Abstract
Non-coding DNA (ncDNA) refers to the portion of the genome that does not code for proteins and accounts for the greatest physical proportion of the human genome. ncDNA includes sequences that are transcribed into RNA molecules, such as ribosomal RNAs (rRNAs), microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and un-transcribed sequences that have regulatory functions, including gene promoters and enhancers. Variation in non-coding regions of the genome have an established role in human disease, with growing evidence from many areas, including several cancers, Parkinson’s disease and autism. Here, we review the features and functions of the regulatory elements that are present in the non-coding genome and the role that these regions have in human disease. We then review the existing research in epilepsy and emphasise the potential value of further exploring non-coding regulatory elements in epilepsy. In addition, we outline the most widely used techniques for recognising regulatory elements throughout the genome, current methodologies for investigating variation and the main challenges associated with research in the field of non-coding DNA.
Keywords: non-coding DNA, regulatory sequences, epilepsy, gene regulation, non-coding mutations
1. Introduction
Why do we not have a better understanding of disease biology? What generates wide phenotypic variation in a disease even when the major components of the disease are readily recognisable? Does this heterogeneity arise from environmental factors or genetic background, or both? In the case of brain disorders, the brain is a complex organ with both environmental and genetic determinants of structure, function and disease. Focusing on internal determinants, the complexity of the brain is intuitively linked to the number and connectivity of differentiated cell types that express cell-specific genes, exhibit unique properties and perform specialised functions (1). At the genomic level, complexity is not simply determined by the expression of cell-specific protein-coding genes but may relate to the way these genes are regulated, with a significant role for non-coding DNA (ncDNA), amongst other influences.
ncDNA refers to the portion of the genome that does not code for proteins and accounts for the greatest proportion of the human genome. Indeed, it is estimated that only about 2% of the human genome encodes proteins, with the rest being non-protein-coding (2). Of this, the exact percentage carrying functional properties is yet to be clarified, with different estimates being proposed (3). Historically, a large percentage of the non-coding genome was referred to as “junk DNA”, due to the prevailing sentiment at the time that it lacked any functional relevance and was essentially useless (4–6). This idea has persisted, predominantly due to the difficulty of investigating such a large and complex field, and the lack of appropriate techniques. In recent years, improvements in sequencing technologies, expression assays and advances in data handling and analysis have made it possible to study the ncDNA and have led to a greater appreciation of its role in human health and disease.
ncDNA includes sequences that are not translated into proteins but are transcribed into RNA molecules, called non-coding RNAs (ncRNAs): these comprise transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), microRNAs (miRNAs), small nucleolar RNAs (snoRNAs), small nuclear RNAs (snRNAs), piwi-interacting RNAs (piRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) (Figure 1). The biology of ncRNA and their roles in disease are not covered in this review (see reviews: (7, 8). Further, the 5’ and 3’ untranslated regions (UTRs), located at either end of the mRNA, also fall into this category of ncDNA. UTRs are crucial for the regulation of protein expression. For example, the translation of upstream open reading frames (uORFs) within the 5’ UTR is known to be a common mechanism for controlling the level of protein production downstream, according to a general model based on competition for ribosome binding (9). Due to their role in mRNA translation initiation, the question arises of whether these regions should be classified as coding DNA rather than non-coding DNA. Therefore, despite the importance of UTRs in the regulation of gene expression, due to the ambiguity in their definition, UTRs will not be discussed further. ncDNA also contains un-transcribed regions that function as regulatory elements, which represent the main focus of this review. These include gene promoters and transcription factor binding sites, enhancers, transposable elements and topologically associating domain (TAD) boundaries (Figure 2). ncDNA and variation in non-coding regions of the genome have an established role in human disease, with evidence in cancers, Parkinson’s disease and autism, amongst others (10–13).
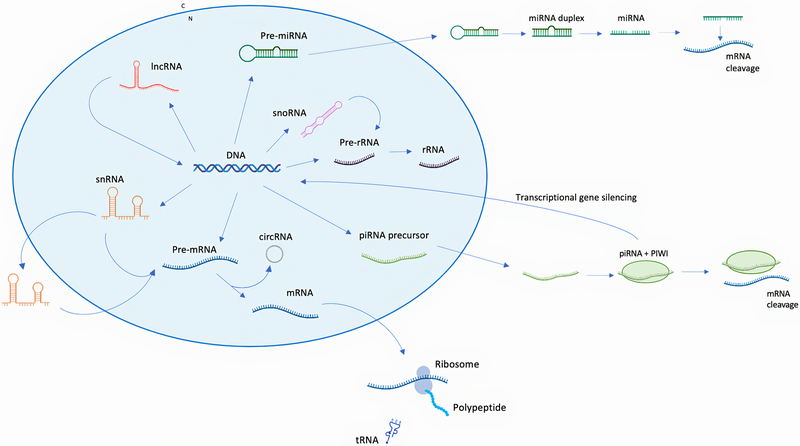
Figure 1:
RNA types and functions. Protein-coding genes are transcribed as pre-mRNAs, which undergo post-transcriptional modifications, becoming mature mRNAs. Among the post-transcriptional modifications of pre-mRNAs is the removal of introns (splicing), which occurs through snRNAs. snRNAs, which guide the splicing process, can be divided into two classes: one class never leaves the nucleus, while another class undergo post-transcriptional modifications in the cytoplasm, before re-entering the nucleus and being functional. The process of splicing may lead to the formation of circRNAs. circRNAs have their 5’ and 3’ ends bound together and are involved in the regulation of alternative splicing of the same genes from which they derive. circRNAs can also interact with miRNAs and inhibit their activity. Mature mRNAs exit the nucleus and reach the cytoplasm, where they are translated into proteins. mRNA translation involves ribosomes, macromolecules composed of proteins and rRNAs. rRNAs are initially transcribed as pre-rRNAs and undergo post-transcriptional modifications that involve snoRNAs. snoRNAs guide the post-transcriptional modifications of rRNAs, snoRNAs and tRNAs. tRNAs are also involved in the mRNA translation process: tRNAs recognise specific mRNA codons and carry the corresponding amino acids to the protein synthesis site. Translation of mRNAs into proteins may be prevented by miRNAs. miRNAs are transcribed in the nucleus as pre-miRNAs, which exit the nucleus and undergo cleavage steps, becoming functional. miRNAs show complementarity with a target mRNA, bind to these target mRNAs and induce their cleavage. Another class of RNA are lncRNAs, which have multiple functions, including interacting with DNA and recruiting regulatory proteins to modulate histone modification. piRNAs are transcribed as precursor molecules, which exit the nucleus and interact with the regulatory proteins PIWI (abbreviation of P-element Induced WImpy testis in Drosophila). The piRNA-PIWI complex is involved in stem cell differentiation and silencing of transposable elements, acting both at the transcription level, by silencing the gene, and post-transcription level, inducing the cleavage of mRNA.
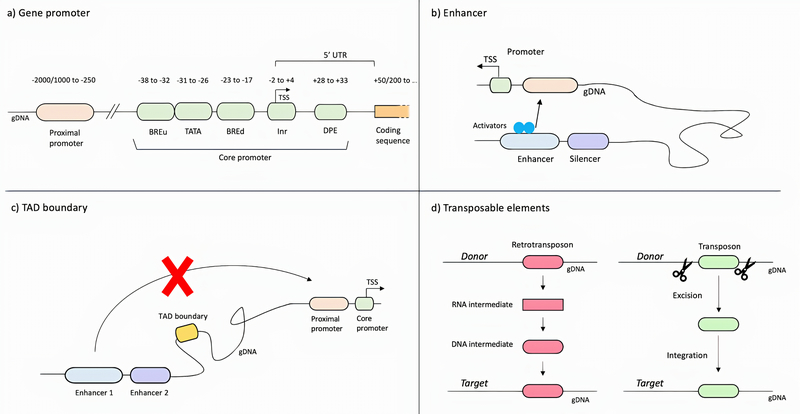
Figure 2:
Schematic representation of non-coding regulatory elements. a) Gene promoters represent the site where the transcriptional machinery assembly occurs, and transcription factors interact to regulate the expression of genes. Gene promoters include the core promoter, the minimal required portion to initiate the transcription, and the proximal promoter. b) Enhancers are regulatory elements that can be bound by activators and repressor and modulate the expression of target genes. c) TAD boundaries may prevent the physical interaction between enhancers and the target genes. d) Transposable elements are regulatory sequences capable of altering their position in the genome. Retrotransposons use a “copy and paste” mechanism and introduce a copy of themselves to a different genetic location; transposons use a “cut and paste” mechanism: they excise themselves from one genetic locus and integrate into a different one. Translocation of TE occurs predominantly in germ cells and during early embryogenesis, when TEs contribute defining the temporal expression of genes. BREu: upstream TFIIB Recognition Element, BREd: downstream TFIIB Recognition Element, TATA box, Inr: Initiator element, DPE: downstream promoter element, TSS: transcription start site, gDNA: genomic DNA.
There is still an open question about the causes of the wide phenotypic variability that characterise many epilepsies with a known genetic cause; ncDNA may contribute to this variability (14). Non-coding regulatory regions may harbour variations that influence gene expression, and have a disease-modifying effect, or influence treatment response. Here, we consider the evidence supporting the role of ncDNA in epilepsy. The regulatory elements that are present in the non-coding genome will be described and the role that these regions have in human disease will be discussed. Examples of the importance that non-coding regulatory regions have in human diseases will be reported, and the existing research in the field of epilepsy will be reviewed. We make the case for further research to appreciate the potential value of non-coding regulatory elements in epilepsy. In addition, this review will also outline the most widely used techniques for recognising regulatory elements throughout the genome, the main methodologies for investigating variation and the main challenges associated with research in the field of ncDNA.
2. Non-coding regulatory regions
One of the most important classes of non-coding regulatory elements of the genome are gene promoters, which are essential for determining the direction of transcription, indicating the sense strand of the DNA and regulating gene expression. The gene promoter is located upstream of, and partially overlapping with, the transcription start site (TSS) of the gene it regulates, thus occupying the first part of the 5’UTR region, as shown in Figure 2a (15). The minimal portion of the promoter required to initiate transcription is called the core promoter; it spans between 60–120 base-pairs and represents the transcriptional machinery assembly site (16–18). The core promoter contains the RNA polymerase binding site, the TSS and optional motifs, including the Goldberg-Hogness box (commonly called TATA box), the Initiator element (Inr), the downstream promoter element (DPE) and the TFIIB recognition element (BRE) (19). Such optional motifs may extend downstream of the TSS: for example the DPE motif, when present, is located 28–33 nucleotides after the TSS (Figure 2a) (20). Beyond the core promoter is the proximal promoter, located approximately 250 bp upstream of the TSS, and usually extending up to 1000–2000bp (21). The proximal promoter contains binding sites for both general and sequence-specific transcription factors (22).
The activity of promoters is influenced by additional regulatory sequences that can modulate the expression of genes from a genomic location even further away. These elements are called enhancers, DNA sequences that range from 50 to 1500 bp in length, to which proteins called activators and repressors can bind. The interaction of the enhancer and the activator/repressor results in the creation of a chromatin loop that can shift the enhancer closer to the gene promoter and allows mediator proteins to be recruited. Mediator proteins either promote or prevent the binding of RNA polymerase, resulting in promotion or repression of the target gene expression. Genes can be modulated by different enhancers, and each enhancer can modulate multiple genes (15). Enhancers may be located thousands of base pairs away from the target gene, either upstream or downstream of the TSS (15). The interaction between enhancer and the target promoter occurs through chromatin loops and is supported by proteins called cohesins. Chromatin loops represent non-random three-dimensional folds of chromatin that generate physical interactions between distantly located genetic sequences, including long-range interactions between regulatory sequences and the corresponding target genes (23). Clusters of multiple enhancers may occur in the genome: these typically exhibit similar activity and regulate the same genes. Such redundancy of enhancers may be crucial, especially during development, to provide robustness in case of loss-of-function mutations and to ensure the correct spatiotemporal expression of target genes, necessary to guide development (24–26). Protein-coding gene redundancy has mostly been lost during evolution, and it is possible that today’s redundancy involves regulatory elements rather than protein-coding genes in order to achieve differential gene expression in various tissues with the least amount of “space” in the genome (27). Enhancer functionality may be limited by TAD boundaries (or insulator elements), which represent another class of DNA regulatory elements that are capable of blocking the physical interaction between enhancer and gene promoter (28). TAD boundaries also function as a chromatin barrier: these regions interact with cohesins and CCCTC-binding factor (CTCF), a transcriptional repressor protein, forming a complex that constitutes a physical impediment to prevent the excessive spread of heterochromatin (28).
Transposable elements (TEs) represent another class of ncDNA elements involved in regulatory control. TEs are capable of altering their position in the genome and can be divided into two different categories, based on the mechanism of transposition. Retrotransposons, TEs of Class 1, use a “copy and paste” mechanism: through reverse-transcription and the production of an RNA intermediate, they introduce a new copy of themselves to a different genetic location. Transposons, Class 2 TEs, use a “cut and paste” mechanism: their sequence includes the genetic code for the transposase enzyme, which they use to excise themselves from one genetic locus and integrate into a different one (29).
Most copies of TEs in our genome have lost the ability to mobilise due to mutations and now have a fixed genetic location. TEs that are still mobile within an individual’s genome mobilise predominantly in germ cells and during early embryogenesis (30, 31). Since TEs often include TSSs and other regulatory sequences in their own sequence, their mobilisation has contributed to the formation of novel tissue-specific promoters and transcription factor binding sites (TFBSs), which now have a role in ensuring the correct spatiotemporal gene expression during development (32, 33). Furthermore, TE mobilisation also occurs in somatic cells. This happens in the brain, particularly in the hippocampus, where the somatic transposition of the human long interspersed nuclear element-1 (LINE-1, also called L1) in neural precursor cells contributes to neuronal hippocampal diversity (31, 34). However, TE insertion into the genome may also have a deleterious effect and cause disease. Examples include Haemophilia A, the first disease in which an association with TEs was proven, and neurofibromatosis type 1 (NF1) (35, 36). Another example is Rett syndrome, which is a neurological condition caused by mutations in the methyl-CpG-binding protein 2 gene (MECP2). MECP2 is a regulator of L1 transposition, and patients with Rett Syndrome, carrying a mutated MECP2, show increased L1 mobilisation, which possibly contributes to the Rett phenotype (37, 38). The regulation and silencing of TE transposition are complex and rely on several elements: piRNAs, which interact with PIWI proteins and drive repressive chromatin marks on the promoter region of TEs, and zinc finger proteins containing the Kruppel-associated box (KRAB-ZFPs), which bind to the TE sequence and recruit additional proteins, ultimately adding repressive chromatin marks to the promoter region of the TE (39, 40).
3. Non-coding variation
The term “non-coding variation” refers to genetic changes that occur within non-coding regions of the genome. Non-coding variation may be represented either by single-nucleotide polymorphisms (SNPs) or structural variations, including short insertions or deletions (collectively called InDels), copy-number variations (CNVs) and repeat expansions.
SNPs, described as single nucleotide substitutions at specific positions of a DNA sequence, represent the most common type of DNA variation. It is estimated that approximately 90% of disease-associated SNPs fall in non-coding sequences of the genome (41). However, for most of the millions of SNPs that have been identified in ncDNA by the Human Genome Project and investigated in GWAS studies, we do not yet understand the functional implications.
Structural variations include small insertions and deletions (defined as the loss and gain of sequences up to 1kb in length), duplications, inversions, translocation and CNVs, which represent duplications and deletions of sequences greater than 1kb in length (42, 43). CNVs have been observed throughout the genome, but interestingly not all chromosomes are affected equally: some chromosomes typically have large numbers of CNVs (such as chromosome 19 and 22), whereas other regions are described as CNV-deserts, often being devoid of CNVs (44, 45).
Another type of structural variations is represented by repeat expansions, which represent the expansion of repeated DNA sequences: the size of the repeat may vary from trinucleotide repeats to 12-nucleotide long sequences, and the number of times this sequence is repeated is also variable (46, 47). Examples of diseases caused by repeat expansions occurring in noncoding sequences include Friedreich’s ataxia, amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (48, 49). Non-coding repeat expansions have also been found in some epilepsies, including progressive myoclonus epilepsy of the Unverricht–Lundborg type (EPM1), associated with the expansion of a dodecamer repeat in the promoter region of the cystatin B gene (CSTB), and benign adult familial myoclonic epilepsy (BAFME), associated with intronic expansions of a five-nucleotide long sequence (TTTCA or TTTTA): expansions of this sequence have been identified in introns of different genes in different patients (50–52).
4. Techniques for identifying regulatory elements
In order to detect and analyse variation in non-coding regulatory elements, it is first necessary to localise these regions in the genome, as summarised in the pipeline in Figure 3. Several strategies, which are outlined in the following sections, can be used to achieve this (Table 1). The most reliable and accurate method for predicting the location of regulatory elements is to integrate data from multiple methods, as will be described in section 3.6 “Online databases”.
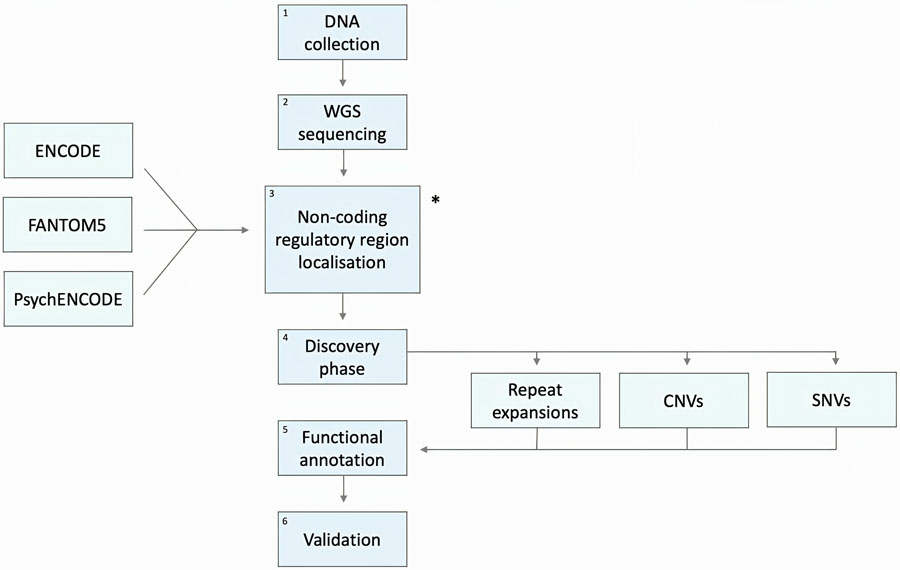
Figure 3:
Flowchart of the steps required to study non-coding genetic variations. After DNA collection and whole-genome sequencing (WGS), is the identification of non-coding regulatory elements, which can be achieved using different approaches, described in Table 1. In the subsequent discovery phase, variant calling tools will be used to identify genetic variations in the cohort of interest. For the functional annotation of variants, the one reliable strategy is to assess the functional consequences of variants using multiple techniques and compare these predictions to provide robustness to the interpretation. Finally, the functional consequences of variants will be evaluated and then validated using wet-lab experiments. WGS: Whole Genome Sequencing, SNVs: Single Nucleotide Variants, CNVs: Copy Number Variations.
Table 1:
Strategies for identifying non-coding regulatory elements.
| Strategy | Methods | Details | Advantages | Disadvantages | Additional Information |
|---|---|---|---|---|---|
| Transcription Factor Binding Site localisation | ChIP-seq | In the ChIP-seq protocol, cells are treated with formaldehyde to crosslink proteins to DNA, followed by DNA fragmentation. Immunoprecipitation with antibodies specific to the target TF allows for the recognition and isolation of the DNA-TF complexes. The precipitated DNA is then purified and sequenced to identify the genetic sequence corresponding to the TFBS. | - Genome-wide coverage - High resolution, even in repetitive sequences |
- Immunoprecipitation with antibodies: the quality of the output data depends on the quality of the antibodies used - Large number of input cells required |
Recent developments increase the resolution close to the single nucleotide level (ChIP-exo, ChIP-nexus) and require lower numbers of input cells (Nano-ChIP-seq). |
| Footprinting | Chromatin accessibility data is required to perform this assay. When chromatin accessibility assays are performed, DNA-bound TFs prevent enzymes used in open-chromatin assays from cleaving DNA in nucleosome-free regions, leaving recognisable footprints. Computational footprinting tools detect the DNA-bound TF footprints, allowing TFBS localisation. | - High sensitivity - Robust protocol for in-vitro and in vivo formed protein–DNA complexes |
- Limitations of open chromatin assays (discussed in the corresponding section) - Need to apply computational bias corrections, which vary depending on the open chromatin assay employed |
||
| Position Weight Matrix | For a given TF, PWM models the predicted sequence of the binding site and assigns a score to each of the four bases at each motif position, indicating the preference for each base at each position. Matching patterns are identified by scanning a PWM against a target DNA sequence, and each potential binding site is assigned a score corresponding to the predicted relative strength of binding. | - Prediction of both the potential binding sites of a TF and the relative strength of binding to each TFBS | - Nucleotide interdependencies and dinucleotide interactions are not considered - Susceptible to false-positive prediction |
Known TFBS motifs resulting from PWM studies are collated in the JASPAR database (http://jaspar.genereg.net) and the TRANSFAC (https://genexplain.com/transfac/) Recent improved algorithms incorporate dinucleotide interactions |
|
| Chromatin accessibility assays | DNase-I hypersensitivity assay | In the DNase-I hypersensitivity assay, cells are lysed and DNA is digested with DNase I endonuclease. Nucleosome-wrapped DNA regions are resistant to endonuclease treatment, whereas nucleosome-free regions are sensitive to cleavage by DNase I endonuclease: these regions are known as DNase I hypersensitive sites (DHSs) and correspond to regulatory elements. The protocol terminates with targeted region amplification and sequencing, which allow for the characterisation of regulatory sequences. | - Gold standard technique for evaluating chromatin accessibility - Genome-wide coverage - High resolution |
- DNase-I cleaves DNA unevenly, depending on sequence composition and cleavage propensities - The preliminary procedures of cell permeabilization and nuclei isolation may affect chromatin status |
|
| ATAC-seq | In the ATAC-seq protocol, cells are lysed, and chromatin is fragmented and tagged with the hyperactive Tn5 transposase, which introduces sequencing adapters into open-chromatin regions. These tagged sequences, corresponding to transcriptionally active and regulatory regions, are then purified and sequenced. | - Assesses chromatin accessibility in single cells and from frozen tissue - Suitable in cases of low amount of sample |
- Prone to sequencing contamination by mitochondrial DNA | Recent improvements reduce mitochondrial reads | |
| FAIRE-seq | In FAIRE seq, nucleosome-DNA interactions are stabilised through formaldehyde. DNA is then fragmented and isolated via phenol-chloroform extraction, which allows the physical separation of the crosslinked DNA (in the organic phase) and protein-free DNA regions (in the aqueous layer). Accessible nucleosome-free DNA regions, which correspond to regulatory sequences, are purified and sequenced. | - Less damaging effect on chromatin than DNAse-I and ATAC-seq | - Low sensitivity for detection in promoter regions - Lower resolution than DNAse-I and ATAC-seq |
||
| Epigenetic Assays | ChIP-seq + MSP | The ChIP-seq protocol performed using histone-specific antibodies identifies genetic regions enriched for histone modification. MSP can be used to selectively amplify methylated sequences. To perform MSP, DNA is bisulphite treated, and PCR is conducted using two pairs of primers. One set of primers amplifies methylated DNA, while the other set amplifies unmethylated DNA. By performing PCR with both pairs of primers, methylated DNA and unmethylated DNA sequences are distinguishable. | - High sensitivity - Small amounts of DNA are required - The protocol can be performed on DNA extracted from paraffin-embedded samples |
- High false-positive rate - Variability of results due to assay conditions (e.g., primer design, annealing temperature, cycle number) |
|
| Whole-genome bisulphite sequencing | Genomic DNA is fragmented and ligated to sequencing adapters, which are characterised by having all cytosines methylated, to allow the subsequent phases of primer binding and amplification after bisulphite conversion. Denatured adapter-ligated DNA fragments undergo bisulphite treatment, which distinguishes methylated and unmethylated cytosines. Methylated sequences are amplified and sequenced. | - Efficient for genome-wide investigations - Ability to assess the methylation state of nearly every CpG site |
- DNA degradation after bisulphite treatment | ||
| Illumina MethylationEPIC [850k] Array | Illumina MethylationEPIC [850k] Array is a DNA methylation microarray that queries the methylation status of more than 850,000 methylation sites, including almost 350,000 CpGs located in regulatory regions identified by the ENCODE and FANTOM5 projects. | - Easy, time-efficient and cost-effective protocol - Increased coverage in enhancer regions compared to the predecessor |
- Only one CpG probe is interrogated per enhancer region | ||
| Chromosome Conformation Capture methods | 5C / Hi-C | 5C (many vs many): Cells are formaldehyde-treated to crosslink interacting loci, then lysed and DNA fragmented. The ends of DNA fragments are re-ligated in diluted conditions to promote ligation between cross-linked interacting fragments. The crosslinking is then reversed, and the samples undergo ligation-mediated amplification (LMA). LMA employs primer pairs that anneal across the ligated junctions of the 3C library and allow for the amplification of the target fragments where chromatin interactions occurred. The target fragments are subsequently sequenced. Hi-C (all vs all): The Hi-C protocol begins with the same steps as 5C. Following the DNA digestion phase, the fragment ends are labelled with biotinylated nucleotides. Then ligation and reversal of the crosslinks occur. Ligated fragments, which contain the internal biotin tag, are purified and sequenced using paired-end sequencing. |
5C: - Cheaper than Hi-C Hi-C: - Genome-wide coverage |
5C: - Complex primer design - Lower quality results than Hi-C - Limited coverage (regions up to 1 Mb) Hi-C: - Limited resolution for short distance interactions |
|
| miniMDS | miniMDS is an algorithm that uses normalised Hi-C data, partitions it, and then performs high-resolution MDS on each partition separately and in parallel, yielding a high-resolution structure for each partition. Using low-resolution data, the partitions are then assembled into a global structure, depicting the three-dimensional organisation of the chromatin. | - Greater speed and lower memory requirements compared to alternative methods - High resolution |
- The output structure sometimes contains densely clustered sites with indistinguishable chromosomal features | http://mahonylab.org/software/minimds/ | |
| Chrom3D | Chrom3D is a software that combines 5C/Hi-C data and lamina-associated domain data to simulate the spatial organisation of chromosome domains with respect to each other and to the nuclear space. | - Chromatin organisation predicted with respect to the nuclear space and nuclear inner membrane location | - A spherical nuclear boundary is assumed | https://github.com/Chrom3D/Chrom3D | |
| Chromosome3D | Chromosome3D is a tool that uses Hi-C data to reconstruct the three-dimensional structure of small genetic regions or chromosomes. Chromosome3D transforms Hi-C contact counts into Euclidean distances between chromosomal regions and produces a structural reconstruction model. | - High speed - Reliable and robust results against noise in Hi-C data |
- Inability to handle long genomic regions - Not suitable for genome-wide investigations |
http://sysbio.rnet.missouri.edu/chromosome3d/ | |
| Comparative genomics tools | BLAST | BLAST compares the nucleotides of a queried sequence with a reference and finds matches and regions of similarity across species. | - Comprehensive collection of genomes available | - Low resolution for highly repetitive sequences | www.ncbi.nlm.nih.gov |
| PhastCons | PhastCons identifies evolutionary conserved sequences by performing multiple alignments of the genome of different species and employing a phylogenetic hidden Markov model. The model assesses evolutionary conservation as the probability of negative selection, with scores ranging from 0 to 1. Multiple versions of PhastCons exist. | - Unsupervised method, resilient to possibly incomplete and erroneous annotations - Suitable to evaluate element-based conservation (identification of conserved elements) - Integrates signals from consecutive nucleotides |
- Fast evolving sequences are not assessed by the model | http://hgdownload.cse.ucsc.edu/goldenPath/hg38/phastCons20way/ | |
| PhyloP | PhyloP measures evolutionary conservation at individual alignment sites, under the null hypothesis of neutral evolution. The algorithm produces conservation/acceleration scores. Positive scores represent evolutionary conservation; negative scores indicate accelerated evolution. Different versions of PhyloP exist. | - Suitable to evaluate base-wise conservation (identification of evolutionary signature at specific classes of nucleotides, eg. third codon positions) - Ability to assess acceleration - Evaluates each base independently |
- A constant evolutionary rate is assumed | http://hgdownload.cse.ucsc.edu/goldenPath/hg38/phyloP20way/ | |
| Online databases |
ENCODE | ENCODE stores a comprehensive catalogue of DNA functional elements, produced by combining multiple assays: DNase-I hypersensitivity assay, DNA methylation assay, ChIP-seq data | https://www.encodeproject.org/ | ||
| FANTOM5 | FANTOM5 uses Cap Analysis of Gene Expression (CAGE) and maps regulatory sequences in multiple cell types and tissues, producing an atlas of TFBSs, promoters and enhancers. Recently, atlases for miRNA and lncRNA have been released | http://fantom.gsc.riken.jp/5/ | |||
| PsychENCODE | PsychENCODE stores a comprehensive catalogue of the gene regulatory landscape of the human brain, produced combining transcriptomic, epigenetic and genomic data from adult and developing human brain tissue | http://www.psychencode.org | |||
| RefMap | By supplying epigenetic and GWAS data from a particular disease, RefMap outputs the active genetic regions in a particular cell type and relevant to that disease. |
All strategies listed contribute to phase number 3 of the pipeline illustrated below in Figure 3. TFBS: transcription factor binding site, TF: transcription factor.
4.1. Transcription Factor Binding Site Localisation
One strategy for identifying non-coding regulatory elements is to localise transcription factor binding sites (TFBSs) across the genome, which can be achieved using Chromatin immunoprecipitation coupled with massively parallel DNA sequencing (ChIP-seq), or by associating open-chromatin assays with computational footprinting methods (53–55). Moreover, all possible binding sites of a particular transcription factor can be examined using position-weight matrix methods (PWM) (53, 56).
4.2. Chromatin accessibility assays
Chromatin accessibility assays enable the prediction of the three-dimensional structure of chromatin, recognising the open and active DNA regions and thus allowing the identification of active regulatory elements. Examples of these assays include the DNase-I hypersensitivity assay, the Assay for Transposase-Accessible Chromatin by sequencing (ATAC-seq) and the formaldehyde-assisted isolation of regulatory elements and sequencing (FAIRE-seq) (55, 57–60).
4.3. Epigenetic assays
Epigenetic modifications represent heritable changes (DNA methylation and histone modification) that influence chromatin structure and DNA accessibility, thus regulating the expression of genes, without altering the genetic sequence (61, 62). Epigenetic assays identify the genetic locations where epigenetic changes have occurred throughout the genome, and this information can be used to locate non-coding regulatory elements. For example, tri-methylation at the lysine-4 residue of histone protein H3 (H3K4me3) is an epigenetic modification that makes chromatin more accessible to transcription factors and is associated with active promoters (63). The most commonly used techniques to track the genome-wide distribution of epigenetic modifications include ChIP-seq, which can be combined with methylation-sensitive PCR (MSP), whole-genome bisulfite sequencing and the Illumina MethylationEPIC [850k] Array (64–66).
4.4. Chromosome Conformation Capture methods
Chromosome Conformation Capture (3C) methods are techniques that predict the physical interactions between genetic loci (67). Since these interactions may reflect interactions between promoters and regulatory elements, 3C methods may help identify non-coding regulatory regions (68). Computational tools, including miniMDS, Chrom3D and Chromosome3D, use chromosome conformation capture data to predict the three-dimensional organisation of the chromatin (69–73).
4.5. Comparative Genomics tools
Comparative genomics tools may be used to identify in silico conserved non-coding sequences, which may correspond to functional regions according to the evolutionary conservation principle (74). The hypothesis is that vertebrates use the same regulatory sequences across phylogeny to control gene expression and, assuming that mutations in such sequences are deleterious or disadvantageous to the organism, these regions are likely to have remained stable and unmutated throughout evolution (75, 76). However, non-coding sequences are known to have a higher evolutionary turnover than protein-coding sequences, such that conservation per se is not a strong indicator of functional relevance but may be useful if combined with other types of data. Examples of comparative genomics and sequence conservation tools are the Basic Local Alignment Search Tool (BLAST), PhastCons and PhyloP (77–81).
4.6. Online databases
The use of online databases is the most comprehensive and convenient method to identify regulatory regions throughout the genome because these integrate data from many of the assays described above to accurately locate functional elements. The main limitation of these databases is that the majority of the regulatory elements for which they provide information, especially enhancers, are simply putative, predicted regulatory regions, of which only a small portion has been experimentally validated (82). Examples include the Encyclopedia of DNA Elements (ENCODE database), the Functional Annotation of the Mammalian Genome project (FANTOM5), the PsychENCODE Consortium and the machine learning tool RefMap (83, 84).
All the methods and strategies described above contribute to phase 3 of the pipeline illustrated in Figure 3.
5. Variant annotation
Once the variants falling in regulatory regions of the genome have been identified, the next and most difficult step is variant annotation, that is assigning a functional impact to each variant. This is a major challenge due to various factors, including the issue of identifying the target gene(s) regulated by a particular regulatory element (85). Regulatory elements may modulate the expression of nearby genes, referred to as cis-regulation, or distantly located genes, called trans-regulation. A further complication is the presence of groups of alleles that co-occur and are co-inherited, a phenomenon known as linkage disequilibrium (LD). LD structure complicates the functional evaluation of variants because, assuming a group of SNPs in LD, it is difficult to determine the actual causal variant that affects the phenotype (85, 86). Additionally, variants falling in regulatory regions are likely to have a small and quantitative functional effect, which is much more difficult to detect and interpret than the large qualitative consequences caused by many deleterious variants in protein-coding genes (87, 88).
There are several methods utilised to score non-coding variants and predict their potential functional impact (Table 2). One of the most widely used prioritisation and functional prediction tool is the Combined Annotation Dependent Depletion (CADD) software. CADD uses a linear kernel support vector machine (SVM) trained to distinguish between variants defined as “fixed”, which represent ancestral variants that have arisen in the distant past and are considered to be beneficial or neutral, and “simulated”, de novo variants, which may be either neutral or deleterious. For any queried variant, CADD combines more than 60 annotations and predicts the functional effect of a variant, based on whether the variant is likely to be an observed or simulated variant (89, 90). One of the limitations of CADD is that the training model used to classify the variants as benign or pathogenic is imperfect, such that an unknown proportion of variants that are classified by CADD as deleterious are actually neutral. To overcome this issue, the newer version of CADD correlates the model predictions with experimentally validated functional effect data to calibrate the parameters and evaluate CADD performance (90). Moreover, to provide robustness to the interpretation, it is useful to integrate CADD prediction scores with additional data, such as independent conservation scores and predictions from other functional annotation software, including the Genome-Wide Annotation of Variants (GWAVA) software, the FATHMM-MKL and the Variant Effect Predictor (VEP) software.
Table 2:
Techniques and resources used to annotate non-coding variants.
| Non-coding variant annotation | |||||
|---|---|---|---|---|---|
| Tool | Data sources | Purpose | Output | Link | Accessibility |
| CADD | • ENCODE • UCSC • Ensembl • VEP • Conservation data |
Annotation of coding and non-coding variants | C-score, indicating pathogenicity | http://cadd.gs.washington.edu/ | Software download Online |
| GWAVA | • ENCODE • Ensembl • Conservation data • GC-content data |
Annotation of non-coding variants | Prediction score (0–1) | https://www.sanger.ac.uk/sanger/StatGen_Gwava | Software download Online |
| FATHMM-MKL | • ChIP-seq data • TFBSs prediction • Conservation data • DNase-I hypersensitivity assay |
Annotation of coding and non-coding variants | p-value, indicating deleteriousness | http://fathmm.biocompute.org.uk | Software download Online |
| FATHMM-XF | • FATHMM-MKL • ENCODE • Roadmap Epigenomics Project data |
Annotation of coding and non-coding variants | p-value, indicating deleteriousness | http://fathmm.biocompute.org.uk/fathmm-xf/ | Software download Online |
| VEP | • Ensembl Regulatory Build • ENCODE • CADD • Conservation data |
Annotation of coding and non-coding variants | Variant location, functional effect | http://www.ensembl.org/info/docs/tools/vep/index.html | Web-interface Command-line tool |
| Other Tools | |||||
| GTEx | • WGS data • RNAseq data |
eQTL analysis | eQTL | https://gtexportal.org/home/ | Web-interface |
| PsychENCODE | • WGS • Epigenomic data Transcriptomic data |
Catalogue of the gene regulatory landscape of the human brain | Genomic elements active in the human brain | http://www.psychencode.org/?page_id=160 | Data download |
| H-MAGMA | • Genomic data • Hi-C data |
Noncoding variant-target gene prediction | Target gene prediction | https://github.com/thewonlab/H-MAGMA | Command-line tool |
GWAVA integrates multiple genomic and epigenomic annotations to predict the functional impact of variants, but can also be applied to a given genetic region, returning all known variants in that region and the corresponding prediction scores (87). FATHMM-MKL is a variant annotation software suitable for both coding and non-coding variants, characterised by the ability to integrate and weight different types of annotations based on relevance and availability: not all annotations are available for all genomic positions, therefore FATHMM-MKL produces a p-value for all positions adjusting the weights relative to the available annotations (91). The improved version of FATHMM-MKL is called FATHMM-XF (FATHMM with extended features), and exhibits greater prediction accuracy than FATHMM-MKL (92, 93). VEP is also frequently used for variant annotation, because it predicts the functional consequences of variants and integrates multiple conservation and functional annotation scores, as well as identifying the location where a variant falls (whether it falls within a protein-coding sequence, non-coding RNA or non-coding regulatory region) (94, 95).
Another useful tool for assessing the implications of non-coding variation is the Genotype-Tissue Expression database (GTEx), which represents a comprehensive public catalogue of tissue-specific gene expression and regulation data (96). The information stored in the GTEx catalogue can be used to determine whether a queried variant functions as an eQTL for specific genes and is capable of modulating the expression of protein-coding genes. In the investigation of non-coding variants, eQTL data, such as those produced by GTEx, may be useful for identifying likely target genes that are affected by a particular non-coding regulatory variant (97). For brain-related data and neurological disorder studies, the same type of information obtained from GTEx can be collected through the PsychENCODE Consortium: a multi-site project that aims at creating a comprehensive catalogue of the gene regulatory landscape of the human brain (98). An additional tool that may be useful to annotate non-coding variants in brain-related studies is Hi-C coupled multimarker analysis of genomic annotation (H-MAGMA) (99).
6. Non-coding variation in disease
The first evidence of direct involvement of non-coding variation in disease dates back to 1982, when a single nucleotide substitution was detected in the promoter region of the haemoglobin subunit beta gene (HBB) (encoding β-globin, a subunit of haemoglobin) and was found to reduce HBB gene expression (100). Subsequently, the variant was demonstrated to alter the binding of a transcription factor. Since then, with the emergence of genomic sequencing and the technical advances, the evidence supporting the involvement of ncDNA in health and disease has grown. Indeed, GWAS studies have shown that more than 88% of disease and trait-associated variants fall within non-coding regions of the genome (101, 102).
Numerous non-coding variants have been shown to contribute in different ways to many diseases; they may function as disease-modifiers, altering disease susceptibility, and in some cases represent the disease-causing variant. One example is Liebenberg syndrome, a rare genetic condition characterised by abnormal development of the arms, carpal bone defects and brachydactyly. This condition is caused by structural changes (deletions and translocations) occurring within an enhancer of the paired-like homeodomain 1 gene (PITX1), which is part of the RIEG/PITX homeobox family. PITX1 is expressed in the lower limbs and is involved in leg development; in Liebenberg syndrome, improper PITX1 activation in the upper limbs leads to the partial transformation of the arms into leg-like limbs (103–105). In autism spectrum disorder (ASD), paternally inherited non-coding structural variations have been shown to be associated with an increased risk of ASD in children (13). In Parkinson’s disease, protective and susceptibility risk variants have been described within an enhancer of SNCA, a key gene in Parkinson’s disease pathogenesis. One of the reported variants lowers the expression of SNCA, leading to lower risk of developing Parkinson’s disease, whereas the other reported variant increases SNCA expression, leading to an increased risk of developing Parkinson’s disease (12).
Furthermore, structural variations affecting TAD boundaries are also of relevance. Many studies have shown that disruptions of TAD boundaries and alterations of CTCF-binding sites result in improper chromosomal contacts and altered gene promoter-enhancer interactions (106–108).
In cancer biology, many non-coding variations have been found in the regulatory regions of cancer-related genes and have been found to function as cancer-drivers (10, 11, 109). One example is the case of the telomerase reverse transcriptase gene (TERT) promoter. TERT encodes the catalytic subunit of telomerase, an enzyme that regulates elongation of telomeres, repeated sequences localised at the ends of chromosomes, the purpose of which is to protect chromosomes from end-to-end fusion and end-degradation (110). End-degradation represents the physiological loss of the terminal portion of the DNA strand that occurs during DNA replication. To prevent the loss of coding sequences, telomeres create a protective cap at the ends of chromosomes that is gradually degraded during DNA replication cycles. Physiologically, telomeres progressively shorten throughout life due to the inactivation of TERT (111). In cancer cells, TERT reactivates and results in minimal telomere shortening, leading to telomere stabilisation and the capacity for indefinite cell proliferation (112). Variations within the TERT promoter were originally identified in melanoma: a highly recurrent promoter variant was found to be involved in the tumorigenic process, as it created a novel binding motif for transcription factors, thus supporting permanent telomerase expression (113). Subsequently, variations of the TERT promoter have been described in glioblastoma, hepatocellular carcinoma, bladder cancer, thyroid cancer and breast cancer, among others, associated with increased telomerase activity and in some cases with poor survival (112, 114–118). TERT promoter mutation status may influence treatment response and can be used to predict how patients will respond to treatments: for example, TERT promoter variants have been associated with resistance to radiotherapy in patients with glioma (119–121). The status of the TERT promoter may also be used to stratify patients and predict prognosis (122–125). Furthermore, the TERT promoter is currently being investigated as a potential therapeutic target: a recently published preclinical study explored the use of programmable CRISPR-based base editing on the TERT promoter to reverse the mutation that activates TERT expression and thereby inhibit tumour growth (110, 112).
7. Evidence of non-coding variation in the epilepsies
Many epilepsies are characterised by significant and usually unexplained phenotypic variability, which could be potentially associated with genetic variation in non-coding sequences, functioning as disease modifiers. Additionally, ncDNA may potentially harbour genetic variants that act as disease risk variants, which could be explored as prognostic biomarkers to stratify patients and identify those with a higher risk of developing comorbidities or experiencing more severe symptoms (126). The investigation of ncDNA in epilepsy has been primarily aimed at non-coding RNAs (127, 128). Additional work has been carried out investigating the methylation state and epimutations in non-coding DNA regions (129, 130); however, overall, the non-coding regulatory regions of the genome remain relatively unexplored.
6.1. Variation in promoter regions
There is some limited evidence of variation in the promoter region of several epilepsy-associated genes possibly modulating gene transcription and contributing to the pathogenesis and phenotypic variability of epilepsy. One example of such variation is in the promoter region of the sodium voltage-gated channel alpha subunit 1 gene (SCN1A), which encodes the voltage-gated sodium channel type I alpha subunit and plays a crucial role in the initiation and propagation of action potentials. Mutations in this gene are associated with a wide spectrum of epilepsies, including Dravet Syndrome and genetic epilepsy with febrile seizures plus (GEFS+) (131, 132). Gao et al. studied the promoter region of SCN1A in a cohort of patients with epilepsy and febrile seizures, who did not carry pathogenic variants in the coding sequence, and detected a heterozygous mutation in the SCN1A promoter. They demonstrated that this variant caused a decrease in promoter activity (of approximately 42%) and was responsible for a mild epilepsy phenotype with incomplete penetrance (133). Furthermore, de Lange et al. have shown that common variations in the promoter of the unaffected SCN1A allele influence the disease severity in patients with a pathogenic SCN1A variant. They assessed the functional consequences of five different SCN1A promoter variants, identifying reduced expression and reduced channel function, leading to a more severe phenotype, thus indicating a disease-modifier effect (134). Neither of these studies has been replicated.
Additionally, evidence exists for a relationship between altered methylation status in the promoter regions of genes and the pathogenesis of epilepsy. Although not confirmed, such methylation alterations may occur because of genetic variations in gene promoter sequences. One example is temporal lobe epilepsy, in which hyper-methylation in the promoter region of the reelin gene (RELN), was reported (135). In addition, Belhedi et al. described an increased methylation status in the promoter region of the carboxypeptidase A6 gene (CPA6) in patients with focal epilepsy and febrile seizures (136). Neither study has been replicated, and the credibility of RELN as a gene of relevance in epilepsy per se has been questioned (137). While still a relatively young field, important research has been carried out on changes in the methylation state of non-coding sequences (129, 130).
7.2. Non-coding structural variations
Structural variations are also known to play a relevant role in epilepsy, and there is evidence of structural variation occurring in non-coding sequences, associated with epilepsy pathogenesis. One example is benign adult familial myoclonic epilepsy (BAFME), also described as familial adult myoclonic epilepsy (FAME), autosomal dominant cortical myoclonus and epilepsy (ADCME) and familial cortical tremor and epilepsy (FCTE), which is associated with the aberrant expansion of TTTCA and TTTTA repeats in intronic regions of multiple genes. For BAFME type 1, multiple studies reported a repeat expansion in intronic regions of the sterile alpha motif domain containing 12 gene (SAMD12) (52, 138). In patients with BAFME type 6, repeat expansions were found in the intronic regions of the trinucleotide repeat containing adaptor 6A gene (TNRC6A), in patients with BAFME7 in introns of the rap guanine nucleotide exchange factor 2 gene (RAPGEF2) and the star related lipid transfer domain containing 7 gene (STARD7), whereas in patients with BAFME4 expansions were identified within an intron of the YEATS domain containing 2 gene (YEATS2) and in patients with BAFME3 in intronic regions of the membrane associated ring-CH-type finger 6 gene (MARCH6) (51, 138–140). Such heterogeneity of culpable genes and the presence of the repeat expansion in all the types of BAFME suggests a correlation between the repeat expansion and the pathogenesis of BAFME, regardless of the gene in which the expansion occurs (51, 138, 139).
Another example is progressive myoclonus epilepsy of the Unverricht–Lundborg type (EPM1), which is associated with expansions of a dodecamer repeat in the promoter region of the cystatin B gene (CSTB) (50, 141). Normal alleles have two to three dodecamer repeats, while more than 30 repeats have been found in people with EPM1. A correlation between the expansion size and disease severity is yet to be clarified, although patients who have compound heterozygosity for the repeat expansion and InDels or point mutations present a more severe phenotype than patients homozygous for the dodecamer expansion (50, 142).
Microdeletions have been found within the promoter region of SCN1A. Nakayama et al. described two cases of Dravet syndrome carrying microdeletions in the SCN1A promoter region that resulted in SCN1A haploinsufficiency and reduced channel protein levels, leading to Dravet Syndrome (143). Additionally, Haigh and colleagues have shown that mice harbouring deletions in the non-canonical promoter region of Scn1a, which include the alternative TSS 1b, exhibit a significant reduction in the expression of Scn1a and an epileptic phenotype (144, 145).
One study reported somatic copy number gains in the enhancer region of the epidermal growth factor receptor gene (EGFR) and the promoter region of the platelet derived growth factor receptor alpha gene (PDGFRA), without alterations in the coding sequence, in brain tissue from patients with focal cortical dysplasia operated for treatment-resistant epilepsy. In addition to the amplification of non-coding regulatory elements, an upregulation of EGFR and PDGFRA was also reported. However, this correlation has not been experimentally confirmed and the mechanism responsible for this association was not addressed in the study (146).
Monlong et al. investigated CNVs in a cohort of 198 patients with epilepsy and found an enrichment of non-coding CNVs close to known epilepsy genes, which likely fall into regulatory sequences (147). However, the functional effect of these non-coding CNVs has not been experimentally verified.
8. Strategy to investigate non-coding variation and challenges
A putative schematic representation of the steps to be taken to investigate non-coding genetic variation is shown in Figure 3. In the preliminary phases of the pipeline, blood is currently the gold standard source for DNA collection, as blood-derived DNA samples are of higher quality and are more likely to pass stringent quality controls (QCs) of DNA integrity and concentration, whereas saliva-derived samples, for example, are more prone to QC failure (148). The major drawback of this approach is the inability to detect somatic genetic variations: in the case of epilepsy, for example, somatic variations may occur in the brain, and might only be detectable with the use of brain-tissue derived DNA samples. Relating to the sequencing approach, although the majority of studies use Whole Exome Sequencing (WES) or targeted sequencing panels, neither of these two methods generate information about non-coding elements; for genome-wide exploration, Whole Genome Sequencing (WGS) is the most appropriate approach.
As shown in the figure, the final phase of the workflow is the functional validation of the candidate variants, which is performed predominantly using wet-lab experimental approaches. The most widely used experimental methods are luciferase reporter assays and the use of CRISPR system to generate viable models. The luciferase reporter assay aims to compare the level of luciferase expression in the presence and absence of a variant of interest (149). The CRISPR-mediated genome editing system can be used to generate both cellular and animal models, such as murine models, carrying the candidate variation, thus allowing evaluation of the functional impact in vivo (150).
However, in the investigation of non-coding variants, multiple challenges need to be highlighted. First, the challenge of predicting the functional consequences of non-coding variation, which can result in discordant predictions from different annotation tools: this is a major limitation that also applies to the investigation of protein-coding variants (151). Furthermore, unlike the investigation of protein-coding variants, another challenge in the study of non-coding variation is the current lack of a variation database, which would allow researchers to quickly determine whether a non-coding variant has been previously observed and linked to disease. Second, the detection of non-coding regulatory regions, such as promoter regions, which is complicated by the existence of overlapping genes: distinct genes that share a genetic region. It is estimated that about one-quarter of human protein-coding genes overlap with each other. Most of these are co-expressed in the same tissue type and are likely to be co-regulated (152). The presence of overlapping genes makes it difficult not only to identify the regulatory sequences flanking a protein-coding gene, but also to understand the functional consequences of variants. Indeed, assuming a pair of genes are overlapping, genetic variations falling in the regulatory sequence upstream of both genes may have an impact on the expression of both genes, while variants falling in the regulatory sequence upstream of the second gene may also fall within the coding sequence of the first gene, thus further complicating the functional interpretation of non-coding variants. Third, the reliability of data on regulatory element localisation: to date, most of the available data, particularly for enhancers, is simply prediction, with only a small portion being experimentally validated data, thus limiting the robustness of results and highlighting the need to produce experimentally-validated enhancer data resources. Recently, non-human model systems have been used to identify and experimentally validate non-coding regulatory elements (153).
9. Conclusions and future perspective
In conclusion, despite the lack of a comprehensive and systematic genome-wide investigation of non-coding regulatory variation in epilepsy, we suggest that ncDNA has the potential to be of relevance in epilepsy research. Non-coding regulatory regions may harbour variations that influence gene expression and contribute to the phenotypic variability of disease, may have a disease-modifying effect, or influence treatment response. Furthermore, the findings of studies in other fields indicate that non-coding variation may also represent the main source of disease causation, and, eventually, may represent potential therapeutic targets for innovative treatment strategies. Overall, the non-coding genome represents an exciting area to investigate.
Acknowledgments
AF and JMM are supported by the Wellcome Trust [Grant number 108749/Z/15/Z], the National Human Genome Research Institute of the National Institutes of Health under award number 2U41HG007234 and the European Molecular Biology Laboratory. For the purpose of open access, the author has applied a CC BY public copyright licence to any Author Accepted Manuscript version arising from this submission. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Ensembl is a registered trademark of EMBL.
List of abbreviations
- 3C
Chromosome Conformation Capture
- 5C
3C-carbon copy
- ADCME
autosomal dominant cortical myoclonus and epilepsy (alternative names are BAFME, FAME, FCTE)
- ALS
amyotrophic lateral sclerosis
- ASD
autism spectrum disorder
- ATAC-seq
assay for transposase-accessible chromatin by sequencing
- BAFME
benign adult familial myoclonic epilepsy (alternative names are ADCME, FAME, FCTE)
- BLAST
Basic Local Alignment Search Tool
- BRE
TFIIB recognition element
- CADD
Combined Annotation Dependent Depletion
- ChIP-seq
chromatin immunoprecipitation coupled with massively parallel DNA sequencing
- circRNA
circular RNA
- CNTNAP2
contactin-associated protein 2 gene
- CNV
copy number variation
- CPA6
carboxypeptidase A6 gene
- CSTB
cystatin B gene
- CTCF
CCCTC-binding factor
- DPE
downstream promoter elements
- EGFR
epidermal growth factor receptor gene
- ENCODE
Encyclopedia of DNA Elements.
- EPM1
progressive myoclonus epilepsy of the Unverricht–Lundborg type
- eQTL
expression Quantitative Trait Locus
- FAIRE-seq
formaldehyde-assisted isolation of regulatory elements and sequencing
- FAME
adult myoclonic epilepsy (alternative names are ADCME, BAFME, FCTE)
- FANTOM5
Functional Annotation of the mammalian genome
- FCTE
familial cortical tremor and epilepsy (alternative names are ADCME, BAFME, FAME)
- FDT
frontotemporal dementia
- GTEx
Genotype-Tissue Expression
- GWAS
genome-wide association study
- GWAVA
Genome-Wide Annotation of Variants
- HBB
haemoglobin beta subunit gene
- Inr
Initiator element
- KRAB-ZFPs
zinc finger proteins containing the Kruppel-associated box
- LD
linkage disequilibrium
- LINE-1/L1
long interspersed nuclear element-1
- lncRNA
long non-coding RNA
- MARCH6
membrane associated ring-CH-type finger 6 gene
- MECP2
methyl-CpG-binding protein 2 gene
- miRNA
microRNA
- MSP
methylation sensitive PCR
- ncDNA
non-coding DNA
- NF1
neurofibromatosis type 1
- PDGFRA
platelet derived growth factor receptor alpha gene
- piRNA
piwi-interacting RNA
- PITX1
paired-like homeodomain 1 gene
- PIWI
P-element Induced WImpy testis in Drosophila
- PWM
position weight matrix
- QC
quality control
- RAPGEF2
rap guanine nucleotide exchange factor 2 gene
- RELN
reelin gene
- rRNA
ribosomal RNA
- SAMD12
sterile alpha motif domain containing 12 gene
- SCN1A
sodium voltage-gated channel alpha subunit 1 gene
- snoRNA
small nucleolar RNA
- SNP
single nucleotide polymorphism
- snRNA
small nuclear RNA
- STARD7
star related lipid transfer domain containing 7 gene
- TAD
topologically associating domain
- TE
transposable element
- TERT
telomerase reverse transcriptase gene
- TF
transcription factor
- TFBS
transcription factor binding site
- TFIIB
Transcription factor II B
- TNRC6A
trinucleotide repeat containing adaptor 6A gene
- tRNA
transfer RNA
- TSS
transcription start site
- VEP
Variant Effect Predictor
- WES
Whole Exome Sequencing
- WGS
Whole Genome Sequencing
- YEATS2
YEATS domain containing 2 gene
10. References
- 1.Vinogradov AE, Anatskaya OV. Organismal complexity, cell differentiation and gene expression: human over mouse. Nucleic Acids Research. 2007;35(19):6350–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ENCODE. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graur D An Upper Limit on the Functional Fraction of the Human Genome. Genome Biol Evol. 2017;9(7):1880–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohno S So much “junk” DNA in our genome. Brookhaven Symp Biol. 1972;23:366–70. [PubMed] [Google Scholar]
- 5.Gardiner K Clonability and gene distribution on human chromosome 21: reflections of junk DNA content? Gene. 1997;205(1):39–46. [DOI] [PubMed] [Google Scholar]
- 6.Makałowski W Genomic scrap yard: how genomes utilize all that junk. Gene. 2000;259(1–2):61–7. [DOI] [PubMed] [Google Scholar]
- 7.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. [DOI] [PubMed] [Google Scholar]
- 8.Hombach S, Kretz M. Non-coding RNAs: Classification, Biology and Functioning. Adv Exp Med Biol. 2016;937:3–17. [DOI] [PubMed] [Google Scholar]
- 9.Silva J, Fernandes R, Romão L. Translational Regulation by Upstream Open Reading Frames and Human Diseases. Adv Exp Med Biol. 2019;1157:99–116. [DOI] [PubMed] [Google Scholar]
- 10.Wadi L, Uusküla-Reimand L, Isaev K, Shuai S, Huang V, Liang M, et al. Candidate cancer driver mutations in superenhancers and long-range chromatin interaction networks. bioRxiv. 2017:236802. [DOI] [PubMed] [Google Scholar]
- 11.Rheinbay E, Nielsen MM, Abascal F, Tiao G, Hornshøj H, Hess JM, et al. Discovery and characterization of coding and non-coding driver mutations in more than 2,500 whole cancer genomes. bioRxiv. 2017:237313. [Google Scholar]
- 12.Soldner F, Stelzer Y, Shivalila CS, Abraham BJ, Latourelle JC, Barrasa MI, et al. Parkinson-associated risk variant in distal enhancer of α-synuclein modulates target gene expression. Nature. 2016;533(7601):95–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandler WM, Antaki D, Gujral M, Kleiber ML, Whitney J, Maile MS, et al. Paternally inherited cis-regulatory structural variants are associated with autism. Science. 2018;360(6386):327–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mei D, Cetica V, Marini C, Guerrini R. Dravet syndrome as part of the clinical and genetic spectrum of sodium channel epilepsies and encephalopathies. Epilepsia. 2019;60 Suppl 3:S2–s7. [DOI] [PubMed] [Google Scholar]
- 15.Wu J, Brown M. Chapter 2 - Epigenetics and Epigenomics. In: Hoffman R, Benz EJ, Silberstein LE, Heslop HE, Weitz JI, Anastasi J, et al. , editors. Hematology (Seventh Edition): Elsevier; 2018. p. 17–24. [Google Scholar]
- 16.Oubounyt M, Louadi Z, Tayara H, Chong KT. DeePromoter: Robust Promoter Predictor Using Deep Learning. Front Genet. 2019;10:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadonaga JT. Perspectives on the RNA polymerase II core promoter. Wiley Interdiscip Rev Dev Biol. 2012;1(1):40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leppek K, Das R, Barna M. Functional 5’ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol. 2018;19(3):158–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu M, Gonzalez-Hurtado E, Martinez E. Core promoter-specific gene regulation: TATA box selectivity and Initiator-dependent bi-directionality of serum response factor-activated transcription. Biochim Biophys Acta. 2016;1859(4):553–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol. 2010;339(2):225–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vo Ngoc L, Kassavetis GA, Kadonaga JT. The RNA Polymerase II Core Promoter in Drosophila. Genetics. 2019;212(1):13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haberle V, Stark A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat Rev Mol Cell Biol. 2018;19(10):621–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao L, Wang S, Cao Z, Ouyang W, Zhang Q, Xie L, et al. Chromatin loops associated with active genes and heterochromatin shape rice genome architecture for transcriptional regulation. Nature Communications. 2019;10(1):3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nature Reviews Molecular Cell Biology. 2015;16(3):144–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, et al. Enhancer redundancy provides phenotypic robustness in mammalian development. Nature. 2018;554(7691):239–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kvon EZ, Waymack R, Elabd MG, Wunderlich Z. Enhancer redundancy in development and disease. Nature Reviews Genetics. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conant GC, Wolfe KH. Functional partitioning of yeast co-expression networks after genome duplication. PLoS Biol. 2006;4(4):e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McArthur E, Capra JA. Topologically associating domain (TAD) boundaries stable across diverse cell types are evolutionarily constrained and enriched for heritability. bioRxiv. 2020:2020.01.10.901967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muñoz-López M, García-Pérez JL. DNA transposons: nature and applications in genomics. Curr Genomics. 2010;11(2):115–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klawitter S, Fuchs NV, Upton KR, Muñoz-Lopez M, Shukla R, Wang J, et al. Reprogramming triggers endogenous L1 and Alu retrotransposition in human induced pluripotent stem cells. Nat Commun. 2016;7:10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161(2):228–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, et al. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24(12):1963–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Playfoot CJ, Duc J, Sheppard S, Dind S, Coudray A, Planet E, et al. Transposable elements and their KZFP controllers are drivers of transcriptional innovation in the developing human brain. Genome Res. 2021;31(9):1531–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435(7044):903–10. [DOI] [PubMed] [Google Scholar]
- 35.Kazazian HH Jr., Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332(6160):164–6. [DOI] [PubMed] [Google Scholar]
- 36.Wimmer K, Callens T, Wernstedt A, Messiaen L. The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLoS Genet. 2011;7(11):e1002371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muotri AR, Marchetto MCN, Coufal NG, Oefner R, Yeo G, Nakashima K, et al. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468(7322):443–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao B, Wu Q, Ye AY, Guo J, Zheng X, Yang X, et al. Somatic LINE-1 retrotransposition in cortical neurons and non-brain tissues of Rett patients and healthy individuals. PLoS Genet. 2019;15(4):e1008043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang P, Wang Y, Macfarlan TS. The Role of KRAB-ZFPs in Transposable Element Repression and Mammalian Evolution. Trends Genet. 2017;33(11):871–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molaro A, Malik HS. Hide and seek: how chromatin-based pathways silence retroelements in the mammalian germline. Curr Opin Genet Dev. 2016;37:51–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hrdlickova B, de Almeida RC, Borek Z, Withoff S. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta. 2014;1842(10):1910–22. [DOI] [PubMed] [Google Scholar]
- 42.Chander V, Gibbs RA, Sedlazeck FJ. Evaluation of computational genotyping of structural variation for clinical diagnoses. GigaScience. 2019;8(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Haraksingh R, Grubert F, Abyzov A, Gerstein M, Weissman S, et al. Child development and structural variation in the human genome. Child Dev. 2013;84(1):34–48. [DOI] [PubMed] [Google Scholar]
- 44.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nature Reviews Genetics. 2015;16(3):172–83. [DOI] [PubMed] [Google Scholar]
- 45.Makino T, McLysaght A, Kawata M. Genome-wide deserts for copy number variation in vertebrates. Nature Communications. 2013;4(1):2283. [DOI] [PubMed] [Google Scholar]
- 46.Jain A, Vale RD. RNA phase transitions in repeat expansion disorders. Nature. 2017;546(7657):243–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ellerby LM. Repeat Expansion Disorders: Mechanisms and Therapeutics. Neurotherapeutics. 2019;16(4):924–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campuzano V, Montermini L, Moltò MD, Pianese L, Cossée M, Cavalcanti F, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271(5254):1423–7. [DOI] [PubMed] [Google Scholar]
- 49.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bosak M, Sułek A, Łukasik M, Żak A, Słowik A, Lasek-Bal A. Genetic testing and the phenotype of Polish patients with Unverricht–Lundborg disease (EPM1) — A cohort study. Epilepsy & Behavior. 2020;112:107439. [DOI] [PubMed] [Google Scholar]
- 51.Florian RT, Kraft F, Leitão E, Kaya S, Klebe S, Magnin E, et al. Unstable TTTTA/TTTCA expansions in MARCH6 are associated with Familial Adult Myoclonic Epilepsy type 3. Nat Commun. 2019;10(1):4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu C, Song Y, Yuan Y, Peng Y, Pang N, Duan R, et al. TTTCA Repeat Expansion of SAMD12 in a New Benign Adult Familial Myoclonic Epilepsy Pedigree. Front Neurol. 2020;11:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jayaram N, Usvyat D, R. Martin AC. Evaluating tools for transcription factor binding site prediction. BMC Bioinformatics. 2016;17(1):547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gusmao EG, Allhoff M, Zenke M, Costa IG. Analysis of computational footprinting methods for DNase sequencing experiments. Nat Methods. 2016;13(4):303–9. [DOI] [PubMed] [Google Scholar]
- 55.Li Z, Schulz MH, Look T, Begemann M, Zenke M, Costa IG. Identification of transcription factor binding sites using ATAC-seq. Genome Biology. 2019;20(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aerts S Chapter five - Computational Strategies for the Genome-Wide Identification of cis-Regulatory Elements and Transcriptional Targets. In: Plaza S, Payre F, editors. Current Topics in Developmental Biology. 98: Academic Press; 2012. p. 121–45. [DOI] [PubMed] [Google Scholar]
- 57.Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010(2):pdb.prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc. 2012;7(2):256–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21 9 1–9 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nature Methods. 2017;14(10):959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bogdanović O, Lister R. DNA methylation and the preservation of cell identity. Current Opinion in Genetics & Development. 2017;46:9–14. [DOI] [PubMed] [Google Scholar]
- 62.Lawrence M, Daujat S, Schneider R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends in Genetics. 2016;32(1):42–56. [DOI] [PubMed] [Google Scholar]
- 63.Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537(7621):553–7. [DOI] [PubMed] [Google Scholar]
- 64.Li J, Li R, Wang Y, Hu X, Zhao Y, Li L, et al. Genome-wide DNA methylome variation in two genetically distinct chicken lines using MethylC-seq. BMC Genomics. 2015;16(1):851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Urich MA, Nery JR, Lister R, Schmitz RJ, Ecker JR. MethylC-seq library preparation for base-resolution whole-genome bisulfite sequencing. Nature Protocols. 2015;10(3):475–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Olova N, Krueger F, Andrews S, Oxley D, Berrens RV, Branco MR, et al. Comparison of whole-genome bisulfite sequencing library preparation strategies identifies sources of biases affecting DNA methylation data. Genome Biology. 2018;19(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. Mapping Chromatin Interactions by Chromosome Conformation Capture. Current Protocols in Molecular Biology. 2006;74(1):21.11.1–21.11.20. [DOI] [PubMed] [Google Scholar]
- 68.Davies JO, Oudelaar AM, Higgs DR, Hughes JR. How best to identify chromosomal interactions: a comparison of approaches. Nat Methods. 2017;14(2):125–34. [DOI] [PubMed] [Google Scholar]
- 69.Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58(3):268–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rieber L, Mahony S. miniMDS: 3D structural inference from high-resolution Hi-C data. Bioinformatics. 2017;33(14):i261–i6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paulsen J, Liyakat Ali TM, Collas P. Computational 3D genome modeling using Chrom3D. Nature Protocols. 2018;13(5):1137–52. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Z, Li G, Toh KC, Sung WK. 3D chromosome modeling with semi-definite programming and Hi-C data. J Comput Biol. 2013;20(11):831–46. [DOI] [PubMed] [Google Scholar]
- 73.Adhikari B, Trieu T, Cheng J. Chromosome3D: reconstructing three-dimensional chromosomal structures from Hi-C interaction frequency data using distance geometry simulated annealing. BMC Genomics. 2016;17(1):886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server issue):W273–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3(1):e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vassalli QA, Anishchenko E, Caputi L, Sordino P, D’Aniello S, Locascio A. Regulatory elements retained during chordate evolution: coming across tunicates. Genesis. 2015;53(1):66–81. [DOI] [PubMed] [Google Scholar]
- 77.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328(5981):1036–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, et al. Rapid Turnover of Long Noncoding RNAs and the Evolution of Gene Expression. PLOS Genetics. 2012;8(7):e1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Quinn JJ, Zhang QC, Georgiev P, Ilik IA, Akhtar A, Chang HY. Rapid evolutionary turnover underlies conserved lncRNA-genome interactions. Genes Dev. 2016;30(2):191–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15(8):1034–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20(1):110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gasperini M, Tome JM, Shendure J. Towards a comprehensive catalogue of validated and target-linked human enhancers. Nature Reviews Genetics. 2020;21(5):292–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507(7493):455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang S, Cooper-Knock J, Weimer AK, Shi M, Moll T, Harvey C, et al. Genome-wide Identification of the Genetic Basis of Amyotrophic Lateral Sclerosis. 2020. [DOI] [PMC free article] [PubMed]
- 85.Elkon R, Agami R. Characterization of noncoding regulatory DNA in the human genome. Nat Biotechnol. 2017;35(8):732–46. [DOI] [PubMed] [Google Scholar]
- 86.Kim SA, Cho CS, Kim SR, Bull SB, Yoo YJ. A new haplotype block detection method for dense genome sequencing data based on interval graph modeling of clusters of highly correlated SNPs. Bioinformatics. 2018;34(3):388–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ritchie GR, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nat Methods. 2014;11(3):294–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Povysil G, Petrovski S, Hostyk J, Aggarwal V, Allen AS, Goldstein DB. Rare-variant collapsing analyses for complex traits: guidelines and applications. Nat Rev Genet. 2019;20(12):747–59. [DOI] [PubMed] [Google Scholar]
- 89.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Research. 2018;47(D1):D886–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day IN, et al. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31(10):1536–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rogers MF, Shihab HA, Mort M, Cooper DN, Gaunt TR, Campbell C. FATHMM-XF: accurate prediction of pathogenic point mutations via extended features. Bioinformatics. 2018;34(3):511–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rojano E, Seoane P, Ranea JAG, Perkins JR. Regulatory variants: from detection to predicting impact. Brief Bioinform. 2019;20(5):1639–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zerbino DR, Wilder SP, Johnson N, Juettemann T, Flicek PR. The ensembl regulatory build. Genome Biol. 2015;16:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gamazon ER, Segrè AV, van de Bunt M, Wen X, Xi HS, Hormozdiari F, et al. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nature Genetics. 2018;50(7):956–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu Z, Ioannidis NM, Zou J. Predicting target genes of non-coding regulatory variants with IRT. Bioinformatics. 2020;36(16):4440–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.PsychENCODE C Revealing the brain’s molecular architecture. Science. 2018;362(6420):1262–3. [DOI] [PubMed] [Google Scholar]
- 99.Sey NYA, Hu B, Mah W, Fauni H, McAfee JC, Rajarajan P, et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nature Neuroscience. 2020;23(4):583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Orkin SH, Kazazian HH, Antonarakis SE, Goff SC, Boehm CD, Sexton JP, et al. Linkage of β-thalassaemia mutations and β-globin gene polymorphisms with DNA polymorphisms in human β-globin gene cluster. Nature. 1982;296(5858):627–31. [DOI] [PubMed] [Google Scholar]
- 101.Edwards Stacey L, Beesley J, French Juliet D, Dunning Alison M. Beyond GWASs: Illuminating the Dark Road from Association to Function. The American Journal of Human Genetics. 2013;93(5):779–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cannon ME, Mohlke KL. Deciphering the Emerging Complexities of Molecular Mechanisms at GWAS Loci. Am J Hum Genet. 2018;103(5):637–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Al-Qattan MM, Al-Thunayan A, AlAbdulkareem I, Al Balwi M. Liebenberg syndrome is caused by a deletion upstream to the PITX1 gene resulting in transformation of the upper limbs to reflect lower limb characteristics. Gene. 2013;524(1):65–71. [DOI] [PubMed] [Google Scholar]
- 104.Spielmann M, Brancati F, Krawitz PM, Robinson PN, Ibrahim DM, Franke M, et al. Homeotic arm-to-leg transformation associated with genomic rearrangements at the PITX1 locus. Am J Hum Genet. 2012;91(4):629–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kragesteen BK, Brancati F, Digilio MC, Mundlos S, Spielmann M. H2AFY promoter deletion causes PITX1 endoactivation and Liebenberg syndrome. J Med Genet. 2019;56(4):246–51. [DOI] [PubMed] [Google Scholar]
- 106.Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin David U, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162(4):900–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163(7):1611–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lupiáñez DG, Spielmann M, Mundlos S. Breaking TADs: How Alterations of Chromatin Domains Result in Disease. Trends Genet. 2016;32(4):225–37. [DOI] [PubMed] [Google Scholar]
- 109.Hornshøj H, Nielsen MM, Sinnott-Armstrong NA, Świtnicki MP, Juul M, Madsen T, et al. Pan-cancer screen for mutations in non-coding elements with conservation and cancer specificity reveals correlations with expression and survival. NPJ Genom Med. 2018;3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jager K, Walter M. Therapeutic Targeting of Telomerase. Genes (Basel). 2016;7(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shay JW. Role of Telomeres and Telomerase in Aging and Cancer. Cancer Discov. 2016;6(6):584–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li X, Qian X, Wang B, Xia Y, Zheng Y, Du L, et al. Programmable base editing of mutated TERT promoter inhibits brain tumour growth. Nature Cell Biology. 2020;22(3):282–8. [DOI] [PubMed] [Google Scholar]
- 113.Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA. Highly Recurrent <em>TERT</em> Promoter Mutations in Human Melanoma. Science. 2013;339(6122):957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proceedings of the National Academy of Sciences. 2013;110(15):6021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pierini T, Nardelli C, Lema Fernandez AG, Pierini V, Pellanera F, Nofrini V, et al. New somatic TERT promoter variants enhance the Telomerase activity in Glioblastoma. Acta Neuropathol Commun. 2020;8(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rachakonda PS, Hosen I, de Verdier PJ, Fallah M, Heidenreich B, Ryk C, et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc Natl Acad Sci U S A. 2013;110(43):17426–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu X, Bishop J, Shan Y, Pai S, Liu D, Murugan AK, et al. Highly prevalent TERT promoter mutations in aggressive thyroid cancers. Endocr Relat Cancer. 2013;20(4):603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shimoi T, Yoshida M, Kitamura Y, Yoshino T, Kawachi A, Shimomura A, et al. TERT promoter hotspot mutations in breast cancer. Breast Cancer. 2018;25(3):292–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trybek T, Walczyk A, Gąsior-Perczak D, Pałyga I, Mikina E, Kowalik A, et al. Impact of BRAF V600E and TERT Promoter Mutations on Response to Therapy in Papillary Thyroid Cancer. Endocrinology. 2019;160(10):2328–38. [DOI] [PubMed] [Google Scholar]
- 120.Shu C, Wang Q, Yan X, Wang J. The TERT promoter mutation status and MGMT promoter methylation status, combined with dichotomized MRI-derived and clinical features, predict adult primary glioblastoma survival. Cancer Med. 2018;7(8):3704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Gao K, Li G, Qu Y, Wang M, Cui B, Ji M, et al. TERT promoter mutations and long telomere length predict poor survival and radiotherapy resistance in gliomas. Oncotarget. 2016;7(8):8712–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Arita H, Yamasaki K, Matsushita Y, Nakamura T, Shimokawa A, Takami H, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med. 2015;372(26):2499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shaughnessy M, Njauw CN, Artomov M, Tsao H. Classifying Melanoma by TERT Promoter Mutational Status. J Invest Dermatol. 2020;140(2):390–4.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hysek M, Paulsson JO, Jatta K, Shabo I, Stenman A, Höög A, et al. Clinical Routine TERT Promoter Mutational Screening of Follicular Thyroid Tumors of Uncertain Malignant Potential (FT-UMPs): A Useful Predictor of Metastatic Disease. Cancers (Basel). 2019;11(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pitkänen A, Löscher W, Vezzani A, Becker AJ, Simonato M, Lukasiuk K, et al. Advances in the development of biomarkers for epilepsy. The Lancet Neurology. 2016;15(8):843–56. [DOI] [PubMed] [Google Scholar]
- 127.Villa C, Lavitrano M, Combi R. Long Non-Coding RNAs and Related Molecular Pathways in the Pathogenesis of Epilepsy. Int J Mol Sci. 2019;20(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shao Y, Chen Y. Pathophysiology and Clinical Utility of Non-coding RNAs in Epilepsy. Front Mol Neurosci. 2017;10:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pathak S, Miller J, Morris EC, Stewart WCL, Greenberg DA. DNA methylation of the BRD2 promoter is associated with juvenile myoclonic epilepsy in Caucasians. Epilepsia. 2018;59(5):1011–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Berger TC, Vigeland MD, Hjorthaug HS, Etholm L, Nome CG, Taubøll E, et al. Neuronal and glial DNA methylation and gene expression changes in early epileptogenesis. PLoS One. 2019;14(12):e0226575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Scheffer IE, Nabbout R. SCN1A-related phenotypes: Epilepsy and beyond. Epilepsia. 2019;60 Suppl 3:S17–s24. [DOI] [PubMed] [Google Scholar]
- 132.Myers KA, Burgess R, Afawi Z, Damiano JA, Berkovic SF, Hildebrand MS, et al. De novo SCN1A pathogenic variants in the GEFS+ spectrum: Not always a familial syndrome. Epilepsia. 2017;58(2):e26–e30. [DOI] [PubMed] [Google Scholar]
- 133.Gao QW, Hua LD, Wang J, Fan CX, Deng WY, Li B, et al. A Point Mutation in SCN1A 5’ Genomic Region Decreases the Promoter Activity and Is Associated with Mild Epilepsy and Seizure Aggravation Induced by Antiepileptic Drug. Mol Neurobiol. 2017;54(4):2428–34. [DOI] [PubMed] [Google Scholar]
- 134.de Lange IM, Weuring W, van ‘t Slot R, Gunning B, Sonsma ACM, McCormack M, et al. Influence of common SCN1A promoter variants on the severity of SCN1A-related phenotypes. Mol Genet Genomic Med. 2019;7(7):e00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kobow K, Jeske I, Hildebrandt M, Hauke J, Hahnen E, Buslei R, et al. Increased Reelin Promoter Methylation Is Associated With Granule Cell Dispersion in Human Temporal Lobe Epilepsy. Journal of Neuropathology & Experimental Neurology. 2009;68(4):356–64. [DOI] [PubMed] [Google Scholar]
- 136.Belhedi N, Perroud N, Karege F, Vessaz M, Malafosse A, Salzmann A. Increased CPA6 promoter methylation in focal epilepsy and in febrile seizures. Epilepsy Research. 2014;108(1):144–8. [DOI] [PubMed] [Google Scholar]
- 137.Machado RA, Benjumea-Cuartas V, Zapata Berruecos JF, Agudelo-Flóres PM, Salazar-Peláez LM. Reelin, tau phosphorylation and psychiatric complications in patients with hippocampal sclerosis and structural abnormalities in temporal lobe epilepsy. Epilepsy & Behavior. 2019;96:192–9. [DOI] [PubMed] [Google Scholar]
- 138.Ishiura H, Doi K, Mitsui J, Yoshimura J, Matsukawa MK, Fujiyama A, et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat Genet. 2018;50(4):581–90. [DOI] [PubMed] [Google Scholar]
- 139.Yeetong P, Pongpanich M, Srichomthong C, Assawapitaksakul A, Shotelersuk V, Tantirukdham N, et al. TTTCA repeat insertions in an intron of YEATS2 in benign adult familial myoclonic epilepsy type 4. Brain. 2019;142(11):3360–6. [DOI] [PubMed] [Google Scholar]
- 140.Corbett MA, Kroes T, Veneziano L, Bennett MF, Florian R, Schneider AL, et al. Intronic ATTTC repeat expansions in STARD7 in familial adult myoclonic epilepsy linked to chromosome 2. Nature Communications. 2019;10(1):4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Di Matteo F, Pipicelli F, Kyrousi C, Tovecci I, Penna E, Crispino M, et al. Cystatin B is essential for proliferation and interneuron migration in individuals with EPM1 epilepsy. EMBO Mol Med. 2020;12(6):e11419–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Assenza G, Benvenga A, Gennaro E, Tombini M, Campana C, Assenza F, et al. A novel c132–134del mutation in Unverricht-Lundborg disease and the review of literature of heterozygous compound patients. Epilepsia. 2017;58(2):e31–e5. [DOI] [PubMed] [Google Scholar]
- 143.Nakayama T, Ogiwara I, Ito K, Kaneda M, Mazaki E, Osaka H, et al. Deletions of SCN1A 5’ genomic region with promoter activity in Dravet syndrome. Hum Mutat. 2010;31(7):820–9. [DOI] [PubMed] [Google Scholar]
- 144.Martin MS, Tang B, Ta N, Escayg A. Characterization of 5′ untranslated regions of the voltage-gated sodium channels SCN1A, SCN2A, and SCN3A and identification of cis-conserved noncoding sequences. Genomics. 2007;90(2):225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Haigh JL, Adhikari A, Copping NA, Stradleigh T, Wade AA, Catta-Preta R, et al. Deletion of a non-canonical promoter regulatory element causes loss of Scn1a expression and epileptic phenotypes in mice. bioRxiv. 2019:766634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Vasudevaraja V, Rodriguez JH, Pelorosso C, Zhu K, Buccoliero AM, Onozato M, et al. Somatic Focal Copy Number Gains of Noncoding Regions of Receptor Tyrosine Kinase Genes in Treatment-Resistant Epilepsy. J Neuropathol Exp Neurol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Monlong J, Girard SL, Meloche C, Cadieux-Dion M, Andrade DM, Lafreniere RG, et al. Global characterization of copy number variants in epilepsy patients from whole genome sequencing. PLoS Genet. 2018;14(4):e1007285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yao RA, Akinrinade O, Chaix M, Mital S. Quality of whole genome sequencing from blood versus saliva derived DNA in cardiac patients. BMC Medical Genomics. 2020;13(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nair AK, Baier LJ. Using Luciferase Reporter Assays to Identify Functional Variants at Disease-Associated Loci. Methods Mol Biol. 2018;1706:303–19. [DOI] [PubMed] [Google Scholar]
- 150.Kweon J, Jang A-H, Shin HR, See J-E, Lee W, Lee JW, et al. A CRISPR-based base-editing screen for the functional assessment of BRCA1 variants. Oncogene. 2020;39(1):30–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yen JL, Garcia S, Montana A, Harris J, Chervitz S, Morra M, et al. A variant by any name: quantifying annotation discordance across tools and clinical databases. Genome Medicine. 2017;9(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen CH, Pan CY, Lin WC. Overlapping protein-coding genes in human genome and their coincidental expression in tissues. Sci Rep. 2019;9(1):13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Vormstein-Schneider D, Lin JD, Pelkey KA, Chittajallu R, Guo B, Arias-Garcia MA, et al. Viral manipulation of functionally distinct interneurons in mice, non-human primates and humans. Nat Neurosci. 2020;23(12):1629–36. [DOI] [PMC free article] [PubMed] [Google Scholar]