Abstract
The malaria hypothesis predicts local, balancing selection of deleterious alleles that confer strong protection from malaria. Three protective variants recently discovered in red cell genes are indeed more common in African than European populations. Still, up to 89% of the heritability of severe malaria is attributed to many genome-wide loci with individually small effects. Recent analyses of hundreds of genome-wide association studies (GWAS) in humans suggest that most functional, polygenic variation is pleiotropic for multiple traits. Interestingly, GWAS alleles and red cell traits associated with small reductions in malaria risk are not enriched in African populations. We propose that other selective and neutral forces, in addition to malaria prevalence, explain the global distribution of most genetic variation impacting malaria risk.
Keywords: malaria, red blood cells, evolution, polygenic, pleiotropy
The malaria hypothesis predicts strong selection on protective variants of large effect
Malaria caused by the parasite Plasmodium falciparum has caused hundreds of millions of deaths over the course of human history. Consequently, even deleterious genetic variants that limit malaria severity are expected to be maintained by balancing selection (see Glossary) within malaria-endemic populations. This “malaria hypothesis” is supported by geographical patterns of monogenic disorders of red blood cells (RBCs) that confer protection against severe malaria (Box 1). For example, the sickle cell allele of hemoglobin beta (HbS) arose and is maintained in sub-Saharan Africa [1], where the vast majority of malaria deaths occur [2]. Sickle cell disease and other RBC disorders (Box 1) provide compelling evidence that malaria has imposed strong selective pressures on human populations throughout the world [3]. Here we focus on selection in Africa from P. falciparum, the deadliest malaria parasite, although RBC disorders and Plasmodium species are found in tropical regions worldwide.
Box 1. History of the malaria hypothesis.
The malaria hypothesis offers an explanation for the high prevalence of hematologic disease in malaria-endemic areas. It is frequently attributed to the population geneticist JBS Haldane, who in the late 1940s, recognized that the prevalence of thalassemia in the Mediterranean was far higher than could be explained by mutation alone [65]. Thalassemia is a life-threatening anemia caused by a homozygous deletion of one of the genes encoding hemoglobin. Haldane proposed that the causal allele could be maintained within a population, despite its fitness cost in homozygotes, by offering a fitness benefit to heterozygotes—a phenomenon now known as balancing selection. RBCs of heterozygous carriers of thalassemia are unusually small and resistant to hypotonic solutions, so Haldane reasoned that they might also be resistant to Plasmodium parasites. Thalassemia has since been associated with reduced clinical malaria [3] and slower P. falciparum growth in vitro [15], although the full mechanism of protection remains unknown.
Also in the mid-twentieth century, the South African scientist AC Allison compiled extensive epidemiological data for sickle cell trait and malaria in Africa [66]. Similar to thalassemia, sickle cell disease is a life-threatening anemia caused by a homozygous point mutation in the hemoglobin beta gene. Heterozygous carriers lack serious symptoms, though they may have clinically abnormal RBCs. Allison recorded a much higher prevalence of sickle cell trait in malaria-hyperendemic areas of East Africa. In 1954, he experimentally confirmed his own malaria hypothesis by infecting 30 native Luo men with P. falciparum parasites and charting the course of their disease for forty days [67]. His study provided strong evidence of the anti-malarial effects of HbS, but it included no record of the informed consent process, if any such process existed [68]. HbS appears to offer protection via hemoglobin polymerization in low oxygen conditions, leading to parasite growth arrest [69].
The malaria hypothesis is also generally accepted to explain the prevalence of Hemoglobin C disease, Hemoglobin E disease, Southeast Asian Ovalocytosis, and G6PD deficiency [70–73]. In the first three diseases, the heterozygous state provides a clinical advantage against malaria while the rare homozygous state is strongly deleterious. G6PD deficiency, in contrast, involves several common alleles with complex effects on malaria and metabolism [71]. Anemia can be induced in G6PD-deficient individuals by fava beans, infection, and other factors, though most carriers have a mild deficiency with no symptoms.
The malaria hypothesis is based on RBC disorders with simple genetic causes (Box 1), but it is sometimes extended to polygenic traits like RBC volume, RBC membrane fragility, and malaria resistance itself [4–9]. Here, we explore whether the simple predictions of the malaria hypothesis can be easily generalized to complex traits. Simultaneous analyses of hundreds of genome-wide association studies (GWAS) have suggested that most functional variants in humans have small effects and are pleiotropic, or associated with more than one phenotype. We hypothesize that complex traits related to malaria may not conform to the malaria hypothesis—even when several large-effect alleles do—because of pleiotropy and selective constraint on genetically correlated traits. Furthermore, because most common variants have a global distribution [10], we expect that most alleles that impact malaria are present in all human populations (Box 2). An overly narrow focus on the malaria hypothesis may distract from progress on the complex biological pathways that contribute to the pathogenesis of severe malaria.
Box 2. Genetic structure and similarity of all human populations.
Most common genetic variation present in humans today is descended from populations living in Africa 100 to 250 thousand years ago (ka) [74]. Between 40 and 60 ka, a population carrying a subset of this African diversity expanded throughout the rest of the world. As they migrated, human populations experienced a series of bottlenecks that reduced their genetic diversity relative to their distance from Africa [75]. Some populations of Homo sapiens mixed with Neanderthals and Denisovans [74], and all populations experienced mutation, recombination, drift, and selection on their unique genetic backgrounds. Genetic variation is clearly structured across human populations (Figure I), primarily by geography and demography [76].
Human population structure allows the regional ancestry of an individual to be inferred from a sufficiently large panel of genetic markers (Figure I). For example, around 1.3 million SNPs are common across Africans but rare or undetected in the rest of the world. Since much of this population structure was created by the loss of SNPs in out-of-Africa bottlenecks, it is unlikely that most of these SNPs are strongly related to malaria.
Furthermore, the majority of common genetic variants in humans are shared among all major populations [10] (Figure I), due to our fairly recent common ancestor. This fact was first demonstrated in 1972 using red blood cell genes [77], including several associated with Plasmodium. In terms of variance partitioning, a large majority (~94%) of human genetic diversity is found within populations, as opposed to across them [75]. In fact, if we were to randomly sample two members of different populations, they would have on average only ~5% more alleles that differ than two members of the same population [75].
Despite sharing many alleles inherited from common ancestors, human populations obviously do exhibit differences in phenotypes and allele frequencies. Understanding the extent of local, balancing selection in shaping these differences requires an appropriate neutral (demographic) model; recognition of environmental factors; and knowledge of genetically correlated traits [41,78].
Figure I. Most common human alleles have a broad geographic distribution.
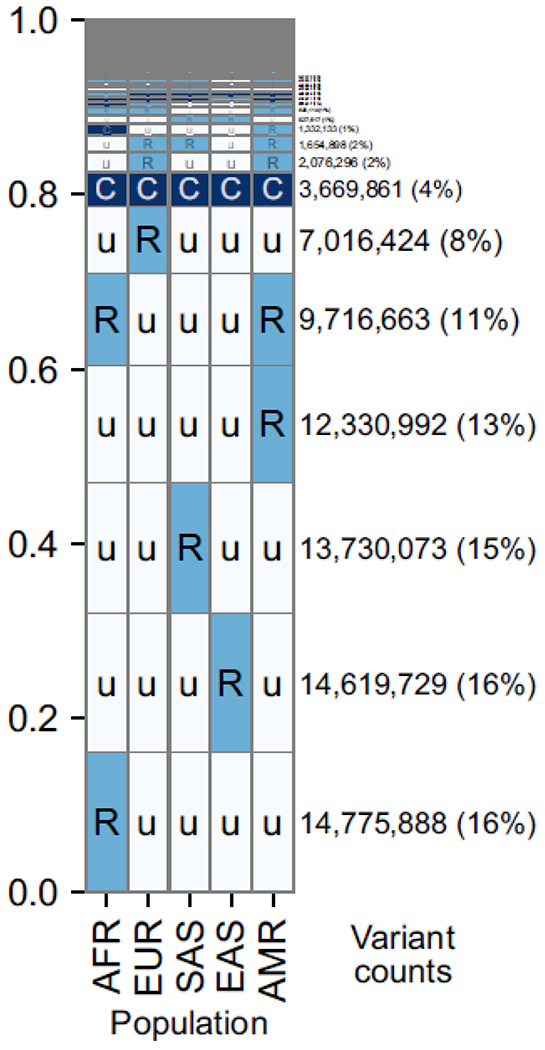
Data are summarized across ~92 million bi-allelic variants from 1000 Genomes. Within each cell, C = common (>5% frequency), R = rare (<5%), u= unobserved (0%). The number and proportion of variants with a given geographical distribution is indicated by the text on the right, as well as the height of the row. AFR=African, EUR=European, SAS=South Asian, EAS=East Asian, AMR=Admixed American. Reproduced from [10].
Three novel alleles with large protective effects conform to the malaria hypothesis
In the last five years, genetic variants at three RBC loci have been associated with strong protection from severe malaria and replicated in independent studies (Table 1). The first locus, ATP2B4, encodes the main calcium transporter of RBCs. An ATP2B4 haplotype associated with decreased risk of severe malaria [7] was recently linked to decreased calcium export and increased dehydration in RBCs [7,11–14]. RBC dehydration may directly contribute to malaria resistance by decreasing P. falciparum growth rate in RBCs [15,16]. Similarly, PIEZO1 encodes a mechanosensitive ion channel implicated in xerocytosis, a rare disease characterized by extreme RBC dehydration [6]. The microsatellite variant PIEZO1 E756del has been associated with strong protection from severe malaria [17], reduced P. falciparum growth [6,15], and mild RBC dehydration [6,18], although these phenotypes have not been replicated in all studies [15,17,19,20]. Genes at the glycophorin locus (GYPA/GYPB) encode red cell surface receptors coated with sialic acid, which facilitate parasite invasion by specific binding to P. falciparum ligands [21]. The Dantu allele at this locus encodes a hybrid protein in which the extracellular domain of GYPB is fused to the transmembrane and intracellular domains of GYPA [22]. Interestingly, the protective effect of Dantu against severe malaria is mediated not through receptor-ligand interactions, but through an increase in RBC membrane tension that inhibits P. falciparum invasion [23]. For all three of these loci, the same polymorphism that is associated with strong protection from clinical malaria [7,17,22] is also associated with RBC trait variation [6,11,14,15,23] and impaired P. falciparum development in vitro [6,15,17,23].
Table 1.
Novel alleles associated with strong protection from severe malaria.
| Gene | Protective variant | Malaria phenotype in vivoa | Possible protective mechanisms | Other associated phenotypes | Geographic distribution of allele frequency | References |
|---|---|---|---|---|---|---|
| ATP2B4/PMC4a (calcium channel) | Haplotype with >50 variants, including one disrupting a GATA binding site | 23-35% protection against severe malaria and placental malaria | Reduction of parasitemia in vivo; reduction of P. falciparum growth in vitro; RBC dehydration; additional pleiotropic effects | Decreased transcript and protein expression; decreased calcium export; increased MCHCb; increased HGBC; decreased RDWd; decreased reticulocytes | African: 35% European: 11% South Asian: 15% East Asian: 3% Admixed American: 11% (chr1:20368 4896-T-G) | [7,11–15,43,79,80] |
| PIEZO1 (mechanose nsitive ion channel) | E756del | 50% protection against severe malaria in a Gabonese population | Variable reduction of P. falciparum growth in vitro. Some studies suggest RBC dehydration, but others find no association. Also associated with reduced var2csa expression | Macrophage-mediated iron overload; increased jumping performance | African: 17% European: <0.06% South Asian: <0.2% East Asian: <0.08% Admixed American: 1.5% (chr16:8873 3964-GTCC-G) | [6,15,17–19,27,28,79,79] |
| GYPA/GYPB hybrid (RBC membrane sialoglycoprotein) | Haplotype of a duplicated chimeric protein known as the Dantu antigen | 33-40% protection against severe malaria | Significant reduction of P. falciparum invasion and growth in vitro; partially mediated by increased RBC membrane tension | Reduced MCVe and MCHf in hetero zygotes and especially homozygotes; substantial changes to expression of RBC membrane proteome | Absent from most of Africa and the rest of the world, but present at 2-13% frequency in parts of East Africa | [22–24,80] |
Based on odds ratio for the broadest category of severe malaria in an additive or recessive association model with one copy of the allele.
mean corpuscular hemoglobin concentration (g/dL red blood cells)
hemoglobin (g/dL whole blood)
red blood cell distribution width (% coefficient of variation)
mean corpuscular volume of red blood cells (fL)
mean cell hemoglobin of red blood cells (pg)
All three protective variants in Table 1 occur at a higher frequency in Africa than Europe, consistent with the malaria hypothesis. PIEZO1 E756del is found in about one third of Africans in 1000 Genomes, which motivated its initial selection for detailed study [6]. The Dantu variant of GYPA/GYPB has a frequency of 2-13% in some parts of East Africa, but is absent from most of Africa and the rest of the world [22,24]. The protective ATP2B4 haplotype, like variation in the ABO blood group, is found at appreciable frequency (~11-15%) in Europe and South Asia and higher frequency (~35%) across Africa.
The degree of protection observed for these variants in vitro has been smaller than in clinical studies, which has been interpreted as evidence of pleiotropic effects through additional tissues or phenotypes [7,15,17]. For example, the protective ATP2B4 haplotype is associated with up to 35% reduction in severe malaria risk, but only ~6% slower growth of P. falciparum in RBCs (Table 1). The same haplotype is associated by GWAS with lymphocyte counts, in addition to RBC dehydration and size [25], which may contribute to its larger effect on severe malaria. However, part of the difference between GWAS and in vitro models might be explained by limitations of current methods of parasite culture. P. falciparum is typically cultured within RBCs in rich media and at low density, independent of immune cells and normal circulation. These conditions necessarily represent only a fraction of the host biology involved in malaria disease. Furthermore, lab culture conditions are known to select for deleterious parasite traits, including the loss of gametocytogenesis [26].
Similar to the pleiotropic effects of ATP2B4, PIEZO1 E756del has been linked to macrophage-mediated iron metabolism and athletic jumping performance [27,28] in addition to RBC phenotypes. Both ATP2B4 and PIEZO1 are widely expressed throughout the body, so variants impacting their function could potentially be pleiotropic through immune cells, the vasculature, or other tissue types. Glycophorin proteins are expressed exclusively on RBCs, but the Dantu variant is associated with altered expression of 100 RBC surface proteins [23] that could be pleiotropic for interactions with immune cells or other issues. The glycophorins and PIEZO1 are both exceptionally polymorphic among RBC genes [15,22,24], with growing GWAS evidence linking multiple variants to multiple traits [25]. Glycophorin loci are also thought to be ancient sites of positive and balancing selection, perhaps because of their roles in several infectious diseases [22,29,30].
Overall, it is plausible that the higher frequencies of protective alleles in ATP2B4, PIEZO1, and GYPA/GYPB in sub-Saharan Africa, compared to the rest of the world, have been driven by selection consistent with the malaria hypothesis (Box 1). Understanding the degree of protection that can be attributed to red cells, as opposed to other tissue types, will require more functional studies. Given the available evidence of pleiotropy (Table 1), it is also possible that selective forces besides malaria have contributed to the observed frequency variation. Discovery and characterization of additional traits linked to these loci may aid in the design of novel treatments and preventative therapies for malaria.
Malaria susceptibility is highly polygenic
The largest GWAS of severe malaria to date sampled more than 17,000 individuals across 11 countries and 3 continents [7]. It successfully identified five genome-wide significant variants—in the genes HBB, GYPA/GYPB, ATP2B4, ABO, and an intergenic region on chromosome 6—that collectively explain ~11 % of the heritability of severe malaria risk. Aside from the identities of these specific variants, then, a major GWAS result is that the majority of heritability cannot yet be attributed to specific genomic loci (Figure 1A, Key Figure).
Figure 1, Key Figure. The main polygenic component of malaria resistance is not enriched in African populations.
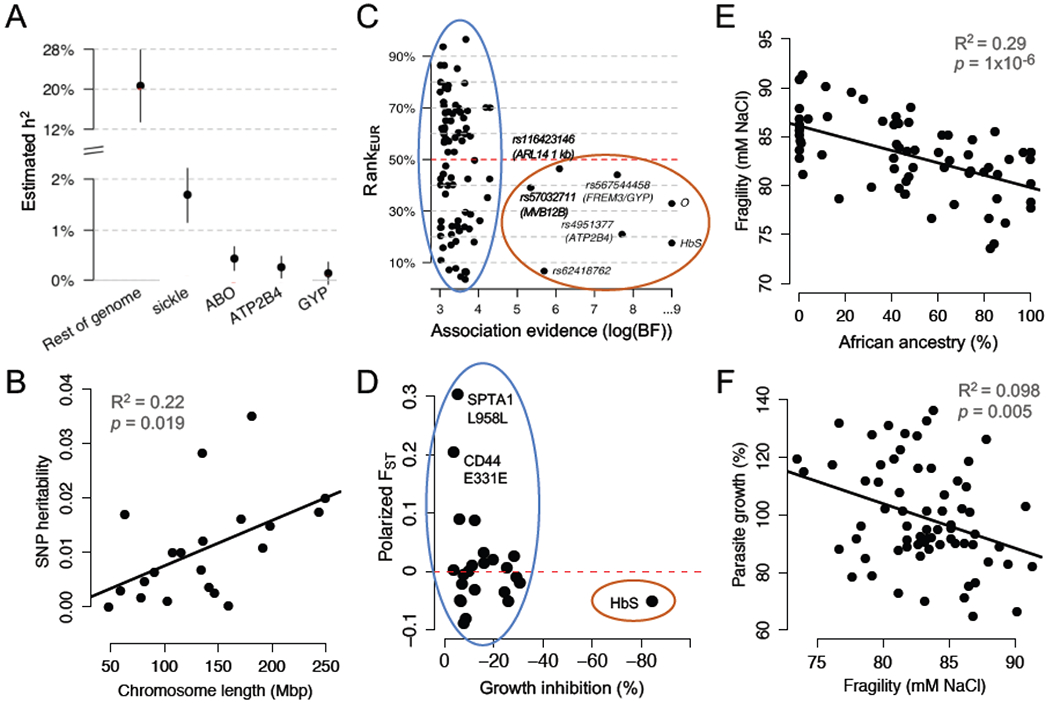
(A) 89% of the estimated heritability of severe malaria risk is attributed to variants with effects too small to be confidently detected by GWAS. Reproduced from [7]. (B) SNP-heritability of severe malaria risk is distributed across chromosomes proportional to their length. Replotted from [36]. (C) Strongly associated (large-effect) alleles from a GWAS for severe malaria risk occur at higher frequencies in African populations, compared to European populations (red oval). Smaller-effect alleles, which are less confidently associated, are equally likely to be more frequent in European populations (blue oval). RankEUR is the conditional rank of the allele in European populations, given African allele count; values greater than/less than 50% indicate that the allele is more common in Europe/Africa. Reproduced from [7]. (D) Alleles from healthy RBCs associated with reduced P. falciparum growth rate in vitro are not enriched for higher frequencies in African populations (blue oval). Polarized FST indicates the differentiation in allele frequencies between African and European populations in 1000 Genomes; values greater than/less than 0 indicate that the allele is more common in Europe/Africa. Replotted from [15]. (E) In donors who lack large-effect alleles for malaria resistance, red cell osmotic fragility is negatively related to exome-wide African ancestry. Replotted from [15]. (F) In donors who lack large-effect alleles for malaria resistance, higher red cell osmotic fragility predicts slower P. falciparum growth in vitro. Replotted from [15]. Panels A and C are reproduced with minor modifications under a Creative Commons license (https://creativecommons.org/licenses/by/4.0/).
This “missing heritability” has a number of potential explanations that have been reviewed recently elsewhere [31,32]. As one example, GWAS have found variable associations between HbS and severe malaria because of variation in technical design (e.g. genotyping method) and environmental factors (e.g. local parasite genotype [33]). Furthermore, because Africans do not comprise a single, homogeneous population, detection of some variants (like Dantu) will depend on their local frequency and population structure. For these reasons, additional loci with large effects may remain to be identified by future studies, particularly those using African-centric SNP arrays or whole-genome sequencing.
Despite these limitations of GWAS, most authors still interpret large fractions of “missing heritability” as evidence that complex traits are highly polygenic [7,34,35]. From this perspective, nearly nine-tenths of the heritability of severe malaria risk could be attributed to variants with individual effects too small to be detected by GWAS of feasible size. Yet these variants appear to be so numerous that the sum of their tiny effects outweighs the much larger effects of the few most dramatic alleles (Figure 1A). A recent heritability analysis [36] demonstrated that the polygenic variation shaping severe malaria risk is distributed throughout the human genome, with each chromosome contributing a share of heritability roughly proportional to its physical size (Figure 1B). This diffuse genetic architecture resembles many other human traits, from red cell volume to type 1 diabetes [34,37]. How and whether polygenic variation may contribute to adaptation is an active area of interest in human population genetics [38–41]. Since severe malaria is well-established as a strong selective pressure (Box 1), it offers a unique opportunity to contrast the evolution of protective alleles with small and large effects.
Polygenic variation linked to malaria protection is not enriched in African populations
The seven SNPs most strongly associated with malaria protection by GWAS (Bayes factor (BF) > 100,000) are all more common in Africans than Europeans [7] (Figure 1C, red oval). Although neither continental group represents a homogenous population, this pattern is consistent with the malaria hypothesis, which predicts local balancing selection of variants with large protective effects (Box 1). In contrast, only 45 of the top 91 SNPs associated with malaria protection (BF > 1,000) are more common in Africans than Europeans [7] (Figure 1C, blue oval). Although these weaker associations likely contain more false positives, this proportion is not meaningfully different from neutral (non-selective) expectations based on demography and population structure (Box 2). Since SNPs with weaker evidence of association also tend to have smaller effect sizes, this result could suggest a distinct mode of evolution for the small-effect, polygenic variation underlying the majority of malaria susceptibility.
Our group recently found a similar result for RBC variants associated with P. falciparum growth in laboratory experiments [15]. Using RBCs sampled from healthy donors with a range of African ancestry, we identified 24 alleles (including PIEZO1 E756del and the ATP2B4 haplotype; Table 1) associated with reduced parasite growth. About half of these protective RBC variants are more frequent in Africans than Europeans (Figure 1D, blue oval), which does not differ from expectations based on random exomic SNPs [15]. Notably, the sickle allele of hemoglobin (HbS) was the only single variant with a dramatic protective effect in these RBC assays (Figure 1D, red oval). Although large-effect alleles from both GWAS and in vitro of studies do conform to the malaria hypothesis, it does not appear that small-effect alleles are, as a group, similarly differentiated between endemic and non-endemic populations.
Protective red cell traits are not enriched in African populations
Red blood cell phenotypes have been a constant focus of malaria research, from the initial formation of the malaria hypothesis (Box 1) to modern GWAS of protective loci (Table 1). Red cell traits related to volume, density, and hemoglobin content—known as red cell indices—are routinely measured in clinical settings around the globe. RBC membrane properties like fragility and deformability are also key to red cell function, blood storage, and diagnosis of hemolytic disease [5,8]. Most red cell traits have high heritability (40-90%), which large GWAS in multiple populations have attributed to polygenic variation [8,12,13,42,43].
Our recent study of healthy variation in RBCs without disease alleles (see Box 1) identified 10 polygenic traits, including RBC volume, that were strongly associated with P. falciparum growth in vitro [15]. Intriguingly, none of these protective traits (e.g. smaller RBC volume) were positively associated with African ancestry in non-carriers of disease alleles. Some RBC phenotypes were related to African ancestry, but not in the direction predicted by the malaria hypothesis. For example, African ancestry and African American ethnicity are strongly associated with decreased osmotic fragility of the RBC membrane [5,15] (Figure 1E). Membrane fragility, similar to “tension” measured by other studies [23], can be understood as an inverse of membrane deformability [15,44]. Our study found that lower osmotic fragility was associated with increased parasite growth [15] (Figure 1F), implying that RBCs with greater African ancestry are more susceptible to malaria parasites. Indeed, greater African ancestry in individuals without disease alleles predicted greater invasion efficiency of a clinical P. falciparum strain from Senegal [15].
An exaggerated protective effect of similar membrane properties has also been observed in African RBCs carrying the rare Dantu variant [23] (Table 1). In that study, high-frame-rate video microscopy was used to simultaneously measure red cell tension and P. falciparum invasion, revealing that parasites preferentially invade RBCs with lower membrane tension. Accordingly, RBCs with higher baseline tension also experienced less membrane deformation when contacted by P. falciparum merozoites. Other recent studies have confirmed that RBC deformability upon merozoite contact is essential for, and correlated with, for P. falciparum invasion success [45,46]. Together, these data suggest that the low membrane fragility characteristic of RBCs with African ancestry makes them relatively more susceptible to P. falciparum parasites. It is possible that decreased membrane fragility serves another functional purpose, unrelated to malaria, that is yet to be discovered. We note that P. falciparum may adapt to the red cell traits that it usually encounters, more quickly than red cell traits themselves can evolve. Indeed, recent work has shown that that the protective effect of HbS varies by parasite genotype, consistent with HbS imposing selection on parasite alleles [33].
Lower red cell volume (MCV) has also been linked to reduced P. falciparum growth and/or invasion in diverse healthy adults [15], anemic African children [4], and chemically dehydrated RBCs [16]. Some earlier studies described lower MCV in African than European Americans, which has been attributed to selection for malaria resistance [4]. However, the deletion allele that causes alpha-thalassemia (see Box 1) strongly diminishes MCV [15,47] and is more common in African-Americans. When carriers of thalassemia are excluded, the apparent association between MCV and ancestry disappears, although the association between MCV and parasite growth remains [15,48]. While environmental variation can complicate inference of population-specific selection [49], Africans and Europeans might be expected to have distinct MCV optima, given differences in malaria prevalence and the protective value of low MCV. This lack of differentiation in a protective trait may suggest that other evolutionary forces are important for shaping polygenic variation in RBC volume, or that environmental factors have obscured the effects of selection.
One phenotype that has repeatedly and robustly been observed to differ between African- and European-Americans is hemoglobin concentration [50]. Polygenic, non-environmental factors appear to drive 6-7% lower blood hemoglobin concentration in African-Americans [50]. In malaria-endemic settings, this difference may be minor compared to anemia from non-genetic factors, which can reduce hemoglobin concentrations to 60% or less of reference values [51]. In 16 sub-Saharan African countries surveyed from 2010-2016, moderate to severe anemia was present in an average of 39.7% of children under 5 years of age [51]. Various recent studies have also found that hemoglobin concentration either improves [4,52], exacerbates [51,53], or has no effect [54] on malaria outcomes. This uncertainty, combined with the much larger impact of environment compared to genetics, raises the possibility that additional factors besides malaria pressure may have contributed to differences in hemoglobin concentration between Africans and Europeans.
Overall, there is currently little evidence that mean RBC trait differences between Africans and Europeans contribute to greater malaria resistance in Africans without known disease alleles. Since the RBC traits discussed here are strongly polygenic, this finding is consistent with the lack of differentiation of detectable, small-effect alleles between populations (Figure 1C–D). How can these conclusions be reconciled with evidence from large-effect alleles (Box 1), which indicates strong selection to reduce P. falciparum growth in African RBCs?
Pervasive pleiotropy may limit polygenic adaptation to malaria
Many functional alleles, including those that confer malaria resistance (Table 1; Box 1), have pleiotropic effects on multiple phenotypes. In recent years, the explosion of GWAS for human traits has enabled direct assessments of the extent of pleiotropy in the human genome. So far, 50-60% of SNPs that have been associated with one human trait are also associated with at least one more [55,56], consistent with widespread pleiotropy. A recent analysis of 558 GWAS traits found that overlapping, associated loci cover over half of the human genome, with over 90% of such loci associated with multiple traits [56]. In many cases, pleiotropic variants impact broadly expressed genes and signaling pathway components that are active in a multitude of cell types [55]. However, since SNPs identified by GWAS are usually in linkage disequilibrium with the true causal SNPs, it is theoretically possible that they tag multiple causal alleles with independent effects on multiple traits. We note that other limitations of GWAS, including technical explanations for missing heritability, also apply to these meta-analyses of multiple traits.
Four years ago, an influential model was proposed in which complex traits may be not just polygenic, but omnigenic: affected by all genetic variants that impact gene expression in relevant cell types [34]. Such a model is plausible because biological networks, including transcriptional networks, are highly interconnected [57]. For example, a genetic variant that changes the transcription of one unit in a protein complex could consequently alter the folding, stability, physical interactions, or other activity of any number of functionally related proteins. The omnigenic model thus predicts extreme pleiotropy, even beyond what is directly observable from current GWAS data. It also helps explain why the heritability of most complex traits is spread so broadly throughout the genome (Figure 1B).
Understanding how abundant, functional variation is maintained within populations remains an ongoing challenge for evolutionary biologists [58,59]. If pleiotropy is so widespread that each functional allele has effects on many traits, then selection on all the traits—not just malaria resistance—could be expected to shape its allele frequency. We therefore propose that widespread pleiotropy may explain the current lack of observable differentiation between malaria-endemic and non-endemic populations for healthy variation that impacts malaria resistance (Figure 1C–D).
The red cell disorders that inspired the malaria hypothesis (Box 1) are testaments to the strength of malaria selection, precisely because their large antimalarial effects outweigh their serious damage to other organismal phenotypes. These examples, however, may be exceptions. Most alleles associated with malaria protection have only small effects on individual risk (Figure 1C–D), but their combined effects far outweigh the handful of large-effect alleles in explaining malaria risk within a population (Figure 1A). Although malaria is a strong selective pressure, and RBC variation appears to impact malaria, we propose that the frequency of any functional allele will depend on its strength and direction of effect on many different traits—not only malaria resistance. Not all alleles that limit malaria should be expected to experience strong, population-specific selection; just as not every allele that is more common in Africa should be expected to impact malaria (Box 2).
Concluding Remarks
Growing data from diverse human populations suggest that the malaria hypothesis may be a misleading heuristic for understanding global patterns of genetic variation, in part because malaria susceptibility is highly polygenic, and widespread pleiotropy may limit polygenic adaptation to malaria. This interpretation may be subject to change, however, as statistical and experimental methodologies improve. Current GWAS data are seriously limited by the large fraction of “missing heritability” for severe malaria risk (Figure 1A), but these limitations are expected to diminish as genetic diversity from non-European populations is discovered and incorporated into GWAS design. Similarly, steady increases in GWAS size; improved analysis of genetic correlations [38,40,41]; and steps toward controlling environmental variation [60] should enhance GWAS performance for small-effect, polygenic variation.
Our conclusions based on in vitro studies of P. falciparum and RBCs may also evolve as more physiologic models of this complex disease are developed, though this will require a substantial commitment of resources. For example, widespread adoption of improved humanized mouse models [61] will allow more cohesive and physiologic studies of severe malaria as a multi-organ disease. Increasingly detailed models of specific organ systems, such as the 3D architecture of the microvasculature [62] and blood-brain barrier [63], may also reveal non-RBC variation that is relevant to the malaria hypothesis. Still, the most direct test of the malaria hypothesis for polygenic variation may be large-scale, controlled infections of humans from diverse genetic backgrounds. At present, we have little knowledge of whether non-African populations contain their own large-effect variants for malaria resistance, nor whether the heritability of severe malaria varies around the world. A similar test could also be performed with genetically diverse non-human models, such as rodents sampled from wild populations.
As we await these developments, it is clear that continuing to explore aspects of malaria biology that do not necessarily adhere to the malaria hypothesis has the potential to advance our understanding of the pathogenesis of this complex disease. For example, RBC membrane dynamics clearly influence P. falciparum fitness [15,23], despite the higher frequency of susceptible phenotypes in African populations (Figure 1E–F). RBC membrane traits may represent a particularly exciting avenue for malaria prevention and therapeutics, given that RBC membrane variation is widespread and apparently well-tolerated in humans.
Casual invocation of the malaria hypothesis is not without costs, as its racial basis is known to reinforce misconceptions of biological heterogeneity within and across human groups [64] (Box 2). Most genetic loci for malaria susceptibility likely have individually small effects, are spread across the genome, are pleiotropic for other traits, and are shared among many human populations. Despite evidence that protective alleles of large effect have experienced balancing selection, it is possible that polygenic variation shaping malaria susceptibility is constrained by correlation with other traits. In our opinion, a major goal of studying genetic susceptibility to disease is to identify novel biological pathways for therapeutic targeting (see Outstanding Questions). Future research that embraces the polygenic, interconnected basis of complex traits will provide new insights into the progression of severe malaria.
Outstanding Questions.
What are the pleiotropic effects of the malaria-protective variants in ATP2B4, PIEZO1, and GYPA/GYPB?
Besides red blood cells, which tissues express genes that contribute to the heritability of severe malaria?
What traits are genetically correlated with severe malaria risk?
How many common variants adhering to the malaria hypothesis remain to be identified, particularly as knowledge of African genetic diversity improves?
Would larger GWAS or in vitro studies yield more robust signals of polygenic adaptation to malaria?
Why are red cell traits so variable within and across human populations?
Is common red cell variation important for P. falciparum parasites in vivo? How does common red cell variation contribute to severe malaria?
Highlights.
In malaria-endemic populations, the malaria hypothesis predicts balancing selection on protective loci that interfere with red blood cell (RBC) function.
Novel variants in the RBC genes PIEZO1, ATP2B4, and GYPA/GYPB are associated with strong protection from severe malaria and are more common in Africans than Europeans, consistent with the malaria hypothesis.
GWAS attribute most of the heritability of severe malaria risk to many loci with small effects spread across the genome.
African populations are not enriched for small-effect alleles associated with protection from severe malaria. Similarly, healthy RBC traits associated with reduced P. falciparum growth in vitro are not positively associated with African ancestry.
Pervasive pleiotropy may limit adaptation in complex traits. Therefore, host variation that impacts malaria may not necessarily conform to the malaria hypothesis.
Acknowledgements
We thank Abigail LaBella, Grant Kinsler, Sophie Walton, and members of the Egan lab for helpful discussion and comments. E.R.E. is supported by an NSF Postdoctoral Research Fellowship in Biology (Grant No. 2109851). This work was supported by NIH grant 1DP2HL13718601 to E.S.E., who is a Tashia and John Morgridge Endowed Faculty Scholar in Pediatric Translational Medicine through the Stanford Maternal Child Health Research Institute.
Glossary
- Anemia
a condition in which red blood cell quantity or hemoglobin concentration is lower than normal.
- Balancing selection
the active maintenance of multiple alleles at one genetic locus.
- Bayes factor (BF)
a ratio of the likelihood of one hypothesis over another. In GWAS, larger values are interpreted as stronger evidence of association between an allele and a trait, compared to a null hypothesis of no association.
- Bottleneck
a sharp decline in population size that reduces genetic diversity.
- Complex trait
a phenotype influenced by many genetic and environmental factors.
- Drift
a change in allele frequency due to chance, such as random sampling of chromosomes during meiosis. Drift has stronger impacts in smaller populations.
- Effect size
when estimated from an association study, the coefficient of an allele in a linear regression model of a trait.
- Fitness
a relative, average measure of the genetic contribution to the next generation from individuals with a certain genotype or phenotype.
- Gametocytogenesis
the production of sexual cells. In P. falciparum, gametocytes produced from asexual blood stages are essential to mosquito transmission.
- Genome-wide significant
an association between a trait and allele with p<5x10−8, based on a Bonferroni correction of all common independent SNPs in the human genome. Alleles with larger effect sizes and higher frequency are more likely to be genome-wide significant.
- GWAS
an observational study of the association between a trait and genome-wide genetic markers, usually conducted in many individuals.
- Heritability (narrow-sense)
the fraction of variance in a trait explained by additive genetic variation, as opposed to environment and measurement error.
- Linkage disequilibrium
correlation between alleles within a population, usually on the same physical chromosome.
- Neutral
not selected; having no impact on fitness.
- Odds ratio (OR)
the likelihood of an outcome for one group relative to another. For example, the likelihood of severe malaria for individuals with a particular allele.
- Omnigenic
influenced by all genes expressed in cell types relevant to a trait. The omnigenic model proposes that expression of a trait’s ‘core’ genes is regulated by many ‘peripheral’ genes in an interconnected network, allowing any variant within that network to have indirect effects on the trait.
- Osmotic fragility
the propensity of a red cell to lyse while absorbing water under hypotonic stress, which reflects biophysical membrane properties.
- Polygenic
traditionally, influenced by more than one gene. Today, this term is commonly used for traits influenced by hundreds or thousands of genetic variants, each with a small effect on total trait variation.
- Population structure
the organization of genetic variation among populations, driven by the evolutionary processes of mutation, recombination, drift, and selection.
- Pleiotropic
(of an allele) influencing two or more traits.
- Red cell indices
standardized blood tests of red cell count, size, hemoglobin content, and variability.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.Shriner D and Rotimi CN (2018) Whole-Genome-Sequence-Based Haplotypes Reveal Single Origin of the Sickle Allele during the Holocene Wet Phase. Am. J. Hum. Genet 102, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization (2020) World malaria report 2020: 20 years of global progress and challenges, World Health Organization [Google Scholar]
- 3.Kariuki SN and Williams TN (2020) Human genetics and malaria resistance. Hum. Genet 139, 801–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goheen MM et al. (2016) Anemia Offers Stronger Protection Than Sickle Cell Trait Against the Erythrocytic Stage of Falciparum Malaria and This Protection Is Reversed by Iron Supplementation. EBioMedicine 14, 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanias T et al. (2017) Ethnicity, sex, and age are determinants of red blood cell storage and stress hemolysis: Results of the REDS-III RBC-Omics study. Blood Adv. 1, 1132–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma S et al. (2018) Common PIEZO1 Allele in African Populations Causes RBC Dehydration and Attenuates Plasmodium Infection. Cell 173, 443–455.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Network MGE (2019) Insights into malaria susceptibility using genome-wide data on 17,000 individuals from Africa, Asia and Oceania. Nat. Commun 10, 5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Page GP et at. (2021) Multiple-ancestry genome-wide association study identifies 27 loci associated with measures of hemolysis following blood storage. J. Clin. Invest 131, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang DL et al. (2018) Erythrocytic ferroportin reduces intracellular iron accumulation, hemolysis, and malaria risk. Science 359, 1520–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biddanda A et al. (2020) A variant-centric perspective on geographic patterns of human allele frequency variation. eLife 9, e60107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zámbó B et al. (2017) Decreased calcium pump expression in human erythrocytes is connected to a minor haplotype in the ATP2B4 gene. Cell Calcium 65, 73–79 [DOI] [PubMed] [Google Scholar]
- 12.Hodonsky CJ et al. (2018) Generalization and fine mapping of red blood cell trait genetic associations to multi-ethnic populations: The PAGE study. Am. J. Hematol 93, 1061–1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Astle WJ et al. (2016) The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell 167, 1415–1429.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lessard S et al. (2017) An erythroid-specific ATP2B4 enhancer mediates red blood cell hydration and malaria susceptibility. J. Clin. Invest 127, 3065–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebel ER et al. (2021) Common host variation drives malaria parasite fitness in healthy human red cells. eLife 10, e69808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tiffert T et al. (2005) The hydration state of human red blood cells and their susceptibility to invasion by Plasmodium falciparum. Blood 105, 4853–4860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguetse CN et al. (2020) A common polymorphism in the mechanosensitive ion channel PIEZO1 is associated with protection from severe malaria in humans. Proc. Natl. Acad. Sci 117, 9074–9081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilboudo Y et al. (2018) A common functional PIEZO1 deletion allele associates with red blood cell density in sickle cell disease patients. Am. J. Hematol 93, E362–E365 [DOI] [PubMed] [Google Scholar]
- 19.Rooks H et al. (2019) A gain of function variant in PIEZO1 (E756del) and sickle cell disease. Haematologica 104, e91–e93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thye T et al. (2021) Human genetic variant E756del in the ion channel PIEZO1 not associated with protection from severe malaria in a large Ghanaian study. J. Hum. Genet DOI: 10.1038/s10038-021-00958-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaskiewicz E et al. (2019) Erythrocyte glycophorins as receptors for Plasmodium merozoites. Parasit. Vectors 12, 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leffler EM et al. (2017) Resistance to malaria through structural variation of red blood cell invasion receptors. Science 356, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kariuki SN et al. (2020) Red blood cell tension protects against severe malaria in the Dantu blood group. Nature 585, 579–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Algady W et al. (2018) The Malaria-Protective Human Glycophorin Structural Variant DUP4 Shows Somatic Mosaicism and Association with Hemoglobin Levels. Am. J. Hum. Genet 103, 769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buniello A et al. (2019) The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campino S et al. (2016) Genomic variation in two gametocyte non-producing Plasmodium falciparum clonal lines. Malar. J 15, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Passini FS et al. (2021) Shear-stress sensing by PIEZO1 regulates tendon stiffness in rodents and influences jumping performance in humans. Nat. Biomed. Eng DOI: 10.1038/s41551-021-00716-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma S et al. (2021) A role of PIEZO1 in iron metabolism in mice and humans. Cell 184, 969–982.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bigham AW et al. (2018) Complex signatures of natural selection at GYPA. Hum. Genet 137, 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson KE and Voight BF (2018) Patterns of shared signatures of recent positive selection across human populations. Nat. Ecol. Evol 2, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Génin E (2020) Missing heritability of complex diseases: case solved? Hum. Genet 139, 103–113 [DOI] [PubMed] [Google Scholar]
- 32.Young AI (2019) Solving the missing heritability problem. PLOS Genet. 15, e1008222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Band G et al. (2021) Malaria protection due to sickle haemoglobin depends on parasite genotype. Nature DOI: 10.1038/s41586-021-04288-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boyle EA et al. (2017) An expanded view of complex traits: from polygenic to omnigenic. Cell 169, 1177–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crouch DJM and Bodmer WF (2020) Polygenic inheritance, GWAS, polygenic risk scores, and the search for functional variants. Proc. Natl. Acad. Sci 117, 18924–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Damena D and Chimusa ER (2020) Genome-wide heritability analysis of severe malaria resistance reveals evidence of polygenic inheritance. Hum. Mol. Genet 29, 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi H et al. (2016) Contrasting the Genetic Architecture of 30 Complex Traits from Summary Association Data. Am. J. Hum. Genet 99, 139–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berg JJ et al. (2019) Reduced signal for polygenic adaptation of height in UK Biobank. eLife 8, e39725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Racimo F et al. (2018) Detecting Polygenic Adaptation in Admixture Graphs. Genetics 208, 1565–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sohail M et al. (2019) Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. eLife 8, e39702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stern AJ et al. (2021) Disentangling selection on genetically correlated polygenic traits via whole-genome genealogies. Am. J. Hum. Genet 108, 219–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen M-FI. et al. (2020) Trans-ethnic and Ancestry-Specific Blood-Cell Genetics in 746,667 Individuals from 5 Global Populations. Cell 182, 1198–1213.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vuckovic D et al. (2020) The Polygenic and Monogenic Basis of Blood Traits and Diseases. Cell 182, 1214–1231.e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews K et al. (2017) Microfluidic analysis of red blood cell deformability as a means to assess hemin-induced oxidative stress resulting from Plasmodium falciparum intraerythrocytic parasitism. Integr. Biol 9, 519–528 [DOI] [PubMed] [Google Scholar]
- 45.Blake TCA et al. (2020) Actomyosin forces and the energetics of red blood cell invasion by the malaria parasite Plasmodium falciparum. PLOS Pathog. 16, e1009007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sisquella X et al. (2017) Plasmodium falciparum ligand binding to erythrocytes induce alterations in deformability essential for invasion. eLife 6, e21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nunchai C et al. (2020) Optimal cutoff of mean corpuscular volume (MCV) for screening of alpha-thalassemia 1 trait. J. Obstet. Gynaecol. Res 46, 774–778 [DOI] [PubMed] [Google Scholar]
- 48.Beutler E and West C (2005) Hematologic differences between African-Americans and whites: the roles of iron deficiency and alpha-thalassemia on hemoglobin levels and mean corpuscular volume. Blood 106, 740–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harpak A and Przeworski M (2021) The evolution of group differences in changing environments. PLOS Biol. 19, e3001072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kang W et al. (2021) Ethnic Differences in Iron Status. Adv. Nutr DOI: 10.1093/advances/nmab035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Papaioannou I et al. (2019) Malaria-anemia comorbidity prevalence as a measure of malaria-related deaths in sub-Saharan Africa. Sci. Rep 9, 11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goheen MM et al. (2017) Host iron status and erythropoietic response to iron supplementation determines susceptibility to the RBC stage of falciparum malaria during pregnancy. Sci. Rep 7, 17674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lombardo P et al. (2017) Hemoglobin Levels and the Risk of Malaria in Papua New Guinean Infants: A Nested Cohort Study. Am. J. Trop. Med. Hyg 97, 1770–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muriuki JM et al. (2019) Iron Status and Associated Malaria Risk Among African Children. Clin. Infect. Dis 68, 1807–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shikov AE et al. (2020) Phenome-wide functional dissection of pleiotropic effects highlights key molecular pathways for human complex traits. Sci. Rep 10, 1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Watanabe K et al. (2019) A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet 51, 1339–1348 [DOI] [PubMed] [Google Scholar]
- 57.Singh RS and Gupta BP (2020) Genes and genomes and unnecessary complexity in precision medicine. Npj Genomic Med. 5, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Llaurens V et al. (2017) Genetic architecture and balancing selection: the life and death of differentiated variants. Mol. Ecol 26, 2430–2448 [DOI] [PubMed] [Google Scholar]
- 59.Bertram J and Masel J (2019) Different mechanisms drive the maintenance of polymorphism at loci subject to strong versus weak fluctuating selection. Evolution 73, 883–896 [DOI] [PubMed] [Google Scholar]
- 60.Watson JA et al. (2021) Improving statistical power in severe malaria genetic association studies by augmenting phenotypic precision. eLife 10, e69698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Minkah NK et al. (2018) Humanized Mouse Models for the Study of Human Malaria Parasite Biology, Pathogenesis, and Immunity. Front. Immunol 9, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernabeu M et al. (2021) Bioengineered 3D Microvessels for Investigating Plasmodium falciparum Pathogenesis. Trends Parasitol. 37, 401–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Long RKM et al. (2021) Understanding parasite–brain microvascular interactions with engineered 3D blood–brain barrier models. Mol. Microbiol 10.1111/mmi.14852 [DOI] [PubMed] [Google Scholar]
- 64.Sparks RA et al. (2020) Using Culturally Relevant Pedagogy to Reconsider the Genetics Canon. J. Microbiol. Biol. Educ 21, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haldane JBS (1949) THE RATE OF MUTATION OF HUMAN GENES. Hereditas 35, 267–273 [Google Scholar]
- 66.Allison AC (1954) The distribution of the sickle-cell trait in East Africa and elsewhere, and its apparent relationship to the incidence of subtertian malaria. Trans. R. Soc. Trop. Med. Hyg 48, 312–318 [DOI] [PubMed] [Google Scholar]
- 67.Allison AC (1954) Protection Afforded by Sickle-cell Trait Against Subtertian Malarial Infection. BMJ 1, 290–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jamrozik E and Selgelid MJ (2021) History of Human Challenge Studies. In Human Challenge Studies In Endemic Settings: Ethical and Regulatory Issues (Jamrozik E and Selgelid MJ, eds), pp. 9–23, Springer International Publishing [Google Scholar]
- 69.Archer NM et al. (2018) Resistance to Plasmodium falciparum in sickle cell trait erythrocytes is driven by oxygen-dependent growth inhibition. Proc. Natl. Acad. Sci 115, 7350–7355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ha J et al. (2019) Hemoglobin E, malaria and natural selection. Evol. Med. Public Health 2019, 232–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clarke GM et al. (2017) Characterisation of the opposing effects of G6PD deficiency on cerebral malaria and severe malarial anaemia. eLife 6, e15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paquette AM et al. (2015) The evolutionary origins of Southeast Asian Ovalocytosis. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis 34, 153–159 [DOI] [PubMed] [Google Scholar]
- 73.Tétard M et al. (2017) Heterozygous HbAC but not HbAS is associated with higher newborn birthweight among women with pregnancy-associated malaria. Sci. Rep 7, 1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bergström A et al. (2021) Origins of modern human ancestry. Nature 590, 229–237 [DOI] [PubMed] [Google Scholar]
- 75.Rosenberg NA (2011) A Population-Genetic Perspective on the Similarities and Differences among Worldwide Human Populations. Hum. Biol 83, 659–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Coop G et al. (2009) The Role of Geography in Human Adaptation. PLoS Genet. 5, e1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lewontin RC (1972) The Apportionment of Human Diversity. In Evolutionary Biology: Volume 6 (Dobzhansky T et al. , eds), pp. 381–398, Springer US [Google Scholar]
- 78.Rosenberg NA et al. (2019) Interpreting polygenic scores, polygenic adaptation, and human phenotypic differences. Evol. Med. Public Health 2019, 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karczewski KJ et al. (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ndila CM et al. (2018) Human candidate gene polymorphisms and risk of severe malaria in children in Kilifi, Kenya: a case-control association study. Lancet Haematol. 5, e333–e345 [DOI] [PMC free article] [PubMed] [Google Scholar]


