Abstract
MicroRNA (miRNA), a noncoding ribonucleic acid, is considered to be important for the progression of gene expression in plants and animals by rupture or translational repression of targeted mRNAs. Many types of miRNA regulate plant metabolism, growth, and response to biotic and abiotic factors. miRNA characterization helps to expose its function in regulating the process of post-transcriptional genetic regulation. There are a lot of factors associated with miRNA function, but the function of miRNA in the organic synthesis of by-products by natural products is not yet fully elucidated. The current review is aimed at observing and characterizing miRNAs and identifying those involved in the functioning of the biosynthesis of secondary metabolites in plants, with their use in controlled manipulation.
1. Introduction
miRNAs are eighteen to twenty-eight nucleotides containing single-stranded RNA molecules which are not transcribed into proteins during transcription. It takes part in post-transcriptional regulation by binding to the messenger RNA and inhibiting the expression of specific genes [1]. These types of molecules are usually expressed in eukaryotes such as animals and plants and some viruses [2, 3].
The first miRNA was reported in Caenorhabditis elegans is Lin-4; then, further studies identify more than 18,226 other types of miRNAs in the same organisms such as 22 (nt) lin-4 and 21 (nt) let-7 [4]. While miRNA-targeted gene interconnections are extensively prolonged, this technique is restrained among domains [5]. miRNAs are disbursed in genetic material as clumps exhibit as a polycistronic segment with features [6]. Most miRNAs in flora are encrypted with their prime transcript and some precedents of miRNA clumps described as miR395. The basic identified hotspots for their beginning are introns [7]. Most of the processes in which they affect are the expansion of time and host-pathogen connection in addition to cell differentiation, proliferation, apoptosis, and tumorigenesis.
In eukaryotes and metazoans, miRNAs are a form of noncoding (22-nucleotide) ribo regulators that control gene expression which has an exciting role in the biosynthesis of plant secondary metabolites like flavonoids, terpenoids, alkaloids, and some other compounds [8, 9]. RNA polymerase II is a precursor RNA also described as the pri-miRNA that helps to stimulate the production of miRNA, processed by DICER-LIKE 1 (DCL1) to fully grown or developed miRNA [8].
In this review, we have updated the knowledge about the present understanding of miRNA-based regulation of biosynthesis and accumulation of secondary metabolites in plants. The data written in English were collected, using scientific search engines such as PubMed/Medline, ScienceDirect, Web of Science, Scopus, and Google Scholar. The search terms used were microRNA, biosynthesis, secondary metabolites, miRNA, flavonoids, alkaloids, and terpenoids.
The scientific names of the plants have been validated according to the Plant List [10, 11].
2. miRNA as a Secondary Metabolite Regulator
The polyadenylated caps and RNAs are the precursor molecules for the synthesis of miRNA both in plants and animals, and RNA polymerase II (RNAPII) transcribed many coding RNAs, although in plants, nucleus RNAase dicer-like 1 (DCL1) and their essential proteins SERRATE (SE) and hyponastic leaves (HYL1) synthesize primary miRNA but the Drosha gene is not found in plants [5, 12]. In Arabidopsis thaliana, the precursor miRNA, dsRNA, is synthesized in the nucleus by the splitting of pri-miRNA by HYL1 and DCL1. The second splitting is done by the activity of HYL1 and DCL1 which results in releasing of miRNA; methylation of two nucleotides 3′ overhangs is performed by methyltransferase HUA enhancer1 (HEN1). Adult single-stranded cytoplasmic miRNA activated by AGO1 is found in the RISC (RNA-induced silencing complex) ribonucleoprotein and by breaking mRNA suppresses its function [13]. The miRNA binds with mRNA at the 3′ untranslated region (3′ UTR) which inhibits the activity of miRNA [14]. The transcriptional factors regulate the expression of miRNA. The transcription of miRNA genes and effective use of DCL1 in the biosynthesis of miRNA is facilitated by At-Negative on TATA-less 2 (NOT2) [15]. Cell division cycle 5 (CDC5) is associated with miRNA which is a positive transcription factor. The drosophila exportin-5 ortholog HASTY (HST) factor exports miRNA duplex to the cytoplasm.
The miRNA-targeted genes may be solitary members of a gene group or synchronize various family groups. However, more than one type of miRNA gene can regulate a single individual with tissues and a single-miRNA gene can control many family individuals. The territorial and worldly articulation of plenitude-developed miRNAs is firmly managed; they differ enormously within various miRNAs, and the bounty likewise fluctuates relying upon the type of tissues or growth and embryonic stages [16].
The miRNAs inhibit the activity of mRNA to control the gene expression at the level of post-transcription. In contrast to animals, plants have an ideal relationship between mRNA and miRNA [17]. For the silencing, a ribonucleoprotein RNA-induced silencing complex (RISC) is formed [18]. Different types of slicers have been reported: AGO10, AGO7, AGO4, AGO2, and AGO1. However, AGO1 is mostly related to miRNA [19]. The RNA splitting is activated by AGO1 by inhibiting specific genes [20].
In plants, the genes coding for transcription, stress response proteins, and other factors which are associated with different biological mechanisms are controlled by miRNA-like conservation of genetic materials, maintenance of metabolism, and enlargement of plants, signal transduction, energy pathway, homeostasis, natural immunity, and adaptive feedback to biotic and abiotic stress [21, 22]. The phytochemicals are peripheral metabolites that play an important role in different processes which are related to the association between plants and its surrounding [23–25]. The mechanism of action of miRNAs in plants is shown in Figure 1.
Figure 1.
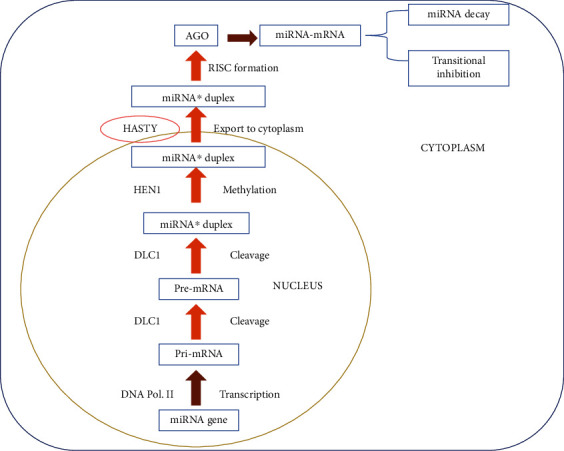
Mechanism of action of miRNAs in plants. miRNA: microRNA; pri-miRNA: primary transcripts; DCL1: RNAase dicer-like 1.
These compounds involve isoprenoids, tannins, alkaloids, phenols, glycosides, and triterpene glycosides, which protect the plants from different living and nonliving stress-causing factors [26, 27]. Such kinds of molecules are produced by herbs for self-protection; these compounds are also used in various industries like nutraceuticals, flavouring F compounds, dyes, and insecticides as they are providing a great response in terms of human health [28, 29]. Materialistic significance has brought about an extraordinary enthusiasm for considering the prospects of improving its creation [30]. It concluded that miRNAs regulate several biological cycles at the stage of post-transcription. Several recent investigations concluded that the miRNA plays an important part in the transduction of the peripheral metabolic series [31]. So, the products of peripheral metabolites can be regulated by miRNAs. The production of any metabolite can be regulated by a positive or negative feedback mechanism, so we can stop the production of undesired products and enhance the production of desired metabolites [32].
Computational evaluation executed in two transcriptomes of Swertia ensured to identify miRNAs connected with secondary metabolites, miR-11320, miR-168, miR-156a, miR-11071, miR-166a, and miR-166b focused on metabolic enzymes consisting of premnaspirodiene oxygenase, aspartate aminotransferase, phosphoglycerate mutase, ribulose-phosphate 3-epimerase, acetyl-CoA, and acetyltransferase. Additionally, it consists of a gene encrypting a homeobox-leucine zipper protein (HD-ZIP) along with secondary metabolite organic production in Swertia chirayita (Roxb.) Buch.-Ham. ex C.B.Clarke [33].
The production of self-protecting metabolites is reduced in diseased plants due to alteration in gene expression so these may be maintained by miRNAs. When Solanum tuberosum L. is exposed to sunlight, brilliance-reactive miRNAs are produced which plays an important role in nicotine metabolism, UMP salvage, fat production, and cellulose catabolism [34].
The stress of cadmium in oil seed rape (Brassica napus L.) described miRNA as a crucial element in the synchronization of transcription factors, living thing stress protection, ion balancing, and peripheral metabolism production [35].
Nicotiana tabacum tainted with tobacco mosaic virus (TMV), at the beginning phase of the disease (5 dpi), shows a bunch of miRNAs with low aggregation, while a large portion of the miRNAs was upregulated at 15 and 22 dpi including both miRNAs and miRNA targets [36, 37].
3. Flavonoid Biosynthesis and miRNA
Flavonoids have a hydroxylated phenolic structure and contain benzo-γ-pyron, which is obtained from phenylpropanoid [38, 39]. Flavonoids have different compounds including chalcones, catechins, flavonols, flavonols, anthocyanins, flavanones, and flavanonols. These compounds are synthesized with the help of others in plants and microorganisms, and their concentration varies as per the environmental conditions [40, 41].
The biological functions of flavonoids vary greatly with the structure and result in various bioactivities such as antimutagenic, antioxidative, anticarcinogenic, anti-inflammatory, and antiplatelet aggregation [42]. These biological activities of flavonoids impart different applications such as medicinal, nutraceutical, cosmetics, and pharmaceutical [43–45].
miRNA-initiated reactions take part in the assemblage of peripheral metabolites (Table 1).
Table 1.
miRNA effect on the synthesis of secondary metabolites.
| Botanical name | Common name | Family | miRNA | Target | Function | References |
|---|---|---|---|---|---|---|
| Helianthus annuus L. | Sunflower | Asteraceae | miR2911 | Gamma-tocopherol methyltransferase | Tocopherol biosynthesis | [46] |
| Salvia sclarea L. | Clary | Lamiaceae | miR828a, miR948a | MYB12 lipoxygenase | Flavonoid biosynthesis | [47] |
| Curcuma longa L. | Turmeric | Zingiberaceae | miR2919 | Flavanone synthase | [48] | |
| miR1168.2, miR156b | Curcumin biosynthesis | |||||
| miR1858 | ||||||
| Ipomoea batatas (L.) Lam. | Sweet potato | Convolvulaceae | ib-miR156 | ibSPL | Anthocyanin biosynthesis | [49] |
| Actinidia arguta (Siebold & Zucc.) Planch. ex Miq. | Kiwifruit | Actinidiaceae | miR858 | AaF3H, AaLDOX | [50] | |
| AaF3GT | ||||||
| Diospyros kaki L. f. | Kaki | Ebenaceae | miR395p-3p | bHLH, MYB | Proanthocyanidin biosynthesis | [51] |
| miR858b | ||||||
| Lonicera japonica Thunb. | Japanese honeysuckle | Caprifoliaceae | miRNAs (U436803, U977315, U805963, U3938865, 4351355) | R2R3-MYB transcription factors | Flavonoid biosynthesis | [2, 3] |
| Halostachys caspica C.A.Mey. | Bail | Amaranthaceae | miR6194 | Flavanone 3-hydroxylase | Flavonol, anthocyanidin, and proanthocyanidin syntheses | [52] |
| miR5655 | ||||||
| Arabidopsis thaliana (L.) Heynh. | Thale cress | Brassicaceae | miR156∗ | SPL9 | Anthocyanin biosynthesis | [53] |
| miR858a∗ | R2R3-MYB transcription factors | Flavonoid biosynthesis | [54] | |||
| miR156 | SPL transcription factor, transcription factors | Flavonoid biosynthesis | [53, 54] | |||
| miRNA858a | ||||||
| R2R3-MYB | ||||||
| Rauvolfia serpentina (L.) Benth. ex Kurz | Indian snakeroot | Apocynaceae | miR396b | Kaempferol 3-O-beta-dgalactosyltransferase | Flavonol glycoside | [55] |
| miR828a | Anthocyanin regulatory C1 protein | Anthocyanin biosynthesis | ||||
| Podophyllum hexandrum Royle | May apple | Berberidaceae | miR1438 | Caffeoyl-CoA O-methyl transferase | Lignin biosynthesis | [56] |
| miR1873 | Dihydroflavonol 4-reductase C | Flavonoid biosynthesis | ||||
| miR1873 | Phenylalanine ammonia lyase (PAL) | Gingerol (phenolic) biosynthesis | [57] | |||
| miR5532 | 2-Hydroxyisoflavanone dehydratase | Isoflavonoid biosynthesis | [56] | |||
| miR1873/miR5532 | Dihydroflavonol | Flavonoid/isoflavonoid biosynthesis | ||||
| 4-Reductase C/-hydroxyisoflavanone dehydratase | ||||||
| miR172i | 4-Coumarate–CoA ligase | Flavonoid biosynthesis | ||||
| miR829.1 | Chalcone synthase | |||||
| Glycine max (L.) Merr. | Soybean | Fabaceae | CHS-siRNA | Chalcone synthase | Flavonoid biosynthesis | [58, 59] |
| Pyrus bretschneideri Rehder | Pear tree | Rosaceae | miR1061-3p | Naringenin 3-dioxygenase | Flavonoid biosynthesis | [60] |
| Helianthus annuus L. | Sunflower | Asteraceae | miR2911 | γ-Tocopherol methyl transferase | α-Tocopherol biosynthesis | [46] |
| miRNAs (U4992168, U2743257) | Isoflavone 20-hydroxylase; dihydroflavonol 4-reductase | Flavones biosynthesis | [2] | |||
| Raphanus sativus L. | Radish | Brassicaceae | miR156 | MYBs, bHLH, WD40, SPLs, ARF, EIN3, WRKY, MADS-box, sucrose synthase, sugar/inositol transporter ABC transporter | Anthocyanin biosynthesis | [61] |
| miR828 | ||||||
| miR858 | ||||||
| Vitis vinifera L. | Grape | Vitaceae | miR828 | MYB114, flavonoid | Flavonol biosynthesis | [62] |
| miR858 | 3′-Monooxygenase (F3MO) | |||||
| Osmanthus fragrans Lour. | Sweet osmanthus | Oleaceae | miR858, miR858a | MYB1- > CHI, CHS, FLS | Flavonoid synthesis | [63] |
| Camellia sinensis (L.) Kuntze | Tea | Theaceae | csn-miR160a | Auxin response factor 18 | Catechin biosynthesis | [64] |
| csn-miR396a | Growth-regulating factor 7, calcium-transporting ATPase 13, DNA-directed RNA polymerase V subunit | |||||
| csn-miR167a | Auxin response factor 6 ARF6, CHI enzyme | |||||
| csn-miR4380a | DFR | |||||
| csn-miR3444b | ||||||
| csn-miR5251 | C4H | |||||
| csn-miR7777-5P.1 | ||||||
| miR2593e | ANR | |||||
| miR166, miR169 | HD-ZIP, NF-YA, GATA | Phytohormone biosynthesis (gallated catechin, caffeine, theanine) | [65] | |||
| miR398 | ||||||
| Mentha spp. | Mint | Lamiaceae | miR156 | Basic helix-loop-helix (bHLH) | Flavone/flavonol biosynthesis | [66] |
| Halostachys caspica C.A.Mey. | Yan Sui Mu | Berberidaceae | miR6194 | Flavanone 3b-hydroxylase (F3H) | Flavonols, anthocyanidins | [52] |
| Biosynthesis |
Scarce studies are present about the importance of miRNAs in the organic synthesis of flavonoids. In Helianthus, 323,318 ESTs had been logically observed for the miRNA recognition and the miR911 group was observed being connected to the biosynthesis of tocopherols. Himalayan May apple (Podophyllum hexandrum Royle), miR1438, affects caffeoyl-CoA O-methyltransferase and is associated with flavonoid biosynthesis; phenylalanine metabolism; stilbenoid, diarylheptanoid, and gingerol biosynthesis; and phenylpropanoid biosynthesis.
Dihydroflavonol 4-reductase C is affected with miR1873 which is associated with flavonoid biosynthesis (Figure 2). Isoflavonoid biosynthesis is associated with the miR5532 2-hydroxyisoflavanone dehydratase (Figure 2; Table 1) [56].
Figure 2.
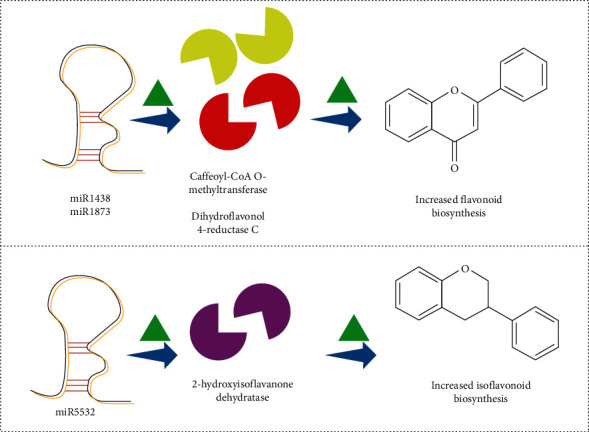
miRNA increases the activity of biosynthetic enzymes involved in flavonoid and isoflavonoid production and resulting in an improved yield of flavonoid and isoflavonoids.
miR2911 regulates tocopherol synthesis genes such as gamma-tocopherol methyltransferase in sunflower plants (Helianthus annuus L.) [46]. Furthermore, mRNA (miR828a and miR948a) helps to accelerate flavonoid biosynthesis by regulating MYB12 lipoxygenase in Clary [47]. Flavonoid's synthesis (curcumin) in turmeric plants (Curcuma longa L.), miR2919, miR1168.2, miR156b, and miR1858 promote the flavanone synthase gene [48].
Anthocyanins are one of the important members of the flavonoid family [67, 68]. In the tuberous root of sweet potato, the expression of ib-miR156 is its target ibSPL, which aids in anthocyanin production [49]. Kiwifruit, miR858, reported accelerating anthocyanin accumulation [69]. High microR156 movement initiates aggregation of anthocyanin along with movement initiated by flavonols. The current study also illustrated that squamosa promoter-binding-like protein 9 SPL9 adversely initiated anthocyanin aggregation with the disruption of an MYB-bHLH-WD40 transcriptional-initiated system. Diospyros kaki L.f. fruit results acquired at tested periods (15 and 20 WAF) confirmed the contrasting appearance of the messenger RNAs, suggesting that those miRNAs throughout the growth and a number of them are miRNAs 858 and 56 which adjust the synthesis of PA. PA production genes are positively initiated, miR858 but negative effects are observed by miR156. Another miR395 and miR858b play their role in biosynthesis proanthocyanidin [51]. Some miRNAs (U4351355, U3938865, U805963, U977315, and U436803) initiate lipid and flavonol organic production in Lonicera japonica Thunb. [2, 3]. Yang et al. [52] imply that salty situations modify miRNAs in which few salt pressure-associated organic channels consist of flavonoid biosynthesis, calcium signaling pathway, and plant hormone signal transduction [52].
Gou and colleagues reported that in the stem of Arabidopsis thaliana anthocyanins accumulate in an acropetal manner, this model is controlled by SPL (squamosa binding protein-like) genes and stimulated by miR156 [53]. The depiction of A. thaliana indicates that miR858a targets myeloblastosis (MYB) transcript elements which are engaged in flavonoid organic production, development, and growth. MYB transcription factors down synchronize with overexpression of miR858a and the redirection of metabolic flux initiated with the excessive appearance of MYBs concerning the composition of flavonoids [54]. The biosynthesis of flavonoids (anthocyanins) is regulated by miR156, miR858a by stimulating the transcription factors R2R3-MYB and SPL9 [53, 54]. Rauvolfia serpentine (Indian snakeroot) has two essential types of miRNA such as miR396b and miR828a, both of which targets kaempferol 3-O-β-D-galactosyltransferase and anthocyanin regulatory C1 protein for synthesizing flavonol glycoside and anthocyanin biosynthesis, respectively [55].
Podophyllum hexandrum Royle (May apple) is an endangered medicinal plant [56]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway study demonstrated that miR1438 and miR1873 regulate various metabolic pathways, especially the biosynthesis of secondary metabolites like lignin and flavonoid via caffeoyl-CoA O-methyl transferase and dihydroflavonol 4-reductase C gene, respectively [56]. Biswas et al. [56] further revealed that miR5532 upregulates the 2-hydroxyisoflavanone dehydratase gene for synthesizing isoflavonoid in the May apple tree. Flavonoid biosynthesis also occurred in May apple through inducing chalcone synthase and 4-coumarate–CoA ligase. Both of them are targeted by miR829.1 and miR172i, respectively [56]. 4-Coumarate-CoA ligase catalyzes the activation of the corresponding thiol esters by 4-coumarate and other 4-hydroxycinnamates. These activated thiol esters are subsequently employed as flavonoid and monolignol precursors [70]. miR5015 has been shown to influence the production of gingerol by suppressing phenyl ammonia-lyase (PAL) which is a basic enzyme in the synthesis [57]. Glycine max (L.) Merr. (soybean) CHS-siRNA targets chalcone synthase enzyme to the biosynthesis of flavonoids [58, 59].
In Pyrus bretschneideri Rehder, miR1061-3p regulates naringenin 3-dioxygenase, which produces flavonoids [60]. Helianthus annuus produces α-tocopherol, and flavones by miRNA and miRNA influence the regulation process via targeting γ-tocopherol methyltransferase, isoflavone 20-hydroxylase, and dihydroflavonol 4-reductase [2]. miR156, miR828, and miR858 are reported to play an important function in regulating anthocyanin biosynthesis in radish through MYBs, bHLH, WD40, SPLs, ARF, EIN3, WRKY, MADS-box, sucrose synthase, sugar/inositol transporter, and ABC transporter genes [61]. Furthermore, MYB114 and flavonoid 3′-monooxygenase (F3MO) are also targeted by miR828 and miR858 in the grape plant for the biosynthesis of flavonol [62]. MYB, an essential step in flavonol biosynthesis, upregulates CHI, CHS, and FLS genes for the production of flavonoids [60, 63, 71].
Sun and coworkers reported that the expression of eight miRNAs (csn-miR160a, csn-miR396a, csn-miR167a, csn-miR4380a, csn-miR3444b, csn-miR5251, csn-miR7777-5P.1, and miR2593e) and their target genes (auxin response factor 18, growth-regulating factor 7, calcium-transporting ATPase 13, DNA-directed RNA polymerase V subunit, auxin response factor 6 ARF6, CHI enzyme, DFR, C4H, and ANR) is involved in catechin biosynthesis [64]. By decreasing the expression of their biosynthesis-related gene transcription, miRNAs could adversely control catechin production.
Another study reported that miR166, miR169, miR398 target HD-ZIP, NF-YA, and GATA gene in the tea plant [65] as well as miR6194 influence flavanone 3β-hydroxylase (F3H) for flavonols and anthocyanidin biosynthesis [52].
4. Terpenoid Biosynthesis and miRNA
Isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) are a C5 predecessor of plant terpenoid secondary metabolites. Terpenoids range in different groups according to the orders of carbon atoms such as polyphenols (>45), carotenoids (C40), triterpenes (C30), diterpenes (C20), sesquiterpenes (C15), and monoterpenes (C10) [72]. Terpenoids also have different applications as biological and industrial issues like flavonoids and alkaloids. The computational study identifies the change of miRNAs into 6 transcriptomes of Picrorhiza kurroa Royle ex Benth. which discovered that miRNA-4995 plays a promoter function in the biosynthesis of terpenoids (Table 2), finally disturbing the manufacturing of picroside-I [73]. The miR-5021 effect on two kinds of enzymes that take part in organic synthesis of GCPE protein, indole alkaloids (IAs), and terpenoid cyclases was first time observed in Catharanthus roseus (L.) G.Don [74]. miRNAs (1134, 5021, and 7539) are probably concerned with promoting terpenoid organic synthesis through the terpenoid channel genes; nontranscriptional factor proteins, like IDS, DXS, IDI, and HMGR, are necessary to bring out DMAPP and IPP, the main predecessor for all kinds of downstream terpenoids [75].
Table 2.
MicroRNA effect on synthesis of terpenoid secondary metabolites.
| Botanical name | Common name | Family | miRNA | Target | Function | References |
|---|---|---|---|---|---|---|
| Picrorhiza kurroa Royle ex Benth. | Kutki | Scrophulariaceae | iRNA-4995 | 3-Deoxy-7-phosphoheptulonate synthase (DAHP synthase) | Terpenoid biosynthesis | [73] |
| Catharanthus roseus (L.) G.Don | Tropical periwinkle | Apocynaceae | mir-5021 | MYB transcription factor, geranyl diphosphate synthase, GCPE protein, UDP-glucose iridoid glucosyltransferase | Isoprenoid/terpenoid biosynthesis | [74] |
| Ferula gummosa Boiss. | Galbanum | Apiaceae | miR2919, miR5251, miR838, miR5021, miR5658 | SPL7, SPL11, ATHB13 TFs | Terpene biosynthesis | [76] |
| Pogostemon cablin (Blanco) Benth. | Patchouli | Lamiaceae | miR156∗ | SPL9 | Sesquiterpenoid, triterpenoid biosynthesis | [77] |
| Mentha spp. | Mint | Lamiaceae | miR156, miR414, and miR5021 | Basic helix-loop-helix (bHLH) geranyl di-phosphate synthase subunit alpha-like protein (NACA) | Terpenoid backbone biosynthesis | [66] |
| miR156∗ | 1-Deoxy-D-xylulose | Terpenoid biosynthesis | [66] | |||
| 5-Phosphate synthase (DXS) | ||||||
| miR414 | Terpene synthase 21 (TPS21) | Sesquiterpenoid, triterpenoid biosynthesis | ||||
| Rauvolfia serpentina (L.) Benth. ex Kurz | Indian snakeroot | Apocynaceae | miR396b | Secologanin synthase | Secologanin | [55] |
| Salvia miltiorrhiza | Red sage | Lamiaceae | miR5072 | Acetyl-CoA C-acetyl transferase | Tanshinones (abietane-type norditerpenoid quinones) | [78] |
| Zingiber officinale Roscoe | Ginger | Zingiberaceae | miR838 | CYP71 | Terpenoid metabolism | [57] |
| Picrorhiza kurroa Royle | Kutki | Plantaginaceae | miR4995 | 3-Deoxy-7-phosphoheptulonate synthase (DAHP synthase) | Picroside biosynthesis | [73] |
| Podophyllum hexandrum Royle | Mayapple | Berberidaceae | miR5538 | Protein-S-isoprenylcysteine | Terpenoid backbone biosynthesis | [56] |
| O-methyltransferase | ||||||
| Xanthium strumarium L. | Rough cocklebur | Asteraceae | miR7539, miR5021, miR1134 | Upstream genes of terpenoid pathway | Terpenoid | [75] |
| miR5491, miR5255 | Beta-amyrin synthase | Xanthanolide (sesquiterpenoids) | ||||
| miR6449, miR5183 | Squalene epoxidase | |||||
| miR7540 | Ent-kaurene synthase | |||||
| Gibberellin 3-oxidase | ||||||
| R-Linalool synthase | ||||||
| miR7540, mmiR5183, miR6449, miR5255, miR5491, miR6435 | R-Linalool synthase, gibberellin 3-oxidase, ent-kaurene synthase, squalene epoxidase, beta-amyrin synthase, germacrene A oxidase | Mono-, sesqui-, di-, and triterpenoid biosynthesis | ||||
| miR1134 | 3-Hydroxy-3-methylglutaryl coenzyme A reductase (HMGR) | Terpenoid backbone biosynthesis | ||||
| miR5183 | Gibberellin 3-oxidase | Diterpenoid | ||||
| miR5255 | Squalene epoxidase | Triterpenoid | ||||
| miR5491 | Beta-amyrin synthase | Triterpenoid | ||||
| miR6435 | Germacrene A oxidase | Sesquiterpenoid | ||||
| miR6449 | Ent-kaurene synthase | Diterpenoid | ||||
| miR7539 | 1-Deoxy-D-xylulose | Terpenoid backbone | ||||
| 5-Phosphate synthase (DXS) | ||||||
| miR7540 | R-Linalool synthase | Monoterpenoid | ||||
| Panax ginseng C.A.Mey. | Asian ginseng | Araliaceae | miR854e miR854b miR854c | Farnesyl diphosphate synthase, squalene epoxidase (SE) | Ginsenosides (triterpene) | [79] |
| miR1439b, miR1439h | Beta amyrin synthase | |||||
| Artemisia annua Pall. | Sweet wormwood | Asteraceae | miR390 | A gene involved in trichome development | Artemisinin (sesquiterpene) | [72] |
| Swertia chirayita (Roxb.) Buch.-Ham. ex C.B.Clarke | Swertia | Gentianaceae | miR-168 | Acetyl-CoA acetyltransferase | PSMs | [33] |
| miR-11320 | Aspartate aminotransferase | |||||
| miR-166a | Premnaspirodiene oxygenase | |||||
| miR-11071 | Ribulose-phosphate3-epimerase | |||||
| miR-156a | Phosphoglycerate mutase, | |||||
| miR-166b | ||||||
| Curcuma longa L. | Turmeric | Zingiberaceae | miR5649a | Geranylgeranyl-diphosphate specific | Terpenoid backbone biosynthesis | [48] |
| miR5021 | Geranyl-diphosphate synthase | |||||
| 1-Deoxy-D-xylulose-5-phosphate synthase (DXS) | ||||||
| Geranylgeranyl-diphosphate synthase | ||||||
| Camellia sinensis (L.) Kuntze | Tea | Theaceae | miR3630 | MYC2 | Terpenoid biosynthesis | [65] |
| miR33418 | MYB. | |||||
| miR156f | SPL9 | |||||
| miR156 | AaERF1/2 | |||||
| miR535 | ||||||
| miR858 | ||||||
| Persicaria minor (Huds.) Opiz | Kesum | Polygonaceae | pmi-miR530 pmimiR6173 | MVD, probable sulphate transporter; Sesquiterpene synthase, HMGR, farnesyl diphosphate synthase 1 | Terpenoid biosynthesis | [80] |
| pmi-miR6300 | ||||||
| Ginkgo biloba L. | Maidenhair tree | Ginkgoaceae | miR_2924, miR_218 | HMGS, MK, MPDC, LPS | Terpene trilactones (TTLs) biosynthesis | [81] |
| miR_2642 | ||||||
| miR159a, miR159, miR_2432 | LPS | Terpene trilactones (TTLs) biosynthesis | ||||
| Artemisia annua Pall. | Sweet wormwood | Asteraceae | miR159, miR172 | Cytochrome P450 reductase; CYP71AV1 catalyzes | Artemisinin (ART) biosynthesis (sesquiterpene) | [82] |
| miR166 | ||||||
| Ferula gummosa Boiss. | Ferula | Apiaceae | miR1533, miR5021, miR5658 | SPL7, SPL11, ATHB13 | Terpene biosynthesis | [76] |
| Stevia rebaudiana (Bertoni) Bertoni | Candyleaf | Asteraceae | miRstv_7∗ | UDP-glycosyl transferase76G1 (UGT76G1) | Steviol glycoside biosynthesis | [83] |
| miRstv_7∗ | Kaurenoic acid hydroxylase (KAH) | Steviol glycoside biosynthesis | ||||
| miRstv_7∗ | Kaurene oxidase (KO) | Steviol glycoside biosynthesis |
miRNAs (6435, 6449, 7540, 5491, 5183, and 5255) affect downward enzymes in the organic synthesis of mono-, sesqui-, di-, and tri-terpenoids; they included germacrene A oxidase, R-linalool synthase, beta-amyrin synthase (bAS), gibberellin 3-oxidase, squalene epoxidase, and ent-kaurene synthase [75]. miRNAs (5658, 5251, 5021, 2919, and 838) were recognized to be included in the terpene organic synthesis channel in Ferula gummosa Boiss.
Squamosa promoter binding and ATHB13 were observed to be promoted by miRNAs (1533, 5021, and 5658) [76]. Sesquiterpene biosynthesis is mediated by miRNA156-connected SPL gene [77]. miR414 is involved in the triterpenoid and sesquiterpenoid organic syntheses while miR5021 also plays a role in terpenoid formation (Table 1) [66]. Investigators revealed that SPL9 specifically connects to and triggers the TPS21 promoter. Due to the great conservation of miR156-SPLs in plants, the findings establish a molecular connection between the developmental time and sesquiterpene synthesis and provide a novel method for engineering plants to grow faster while producing more terpenoids [77]. In all, 130 unique candidate transcripts for eight miRNA families were anticipated. All anticipated miRNA targets govern development, reproduction, stimulus response, signaling, and various metabolic processes. Additionally, a network of miRNA-mediated gene regulation was built using the hybridized lowest free energy of discovered miRNAs and their targets. The study discovered that the mint family's miR414, miR156, and miR5021 might regulate the essential oil biosynthesis gene control mechanism. Additionally, three miRNA candidates were identified as important in trichome formation (miR5021, miR156, and miR5015b) [66]. However, apart from providing first-hand knowledge on Rauvolfia serpentina (L.) Benth. ex Kurz miRNAs and their objectives, the outcomes in this work help to a profound comprehension of miRNA-mediated gene regulation mechanisms in plants [55].
Another study demonstrated that among 452 recognized miRNAs, which equate to 589 precursor miRNAs, 62 miRNAs are expressed exclusively in the base, 95 miRNAs are expressed exclusively in the stem, 19 miRNAs are expressed exclusively in the leaf, and 71 miRNAs are expressed exclusively in flower. The degradome study found 69 targets that may be cleaved by 25 miRNAs. Among these, miR5072 cleaved acetyl-CoA C-acetyltransferase, which is implicated in the production of tanshinones. This investigation expanded significantly to our comprehension of the tissue-specific expression patterns of miRNAs in Salvia miltiorrhiza Bunge and laid the groundwork for future exploration on the production of tanshinone mediated by miRNAs [78].
Moreover, recent findings indicated that most anticipated miRNAs were engaged in rhizome formation control. miR854, miR5021, and miR838 have been revealed as miRNAs that control ginger rhizome growth and essential oil production [57]. It is demonstrated that miRNA-4995 performs a key role in regulating terpenoid biosynthesis, subsequently influencing the development of picroside-I. miR-5532 and miR-5368 expression rates were significantly lower in field-grown specimens than in vitro-cultured samples, implying a role in controlling Picrorhiza kurroa Royle ex Benth. development under culture environments [73]. Additionally, an investigation concluded 60 mature miRNAs and 6 precursor miRNAs in Podophyllum hexandrum Royle using 454 pyrosequencing. The finding contributes fundamental knowledge regarding the control of secondary metabolite production in Podophyllum hexandrum Royle by miRNAs [56].
A study demonstrated to pinpoint the trichome-specific miRNAs and mRNA targets. The findings from the experiment give a basis for additional research in Xanthium strumarium L. glandular cells to elucidate the governing strategy of miRNAs underpinning the production of secondary, in particular, terpenoids [75]. Researchers discovered that a large number of the miRNAs pinpointed had tissue-specific expression. By scanning the ginseng EST database for anticipated objectives of the estimated 69 conserved miRNAs, 346 promising targets were recognized. The anticipated targets were primarily engaged in secondary metabolic processes responding to transcription regulator activities and biotic and abiotic stress, among other metabolic pathways [79]. Conversely, miR390 was expected to engage a gene associated with trichome formation, the location of artemisinin production, and therefore may be possible for genetic transformation to increase the artemisinin content [72]. Also, a new study demonstrated insight into the role of miRNAs in the control of secondary metabolite production in the herbal plant Swertia chirayita (Roxb.) Buch.-Ham. ex C.B.Clarke. Additionally, the putatively discovered miRNAs can increase secondary metabolite production in S. chirayita by genetic modification [33].
Another study revealed that three miRNAs (miR156b, miR1168.2, and miR1858) were involved in curcumin production. Other miRNAs were discovered to be engaged in turmeric's development and growth. Additionally, a phylogenetic analysis of selected miRNAs was done in this study [48]. Moreover, another investigation demonstrated the identification of miRNA associated with control gene channels regulating terpenoid biosynthesis in Camellia sinensis (L.) Kuntze at various growth stages [65]. Investigating the regulation of terpenoid biosynthesis by miRNA outcomes indicates that six miRNAs examined control terpenoid production in P. minor post-transcriptionally. This governmental function of miRNAs suggests that they may be used as a genetic tool to control terpenoid production in Persicaria minor (Huds.) Opiz [80]. Another study also evaluated miRNAs and their presumed target genes involved in transthyretin-like (TTL) biosynthesis, and the results showed the existence of a complex miRNA-mRNA regulatory network involved in Ginkgo biloba L. TTL metabolism [81]. Figure 3 shows few examples in which miRNA assists in the improved production of terpenoid compounds.
Figure 3.
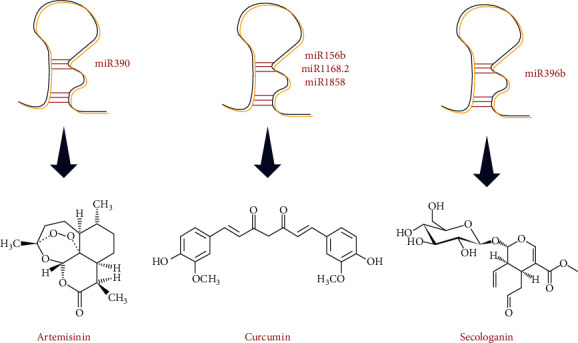
miRNA improves the production of terpenoid compounds.
The investigation uncovered multiple previously unknown miRNA families in Artemisia annua L., including miR399, miR396, miR319 miR858, miR6111, and miR5083. The expression patterns and correlations between miRNAs and their associated targets in A. annua plant are different, depending on its developmental phases [82]. Saifi and coworkers found nine miRNAs (miR319c, miR319a, miR319f, miR319b, miR319h, miR319d, miRstv-7, miR319e, and miRstv-9) and authors reported that these miRNAs are correspondingly connected to the expression thresholds of their target mRNAs and steviol glycosides were detected to be directly connected to the expression stages of their target mRNAs. This work established a foundation for a better knowledge of the steviol glycoside biosynthetic process, and these miRNAs may be used to control the production of these metabolites in Stevia rebaudiana (Bertoni) Bertoni to increase their concentration and productivity [83].
5. Alkaloid and Other N-Containing Metabolites and miRNA
Alkaloids are naturally occurring organic compounds that contain one or more nitrogen atoms in their heterocyclic ring structure, have alkali-like properties [84], have a variety of biological effects such as antimalarial (e.g., quinine), antiasthma (e.g., ephedrine), anticancer (e.g., homoharringtonine) [85], cholinomimetic (e.g., galantamine) [86], vasodilatory (e.g., vincamine), antiarrhythmic (e.g., quinidine), analgesic (e.g., morphine) [87], antibacterial (e.g., chelerythrine) [88], and antihyperglycemic activities (e.g., piperine) [63], and play significant roles in the defence against herbivores [89].
In plants, a myriad of bioactive compounds is synthesized naturally by several pathways. Among them, the involvement of miRNA is the most common way used for the biosynthesis of naturally occurring alkaloids. Pyrrolidine, purine, imidazole, indole, quinolizidine, pyrrolizidine, isoquinoline, and tropane are among the various types of alkaloids listed [90]. Disparate to other bioactive compounds, this group is extremely diverse and heterogeneous with an estimated 12,000 alkaloids in nature [89]. Alkaloids according to their toxic nature react as resistance compounds against many kinds of herbivores and pathogens. The understanding of alkaloid biosynthesis balancing is critical for its production.
Tobacco (Nicotiana tabacum L.) belongs to the Solanaceae family; a study revealed six different kinds of distinctive tobacco miRNAs, namely, miRX13, miRX17, miRX19, miRX20, miRX27 [50], and miR164 [91], that have been anticipated to target important genes of the nicotine organic biosynthesis and dissimilation channel, cytochrome P450 monooxygenase (CYP82E4), quinolinate phosphoribosyl-transferase 1 (QPT1), quinolinate phosphoribosyl-transferase 2 (QPT2), putrescine methyltransferase 2 (PMT2), and NtNAC-R1 genes [50].
In the opium poppy, the botanical name of Papaver somniferum L., a member of the Papaveraceae family, it has been identified that transcription of pso-miR408, pso-miR13 and pso-miR2161 (Table 1) has an important role in the biosynthesis of organic benzylisoquinoline alkaloids (BIA) [92]. pso-miR13, pso-miR2161, and pso-miR408 would possibly break 7-O-methyltransferase, S-adenosyl-l-methionine:3′-hydroxy-N-methylcoclaurine, 4′-O-methyltransferase 2/4′-O-methyltransferase 2 (4-OMT)/FAD-binding, and BBE domain-containing protein, respectively, transcript concerned with inside the transformation of S-reticuline to morphinan alkaloids (BIA). 4-OMT is the target of pso-miR216 and moderates the assembly of S-reticuline; this is additionally an intermediary particle in BIA organic production. On the opposite side, pso-microR408 feasibly reacts with messenger RNA from reticuline oxidase-like protein in the rate of transformation of S-reticuline to (S)-scoulerine within the BIA channel [92]. Figure 4 showed the role of miRNA in the synthesis of alkaloid and other N-containing metabolites.
Figure 4.
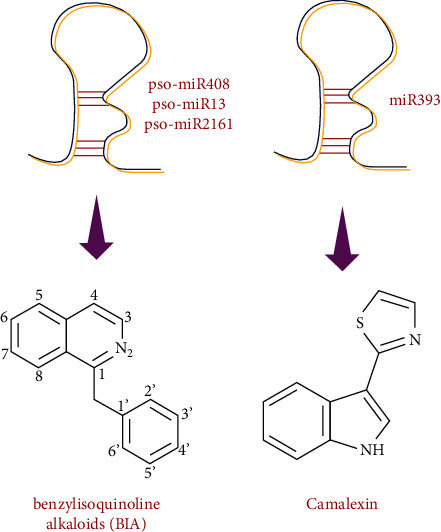
Role of miRNA in the synthesis of alkaloid and other N-containing metabolites.
Two kinds of two paclitaxel biosynthetic genes in Taxus baccata L., namely, taxane 2-α-O-benzoyl transferase and taxane 13-α-hydroxylase, are the breaking objects of microR164 and 171 [93, 94]. In silico inspection identified that microR396b in Rauvolfia serpentina (L.) Benth. ex Kurz hits kaempferol 3-O-beta-D-galactosyltransferase whose interest as transferase shifted the hexosyl class for the development of flavonoid glycosides [55]. miR-5021 in Madagascar periwinkle (Catharanthus roseus (L.) G.Don) regulates the enzyme involved in the biosynthesis of TIAs which includes UDP-glucose iridoid glucosyltransferase [74].
In Madagascar periwinkle, there are some other miRNAs such as cro-miR160, cro-miR164a, cro-miR164b, and cro-miR393d, which have a regulatory role in targeting CrARF16, ORCA3, ORCA4, and BIS1 genes in the TIA biosynthetic pathway and synthesis indole alkaloids [95]. In addition, Robert-Seilaniantz et al. [96] demonstrated that miR393 with its related auxin receptor genes is involved in alkaloid (camalexin) biosynthesis in Thale cress (Arabidopsis thaliana (L.) Heynh.) and in turmeric also named as Curcuma longa; miR5021 is illustrated to have a role to synthesize isoquinoline alkaloid interacting with two enzyme genes like aspartate transaminase and aromatic-amino-acid transaminase [48]. Furthermore, in sunflower plants, U3938865 and U4351355 involved glucosinolates and camalexin (indole alkaloid) biosynthesis through regulating the MYB34 gene [2] (Table 3).
Table 3.
Alkaloid and other N-containing metabolites and miRNA.
| Botanical name | Common name | Family | miRNA | Target | Function | References |
|---|---|---|---|---|---|---|
| Nicotiana tabacum L. | Tobacco | Solanaceae | miRX13∗ | Putrescine methyltransferase 2 (PMT2) | Nicotine biosynthesis | [50] |
| miRX17∗ | Quinolinate phosphoribosyl transferase 1 (QPT1) | |||||
| miRX20∗ | Cytochrome P450 monooxygenase (CYP82E4) | |||||
| miRX27∗ | Quinolinate phosphoribosyl-transferase 2 (QPT2) | |||||
| miR164 | NtNAC-R1 | [91] | ||||
| miRX17, miRX27 | QPT1, QPT2, CYP82E4, PMT2 | [50] | ||||
| miRX20, miRX19 | ||||||
| Papaver somniferum L. | Opium poppy | Papaveraceae | miR13 | 7-O-Methyltransferase (7-OMT) | BIA biosynthesis | [92] |
| miR408 | Reticuline oxidase | |||||
| miR2161 | 4′-O-ethyltransferase 2 (4-OMT) | |||||
| pso-miR13 | 7-O-methyltransferase, S-adenosyl-l-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase 2/4′-O-methyltransferase2 (4-OMT)/FAD-binding and BBE domain-containing protein | |||||
| pso-miR2161 | ||||||
| pso-miR408 | ||||||
| pso-miR13 | 7-OMT | Morphinan | ||||
| Taxus baccata L. | English yew | Taxaceae | miR164 | Taxane 13α hydroxylase | Taxol | [93] |
| Catharanthus roseus (L.) G.Don | Madagascar periwinkle | Apocynaceae | miR-5021 | Two enzymes involved in biosynthesis of TIAs | Indole alkaloids | [74] |
| UDP-glucose iridoid glucosyltransferase | ||||||
| cro-miR160, cro-miR164a, cro-miR164b | CrARF16, ORCA3, ORCA4, and BIS1, in TIA biosynthetic pathway | Indole | [95] | |||
| cro-miR393d | Alkaloid biosynthesis | |||||
| Arabidopsis thaliana (L.) Heynh. | Thale cress | Brassicaceae | miR393 | Auxin receptor | Camalexin | [96] |
| Curcuma longa L. | Turmeric | Zingiberaceae | miR5021 | Aspartate transaminase, aromatic-amino-acid transaminase | Isoquinoline alkaloid biosynthesis | [48] |
| Helianthus annuus L. | Sunflower | Asteraceae | U3938865 and U4351355 | MYB34 | Glucosinolates and camalexin (indole alkaloid) biosynthesis | [2] |
6. Fatty Acids and Other Compound Biosynthesis and miRNA
miRNA plays an important role in fatty acid secondary metabolite biosynthesis [97]. Lonicera japonica targets regulatory genes which help in the process of synthesis such as acyl-CoA synthetase, acyl carrier protein, and fatty acid hydroxylase. miRNAs involved in the fatty acid synthesis are U436803, U977315, and U805963 [2]. Besides this, in Paeonia × suffruticosa Andrews (tree peony) plant, fatty acids are synthesized by miR156b and miR7826 via regulating 1-acyl-sn-glycerol-3-phosphate acyltransferase and ACCase gene activities [98].
In Corynespora cassiicola plant, some novel miRNA was found that positively or negatively regulates secondary metabolite production in plants. These novel types of miRNAs are novel-miR1 to novel-miR7 which targets genes in phenylpropanoid synthesis [15]. Furthermore, phenylpropanoid biosynthesis is also regulated by miRNA (miR2673a and miR396b) in Podophyllum hexandrum Royle [99].
Withanolide is a class of active secondary metabolites in plants with medicinal interest. miR5140, miR159, miR477, and miR530 upregulated cycloartenol synthase (CAS1) sterol delta-7 reductase 1, CYP82G, zeatin o-glycosyl transferase (UGTs), and secoisolariciresinol dehydrogenase (ABA2) for synthesizing withanolide in ashwagandha plant [100]. Moreover, the tobacco plant synthesized glucosinolate with the help of miRNAs [53]. In Arabidopsis thaliana (L.) Heynh., miR5090 and miR826 proportion target AOP2 encrypting a 2-oxoglutarate-structured-dioxygenase, which is concerned in sulfur and nitrogen organic production [49, 101].
miRNAs had been observed from laboratory subculture of plant essential sections of the transcriptome of Withania somnifera (L.) Dunal in which miRNAs 5303, 159, 5140, and 172 are found in root tissues and miRNAs 5079, 530, 477, and 1426 are found in leaf tissues. Such miRNA is connected to the elongation of peripheral metabolites. Endogenetic mRNAs from roots (159 and 5140) and leaves (477 and 530) may be involved in the increase of metabolite production, although miR530 from leaves and miR172 and miR159 from roots were entangled in the balancing of peripheral metabolites connected with mRNAs [100]. Arabidopsis thaliana (L.) Heynh., Oryza sativa L., and Chlorophytum borivilianum Santapau & R.R.Fern.-targeted gene forecast imply that miR166, miR172, miR894, and miR9662 are probably concerned in enhancing the organic production of saponin [102]. miRNAs (5298b and 8154) raise phenylpropanoid, taxol, and flavonol organic synthesis in subcultured Taxus cells [103]. Salvia miltiorrhiza Bunge miRNA5072 targets acetyl-CoA which is concerned with the organic synthesis of tanshinone [78] (Table 4). miRNA826 targets hydroxyalkyl generating 2 oxoglutarate dioxygenases that is concerned with sulfur and nitrogen production [101].
Table 4.
Fatty acids, other compound biosynthesis, and miRNA.
| Botanical name | Common name | Family | miRNA | Target | Function | References |
|---|---|---|---|---|---|---|
| Lonicera japonica Thunb. | Honeysuckle | Caprifoliaceae | U436803 | Acyl-CoA synthetase, acyl carrier protein, fatty acid hydroxylase | Fatty acid biosynthesis | [2] |
| U977315 | ||||||
| U805963 | ||||||
| Paeonia × suffruticosa Andrews | Tree peony | Paeoniaceae | miR156 b | 1-Acyl-sn-glycerol-3-phosphate acyltransferase | Fatty acid biosynthesis | [98] |
| Corynespora cassiicola | Cucumber | Corynesporascaceae | Novel-miR1 | 4-Coumarate: CoA ligase, | Phenylpropanoid synthesis | [15] |
| Novel-miR1-novel-miR7 | Novel-miR2 | S-Adenosylmethionine decarboxylase | ||||
| Novel-miR3 | Peroxidase | |||||
| Novel-miR4 | Phosphoenolpyruvate carboxylase | |||||
| Novel-miR5 | Pleiotropic drug resistance protein | |||||
| Novel-miR6 | Xyloglucan endotransglucosylase/hydrolase family | |||||
| Novel-miR7 | Phenylalanine ammonia-lyase | |||||
| Podophyllum hexandrum Royle | May apple | Berberidaceae | miR2673a | MYB F1, WRKY37 | [99] | |
| miR396b | Flavonol synthase | |||||
| UDP glycosyltransferase | ||||||
| Withania somnifera (L.) Dunal | Ashwagandha | Solanaceae | miR5140 | Cycloartenol synthase (CAS1), sterol delta-7 reductase 1 | Withanolide biosynthesis | [100] |
| miR159 | CYP82G | |||||
| miR477 | Zeatin o-glycosyl transferase (UGTs) | |||||
| miR530 | Secoisolariciresinol dehydrogenase (ABA2) | |||||
| Nicotiana tabacum L. | Tobacco | Solanaceae | miRn24 | Branched-chain amino acid transaminase 3 (BCAT3) | Glucosinolate biosynthesis | [53] |
| Arabidopsis thaliana (L.) Heynh. | Thale cress | Brassicaceae | miR826 | Alkenyl hydroxyalkyl producing 2 (AOP2) | Transgenic approach | [49, 101] |
| miR5090∗ | ||||||
| Swertia chirayita (Roxb.) Buch.-Ham. ex C.B.Clarke | Chiretta | Gentianaceae | miR-168 | Acetyl-CoA acetyltransferase (AACT), aspartate aminotransferase (PHAT) premnaspirodiene oxygenase (PSO) | Secondary metabolites biosynthesis | [23] |
| miR-11320 miR166a | Ribulose-phosphate 3-epimerase (RPE) | |||||
| miR-11071 miR-156a miR-166b | Phosphoglycerate mutase (PGM) | |||||
| Gene encoding homeobox-leucine zipper protein (HD-ZIP) | ||||||
| Arabidopsis thaliana (L.) Heynh. | Thale cress | Brassicaceae | miR826 | Alkenyl hydroxyalkyl producing 2 (AOP2) | Glucosinolate biosynthesis | [49] |
| miR5090 | ||||||
| Solanum tuberosum L. | Potato | Solanaceae | miR6023, miR6024 and miR6027 | Alkaloid metabolism, UMP salvage, lipid biosynthesis, and cellulose catabolism; glycoalkaloid metabolism via JA signaling pathway | UDP-glucose biosynthesis | [34] |
| Salvia miltiorrhiza Bunge | Red sage | Lamiaceae | miR5072 | Acetyl-CoA | Tanshinones biosynthesis | [78] |
| C-acetyl transferase | ||||||
| Stevia rebaudiana (Bertoni) Bertoni | Candyleaf | Asteraceae | miR319g miRstv_11 | KO, UGT85C2, KS, KAH | Steviol glycoside biosynthesis | [83] |
| Picrorhiza kurroa Royle ex Benth. | Kutki | Plantaginaceae | miRNA-4995 | One enzyme involved in biosynthesis of terpenoids | Picroside-I (iridoid glycoside) | [73] |
7. Conclusion
miRNAs are microscopic particles related to growing functions that direct gene expression. The method assumed post-transcriptional and transductional procedures. miRNA secondary metabolism dominance is the latest kind of field and better information about the process of peripheral metabolism in plants will play a critical role in achieving new kinds of outcomes in controlled systems. These outcome by-products have good economic values due to their utilization in cosmetics product, food, agronomy, and other industries. Further investigations for the production of secondary metabolites by maneuvering the role of miRNA in various other crops such as spices, flowering plants, and medicinal plants may result in improving the profitability of the food and pharmaceutical industries.
Acknowledgments
We are very much thankful to the International Centre for Empirical Research and Development (ICERD), Bangladesh. We acknowledge the Bioinformatics Centre, Banasthali Vidyapith supported by DBT and DST for providing computation and networking support through the FIST and CURIE programs at the Department of Bioscience and Biotechnology, Banasthali Vidyapith, Rajasthan.
Abbreviations
- RISC:
Ribonucleoprotein RNA-induced silencing complex
- AACT:
Acetyl-CoA acetyltransferase
- PSO:
Premnaspirodiene oxygenase
- RPE:
Ribulose-phosphate 3-epimerase
- PGM:
Phosphoglycerate mutase
- HD-ZIP:
Omeobox-leucine zipper protein
- TMV:
Tobacco mosaic virus
- DCL1:
RNAase dicer-like 1
- HYL1:
Hyponastic leaves
- miRNAs:
MicroRNAs
- SE:
Serrate
- RNAPII:
RNA polymerase II
- HEN1:
HUA enhancer 1
- 3′ UTR:
3′ Untranslated region
- CDC5:
Cell division cycle 5
- SPL:
Squamosa promoter targeting protein like
- MyB:
Myeloblastosis
- DMAPP:
Dimethylallyl diphosphate
- IPP:
Isopentenyl diphosphate
- bAS:
Beta-amyrin synthase
- CASI:
Cycloartenol synthase
- QPT2:
Quinolinate phosphoribosyl-transferase 2
- PMT2:
Putrescine methyltransferase 2
- BIA:
Benzyliso quinoline alkaloids
- CyP82E4:
Cytochrome P450 monooxygenase
- QPT1:
Quinolinate Phosphoribosyltransferase 1.
Contributor Information
Javad Sharifi-Rad, Email: javad.sharifirad@gmail.com.
Daniela Calina, Email: calinadaniela@gmail.com.
Data Availability
The data supporting this review are from previously reported studies and datasets, which have been cited. The processed data are available from the corresponding author upon request.
Conflicts of Interest
The authors declared no conflict of interest.
Authors' Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis, and interpretation or in all these areas, that is, revising or critically reviewing the article; giving final approval of the version to be published one; agreeing on the journal to which the article has been submitted; and confirming to be accountable for all aspects of the work. All authors have read and agreed to the published version of the manuscript. Rajib Hossain and Divya Jain have contributed equally to this work.
References
- 1.Treiber T., Treiber N., Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nature Reviews Molecular Cell Biology . 2019;20(1):5–20. doi: 10.1038/s41580-018-0059-1. [DOI] [PubMed] [Google Scholar]
- 2.Liu J., Yuan Y., Wang Y., et al. Regulation of fatty acid and flavonoid biosynthesis by miRNAs in Lonicera japonica. RSC Advances . 2017;7(56):35426–35437. doi: 10.1039/C7RA05800D. [DOI] [Google Scholar]
- 3.Liu S.-R., Zhou J.-J., Hu C.-G., Wei C.-L., Zhang J.-Z. MicroRNA-mediated gene silencing in plant defense and viral counter-defense. Frontiers in Microbiology . 2017;8 doi: 10.3389/fmicb.2017.01801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melo C. A., Melo S. A. RNA Mapping . Springer; 2014. MicroRNA biogenesis: dicing assay; pp. 219–226. [DOI] [PubMed] [Google Scholar]
- 5.Millar A. A., Waterhouse P. M. Plant and animal microRNAs: similarities and differences. Functional & Integrative Genomics . 2005;5(3):129–135. doi: 10.1007/s10142-005-0145-2. [DOI] [PubMed] [Google Scholar]
- 6.Bruscella P., Bottini S., Baudesson C., Pawlotsky J.-M., Feray C., Trabucchi M. Viruses and miRNAs: more friends than foes. Frontiers in Microbiology . 2017;8:p. 824. doi: 10.3389/fmicb.2017.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.França G. S., Hinske L. C., Galante P. A., Vibranovski M. D. Unveiling the impact of the genomic architecture on the evolution of vertebrate microRNAs. Frontiers in Genetics . 2017;8:p. 34. doi: 10.3389/fgene.2017.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones-Rhoades M. W., Bartel D. P., Bartel B. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology . 2006;57(1):19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 9.Reinhart B. J., Weinstein E. G., Rhoades M. W., Bartel B., Bartel D. P. MicroRNAs in plants. Genes & Development . 2002;16(13):1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinrich M., Lardos A., Leonti M., et al. Best practice in research: consensus statement on ethnopharmacological field studies - ConSEFS. Journal of Ethnopharmacology . 2018;211:329–339. doi: 10.1016/j.jep.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 11.The Plant List. 2020. http://www.theplantlist.org/
- 12.Achkar N. P., Cambiagno D. A., Manavella P. A. miRNA biogenesis: a dynamic pathway. Trends in Plant Science . 2016;21(12):1034–1044. doi: 10.1016/j.tplants.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Eamens A., Wang M.-B., Smith N. A., Waterhouse P. M. RNA silencing in plants: yesterday, today, and tomorrow. Plant Physiology . 2008;147(2):456–468. doi: 10.1104/pp.108.117275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Brien J., Hayder H., Zayed Y., Peng C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Frontiers in Endocrinology . 2018;9:p. 402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Song X., Gu L., et al. NOT2 proteins promote polymerase II–dependent transcription and interact with multiple microRNA biogenesis factors in Arabidopsis. The Plant Cell . 2013;25(2):715–727. doi: 10.1105/tpc.112.105882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jagadeeswaran G., Zheng Y., Sumathipala N., et al. Deep sequencing of small RNA libraries reveals dynamic regulation of conserved and novel microRNAs and microRNA-stars during silkworm development. BMC Genomics . 2010;11:1–18. doi: 10.1186/1471-2164-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catalanotto C., Cogoni C., Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. International Journal of Molecular Sciences . 2016;17(10):p. 1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwakawa H.-O., Tomari Y. The functions of microRNAs: mRNA decay and translational repression. Trends in Cell Biology . 2015;25(11):651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Singh R. K., Gase K., Baldwin I. T., Pandey S. P. Molecular evolution and diversification of the Argonaute family of proteins in plants. BMC Plant Biology . 2015;15:1–16. doi: 10.1186/s12870-014-0364-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arribas-Hernández L., Marchais A., Poulsen C., et al. The slicer activity of ARGONAUTE1 is required specifically for the phasing, not production, of trans-acting short interfering RNAs in Arabidopsis. The Plant Cell . 2016;28(7):1563–1580. doi: 10.1105/tpc.16.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Djami-Tchatchou A. T., Sanan-Mishra N., Ntushelo K., Dubery I. A. Functional roles of microRNAs in agronomically important plants—potential as targets for crop improvement and protection. Frontiers in Plant Science . 2017;8:p. 378. doi: 10.3389/fpls.2017.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferrari C., Balandras F., Guedon E., Olmos E., Chevalot I., Marc A. Limiting cell aggregation during mesenchymal stem cell expansion on microcarriers. Biotechnology Progress . 2012;28(3):780–787. doi: 10.1002/btpr.1527. [DOI] [PubMed] [Google Scholar]
- 23.Sharifi-Rad J., Dey A., Koirala N., et al. Cinnamomum species: bridging phytochemistry knowledge, pharmacological properties and toxicological safety for health benefits. Frontiers in Pharmacology . 2021;12 doi: 10.3389/fphar.2021.600139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mahajan V., Mahajan A., Pagoch S., Bedi Y., Gandhi S. microRNA mediated regulation of plant secondary metabolism: an in silico analysis. Journal of Natural Science, Biology and Medicine . 2011;2 [Google Scholar]
- 25.Aljawish A., Chevalot I., Paris C., Muniglia L. Green synthesis of glyco-phenol by enzymatic coupling between ferulic acid and glucosamine: An ecofriendly procedure. Biotechnology and Applied Biochemistry . 2021 doi: 10.1002/bab.2215. [DOI] [PubMed] [Google Scholar]
- 26.Painuli S., Quispe C., Herrera-Bravo J., et al. Nutraceutical profiling, bioactive composition, and biological applications of Lepidium sativum L. Oxidative Medicine and Cellular Longevity . 2022;2022:20. doi: 10.1155/2022/2910411.2910411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hossain R., Quispe C., Herrera-Bravo J., et al. Lasia spinosa chemical composition and therapeutic potential: a literature-based review. Oxidative Medicine and Cellular Longevity . 2021;2021:12. doi: 10.1155/2021/1602437.1602437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharifi-Rad J., Quispe C., Herrera-Bravo J., et al. Phytochemical constituents, biological activities, and health-promoting effects of the Melissa officinalis. Oxidative Medicine and Cellular Longevity . 2021;2021:20. doi: 10.1155/2021/6584693.6584693 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Sharifi-Rad J., Quispe C., Imran M., et al. Genistein: an integrative overview of its mode of action, pharmacological properties, and health benefits. Oxidative Medicine and Cellular Longevity . 2021;2021:36. doi: 10.1155/2021/3268136.3268136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiwari R., Rana C. Oil price and exchange rate in Malaysia: a time-frequency analysis. International Journal of Engineering Research and General Science . 2015;5(4):661–670. doi: 10.18488/journal.aefr/2015.5.4/102.4.661.670. [DOI] [Google Scholar]
- 31.Gupta O. P., Karkute S. G., Banerjee S., Meena N. L., Dahuja A. Contemporary understanding of miRNA-based regulation of secondary metabolites biosynthesis in plants. Frontiers in Plant Science . 2017;8:p. 374. doi: 10.3389/fpls.2017.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sabzehzari M., Naghavi M. Phyto-miRNAs-based regulation of metabolites biosynthesis in medicinal plants. Gene . 2019;682:13–24. doi: 10.1016/j.gene.2018.09.049. [DOI] [PubMed] [Google Scholar]
- 33.Padhan J. K., Kumar P., Sood H., Chauhan R. S. Prospecting NGS-transcriptomes to assess regulation of miRNA-mediated secondary metabolites biosynthesis inSwertia chirayita, a medicinal herb of the North-Western Himalayas. Medicinal Plants-International Journal of Phytomedicines and Related Industries . 2016;8(3):219–228. doi: 10.5958/0975-6892.2016.00029.0. [DOI] [Google Scholar]
- 34.Qiao Y., Zhang J., Zhang J., et al. Integrated RNA-seq and sRNA-seq analysis reveals miRNA effects on secondary metabolism in Solanum tuberosum L. Molecular Genetics and Genomics . 2017;292(1):37–52. doi: 10.1007/s00438-016-1253-5. [DOI] [PubMed] [Google Scholar]
- 35.Jian H., Yang B., Zhang A., et al. Genome-wide identification of microRNAs in response to cadmium stress in oilseed rape (Brassica napus L.) using high-throughput sequencing. International Journal of Molecular Sciences . 2018;19(5):p. 1431. doi: 10.3390/ijms19051431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bazzini A. A., Manacorda C. A., Tohge T., et al. Metabolic and miRNA profiling of TMV infected plants reveals biphasic temporal changes. PLoS One . 2011;6(12, article e28466) doi: 10.1371/journal.pone.0028466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain D., Chaudhary P., Varshney N., et al. Tobacco smoking and liver cancer risk: potential avenues for carcinogenesis. Journal of Oncology . 2021;2021:11. doi: 10.1155/2021/5905357.5905357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salehi B., Quispe C., Chamkhi I., et al. Pharmacological properties of chalcones: a review of preclinical including molecular mechanisms and clinical evidence. Frontiers in Pharmacology . 2021;11 doi: 10.3389/fphar.2020.592654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Islam M. S., Quispe C., Hossain R., et al. Neuropharmacological effects of quercetin: a literature-based review. Frontiers in Pharmacology . 2021;12 doi: 10.3389/fphar.2021.665031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salehi B., Lopez-Jornet P., Pons-Fuster López E., et al. Plant-derived bioactives in oral mucosal lesions: a key emphasis to curcumin, lycopene, chamomile, aloe vera, green tea and coffee properties. Biomolecules . 2019;9(3):p. 106. doi: 10.3390/biom9030106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salehi B., Rescigno A., Dettori T., et al. Avocado–soybean unsaponifiables: a panoply of potentialities to be exploited. Biomolecules . 2020;10(1):p. 130. doi: 10.3390/biom10010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharifi-Rad J., Quispe C., Shaheen S., et al. Flavonoids as potential anti-platelet aggregation agents: from biochemistry to health promoting abilities. Critical Reviews in Food Science and Nutrition . 2021;13:1–14. doi: 10.1080/10408398.2021.1924612. [DOI] [PubMed] [Google Scholar]
- 43.Salehi B., Shivaprasad S. M. V. A. K. N., V Anil Kumar N., et al. Veronica plants-drifting from farm to traditional healing, food application, and Phytopharmacology. Molecules . 2019;24(13):p. 2454. doi: 10.3390/molecules24132454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salehi B., Sharifi-Rad J., Capanoglu E., et al. Cucurbita plants: from farm to industry. Applied Sciences . 2019;9(16):p. 3387. doi: 10.3390/app9163387. [DOI] [Google Scholar]
- 45.Diwan A., Ninawe A., Harke S. Gene editing (CRISPR-Cas) technology and fisheries sector. Canadian Journal of Biotechnology . 2017;1(2):65–72. doi: 10.24870/cjb.2017-000108. [DOI] [Google Scholar]
- 46.Barozai M. Y. K., Baloch I. A., Din M. Identification of microRNAs and their targets in helianthus. Molecular Biology Reports . 2012;39(3):2523–2532. doi: 10.1007/s11033-011-1004-y. [DOI] [PubMed] [Google Scholar]
- 47.Legrand S., Valot N., Nicolé F., et al. One-step identification of conserved miRNAs, their targets, potential transcription factors and effector genes of complete secondary metabolism pathways after 454 pyrosequencing of calyx cDNAs from the Labiate Salvia sclarea L. Gene . 2010;450(1-2):55–62. doi: 10.1016/j.gene.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 48.Singh N., Sharma A. Turmeric (Curcuma longa): miRNAs and their regulating targets are involved in development and secondary metabolite pathways. Comptes Rendus Biologies . 2017;340(11-12):481–491. doi: 10.1016/j.crvi.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 49.He H., Liang G., Li Y., Wang F., Yu D. Two young microRNAs originating from target duplication mediate nitrogen starvation adaptation via regulation of glucosinolate synthesis in Arabidopsis thaliana. Plant Physiology . 2014;164(2):853–865. doi: 10.1104/pp.113.228635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li F., Wang W., Zhao N., et al. Regulation of nicotine biosynthesis by an endogenous target mimicry of microRNA in tobacco. Plant Physiology . 2015;169(2):1062–1071. doi: 10.1104/pp.15.00649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo Y., Zhang X., Luo Z., Zhang Q., Liu J. Identification and characterization of microRNAs from Chinese pollination constant non-astringent persimmon using high-throughput sequencing. BMC Plant Biology . 2015;15:1–18. doi: 10.1186/s12870-014-0400-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang R., Zeng Y., Yi X., Zhao L., Zhang Y. Small RNA deep sequencing reveals the important role of microRNAs in the halophyte Halostachys caspica. Plant Biotechnology Journal . 2015;13(3):395–408. doi: 10.1111/pbi.12337. [DOI] [PubMed] [Google Scholar]
- 53.Gou J.-Y., Felippes F. F., Liu C.-J., Weigel D., Wang J.-W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. The Plant Cell . 2011;23(4):1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma D., Tiwari M., Pandey A., Bhatia C., Sharma A., Trivedi P. K. MicroRNA858 is a potential regulator of phenylpropanoid pathway and plant development. Plant Physiology . 2016;171(2):944–959. doi: 10.1104/pp.15.01831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prakash P., Rajakani R., Gupta V. Transcriptome-wide identification of Rauvolfia serpentina microRNAs and prediction of their potential targets. Computational Biology and Chemistry . 2016;61:62–74. doi: 10.1016/j.compbiolchem.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 56.Biswas S., Hazra S., Chattopadhyay S. Identification of conserved miRNAs and their putative target genes in Podophyllum hexandrum (Himalayan Mayapple) Plant Gene . 2016;6:82–89. doi: 10.1016/j.plgene.2016.04.002. [DOI] [Google Scholar]
- 57.Singh N., Srivastava S., Sharma A. Identification and analysis of miRNAs and their targets in ginger using bioinformatics approach. Gene . 2016;575(2):570–576. doi: 10.1016/j.gene.2015.09.036. [DOI] [PubMed] [Google Scholar]
- 58.Tuteja J. H., Zabala G., Varala K., Hudson M., Vodkin L. O. Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in Glycine max seed coats. The Plant Cell . 2009;21(10):3063–3077. doi: 10.1105/tpc.109.069856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho Y. B., Jones S. I., Vodkin L. The transition from primary siRNAs to amplified secondary siRNAs that regulate chalcone synthase during development of Glycine max seed coats. PLoS One . 2013;8(10, article e76954) doi: 10.1371/journal.pone.0076954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu J., Wang D., Liu Y., Wang L., Qiao X., Zhang S. Identification of miRNAs involved in pear fruit development and quality. BMC Genomics . 2014;15:1–19. doi: 10.1186/1471-2164-15-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun Y., Qiu Y., Duan M., et al. Identification of anthocyanin biosynthesis related microRNAs in a distinctive Chinese radish (Raphanus sativus L.) by high-throughput sequencing. Molecular Genetics and Genomics . 2017;292(1):215–229. doi: 10.1007/s00438-016-1268-y. [DOI] [PubMed] [Google Scholar]
- 62.Tirumalai V., Swetha C., Nair A., Pandit A., Shivaprasad P. V. miR828 and miR858 regulate VvMYB114 to promote anthocyanin and flavonol accumulation in grapes. Journal of Experimental Botany . 2019;70(18):4775–4792. doi: 10.1093/jxb/erz264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shi Q., Hui S., Zhang A.-H., et al. Natural alkaloids: basic aspects, biological roles, and future perspectives. Chinese Journal of Natural Medicines . 2014;12:401–406. doi: 10.1016/S1875-5364(14)60063-7. [DOI] [PubMed] [Google Scholar]
- 64.Sun P., Cheng C., Lin Y., Zhu Q., Lin J., Lai Z. Combined small RNA and degradome sequencing reveals complex microRNA regulation of catechin biosynthesis in tea (Camellia sinensis) PLoS One . 2017;12(2, article e0171173) doi: 10.1371/journal.pone.0171173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao S., Wang X., Yan X., et al. Revealing of microRNA involved regulatory gene networks on terpenoid biosynthesis in Camellia sinensis in different growing time points. Journal of Agricultural and Food Chemistry . 2018;66(47):12604–12616. doi: 10.1021/acs.jafc.8b05345. [DOI] [PubMed] [Google Scholar]
- 66.Singh N., Srivastava S., Shasany A. K., Sharma A. Identification of miRNAs and their targets involved in the secondary metabolic pathways of Mentha spp. Computational Biology and Chemistry . 2016;64:154–162. doi: 10.1016/j.compbiolchem.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 67.Salehi B., Sharifi-Rad J., Cappellini F., et al. The therapeutic potential of anthocyanins: current approaches based on their molecular mechanism of action. Frontiers in Pharmacology . 2020;11 doi: 10.3389/fphar.2020.01300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharifi-Rad J., Quispe C., Herrera-Bravo J., et al. A pharmacological perspective on plant-derived bioactive molecules for epilepsy. Neurochemical Research . 2021;46(9):2205–2225. doi: 10.1007/s11064-021-03376-0. [DOI] [PubMed] [Google Scholar]
- 69.Li Y., Cui W., Wang R., et al. MicroRNA858-mediated regulation of anthocyanin biosynthesis in kiwifruit (Actinidia arguta) based on small RNA sequencing. PLoS One . 2019;14(5, article e0217480) doi: 10.1371/journal.pone.0217480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gross G. G., Zenk M. H. Isolation and properties of hydroxycinnamate: CoA ligase from lignifying tissue of Forsthia. European Journal of Biochemistry . 1974;42(2):453–459. doi: 10.1111/j.1432-1033.1974.tb03359.x. [DOI] [PubMed] [Google Scholar]
- 71.Zhang Y., Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evolutionary Biology . 2005;5(1):1–12. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pérez-Quintero Á. L., Sablok G., Tatarinova T. V., Conesa A., Kuo J., López C. Mining of miRNAs and potential targets from gene oriented clusters of transcripts sequences of the anti-malarial plant, Artemisia annua. Biotechnology Letters . 2012;34(4):737–745. doi: 10.1007/s10529-011-0808-0. [DOI] [PubMed] [Google Scholar]
- 73.Vashisht I., Mishra P., Pal T., Chanumolu S., Singh T. R., Chauhan R. S. Mining NGS transcriptomes for miRNAs and dissecting their role in regulating growth, development, and secondary metabolites production in different organs of a medicinal herb, Picrorhiza kurroa. Picrorhiza kurroa. Planta . 2015;241(5):1255–1268. doi: 10.1007/s00425-015-2255-y. [DOI] [PubMed] [Google Scholar]
- 74.Pani A., Mahapatra R. K. Computational identification of microRNAs and their targets in Catharanthus roseus expressed sequence tags. Genomics Data . 2013;1:2–6. doi: 10.1016/j.gdata.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fan R., Li Y., Li C., Zhang Y. Differential microRNA analysis of glandular trichomes and young leaves in Xanthium strumarium L. reveals their putative roles in regulating terpenoid biosynthesis. PLoS One . 2015;10(9, article e0139002) doi: 10.1371/journal.pone.0139002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Najafabadi A. S., Naghavi M. R. Mining Ferula gummosa transcriptome to identify miRNAs involved in the regulation and biosynthesis of terpenes. Gene . 2018;645:41–47. doi: 10.1016/j.gene.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 77.Yu Z.-X., Wang L.-J., Zhao B., et al. Progressive regulation of sesquiterpene biosynthesis in Arabidopsis and Patchouli (Pogostemon cablin) by the miR156-targeted SPL transcription factors. Molecular Plant . 2015;8(1):98–110. doi: 10.1016/j.molp.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 78.Xu X., Jiang Q., Ma X., et al. Deep sequencing identifies tissue-specific microRNAs and their target genes involving in the biosynthesis of tanshinones in Salvia miltiorrhiza. PLoS One . 2014;9(11, article e111679) doi: 10.1371/journal.pone.0111679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mathiyalagan R., Subramaniyam S., Natarajan S., et al. Insilico profiling of microRNAs in Korean ginseng (Panax ginsenggenes involving in the biosynthesis of ta Meyer) Journal of Ginseng Research . 2013;37(2):227–247. doi: 10.5142/jgr.2013.37.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Samad A. F. A., Rahnamaie-Tajadod R., Sajad M., et al. Regulation of terpenoid biosynthesis by miRNA in Persicaria minor induced by Fusarium oxysporum. BMC Genomics . 2019;20:1–22. doi: 10.1186/s12864-019-5954-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ye J., Zhang X., Tan J., et al. Global identification of Ginkgo biloba microRNAs and insight into their role in metabolism regulatory network of terpene trilactones by high-throughput sequencing and degradome analysis. Industrial Crops and Products . 2020;148:p. 112289. doi: 10.1016/j.indcrop.2020.112289. [DOI] [Google Scholar]
- 82.Khan S., Ali A., Saifi M., Saxena P., Ahlawat S., Abdin M. Z. Identification and the potential involvement of miRNAs in the regulation of artemisinin biosynthesis in A. annua. Scientific Reports . 2020;10:1–11. doi: 10.1038/s41598-020-69707-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saifi M., Nasrullah N., Ahmad M. M., Ali A., Khan J. A., Abdin M. In silico analysis and expression profiling of miRNAs targeting genes of steviol glycosides biosynthetic pathway and their relationship with steviol glycosides content in different tissues of Stevia rebaudiana. Plant Physiology and Biochemistry . 2015;94:57–64. doi: 10.1016/j.plaphy.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 84.Sharifi-Rad J., Kamiloglu S., Yeskaliyeva B., et al. Pharmacological activities of Psoralidin: a comprehensive review of the molecular mechanisms of action. Frontiers in Pharmacology . 2020;11 doi: 10.3389/fphar.2020.571459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kittakoop P., Mahidol C., Ruchirawat S. Alkaloids as important scaffolds in therapeutic drugs for the treatments of cancer, tuberculosis, and smoking cessation. Current Topics in Medicinal Chemistry . 2014;14(2):239–252. doi: 10.2174/1568026613666131216105049. [DOI] [PubMed] [Google Scholar]
- 86.Russo P., Frustaci A., Del Bufalo A., Fini M., Cesario A. Multitarget drugs of plants origin acting on Alzheimer’s disease. Current Medicinal Chemistry . 2013;20(13):1686–1693. doi: 10.2174/0929867311320130008. [DOI] [PubMed] [Google Scholar]
- 87.Sinatra R. S., Jahr J. S., Watkins-Pitchford J. M. The essence of analgesia and analgesics . Cambridge University Press; 2010. [DOI] [Google Scholar]
- 88.Cushnie T. T., Cushnie B., Lamb A. J. Alkaloids: an overview of their antibacterial, antibiotic-enhancing and antivirulence activities. International Journal of Antimicrobial Agents . 2014;44(5):377–386. doi: 10.1016/j.ijantimicag.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 89.Ziegler J., Facchini P. J. Alkaloid biosynthesis: metabolism and trafficking. Annual Review of Plant Biology . 2008;59(1):735–769. doi: 10.1146/annurev.arplant.59.032607.092730. [DOI] [PubMed] [Google Scholar]
- 90.Roy A. A review on the alkaloids an important therapeutic compound from plants. IJPB . 2017;3:1–9. [Google Scholar]
- 91.Fu Y., Guo H., Cheng Z., et al. NtNAC-R1, a novel NAC transcription factor gene in tobacco roots, responds to mechanical damage of shoot meristem. Plant Physiology and Biochemistry . 2013;69:74–81. doi: 10.1016/j.plaphy.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 92.Boke H., Ozhuner E., Turktas M., Parmaksiz I., Ozcan S., Unver T. Regulation of the alkaloid biosynthesis by miRNA in opium poppy. Plant Biotechnology Journal . 2015;13(3):409–420. doi: 10.1111/pbi.12346. [DOI] [PubMed] [Google Scholar]
- 93.Hao D. C., Yang L., Xiao P. G., Liu M. Identification of Taxus microRNAs and their targets with high-throughput sequencing and degradome analysis. Physiologia Plantarum . 2012;146(4):388–403. doi: 10.1111/j.1399-3054.2012.01668.x. [DOI] [PubMed] [Google Scholar]
- 94.Sharifi-Rad J., Quispe C., Patra J. K., et al. Paclitaxel: application in modern oncology and nanomedicine-based cancer therapy. Oxidative Medicine and Cellular Longevity . 2021;2021:24. doi: 10.1155/2021/3687700.3687700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shen E. M., Singh S. K., Ghosh J. S., et al. The miRNAome of Catharanthus roseus: identification, expression analysis, and potential roles of microRNAs in regulation of terpenoid indole alkaloid biosynthesis. Scientific Reports . 2017;7:1–13. doi: 10.1038/srep43027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robert-Seilaniantz A., MacLean D., Jikumaru Y., et al. The microRNA miR393 re-directs secondary metabolite biosynthesis away from camalexin and towards glucosinolates. The Plant Journal . 2011;67(2):218–231. doi: 10.1111/j.1365-313X.2011.04591.x. [DOI] [PubMed] [Google Scholar]
- 97.Salehi B., Prakash Mishra A., Nigam M., et al. Ficusravel candidate miRNAs and their a plants: state of the art from a phytochemical, pharmacological, and toxicological perspective. Phytotherapy Research . 2021;35(3):1187–1217. doi: 10.1002/ptr.6884. [DOI] [PubMed] [Google Scholar]
- 98.Yin D.-D., Li S.-S., Shu Q.-Y., et al. Identification of microRNAs and long non-coding RNAs involved in fatty acid biosynthesis in tree peony seeds. Gene . 2018;666:72–82. doi: 10.1016/j.gene.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 99.Kumar P., Padhan J. K., Kumar A., Chauhan R. S. Transcriptomes of Podophyllum hexandrum unravel candidate miRNAs and their association with the biosynthesis of secondary metabolites. Journal of Plant Biochemistry and Biotechnology . 2018;27(1):46–54. doi: 10.1007/s13562-017-0414-x. [DOI] [Google Scholar]
- 100.Srivastava S., Singh R., Srivastava G., Sharma A. Comparative study of Withanolide biosynthesis-related miRNAs in root and leaf tissues of Withania somnifera. Applied Biochemistry and Biotechnology . 2018;185(4):1145–1159. doi: 10.1007/s12010-018-2702-x. [DOI] [PubMed] [Google Scholar]
- 101.Liang G., He H., Yu D. Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS One . 2012;7(11, article e48951) doi: 10.1371/journal.pone.0048951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kajal M., Singh K. Small RNA profiling for identification of miRNAs involved in regulation of saponins biosynthesis in Chlorophytum borivilianum. BMC Plant Biology . 2017;17(1):1–14. doi: 10.1186/s12870-017-1214-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang M., Dong Y., Nie L., Lu M., Fu C., Yu L. High-throughput sequencing reveals miRNA effects on the primary and secondary production properties in long-term subcultured Taxus cells. Frontiers in Plant Science . 2015;6:p. 604. doi: 10.3389/fpls.2015.00604. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting this review are from previously reported studies and datasets, which have been cited. The processed data are available from the corresponding author upon request.


