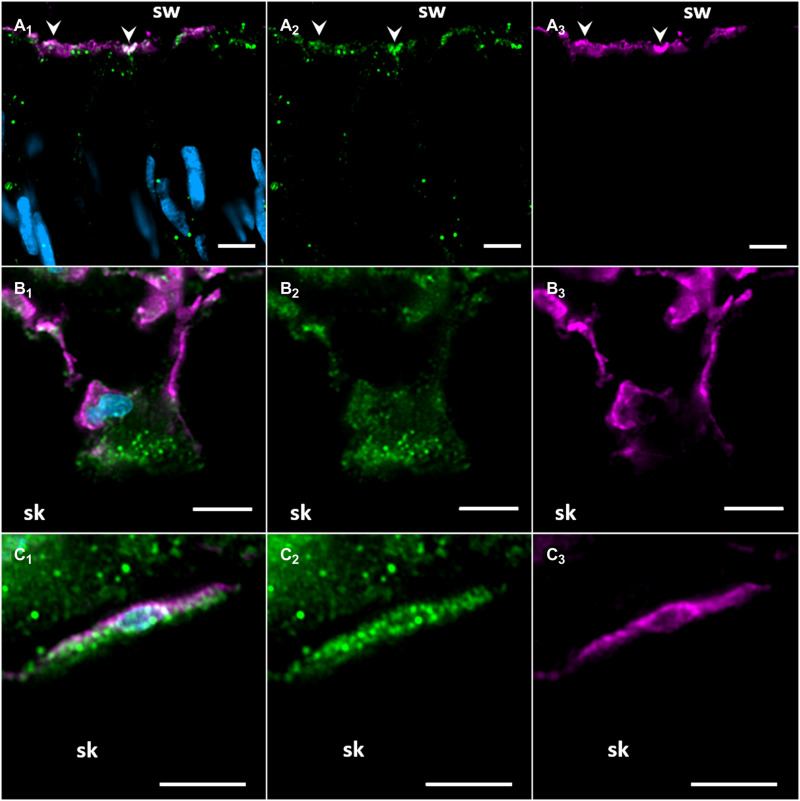
Fig. 3. Confocal Airyscan immunolocalization of Acropora yongei Rhesus protein (ayRhp1) in the oral epidermis, desmocytes, and calcifying cells.
(A1–3) ayRhp1 on the apical membrane of columnar cells in the oral epidermis. (B1–3) Desmocyte with intense ayRhp1 signal in its apical region. (C1–3) Calcifying cells displaying ayRhp1 signal on membranes and in the cytosol. Corresponding areas between panels are marked with arrowheads. [(A1) to (C1)], [(A2) to (C2)], and [(A3) to (C3)] show ayRhp1, Na+/K+-ATPase (NKA), and 4′,6-diamidino-2-phenylindole (DAPI) signals, ayRhp1 signal alone, or NKA signal alone, respectively. Nuclei (DAPI) are shown in blue, ayRhp1 in green, and the NKA in purple. Scale bars, 5 μm.
In the calicodermis, ayRhp1 was expressed in both calcifying cells that deliver dissolved inorganic carbon (DIC), Ca2+, and matrix proteins for skeletal formation and in desmocytes that anchor living coral tissue to the skeleton. The ayRhp1 signal in desmocytes was very intense, especially at the apical membrane adjacent to the skeleton (Figs. 2C1 and 3B and fig. S4A1). Previous studies have provided morphological descriptions of coral desmocytes (41, 42); however, to our knowledge, this is the first description of any protein specifically expressed in this cell type. As an NH3 channel, ayRhp1 may contribute to coral calcification by enhancing NH3 diffusion to buffer the pH of the extracellular calcifying medium (ECM) and maintain conditions favorable for calcification. Metabolic NH3 has been proposed to promote biological calcification of avian egg shells (43), land snail shells (44), and coral skeletons (45) by buffering H+ produced during CaCO3 precipitation as NH4+. As a CO2 channel, ayRhp1 could help deliver DIC to the ECM following the outwardly directed pCO2 gradient favoring CO2 diffusion from calicodermis cells into the ECM, which is an important source of DIC for calcification in multiple coral species (46, 47). In the gastroderm, ayRhp1 was highly expressed in alga-hosting coral cells surrounding the symbiotic algae (Fig. 2D1), in a pattern that resembled that of VHA in the symbiosome membrane (4). This was explored in further detail.
ayRhp1 is present in the symbiosome membrane
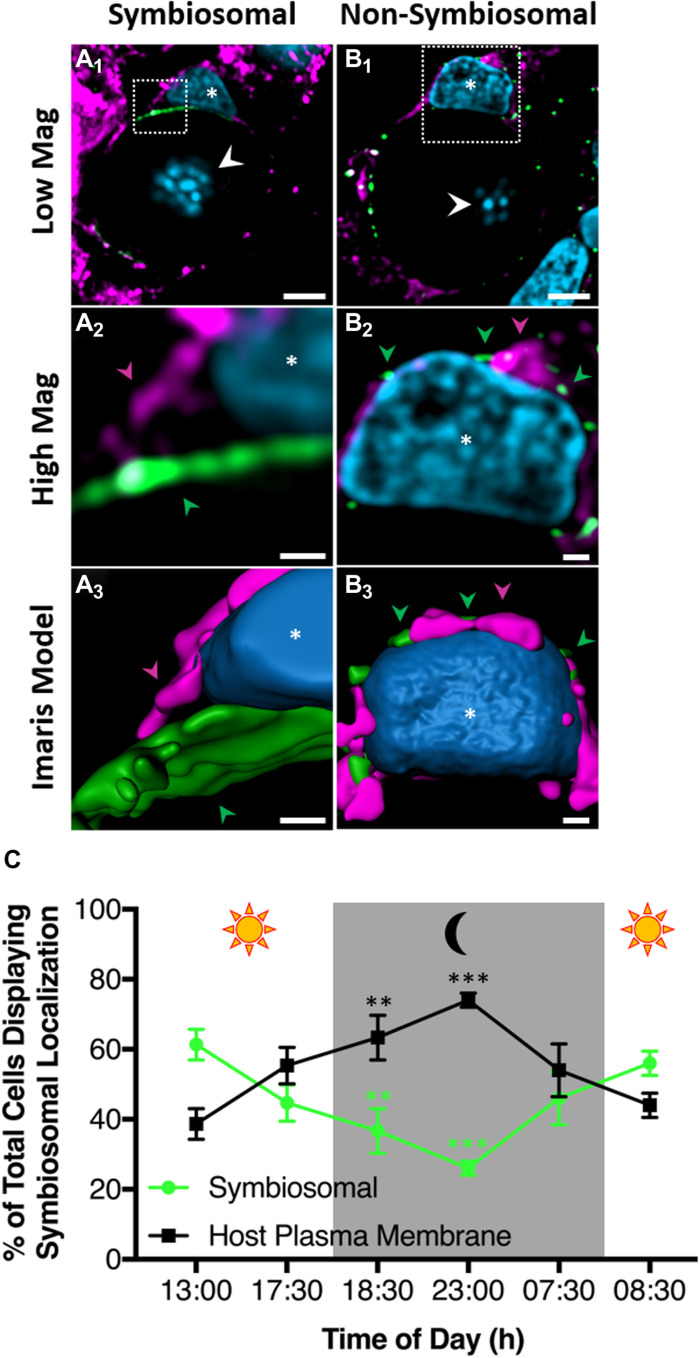
Confocal Airyscan microscopy and coimmunostaining of ayRhp1 and Na+/K+-ATPase (NKA) allowed us to definitively establish ayRhp1’s subcellular localization within the tightly packaged host cells in coral tissues. Consistent with its universal presence in the plasma membrane (48), NKA outlined the perimeter of all alga-containing host cells (Fig. 4, A and B). The ayRhp1 signal was also present around the algae; but in most cells, it was internal to that of NKA and also present in the thin region between the host cell nucleus and the alga (Fig. 4 A1). Since these cells are tightly packed, this region is occupied by the symbiosome membrane (4, 49). This was most readily evident in the region adjacent to the coral nucleus where a region of cytosol separates the external NKA and internal ayRhp1 signals, further indicating their respective presence in the plasma and symbiosome membranes (Figs. 4A2 and 3). The symbiosomal localization of ayRhp1 was further confirmed in host cells containing two algae (fig. S5C), which are rather scarce but contain a larger cytoplasmic region that allows for better visualization of subcellular compartments. In addition, a minority of cells lacked ayRhp1 in the region between the host nucleus and the algae (Fig. 4B), which instead colocalized with or appeared slightly internal to NKA around the nuclear periphery, indicating ayRhp1’s presence in the host plasma membrane, cytosolic vesicles, or both. We attempted to quantify the two ayRhp1 subcellular localization patterns using tissue sections; however, the high density of tightly packed gastrodermal cells coupled with their intense NKA signal confounded imaging and prevented an unbiased approach. To circumvent these limitations, we immunostained isolated coral cells, an approach we previously used to confirm the presence of VHA in the symbiosome membrane (4). Similar to tissue sections, a majority of isolated cells displayed ayRhp1 signal in the thin region between the host cell nucleus and the alga, indicative of ayRhp1 symbiosomal localization (fig. S5, B, C, and E to I). We also observed a minority of cells with ayRhp1 signal around the host cell nucleus (fig. S5, D and J to N), indicative of ayRhp1’s presence in the cytoplasm or plasma membrane of the host cell. Free algal cells released during the isolation procedure, identified by the lack of an adjacent host nucleus, did not have ayRhp1 signal (fig. S5A).
Fig. 4. Confocal Airyscan immunolocalization of Acropora yongei Rhesus protein (ayRhp1) in alga-containing coral cells.
(A1) Cells displaying ayRhp1 in the symbiosome membrane of tissue sections. (B1) Cells displaying nonsymbiosomal ayRhp1 of tissue sections. (A2 and B2) Higher magnification of the region denoted by the white boxes in (A1) and (B1). (A3 and B3) Three-dimensional renderings of (A2) and (B2). Corresponding areas between (A2) and (B2) and (A3) and (B3) are marked. Nuclei are shown in blue, ayRhp1 in green, and the NKA in purple. Notice the separation (A1–3) or colocalization (B1–3) of ayRhp1 and NKA corresponding to symbiosomal or nonsymbiosomal ayRhp1 localizations, respectively. Host nuclei are denoted with an asterisk, and algal nuclei with arrowheads. Scale bars, 5 μm (A1 and B1), 0.5 μm (A2 and B2), and 0.5 μm (A3 and B3). (C) Percentage of total alga-containing A. yongei host cells with symbiosomal ayRhp1 over a diel cycle. Data show means ± SEM. n = 3 per time point, 50 cells per n, 900 cells total. The asterisks indicate significant differences with the 1300-hour time point (two-way repeated-measures ANOVA followed by Dunnett’s posttest; **P < 0.01; ***P < 0.0001).
Diel trafficking of ayRhp1
On the basis of established patterns of nitrogen delivery to coral algal symbionts (9, 11), we hypothesized that the ayRhp1 subcellular localization would change in a diel fashion. We therefore quantified ayRhp1 subcellular localization patterns using epifluorescence microscopy on isolated alga-containing coral cells over a diel cycle. This allowed us to achieve sufficient replication (50 cells from 18 coral branches, 3 coral branches at each of six time points, for a total of 900 cells observed in blind fashion). Once an algal-containing coral host cell was identified, the observer rapidly and continuously shifted the focal plane and alternated between fluorescence and bright-field DIC while looking through the microscope eyepiece. This technique allowed the observer to determine whether the ayRhp1 signal was present in between the host cell nucleus and the alga (classified as “symbiosomal localization”; Fig. 4A and fig. S5, B, C, and E to I), or around the periphery of the host cell’s nucleus (“nonsymbiosomal localization”; Fig. 4B and fig. S5, D and J to N). The percentage of cells displaying ayRhp1 symbiosomal localization was significantly higher during the day, with a maximum of 61.3 ± 4.4% cells displaying this pattern at 1300 hours in contrast to only 26.0 ± 2.0% of cells at 2300 hours (P < 0.001) (Fig. 4C). These results indicate that ayRhp1 is preferentially present in the symbiosome membrane during the day. To our knowledge, this is the first report of diel changes in proteomic makeup of the cnidarian symbiosome membrane and furthers the notion that this interface that separates symbiotic partners can be dynamically modified by the host cell to control the physiology of the alga.
A putative host-controlled nitrogen concentrating mechanism
The pKa for Tamm combined with the pH difference between the host cell’s cytosol and the symbiosome dictates >2000 higher pNH3 in the former. Although this establishes a steep partial pressure gradient favoring NH3 diffusion into the symbiosome, diffusion across lipidic membranes is generally limited [reviewed in (16)]. The presence of ayRhp1 in the symbiosome membrane is poised to overcome this limitation, thus enhancing NH3 delivery to the algal symbionts in an analogous manner to nodulin-intrinsic proteins in plant-Rhizobium symbioses (17, 18). Once inside the highly acidic symbiosome, NH3 will be immediately converted into NH4+, which cannot move across the plasma membrane, or through ayRhp1. This mechanism is known as “NH4+ acid trapping” and is well documented in diverse excretory epithelia from humans (20), teleost fishes (39), cephalopod and bivalve mollusks (38, 50), and crustaceans [reviewed in (26); (20, 38, 39, 50)]. Moreover, the uptake of NH4+ by the alga will ensure the continuous conversion of NH3 into NH4+ in the symbiosome, which in turn will maintain NH3 diffusion from the host cytosol. By analogy to the CCM that facilitates symbiont photosynthesis (4), NH3 transport by ayRhp1 coupled to acid trapping of NH4+ in the symbiosome can be considered a host-controlled nitrogen concentrating mechanism (NCM). The high degree of conservation among cnidarian Rh channels (fig. S1) and the presence of an acidic symbiosome in anemones and corals from both the complex and robust clades (4) suggest that Rh-mediated NCMs are widespread in cnidarians. However, species- and environment-specific differences in the NCM contribution to Tamm transport may exist and must be explored.
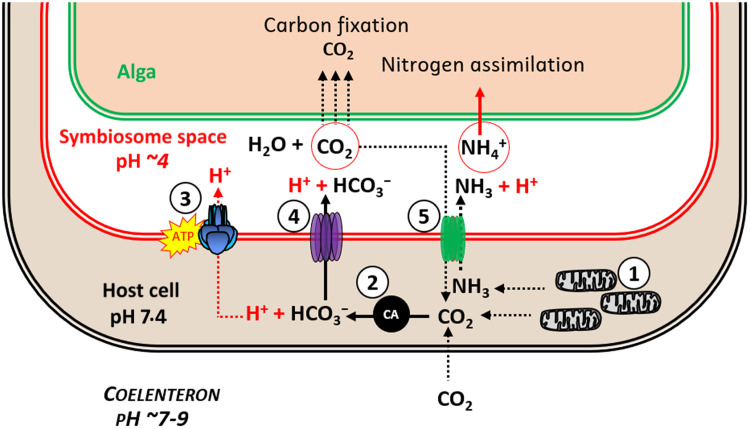
In addition, CO2-facilitated diffusion by ayRhp1 has implications for the host-controlled symbiosomal CCM (4). In this model, H+ transport by VHA generates an acidic symbiosome that drives the DIC equilibrium toward CO2 accumulation in the symbiosome space, which then diffuses into the alga where it is fixed by algal Rubisco. The presence of ayRhp1 in the symbiosome membrane may initially seem counterproductive for the CCM, as it provides a pathway for CO2 to leak back into the host cell cytoplasm. Continued carbon fixation by the algae, however, ensures a more favorable gradient for CO2 diffusion into the algae compared with the host cell. The CO2 that backflows into the coral host cell would be hydrated by cytosolic carbonic anhydrases (CA) into H+ and HCO3− (51), which can then be used as substrates for VHA and HCO3− transporters in the symbiosome membrane. Furthermore, the fluid in the coelenteron and mitochondria in the gastrodermal cells (4) are additional sources of CO2 that can fuel symbiosome acidification (Fig. 5). This mechanism is akin to the human kidney collecting duct, where VHA, CAs, HCO3− transporters, and Rh channels interact with each other for the purposes of HCO3− reabsorption and Tamm excretion (52).
Fig. 5. Model of the coral nitrogen concentrating mechanism in alga-containing coral host cells.
(1) Coral mitochondria produce NH3 and CO2. (2) Cytosolic CA catalyzes CO2 hydration into H+ and HCO3−. (3) H+ and (4) HCO3− are moved into the symbiosome space by VHA and an unidentified HCO3− transporter, respectively. Inside the symbiosome, H+ and HCO3− dehydrate into CO2, which diffuses into and is photosynthetically fixed by the alga. (5) NH3 diffuses via the A. yongei Rhesus protein (ayRhp1) into the symbiosome space, where it is immediately protonated and trapped as NH4+. An unidentified algal transporter imports NH4+ into the alga, where it is assimilated. Some CO2 also diffuses via ayRhp1 back into the host cell’s cytosol, where it is rehydrated and transported back to the symbiosome space. ATP, adenosine 5′-triphosphate.
The increased proportion of cells displaying symbiosomal ayRhp1 localization during the day matches established patterns of increased Tamm delivery from host to symbiont (53) and symbiont nitrogen assimilation (9) during light conditions. The diurnal nitrogen supply is primarily used not to advance growth but to sustain a high turnover of photosystem proteins and pigments damaged by ultraviolet radiation and electron transfer, which is essential for continued and efficient photosynthesis (9, 54, 55). This situation highlights the need for unique regulatory mechanisms in photosymbiotic associations compared with symbioses with nonphotosymbiotic microbes, such as those in plant roots. Conversely, the removal of ayRhp1 from the symbiosome membrane at night would serve to restrict nitrogen supply to symbiotic algae, thus limiting the synthesis of nonphotosynthetic proteins that would be essential to sustain their growth and reproduction (55). This mechanism gains additional significance when we consider that the coral symbiosome is highly acidic in both light and dark conditions (4), and this implies a continued steep inwardly directed pNH3 gradient. Moreover, alga-containing gastrodermal cells are in contact with the gastrovascular cavity or coelenteron. This compartment contains Tamm at concentrations that can be several hundred-fold higher compared with seawater (56) and experiences steep diel pH fluctuations that can reach pH values as high as 9 during the day and as low as 6.75 at night (56, 57). The presence of ayRhp1 channels in the host plasma membrane at night would facilitate the removal of NH3 from the host cell into the coelenteron, where it would be trapped as NH4, further restricting nitrogen supply to the algae at night.
The regulation of nitrogen delivery via changes in ayRhp1 subcellular localization is not mutually exclusive with regulation via the GS/GDH/GOGAT pathway (11, 58, 59), and they complement each other. The involvement of this pathway is largely based on changes in gene expression or enzyme activity upon transitioning from symbiotic to aposymbiotic stages (22) or during long-term environmental disturbances (60, 61). But despite diel mRNA expression patterns (62), the abundance of most metabolic enzymes in coral cells, including that of GS, does not seem to change on a diel basis (63). However, this does not preclude diel regulation of enzyme activity by posttranslational modifications or substrate availability that could act synergistically with ayRhp1 to control nitrogen to symbiotic algae. Moreover, ayRhp1 subcellular localization was not identical in all cells at any time period, indicating finer regulation based on position on the coral colony, symbiotic stage, or some other unidentified factors.
Perspectives and limitations
The ayRhp1- and VHA-dependent NCM identified here together with diel changes in ayRhp1 subcellular distribution provide a potential mechanism whereby coral host cells can supply nitrogen to their algal symbionts while still maintaining them in a nitrogen-limited state to control their growth under oligotrophic conditions. Alterations in nitrogen delivery to coral symbiotic algae have been linked to eutrophication and other environmental stressors that result in disruption of the symbiosis at the colony level, commonly known as coral bleaching (64–69). For example, heat stress may promote coral amino acid catabolism as a means to meet increased metabolic demand in a warmed environment, which has been suggested to trigger a feedback loop that releases symbionts from nitrogen limitation, uncouples the symbiotic relationship, and leads to bleaching (70). In addition, future studies must take into account that ayRhp1 is present in multiple cell types throughout coral tissues, which cannot be discerned using transcriptomics, proteomics, or metabolomics assays on bulk coral colony samples. Specifically, changes in ayRhp1 abundance could be driven by ayRhp1 in cells in the epidermis, gastrodermis, calicodermis, or combinations, and each of these conditions would reflect a unique coral response, in some cases with opposite implications for coral health. With this in mind, techniques that allow the investigation of coral biology at the cellular and molecular levels such as nanoscale secondary ion mass spectrometry (“nano-SIMS”) (11, 70) and confocal microscopy (71–73) are essential complements to “-omics” techniques. In particular, confocal Airyscan microscopy will allow studying physiological processes in calcifying cells and at the symbiosome membrane in unprecedent detail. Further work is required to determine whether the observed ayRhp1 symbiosomal localization is widespread among scleractinian corals. In addition, future studies should examine wild corals to ascertain the importance of this putative Rh channel–dependent NCM in the field. Future studies could also explore the role of other environmental nitrogen sources (i.e., urea and NO3−) on host-symbiont metabolism. These are time- and materials-intensive tasks but are necessary to fully contextualize the coral NCM at the ecophysiological level.
METHODS
Organisms
A. yongei colonies were maintained in flow-through seawater at 26°C with a 10/14-hour light/dark cycle with sunrise at 0800 and sunset at 1800. These coral colonies predominantly contain Cladocopium species (formerly Symbiodinium clade C). See the Supplementary Materials and Methods for additional information on coral husbandry.
Cloning of ayRhp1
Following the methods of (15), total RNA was collected by flash-freezing a 2-cm A. yongei nubbin in liquid nitrogen and crushing with a mortar and pestle into a fine powder. Powdered tissue was resuspended in TRIzol reagent (Invitrogen, Carlsbad, CA, USA), and total RNA was extracted following the manufacturer’s protocol. Total RNA was cleaned and concentrated using an RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen) and Oligo(dT) primers according to the manufacturer’s protocol. The resulting cDNA was used as template for all RT-PCRs. The full-length ayRhp1 sequence was obtained and can be found on GenBank (MH025799).
Protein sequences for the phylogenetic analysis were sourced from (74) and GenBank via BLASTtn search. Accession numbers of all Rh sequences used in this analysis can be found in the supporting information file (Table S1).
Plasmid preparation and cDNA synthesis for Xenopus laevis expression of ayRhp1
The open reading frame of ayRhp1 was amplified from pCR2.1 TOPO-ayRhp1 vector (see cloning of ayRhp1) using Q5 high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA, USA) and the restriction site–containing primers (forward primer, 5′-ATACCCGGGATGTCTACTCGACCTCCTACG-3′; reverse primer: 5′-GGCAAGCTTTTACACTTTATCATCTCCGAC-3′), subcloned by sticky-end ligation with T4 ligase (New England Biolabs) into Xma I and Hind III restriction sites of a pGEM-HE vector containing Xenopus beta globin 5′- and 3′-UTR sequences flanking the cloning site. Proper insertion was verified by restriction digest using Eco R1, Bam HI, and Sph I and visualized by gel electrophoresis to confirm ayRhp1 fragments produced were of the expected size. The pGEM-HE vector containing the ayRhp1 insert was linearized using Sph I; the restriction enzyme was heat inactivated at 65°C and linear plasmid column purified (GeneJet PCR Purification Kit; Thermo Fisher Scientific, Waltham, MA, USA). The in vitro transcription of ayRhp1 capped mRNA (cRNA) was performed with HiScribe T7 ARCA mRNA kit (New England Biolabs) on Sph I linearized pGEM-HE-ayRhp1 vector followed by column purification (RNeasy MiniElute Cleanup Kit; Qiagen). The cRNA was quantified spectrophotometrically (NanoDrop, ND-1000; Thermo Fisher Scientific), and its integrity was assessed on a denaturing MOPS agarose gel.
Oocyte microinjection
Stage VI–V oocytes were collected from mature female X. laevis (75). Briefly, the frogs were euthanized via decapitation, and the ovary was dissected and placed in Ca2+-free oocyte ringer (OR2) solution (82.5 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 1 mM Na2HPO4, 5 mM HEPES, pH 7.5) containing collagenase type VI (1 mg ml−1; Thermo Fisher Scientific). After incubation under gentle agitation for 90 min at room temperature, collagenase activity was terminated by rinsing the oocytes three times in OR2 containing 1 mM CaCl2. Oocytes were then manually sorted, rinsed, and allowed to recover in OR2 sterilized using vacuum bottle-top filters (EMD Millipore Steritop) overnight at 16°C (Fisherbrand Mini Refrigerated Incubator). Oocytes were injected with 18.4 ng of ayRhp1 cRNA (36.8 nl with 0.5 ng nl−1) (ayRhp1) or equivalent volume of nuclease-free water (control) using a Nanoject II or III auto-nanoliter injector (Drummond Scientific, Broomall, PA, USA). Experiments were conducted 3 days postinjection; during this time, the oocytes were stored in OR2 supplemented with 2.5 mM sodium pyruvate, penicillin-streptomycin (1 mg ml−1), and gentamicin (50 μg ml−1). Oocytes that died during experiments were discounted from analyses. All procedures followed the Guidelines of the Canadian Council on Animal Care and were approved by the University of Manitoba Animal Research Ethics Board.
Oocyte Tamm uptake rates
Groups of control (water-injected) or ayRhp1 (ayRhp1 cRNA-injected) oocytes (24 oocytes = 1 replicate; n = 6 to 8) were placed in 15-ml tubes and incubated for 1 hour at room temperature in OR2 solutions containing (a) 0 mM NH4Cl, pH 7.5; (b) 1 mM NH4Cl, pH 6.5; (c) 1 mM NH4Cl, pH 7.5; (d) 1 mM NH4Cl, pH 8.5; or (e) 10 mM NH4Cl, pH 7.5. Osmolarity was maintained by substituting NaCl with NH4Cl, and pH was adjusted by adding NaOH or HCl. Following incubation, oocytes were washed in ice-cold OR2 to remove excess NH4Cl and placed in groups of three oocytes in 27 μl of 6% perchloric acid to deproteinize samples (76). After pH neutralization with 3 M KOH, samples were diluted 1:10–1:40 with MilliQ water, and Tamm was measured using a hypochlorite-salicylate-nitroprusside–based assay (77). Tamm uptake rate was calculated according to the formula
where [Tamm]t = 1h is the Tamm measured in oocytes after incubation in OR2 (b) to (e) for 1 hour, [Tamm]t = 0h is Tamm measured in oocytes in OR2 (a) before the start of the incubations, 1 μl is the average oocyte volume, and 60 min was used to calculate rates on a per minute basis. The average Tamm uptake rate of control oocytes was subtracted from that of ayRhp1 oocytes before statistical analysis (fig. S3). Tamm uptake kinetics were calculated using a nonlinear regression to fit the Michaelis-Menten equation.
Oocyte CO2 release rates
Hypercapnic OR2 was generated by aeration with 5% CO2 until pH reached equilibrium. Groups of control or ayRhp1 oocytes (25 oocytes = 1 replicate; n = 8) were incubated in 40 ml of hypercapnic OR2 for 1.5 hours at 16°C. Oocytes were transferred along with hypercapnic OR2 into 2-ml septum capped gas tight vials. Hypercapnic OR2 was sequentially removed from each vial, replaced with normocapnic OR2, and sealed while submerged in OR2 to prevent air bubbles. Rapid transfer from hypercapnic to normocapnic OR2 was used to generate an oocyte-to-OR2 pCO2 gradient. One minute after sealing the chambers, three replicate 5-μl samples were taken with a gas tight Hamilton syringe from the vial by piercing the septum on the cap and injected into a custom-built total CO2 analyzer using a Licor 850 (LI-COR Biosciences, Lincoln, NE, USA) for CO2 detection as previously described (36). Standards of 0, 0.2, 0.3, 0.4, and 0.5 mmol l−1 NaHCO3 were used to calibrate the CO2 analyzer and produced an R2 of 0.99. CO2 release rate was calculated according to the formula
where CO2 releaset = 1min is the CO2 measured in the sample after 1 min of incubation in normocapnic OR2, 0.04644 and 0.1960 are corrections calculated from the standard curve, 0.002 L is the chamber volume, 25 oocytes are the number of oocytes per chamber, and 1 min is the CO2 flux period, The integrity of oocytes in all vials was confirmed under a microscope at the end of the sampling period.
Antibodies
Custom-made, affinity-purified anti-ayRhp1 rabbit polyclonal antibodies were developed (GenScript USA Inc., Piscataway, NJ, USA) against the peptide CHNKDAHGSHKEGSN, which is present in a putative Rhp1 protein predicted from the Acropora digitifera genome (XP_015769291.1) (78). This epitope has just one amino acid difference in ayRhp1 (CHNKDAHGSPKEGSN). NKA was immunolocalized with a commercially available monoclonal antibody (SC-48345, Santa Cruz Biotechnology, Dallas, TX, USA).
ayRhp1 protein expression and antibody validation
Using methods adapted from (4), A. yongei tissue was removed from the skeleton using an airbrush loaded with homogenization buffer. Briefly, homogenate was sonicated on ice and centrifuged to pellet down debris; the supernatant was kept on ice. Sample protein concentrations were determined using a Bradford Protein Assay (Bio-Rad, Hercules, CA, USA) with a bovine serum albumin standard curve. Samples were then incubated in 4× Laemmli sample buffer (Bio-Rad) and 10% β-mercaptoethanol before heating at 90°C for 3 min and loaded onto an SDS–polyacrylamide gel electrophoresis (PAGE) gel. Following electrophoresis, proteins were transferred from the gel onto a polyvinylidene difluoride (PVDF) membrane using a Mini Tans-Blot Cell (Bio-Rad) overnight. The membrane was blocked with 5% powdered fat-free milk in TBS-T for 1 hour on a shaker at room temperature before overnight incubation on a shaker (4°C) with anti-ayRhp1 primary antibody (0.216 μg ml–1), primary antibody with 400× excess peptide on a molar base (“preabsorption control”), or preimmune serum (0.216 μg ml–1) diluted in blocking buffer. Membranes were washed with 4 × 15 min TBS-T washes before incubation with secondary antibody [goat anti–rabbit–horseradish peroxidase (HRP) diluted 1:10,000, Bio-Rad] for 1 hour on a shaker at room temperature. Membranes were again washed with 4 × 15 min TBS-T washes and a final 15-min TBS wash before band development with an ECL Prime Western blot Detection Kit (GE Healthcare, Chicago, IL, USA) and imaged using a Chemidoc Imaging system (Bio-Rad) (fig. S4D). See the Supplementary Materials and Methods for additional information.
Immunofluorescence
Some A. yongei nubbins were fixed and decalcified, and others were brushed with a toothbrush to isolate cells prior to fixing following previously described methods (4, 15, 49, 71, 73) (see the Supplementary Materials and Methods for details). Tissue sections and isolated cells were incubated for 1 hour at room temperature in blocking buffer [4 ml of phosphate-buffered saline with Triton X-100 (PBS-TX), 80 μl of normal goat serum, and 0.8 μl of keyhole limpet hemocyanin solution], followed by overnight incubation (4°C) with anti-ayRhp1 antibodies (2.16 μg ml–1), anti-ayRhp1 antibodies preabsorbed with excess peptide (8.64 μg ml–1), or preimmune serum (2.16 μg ml–1) alone or in combination with the anti-NKA antibody (2.00 μg ml–1) (all in blocking buffer) (fig. S4, A to C).
Slides were washed in PBS-TX to remove unbound primary antibodies (3 × 5 min). Secondary antibodies (goat anti–rabbit–Alexa Fluor 555, goat anti–rabbit–Alexa Fluor 488, and/or goat anti–mouse–Alexa Fluor568, 4 μg/ml in blocking buffer; Invitrogen) were then added for 1 hour at room temperature followed by 4′,6-diamidino-2-phenylindole (DAPI) DNA stain (1 μg ml–1 in blocking buffer; Invitrogen) for 5 min at room temperature and washed again in PBS-TX to remove unbound secondary antibodies and DAPI (3 × 5 min).
Epifluorescence microscopy was performed on a Zeiss AxioObserver Z1 (Carl Zeiss AG, Oberkochen, Germany) connected to a metal halide lamp. ayRhp1 and DAPI signals in tissue sections (Fig. 2 and fig. S4, A to C1) were visualized using HE Cy3 [excitation (ex): 550 nm, emission (em): 570 nm] and DAPI (ex: 359 nm, em: 461 nm) filters, respectively. ayRhp1 and DAPI signals in isolated cells (fig. S5) were visualized using HE DsRed (ex: 538 to 562 nm, em: 570 to 640 nm) and FURA (ex: 335 to 345 and 375 to 385 nm, em: 505 to 530 nm) filters, respectively.
Confocal Airyscan microscopy was performed on a Zeiss AxioObserver Z1 connected to a laser scanner equipped with 405-, 488-, 561-, and 640-nm laser lines (Zeiss LSM 800 with Airyscan, Carl Zeiss AG). This device uses a 32-channel photomultiplier detector and linear deconvolution to obtain 140-nm lateral (X-Y) and 400-nm axial (Z) resolution. ayRhp1, NKA, and DAPI signals in tissue sections (Figs. 4, A and B, and 3) were visualized using goat anti–rabbit–Alexa Fluor 488 and goat anti–mouse–Alexa Fluor568 secondary antibodies (Invitrogen) and DAPI stain (Invitrogen), respectively (Alexa Fluor 488—ex: 517 nm, em: 497 to 574 nm; Alexa Fluor 568—ex: 577 nm, em: 560 to 643 nm; DAPI—ex: 465 nm, em: 400 to 484 nm). Three-dimensional reconstructions of z-stacks were generated using Imaris 9.0 (Bitplane, Zurich, Switzerland). To facilitate visualization by color-blind readers, NKA, ayRhp1, and DAPI signals are presented using the false colors violet, green, and blue, respectively, in all figures.
Assessment of ayRhp1 subcellular localization over day-night cycles
Cell isolations were prepared from coral nubbins randomly selected from three separate tanks. Nubbins were sampled 30 min before and after sunrise and sunset (0730, 0830, 1730, and 1830 hours) as well as halfway between lighting condition changes (1300 and 2300 hours). Samples taken during the day were continually illuminated during cell isolation and fixation, while those taken during the night were kept in the dark. At each time point, cells were immunostained for ayRhp1 and imaged as described above. Starting from the upper-right corner of the field of view, the first 50 intact alga-hosting A. yongei cells displaying ayRhp1 signal were counted and classified into one of two subcellular localization patterns: “symbiosomal ayRhp1 localization” (the ayRhp1 signal clearly traversed the region between the nuclei of the coral host cell and the algae) or nonsymbiosomal localization (ayRhp1 signal was absent from this region but present exterior and adjacent to the host nucleus). Cells were counted and classified in a double-blind manner: Slides were named with random identifiers by an independent person before being observed on the fluorescence microscope by another person. Classification was conducted during observation through the microscope eyepiece, as this allowed a better determination of ayRhp1 subcellular localization by rapid and repetitive adjustments to the fine focus and alternation between the Alexa Fluor 555 and DAPI channels. Cells from three separate branches were classified at each time point, resulting in a total of 150 cells per time point and 900 cells in total. Time points were matched with random slide names only once all 900 cells were classified. Raw count data are presented in data S1.
Statistical analysis
All statistical tests were run in GraphPad Prism 7 (San Diego, CA, USA). Tamm uptake, CO2 release, and ayRhp1 localization data were tested for normality and homogeneity of variance using D’Agostino and Pearson or Shapiro-Wilk normality tests and Brown-Forsythe tests. Tamm uptake data were analyzed using one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons. CO2 release data were analyzed using a Welch’s t test (two-tailed, unequal variance). Data from ayRhp1 localization in isolated cells were analyzed using two-way repeated-measures ANOVA followed by Dunnett’s posttest using the data from 1300 hours as control. Alpha was set at 0.05 for all statistical tests.
Acknowledgments
We thank M. Romero (Mayo Clinic) for the gift of Xenopus expression vector; M. Ortega, S. Nöel, A. Serna, C. Hassabi, D. Jio, and P. Zerofski (Scripps Institution of Oceanography) for help in maintaining coral cultures; and M. Nash (University of Manitoba) for support with oocyte sorting and maintenance.
Funding: This work was partially supported by the National Science Foundation (NSF) EF #1220641 to M.T., NSF Graduate Research (GRFP 2019271478) and SIO Doctoral Scholar Fellowships to A.B.T., Natural Sciences and Engineering Research Council of Canada (NSERC) Postgraduate Doctoral Scholarship-Doctoral to A.R.Q.-R., University of Manitoba Graduate Fellowship to H.Z., and NSERC Discovery grant (RGPIN/5013-2018) to D.W.
Author contributions: M.T. and A.B.T. conceived the project. M.T. and D.W. directed the research. A.B.T. performed all cloning, antibody validation, and microscopy experiments. A.R.Q.-R. and H.Z. performed oocyte experiments. A.B.T., A.R.Q.-R., D.W., and M.T. analyzed the data. A.B.T. and M.T. wrote the manuscript. All authors read and edited the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S6
Table S1
References
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.van Oppen M. J. H., Medina M., Coral evolutionary responses to microbial symbioses. Philos. Trans. R. Soc. Lond. B Biol. Sci. 375, 20190591 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanaka Y., Suzuki A., Sakai K., The stoichiometry of coral-dinoflagellate symbiosis: Carbon and nitrogen cycles are balanced in the recycling and double translocation system. ISME J. 12, 860–868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang B. L., Thoughts on a very acidic symbiosome. Front. Microbiol. 6, 816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barott K. L., Venn A. A., Perez S. O., Tambutté S., Tresguerres M., Coral host cells acidify symbiotic algal microenvironment to promote photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 112, 607–612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahav O., Dubinsky Z., Achituv Y., Falkowski P. G., Ammonium metabolism in the zooxanthellate coral, Stylophora pistillata. Proc. R. Soc. Lond. B. 236, 325–337 (1989). [Google Scholar]
- 6.Wright P. A., Nitrogen excretion: Three end products, many physiological roles. J. Exp. Biol. 198, 273–281 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Szmant A. M., Ferrer L. M., FitzGerald L. M., Nitrogen excretion and O:N ratios in reef corals: Evidence for conservation of nitrogen. Mar. Biol. 104, 119–127 (1990). [Google Scholar]
- 8.Cui G., Liew Y. J., Li Y., Kharbatia N., Zahran N. I., Emwas A. H., Eguiluz V. M., Aranda M., Host-dependent nitrogen recycling as a mechanism of symbiont control in Aiptasia. PLOS Genet. 15, e1008189 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopp C., Pernice M., Domart-Coulon I., Djediat C., Spangenberg J. E., Alexander D. T. L., Hignette M., Meziane T., Meibom A., Highly dynamic cellular-level response of symbiotic coral to a sudden increase in environmental nitrogen. mBio 4, e00052-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Elia C., Domotor S., Webb K., Nutrient uptake kinetics of freshly isolated zooxanthellae*. Mar. Biol. 167, 157–167 (1983). [Google Scholar]
- 11.Pernice M., Meibom A., Van Den Heuvel A., Kopp C., Domart-Coulon I., Hoegh-Guldberg O., Dove S., A single-cell view of ammonium assimilation in coral-dinoflagellate symbiosis. ISME J. 6, 1314–1324 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rees T. A. V., Are symbiotic algae nutrient deficient? Proc. R. Soc. B Biol. Sci. 243, 227–233 (1991). [Google Scholar]
- 13.Krueger T., Horwitz N., Bodin J., Giovani M. E., Escrig S., Fine M., Meibom A., Intracellular competition for nitrogen controls dinoflagellate population density in corals. Proc. Biol. Sci. 287, 20200049 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sproles A. E., Kirk N. L., Kitchen S. A., Oakley C. A., Grossman A. R., Weis V. M., Davy S. K., Phylogenetic characterization of transporter proteins in the cnidarian-dinoflagellate symbiosis. Mol. Phylogenet. Evol. 120, 307–320 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Barott K. L., Barron M. E., Tresguerres M., Identification of a molecular pH sensor in coral. Proc. R. Soc. B 284, 20171769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiner I. D., Verlander J. W., Ammonia transporters and their role in acid-base balance. Physiol. Rev. 97, 465–494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallace I. S., Choi W.-G., Roberts D. M., The structure, function and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochim. Biophys. Acta 1758, 1165–1175 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Niemietz C. M., Tyerman S. D., Channel-mediated permeation of ammonia gas through the peribacteroid membrane of soybean nodules. FEBS Lett. 465, 110–114 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Geyer R. R., Parker M. D., Toye A. M., Boron W. F., Musa-Aziz R., Relative CO2/NH3 permeabilities of human RhAG, RhBG and RhCG. J. Membr. Biol. 246, 915–926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruswitz F., Chaudhary S., Ho J. D., Schlessinger A., Pezeshki B., Ho C.-M., Sali A., Westhoff C. M., Stroud R. M., Function of human Rh based on structure of RhCG at 2.1 A. Proc. Natl. Acad. Sci. U.S.A. 107, 9638–9643 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baday S., Orabi E. A., Wang S., Lamoureux G., Bernèche S., Mechanism of NH4+ recruitment and NH3 transport in Rh proteins. Structure 23, 1550–1557 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Lehnert E. M., Mouchka M. E., Burriesci M. S., Gallo N. D., Schwarz J. A., Pringle J. R., Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3 4, 277–295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii Y., Maruyama S., Takahashi H., Aihara Y., Yamaguchi T., Yamaguchi K., Shigenobu S., Kawata M., Ueno N., Minagawa J., Global shifts in gene expression profiles accompanied with environmental changes in cnidarian-dinoflagellate endosymbiosis. G3 9, 2337–2347 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganot P., Moya A., Magnone V., Allemand D., Furla P., Sabourault C., Adaptations to endosymbiosis in a Cnidarian-Dinoflagellate association: Differential gene expression and specific gene duplications. PLOS Genet. 7, e1002187 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu M., Zheng X., Fan C.-M., Zheng Y., Lineage dynamics of the endosymbiotic cell type in the soft coral Xenia. Nature 582, 534–538 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D. Weihrauch, M. O’Donnell, Acid-Base Balance and Nitrogen Excretion in Invertebrates (Springer International Publishing, 2017); 10.1007/978-3-319-39617-0. [DOI]
- 27.Benghezal M., Gotthardt D., Cornillon S., Cosson P., Localization of the Rh50-like protein to the contractile vacuole in Dictyostelium. Immunogenetics 52, 284–288 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Han K. H., Mekala K., Babida V., Kim H. Y., Handlogten M. E., Verlander J. W., Weiner I. D., Expression of the gas-transporting proteins, Rh B glycoprotein and Rh C glycoprotein, in the murine lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 297, L153–L163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry S. F., Braun M. H., Noland M., Dawdy J., Walsh P. J., Do zebrafish Rh proteins act as dual ammonia-CO2 channels? J. Exp. Zool. A Ecol. Genet. Physiol. 313, 618–621 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Endeward V., Cartron J. P., Ripoche P., Gros G., RhAG protein of the Rhesus complex is a CO2 channel in the human red cell membrane. FASEB J. 22, 64–73 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Nawata C. M., Wood C. M., O’Donnell M. J., Functional characterization of Rhesus glycoproteins from an ammoniotelic teleost, the rainbow trout, using oocyte expression and SIET analysis. J. Exp. Biol. 213, 1049–1059 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Li X.-D., Lupo D., Zheng L., Winkler F., Structural and functional insights into the AmtB/Mep/Rh protein family. Transfus. Clin. Biol. 13, 65–69 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Stolarski J., Kitahara M. V., Miller D. J., Cairns S. D., Mazur M., Meibom A., The ancient evolutionary origins of Scleractinia revealed by azooxanthellate corals. BMC Evol. Biol. 11, 316 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marini A. M., Boeckstaens M., Benjelloun F., Chérif-Zahar B., André B., Structural involvement in substrate recognition of an essential aspartate residue conserved in Mep/Amt and Rh-type ammonium transporters. Curr. Genet. 49, 364–374 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Li X., Jayachandran S., Nguyen H.-H. H. T., Chan M. K., Structure of the Nitrosomonas europaea Rh protein. Proc. Natl. Acad. Sci. U.S.A. 104, 19279–19284 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee D. J., Gutbrod M., Ferreras F. M., Matthews P. G. D., Changes in hemolymph total CO2 content during the water-to-air respiratory transition of amphibiotic dragonflies. J. Exp. Biol. 221, jeb181438 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Sasaki S., Ishibashi K., Nagai T., Marumo F., Regulation mechanisms of intracellular pH of Xenopus laevis oocyte. Biochim. Biophys. Acta 1137, 45–51 (1992). [DOI] [PubMed] [Google Scholar]
- 38.Thomsen J., Himmerkus N., Holland N., Sartoris F. J., Bleich M., Tresguerres M., Ammonia excretion in mytilid mussels is facilitated by ciliary beating. J. Exp. Biol. 219, 2300–2310 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Wright P. A., Wood C. M., A new paradigm for ammonia excretion in aquatic animals: Role of Rhesus (Rh) glycoproteins. J. Exp. Biol. 212, 2303–2312 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Thiel D., Hugenschutt M., Meyer H., Paululat A., Quijada-Rodriguez A. R., Purschke G., Weihrauch D., Ammonia excretion in the marine polychaete Eurythoe complanata (Annelida). J. Exp. Biol. 220, 425–436 (2017). [DOI] [PubMed] [Google Scholar]
- 41.Tidball J. G., Fine structural aspects of anthozoan desmocyte development (Phylum Cnidaria). Tissue Cell 14, 85–96 (1982). [DOI] [PubMed] [Google Scholar]
- 42.Muscatine L., Tambutte E., Allemand D., Morphology of coral desmocytes, cells that anchor the calicoblastic epithelium to the skeleton. Coral Reefs 16, 205–213 (1997). [Google Scholar]
- 43.Campbell J. W., Speeg K. V., Ammonia and biological deposition of calcium carbonate. Nature 224, 725–726 (1969). [Google Scholar]
- 44.Loest R. A., Ammonia-forming enzymes and calcium-carbonate deposition in terrestrial pulmonates. Physiol. Zool. 52, 470–483 (1979). [Google Scholar]
- 45.Crossland C. J., Barnes D. J., The role of metabolic nitrogen in coral calcification. Mar. Biol. 28, 325–332 (1974). [Google Scholar]
- 46.Cai W. J., Ma Y., Hopkinson B. M., Grottoli A. G., Warner M. E., Ding Q., Hu X., Yuan X., Schoepf V., Xu H., Han C., Melman T. F., Hoadley K. D., Pettay D. T., Matsui Y., Baumann J. H., Levas S., Ying Y., Wang Y., Microelectrode characterization of coral daytime interior pH and carbonate chemistry. Nat. Commun. 7, 11144 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allison N., Cohen I., Finch A. A., Erez J., Tudhope A. W., Corals concentrate dissolved inorganic carbon to facilitate calcification. Nat. Commun. 5, 5741 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Kaplan J. H., Biochemistry of Na,K-ATPase. Annu. Rev. Biochem. 71, 511–535 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Venn A. A., Tambutté E., Lotto S., Zoccola D., Allemand D., Tambutté S., Imaging intracellular pH in a reef coral and symbiotic anemone. Proc. Natl. Acad. Sci. U.S.A. 106, 16574–16579 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu M. Y., Guh Y. J., Stumpp M., Lee J. R., Chen R. D., Sung P. H., Chen Y. C., Hwang P. P., Tseng Y. C., Branchial NH4+-dependent acid-base transport mechanisms and energy metabolism of squid (Sepioteuthis lessoniana) affected by seawater acidification. Front. Zool. 11, 55 (2014). [Google Scholar]
- 51.Bertucci A., Moya A., Tambutté S., Allemand D., Supuran C. T., Zoccola D., Carbonic anhydrases in anthozoan corals - a review. Bioorg. Med. Chem. 21, 1437–1450 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Weiner I. D., Verlander J. W., Renal ammonia metabolism and transport. Compr. Physiol. 3, 201–220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grover R., Maguer J.-F., Reynaud-vaganay S., Ferrier-Pagès C., Uptake of ammonium by the scleractinian coral Stylophora pistillata: Effect of feeding, light, and ammonium concentrations. Limnol. Oceanogr. 47, 782–790 (2002). [Google Scholar]
- 54.Beraud E., Gevaert F., Rottier C., Ferrier-Pages C., The response of the scleractinian coral Turbinaria reniformis to thermal stress depends on the nitrogen status of the coral holobiont. J. Exp. Biol. 216, 2665–2674 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Rosic N., Kaniewska P., Chan C.-K., Ling E. Y., Edwards D., Dove S., Hoegh-Guldberg O., Early transcriptional changes in the reef-building coral Acropora aspera in response to thermal and nutrient stress. BMC Genomics 15, 1052 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agostini S., Suzuki Y., Higuchi T., Casareto B. E., Yoshinaga K., Nakano Y., Fujimura H., Biological and chemical characteristics of the coral gastric cavity. Coral Reefs 31, 147–156 (2012). [Google Scholar]
- 57.Furla P., Galgani I., Durand I., Allemand D., Sources and mechanisms of inorganic carbon transport for coral calcification and photosynthesis. J. Exp. Biol. 203, 3445–3457 (2000). [DOI] [PubMed] [Google Scholar]
- 58.Yellowlees D., Rees T. A. V., Leggat W., Metabolic interactions between algal symbionts and invertebrate hosts. Plant. Cell Environ. 31, 679–694 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Oakley C. A., Ameismeier M. F., Peng L., Weis V. M., Grossman A. R., Davy S. K., Symbiosis induces widespread changes in the proteome of the model cnidarian Aiptasia. Cell. Microbiol. 18, 1009–1023 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Yellowlees D., Rees T., Fitt W., Effect of ammonium-supplemented seawater on glutamine synthetase and glutamate dehydrogenase activities in host tissue and zooxanthellae of Pocillopora damicornis and on ammonium uptake rates of the zooxanthellae. Pacific Sci. 48, 291–295 (1994). [Google Scholar]
- 61.Tang J., Ni X., Wen J., Wang L., Luo J., Zhou Z., Increased ammonium assimilation activity in the scleractinian coral Pocillopora damicornis but not its symbiont after acute heat stress. Front. Mar. Sci. 7, 565068 (2020). [Google Scholar]
- 62.Levy O., Kaniewska P., Alon S., Eisenberg E., Karako-Lampert S., Bay L. K., Reef R., Rodriguez-Lanetty M., Miller D. J., Hoegh-Guldberg O., Complex diel cycles of gene expression in coral-algal symbiosis. Science 331, 175 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Linsmayer L. B., Deheyn D. D., Tomanek L., Tresguerres M., Dynamic regulation of coral energy metabolism throughout the diel cycle. Sci. Rep. 10, 19881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuntz N. M., Kline D. I., Sandin S. A., Rohwer F., Pathologies and mortality rates caused by organic carbon and nutrient stressors in three Caribbean coral species. Mar. Ecol. Prog. Ser. 294, 173–180 (2005). [Google Scholar]
- 65.Schlöder C., D’Croz L., Responses of massive and branching coral species to the combined effects of water temperature and nitrate enrichment. J. Exp. Mar. Biol. Ecol. 313, 255–268 (2004). [Google Scholar]
- 66.Wooldridge S. A., Done T. J., Thomas C. R., Gordon I. I., Marshall P. A., Jones R. N., Safeguarding coastal coral communities on the central Great Barrier Reef (Australia) against climate change: Realizable local and global actions. Clim. Change 112, 945–961 (2012). [Google Scholar]
- 67.Cleves P. A., Krediet C. J., Lehnert E. M., Onishi M., Pringle J. R., Insights into coral bleaching under heat stress from analysis of gene expression in a sea anemone model system. Proc. Natl. Acad. Sci. U.S.A. 117, 28906–28917 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morris L. A., Voolstra C. R., Quigley K. M., Bourne D. G., Bay L. K., Nutrient availability and metabolism affect the stability of coral–symbiodiniaceae symbioses. Trends Microbiol. 27, 678–689 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Wiedenmann J., D’Angelo C., Smith E. G., Hunt A. N., Legiret F. E., Postle A. D., Achterberg E. P., Nutrient enrichment can increase the susceptibility of reef corals to bleaching. Nat. Clim. Chang. 3, 160–164 (2013). [Google Scholar]
- 70.Rädecker N., Pogoreutz C., Gegner H. M., Cárdenas A., Roth F., Bougoure J., Guagliardo P., Wild C., Pernice M., Raina J. B., Meibom A., Voolstra C. R., Heat stress destabilizes symbiotic nutrient cycling in corals. Proc. Natl. Acad. Sci. U.S.A. 118, e2022653118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barron M. E., Thies A. B., Espinoza J. A., Barott K. L., Hamdoun A., Tresguerres M., A vesicular Na+/Ca2+ exchanger in coral calcifying cells. PLOS ONE 13, e0205367 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ganot P., Tambutté E., Caminiti-Segonds N., Toullec G., Allemand D., Tambutté S., Ubiquitous macropinocytosis in anthozoans. eLife 9, e50022 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barott K. L., Venn A. A., Thies A. B., Tambutté S., Tresguerres M., Regulation of coral calcification by the acid-base sensing enzyme soluble adenylyl cyclase. Biochem. Biophys. Res. Commun. 525, 576–580 (2020). [DOI] [PubMed] [Google Scholar]
- 74.Huang C.-H., Peng J., Evolutionary conservation and diversification of Rh family genes and proteins. Proc. Natl. Acad. Sci. 102, 15512–15517 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soreq H., Seidman S., Xenopus oocyte microinjection: From gene to protein. Methods Enzymol. 207, 225–265 (1992). [DOI] [PubMed] [Google Scholar]
- 76.Veauvy C. M., Walsh P. J., Mcdonald M. D., Effect of elevated ammonia on tissue nitrogen metabolites in the ureotelic gulf toadfish (Opsanus beta) and the ammoniotelic midshipman (Porichthys notatus). Physiol. Biochem. Zool. 82, 345–352 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Kempers A. J., Kok C. J., Re-examination of the determination of ammonium as the indophenol blue complex using salicylate. Anal. Chim. Acta 221, 147–155 (1989). [Google Scholar]
- 78.Shinzato C., Shoguchi E., Kawashima T., Hamada M., Hisata K., Tanaka M., Fujie M., Fujiwara M., Koyanagi R., Ikuta T., Fujiyama A., Miller D. J., Satoh N., Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476, 320–323 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Edgar R. C., MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stamatakis A., RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sonnhammer E. L., von Heijne G., Krogh A., A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 (1998). [PubMed] [Google Scholar]
- 82.Krogh A., Larsson B., Von Heijne G., Sonnhammer E. L. L., Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 83.M. E. Jørgensen, H. H. Nour-Eldin, B. A. Halkier, in Biotechnology of Plant Secondary Metabolism: Methods and Protocols, A. G. Fett-Neto, Ed. (Springer Science & Business Media, 2016); 10.1007/978-1-4939-3393-8, vol. 1405, pp. 99–107. [DOI]
- 84.Barott K., Tresguerres M., Immunolocalization of proteins in corals: The V-type H+-ATPase proton pump. Bio-protocol 5, e1573 (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S6
Table S1
References
Data S1