Abstract
Smoking-associated DNA methylation (DNAm) signatures are reproducible among studies of mostly European descent, with mixed evidence if smoking accelerates epigenetic aging and its relationship to longevity. We evaluated smoking-associated DNAm signatures in the Costa Rican Study on Longevity and Healthy Aging (CRELES), including participants from the high longevity region of Nicoya. We measured genome-wide DNAm in leukocytes, tested Epigenetic Age Acceleration (EAA) from five clocks and estimates of telomere length (DNAmTL), and examined effect modification by the high longevity region. 489 participants had a mean (SD) age of 79.4 (10.8) years, and 18% were from Nicoya. Overall, 7.6% reported currently smoking, 35% were former smokers, and 57.4% never smoked. 46 CpGs and five regions (e.g. AHRR, SCARNA6/SNORD39, SNORA20, and F2RL3) were differentially methylated for current smokers. Former smokers had increased Horvath’s EAA (1.69-years; 95% CI 0.72, 2.67), Hannum’s EAA (0.77-years; 95% CI 0.01, 1.52), GrimAge (2.34-years; 95% CI1.66, 3.02), extrinsic EAA (1.27-years; 95% CI 0.34, 2.21), intrinsic EAA (1.03-years; 95% CI 0.12, 1.94) and shorter DNAmTL (− 0.04-kb; 95% CI − 0.08, − 0.01) relative to non-smokers. There was no evidence of effect modification among residents of Nicoya. Our findings recapitulate previously reported and novel smoking-associated DNAm changes in a Latino cohort.
Subject terms: DNA methylation, Epidemiology
Introduction
Cigarette smoke causes adverse health outcomes; however, smoking cigarettes remains a common behavior around the world1. The smoke is comprised of a complex chemical mixture of over 7000 compounds, including multiple known human carcinogens2. Smoking is a major environmental risk factor for the development of respiratory and cardiovascular illnesses, including chronic obstructive pulmonary disease and coronary heart disease3,4. Smoking cigarettes has deleterious effects on multiple organs throughout the body and is both the largest preventable cause of cancer-related deaths5–7 and the leading intervenable cause of death in the United States, greater than diet and physical activity patterns combined8,9. Biological pathways implicated in the smoking-induced pathogenesis of diseases include inflammation, oxidative stress, cellular apoptosis, extracellular matrix destruction, and impaired cellular signaling10,11. Growing evidence suggests that both direct DNA damage and epigenetic mechanisms play major roles in smoking-associated diseases.
A common epigenetic modification is DNA methylation, the addition of a methyl group to a cytosine nucleotide followed by a guanine (CpG), which can regulate gene expression. Multiple epigenome-wide association studies (EWAS) conducted in blood samples have shown that adult cigarette smoking is associated with altered DNA methylation patterns of leukocytes across cohorts of mostly European descent12–18. These smoking-associated changes in DNA methylation may contribute to an increased risk for poor health outcomes among smokers. For example, studies have consistently found hypomethylation of the F2RL3 (coagulation factor II receptor-like 3) and AHRR (aryl hydrocarbon receptor repressor) genes in smokers compared to non-smokers, which, in turn, has been associated with reduced lung function and increased mortality13,19–22. Most studies and meta-analyses have tested DNA methylation changes with the 450K methylation array (Infinium HumanMethylation450 BeadChip; Illumina), and only few have used the newer and more comprehensive 850K methylation array (Infinium HumanMethylation EPIC BeadChip; Illumina), which may provide further insights23. Furthermore, the reproducibility of smoking-associated DNA methylation signatures in non-Caucasian populations is not well established.
Other important biomarkers to elucidate the effects of environmental exposures, specifically on the aging process and age-related diseases, are epigenetic clocks. Epigenetic clocks use DNA methylation levels of age-associated CpG sites to estimate an epigenetic age that can then be compared to chronological age in order to determine age acceleration, a measure of biological aging24. Among adults, increased epigenetic age acceleration is associated with factors like an unhealthy diet, lack of exercise, and lifetime stress and has been shown to help evaluate susceptibility to diseases like lung cancer25–28. Several epigenetic clocks have been developed in order to include epigenetic markers in specific or multiple tissues and improve predictive performance for specific aging measures, morbidity, or mortality. These include the Hannum blood29, Horvath Pan Tissue24, Skin-Blood30, PhenoAge31, and GrimAge32 clocks. Additionally, telomere length can be estimated from DNA methylation (DNAmTL), which more closely correlates with chronological age relative to measured telomere length33. The GrimAge clock is particularly successful at predicting mortality associated with factors like smoking and obesity32. Unlike the other clocks, GrimAge was trained on smoking pack-years and includes chronological age in its input. Cigarette smoking has recently been shown to be associated with increased age acceleration in respiratory tissues, which may be reversed by smoking cessation34.
Overall, there is a lack of EWAS data and epigenetic age acceleration studies on smoking conducted with diverse populations, including Latinos, in which three of the top five populations-specific causes of death—cancer, stroke, and heart disease—are all associated with smoking35. Studying the epigenetic effects of smoking in a Latino population could contribute to characterizing health disparities this group experiences36,37 as well as assess the generalizability of findings from other studies, as epigenetic analyses on the effects of smoking have been found to differ by ethnic groups14. In this study, we investigated DNA methylation patterns of current and former smokers compared to non-smokers in people living in Costa Rica in order to address the lack of studies in Latino populations. The study population includes participants from the Nicoya peninsula: a “Blue Zone” characterized by exceptionally high longevity compared to the rest of Costa Rica and the world38,39. We also use multiple epigenetic aging biomarkers to understand how smoking may impact biological aging of participants. We hypothesized that most previously identified smoking signatures would be generalizable to this Latino cohort and that study participants from the high longevity region would exhibit epigenomic resiliency to smoking-associated epigenetic changes and epigenetic age acceleration.
Methods
Data collection and sample preparation
The study participants were selected from The Costa Rican Study on Longevity and Healthy Aging (CRELES) cohort; the study protocol has been previously described40–42. Briefly, CRELES is a prospective longitudinal study of a nationally representative sample of 2827 residents of Costa Rica who were age 60 years and older at baseline in 2004–2006, with a second wave of interviews and data collection in 2006–2008. Information from a CRELES-complementary sample of Nicoyan quasi-centenarians (age 95 and above) were also collected. All data, examinations, and specimens were taken in the participants’ homes, and details about sample, field, and laboratory procedures have been previously reported40,41. The Ethical Science Committee of the University of Costa Rica granted human subjects approval to CRELES (VI-763-CEC-23-04). All participants granted written informed consent by means of their signature and the study was conducted according to the guidelines laid down in the Declaration of Helsinki.
We randomly selected 512 individual samples from both wave 1 and wave 2 blood samples for DNA methylation (DNAm) analysis. We ascertained smoking behavior by interviews and classified participants as current smokers if they reported smoking > 100 cigarettes in their lifetime as well as currently smoking at the study visit. Former smokers reported smoking > 100 cigarettes in their lifetime but not currently smoking at the study visit, while non-smokers reported not smoking over > 100 cigarettes in their lifetime and currently not smoking. We also investigated ever smoking (> 100 cigarettes in their lifetime) vs. non-smokers for comparability with previous studies.
DNA methylation measurements
Whole blood samples were collected via venipuncture and processed at the University of Costa Rica, as previously described43. Genomic DNA was extracted from 2 mL of frozen whole blood using the phenolchloroform method. DNA was bisulfite converted with the Zymo Research EZ DNA Methylation™ Kit (Irvine, CA, USA). Bisulfite-converted DNA from each sample was randomized across Infinium MethylationEPIC BeadChips as well as sentrix row and run in one batch according to the manufacturer’s protocol (San Diego, CA, USA)42.
We processed raw DNA methylation image files using the R statistical software (www.r-project.org/) and several Bioconductor packages44 including the minfi pipeline for quality control45. All samples had median methylated and unmethylated log-intensities above a threshold considered to be of good quality (> 10.5). We used functional normalization with 3 principal components capturing > 90% of the variation in the control probes to normalize samples. We chose this normalization method given that participants came from two geographic regions in Costa Rica, and we expected genetic ancestry differences that could influence DNA methylation by region. A total of 512 samples were analyzed, and 12 samples were identified as outliers based on principal component analyses29 or by having ≥ 5% of CpGs with non-significant detection (P > 1 × 10–16). We also removed 3 samples mismatched on recorded sex and 8 technical replicates, leaving a total of 489 samples. Data from the 489 participants from waves 1 (n = 274) and 2 (n = 215) were included after quality control. We removed 59 SNP probes, probes with < 3 beads or with a non-significant detection (P > 1 × 10–16) in ≥ 1% of samples, removing a total of 22,261 non-reliable probes. We further removed 18,474 probes in XY chromosomes and 25,395 polymorphic probes containing a SNP at the CpG site or single base extension with a minor allele frequency > 1%. We removed 9671 autosomal probes that cross-hybridize to sex chromosomes46. A total of 790,058 high quality CpGs were used in statistical analyses. Finally, we corrected for sample plate, row (position within the array), and chip using ComBat47. We visualized the density distributions for samples at all processing steps and performed principal components (PC) analyses to examine the associations of methylation differences with technical, biological, and measured traits with global DNA methylation variation. For each CpG site, methylation is reported as the average β-value, corresponding to an interval scaled quantity between zero and one interpreted as the fraction of DNA molecules whose target CpG is methylated. All results are presented on the β-value scale multiplied by 100 to ease interpretability as percent change in DNA methylation. To estimate leukocyte composition as proportions, we used a reference panel of isolated leukocytes with the IDOL projection and Houseman method48 to estimate cell-type proportions (CD8 + T cells, CD4 + T cells, NK, B-cells, monocytes, and neutrophils)49.
Genotyping of samples
Genotyping data was measured at 618,540 single nucleotide polymorphism (SNP) sites using the Infinium Global Screening Array (GSA) BeadChips according to the Illumina’s standard protocol (Illumina). GenomeStudio 2.0 Genotyping software was used to transform the raw intensity files into clusters, and subsequently genotype calls by producing cohort-specific clustering files and manifest GSA-24v1-0_C1 (Version 1 A2, Illumina). We applied standard quality control procedures to the array, and SNPs with a MAF ≤ 5% were removed prior to performing PCA using pca (PCAtools). Horn’s analysis was used to determine how many PCs to retain (n = 2) using the paran function (MASS)50. Participants were ascribed the rotated PC1 and PC2 loadings to represent and control for genetic ancestry differences in subsequent analyses. After quality control, a total of 465 participants had complete DNA methylation and genetic ancestry data. EWAS of smoking adjusting for genotype PCs was restricted to the 465 participants with high quality DNA methylation and genotyping data.
Calculation of epigenetic aging biomarkers
Epigenetic age was calculated using five clocks: the Horvath Pan Tissue, Horvath Skin-Blood, Hannum Blood, PhenoAge, and GrimAge clocks. We calculated all epigenetic aging biomarkers utilizing the online Horvath calculator (http://dnamage.genetics.ucla.edu/) with the advanced analysis option. The outcome of interest was the “AgeAccelerationResidual” or residuals resulting from a linear regression model where each DNA methylation clock is regressed on chronological age of each participant. We refer to all acceleration measures as Epigenetic Age Acceleration (EAA) for the specific clock and defined the residuals of the DNA methylation estimate of telomere length linearly regressed on chronological age as DNAmTL adjusted for age. A positive EAA indicates that the estimated epigenetic age is higher than the chronological age (increased biological aging) and a negative DNAmTL adjusted for age reflects a shorter telomere length. In addition, we tested Extrinsic EAA (EEAA) and Intrinsic EAA (IEAA) for Hannum’s and Horvath’s clocks, respectively51. The EEAA measure is associated with age-related changes in blood cell counts due to immune system aging and is calculated by upweighting the contributions of age-associated blood cell counts (naive cytotoxic T cells, exhausted cytotoxic T cells, and plasmablasts). The IEAA measure is independent of blood cell counts, represents intrinsic cellular aging, and is calculated by adding immune cell counts in addition to chronological age when calculating regression residuals. Analyses of epigenetic clocks included the 489 participants with high quality DNA methylation data.
Statistical analysis
We described our study sample using means and proportions for the variables analyzed and evaluated accuracy of all epigenetic aging biomarkers via their empirical correlation with chronological age as well as scatterplots. In a linear EWAS model, we compared current and former smokers to non-smokers via limma52, with each individual CpG on the beta value scale while adjusting for sex, chronological age, BMI, education, household assets, the first two principal components from genetic data, and estimated cell-type composition. We report statistically significant results adjusted for multiple comparisons using both a Bonferroni correction of α = 0.05/790,058, or P < 6.33 × 10–8 and by controlling the False Discovery Rate at 5% (FDR < 0.05). Additionally, we tested for Differentially Methylated Regions (DMRs) using DMRcate53 for the comparison of current to non-smokers as well as current to former smokers while adjusting for the same covariates as individual linear models. To test for effect modification by region, we fitted a linear model that incorporated interactions between smoking status (current, former, and non-smokers) and residence in the Nicoya Peninsula as binary. We evaluated EWAS model fit by visualizing quantile–quantile plots of the observed vs. expected P-values and estimated the genomic inflation factor (λ). We used missMethyl54 to test for enrichment of differentially methylated sites across KEGG biological pathways. We summarized results using Manhattan and volcano plots of EWAS as well as a circular genomic plot. To test for EAA, we used linear regression to estimate mean differences between smoking groups across acceleration measures of epigenetic aging as the outcome. Namely, we tested the residuals of regressing each epigenetic clock on chronological age against self-reported smoking behavior (current, former and non-smokers). We also tested associations between ever smoking vs. never smoking for comparability with other studies of EAA. Effect modification was evaluated by fitting a multiplicative term between smoking behavior and Nicoya residency in linear regression models of EAA. We report estimates and 95% confidence intervals (95% CI) as well as unadjusted P-values for EAA analyses.
Results
A total of 489 CRELES study participants had complete data and DNA methylation measurements (Table 1); they had a mean (SD) age of 79.4 years (10.8 years), and 90 (18.4%) lived in the high longevity region of the Nicoya Peninsula. Of all participants, 278 were female (57%), 37 (7.6%) reported currently smoking, 171 (35%) were former smokers, and 281 (57.4%) reported never smoking. The majority of participants only had an elementary school education (69%), and 97 participants (20%) had no formal education. To control for population stratification, we adjusted EWAS models for two principal components from genome-wide SNP arrays, limiting these analyses to 465 study participants. These two principal components significantly differed between the regions (P < 1 × 10–7), with region of residence explaining 28% and 8% of the variance for the first and second genetic principal components, respectively.
Table 1.
Characteristics of participants from the Costa Rican Study on Longevity and Healthy Aging (CRELES) included in this study, overall and stratified by residents from the Nicoya Peninsula (Nicoyans) or non-residents (non-Nicoyans).
| Participant characteristics | Overall (N = 489) | Non-Nicoyans (N = 399) | Nicoyans (N = 90) |
|---|---|---|---|
| Sex | |||
| Male | 211 (43%) | 165 (41%) | 46 (51%) |
| Female | 278 (57%) | 234 (59%) | 44 (49%) |
| Level of education | |||
| None | 97 (20%) | 78 (20%) | 19 (21%) |
| Elementary (1–6 years) | 338 (69%) | 270 (68%) | 68 (76%) |
| Secondary (7–11 years) | 27 (6%) | 27 (7%) | – |
| Post-Secondary (> = 12 years) | 27 (6%) | 24 (6%) | 3 (3%) |
| Household assets (0–10) | 7.99 (1.76) | 8.13 (1.69) | 7.36 (1.93) |
| BMI (kg/m^2) | 25.2 (5.5) | 25.7 (5.6) | 23.4 (4.7) |
| Age, (years) | 79.4 (10.8) | 78 (10.4) | 85.3 (10.9) |
| Smoking status | |||
| Current | 37 (7.6%) | 28 (7%) | 9 (10%) |
| Former | 171 (35%) | 138 (34.6%) | 33 (36.7%) |
| Never | 281 (57.4%) | 233 (58.4%) | 48 (53.3%) |
| Genetic principal components (PCs)* | |||
| PC1 | − 0.55 (32.69) | − 8.51 (27.17) | 37.11 (31.23) |
| PC2 | 0.11 (22.32) | 3.11 (20.51) | − 12.08 (23.06) |
| Missing | – | 15 (3.8%) | 9 (10%) |
| Estimated leukocyte composition (%) | |||
| Neutrophils | 54.5 (10.6) | 54.7 (10.3) | 53.9 (12) |
| NK | 8.1 (3.3) | 8 (3.2) | 8.6 (3.3) |
| CD4 + -T | 14.1 (5.2) | 14.1 (5.3) | 14.2 (5.1) |
| CD8 + -T | 10.9 (5.0) | 10.9 (5) | 10.6 (4.9) |
| Monocytes | 8.3 (2.4) | 8.2 (2.2) | 8.7 (2.9) |
| B-cells | 5.3 (2.5) | 5.3 (2.5) | 5.4 (2.5) |
Statistics presented are n (%) or mean (SD).
*Missing genetic data for 24 study participants.
Epigenome-wide association analyses (EWAS) of current smoking
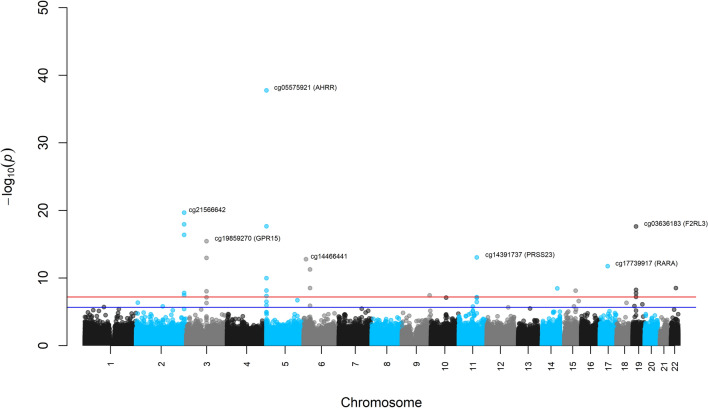
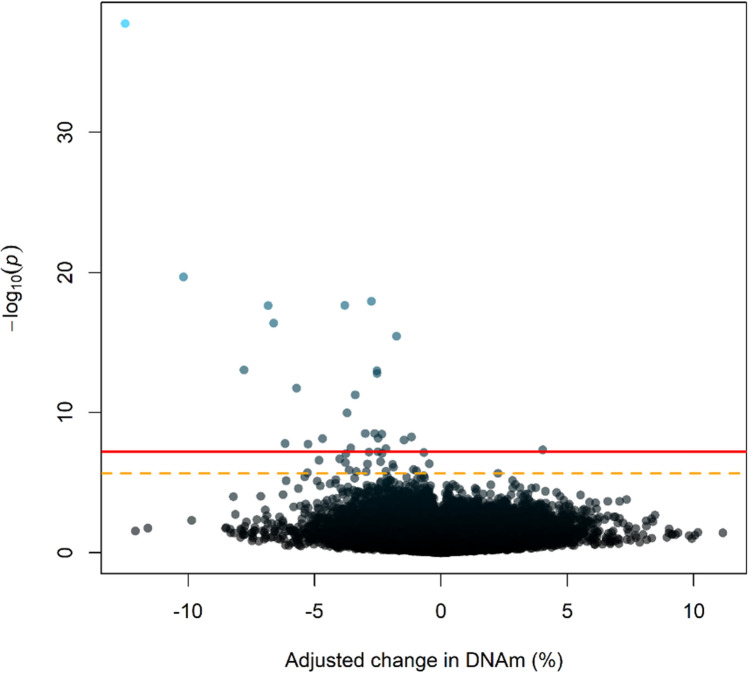
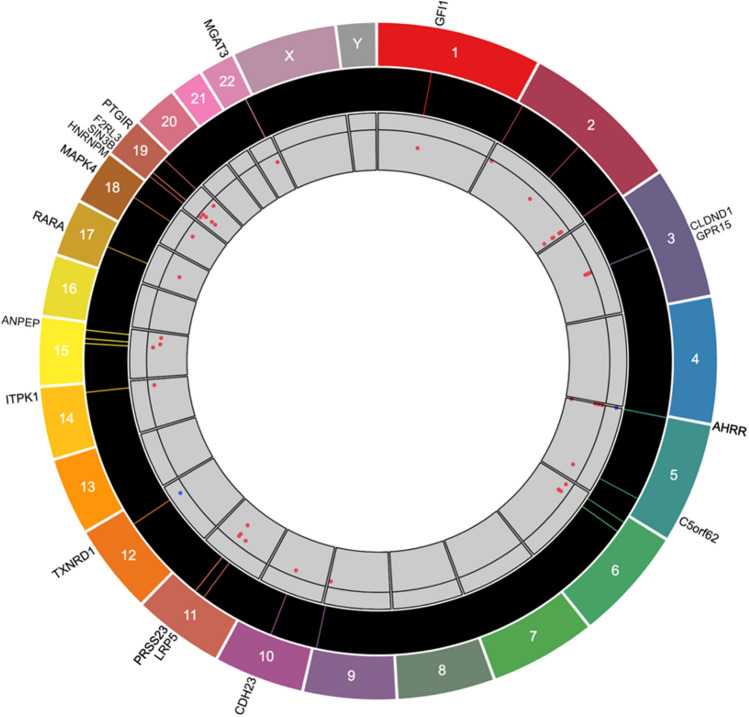
In EWAS adjusted for sex, age, BMI, education, household assets, two principal components of genotypes, and estimated leukocyte composition, a total of 46 CpGs were differentially methylated when comparing current smokers to non-smokers, while adjusting for former smokers. Among the 46 CpGs with a FDR < 0.05, 26 CpGs were statistically significant using a Bonferroni adjusted threshold of P < 6.33 × 10–8 (α = 0.5/790,058). A Manhattan plot, volcano plot, and circular genomic plot of EWAS results for current vs. non-smokers displaying genomic position, effect sizes and gene annotations are shown in Figs. 1, 2 and 3. Multiple of the top 30 differentially methylated CpGs from this analysis annotated to the same genes (AHRR, PRSS23, SIN3B, and F2RL3) and a region in chromosome 2 (chr2: 233,283,010–233,286,229), and almost all were hypomethylated, as shown in Table 2. Among all 46 differentially methylated CpGs found with a FDR < 0.05, 44 (96%) were hypomethylated (Table S1 in Supplementary File 1). The CpGs with the greatest increase (cg07943658; 4.0%) and decrease (cg05575921; − 12.49%) in methylation among current smokers were both annotated to the AHRR gene. A comparison of the CpGs found in our study across many others is available in Table S2 in Supplementary File 1. Five CpGs (cg04706667, cg06991517, cg13511253, cg18234441, and cg22812571) have not been previously reported to be differentially methylated in other smoking-related studies, including large meta-analyses (Table S2 in Supplementary File 1). Four of these novel CpGs were hypomethylated and annotated to the mitogen-activated protein kinase 4 (MAPK4), the heterogeneous nuclear ribonucleoprotein M (HNRNPM), and the prostaglandin I2 receptor (PTGIR) genes. The remaining CpG was hypermethylated and annotated to the thioredoxin reductase 1 (TXNRD1) gene. The directionality of methylation—increased or decreased—for the remaining 41 sites is in alignment with prior findings in the literature, such as the large meta-analysis of Joehanes et al.55 and a recent analysis of samples evaluated with the EPIC array from Domingo-Relloso et al. (2020) study55,56. The genomic inflation factor (λ = 1.05) and quantile–quantile plot of observed vs. expected P-value distribution show no major concerns for the analyses (Supplementary Figure S1).
Figure 1.
Manhattan plot of epigenome-wide associations among current smokers relative to non-smokers (red line represents a Bonferroni corrected level of significance; blue line represents a false discovery rate of 5%).
Figure 2.
Volcano plot of the -log10(p-values) against adjusted regression coefficients for each CpG for the Epigenome-Wide Association Study comparing current smokers to non-smokers (red solid line represents a Bonferroni corrected level of significance; orange dotted line represents a false discovery rate of 5%).
Figure 3.
Circular genomics plot for current vs. never smokers. From the outermost ring to the center: gene names, chromosome numbers, locations of CpG sites within each chromosome, and effect sizes of DNA methylation (blue points above the horizontal line of 0 represent hypermethylation; red points below the line represent hypomethylation).
Table 2.
Top 30 differentially methylated CpGs for current smokers relative to non-smokers, ranked by smallest P-value and sorted by chromosome and position.
| CpG | CHR | Position | UCSC Gene Name | Adjusted %-DNA methylation differencea | FDRP-value | Relation to Island | Gene region |
|---|---|---|---|---|---|---|---|
| cg17087741 | 2 | 233283010 | − 2.75 | 2.92E−13 | N_Shore | ||
| cg03329539 | 2 | 233283329 | − 3.56 | 1.15E−03 | N_Shore | ||
| cg21566642 | 2 | 233284661 | − 10.18 | 8.24E−15 | Island | ||
| cg01940273 | 2 | 233284934 | − 6.61 | 5.29E−12 | Island | ||
| cg22812571 | 2 | 233286229 | − 6.17 | 6.05E−04 | S_Shore | ||
| cg08064403 | 3 | 98240258 | CLDND1 | − 1.46 | 3.58E−04 | N_Shore | Body |
| cg19859270 | 3 | 98251294 | GPR15 | − 1.76 | 3.89E−11 | OpenSea | 1st Exon |
| cg04180924 | 3 | 98272064 | − 0.67 | 1.96E−03 | OpenSea | ||
| cg02978227 | 3 | 98292027 | − 2.54 | 8.86E−09 | OpenSea | ||
| cg07943658 | 5 | 352001 | AHRR | 4.03 | 1.43E−03 | OpenSea | Body |
| cg05575921 | 5 | 373378 | AHRR | − 12.49 | 1.35E−32 | N_Shore | Body |
| cg04551776 | 5 | 393366 | AHRR | − 2.48 | 2.94E−04 | OpenSea | Body |
| cg25648203 | 5 | 395444 | AHRR | − 3.81 | 3.54E−13 | OpenSea | Body |
| cg21161138 | 5 | 399360 | AHRR | − 3.71 | 6.23E−06 | OpenSea | Body |
| cg14466441 | 6 | 11392193 | − 2.52 | 1.26E−08 | OpenSea | ||
| cg24859433 | 6 | 30720203 | − 3.39 | 3.54E−07 | OpenSea | ||
| cg15342087 | 6 | 30720209 | − 2.62 | 1.63E−04 | OpenSea | ||
| cg26768182 | 9 | 134272679 | − 2.18 | 1.21E−03 | S_Shelf | ||
| cg10750182 | 10 | 73497514 | C10orf105 | − 2.33 | 2.16E−03 | OpenSea | 5' UTR |
| cg11660018 | 11 | 86510915 | PRSS23 | − 3.76 | 2.22E−03 | N_Shore | TSS1500 |
| cg14391737 | 11 | 86513429 | PRSS23 | − 7.79 | 8.86E−09 | S_Shore | 5' UTR |
| cg00475490 | 11 | 86517110 | PRSS23 | − 2.85 | 1.91E−03 | OpenSea | 5' UTR |
| cg05284742 | 14 | 93552128 | ITPK1 | − 2.34 | 1.65E−04 | OpenSea | Body |
| cg18110140 | 15 | 75350380 | − 4.69 | 2.98E−04 | OpenSea | ||
| cg17739917 | 17 | 38477572 | RARA | − 5.71 | 1.26E−07 | S_Shelf | 5' UTR |
| cg03384915 | 19 | 16986822 | SIN3B | − 1.16 | 2.50E−04 | Island | Body |
| cg14712058 | 19 | 16988083 | SIN3B | − 2.49 | 1.86E−03 | N_Shore | Body |
| cg21911711 | 19 | 16998668 | F2RL3 | − 5.26 | 6.36E−04 | N_Shore | TSS1500 |
| cg03636183 | 19 | 17000585 | F2RL3 | − 6.84 | 3.54E−13 | N_Shore | Body |
| cg05086879 | 22 | 39861490 | MGAT3 | − 3 | 1.63E−04 | OpenSea | 5'UTR |
aLinear regression models adjusted for sex, age, BMI, education, household assets, two principal components from genetic data, and estimated cell-type composition (CD8 + -T, CD4 + -T, NK, B-cell, monocytes, and neutrophils).
EWAS of former smoking
For the adjusted EWAS comparing former to non-smokers while adjusting for current smoking, only one CpG site (cg05575921; AHRR) was found to have a significant association with smoking, with a smaller magnitude of effect (− 2.93%). The quantile–quantile P-value and Manhattan plots for the EWAS of former smokers relative to non-smokers are shown in Supplementary Figures S2 and S3, respectively.
DMRs among current smokers and modification of associations by longevity region
Testing for DMRs yielded five DMRs hypomethylated among current smokers relative to non-smokers. Two DMRs annotated to the AHRR gene and a single DMR was observed for the F2RL3, the Small Nucleolar RNA, H/ACA Box 20 (SNORA20), and Small Cajal Body-Specific RNA 6 (SCARNA6)/Small nucleolar RNA (SNORD55/SNORD39) genes. Results of DMR analyses are shown in Table 3. No DMRs were found for former smokers relative to non-smokers. Among the differentially methylated CpGs for current smokers relative to non-smokers, three KEGG biological pathways were marginally enriched or overrepresented (Punadjusted < 0.05), with more than one gene differentially methylated: hsa05200 or “Pathways in cancer”, hsa04611 “Platelet activation,” and hsa04080 “Neuroactive ligand-receptor interaction.” However, these results did not survive multiple testing adjustments.
Table 3.
Differentially methylated regions (DMRs) associated with current smoking status relative to non-smokers.
| Chromosome | Start | End | Width | CpGs | Stouffer P-value | Mean %-DNAm difference at DMR | Overlapping genes |
|---|---|---|---|---|---|---|---|
| chr2 | 233283010 | 233286229 | 3220 | 12 | 3.23 × 10–13 | − 3.29 | SCARNA6, SNORD39 |
| chr5 | 373378 | 374093 | 716 | 3 | 5.38 × 10–7 | − 4.34 | AHRR |
| chr5 | 395444 | 395717 | 274 | 2 | 9.30 × 10–5 | − 2.08 | AHRR |
| chr6 | 30719807 | 30720484 | 678 | 6 | 0.017 | − 1.64 | SNORA20 |
| chr19 | 17000585 | 17001002 | 418 | 3 | 0.022 | − 2.29 | F2RL3 |
In EWAS analyses with interactions between smoking (current, former, and never) and longevity region residency, there was no statistical evidence that smoking related DNAm signatures differed between Nicoyan and non-Nicoyan smokers after adjusting for multiple testing (Supplementary Figure S4).
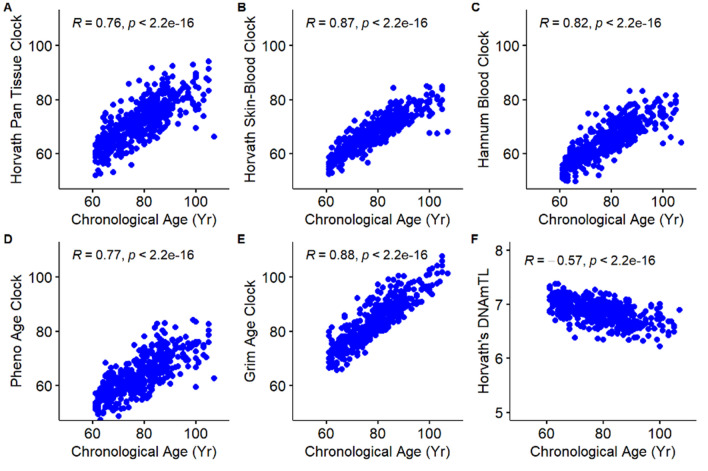
Correlation plots comparing each study participant’s epigenetic age, as determined by five different epigenetic clocks, DNAmTL biomarker, and chronological age are displayed in Fig. 4. Overall, there were strong, positive, and significant correlations between chronological age and epigenetic age for the Horvath Pan Tissue (r = 0.76), Skin-Blood (r = 0.87), Hannum Blood (r = 0.82), PhenoAge (r = 0.77), and the GrimAge (r = 0.88) clocks. Of note, the GrimAge clock includes chronological age as an input. A moderate negative correlation was observed between DNAmTL estimates and chronological age (r = − 0.57). As expected, the EAA measures were uncorrelated with chronological ages.
Figure 4.
Scatterplots and correlation coefficients of DNA methylation aging biomarkers (epigenetic clocks) and chronological age in years: (A) Horvath’s Clock, (B) Horvath’s Skin-Blood Clock, (C) Hannum’s Clock, (D) PhenoAge Clock, (E) GrimAge Clock, and (F) Horvath’s DNAm telomere length (DNAmTL).
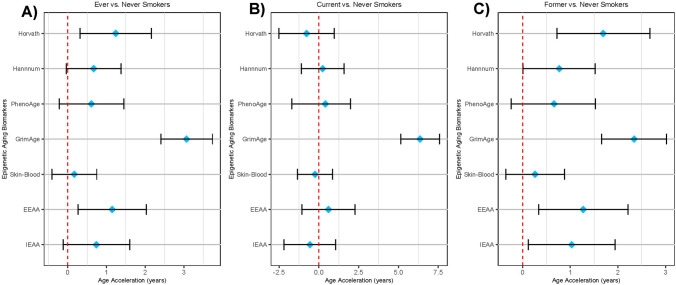
Epigenetic age acceleration for ever versus never smokers
To compare our data to previous reports, we first tested differences in epigenetic age acceleration when comparing ever smokers, that included current and former smokers, to never-smokers (Fig. 5A); the data are also displayed in Supplementary Table S3. The highest epigenetic age acceleration among ever smokers was found for the GrimAge clock (3.07 years, 95% CI 2.41, 3.74), which incorporates DNA methylation estimated smoking pack-years in its calculation, so this was expected. All clocks showed the same trend of increased age acceleration among smokers, with the Horvath Clock and Extrinsic EAA measure demonstrating significantly increased EAA: 1.24 years (95% CI 0.32, 2.16) and 1.15 years (95% CI 0.27, 2.03), respectively. Additionally, smokers had on average 0.04 kb shorter (95% CI − 0.07, − 0.01) DNA methylation estimates of telomere length residuals after adjusting for chronological age. Similar results were observed when stratifying models with participants from the Nicoya region, with the exception that PhenoAge was significantly accelerated among ever smokers compared to non-smokers from the Nicoya Peninsula (2.11 years; 95% CI 0.14, 4.08) but not in the other region (P = 0.44). However, no effect modification of smoking on epigenetic aging was observed for the longevity region in multiplicative interaction models (P > 0.05).
Figure 5.
Estimated differences in years of epigenetic age acceleration when comparing: (A) ever smokers to never smokers, (B) current smokers to never smokers, and (C) former smokers to never smokers. Diamonds represent point estimates, error bars represent 95% confidence intervals, and red dashed line represent the null hypothesis. The telomere length biomarker (DNAmTL) was excluded due to scaling.
Epigenetic age acceleration for current and former smokers
Furthermore, we tested differences between current and former smokers relative to non-smokers across all age acceleration measures (Fig. 5B,C) and stratified by Nicoya residence (Table S4). Epigenetic age acceleration estimates among current smokers relative to non-smokers were not statistically significant, inconsistent in directionality, and relatively weak in strength, except for GrimAge, which was expected due to the incorporation of pack-years in its calculation. For former smokers, acceleration measures for the Horvath Pan Tissue, Hannum Blood, EEAA, and IEAA measures were positive and statistically significant: 1.69 years (95% CI 0.72, 2.67), 0.77 years (95% CI 0.01, 1.52), 1.27 years (95% CI 0.34, 2.21), and 1.03 (95% CI 0.12, 1.94), respectively. Overall, the greatest age acceleration was again observed for the GrimAge clock: 6.36 years (95% CI 5.14, 7.58) for current smokers and 2.34 years (95% CI 1.66, 3.02) for former smokers. After regressing age out from DNAmTL, a 0.04 kb lower (95% CI − 0.08, − 0.01) estimate was observed among former smokers compared to current smokers. No significant effect modification was observed for the relationship between smoking and epigenetic aging markers by longevity region residency (P > 0.05).
Overall, stratified results were consistent between Nicoyans and non-Nicoyans despite the sample size of the former group. However, the EAA for the PhenoAge clock was significantly accelerated, 2.28 years (95% CI 0.11, 4.44) among former smokers in Nicoya compared to 0.39 years (95% CI − 0.58, 1.36) for former smokers not from Nicoya. Some prior studies have found negative and/or insignificant differences in age acceleration when using whole blood samples from former, current, and never smokers, as summarized in Supplementary Table S5.
Discussion
In this study of a Latino adult population living in Costa Rica, including residents from the high longevity region of the Nicoya Peninsula, we investigated associations between current, former, and never smoking status with DNA methylation signatures and epigenetic age acceleration. Our findings replicated previously reported associations within the AHRR, PRSS23, SIN3B, and F2RL3 genes and found 5 novel signatures, which annotated to the MAPK4, HNRNPM, PTGIR, and TXNRD1 genes. Lastly, our results provided strong support that former smokers have accelerated epigenetic aging for Horvath’s and Hannum’s epigenetic clocks as well as extrinsic and intrinsic measures of aging. Consistently, former smokers had shorter DNA methylation estimates of telomere length adjusted for age. In addition, we did not observe significant epigenetic age acceleration among current smokers, except for GrimAge, which could be due to small sample size and suggest importance of differentiating current and past smoking habits to test associations.
In the EWAS comparing current smokers to non-smokers, we found 41 CpG sites that were replicated in previous studies as well as five novel CpGs. The directionality of methylation for the overlapping CpG sites aligns with findings from previous studies55–61. The majority of the significant CpG sites in this study overlapped with findings from a study on cigarette smoking among American Indian adults56, and fewer overlapped with findings from a study of African American women58, both using the EPIC array. The five novel sites that we found have not been reported in other studies, even those that similarly used the 850K EPIC Illumina BeadChip and included participants from a racial minority population56,58,62. Four of these sites were hypomethylated and annotated to the MAPK4, HNRNPM, and PTGIR genes. MAPK4 is an atypical kinase involved with the AKT/mTOR signaling pathway, and overexpression is associated with acute lung injury and cancers63,64. Similarly, the HNRNPM and PTGIR proteins have been shown to promote cancerous cell growth and be associated with poorer oncogenic outcomes65,66. The latter is also involved with vascular remodeling, and its loss of function may increase risk for vessel stenosis and dissection67. If MAPK4 (TSS1500), HNRNPM, and PTGIR hypomethylation among smokers—what we observed—leads to greater protein expression, this could partially explain their increased risk for lung and heart diseases and several cancers. The remaining novel CpG site was hypermethylated and annotated to the TXNRD1 gene. The TXNRD1 protein is involved in protecting cells from reactive oxygen species and also promotes tumor growth and DNA replication68. Further study of changes to gene expression arising from epigenetic modifications among smokers can elucidate how environmental tobacco exposure leads to the development of diseases.
Our analysis of epigenetic age acceleration demonstrated that for several biological aging biomarkers, ever smokers experience accelerated aging compared to non-smokers. The varied age acceleration results may be because some clocks are better at capturing adverse health impacts from specific environmental stimuli than other clocks. For example, the largest age acceleration associated with smoking was found for the GrimAge clock, which is in alignment with previous studies demonstrating the clock’s success at predicting mortality associated with smoking exposure32,69. We expected this result, as pack-years is used in the estimation of GrimAge years32. In all other clocks, age acceleration was positive and statistically significant for former smokers but not current smokers. This might be explained by delayed effects of smoking on the development of negative health outcomes or active compensation in the epigenome of current smokers for the toxic exposure of cigarette smoke, which could explain the mostly null age accelerations. Alternately, current smokers that made it into the study at the older ages of recruitment might be uniquely unaffected by smoking. For example, former smokers might have ceased to smoke due to declining health while current smokers continued as they were unaffected. This might be partially supported by genetic findings showing that long-lived smokers carry variants that may confer protection70. This hypothesis warrants further testing in the context of epigenetic clocks. Interestingly, the opposite was true in site-by-site analyses for EWAS, where we observed strong associations among current smokers compared to former smokers.
Overall, there are inconsistent results across existing studies that have used a variety of epigenetic clocks on whole blood samples to assess the effects of smoking status on epigenetic age acceleration. Many studies that found null age acceleration results comparing smokers to non-smokers did not stratify smokers into current and former smokers25,71,72. This may lead to results that misrepresent the epigenetic effects of cigarette smoke exposure. Also, inconsistencies in those studies and this one may be due to participants’ duration and intensity of smoking as well as time since smoking cessation, which are individual-level factors that can affect epigenetic age acceleration outcomes73–75. Among ever smokers, some studies found positive results that are not statistically significant or slightly negative epigenetic age acceleration25,71,73,76,77. We also observed non-significant epigenetic age deacceleration among current smokers for Horvath’s Clock, the Skin-Blood Clock, and the IEAA measure. In contrast to our study, previous analyses of smoking have found no associations with IEAA25,28. Our analysis builds on previous evidence by including a comprehensive set of epigenetic age acceleration outcomes to assess the consistency of results across different epigenetic clocks. While we did not find evidence of effect modification by the high longevity region, future research should evaluate associations between epigenetic aging and plant-based diets, consistent physical activity, and sociocultural connectedness—factors found to be increased in Blue Zones78. For example, residents from Nicoya report higher levels of physical activity and greater intake of fruits and vegetables, black beans, corn tortillas and rice79.
This study has some limitations. Due to the study design, we were unable to determine temporality and results might be influenced by recall bias. Also, the associations that we found could be explained by a common factor that was unaccounted for, as residual confounding is a common source of bias in observational studies. However, given the replication of previous smoking signatures, we think this is less likely. To mitigate the chance that this may occur, we controlled for sociodemographic characteristics, genetic principal components, and estimated cell-type composition in EWAS models. Another limitation is that only 90 of the study participants lived in the Nicoya region and only 9 were current smokers, which reduces the statistical power of the study to detect differences in associations among smokers and non-smokers living inside and outside the region. The sample from Nicoya was also older, which might introduce survivor bias regarding the smokers included in this study. Importantly, cigarette smoking data was self-reported by participants, which may introduce bias in the exposure assessment, but we expect this to be non-differential relative to DNA methylation or epigenetic aging measures.
In this EWAS of smoking conducted with a Latino cohort, we found five novel differentially methylated CpG sites among smokers. It also replicated several DNA methylation signatures of current smoking found in previous studies, such as hypomethylation of CpG sites annotated to the AHRR, F2RL3, SIN3B, and PRSS23 genes. In our study, former smokers exhibited consistent increased epigenetic age acceleration for several epigenetic clocks. Future studies with diverse populations and transcriptome analysis would assist in determining how environmental factors increase health risks among smokers and affect health disparities. Importantly, addressing factors that might promote resilient epigenomes even in the presence of harmful exposure can help optimize public health interventions.
Supplementary Information
Acknowledgements
We are grateful to the participants of the study for providing valuable information and biological specimens without payment or compensation. We are also grateful to the staff and fieldworkers at the Universidad de Costa Rica, Centro Centroamericano de Población, and Instituto de Investigaciones en Salud who made the CRELES study possible.
Author contributions
A.C., S.E., N.G., K.H. and H.R.E. contributed to the data analyses and processing of the data. W.H.D. and L.R.B. conceived the initial CRELES Study. R.P.F., A.O., L.M.W. and M.S.K. were major contributors to the analysis, data interpretation, and writing of the manuscript. All authors read and approved of the final manuscript.
Funding
This work was supported by the UC Berkeley Center on the Economics and Demography of Aging and the United States National Institutes of Health grants 2P30AG012839, R01ES031259 (AC) and R03AG067064 (AC) and R21MD013296 (DHR), R01MD011721 (DHR).
Data availability
Public-use version of the CRELES data is available from the Inter-University Consortium for Political and Social Research (ICPSR) repository (http://doi.org/10.3886/ICPSR31263.v1). Since data DNA methylation and the complementary sample of centenarians in Nicoya are not currently part of the public-use, requests for restricted access to data can be submitted at http://www.creles.berkeley.edu/ following institutional review approval.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-08160-w.
References
- 1.Rodgman A, Smith CJ, Perfetti TA. The composition of cigarette smoke: A retrospective, with emphasis on polycyclic components. Hum. Exp. Toxicol. 2016 doi: 10.1191/096032700701546514. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (US), National Center for Chronic Disease Prevention and Health Promotion (US), & Office on Smoking and Health (US). How Tobacco Smoke Causes Disease: The Biology and Behavioral Basis for Smoking-Attributable Disease: A Report of the Surgeon General. (Centers for Disease Control and Prevention (US), 2010). [PubMed]
- 3.Morris PB, et al. Cardiovascular effects of exposure to cigarette smoke and electronic cigarettes: Clinical perspectives from the prevention of cardiovascular disease section leadership council and early career councils of the American college of cardiology. J. Am. Coll. Cardiol. 2015;66:1378–1391. doi: 10.1016/j.jacc.2015.07.037. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida T, Tuder RM. Pathobiology of cigarette smoke-induced chronic obstructive pulmonary disease. Physiol. Rev. 2007;87:1047–1082. doi: 10.1152/physrev.00048.2006. [DOI] [PubMed] [Google Scholar]
- 5.Onor IO, et al. Clinical effects of cigarette smoking: Epidemiologic impact and review of pharmacotherapy options. Int. J. Environ. Res. Public Health. 2017;14:1147. doi: 10.3390/ijerph14101147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jha P. The hazards of smoking and the benefits of cessation: A critical summation of the epidemiological evidence in high-income countries. Elife. 2020;9:e49979. doi: 10.7554/eLife.49979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saha SP, Bhalla DK, Whayne TF, Gairola C. Cigarette smoke and adverse health effects: An overview of research trends and future needs. Int. J. Angiol. 2007;16:77–83. doi: 10.1055/s-0031-1278254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 9.McGinnis JM, Foege WH. Actual causes of death in the United States. JAMA. 1993;270:2207–2212. [PubMed] [Google Scholar]
- 10.Ambrose JA, Barua RS. The pathophysiology of cigarette smoking and cardiovascular disease: An update. J. Am. Coll. Cardiol. 2004;43:1731–1737. doi: 10.1016/j.jacc.2003.12.047. [DOI] [PubMed] [Google Scholar]
- 11.Salahuddin S, Prabhakaran D, Roy A. Pathophysiological mechanisms of tobacco-related CVD. Glob. Heart. 2012;7:113–120. doi: 10.1016/j.gheart.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Besingi W, Johansson A. Smoke-related DNA methylation changes in the etiology of human disease. Hum. Mol. Genet. 2014;23:2290–2297. doi: 10.1093/hmg/ddt621. [DOI] [PubMed] [Google Scholar]
- 13.Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am. J. Hum. Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott HR, et al. Differences in smoking associated DNA methylation patterns in South Asians and Europeans. Clin. Epigenetics. 2014;6:4. doi: 10.1186/1868-7083-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monick MM, et al. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;159B:141–151. doi: 10.1002/ajmg.b.32021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shenker NS, et al. DNA methylation as a long-term biomarker of exposure to tobacco smoke. Epidemiology. 2013;24:712–716. doi: 10.1097/EDE.0b013e31829d5cb3. [DOI] [PubMed] [Google Scholar]
- 17.Zeilinger S, et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlid S, Xu Z, Panduri V, Sandler DP, Taylor JA. CpG sites associated with cigarette smoking: Analysis of epigenome-wide data from the sister study. Environ. Health Persp. 2014;122:673–678. doi: 10.1289/ehp.1307480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philibert R, Dogan M, Beach SRH, Mills JA, Long JD. AHRR methylation predicts smoking status and smoking intensity in both saliva and blood DNA. Am. J. Med. Genet. Part B Neuropsychiatric Genet. Off. Publ. Int. Soc. Psychiat. Genet. 2020;183:51–60. doi: 10.1002/ajmg.b.32760. [DOI] [PubMed] [Google Scholar]
- 20.Cole JW, Xu H. Aryl hydrocarbon receptor repressor methylation: A link between smoking and atherosclerosis. Circ. Cardiovasc. Genet. 2015;8:640–642. doi: 10.1161/CIRCGENETICS.115.001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, et al. F2RL3 methylation in blood DNA is a strong predictor of mortality. Int. J. Epidemiol. 2014;43:1215–1225. doi: 10.1093/ije/dyu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kodal, J. B., Kobylecki, C. J., Vedel-Krogh, S., Nordestgaard, B. G. & Bojesen, S. E. AHRR hypomethylation, lung function, lung function decline and respiratory symptoms. Eur. Respirat. J.51, (2018). [DOI] [PubMed]
- 23.Pidsley R, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 2016;17:208. doi: 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:3156. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quach A, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging. 2017;9:419–446. doi: 10.18632/aging.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horvath S, et al. Obesity accelerates epigenetic aging of human liver. PNAS. 2014;111:15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zannas AS, et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: Relevance of glucocorticoid signaling. Genome Biol. 2015;16:266. doi: 10.1186/s13059-015-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levine ME, et al. DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging. 2015;7:690–700. doi: 10.18632/aging.100809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horvath S, et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging. 2018;10:1758–1775. doi: 10.18632/aging.101508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine ME, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10:573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu AT, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11:303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu AT, et al. DNA methylation-based estimator of telomere length. Aging (Albany NY) 2019;11:5895–5923. doi: 10.18632/aging.102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X, et al. Effect of tobacco smoking on the epigenetic age of human respiratory organs. Clin. Epigenetics. 2019;11:183. doi: 10.1186/s13148-019-0777-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dominguez K, et al. Vital signs: Leading causes of death, prevalence of diseases and risk factors, and use of health services among Hispanics in the United States—2009–2013. MMWR Morb. Mortal. Wkly Rep. 2015;64:469. [PMC free article] [PubMed] [Google Scholar]
- 36.Velasco-Mondragon E, Jimenez A, Palladino-Davis AG, Davis D, Escamilla-Cejudo JA. Hispanic health in the USA: A scoping review of the literature. Public Health Rev. 2016;37:31. doi: 10.1186/s40985-016-0043-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vega WA, Rodriguez MA, Gruskin E. Health disparities in the latino population. Epidemiol. Rev. 2009;31:99–112. doi: 10.1093/epirev/mxp008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosero-Bixby L, Dow WH, Rehkopf DH. The Nicoya region of Costa Rica: A high longevity island for elderly males. Vienna Yearb. Popul. Res. 2013;11:109–136. doi: 10.1553/populationyearbook2013s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buettner D, Skemp S. Blue zones: Lessons from the world’s longest lived. Am. J. Lifestyle Med. 2016;10:318–321. doi: 10.1177/1559827616637066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rehkopf DH, et al. Longer leukocyte telomere length in Costa Rica’s Nicoyan Peninsula: A population-based study. Exp. Gerontol. 2013;48:1266. doi: 10.1016/j.exger.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosero-Bixby L, et al. Correlates of longitudinal leukocyte telomere length in the Costa Rican Longevity Study of Healthy Aging (CRELES): On the importance of DNA collection and storage procedures. PLoS ONE. 2019;14:e0223766. doi: 10.1371/journal.pone.0223766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McEwen LM, et al. Differential DNA methylation and lymphocyte proportions in a Costa Rican high longevity region. Epigenetics Chromatin. 2017;10:21. doi: 10.1186/s13072-017-0128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosero-Bixby L, Dow WH. Predicting mortality with biomarkers: A population-based prospective cohort study for elderly Costa Ricans. Popul. Health Metr. 2012;10:11. doi: 10.1186/1478-7954-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:1–16. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aryee MJ, et al. Minfi: A flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–1369. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price EM, et al. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6:1–15. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 48.Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salas LA, et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol. 2018;19:1–14. doi: 10.1186/s13059-018-1448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venables, B. & Ripley, B. D. Modern Applied Statistics with S. (2002). 10.1007/978-0-387-21706-2.
- 51.Chen BH, et al. DNA methylation-based measures of biological age: Meta-analysis predicting time to death. Aging. 2016;8:1844. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peters TJ, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Phipson B, Maksimovic J, Oshlack A. missMethyl: An R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics. 2016;32:286–288. doi: 10.1093/bioinformatics/btv560. [DOI] [PubMed] [Google Scholar]
- 55.Joehanes R, et al. Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet. 2016;9:436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Domingo-Relloso A, et al. Cadmium, smoking, and human blood DNA methylation profiles in adults from the strong heart study. Environ. Health Persp. 2020;128:067005. doi: 10.1289/EHP6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S, et al. Causal effect of smoking on DNA methylation in peripheral blood: A twin and family study. Clin. Epigenetics. 2018;10:18. doi: 10.1186/s13148-018-0452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barcelona V, et al. Novel DNA methylation sites associated with cigarette smoking among African Americans. Epigenetics. 2019;14:383–391. doi: 10.1080/15592294.2019.1588683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: A systematic review of DNA methylation studies. Clin. Epigenetics. 2015;7:113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ringh MV, et al. Tobacco smoking induces changes in true DNA methylation, hydroxymethylation and gene expression in bronchoalveolar lavage cells. EBioMedicine. 2019;46:290–304. doi: 10.1016/j.ebiom.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee MK, Hong Y, Kim S-Y, London SJ, Kim WJ. DNA methylation and smoking in Korean adults: Epigenome-wide association study. Clin. Epigenetics. 2016;8:103. doi: 10.1186/s13148-016-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Christiansen C, et al. Novel DNA methylation signatures of tobacco smoking with trans-ethnic effects. Clin. Epigenetics. 2021;13:36. doi: 10.1186/s13148-021-01018-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mao L, et al. Identification of atypical mitogen-activated protein kinase MAPK4 as a novel regulator in acute lung injury. Cell Biosci. 2020;10:121. doi: 10.1186/s13578-020-00484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W, et al. MAPK4 overexpression promotes tumor progression via noncanonical activation of AKT/mTOR signaling. J. Clin. Invest. 2019;129:1015–1029. doi: 10.1172/JCI97712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Misawa K, et al. G protein-coupled receptor genes, PTGDR1, PTGDR2, and PTGIR, are candidate epigenetic biomarkers and predictors for treated patients with HPV-associated oropharyngeal cancer. Microorganisms. 2020;8:E1504. doi: 10.3390/microorganisms8101504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho JS, et al. HNRNPM controls circRNA biogenesis and splicing fidelity to sustain cancer cell fitness. Elife. 2021;10:e59654. doi: 10.7554/eLife.59654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Georges A, et al. Rare loss-of-function mutations of PTGIR are enriched in fibromuscular dysplasia. Cardiovasc. Res. 2021;117:1154–1165. doi: 10.1093/cvr/cvaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu B, et al. TXNRD1 is an unfavorable prognostic factor for patients with hepatocellular carcinoma. BioMed. Res. Int. 2017;2017:e4698167. doi: 10.1155/2017/4698167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao W, et al. Education and lifestyle factors are associated with DNA methylation clocks in older african americans. Int. J. Environ. Res. Public Health. 2019;16:3141. doi: 10.3390/ijerph16173141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine ME, Crimmins EM. A genetic network associated with stress resistance, longevity, and cancer in humans. J. Gerontol. Ser. A Biomed. Sci. Med. Sci. 2016;71:703–712. doi: 10.1093/gerona/glv141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simons RL, et al. Economic hardship and biological weathering: The epigenetics of aging in a U.S. sample of black women. Soc. Sci. Med. 2016;150:192–200. doi: 10.1016/j.socscimed.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luo A, et al. Epigenetic aging is accelerated in alcohol use disorder and regulated by genetic variation in APOL2. Neuropsychopharmacology. 2020;45:327–336. doi: 10.1038/s41386-019-0500-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang Y, et al. Smoking-related DNA methylation is associated with DNA methylation phenotypic age acceleration: The veterans affairs normative aging study. Int. J. Environ. Res. Public Health. 2019;16:2356. doi: 10.3390/ijerph16132356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dugué P-A, et al. Association of DNA methylation-based biological age with health risk factors and overall and cause-specific mortality. Am. J. Epidemiol. 2018;187:529–538. doi: 10.1093/aje/kwx291. [DOI] [PubMed] [Google Scholar]
- 75.McCartney DL, et al. Investigating the relationship between DNA methylation age acceleration and risk factors for Alzheimer’s disease. Alzheimers Dement. (Amst) 2018;10:429–437. doi: 10.1016/j.dadm.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao X, Zhang Y, Breitling LP, Brenner H. Relationship of tobacco smoking and smoking-related DNA methylation with epigenetic age acceleration. Oncotarget. 2016;7:46878–46889. doi: 10.18632/oncotarget.9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horvath S, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17:171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nieddu A, et al. Dietary habits, anthropometric features and daily performance in two independent long-lived populations from Nicoya Peninsula (Costa Rica) and Ogliastra (Sardinia) Nutrients. 2020;12:1621. doi: 10.3390/nu12061621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chacón AM, Jiménez CC, Campos H. Dietary habits and lifestyle among long-lived residents from the Nicoya Peninsula of Costa Rica. Revista Hispanoamericana de Ciencias de la Salud (RHCS) 2017;3:53–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Public-use version of the CRELES data is available from the Inter-University Consortium for Political and Social Research (ICPSR) repository (http://doi.org/10.3886/ICPSR31263.v1). Since data DNA methylation and the complementary sample of centenarians in Nicoya are not currently part of the public-use, requests for restricted access to data can be submitted at http://www.creles.berkeley.edu/ following institutional review approval.