Abstract
Purpose:
To reduce workload and inconsistencies in organ segmentation for radiation treatment planning, we developed and evaluated general and custom auto-segmentation models on CT for three major tumor sites using a well-established deep convolutional neural network (DCNN).
Methods and Materials:
Five CT-based auto-segmentation models for 42 organs at risk (OARs) in head and neck (HN), abdomen (ABD) and male pelvis (MP) were developed using a full 3D DCNN architecture. Two types of DL models were separately trained using either general diversified multi-institutional datasets or custom well controlled single institution datasets. To improve segmentation accuracy, an adaptive spatial resolution approach for small and/or narrow OARs and a pseudo scan extension approach, when CT scan length is too short to cover entire organs, were implemented. The performance of the obtained models was evaluated based on accuracy and clinical applicability of the auto-segmented contours using qualitative visual inspection and quantitative calculation of dice similarity coefficient (DSC), mean distance to agreement (MDA), and time efficiency.
Results:
The five DL auto-segmentation models developed for the three anatomical sites were found to have high accuracy (DSC ranging from 0.8 – 0.98) for 74% OARs and marginally acceptable for 26% OARs. The custom models performed slightly better than the general models, even with smaller custom datasets used for the custom model training. The organ-based approaches improved auto-segmentation accuracy for small or complex organs (e.g., eye lens, optic nerves, inner ears, and bowels). Compared with traditional manual contouring times, the auto-segmentation times, including subsequent manual editing, if necessary, were substantially reduced by 88% for MP, 80% for HN, and 65% for ABD models.
Conclusions:
The obtained auto-segmentation models, incorporating organ-based approaches were found to be effective and accurate for most OARs in the male pelvis, head and neck and abdomen. We have demonstrated that our multi-anatomical deep learning auto-segmentation models are clinically useful for radiation treatment planning.
Keywords: CT-based auto-segmentation, deep learning, radiation therapy planning, adaptive radiation therapy
Introduction
The goal of radiation therapy (RT) is to deliver a precise and effective dose to the tumor while maximally sparing adjacent organs at risk (OARs)1,2. As a contributing factor for this goal, accurate, fast, and efficient delineation of the tumor and OARs is necessary. In current standard RT practice, the delineation of organs is done by manual or semi-automated (e.g., atlas-based) methods with manual editing on planning images (e.g., computed tomography (CT), magnetic resonance imaging (MRI))3,4. These processes are labor intensive and time consuming and inevitably introduce inter- and intra-observer variations during RT planning and delivery5–6. Recently, online adaptive replanning (OLAR) has been introduced into the clinic to account for inter-fractional anatomic and physiological changes7–9. During OLAR, with the patient lying on the table waiting to be treated, manual segmentation on the image of the day can be slow and strongly subjective, thus introducing inconsistencies between fractions10. Hence, a fast, robust, and consistent segmentation of tumors and OARs is a requirement. Deep learning (DL), such as deep convolutional neural network (DCNN) has posed itself as a potential solution for the aforementioned goal11–14.
Substantial efforts have been reported in the literature for using DCNN for auto-segmentation15–18. To name a few, Kazemifar et al. developed a two-dimensional (2D) U-Net (a convolutional neural network) model to delineate OARs in male pelvis CT using a mapping function and 2D convolution layers with variable kernel sizes, channel number, and rectified linear unit (ReLu) as activation function. They reported an average dice similarity coefficient (DSC) of 0.88, 0.95, and 0.92 for the prostate, bladder, and rectum, respectively19. Gibson et al. reported a dense virtual network (VNet) model featuring high resolution activation maps with a batch-wise spatial dropout scheme. The model proved to be more accurate and robust than existing multi-atlas-based methods on abdominal CT20. Dong et al. reported an advanced U-Net-generative adversarial network (U-Net-GAN) to segment a thoracic CT, with high accuracy21. Ye et al. designed a dense connectivity embedding U-net (DEU) based on CNN for auto-segmentation on MRI for nasopharyngeal carcinoma22.
Until recently, the DL auto-segmentation studies have been mostly focused on singular anatomical site. Recently, Chen et al. reported auto-segmentation of whole-body CT with a total of 50 OARs using site-specific networks, e.g., Ua-Net for head and neck, 2.5D U-Net for thorax, and 3D U-net for abdomen and male pelvis. They reported an average DSC of 0.84 and 0.81, respectively, for the models trained with in-house and public datasets23. Similarly, Isensee et al. used a self-configuring network, nn-Unet, to develop auto-segmentation models for multiple anatomical sites on multimodality images (CT, MRI). Their algorithm smartly channeled the knowledge in three primary steps: fixed, rule-based, and empirical parameters, to segment 53 organs.24 Besides these DL auto-segmentation studies for large numbers of organs with traditional encoder-decoder networks, the attention-mechanism combined with DCNN and transformers were also recently introduced for image segmentation tasks with some level of success25–27.
Despite these considerable efforts, DL auto-segmentation has not been routinely used in radiation therapy clinics primarily because the auto-segmented contours are not always acceptable, thus, manual editing is often required. Limitations on performance of CT-based auto-segmentation include intrinsic imaging quality (e.g., low soft tissue contrast, artifacts caused by metal implants or organ motion, inhomogeneous image intensities, and insufficient scan range) and DL algorithms and model building strategies (e.g., overfitting, over parametrization, limited quality, size and consistency of training datasets, challenges with small or complex organs)28. Additionally, comprehensive, and quantitative assessments of DL auto-segmentations based on clinically relevant criteria are generally lacking29–33.
In this work, we developed five DL auto-segmentation models for three major tumor sites, male pelvis (MP), head and neck (HN) and abdomen (ABD), that consist of a comprehensive list of organs, including those that are generally challenging in auto-segmentation. We employed a representative well-established DCNN network and incorporated organ-based approaches to specifically address the issues with small and complex organs. We compared our DL models trained with diversified multi-institutional datasets versus well-controlled single institutional datasets. In addition, we qualitatively and quantitatively evaluated the performance of the obtained models using various clinically relevant metrics including time efficiency, visual inspection, dice similarity coefficient (DSC) and mean distance to agreement (MDA) and compared our results with those from recent publications. To our knowledge, this is one of a few studies reporting DL auto-segmentation for large numbers of OARs for multiple anatomic sites.
Materials and Methods
DCNN auto-segmentation algorithm
A well-established 3D U-Net architecture, named ResUnet3D network34, was modified and used to build the auto-segmentation models. The algorithm included classic encoding and decoding structures35,36, in which the encoding part was responsible for generating/learning multi-scale multi-dimensional image features in multiple levels, while the decoding structure was used to produce the same size label map to the corresponding input image driven by the learned features. In addition, short-range residual connections in residual block as introduced by the Residual Networks37 were used to suppress the gradient vanish behavior and to shorten iteration numbers during the model building process. Besides flexibility, this long and short connection combination allowed the decoding part to preserve the high-resolution features with potential localization of information, which could be potentially lost at the bottom of the encoding part of the network. Through these residual connections, information was better transmitted across different feature levels, resulting in improved model convergence and performance by suppressing the saturation of gradient backpropagation. This is especially important and useful if training is conducted from scratch without initializing models from some early training data. A schematic of the algorithm is shown in Figure 1. In this work, the building of an auto-segmentation model using this algorithm was implemented in an iterative process, consisting of model training, model validation, and algorithm refinement based on quantitative evaluation feedback.
Figure 1.
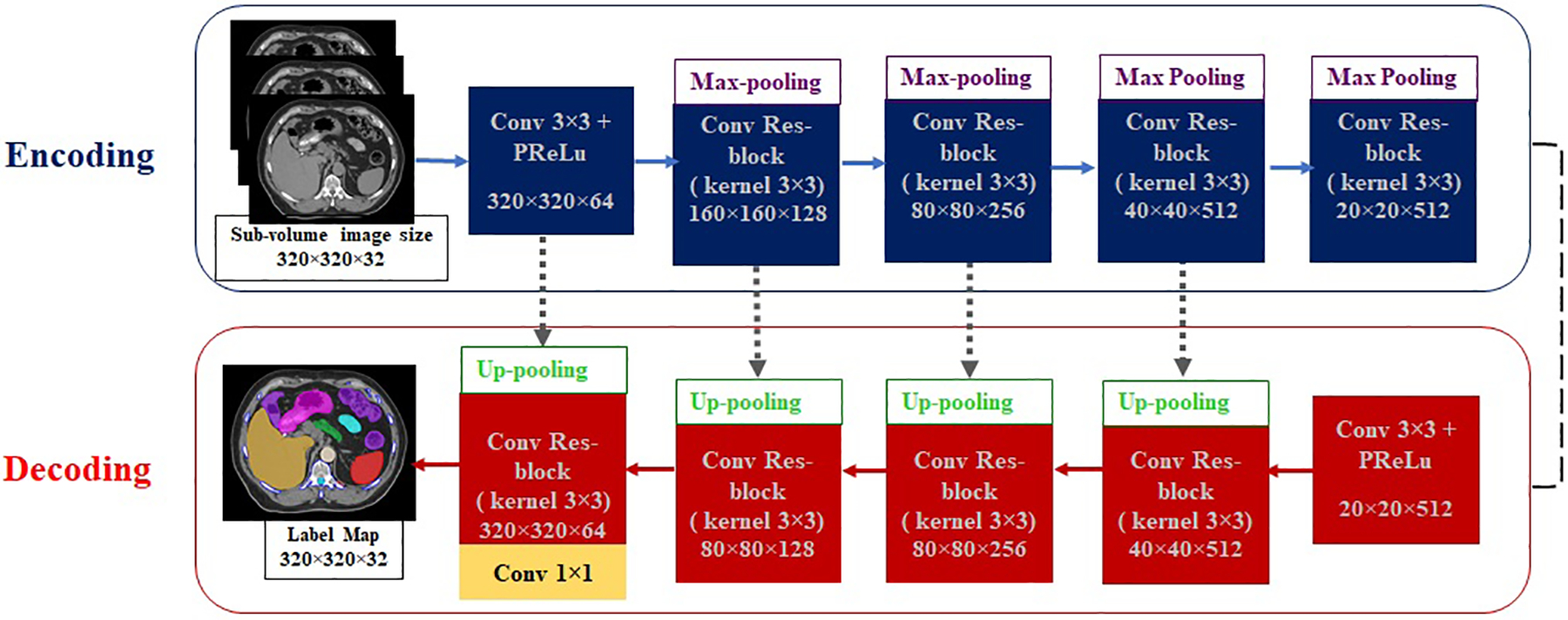
A schematic of ResUnet3D network with encoding and decoding structures. Respective convolution layers of 3×3 kernel size is showed. The batch normalization layer is adopted between layers but is not shown explicitly in this scheme. The gray dotted arrows represent the long-range direct connection, and short skip connections and addition operations are embedded in the convolutional residual blocks. The array shape, for example, 320×320×64 represents the resulting tensor sizes in each layer with ignoring the batch size = 1 and the depth of 3D volume. The last dimension number 64 represents the filter number (out-channel number).
Improvements with organ-based approaches
The default ResUnet3D network employed the same spatial resolution in the entire dataset. As organ volume can vary drastically for the large number of OARs studied in this work, hence, the algorithm convergence for different organs can be quite different, resulting in different accuracy and efficiency in using the obtained model. To balance the accuracy and efficiency, a compromise between image input size and spatial resolution needed to be made. For example, an input image size of 320×320×32 with a default spatial resolution of 2×2×2 mm3 may be used. However, this resolution may be too coarse for small and/or narrow organs such as the inner ear or optic nerve in the HN region. To overcome this problem, we adopted an adaptive spatial resolution (ASR) approach, allowing the use of a higher spatial resolution of 1×1×2 mm3 for a subregion where a small and/or narrow OAR is present38,39. This approach was implemented in four steps, as shown in Figure 2.
Figure 2.

A schematic of adaptive spatial resolution (ASR) approach for head and neck small and/or narrow organs.
Another issue that can negatively affect the auto-segmentation performance is the missing superior and/or inferior slices due to inconsistent and/or insufficient CT scan lengths required to cover entire large organs, such as bowels in abdomen. To address this issue, we implemented a pseudo scan extension (PSE) approach to take into account the missing superior and/or inferior slices. In the PSE approach, the end slice with contours in the scan was repeated with eight-fold elastic contour transformation, mimicking the possible deformation beyond the end slice, thus, reconstructing the missing contours considering their shape changes.
Datasets
The datasets used to train and test the DL models in this study were the CTs along with manually drawn contours (ground truth) collected for three tumor sites, MP, HN and ABD. To investigate the effect of variation in training datasets, two types of datasets: (1) general diversified datasets obtained from multiple institutions/sources that included a wide variability of scanners and protocols (204 CTs for MP, and 65 CTs for HN, including 41 from the MICCAI 201540 and 24 from multiple institutions) and (2) custom well-controlled datasets acquired with standard clinical protocols from a single institution (50 CTs for MP and 47 for HN), were used for the HN and MP model training. For abdomen, only a custom dataset (58 cases) was used for the training as there was no general dataset for ABD available to us. For the custom datasets, ground truths were carefully created manually using available contouring consensus and nomenclature guidelines41–45 and were independently checked by two experienced clinicians per anatomical site. To avoid potential bias and to improve robustness in both model training and testing, the datasets for each tumor site were purposely chosen to include a broad range of variations in image quality and anatomy.
Model training
For MP and HN, two DL models for each were trained: the general models (general male pelvis (G-MP) and general head and neck (G-HN)) trained with the general datasets, and the custom models (custom male pelvis (C-MP) and custom head and neck (C-HN)) trained with the custom datasets. For abdomen, only custom abdomen (C-ABD) model was trained. The G-MP and C-MP models were trained for 8 and 11 OARs, while the G-HN and C-HN were trained for 11 and 19 OARs, respectively. The C-ABD model was trained for 12 OARs.
For all models, CT numbers were normalized to categorize into specific ranges and were linearly transformed into [−1 1] range before they were used for model training and testing. The CT number ranges were [−400,400] HU for MP, [−1000,4000] HU for HN, and [−1000 4000] HU for ABD. During model training, the objective function was set to minimize a weighted cross-entropy loss function and ADAM’s method was adopted for this optimization46. The weight decay was set to be 0.0005 and the momentum parameter was set to 0.9. Initially, the learning rate was set to 0.002 and linear decay was set after half the number of training epochs. The models were trained for 200–400 epochs with online data augmentation that was accomplished by applying an online random 3D elastic transform/deformation on each dataset47,48. All the models were trained on a single NVIDIA V100 GPU, DGX-1 system.
Model evaluation
The trained models were integrated in a research tool (ADMIRE, Elekta Inc) for execution. All models testing was performed on an Intel ® Xeon® Gold 6132 CPU @ 2.6GHz 128GB RAM hardware. The performances of G-MP, C-MP, G-HN, C-HN and C-ABD models were tested with 50, 10, 50, 10, and 13 independent datasets, respectively. To avoid a predisposed evaluation, the testing cases for each model were chosen to imitate the imaging and anatomical variability of the training data. The model performance was evaluated for common OARs in RT planning. A summary of the models, numbers of training and testing datasets used, numbers and names of OARs in each model along with those evaluated for each tumor site is shown in Table 1.
TABLE 1.
Summary of presented models; including training/testing datasets for each and trained and evaluated (underlined) OARs.
| # of training / testing CT datasets | # of trained OARs / nomenclature of OARs | |
|---|---|---|
| General male pelvis (G-MP) | 204 / 50 | 8 / Bladder, Prostate, Rectum, Femure_L, Femure_R, Pelvis_L, Pelvis_R, patient external |
| General head and neck (G-HN) | 65 / 50 | 8 / Brainstem, Bone_Mandible, Parotid_(L/R), Glnd_Submand_(L/R), SpinalCord, patient external |
| Custom male pelvis (C-MP) | 50 / 10 | 11 / Bladder, PenileBulb, Prostate, Rectum, SeminalVes, Sigmoid, Femure_L, Femure_R, Pelvis_L, Pelvis_R, patient external |
| Custom head and neck (C-HN) | 47 / 10 | 19 / Brain, Brainstem, Bone_Mandible, Esophagus, Eye_(L/R), Inner Ear_(L/R), Larynx, Lens_(L/R), OpticNerve_(L/R), Parotid_(L/R), Glnd_Submand_(L/R), SpinalCord, patient external |
| Custom abdomen (C-ABD) | 58 / 13 | 12 / A_Aorta, Bowel_Large, Bowel_Small, Duodenum, Kidney_(L/R), Liver, Pancreas, SpinalCord, Spleen, Stomach, patient external |
Performance of the auto-segmentation models was qualitatively and quantitively assessed in terms of segmentation accuracy, reproducibility, and time. A qualitative assessment of auto-segmented contours was performed by reviewing the obtained contours slice-by-slice. The regions with optimal and sub-optimal contour accuracy were recorded. The 3D average DSC and MDA, as calculated with respect to the ground truth, were used to quantitatively measure the auto-segmentation accuracy. The clinical acceptability of the auto-segmentation was assessed based on AAPM TG-132 report, e.g., 3D average DSC > 0.8 and MDA < 2 mm49.
Computing time for generating all OAR contours using a model for each test case was recorded and the average segmentation time for each model was documented. As the auto-segmented contours are often reviewed and manually edited for inaccuracy, the segmentation time with a DL model should include the time for reviewing and manual editing. We conducted a time study to estimate the reviewing and editing time required to fix the inaccuracies in the auto-segmented contours. Summation of the auto-segmentation time (computer time) and the subsequent manual reviewing and editing time was compared with the time for conventional manual contouring using a commercial clinical contouring tool (MIM version 6.8.6, MIM Software Inc.; Beachwood, OH).
Results
Auto-segmentation accuracy
The accuracy of the auto-segmented contours, as measured by the average 3D DSC and MDA, for the 5 models is presented in Figure 3 using a box plot technique50. The average DSC and MDA values with standard deviation for all the OARs in the 5 models are shown in Table 2. Comparisons of representative auto-segmented contours and the ground truth on the test CT for each of the 5 models are presented in Figure 4.
Figure 3.
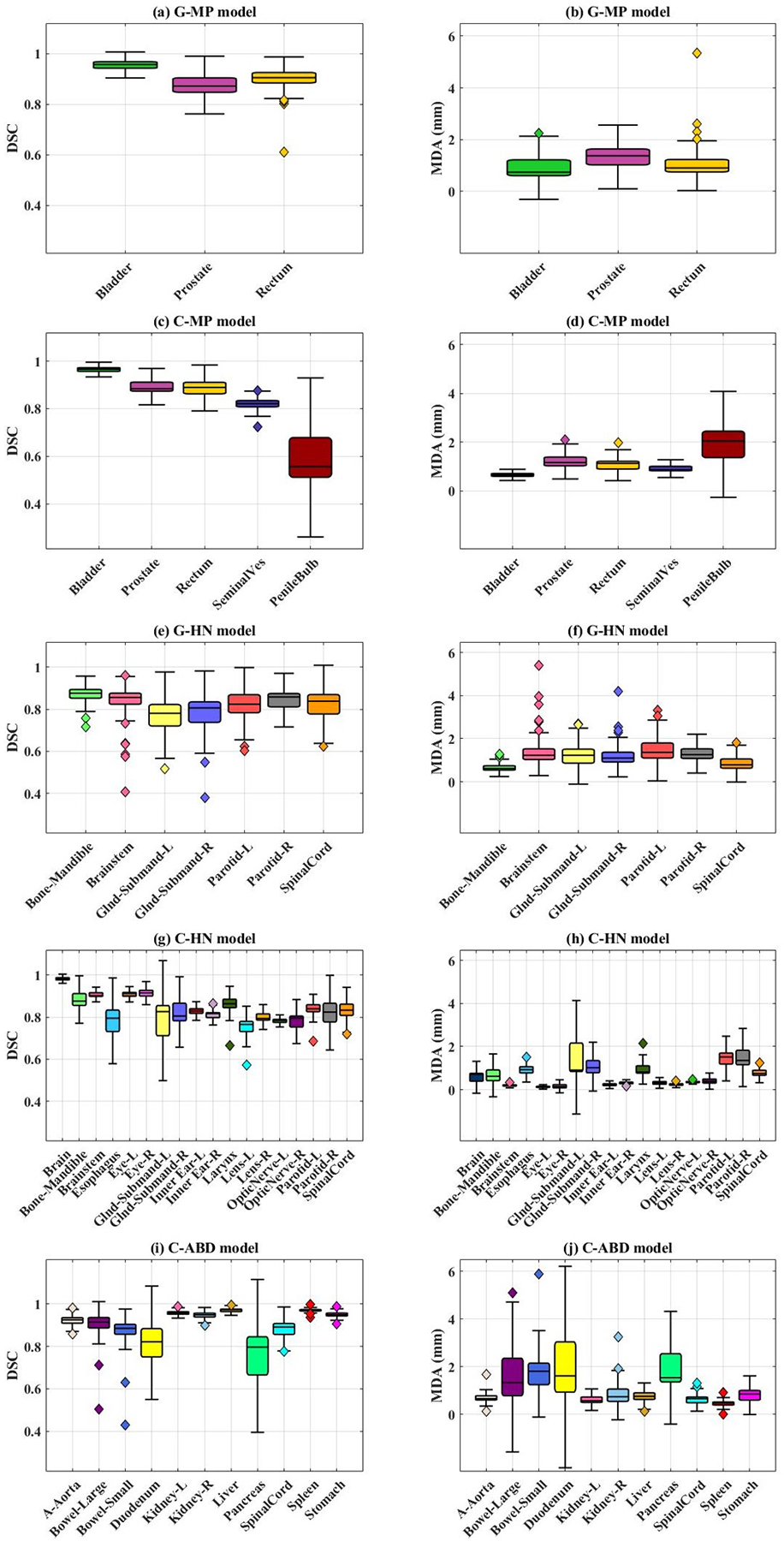
Accuracy metrics, 3D average dice similarity coefficient (DSC) and mean distance to agreement (MDA) for all 5 models are shown. a-b) General male pelvis (G-MP) model; c-d) general head and neck (G-HN) model; e-f) custom male pelvis (C-MP) model; g-h) custom head and neck (C-HN) model; i-j) custom abdomen (C-ABD) model. The whiskers in the box plot represent variability and individual data points represent outliers.
TABLE 2.
Summary of average accuracy metrics dice similarity coefficient (DSC) and mean distance to agreement (MDA) calculated for all models. The custom models shown in bold are the final results.
| DSC ± std | MDA ± std | |
|---|---|---|
| General male pelvis (G-MP) model OARs | ||
| Bladder | 0.95 ± 0.02 | 0.91 ± 0.41 |
| Prostate | 0.87 ± 0.04 | 1.38 ± 0.39 |
| Rectum | 0.89 ± 0.05 | 1.13 ± 0.75 |
| General head and neck (G-HN) model OARs | ||
| Bone_Mandible | 0.87 ± 0.04 | 0.68 ± 0.19 |
| Brainstem | 0.83 ± 0.10 | 1.46 ± 0.90 |
| Gland_Submand_L | 0.75 ± 0.10 | 1.39 ± 0.67 |
| Gland_Submand_R | 0.78 ± 0.10 | 1.27 ± 0.65 |
| Parotid_L | 0.82 ± 0.06 | 1.50 ± 0.62 |
| Parotid_R | 0.84 ± 0.04 | 1.36 ± 0.48 |
| SpinalCord | 0.81 ± 0.08 | 0.91 ± 0.35 |
| Custom male pelvis (C-MP) model OARs | ||
| Bladder | 0.96 ± 0.01 | 0.67 ± 0.08 |
| PenileBulb | 0.62 ± 0.14 | 1.82 ± 0.81 |
| Prostate | 0.88 ± 0.03 | 1.26 ± 0.36 |
| Rectum | 0.89 ± 0.03 | 1.13 ± 0.39 |
| SeminalVes | 0.82 ± 0.04 | 0.91 ± 0.13 |
| Custom head and neck (C-HN) model OARs | ||
| Brain | 0.98 ± 0.01 | 0.58 ± 0.21 |
| Bone_Mandible | 0.88 ± 0.03 | 0.68 ± 0.30 |
| Brainstem | 0.90 ± 0.01 | 0.20 ± 0.06 |
| Esophagus | 0.78 ± 0.06 | 0.98 ± 0.27 |
| Eye_L | 0.91 ± 0.01 | 0.13 ± 0.05 |
| Eye_R | 0.91 ± 0.01 | 0.15 ± 0.07 |
| Gland_Submand_L | 0.77 ± 0.10 | 1.30 ± 0.71 |
| Gland_Submand_R | 0.80 ± 0.07 | 1.10 ± 0.36 |
| InnerEar_L | 0.83 ± 0.01 | 0.24 ± 0.05 |
| InnerEar_R | 0.82 ± 0.02 | 0.32 ± 0.08 |
| Larynx | 0.85 ± 0.07 | 1.03 ± 0.42 |
| Lens_L | 0.74 ± 0.06 | 0.32 ± 0.07 |
| Lens_R | 0.80 ± 0.02 | 0.25 ± 0.07 |
| OpticNerve_L | 0.78 ± 0.01 | 0.36 ± 0.06 |
| OpticNerve_R | 0.79 ± 0.04 | 0.38 ± 0.14 |
| Parotid_L | 0.82 ± 0.05 | 1.52 ± 0.38 |
| Parotid_R | 0.82 ± 0.04 | 1.48 ± 0.40 |
| SpinalCord | 0.83 ± 0.06 | 2.27 ± 4.90 |
| Custom abdomen (C-ABD) model OARs | ||
| A_Aorta | 0.92 ± 0.03 | 0.71 ± 0.34 |
| Bowel_Large | 0.87 ± 0.13 | 2.37 ± 2.80 |
| Bowel_Small | 0.84 ± 0.15 | 3.80 ± 6.88 |
| Duodenum | 0.82 ± 0.09 | 2.10 ± 1.64 |
| Kidney_L | 0.96 ± 0.01 | 0.60 ± 0.20 |
| Kidney_R | 0.94 ± 0.02 | 0.97 ± 0.80 |
| Liver | 0.97 ± 0.01 | 0.76 ± 0.27 |
| Pancreas | 0.76 ± 0.09 | 1.92 ± 0.73 |
| SpinalCord | 0.88 ± 0.05 | 0.68 ± 0.29 |
| Spleen | 0.97 ± 0.01 | 0.50 ± 0.20 |
| Stomach | 0.95 ± 0.02 | 0.85 ± 0.37 |
Figure 4.
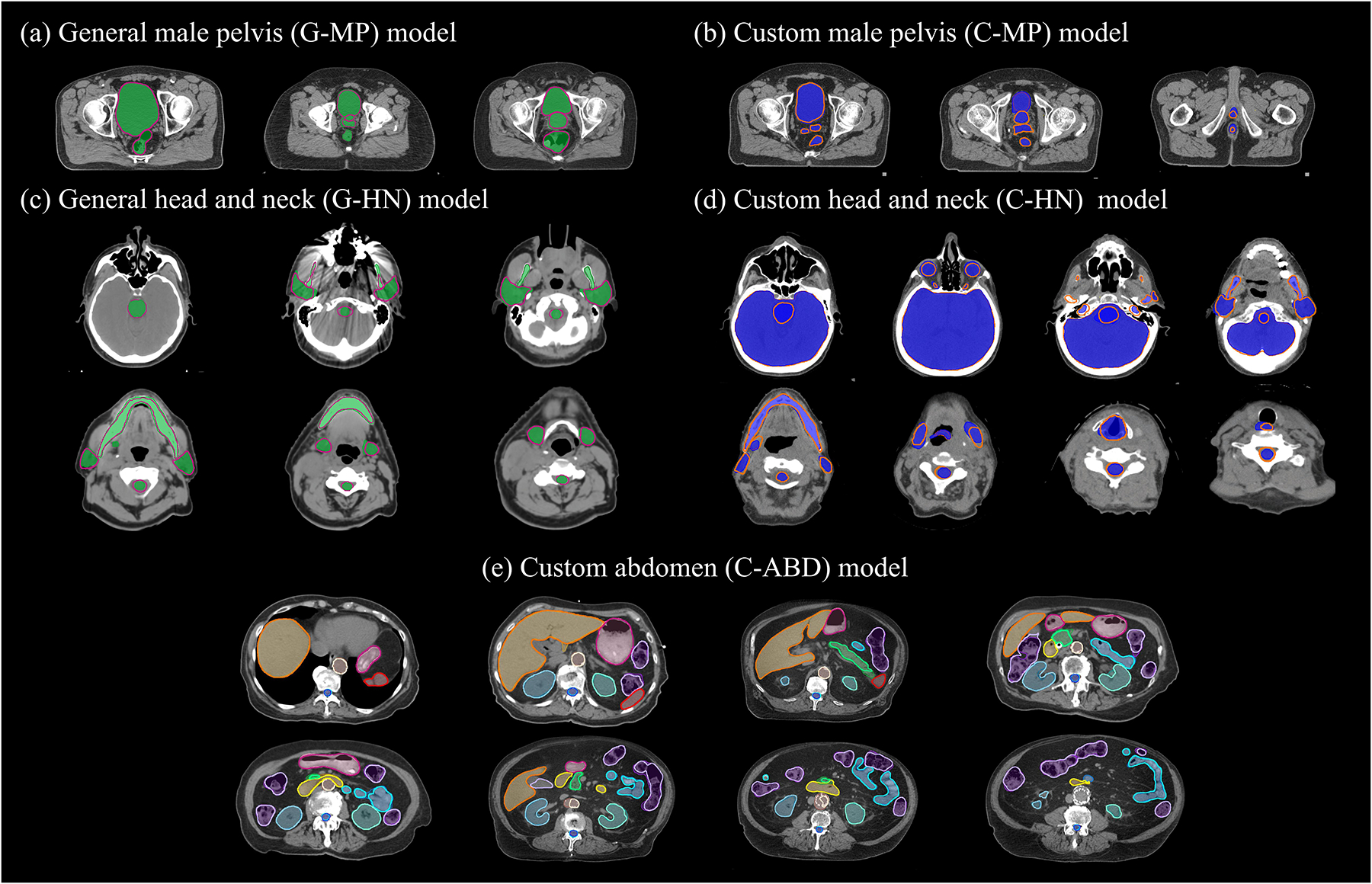
Comparison of representative ground truth (shaded) and auto-segmented (thick lines) contours on axial slices of test CTs for the five models.
Male Pelvis Models
For the G-MP model, the best concordance between the auto-segmented contours and the ground truth on the test data was demonstrated for bladder, with an average DSC of 0.95 ± 0.02, whereas a broader DSC range was observed for rectum with an average DSC of 0.89 ± 0.05, as shown in Figure 3a. Interestingly, high performance was also observed for prostate with an average DSC of 0.87 ± 0.04. Compared to the G-MP model, the C-MP model, trained with smaller dataset than that for the G-MP model, performed slightly better as seen in Figure 3(c, d) (e.g., reduced standard deviation of DSC for the prostate, rectum, and bladder segmentations). With the C-MP model, the average DSC values for the seminal vesicle and penile bulb were 0.82 and 0.62, respectively. The low DSC for penile bulb is expected as the anatomical markers or the necessary soft tissue contrast for the penile bulb is generally lacking on CT. For bladder, rectum, and prostate, as shown in Figure 5(a, b), the auto-segmented contours by the G-MP and C-MP models were acceptable in 46 out of 50 (92%) and 9 out of 10 (90%) test cases respectively. Filled green circles represent individual test cases for which the auto-segmentation accuracy met TG-132 clinical acceptability criteria49, while filled magenta squares indicate unacceptable cases. It was observed that the variability in accurately identifying and segmenting the recto-sigmoidal junction, commonly encountered in manual contouring, was the primary contributor for the broad range of accuracy for the rectum auto-segmentation.
Figure 5.
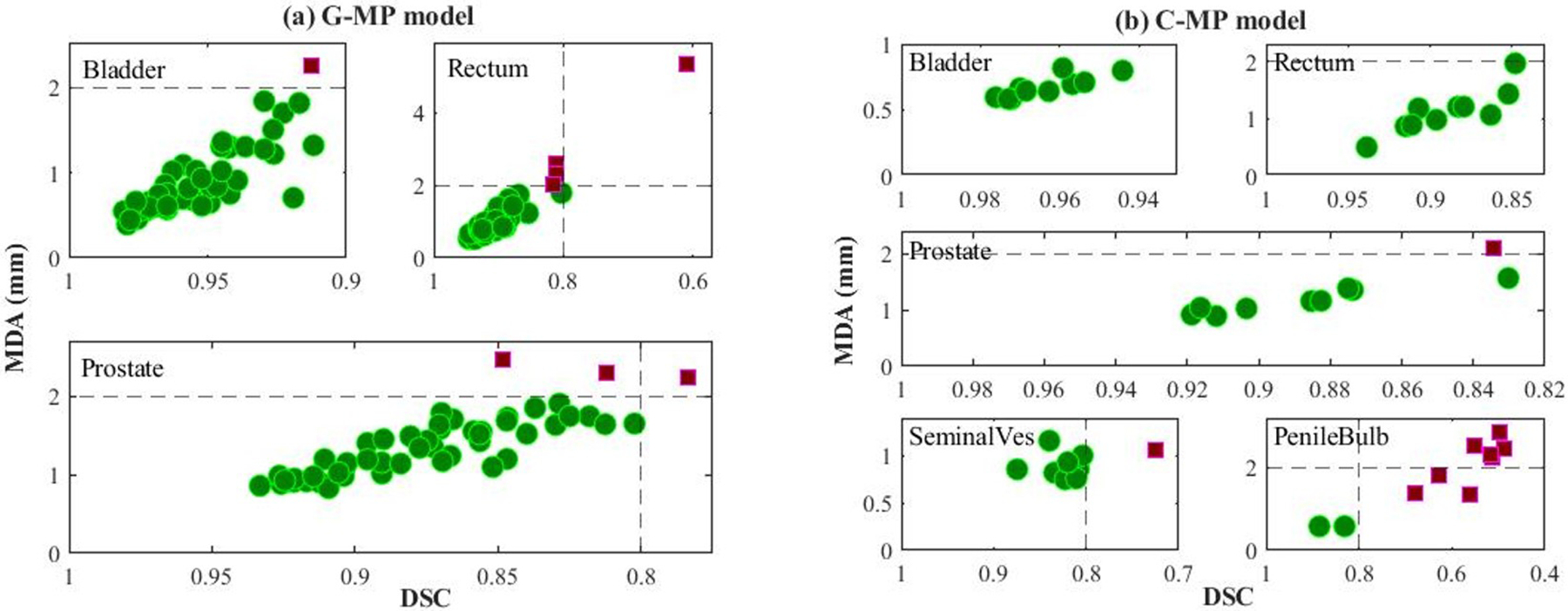
Depiction of male pelvis model performance of all test cases for (a) General male pelvis (G-MP) model and (b) custom male pelvis (C-MP) model. For each subplot, x-axis is dice similarity coefficient (DSC) and y-axis is mean distance to agreement (MDA). Green circles and violet-red squares represent clinically acceptable and unacceptable cases respectively. The dashed lines indicate the TG-132 metrics tolerances (DSC > 0.8, MDA < 2mm).
Head and Neck Models
A reasonably accurate auto-segmentation was observed with both G-HN and C-HN models with an average DSC of > 0.8 for all organs, except submandibular glands, as shown in Figure 3(e–h). As shown in Figure 6(a, b), the case-based analysis confirmed that both models delivered clinically acceptable contours for most OARs with some exceptions. Compared to the G-HN model, the C-HN performed better, e.g., less unacceptable cases for the C-HN models (Figure 6(b)). High auto-segmentation accuracies in 70–100% of the test cases was observed for larynx, eyes, brain, brainstem, and mandible with the C-HN model. Sub-optimal segmentation was observed for submandibular glands. The presence of vessels, the proximity to sternocleidomastoid muscle, and the poor contrast in the region make the submandibular gland a difficult organ to delineate. Moreover, in the initial model iterations, poor and incomplete segmentation was observed for small organs including lens, optic nerves, and inner ears (Table 3). As shown in Table 3, and Figure 3(g–h), the use of the ASR approach allowed a higher resolution of 1×1×2 mm3 in the subregions containing six small (lens, optic nerves, and inner ears) and three medium (brainstem, eyes) sized OARs, and yielded substantially improved auto-segmentation of these small and/or narrow OARs.
Figure 6.
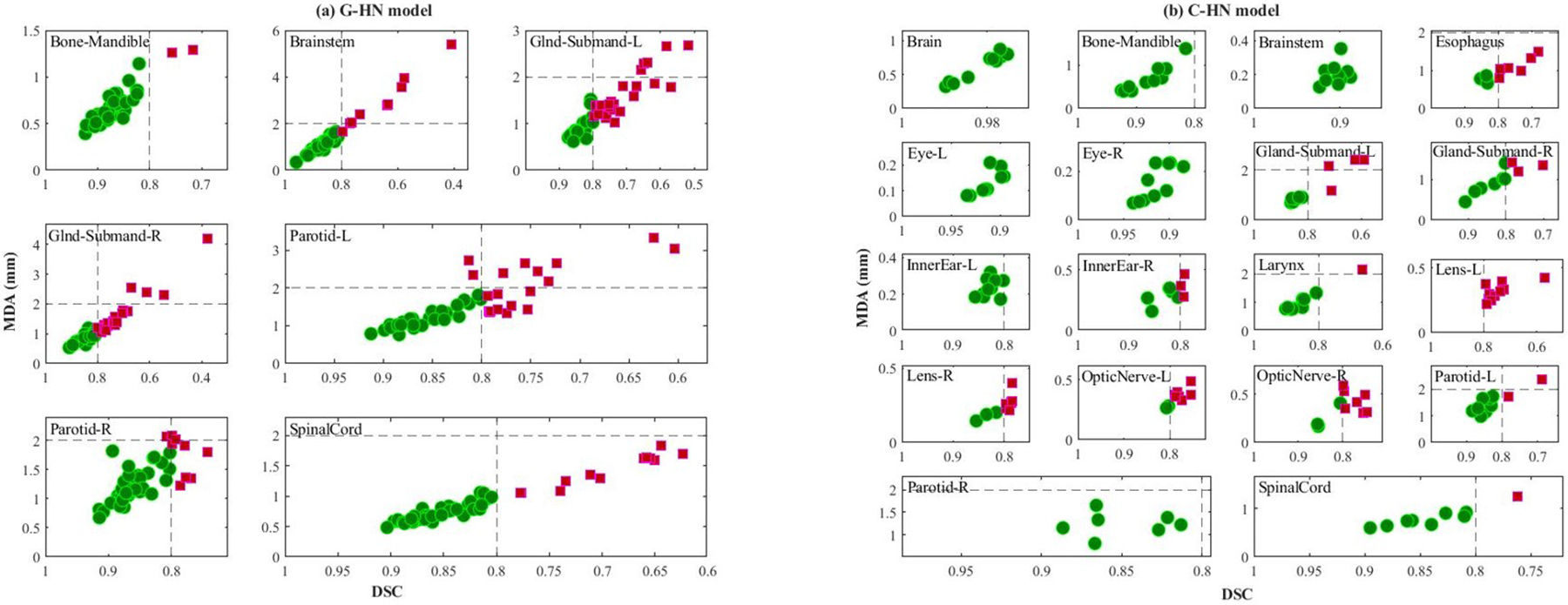
Clinical acceptability of all test cases for (a) general head and neck (G-HN) model and (b) custom head and neck (C-HN) model. For each subplot, x-axis is dice similarity coefficient (DSC) and y-axis is mean distance to agreement (MDA). Green circles and violet-red squares represent clinically acceptable and unacceptable cases respectively. The dashed lines indicate the TG-132 metrics tolerances (DSC > 0.8, MDA < 2mm).
TABLE 3.
Improved segmentation with the custom head and neck (C-HN) model of small and mid-sized organs with the implementation of adaptive spatial resolution (ASR) approach. The best results with ASR are shown in bold.
| Modified ResUnet3D | Modified ResUnet3D with ASR | |||
|---|---|---|---|---|
| DSC | MDA | DSC | MDA | |
| Brainstem | 0.842 | 1.293 | 0.904 | 0.202 |
| Eye_L | 0.891 | 0.810 | 0.913 | 0.130 |
| Eye_R | 0.909 | 0.589 | 0.914 | 0.153 |
| Innerear_L | 0.758 | 0.661 | 0.828 | 0.235 |
| Innerear_R | 0.720 | 0.732 | 0.818 | 0.315 |
| Lens_L | 0.240 | 1.567 | 0.743 | 0.316 |
| Lens_R | 0.191 | 1.846 | 0.803 | 0.246 |
| Opticnerve_L | 0.442 | 1.710 | 0.782 | 0.360 |
| Opticnerve_R | 0.268 | 2.390 | 0.792 | 0.377 |
Abdomen Model
While abdominal organs can have inherent geometrical distortions and organ-based, observer-dependent segmentation imperfections, accurate auto-segmentation was observed for most of the OARs with the C-ABD model, as shown in Figure 3(i, j). A high DSC of 0.92 to 0.97 was observed for aorta, kidneys, liver, stomach, and spleen. While DSC values of > 0.8 were observed for bowels and duodenum, their MDA values were outside the clinically acceptable tolerance range (> 2mm). This suboptimal auto-segmentation for the bowels, duodenum and pancreas is consistent with the difficulty in manual delineation of these organs on CT owing to the drastic changes in their sizes, shapes, and motions between slices. Proximity of vessels to the pancreatic body and tail was found to be the leading cause of inaccuracies, thus resulting in suboptimal performance (DSC 0.76), as shown in Figure 7(a, b). For the bowels, the relatively high DSC values (0.84 – 0.87) was due in part to the use of the PSE approach. The auto-segmentation accuracy of bowels was found to be strongly dependent on the complexity of the anatomy, as shown by long whiskers in the box plots of Figure 3(i, j), indicating large variations and inaccuracies, e.g., mislabeling of bowels in presence of large air pockets, as shown in Figure 7c.
Figure 7.
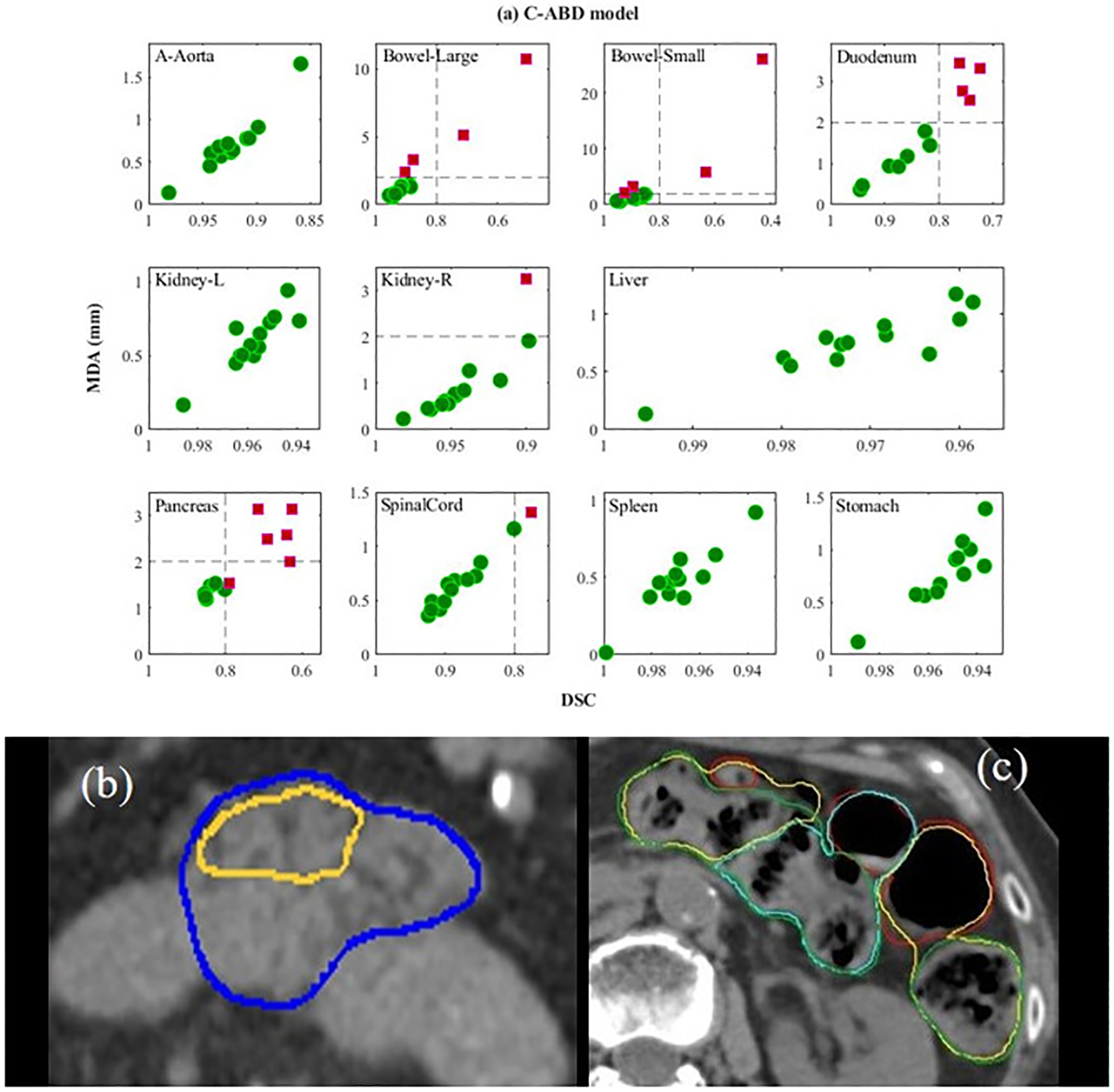
a) Performance of the custom abdomen model (C-ABD) of all test cases. For each subplot, x-axis is dice similarity coefficient (DSC) and y-axis is mean distance to agreement (MDA). Green circles and violet-red squares represent clinically acceptable and unacceptable cases respectively. The dashed lines indicate the TG-132 metrics tolerances (DSC > 0.8, MDA < 2mm); b) the ground truth (blue) vs. inaccurate auto-segmentation (yellow) for normal pancreas; c) ground truth (green) vs. inaccurate auto-segmentation (yellow) (mislabeled air pockets) for the large bowel, and ground truth (red) vs. inaccurate auto-segmentation (cyan) for the small bowel.
Auto-Segmentation Time
On average, the execution of the auto-segmentation for all the OARs on a testing CT set took approximately 30 seconds for male pelvis, 120 seconds for head and neck, and 70 seconds for abdomen. The auto-segmentation time was found to depend on the numbers of CT slices and OARs. The average time for visually reviewing the auto-segmented contours and the manual editing of inaccurate contours for complex cases took up to 5 minutes for the C-MP, 15 minutes for C-HN, and 30 minutes for the C-ABD model. The longer time for ABD was primarily due to the complicated anatomy. In contrast, the average manual contouring from scratch ranged from 30 minutes (for MP) to up to 90 minutes (for HN and ABD). Average reductions on contouring times with using the auto-segmentation models as compared to the full manual contouring were calculated to be 88% for MP, 80% for HN, and 65% for ABD.
Comparison to recent published data
The performance of the present models was compared to three types of results recently published on CT-based DL auto-segmentation for large numbers of organs in multiple anatomic sites similar to this study, (1) the data for HN, MP and ABD by Chen et al using a WBNet algorithm23, (2) the data for the three sites by Isensee et al using a nnUNET network24, and (3) combined data of the MRI-aided DL auto-segmentation on CT for MP51 and NH52,53 and the data of using a self-spaced DenseNet algorithm for ABD54. To our knowledge, the studies (1)23 and (2)24 are the only available studies so far involving all the similar OARs in MP, HN and ABD as in the present work. Table 4 compares DSC values obtained in this work to those reported in the three types of studies for 32 OARs in the three anatomic sites. It is seen that the present work performed slightly better in 17 out of 32 OARs compared to all the published studies included, with DSC values greater than or equal to 0.8 for 26 out of 32 OARs. The current MP and ABD models outperformed the previously published results, except for right kidney and pancreas. The average DSCs for all OARs were 0.863, 0.854 and 0.833 for the current work, the study by Chen et al23 and the study by Isensee et al24, respectively.
TABLE 4.
Comparison of dice similarity coefficients for selected OARs from the current work, the study by Chen et al23, the study by Isensee et al24, and the combined datasets51–54. Bold numbers represent the best results.
| Current work | Study by Chen23 | Study by Isensee24 | Combined datasets51–54 | |
|---|---|---|---|---|
| Bladder | 0.96 | 0.93 | 0.92 | 0.95 |
| Prostate | 0.88 | 0.87 | ||
| Rectum | 0.89 | 0.8 | 0.83 | 0.89 |
| Brain | 0.98 | 0.95 | ||
| Bone_Mandible | 0.88 | 0.94 | 0.94 | 0.89 |
| Brainstem | 0.90 | 0.87 | 0.9 | 0.9 |
| Esophagus | 0.78 | 0.76 | 0.81 | 0.85 |
| Eye_L | 0.91 | 0.92 | 0.84 | 0.89 |
| Eye_R | 0.91 | 0.93 | 0.86 | 0.87 |
| Gland_Submand_L | 0.77 | 0.82 | 0.79 | |
| Gland_Submand_R | 0.80 | 0.82 | 0.8 | |
| InnerEar_L | 0.83 | 0.74 | 0.73 | 0.76 |
| InnerEar_R | 0.82 | 0.77 | 0.73 | 0.75 |
| Larynx | 0.85 | 0.9 | 0.79 | 0.9 |
| Lens_L | 0.74 | 0.83 | 0.77 | 0.79 |
| Lens_R | 0.80 | 0.84 | 0.79 | 0.77 |
| OpticNerve_L | 0.78 | 0.76 | 0.72 | 0.72 |
| OpticNerve_R | 0.79 | 0.75 | 0.75 | 0.72 |
| Parotid_L | 0.82 | 0.85 | 0.8 | 0.89 |
| Parotid_R | 0.82 | 0.85 | 0.78 | 0.88 |
| SpinalCord | 0.83 | 0.86 | 0.86 | 0.86 |
| A_Aorta | 0.92 | 0.94 | ||
| Bowel_Large | 0.87 | 0.8 | 0.82 | |
| Bowel_Small | 0.84 | 0.82 | 0.81 | |
| Duodenum | 0.82 | 0.77 | 0.74 | 0.69 |
| Kidney_L | 0.96 | 0.96 | 0.88 | 0.94 |
| Kidney_R | 0.94 | 0.96 | 0.89 | 0.95 |
| Liver | 0.97 | 0.94 | 0.96 | 0.96 |
| Pancreas | 0.76 | 0.84 | 0.83 | 0.79 |
| SpinalCord | 0.88 | 0.88 | 0.85 | |
| Spleen | 0.97 | 0.96 | 0.95 | 0.95 |
| Stomach | 0.95 | 0.9 | 0.91 | 0.88 |
Discussion
In this work, we have developed clinically applicable auto-segmentation models for 42 OARs in three major tumor sites: head and neck, abdomen and male pelvis using a well-established DL network. Two organ-based approaches, the adaptive spatial resolution, and the pseudo scan extension, were implemented to improve the model performance of small/elongated (e.g., inner ear) and large (e.g., bowels) organs. Separate DL models were trained with using diversified multi-institutional datasets and using well-controlled single institutional datasets and were evaluated based on clinically relevant criteria (e.g., required contour editing effort). We have found that the obtained DL models can quickly generate clinically acceptable contours for most of the organs in the three tumor sites. Performance of the models trained with the single-institutional datasets is generally better than those trained with the multi-institutional datasets. Performance of our models are generally better or comparable to those reported recently on DL auto-segmentation for multi anatomic sites.
Challenges in the DL auto-segmentation were observed in all three tumor sites. For male pelvis, the poor soft tissue contrast at the apex of the prostate and the seminal vesicles and the apex of the prostate and the rectum, led to residual auto-segmentation inaccuracies, consistent with the observations reported previously for manual contouring55. For example, in 76% of the cases tested, the G-MP model generated prostate contours incorporated part of the seminal vesicles (Figure 8a, violet-red contour). Inaccuracies for rectum auto-segmentation were observed in 62% of the test cases at the recto-sigmoidal junction, concurring with uncertainties existing with manual segmentation. These issues were addressed by including the seminal vesicles and sigmoid in the C-MP model, as shown in Figure 8(a,b), resulting in improved segmentation. Even with the challenges at the recto-sigmoidal junction, the rectum segmentation was generally improved (shorter DSC range, and better overlap with ground truth) with the C-MP model (Figure 8b). Additionally, upon introducing abnormal anatomy in the testing data, the robustness of the model was probed. Figure 8c shows an example, where the auto-segmentation model correctly identified the median lobe and excluded it from the bladder contour. Taking in account the aforementioned challenges and the anatomical variations in the testing data, the accuracy comparison of the C-MP model with the recent publications (Table 4) clearly showcased superior accuracy of our bladder, rectum and prostate auto-segmentation.
Figure 8.
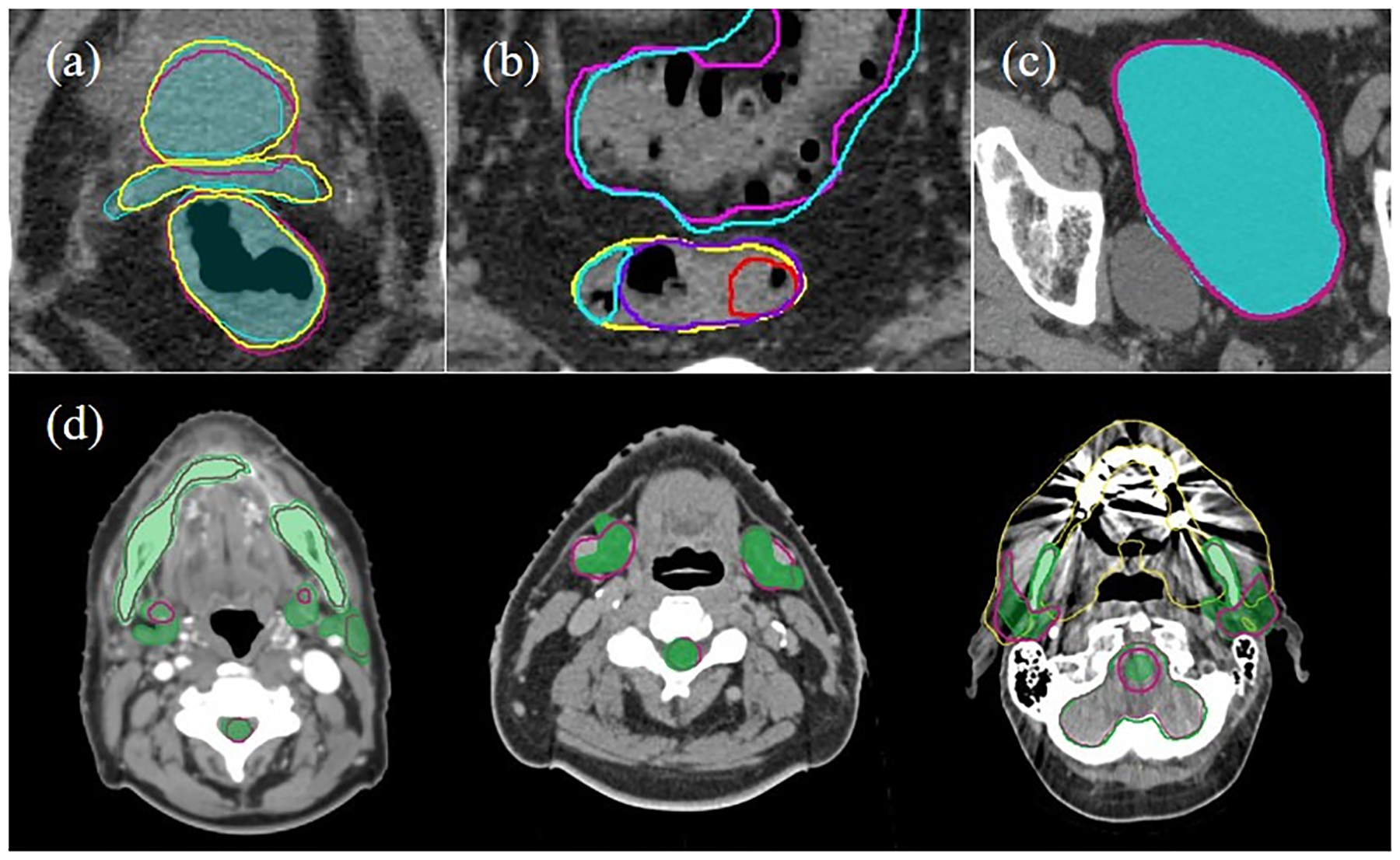
a) A comparison of three contour sets: ground truth (cyan filled), general male pelvis (G-MP) segmentation (violet-red) and custom male pelvis (C-MP) segmentation (yellow) for prostate, seminal vesicle and rectum on an axial slice; b) a comparison of general male pelvis (G-MP) segmented rectum (red) with custom male pelvis (C-MP) segmented sigmoid (cyan) and rectum (purple) and ground truth (yellow); c) accurate segmentation of bladder is observed with exclusion of the median lobe, as observed by a good overlap of the general male pelvis (G-MP) segmentation (violet-red line) on the ground truth (cyan filled); and d) examples of the sub-optimal performance of general head and neck (G-HN) auto-segmentation (violet-red line) as compared with the ground truth (green filled) in the presence of artifacts and anatomical uncertainties. Showing from left-to-right are poor boundary delineation due to the low contrast, vessels in vicinity of submandibular glands, and image artifact from dental implants.
For head and neck, the CT auto-segmentation and the manual editing can be tedious and time-consuming owing to the large number of OARs, small organ volumes, poor soft tissue contrast, and image artifacts (e.g., from dental implants)56. For the G-HN model, the dental artifacts resulted in incomplete delineation of the mandible in 2 out of 50 test cases. The auto-segmentation of the submandibular gland and left parotid gland was imperfect in nearly 50% of the cases (Figure 6a). These substantial inaccuracies in these OARs were due to the high variability in their shapes and locations, as well as their proximity to vessels and/or the disease sites. These specific anatomic situations may not be fully represented in the training datasets, and consequently, the obtained model would result in inaccurate delineation in such situations (Figure 8d middle). The C-HN model was developed to incorporate additional relevant OARs and specifically address the issues of vessels in proximity of glands in low contrast regions. As shown in Figure 6b, the auto-segmentation for parotid glands, and submandibular gland somewhat improved, a consequence of using a consistent cohort of site-specific training datasets. Another challenge in HN DL auto-segmentation is the presence of small organs39. In the earlier iterations of the C-HN model training process, we observed extremely poor segmentation for small organs, e.g., inner ears, eye lens and optic nerves. By using the ASR approach, the auto-segmentation accuracy for the small organs was substantially improved (Table 4). Recently, Gao et al., reported their solution to the auto-segmentation of small organs by using a two-stage deep neural network, FocusNetv with adversarial shape constraint57. Their average DSC for the inner ears is comparable with our result, 0.85 versus 0.83.
Abdomen is one of the most challenging tumor sites for CT auto-segmentation due primarily to the complex anatomy and poor soft tissue contrast. There are limited studies reporting DL auto-segmentation in abdomen in the literature. Mostly these studies focused on either relatively large or stable organs like liver, kidneys, and spleen33,58. To our knowledge, the current work is the first attempt to investigate DL auto-segmentation for 11 abdominal organs, including complex organs such as bowels. The gallbladder was not included in this study as it is often not a major concern in RT planning. Compared to other recently reported DL auto-segmentation work23,24,54, our DL models outperformed for 8 out of 11 abdomen OARs. For the remaining three (aorta, right kidney, and pancreas), comparable results were seen for aorta and right kidney, whereas our model performed worst for the pancreas. The previous study using WBnet23 showed the higher accuracy with a DSC of 0.84 for pancreas. It is generally challenging to auto-segment pancreas on CT due to multiple factors including the lack of contrast with surrounding tissues59, heterogeneity inside the pancreas, and a potential bias in the training data. Moreover, in this study, the training data includes both diseased and normal pancreas cases, with more diseased cases used. Consequently, the auto-segmentation for healthy pancreas may become inaccurate as shown in Figure 7b, a liver cancer case. Moreover, in abdomen OAR segmentation, inaccuracies occur primarily at transition junctions like duodenojejunal and gastroesophageal. These inaccuracies are consistent with challenges faced in manual contouring. The most challenging organ segmentation is the bowels, separately labelled as large and small. In our results, a large variability in accuracy was observed due to anatomic complexity of the bowels, i.e., random changes in shape, location and volume variations owing to distensibility and contractility60. In addition, variable lengths of the CT scans also contributed to segmentation challenges. For DL auto-segmentation, these challenges lead to two scenarios; 1) incomplete segmentation of one or more loops of large bowel, e.g., missing descending colon, 2) mislabeling between the two bowels. To address some of these challenges, we introduced the PSE approach that led to increased DSC/MDA values for small and large bowels from 0.71/7.9 mm and 0.7/14 mm to 0.84/3.8 mm and 0.87/2.37 mm, respectively. Representative image slices are shown in Figure S-1, where the auto-segmentation of three regions of bowels is compared with the ground truth, showing incomplete or mislabeled bowels, and improved bowel segmentation with the PSE approach.
Parallel to the quantitative evaluation of auto-segmentation accuracy, evaluation based on its time efficacy is relevant for clinical application of the DL auto-segmentation in RT planning. Other semi-automatic segmentation (e.g., atlas-based method) is known to save time, however, time efficacy assessment of DL auto-segmentation is almost nonexistent29,61–63. Although DL auto-segmentation can be completed in a matter of seconds or minutes for all relevant OARs in a tumor site, manual edit for the inaccurate auto-segmented contours is required with the currently achievable performance of DL auto-segmentation and can take substantial time. In this study, we have estimated realistic time efficacy of the DL auto-segmentation for all the three anatomical sites. Increased efficiency with our DL auto-segmentation was demonstrated by substantial reduction of the time needed to auto-segment, review, and manually edit versus the time required for the conventional manual contouring. The average time reductions were in the range of 65%−80% for the three tumor sites. Note that van der Veen et al. reported an average time saving of 33% for their DL auto-segmentation when compared to manual delineation for 16 OARs in the head and neck site64. In our study, we observed that, with our DL auto-segmentation techniques, the manual editing was not necessary for most of the OARs, except for the organs that are also considered challenging with manual contouring, e.g., pancreas, bowels, penile bulb, and inferior slices of glands.
Additionally, it was observed that the auto-segmented contours adhered closely to the contouring guidelines used in the training datasets. For example, the exclusion of the hilum of liver and kidneys in the training datasets was observed in the auto-segmented contours, indicating the robustness and reproducibility of our DL models (Figure 4e). For all the OARs, including those with inherently poor CT contrast (e.g., submandibular glands), the models trained using custom training datasets with the additional proximal organs and the consistent contouring guidelines showed slight improvement when compared with the general models.
This study has successfully contributed to the ongoing efforts to bring DL auto-segmentation into routine clinical practice, replacing the current labor-intensive and time-consuming manual contouring in RT planning process. There is still a large room for improvement65, motivating considerable on-going efforts including incorporating high soft tissue contrast imaging (e.g., MRI)51,53,66, developing robust algorithms (e.g., continual learning67), and using large and high-quality datasets.
Conclusions
We have successfully developed clinically applicable deep learning-based auto-segmentation models for 42 OARs in three major tumor sites by using a well-established DNCC network and incorporating organ-based approaches to improve model performance. The DL models trained with using either diversified multi-institutional datasets or well-controlled single institutional datasets can quickly generate clinically acceptable contours for most of the OARs, except for several complex organs that are also considered challenging in manual contouring. Performance of our DL models are generally better or comparable to those from the recent published studies with similar study scopes. The use of the DL models substantially reduced segmentation time and effort as compared to the manual contouring in radiation therapy planning process.
Supplementary Material
Acknowledgements
The authors thank Laura Buchanan PhD, Ivy Krystal Jones PhD, Xiaohuan Zhu, MSc for their help in the abdominal data preparation.
Funding Statement
The work was partially supported by the Medical College of Wisconsin (MCW) Cancer Center and Froedtert Hospital Foundation, the MCW Fotsch Foundation, Elekta AB, and the National Cancer Institute of the National Institutes of Health under award number R01CA247960. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of Interest Statement for All Authors
MCW received institutional research support from Elekta AB. JX and DT are employees of Elekta AB.
Data Availability Statement for this Work
Research data are stored in an institutional repository and selective data will be shared upon request to the corresponding author.
References
- 1.Atun R, Jaffray DA, Barton MB, et al. Expanding global access to radiotherapy. Lancet Oncol 2015;16(10):1153–1186. [DOI] [PubMed] [Google Scholar]
- 2.Citrin DE, Recent Developments in Radiotherapy. N Engl J Med 2017;377:1065–1075. [DOI] [PubMed] [Google Scholar]
- 3.Sharma N, Aggarwal LM. Automated medical image segmentation techniques. J Med Phys 2010;25(1):3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rigaud B, Simon A, Castelli J, et al. Deformable image registration for radiation therapy: principle, methods, applications and evaluation. Acta Oncol 2019;58(9):1225–1237. [DOI] [PubMed] [Google Scholar]
- 5.Yamazaki H, Nishiyama K, Tanaka E, et al. Dummy run for a phase II multi-institute trial of chemoradiotherapy for unresectable pancreatic cancer: inter-observer variance in contour delineation. Anticancer Res 2007;27(4C):2965–2971. [PubMed] [Google Scholar]
- 6.Vinod SK, Jameson MG, Min M, Holloway LC. Uncertainties in volume delineation in radiation oncology: A systematic review and recommendations for future studies. Radiother Oncol 2016;121(2):169–179. [DOI] [PubMed] [Google Scholar]
- 7.Lim-Reinders S, Keller BM, Al-Ward S, Sahgal A, Kim A. Online Adaptive Radiation Therapy. Int J Radiat Oncol Biol Phys 2017;99(4):994–1003. [DOI] [PubMed] [Google Scholar]
- 8.Ahunbay EE, Peng C, Chen GP, et al. An on-line replanning scheme for interfractional variations. Med Phys 2008;35(8):3607–3615. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Hoisak JDP, Li N, et al. Dosimetric benefit of adaptive re-planning in pancreatic cancer stereotactic body radiotherapy. Med Dosim 2015;40(4):318–324. [DOI] [PubMed] [Google Scholar]
- 10.Gupta V, Wang Y, Méndez Romero AM, et al. Fast and robust adaptation of organs-at-risk delineations from planning scans to match daily anatomy in pre-treatment scans for online-adaptive radiotherapy of abdominal tumors. Radiother Oncol 2018;127(2):332–338. [DOI] [PubMed] [Google Scholar]
- 11.Sharp G, Fritscher KD, Pekar V, et al. Vision 20/20: Perspectives on automated image segmentation for radiotherapy. Med Phys 2014;41(5):050902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardenas CE, Yang J, Anderson BM, Court LE, Brock KB. Advances in Auto-Segmentation. Semin Radiat Oncol 2019;29(3):185–197. [DOI] [PubMed] [Google Scholar]
- 13.Thor M, Apte A, Haq R, Iyer A, LoCastro E, Deasy JO. Using Auto-Segmentation to Reduce Contouring and Dose Inconsistency in Clinical Trials: The Simulated Impact on RTOG 0617. Int J Radiat Oncol Biol Phys 2021;109(5):1619–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521(7553): 436–444. [DOI] [PubMed] [Google Scholar]
- 15.Greenspan H, van Ginneken B, Summers RM. Guest Editorial Deep Learning in Medical Imaging: Overview and Future Promise of an Exciting New Technique. IEEE Trans Med Imaging 2016;35(5):1153–1159. [Google Scholar]
- 16.Zhou SK, Greenspan H, Davatzikos C, et al. A review of deep learning in medical imaging: Image traits, technology trends, case studies with progress highlights, and future promises. 2020. arXiv:2008.09104v2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- 18.Wong J, Huang V, Wells D, et al. Implementation of deep learning-based auto-segmentation for radiotherapy planning structures: a workflow study at two cancer centers. Radiat Oncol 2021;16:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazemifar S, Balagopal A, Nguyen D, et al. Segmentation of the prostate and organs at risk in male pelvic CT images using deep learning. Biomed Phys Eng Express 2018;4(5);055003. [Google Scholar]
- 20.Gibson E, Giganti F, Hu Y, et al. Automatic Multi-Organ Segmentation on Abdominal CT with Dense V-Networks. IEEE Trans Med Imaging 2018;37(8):1822–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong X, Lei Y, Wang T, et al. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys 2019;46(5):2157–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Y, Cai Z, Huang B, et al. Fully-Automated Segmentation of Nasopharyngeal Carcinoma on Dual-Sequence MRI Using Convolutional Neural Networks. Front Oncol 2020;10:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen X, Sun S, Bai N, Han K, et al. A deep learning-based auto-segmentation system for organs-at-risk on whole-body computed tomography images for radiation therapy. Radiother Oncol 2021;160:175–184. [DOI] [PubMed] [Google Scholar]
- 24.Isensee F, Jaeger PF, Kohl SAA, Petersen J, et al. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods 2021;18:203–211. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Lu Y, Yu Q, Luo X, et al. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. 2021. arXiv:2102.04306 [Google Scholar]
- 26.Valanarasu JMJ, Oza P, Hacihaliloglu I and Patel VM. Medical Transformer: Gated Axial-Attention for Medical Image Segmentation. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021:36–46. MICCAI 2021. Lecture Notes in Computer Science, vol 12901. Springer, Cham. [Google Scholar]
- 27.Gao Y, Zhou M and Metaxas DN. UTNet: A Hybrid Transformer Architecture for Medical Image Segmentation. 2021. arXiv:2107.00781 [Google Scholar]
- 28.Boldrini L, Bibault J-E, Masciocchi C, Shen Y, Bittner M-I. Deep Learning: A review for the Radiation Oncologist. Front Oncol 2019;9(977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong J, Fong A, McVicar N, et al. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother Oncol 2020;144:152–158. [DOI] [PubMed] [Google Scholar]
- 30.Zhong Y, Yang Y, Fang Y, Wang J, et al. A Preliminary Experience of Implementing Deep-Learning Based Auto-Segmentation in Head and Neck Cancer: A Study on Real-World Clinical Cases. Front Oncol 2021;11:638197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cramer JD, Burtness B, Le QT, Ferris RL. The changing therapeutic landscape of head and neck cancer. Nat Rev Clin Oncol 2019;16:669–683. [DOI] [PubMed] [Google Scholar]
- 32.Feng X, Qing K, Tustison NJ, Meyer CH, Chen Q. Deep convolutional neural network for segmentation of thoracic organs-at-risk using cropped 3D images. Med Phys 2019;46(5):2169–2180. [DOI] [PubMed] [Google Scholar]
- 33.Ma J, Zhang Y, Gu S, et al. Abdomen CT-1K: Is Abdominal Organ Segmentation A Solved Problem? 2020. arXiv:2010.14808v1 [Google Scholar]
- 34.Yu L, Yang X, Chen H, Qin J, Heng PA. Volumetric ConvNets with Mixed Residual Connections for Automated Prostate Segmentation from 3D MR Images. 31st AAAI conference on artificial Intelligence, 2017. [Google Scholar]
- 35.Long J, Shelhamer E, Darrell T. Fully Convolutional Networks for Semantic Segmentation. PROC CVPR IEEE 2015:3431–3440. [DOI] [PubMed] [Google Scholar]
- 36.Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A. (eds) Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015:234–241. MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer, Cham. [Google Scholar]
- 37.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. CVPR IEEE 2016;770–778. [Google Scholar]
- 38.Zhou Y, Xie L, Shen W, Wang Y, Fishman EK, Yuille AL. A fixed-point model for pancreas segmentation in abdominal CT scans. MICCAI 2017. LNCS;10433:693–701. [Google Scholar]
- 39.Roth H, Oda H, Hayashi Y, et al. Hierarchical 3D fully convolutional networks for multi-organ segmentation. 2017. arxiv:1704.06382v1 [Google Scholar]
- 40.Raudaschl PF, Zaffino P, Sharp GC, et al. Evaluation of segmentation methods on head and neck CT: Auto-segmentation challenge 2015. Med. Phys 2015;44(5):2020–2036. [DOI] [PubMed] [Google Scholar]
- 41.Gay HA, Barthold HJ, O’Meara E, et al. Pelvic normal tissue contouring guidelines for radiation therapy: A Radiation Therapy Oncology Group consensus panel atlas. Int J Radiat Oncol Biol Phys 2012;83(3):e353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brouwer CL, Steenbakkers RJHM, Bourhis J, et al. CT-based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines. Radiother Oncol 2015;117(1):83–90. [DOI] [PubMed] [Google Scholar]
- 43.Sun Y, Yu XL, Luo W, et al. Recommendation for a contouring method and atlas of organs at risk in nasopharyngeal carcinoma patients receiving intensity-modulated radiotherapy. Radiother Oncol 2014;110(3):390–397. [DOI] [PubMed] [Google Scholar]
- 44.Jabbour SK, Hashem SA, Bosch W, et al. Upper abdominal normal organ contouring guidelines and atlas: A Radiation Therapy Oncology Group consensus. Pract Radiat Oncol 2014;4(2):82–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright JL, Yom SS, Awan MJ, et al. Standardizing Normal Tissue Contouring for Radiation Therapy Treatment Planning: An ASTRO Consensus Paper. Pract Radiat Oncol 2019;9(2):65–72. [DOI] [PubMed] [Google Scholar]
- 46.Abadi M, Agarwal A, Barham P, et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Distributed Systems. 2016. arxiv:1603.04467v2 [Google Scholar]
- 47.Simard PY, Steinkraus D, Platt JC. Best Practices for Convolutional Neural Networks Applied to Visual Document Analysis. ICDAR 2003: Proceedings of the Seventh International Conference on Document Analysis and Recognition - Volume 2. [Google Scholar]
- 48.Shorten C and Khoshgoftaar TM. A survey on Image Data Augmentation for Deep Learning. J Big Data 2019;6(60). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brock KK, Mutic S, McNutt TR, Li H, Kessler ML. Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132. Med Phys 2017;44(7):e43–e76. [DOI] [PubMed] [Google Scholar]
- 50.Hummersone C. Alternative box plot (https://github.com/IoSR-Surrey/MatlabToolbox), GitHub; 2020. [Google Scholar]
- 51.Dong X, Lei Y, Tian S, Wang T, et al. Synthetic MRI-aided multi-organ segmentation on male pelvic CT using cycle consistent deep attention network. Radiother Oncol 2019;141:192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Lei Y, Fu Y, Wang Y, et al. Head and neck multi-organ auto-segmentation on CT images aided by synthetic MRI. Med Phys 2020;47(9):4294–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai X, Lei Y, Wang T, Zhou J, et al. Automated delineation of head and neck organs at risk using synthetic MRI-aided mask scoring regional convolutional neural network. Med Phys 2021;48(10):5862–5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tong N, Gou S, Niu T, Yang S, et al. Self-paced DenseNet with boundary constraint for automated multi-organ segmentation on abdominal CT images. Phys Med Biol 2020;65:135011. [DOI] [PubMed] [Google Scholar]
- 55.Fiorino C, Reni M, Bolognesi A, Cattaneo GM, Calandrino R. Intra- and inter-observer variability in contouring prostate and seminal vesicles: Implications for conformal treatment planning. Radiother Oncol 1998;47(3):285–292. [DOI] [PubMed] [Google Scholar]
- 56.O’ Daniel JC, Rosenthal DI, Garden AS, et al. The effect of dental artifacts, contrast media, and experience on interobserver contouring variations in head and neck anatomy. Am J Clin Oncol 2007;30:191–198. [DOI] [PubMed] [Google Scholar]
- 57.Gao Y, Huang R, Yang Y, Zhang J, et al. FocusNetv2: Imbalanced large and small organ segmentation with adversarial shape constraint for head and neck CT image. Medical Image Analysis 2021;67:101831. [DOI] [PubMed] [Google Scholar]
- 58.Kavur AE, Gezer NS, Barı M, Aslan S et al. , CHAOS Challenge - combined (CT-MR) healthy abdominal organ segmentation. Med Image Anal 2021;69:101950. [DOI] [PubMed] [Google Scholar]
- 59.Yao X, Song Y, Liu Z. Advances on pancreas segmentation: a review. Multimed Tools Appl 2020;79, 6799–6821. [Google Scholar]
- 60.Devi KG, Radhakrishnan R. Automatic Segmentation of Colon in 3D CT Images and Removal of Opacified Fluid Using Cascade Feed Forward Neural Network, Comput Math Method M 2015, 670739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaa E, Elguindib S, Onochiea I, Gorovetsa D, et al. Clinical implementation of deep learning contour autosegmentation for prostate radiotherapy. Radiother Oncol 2021;159:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kendall JK, Barman A, Stieb S, Fuller CD et al. , Novel Autosegmentation Spatial Similarity Metrics Capture the Time Required to Correct Segmentations Better than Traditional Metrics in a Thoracic Cavity Segmentation Workflow. J Digit Imaging 2021;34(3):541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lustberg T, van Soest J, Gooding M, Peressutti D et al. , Clinical evaluation of atlas and deep learning based automatic contouring for lung cancer. Radiother Oncol 2018;126(2):312–317. [DOI] [PubMed] [Google Scholar]
- 64.van der Veen J, Willems S, Deschuymer S, Robben D, et al. Benefits of deep learning for delineation of organs at risk in head and neck cancer. Radiother Oncol 2019;138:68–74. [DOI] [PubMed] [Google Scholar]
- 65.Tajbakhsh N, Jeyaseelan L, Li Q, Chiang JN, Wu Z, Ding X. Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation. Med Image Anal 2020;63:101693. [DOI] [PubMed] [Google Scholar]
- 66.Liu Y, Lei Y, Fu Y, Wang Y, et al. Head and neck multi-organ auto-segmentation on CT images aided by synthetic MRI. Med Phys 2020;47(9):4294–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim N, Chun J, Chang JS, Le CG et al. Feasibility of Continual Deep Learning-Based Segmentation for Personalized Adaptive Radiation Therapy in Head and Neck Area. Cancers (Basel) 2021;13(4):702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Research data are stored in an institutional repository and selective data will be shared upon request to the corresponding author.


