Abstract
Previous researches have demonstrated the meaning of CTSB for the progress of several tumors, whereas few clues about its immunological characteristic in gliomas. Here we systematically explored its biologic features and clinical significance for gliomas. 699 glioma cases of TCGA and 325 glioma cases of CGGA were respectively included as training and validating cohorts. R software was used for data analysis and mapping. We found that CTSB was remarkably highly-expressed for HGG, IDH wild type, 1p19q non-codeletion type, MGMT promoter unmethylation type and mesenchymal gliomas. CTSB could specifically and sensitively indicate mesenchymal glioma. Upregulated CTSB was an independent hazard correlated with poor survival. CTSB-related biological processes in gliomas chiefly concentrated on immunoreaction and inflammation response. Then we proved that CTSB positively related to most inflammatory metagenes except IgG, including HCK, LCK, MHC II, STAT1 and IFN. More importantly, the levels of glioma-infiltrating immune cells were positively associated with the expression of CTSB, especially for TAMs, MDSCs and Tregs. In conclusion, CTSB is closely related to the malignant pathological subtypes, worse prognosis, immune cells infiltration and immunosuppression of gliomas, which make it a promising biomarker and potential target in the diagnosis, treatment and prognostic assessment of gliomas.
Subject terms: Diagnostic markers, Prognostic markers, Cancer, Diagnosis, Prognosis, Therapeutics, Cancer, Surgical oncology, Cancer therapy, CNS cancer, Tumour biomarkers, Transcription
Introduction
Now gliomas have become the most common type of primary tumors in central nervous system (CNS), comprising 80.8% of the primary malignant tumors and resulting in 88.10% of the deaths in CNS tumors1. The current therapies for gliomas include surgical resection, chemotherapy, radiation therapy and some other emerging treatments such as molecular targeted therapy, immunotherapy and tumor treating fields (TTF)2,3. Yet the holistic treatment status of gliomas has achieved limited improvements with these therapies, and the therapeutic response of high-grade gliomas (HGG) is still poor3–5. More recently, WHO CNS5 has highlight the significance of molecular biomarkers in providing meaningful diagnostic and therapeutic information for gliomas6. Enrichment strategies using precise biomarkers will help to improve the current dilemma of glioma treatment4. Thus, it is urgent to find more specific molecular biomarkers with clinicopathologic utility closely related to the malignant behavior and immune microenvironment of gliomas.
Cathepsin B (CTSB) is an exclusive multifunctional protein of cysteine protease family in that it has an additional pH sensitive occluding loop which allows it to act as an endopeptidase/exopeptidase depending upon pH7. There are three major subtypes of CTSB8: main transcript, main transcript lacking exon 2 or main transcript lacking exon 2 and 3. The latter two isoforms were proved as a result of alternative splicing9,10, which directly influences the translation rate, sorting and catalytical activity of CTSB11–14. CTSB is tightly regulated transcriptionally, post-transcriptionally and post-translationally from biosynthesis to lysosomes15, such as transcription level increased, transcription start sites alterations, splice variants adjustments and post-transcriptional modifications via proteolytic processing, glycosylation, inhibition and trafficking16. Different subtypes of CTSB have different subcellular localizations, which determine its multiple biological functions8. CTSB locates mainly in secretory vesicles, cytoplasm, nucleus, and also binds to plasma membrane or is secreted outside into the extracellular space15,17. It can also be secreted without entering the endosomal-lysosomal system8. Translocation of CTSB may occur under pathological conditions, for example, it can locate on the cell membrane of cancer cells via binding to annexin A2(ANXA2) tetramers or the caveolae site18. The physiological function of CTSB mainly associates with its carboxypeptidase activity, and the endopeptidase activity of CTSB is related to its pathological role in the degradation of extracellular matrix (ECM)8. The principal physiological function of CTSB is the lysosomal degradation of different proteins to maintain the stable state of intracellular proteome15. CTSB has also been proved to participate in ECM remodeling by degrading the structural components, such as collagen and elastin17. It also can affect various intracellular signaling pathways with the processing of several cytokines and chemokines17. CTSB plays a key role in many physiological and pathological processes such as cell proliferation, migration, autophagy, antigen presentation, apoptosis19, hippocampal dependent memory function20, cell differentiation and tumorigenesis17.
In terms of pathological processes, CTSB related to arthritis and atherosclerotic vascular diseases17, and has also been confirmed to be involved in inflammatory brain diseases and brain aging21, memory and psychiatric disorders and neuronal cascade death response after cerebral infarction22. CTSB has been regarded as an important biomarker and potential therapeutic target for many cancers15, such as pancreatic cancer23, gastric cancer24, colon cancer25, breast cancer8, acute myelogenous leukemia26, oral squamous cell carcinoma and thyroid cancer27. Recent studies have confirmed that CTSB is rarely expressed in normal brain tissues but overexpressed in glioblastoma (GBM) and glioblastoma stem cells (GSCs)28,29. The expression level of CTSB in GBM was 6 times higher than that in normal brain tissues30. For GBM, CTSB mainly locates in the invasive margins of tumor infiltration and neovascularization31. Additionally, it was shown that highly-expressed CTSB can promote the proliferation32, migration33, adhesion15, invasion18, anti-apoptosis34, tumor angiogenesis8 and drug resistance of GBM tumor cells35. These make CTSB a possible therapeutic target for gliomas. Yet previous researches about CTSB in gliomas mainly focused on GBM, mostly in vitro, and lacked of comprehensive large-sample clinical analysis, which are inadequate in reflecting the actual role of CTSB regarding the immune microenvironment and tumor heterogeneity of gliomas. More importantly, there were few clues about the role of CTSB in glioma-related immune activities, which greatly limits the understanding of the function and mechanism of CTSB in gliomas and restricts the clinical transformation of CTSB-targeted therapy for gliomas.
In this study, we aimed to comprehensively explore the expression pattern, biological functions and prognostic value of CTSB for gliomas, especially focusing on its role in glioma-related immune responses and immune cells infiltration. So, we chose The Cancer Genome Atlas (TCGA) dataset as training cohort and the Chinese Glioma Genome Atlas (CGGA) dataset as validating cohort to analyze the RNA sequencing (RNA-seq) data of 1024 glioma cases. This study provides a powerful theoretical basis for the design of CTSB-related therapeutics for gliomas.
Results
CTSB is remarkably highly-expressed in HGG, isocitrate dehydrogenase (IDH) wildtype, 1p19q non-codeletion type and O6-methylguanine-DNA methyltransferase (MGMT) promoter unmethylation gliomas
To explore the expression pattern of CTSB in gliomas, we analyzed the RNA-seq data of glioma samples from TCGA and CGGA databases. We found that CTSB was significantly highly-expressed in HGG especially in WHO grade IV gliomas (P < 0.0001) (Fig. 1A,B). This result was consistent in both databases. Meanwhile, the different types of IDH status, 1p/19q-codeleted status and MGMT status has been proved to be crucial to clarify the subtypes of gliomas and predict the prognosis of patients6. Therefore, we analyzed the difference of the expression level of CTSB between these glioma types. It was showed that the expression of CTSB in IDH wide-type (Fig. 1C,D overall grade; E, F low-grade gliomas (LGG); G, H HGG), 1p19q non-codeletion type (Fig. 1I,J) and MGMT promoter unmethylation type (Fig. 1K,L) were significantly higher than those in IDH mutant-type (especially for HGG), 1p19q codeletion type and MGMT promoter methylation type of gliomas respectively. Among these variables, IDH status was proved to be most strongly associated with CTSB expression via multivariate correlation analysis of TCGA (Fig. S1A) and CGGA (Fig. S1B) cohorts. These results reveal that CTSB is highly-expressed in these malignant molecular types which are negatively markers of therapeutic reactivity and prognosis of gliomas.
Figure 1.
Comparison of the expression of CTSB in gliomas of different WHO grades, isocitrate dehydrogenase (IDH) status, 1p/19q-codeleted status and O6-methylguanine-DNA methyltransferase (MGMT) promoter status. CTSB was significantly highly-expressed in high-grade gliomas (HGG), IDH-wildtype, 1p19q non- codeletion and MGMT promoter unmethylation gliomas in TCGA and CGGA datasets. The expression of CTSB in TCGA dataset according to WHO grade (A), IDH status (overall WHO grades (C), LGG (E), HGG(G)), 1p/19q-codeleted status (I) and MGMT status (K). The expression of CTSB in CGGA dataset according to WHO grades (B) and IDH status (overall WHO grades (D), LGG (F), HGG(H)), 1p/19q-codeleted status (J) and MGMT status (L). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The upregulated CTSB is correlated with the poor prognosis of gliomas
The results above suggested that CTSB may be a potential biomarker of malignant gliomas. Then we analyzed the prognostic value of CTSB for gliomas. Firstly, we drew Kaplan Meier curve with the survival data of glioma samples in the two databases (Fig. 2A,B), we found that patients with higher expression of CTSB would have shorter OS time (P < 0.0001). To avoid the influence of tumor heterogeneity, we also compared the effect that CTSB have on the OS time of LGG (Fig. 2C,D) and HGG (Fig. 2E,F) respectively, and this phenomenon was more obvious in HGG. Secondly, we further explored the role that CTSB played in the prognosis of gliomas together with other factors such as gender, age, WHO grade, IDH mutant status, MGMT promoter methylation type and 1p19q codeletion status (Fig. 3A,B overall grade; Fig. S2A, B LGG; Fig. S2C, D HGG). The multi-factors COX analysis also come to the same conclusion that high-expressed CTSB is a risk factor and a negative biomarker related with the poor prognosis of HGG.
Figure 2.
Survival analysis of glioma patients based on CTSB expression in TCGA and CGGA cohorts. Kaplan–Meier analysis indicated that high expression of CTSB was related to significantly worse prognosis overall in overall glioma (A,B), low-grade gliomas (LGG) (C,D) and HGG (E,F) patients.
Figure 3.
Forest plot of hazard ratios for overall survival rates assessed by CTSB and clinicopathological factors. CTSB was an independent prognostic factor after adjusting for age, WHO grade, IDH status, 1p19q status and MGMT status in TCGA (A) and CGGA (B) datasets.
CTSB is highly-expressed in mesenchymal subtype of gliomas and is specific and sensitive in predicting mesenchymal subtype
In the last few years, TCGA network have divided gliomas into four molecular subtypes: proneural, neural, classical and mesenchymal type36. This classification has been proved to be meaningful for patients’ survival status, especially mesenchymal subtype which is representing for invasion and poor prognosis5. So, we analyzed the relationship between CTSB and TCGA subtypes. We found that CTSB was remarkably highly-expressed in mesenchymal subtype both in TCGA and CGGA databases (Fig. 4A,C). Then we performed receiver operator characteristic curve (ROC) to evaluate the specificity and sensitivity of CTSB in predicting mesenchymal subtype gliomas. The area under the curve (AUC) of the TCGA dataset was 0.937, and the specificity and sensitivity were 88.6% and 87.5% respectively at the optimal cut-off value of 14.173 (Fig. 4B). The AUC of the CGGA dataset was 0.894, and the specificity and sensitivity were 82.4% and 85.2%, respectively at the optimal cut-off value of 1.919 (Fig. 4D). These results indicate that CTSB is highly specific to the mesenchymal gliomas and can be used to predict this subtype.
Figure 4.
Comparison of CTSB expression levels in different TCGA molecular subtypes. CTSB was significantly enriched in the mesenchymal subtype in TCGA (A) and CGGA (C) cohorts (P < 0.0001). Receiver operator characteristic curve (ROC) curve analysis showed the predictive value of CTSB for mesenchymal subtype in the TCGA and CGGA cohorts (B,D). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
CTSB is closely related to the immune activities in gliomas
To further study the feature and biological function of CTSB, we ranked the genes related with CTSB via Spearman’s correlation analysis, finally we respectively screened 162 related genes from TCGA database (151 positively corelated genes, 11 negatively corelated genes, |R|> 0.7 and P < 0.05) and 170 related genes from CGGA database (165 positively corelated genes, 5 negatively corelated genes, |R|> 0.6 and P < 0.05), as shown in Supplementary Table S1. Then we used the DAVID website to perform Gene ontology (GO) functional analysis of these genes. The biological processes of CTSB mainly contain signal transduction, immune response, inflammatory response, innate immune response, regulation of immune response, interferon-gamma-mediated signaling pathway, leukocyte migration, positive regulation of T cell proliferation, positive regulation of tumor necrosis factor and interferon-gamma production, antigen processing and presentation, positive regulation of phagocytosis and so on. In terms of cellular components, CTSB mainly act as extracellular exosome, membrane raft, actin filament and lysosome, which located on cell surface, phagocytic vesicle membrane, plasma membrane and in extracellular space. The molecular function of CTSB mainly contains protein binding, serine-type and cysteine-type endopeptidase activity, hydrolase activity, protein homodimerization activity, S100 protein and MHC class II protein complex binding and so on. The above results were consistent among TCGA and CGGA databases (Fig. 5A,B). Meanwhile, we also did GO functional analysis on the overlapped 64 related genes (Supplementary Table S1) of the two databases for validation (Fig. 5C,D). Our study showed that CTSB is correlated with many immune activities and inflammatory response processes in gliomas, and mainly act as extracellular exosome, lysosome and membrane raft in extracellular space or on cell membrane.
Figure 5.

Gene ontology (GO) analysis of CTSB-related characteristics in gliomas. The results revealed that CTSB is related to some important biological processes in glioma based on TCGA (A) and CGGA (B) datasets. GO analysis of 64 genes common to both datasets was used for validation (C,D).
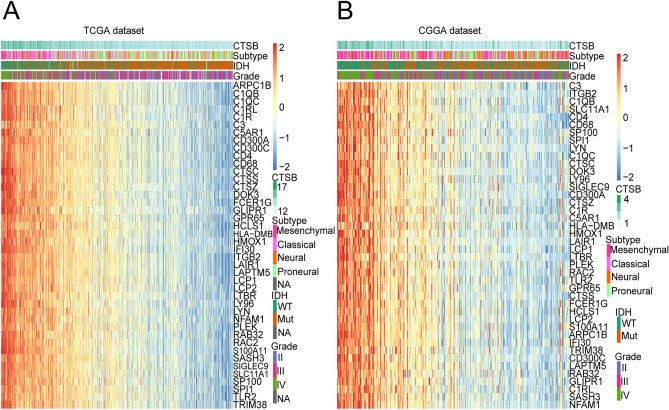
Then for further exploration of the function in glioma-related immune activities, we download the immune genes sets from AmiGO2 website. According to this dataset, we chose 104 genes from TCGA database (|R|> 0.7, P < 0.05) and 94 genes from CGGA database (|R|> 0.6, P < 0.05) which was significantly correlated with CTSB (Supplementary Table S2). Then the overlapped 42 immune-related genes of them (Supplementary Table S2) were used to draw the heat map (Fig. 6A,B). Then we found that most of these genes are positively related to the expression of CTSB in both databases, which further reveals the role that CTSB plays in glioma-related immune activities.
Figure 6.
Heatmap analysis of the relationship between CTSB and immune function-related genes in gliomas. The results showed that CTSB had a markedly positive correlation with most immune genes in both TCGA (A) and CGGA (B) cohorts.
CTSB is closely related to the inflammatory response of gliomas
In the results above, it is suggested that CTSB is related with the inflammatory response of gliomas, so in order to explore the function of CTSB, we analyzed 104 inflammatory genes which could be divided into seven metagenes5. Supplementary Table S3 summarizes the detailed list of these genes. In TCGA and CGGA datasets, we performed heatmap clustering analysis on the above genes and found that all of the gene metagenes are positively associated with CTSB except IgG (Fig. 7A,B; Fig. S3A, B). To validate this result, we applied gene set variation analysis (GSVA) to draw correlograms according to the Pearson correlation between CTSB and the seven gene metagenes (Fig. 7C,D). Finally, the results of the two cohorts were highly consistent with the heatmap. The expression of CTSB is remarkably positively related with HCK, MHC-II, STAT1, LCK, MHC-I and interferon (IFN) metagenes, but negatively associated with IgG metagene.
Figure 7.
CTSB-related inflammatory activities in gliomas. The relationship between CTSB and WHO grade, IDH status, molecular subtypes and inflammatory metagenes are presented as a heatmap in TCGA and CGGA databases (A,B). Correlograms showed the correlation between CTSB and inflammatory metagenes (C,D). Blue represents positive correlations, and red represents negative correlations. Color intensity and the size of the circle in pie charts are proportional to the correlation coefficients. The results indicated that CTSB was significantly positively correlated with most inflammatory activities.
Relationship between CTSB and tumor-infiltrating immune cells in gliomas
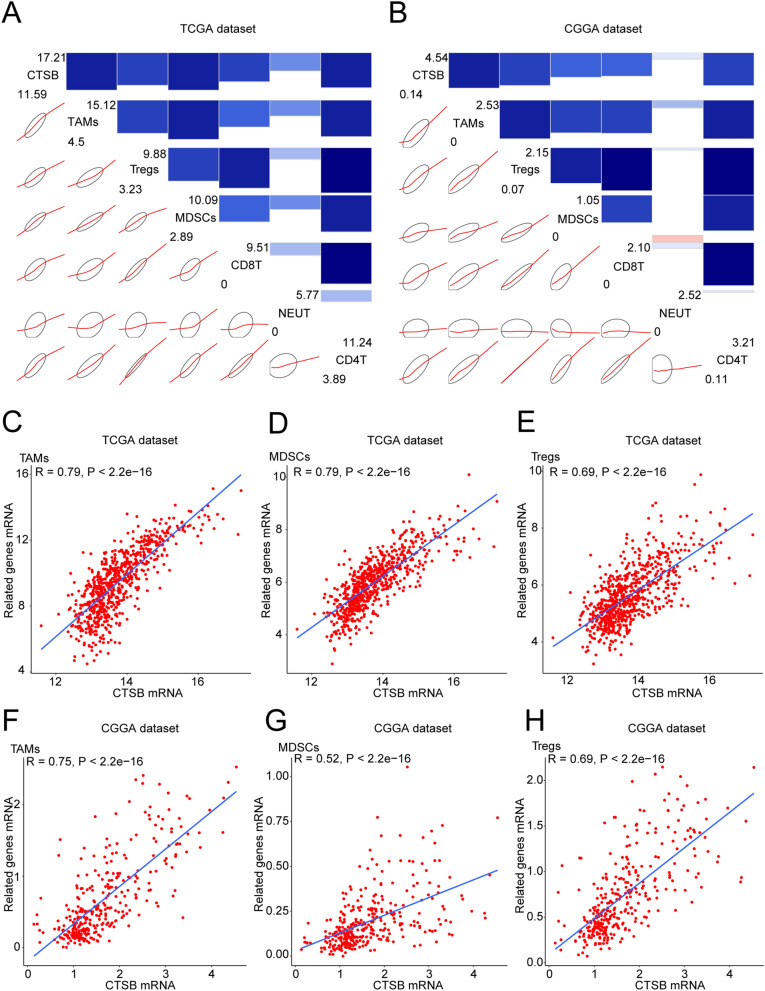
Tumor-infiltrating immune cells were proved to be essential in the invasion progress of gliomas37. Therefore, it is necessary to explore the relationship between CTSB and infiltrating immune cells in gliomas. We analyzed the most common 6 types of immune cells such as CD4 + T cells, regulatory T cells (Tregs), CD8 + T cells, tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs) and neutrophils (NEUT), and the detailed information of the markers of these 6 cell types is summarized in Supplementary Table S4. Then we drew corrgrams to show the correlation between CTSB and above cell types in the two databases. It was shown that these infiltrating immune cells were positively correlated with the expression of CTSB (Fig. 8A,B). We also performed Pearson correlation analysis, and CTSB was significantly correlated with TAMs, MDSCs and Tregs in both databases: TAMs (r = 0.79 in TCGA dataset, r = 0.75 in CGGA dataset; Fig. 8C,F), MDSCs (r = 0.79 in TCGA dataset, r = 0.52 in CGGA dataset; Fig. 8D,G), and Tregs (r = 0.69 in TCGA and CGGA datasets; Fig. 8E,H).
Figure 8.
CTSB-related infiltrating immune cells in gliomas. (A,B) Correlograms showing the correlation between CTSB and immune cell infiltration level based on both datasets. Blue represents positive correlations, and red represents negative correlations. Color intensity and the size of the bars and ellipses are proportional to the correlation coefficients. The leading diagonal contains the minimum and maximum values of variables. (C–H) CTSB was significantly positively correlated with tumor-related macrophages (TAMS), myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs) in both datasets. Each point represents a glioma sample. A regression line was fitted to the dot plot.
Discussion
To date, even lots of trials of targeted therapy for gliomas have not shown anticipated efficacy, enrichment strategies with more precise biomarkers of gliomas will greatly increase the chances of success in the future4,5. Our study indicates that highly-expressed CTSB in the immune microenvironment is closely correlated with the malignant molecular features, worse prognosis, immune cells infiltration and immunosuppression processes of gliomas, which can be regard as a new prognostic biomarker and potential therapeutic target.
First, we found that CTSB is significantly highly-expressed with increasing WHO grade, especially for HGG, which was consistent with previous studies28. We also demonstrated that CTSB is highly-expressed in known malignant glioma molecular phenotypes such as IDH wild-type, MGMT promoter unmethylation, 1p19q non-codeletion and mesenchymal subtype. Meanwhile, CTSB may be a specific and sensitive predictor of mesenchymal subtype gliomas. According to the clinical meaning of these molecular phenotypes in gliomas, patients with highly-expressed CTSB may have a greater risk of tumor recurrence, progression, epithelial mesenchymal transformation (EMT) and therapeutic resistance. The survival analysis showed that the highly-expressed CTSB means shorter OS time of glioma patients, which is also an independent risk factor for the prognosis, and the similar result was reported before in GBM38. The overall expression and enzymatic activity of CTSB in GBM could be regulated through the alternative splicing of the pre-mRNA of CTSB39,40, which determined by the differentiation and microenvironment of tumor cells and in return influence the malignant behavior of GBM39. We preliminarily profiled the splicing patterns of CTSB for gliomas via TCGA SpliceSeq41, and the exon 8 skip is the most common splicing type of CTSB in gliomas (Fig. S4A-E). CTSB also acts as a mediator of EMT process upregulating EMT-activated transcription factors through the Wnt/β-catenin pathway15. As the most invasive lysosomal cathepsin in GBM, CTSB remodels ECM to create prerequisite niches for invasion33 by degrading ECM components(such as laminin, collagen, fibrin, tenascin-C)42, triggering the uPA/plasminogen/plasmin proteolytic cascade42, activating other proteolytic enzyme systems32, releasing cytokines and growth factors, aggravating peritumoral edema and acidic microenvironment43. It also impairs the basement membrane of blood brain barrier (BBB) to promote glioma cells invasion33. CTSB also induces cell senescence mediated malignant transformation via CCNB2/SASP/CTSB & PGE2 axis44. It was also shown that CTSB can promote proliferation by upregulating the expression of p-ERK and c-Myc and reducing the level of Cellular Repressor of E1A Stimulated Genes 1 in GBM45. CTSB can increases the resistance of glioma cells to apoptosis and ferroptosis via lysosome-nuclear pathway of communication15,46. Highly-expressed CTSB interacts with the ANXA2 tetramer and induces the expression of VEGF-C, TGF-β and matrix metalloprotein (MMP)-9 to promote high permeability angiogenesis in gliomas8,47. CTSB promotes the generation and proliferation of GSCs via renin-angiotensin system (RAS)28, and regulates the self-renewal of GSCs through hedgehog components, Bmi1 and Sox229. These studies strongly support our conclusion. Therefore, CTSB can be used for the molecular pathology diagnosis and prognosis evaluation of gliomas as a promising biomarker.
Regarding the role and biological processes of CTSB in gliomas, we found that CTSB is mainly related to signal transduction, immune and inflammatory response, regulation of immune response, leukocyte migration, interferon-gamma-mediated signaling pathway, antigen processing and presentation. It also associates with the positive regulation of T cell proliferation, tumor necrosis factor and interferon-gamma production. Correlation analysis confirmed that glioma-associated immune response and inflammatory activity are significantly correlated with CTSB. Immune cell infiltration and immunosuppression play a vital role in the progression and treatment resistance of gliomas3,5. But the exact function of CTSB in these processes remains unclear. Our data showed that the degree of immune cell infiltration is significantly related to the expression level of CTSB, especially the immunosuppression-related immune cells TAMs, MDSCs and Tregs. This new finding suggests that CTSB in TME may associate with the immunosuppression and progression of gliomas. Previous studies reported that CTSB is actively correlated with T lymphocytes apoptosis in antigenic immune response48 and delayed type hypersensitivity as a regular factor of lysosomal biogenesis and autophagy49. It induces cognitive impairment through inflammatory response and drives inflammatory brain disease via regulating the production and secretion of IL-1β21. CTSB can activate NLRP3 inflammasome to adjust IL-1β and IL-18 production7, and it also controls the secretion of TNF-α in monocytes and IL-12 in macrophages50. It may cause anti-inflammatory response via adjusting autophagy or mitochondrial dynamics of macrophages51. CTSB is highly-expressed in lung cancer and recruits monocytes into tumor to become TAMs, which promotes immunosuppression and tumor progression52. CTSB knock-down significantly decreases MDSCs infiltration of premalignant intestinal polyps53. CTSB-mediated CD18 flaking adjusts the extravasation, transmigration and recruitment of leukocyte from tumor angiogenic vessels47. CTSB is related to infiltrating immune cells with the activation of ELR chemokines and inactivation of Non-ELR chemokines54. CTSB cracks Rip1 kinase to inhibit the necrotic apoptosis of TAMs and promotes macrophage-assisted pro-metastatic processes55. CTSB in TAMs creates an advantageous microenvironment for tumor invasion19 and protects tumor cells from apoptosis induced by etoposide, paclitaxel and doxorubicin56. As proved in mouse model, CTSB in MDSCs is crucial for the tumorigenesis of pancreatic neuroendocrine carcinoma57. Interplay between CTSB and NLRP3 inflammasome in tumor-infiltrating MDSCs results in IL-1 β production and pro-tumor immune response58. CTSB also participates in antigen-presenting, immune cells differentiation and homeostasis48. CTSB is closely related to immunosuppression of cervical cancer59. The CTSB on tumor cells surface can remove the autoreactive lymphocytes and degrade these cytotoxic effector molecules (such as IgG and chemokines CXCR3, CXCL9, CXCL10) synthesized from tumor-suppressive infiltrating immune cells7,60. CTSB promotes the apoptosis of CD8 + T lymphocytes to prevent them from being memory CD8 + T cells and decrease the lasting maintenance of them48. It also induces the programmed death of pro-B cells mediated with CpG TLR-9 and the death of B cells from antigen-dependent germinal center61. Therefore, the role of CTSB in glioma-related immune response and immunosuppression makes it possible to use CTSB as a therapeutic target enhancing the host immune system to interfere with immune evasion of glioma cells, which is a promising prerequisite in improving the reactivity of targeted therapy for gliomas62.
According to the expression patterns, prognostic significance and the biological processes of CTSB in gliomas, we deem that CTSB-based therapy might be an important strategy to improve the overall prognosis and the therapeutic reactivity of glioma patients. BBB-permeable CTSB inhibitors have been used in many neurological diseases such as stroke, Alzheimer’s disease and Parkinson’s disease21. Recently, many researchers explored the therapeutic application of small molecule inhibitors for CTSB in many cancers7, and CTSB-based visible-light-triggered nanoparticles of prodrug can enhance the effect of immune checkpoint blocking therapy, chemotherapy and photodynamic therapy63. Moreover, previous studies have preliminarily investigated the potential clinical value of CTSB for gliomas in vitro and vivo8,28,31,64. Down-regulation of CTSB significantly inhibited the proliferation, invasion and tumor angiogenesis of GBM8. Auranofin and Tivozanib can reduce the invasion of GBM cells through inhibiting the activity of CTSB and blocking the CTSB/uPA/MMP-2 proteolytic cascade respectively31,64. Considering the interaction between CBST and RAS in promoting GSCs invasion, targeted therapy combined CTSB with RAS remarkably reduces the growth of GBM in vivo28. For treatment resistance, overexpressed CTSB promoted EMT, decreased the cytotoxicity of TMZ and reduced TMZ-induced cell death in GBM35. High expression of CTSB also facilitates radioresistance of GBM and paediatric glioma via increasing homology recombination65. Inhibition of CTSB will upregulate the expression of γH2AX and H2AX by inhibiting C-Met signal transduction, resulting in the transcriptional arrest of cells that makes them sensitive to apoptosis induced by radiation, thus improving the radioresistance of gliomas66. Furthermore, the specific expression of CTSB in GBM has been attempted to apply in the design of surgical protocols67. CTSB-based small molecular probes of suicide inhibitors with fluorescently quenched activity have been demonstrated in mouse models that they can quickly recognize the core and edge of GBM, rapidly visualizing marginal infiltrating tumor cells during operation, which provides an innovative imaging method of potential application value for surgical directional resection of gliomas67. Based on our findings and the above studies, CTSB-targeted therapy alone or combined with immunotherapy and vascular targeting therapies may complement current treatment strategies for gliomas. Nevertheless, further studies with single-cell RNA sequencing are currently in progress to optimize the limitations of multicellular level analysis for TCGA and CGGA datasets, which can also be used to furtherly explore splice variant expression and other regulation ways to reveal the specific mechanism of CTSB in the immunosuppressive microenvironment of gliomas. More evidences of preclinical studies are also needed to support the clinical application of CTSB. In the future, CTSB may clinically acts as a potential biomarker and practical target for the precision diagnosis, treatment, image-guided surgery, cellular modification agent and targeted drug delivery of gliomas17.
In brief, this research mainly focused on the expression pattern, biological function and clinical significance of CTSB in gliomas. Our study suggested that highly-expressed CTSB in gliomas was closely associated with the malignant pathological subtypes, poor prognosis, tumor-infiltrating immune cells and immunosuppression, which made it a promising biomarker and potential target in the diagnosis, treatment and prognostic assessment of gliomas. We believe that in the future, CTSB-based therapies alone or in combination with other treatments will be a meaningful strategy in the combination and individualized precision therapies for gliomas.
Methods
Patients and samples
All the selected cases of glioma ranging from WHO grade II to grade IV were collected from the TCGA database (699 cases) (http://cancergenome.nih.gov/) and the CGGA database (325 cases) (http://www.cgga.org.cn). The CGGA database was mainly used to avoid the limitation of single-dataset research. We obtained all the data of RNA-seq, molecular pathological materials and the overall survival (OS) rate information from above cases. We sifted through and removed 63 cases and 15 cases without available above materials from the TCGA and CGGA datasets respectively. This research was approved by the Ethics Committee of the Peking University Third Hospital (S2020018).
Statistical analysis
We performed all the statistical analyses and figures with R software for MacOS, version 4.0.3 (http://www.r-project.org). All the gene expression profiling data in this study were log-transformed for further analysis. Kaplan–Meier survival curves were used to show the overall survival rate difference of glioma patients. Multivariate Cox regression analysis was performed with the survival package in R. Other R packages such as ggplot2, pROC, pheatmap, devtools, corrplot, ggpubr and corrgram package were also used to visualize the analyzed data. We filtered out these genes which were significantly corelated with CTSB via Spearman’s correlation analysis. GO functional analysis was performed using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/) to explore the enriched biological process, molecular function and cellular component. The immune system gene sets were downloaded from the AmiGO 2 website (http://amigo.geneontology.org/amigo) to analyze the role that CTSB plays in glioma-related immune activities. The Pearson’s correlation analysis was also used to evaluate the strength of correlation. One-way ANOVA was used to test the differences among at least 3 groups. Student’s t-test was used to test the differences of each 2-group comparison. The statistically significant differences were defined at the level of P < 0.05.
Ethics approval
This research was approved by the Ethics Committee of the Peking University Third Hospital (S2020018).
Supplementary Information
Acknowledgements
This work was supported by the “National Natural Science Foundation of China” (Grant Number: 82072774 and 81872051), “Peking University Clinical Scientist Program” (Grant Number: BMU2019LCKXJ007), “Key Clinical Projects of Peking University Third Hospital” (Grant Number: BYSY2018060), “China Postdoctoral Science Foundation” (Grant Number: 2020M670064) and “Beijing Natural Science Foundation” (Grant Number: 7214271).
Abbreviations
- CNS
Central nervous system
- TTF
Tumor treating fields
- HGG
High-grade glioma
- CTSB
Cathepsin B
- ANXA2
Annexin A2
- ECM
Extracellular matrix
- GBM
Glioblastoma
- GSCs
Glioblastoma stem cells
- TCGA
The Cancer Genome Atlas
- CGGA
Chinese Glioma Genome Atlas
- RNA-seq
RNA sequencing
- IDH
Isocitrate dehydrogenase
- MGMT
O6-methylguanine-DNA methyltransferase
- LGG
Low-grade glioma
- ROC
Receiver operating characteristic
- AUC
Area under the curve
- GO
Gene ontology
- GSVA
Gene set variation analysis
- IFN
Interferon
- Tregs
Regulatory T cells
- TAMs
Tumor-associated macrophages
- MDSCs
Myeloid-derived suppressor cells
- NEUT
Neutrophils
- EMT
Epithelial mesenchymal transformation
- BBB
Blood brain barrier
- MMP
Matrix metalloprotein
- RAS
Renin-angiotensin system
- OS
Overall survival
Author contributions
K.M. and J.Y. conceived and designed the project; X.C. and W.L. collected the data; K.M and X.C. analyzed the data; S.C. and C.Y. participated in constructing figures; W.L. and S.C. manuscript revision; K.M and J.Y wrote the paper. All authors read and approved the final manuscript.
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Code availability
Custom code used to analyse the RNA-seq data and datasets generated and/or analysed in the current study are available from the corresponding authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-08346-2.
References
- 1.Ostrom, Q. T. et al. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro Oncol. 22, iv1–iv96 (2020). [DOI] [PMC free article] [PubMed]
- 2.Nabors, L. B. et al. Central nervous system cancers, Version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 18, 1537–1570(2020). [DOI] [PubMed]
- 3.Xu S, Tang L, Li X, Fan F, Liu Z. Immunotherapy for glioma: Current management and future application. Cancer Lett. 2020;476:1–12. doi: 10.1016/j.canlet.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Tan AC, et al. Management of glioblastoma: State of the art and future directions. CA Cancer J. Clin. 2020;70:299–312. doi: 10.3322/caac.21613. [DOI] [PubMed] [Google Scholar]
- 5.Ma K, et al. ANXA2 is correlated with the molecular features and clinical prognosis of glioma, and acts as a potential marker of immunosuppression. Sci. Rep. 2021;11:20839. doi: 10.1038/s41598-021-00366-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis, D. N. et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol (2021). [DOI] [PMC free article] [PubMed]
- 7.Jakoš T, Pišlar A, Jewett A, Kos J. Cysteine cathepsins in tumor-associated immune cells. Front. Immunol. 2019;10:2037. doi: 10.3389/fimmu.2019.02037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mijanović O, et al. Cathepsin B: A sellsword of cancer progression. Cancer Lett. 2019;449:207–214. doi: 10.1016/j.canlet.2019.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong Q, Chan SJ, Bajkowski AS, Steiner DF, Frankfater A. Characterization of the cathepsin B gene and multiple mRNAs in human tissues: Evidence for alternative splicing of cathepsin B pre-mRNA. DNA Cell Biol. 1993;12:299–309. doi: 10.1089/dna.1993.12.299. [DOI] [PubMed] [Google Scholar]
- 10.Berardi S, et al. Alternative messenger RNA splicing and enzyme forms of cathepsin B in human osteoarthritic cartilage and cultured chondrocytes. Arthritis Rheum. 2001;44:1819–1831. doi: 10.1002/1529-0131(200108)44:8<1819::AID-ART319>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 11.Müntener K, Zwicky R, Csucs G, Baici A. The alternative use of exons 2 and 3 in cathepsin B mRNA controls enzyme trafficking and triggers nuclear fragmentation in human cells. Histochem. Cell Biol. 2003;119:93–101. doi: 10.1007/s00418-002-0487-y. [DOI] [PubMed] [Google Scholar]
- 12.Baici A, Müntener K, Willimann A, Zwicky R. Regulation of human cathepsin B by alternative mRNA splicing: Homeostasis, fatal errors and cell death. Biol. Chem. 2006;387:1017–1021. doi: 10.1515/BC.2006.125. [DOI] [PubMed] [Google Scholar]
- 13.Müntener K, Zwicky R, Csucs G, Rohrer J, Baici A. Exon skipping of cathepsin B: Mitochondrial targeting of a lysosomal peptidase provokes cell death. J. Biol. Chem. 2004;279:41012–41017. doi: 10.1074/jbc.M405333200. [DOI] [PubMed] [Google Scholar]
- 14.Müntener K, et al. Folding competence of N-terminally truncated forms of human procathepsin B. J. Biol. Chem. 2005;280:11973–11980. doi: 10.1074/jbc.M413052200. [DOI] [PubMed] [Google Scholar]
- 15.Pišlar A, Jewett A, Kos J. Cysteine cathepsins: Their biological and molecular significance in cancer stem cells. Semin. Cancer Biol. 2018;53:168–177. doi: 10.1016/j.semcancer.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Schleyer KA, Cui L. Molecular probes for selective detection of cysteine cathepsins. Org. Biomol. Chem. 2021;19:6182–6205. doi: 10.1039/d1ob00225b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vidak, E., Javoršek, U., Vizovišek, M. & Turk, B. Cysteine cathepsins and their extracellular roles: Shaping the microenvironment. Cells. 8 (2019). [DOI] [PMC free article] [PubMed]
- 18.Li Z, et al. Advances in cancer treatment: A new therapeutic target, Annexin A2. J. Cancer. 2021;12:3587–3596. doi: 10.7150/jca.55173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aggarwal N, Sloane BF. Cathepsin B: Multiple roles in cancer. Proteom. Clin. Appl. 2014;8:427–437. doi: 10.1002/prca.201300105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niemeyer C, Matosin N, Kaul D, Philipsen A, Gassen NC. The role of cathepsins in memory functions and the pathophysiology of psychiatric disorders. Front. Psychiatry. 2020;11:718. doi: 10.3389/fpsyt.2020.00718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakanishi H. Microglial cathepsin B as a key driver of inflammatory brain diseases and brain aging. Neural Regen. Res. 2020;15:25–29. doi: 10.4103/1673-5374.264444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuan D, et al. Interruption of endolysosomal trafficking leads to stroke brain injury. Exp. Neurol. 2021;345:113827. doi: 10.1016/j.expneurol.2021.113827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujimoto T, et al. Cathepsin B is highly expressed in pancreatic cancer stem-like cells and is associated with patients' surgical outcomes. Oncol. Lett. 2021;21:30. doi: 10.3892/ol.2020.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma X, et al. Genetic polymorphisms of Cathepsin B are associated with gastric cancer risk and prognosis in a Chinese population. Cancer Biomark. 2021;32:189–198. doi: 10.3233/CBM-203208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munro MJ, Peng L, Wickremesekera SK, Tan ST. Colon adenocarcinoma-derived cells possessing stem cell function can be modulated using renin-angiotensin system inhibitors. PLoS ONE. 2021;16:e0256280. doi: 10.1371/journal.pone.0256280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng S, et al. CTSB knockdown inhibits proliferation and tumorigenesis in HL-60 cells. Int. J. Med. Sci. 2021;18:1484–1491. doi: 10.7150/ijms.54206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, E. K., Song, M. J., Jang, H. H. & Chung, Y. S. Clinicopathologic Analysis of Cathepsin B as a Prognostic Marker of Thyroid Cancer. Int. J. Mol. Sci. 21 (2020). [DOI] [PMC free article] [PubMed]
- 28.Tan, D. C. et al. Therapeutic targeting of cancer stem cells in human glioblastoma by manipulating the renin-angiotensin system. Cells. 8 (2019). [DOI] [PMC free article] [PubMed]
- 29.Bischof J, et al. Cancer stem cells: The potential role of autophagy, proteolysis, and cathepsins in glioblastoma stem cells. Tumour Biol. 2017;39:1010428317692227. doi: 10.1177/1010428317692227. [DOI] [PubMed] [Google Scholar]
- 30.Rempel SA, et al. Cathepsin B expression and localization in glioma progression and invasion. Cancer Res. 1994;54:6027–6031. [PubMed] [Google Scholar]
- 31.Kast RE. Glioblastoma invasion, cathepsin B, and the potential for both to be inhibited by auranofin, an old anti-rheumatoid arthritis drug. Cent. Eur. Neurosurg. 2010;71:139–142. doi: 10.1055/s-0029-1242756. [DOI] [PubMed] [Google Scholar]
- 32.Cheng YC, Ding YM, Hueng DY, Chen JY, Chen Y. Caffeine suppresses the progression of human glioblastoma via cathepsin B and MAPK signaling pathway. J. Nutr. Biochem. 2016;33:63–72. doi: 10.1016/j.jnutbio.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 33.Pramanik D, Jolly MK, Bhat R. Matrix adhesion and remodeling diversifies modes of cancer invasion across spatial scales. J. Theor. Biol. 2021;524:110. doi: 10.1016/j.jtbi.2021.110733. [DOI] [PubMed] [Google Scholar]
- 34.Huang, R. et al. Adaptive changes allow targeting of ferroptosis for glioma treatment. Cell Mol. Neurobiol. (2021). [DOI] [PMC free article] [PubMed]
- 35.Ho KH, et al. miR-140 targeting CTSB signaling suppresses the mesenchymal transition and enhances temozolomide cytotoxicity in glioblastoma multiforme. Pharmacol. Res. 2019;147:104. doi: 10.1016/j.phrs.2019.104390. [DOI] [PubMed] [Google Scholar]
- 36.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim AR, et al. Absolute quantification of tumor-infiltrating immune cells in high-grade glioma identifies prognostic and radiomics values. Cancer Immunol. Immunother. 2021;70:1995–2008. doi: 10.1007/s00262-020-02836-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Breznik B, et al. Cysteine cathepsins B, X and K expression in peri-arteriolar glioblastoma stem cell niches. J. Mol. Histol. 2018;49:481–497. doi: 10.1007/s10735-018-9787-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berquin IM, Cao L, Fong D, Sloane BF. Identification of two new exons and multiple transcription start points in the 5'-untranslated region of the human cathepsin-B-encoding gene. Gene. 1995;159:143–149. doi: 10.1016/0378-1119(95)00072-e. [DOI] [PubMed] [Google Scholar]
- 40.Berquin IM, Sloane BF. Cathepsin B expression in human tumors. Adv Exp Med Biol. 1996;389:281–294. doi: 10.1007/978-1-4613-0335-0_35. [DOI] [PubMed] [Google Scholar]
- 41.Ryan M, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 2016;44:D1018–1022. doi: 10.1093/nar/gkv1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nettesheim, A. et al. Cathepsin B localizes in the caveolae and participates in the proteolytic cascade in trabecular meshwork cells. Potential new drug target for the treatment of glaucoma. J. Clin. Med. 10 (2020). [DOI] [PMC free article] [PubMed]
- 43.Oishi M, et al. Aquaporin 1 elicits cell motility and coordinates vascular bed formation by downregulating thrombospondin type-1 domain-containing 7A in glioblastoma. Cancer Med. 2020;9:3904–3917. doi: 10.1002/cam4.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, et al. CCNB2/SASP/Cathepsin B & PGE2 axis induce cell senescence mediated malignant transformation. Int. J. Biol. Sci. 2021;17:3538–3553. doi: 10.7150/ijbs.63430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomez-Auli A, et al. The secreted inhibitor of invasive cell growth CREG1 is negatively regulated by cathepsin proteases. Cell Mol. Life Sci. 2021;78:733–755. doi: 10.1007/s00018-020-03528-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Yu C, Kang R, Kroemer G, Tang D. Cellular degradation systems in ferroptosis. Cell Death Differ. 2021;28:1135–1148. doi: 10.1038/s41418-020-00728-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakao S, Zandi S, Sun D, Hafezi-Moghadam A. Cathepsin B-mediated CD18 shedding regulates leukocyte recruitment from angiogenic vessels. FASEB J. 2018;32:143–154. doi: 10.1096/fj.201601229R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perišić Nanut M, Pečar Fonović U, Jakoš T, Kos J. The role of cysteine peptidases in hematopoietic stem cell differentiation and modulation of immune system function. Front. Immunol. 2021;12:680279. doi: 10.3389/fimmu.2021.680279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke AJ, Simon AK. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat. Rev. Immunol. 2019;19:170–183. doi: 10.1038/s41577-018-0095-2. [DOI] [PubMed] [Google Scholar]
- 50.Gonzalez-Leal IJ, et al. Cathepsin B in antigen-presenting cells controls mediators of the Th1 immune response during Leishmania major infection. PLoS Negl. Trop. Dis. 2014;8:e3194. doi: 10.1371/journal.pntd.0003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss-Sadan T, et al. Cathepsins drive anti-inflammatory activity by regulating autophagy and mitochondrial dynamics in macrophage foam cells. Cell Physiol. Biochem. 2019;53:550–572. doi: 10.33594/000000157. [DOI] [PubMed] [Google Scholar]
- 52.Wang R, et al. Increased IL-10 mRNA expression in tumor-associated macrophage correlated with late stage of lung cancer. J. Exp. Clin. Cancer Res. 2011;30:62. doi: 10.1186/1756-9966-30-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gounaris E, et al. Live imaging of cysteine-cathepsin activity reveals dynamics of focal inflammation, angiogenesis, and polyp growth. PLoS ONE. 2008;3:e2916. doi: 10.1371/journal.pone.0002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Repnik U, Starr AE, Overall CM, Turk B. Cysteine cathepsins activate ELR chemokines and inactivate non-ELR chemokines. J. Biol. Chem. 2015;290:13800–13811. doi: 10.1074/jbc.M115.638395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jakoš T, Pišlar A, Pečar Fonović U, Kos J. Lysosomal peptidases in innate immune cells: implications for cancer immunity. Cancer Immunol. Immunother. 2020;69:275–283. doi: 10.1007/s00262-019-02447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shree T, et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011;25:2465–2479. doi: 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shchors K, et al. Increased invasiveness of MMP-9-deficient tumors in two mouse models of neuroendocrine tumorigenesis. Oncogene. 2013;32:502–513. doi: 10.1038/onc.2012.60. [DOI] [PubMed] [Google Scholar]
- 58.Bruchard M, Ghiringhelli F. Impact of chemotherapies on immunosuppression and discovery of new therapeutic targets. Bull Cancer. 2014;101:605–607. doi: 10.1684/bdc.2014.1936. [DOI] [PubMed] [Google Scholar]
- 59.Daneri-Navarro A, et al. Immunosuppressive activity of proteases in cervical carcinoma. Gynecol. Oncol. 2005;98:111–117. doi: 10.1016/j.ygyno.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 60.Høglund, R. A. et al. Human cysteine cathepsins degrade immunoglobulin G in vitro in a predictable manner. Int. J. Mol. Sci. 20 (2019). [DOI] [PMC free article] [PubMed]
- 61.Moshe A, et al. Cystatin C takes part in melanoma-microglia cross-talk: possible implications for brain metastasis. Clin. Exp. Metastasis. 2018;35:369–378. doi: 10.1007/s10585-018-9891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burster T, et al. critical view of novel treatment strategies for glioblastoma: failure and success of resistance mechanisms by glioblastoma cells. Front. Cell Dev. Biol. 2021;9:695325. doi: 10.3389/fcell.2021.695325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi, J. et al. Visible-light-triggered prodrug nanoparticles combine chemotherapy and photodynamic therapy to potentiate checkpoint blockade cancer immunotherapy. ACS Nano (2021). [DOI] [PubMed]
- 64.Momeny M, et al. Blockade of vascular endothelial growth factor receptors by tivozanib has potential anti-tumour effects on human glioblastoma cells. Sci Rep. 2017;7:44075. doi: 10.1038/srep44075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X, Wang X, Xu S, Li X, Ma X. Cathepsin B contributes to radioresistance by enhancing homologous recombination in glioblastoma. Biomed. Pharmacother. 2018;107:390–396. doi: 10.1016/j.biopha.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 66.Malla RR, et al. uPAR and cathepsin B inhibition enhanced radiation-induced apoptosis in gliomainitiating cells. Neuro Oncol. 2012;14:745–760. doi: 10.1093/neuonc/nos088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cutter JL, et al. Topical application of activity-based probes for visualization of brain tumor tissue. PLoS ONE. 2012;7:e33060. doi: 10.1371/journal.pone.0033060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Custom code used to analyse the RNA-seq data and datasets generated and/or analysed in the current study are available from the corresponding authors upon request.