Significance
IL-33R mediates local inflammatory responses and plays crucial roles in the pathogenesis of immune diseases. In this study, we identified USP38, which negatively regulates IL-33-triggered signaling by mediating K27-linked deubiquitination of IL-33R at K511 and its autophagic degradation. USP38 deficiency aggravates IL-33–induced lung inflammatory response and bleomycin-induced pulmonary fibrosis. We further show that the E3 ubiquitin ligase TRAF6 catalyzes K27-linked polyubiquitination of IL-33R at K511, and that deficiency of TRAF6 inhibits IL-33–mediated signaling. Our findings reveal an important mechanism regarding how IL-33R is precisely regulated to ensure its inactivation in rest cells and proper activation following IL-33 stimulation.
Keywords: IL-33R, USP38, polyubiquitination, inflammatory response, pulmonary fibrosis
Abstract
The warning cytokine interleukin-33 receptor (IL-33R) mediates local inflammatory responses and plays crucial roles in the pathogenesis of immune diseases such as pulmonary fibrosis and rheumatoid arthritis. Whether and how IL-33R is regulated remain enigmatic. Here, we identified ubiquitin-specific protease 38 (USP38) as a negative regulator of IL-33R–mediated signaling. USP38 deficiency promotes interleukin-33 (IL-33)–induced downstream proinflammatory responses in vitro and in vivo. Usp38−/− mice are more susceptible to inflammatory damage and death and developed more serious pulmonary fibrosis after bleomycin treatment. USP38 is constitutively associated with IL-33R and deconjugates its K27-linked polyubiquitination at K511, resulting in its autophagic degradation. We further show that the E3 ubiquitin ligase tumor necrosis factor receptor–associated factor 6 (TRAF6) catalyzes K27-linked polyubiquitination of IL-33R at K511, and that deficiency of TRAF6 inhibits IL-33–mediated signaling. Our findings suggest that K27-linked polyubiquitination and deubiquitination of IL-33R by TRAF6 and USP38 reciprocally regulate IL-33R level and signaling, which represents a critical mechanism in the regulation of IL-33–triggered lung inflammatory response and pulmonary fibrosis.
Interleukin-33 (IL-33) is a member of the interleukin-1 (IL-1) family, which plays crucial roles in innate and adaptive immunity, contributing to tissue homeostasis and responses to environmental stresses (1, 2). IL-33 is expressed constitutively in a variety of cells, including human epithelial and pulmonary endothelial cells, as a chromatin-associated nuclear cytokine in the steady state. IL-33 is released as an alarm after cell injury to alert the immune system of tissue damage during trauma or infection (1, 3–5). It has been shown that excessive activation of IL-33 causes severe local inflammation as well as autoimmune diseases or immune-related diseases, such as substantial lung damage (6–8). Overexpression of IL-33 is observed in patients with chronic fibrosis, including idiopathic pulmonary fibrosis (9, 10). IL-33 promotes interleukin-33 receptor (IL-33R)–dependent pulmonary fibrosis by inducing alternate activation of macrophages and innate lymphoid cells in mice (11). Anti–IL-33 antibody treatment attenuates airway inflammation in a murine model of allergic asthma (12).
IL-33R, also known as suppression of tumorigenicity 2 (ST2), is a classical type I transmembrane receptor of IL-33 (13). Binding of IL-33 to IL-33R triggers a cascade of signaling events, leading to local immune responses. The initial step in IL-33 signal transduction is ligand-induced conformational changes in IL-33R, which facilitate recruitment of interleukin-1 receptor accessory protein (IL-1RAP). The activated heterodimer complex recruits downstream signaling components, including myeloid differentiation primary response protein 88 (MyD88), IL-1 receptor (IL-1R)–associated kinase, tumor necrosis factor (TNF) receptor–associated factor 6 (TRAF6), and transforming growth factor (TGF)-β–activated kinase 1 (TAK1) complex, resulting in TAK1 activation. TAK1 subsequently activates downstream kinases inhibitor of nuclear factor kappa-B kinase subunit alpha (IKKα) and IKKβ, which phosphorylate nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) inhibitor (IκB) proteins. These events ultimately lead to activation of the transcription factor NF-κB and induction of downstream effector genes (1, 14–16).
While excessive activation of IL-33–triggered signaling is associated with a variety of diseases, the IL-33/IL-33R axis is tightly regulated under physiological conditions. sST2, a soluble and secreted form of IL-33R/ST2, inhibits IL-33–triggered signaling by competing with IL-33R for IL-33 binding (17, 18). IL-33–triggered signaling is inhibited by the single immunoglobulin domain interleukin-1 receptor–associated molecule (SIGIRR; also known as TIR8), which disrupts dimerization of IL-33R and IL-1RAP (19). Previous studies have demonstrated that posttranslational modifications play critical roles in the regulation of the IL-1R family. For example, the E3 ubiquitin ligases MARCH3 and MARCH8 mediate K48-linked polyubiquitination and degradation of IL-1R and IL-1RAP, respectively, leading to negative regulation of IL-1–triggered inflammatory responses (20–22). It has been previously reported that the E3 ubiquitin ligase FBXL19 mediates polyubiquitination of IL-33R, and overexpression of FBXL19 attenuates IL-33–triggered inflammatory response (23).
Deubiquitinating enzymes remove ubiquitin conjugates from their target proteins, resulting in regulation of their stability, localization, or activity (24–27). Ubiquitin-specific protease 38 (USP38) was originally identified as a negative regulator of type I interferon signaling by its regulation of TANK-bound kinase 1 ubiquitination (28). In addition, it has been shown that USP38 is down-regulated in colorectal cancers and acts as a tumor suppressor by deubiquitinating HDAC3, thereby controlling expression of tumor stem cell–related genes (29).
In this study, we identified USP38 as a negative regulator of IL-33–triggered signaling. USP38 deficiency potentiates IL-33–induced lung inflammatory response and bleomycin-induced pulmonary fibrosis. Mechanistically, we demonstrate that K27-linked polyubiquitination of IL-33RK511 is critical for its stability. USP38 is constitutively associated with IL-33R, resulting in deconjugation of K27-linked polyubiquitin moieties from IL-33RK511 as well as its autophagic degradation. We also identified TRAF6 as the E3 ubiquitin ligase that catalyzes K27-linked polyubiquitination of IL-33RK511. These findings suggest that K27-linked polyubiquitination and deubiquitination of IL-33R by TRAF6 and USP38, respectively, serve as a critical regulatory mechanism for fine-tuning IL-33R–mediated inflammatory response.
Results
USP38 Negatively Regulates IL-33–Triggered Signaling by Down-Regulating IL-33R.
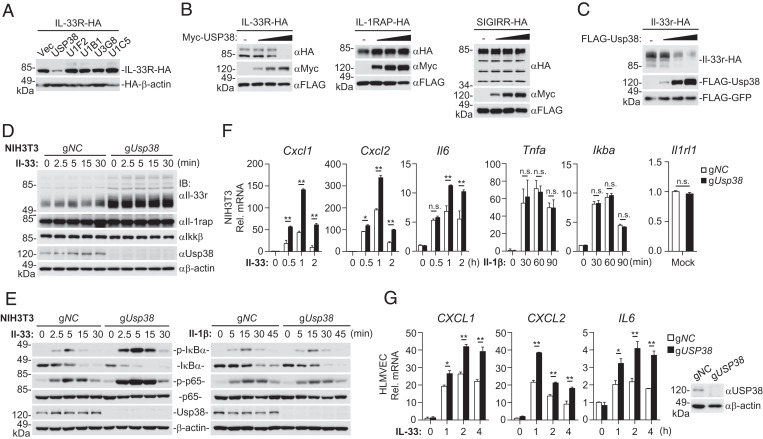
Whether and how IL-33R is regulated by posttranslational modifications are enigmatic. In an attempt to identify proteins that regulate IL-33R–mediated signaling by posttranslational modifications, we screened 247 ubiquitin-related proteins for their abilities to regulate IL-33R level by cotransfection experiments in HEK293 cells. These efforts led to the identification of human USP38, which caused down-regulation of human IL-33R (Fig. 1A). Overexpression of USP38 down-regulated the level of IL-33R but not IL-1RAP or SIGIRR in a dose-dependent manner (Fig. 1B). We further examined whether murine Usp38 (throughout the text, the designations of Homo sapiens genes and proteins are all capitalized, while only the first letters of murine genes and proteins are capitalized) can down-regulate Il-33r. The results showed that Usp38 down-regulated Il-33r in a dose-dependent manner (Fig. 1C), pointing to a conserved function of Usp38 in down-regulating Il-33r. We next generated Usp38-deficient murine embryonic fibroblast NIH 3T3 cells using the CRISPR-Cas9 system. We found that the level of Il-33r but not of Il-1rap and Ikkβ was increased in Usp38-deficient NIH 3T3 cells (Fig. 1D). Consistently, Usp38 deficiency enhanced Il-33– but not Il-1β–induced phosphorylation of IκBα and p65 in NIH 3T3 cells (Fig. 1E), which are hallmarks of Il-33–induced activation of downstream components. qPCR experiments indicated that Usp38 deficiency potentiated Il-33–induced transcription of Cxcl1, Cxcl2, and Il6 genes (Fig. 1F). In contrast, Il-1β–induced transcription of Tnfa and Ikba genes was comparable between Usp38-deficient and control NIH 3T3 cells (Fig. 1F). In these experiments, Usp38 deficiency did not affect IL-33R messenger RNA (mRNA) levels (Fig. 1F). Similarly, USP38 deficiency also potentiated IL-33–induced transcription of CXCL1, CXCL2, and IL6 genes in human pulmonary microvascular endothelial cells (HLMVECs) (Fig. 1G). Reconstitution experiments indicated that ectopic expression of Usp38 in Usp38-deficient NIH 3T3 cells down-regulated Il-33r in a dose-dependent manner (SI Appendix, Fig. S1A) and reversed the potentiation of IL-33–induced transcription of downstream genes by Usp38 deficiency (SI Appendix, Fig. S1B). Moreover, knockdown of Il-33r inhibited the potentiation of IL-33–induced transcription of downstream genes by Usp38 deficiency (SI Appendix, Fig. S1C).These data suggest that endogenous USP38 negatively regulates IL-33–triggered signaling in various mammalian cells through regulation of IL-33R levels.
Fig. 1.
USP38 inhibits IL-33–triggered signaling. (A) Screening of ubiquitin-related enzymes that regulate the stability of IL-33R. HEK293 cells were transfected with IL-33R-HA, HA-β-actin, and the plasmids encoding ubiquitin-related proteins for 24 h before immunoblotting analysis with the indicated antibodies. Data shown are from five representative ubiquitin-related enzyme clones. (B) USP38 promotes degradation of IL-33R in a dose-dependent manner. HEK293 cells (5 × 105) were transfected with IL-33R-HA (0.1 μg), IL-1RAP-HA (0.1 μg), or SIGRR-HA (0.1 μg) together with Flag-GFP (0.1 μg) and Myc-USP38 (0, 0.2, 0.4, and 0.8 μg) for 24 h before immunoblotting analysis with the indicated antibodies. An empty vector was added to ensure that each transfection received the same amount of total DNA. (C) Usp38 destabilizes Il-33r in a dose-dependent manner. HEK293 cells (5 × 105) were transfected with Flag-GFP (0.1 μg), Il-33r-HA (0.1 μg), and Flag-Usp38 (0, 0.3, 0.9, and 2.7 μg) plasmids for 24 h before immunoblotting analysis with the indicated antibodies. An empty vector was added to ensure that each transfection receives the same amount of total DNA. (D) Usp38 deficiency specifically up-regulates the levels of Il-33r in NIH 3T3 cells. The control (gNC) and Usp38-deficient (gUsp38) NIH 3T3 cells (1 × 106) were left untreated or treated with Il-33 (5 ng/mL) for the indicated time before immunoblotting analysis with the indicated antibodies. (E) Effects of Usp38 deficiency on phosphorylation of IκBα and p65 induced by Il-33 or Il-1β. The control and Usp38-deficient NIH 3T3 cells (1 × 106) were left untreated or treated with Il-33 (5 ng/mL) or Il-1β (100 ng/mL) for the indicated time before immunoblotting analysis with the indicated antibodies. (F) Effects of Usp38 deficiency on the transcription of downstream genes induced by Il-33 or Il-1β. The control and Usp38-deficient NIH 3T3 cells (1 × 106) were left untreated or treated with Il-33 (5 ng/mL) or Il-1β (100 ng/mL) for the indicated time before qPCR analysis. (G) Effects of USP38 deficiency on transcription of downstream genes induced by IL-33. The control (gNC) and USP38-deficient (gUSP38) HLMVEC cells (1 × 106) were left untreated or treated with IL-33 (50 ng/mL) for the indicated time before qPCR (histographs in Left) and immunoblotting analysis with the indicated antibodies (blots in Right). Data in F and G show mean ± SEM. n = 3 from one representative experiment. All experiments were repeated at least two times with similar results. n.s., not significant. *P < 0.05; **P < 0.01.
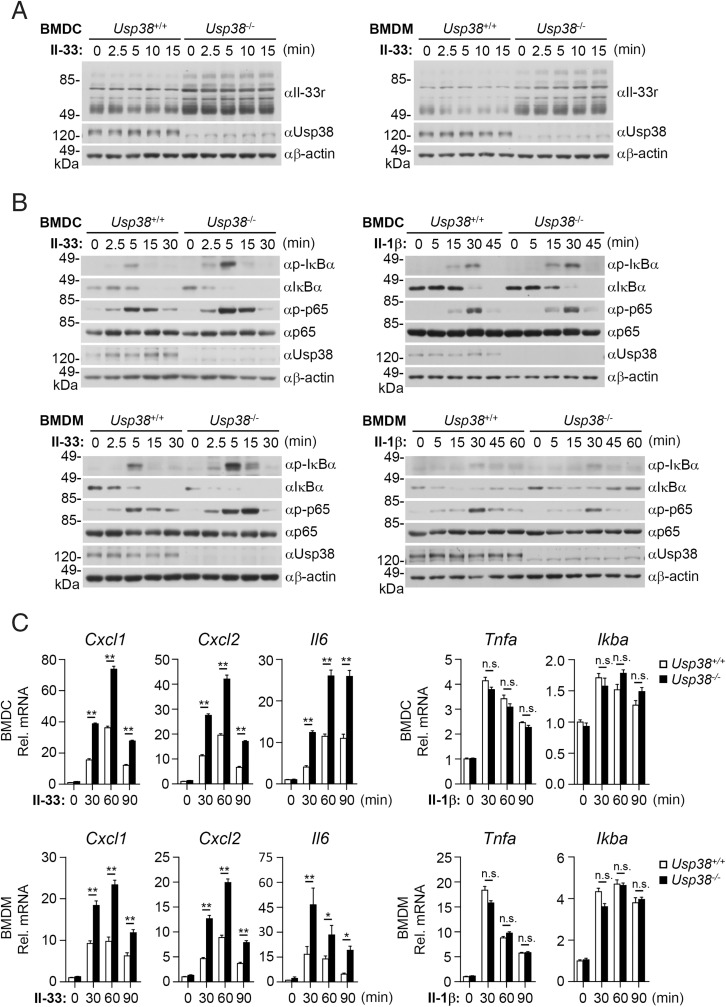
To further investigate the roles of Usp38 in vivo, we prepared Usp38-deficient mice generated by the CRISPR-Cas9 method (30). We found that the levels of Il-33r were increased in Usp38-deficient bone marrow–derived dendritic cells (BMDCs) and bone marrow–derived macrophages (BMDMs) compared with their wild-type counterparts (Fig. 2A). Consistently, Usp38 deficiency potentiated Il-33– but not Il-1β–induced phosphorylation of IκBα and p65 in mouse BMDCs and BMDMs (Fig. 2B). Usp38 deficiency also potentiated Il-33–induced transcription of Cxcl1, Cxcl2, and Il6 genes but not Il-1β–induced transcription of Tnfa and Ikba genes in BMDCs and BMDMs (Fig. 2C). These results suggest that Usp38 negatively regulates Il-33–triggered signaling in primary immune cells.
Fig. 2.
Usp38 negatively regulates Il-33–triggered signaling in primary immune cells. Usp38+/+ or Usp38−/− BMDCs or BMDMs (1 × 106) were left untreated or treated with Il-33 (5 ng/mL) or Il-1β (100 ng/mL) for the indicated time before immunoblotting analysis with the indicated antibodies (A and B) or qPCR analysis for mRNA levels of the indicated genes (C). Graphs show mean ± SEM. n = 3 from one representative experiment. All experiments were repeated at least two times with similar results. n.s., not significant. *P < 0.05; **P < 0.01.
Usp38 Deficiency Promotes Il-33–Induced Lung Inflammatory Response and Bleomycin-Induced Pulmonary Fibrosis.
To investigate the roles of Usp38 in Il-33–induced signaling in vivo, we treated mice with recombinant murine Il-33 intranasally to induce lung inflammatory response. We found that Usp38 deficiency markedly increased eosinophilia and Il-5 levels in the lung following recombinant Il-33 stimulation (SI Appendix, Fig. S2). These data suggest that Usp38 deficiency potentiates Il-33–induced lung inflammatory response in vivo.
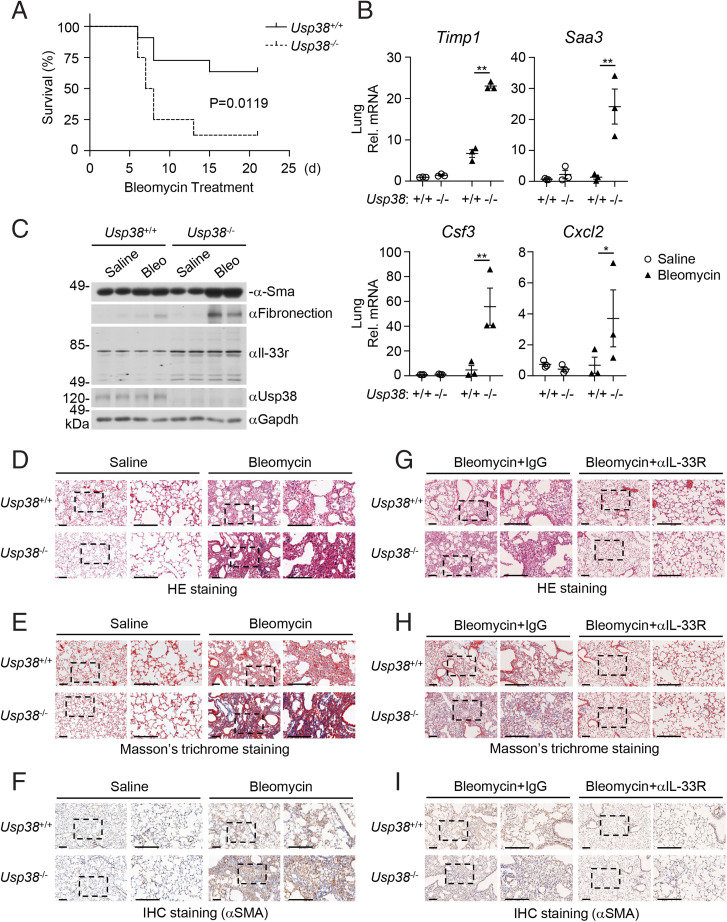
Pulmonary fibrosis is a type of end-stage change of lung diseases characterized by fibroblast proliferation and a large amount of extracellular matrix aggregation, accompanied by inflammatory injury and structural destruction (31, 32). Bleomycin is an important chemotherapeutic agent for cancer. Its cytotoxic activity associated with DNA strand splicing and induction of reactive oxygen species can lead to serious side effects, including lung fibrosis (32, 33). Bleomycin-induced fibrosis in susceptible C57BL/6 mice has been widely used as a model for studying pulmonary fibrosis (31, 32). It has been shown that Il-33r is critical for lung fibrotic pathogenesis (10). IL-33 is induced by bleomycin in lung tissue. ST2 deficiency or anti–IL-33 antibody treatment attenuates bleomycin-induced lung inflammation and fibrosis (11). We, therefore, investigated whether USP38 plays a role in severe pulmonary fibrosis induced by high doses of bleomycin. Compared with the Usp38+/+ littermates, Usp38−/− mice were more susceptible to death induced by bleomycin (10 mg/kg) (Fig. 3A). The mRNA levels of lung inflammatory genes (Cxcl2, Csf3) and fibrosis-related genes (Timp1, Saa3) induced by bleomycin were significantly increased in Usp38−/− mice compared with the wild-type littermates (Fig. 3B). Usp38 deficiency markedly increased the levels of fibronectin and α-smooth muscle actin (α-SMA), which are fibrotic markers induced by bleomycin. Consistently, the protein levels of Il-33r in the lungs of Usp38−/− mice were also up-regulated compared with that in the wild-type littermates (Fig. 3C). Hematoxylin-eosin (HE) staining analysis indicated that Usp38−/− mice showed more serious inflammatory damage of lung after bleomycin administration (Fig. 3D). Moreover, Usp38−/− mice showed more severe collagen deposition (Fig. 3E) and increased α-SMA (Fig. 3F) as illustrated by trichrome and immunohistochemical (IHC) staining, respectively, in the lungs following bleomycin administration. Furthermore, we treated mice with anti–Il-33r neutralizing antibody or control mouse immunoglobulin G (IgG) following bleomycin administration. HE staining analysis indicated that bleomycin-induced airway inflammatory damages were attenuated following treatment with Il-33r neutralizing antibody in the lungs of both wild-type and Usp38−/− mice (Fig. 3G). Consistently, collagen deposition and α-SMA protein in wild-type and Usp38−/− mice were comparable following treatment of Il-33r neutralizing antibody (Fig. 3 H and I). These data suggest that Usp38 deficiency potentiates bleomycin-induced pulmonary fibrosis in mice through regulation of Il-33r stability.
Fig. 3.
Usp38 deficiency promotes bleomycin-induced pulmonary fibrosis. (A) Measurement of survival of Usp38+/+ and Usp38−/− mice following bleomycin treatment. Sex- and age-matched Usp38+/+ (n = 11) and Usp38−/− (n = 8) mice were intranasally injected with bleomycin (10 mg/kg). Mouse survival was monitored daily for a period of 21 d. The survival curve was generated by Kaplan–Meier methods followed by log-rank test analysis. (B) Effects of Usp38 deficiency on transcription of lung inflammatory and fibrotic genes induced by bleomycin. Sex- and age-matched Usp38+/+ (n = 3) and Usp38−/− (n = 3) mice were intranasally injected with saline or bleomycin (20 mg/kg), and lung tissues were taken for qPCR analysis 5 d after injection. Data shown are mean ± SEM. *P < 0.05; **P < 0.01. (C) Effects of Usp38 deficiency on levels of Il-33r and pulmonary fibrosis–related proteins in the lung following bleomycin treatment. Sex- and age-matched Usp38+/+ and Usp38−/− mice were intranasally injected with saline or bleomycin (10 mg/kg), and lung tissues were taken for immunoblotting analysis with the indicated antibodies 14 d after injection. Two biological repeats were performed with similar results. (D–I) Pathological analysis of lung tissues. Sex- and age-matched Usp38+/+ and Usp38−/− mice were intranasally injected with saline or bleomycin (10 mg/kg), and lung tissues were taken for HE staining (D), Masson’s trichrome (E), and IHC (F) staining 14 d after injection. For G–I, the mice were also treated intraperitoneally with anti–Il-33r neutralizing antibody or control rat IgG (R&D; 100 μg per mouse) on days 0, 5, and 10 after bleomycin administration, and lung tissues were taken for HE staining (G), Masson’s trichrome (H), and IHC (I) staining 14 d after bleomycin administration. Three biological repeats were performed with similar results. (Scale bars, 300 μm).
USP38 Removes K27-Linked Polyubiquitin Moieties from IL-33RK511 and Causes Its Autophagic Degradation.
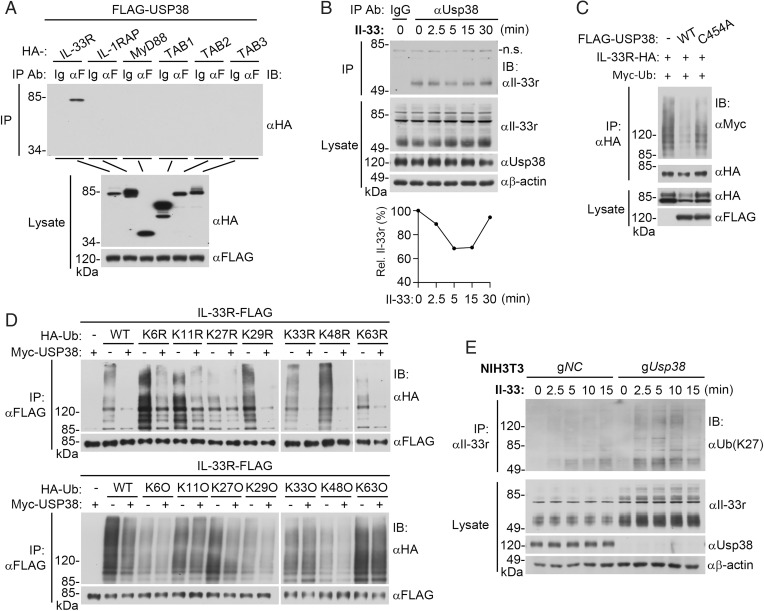
To investigate the molecular mechanisms responsible for the inhibitory effects of USP38 on IL-33–mediated inflammatory response, we examined whether USP38 is associated with components in IL-33–triggered signaling pathways. Mammalian overexpression and coimmunoprecipitation experiments indicated that USP38 was specifically associated with IL-33R but not IL-1RAP or their downstream components, including MyD88, TAB1, TAB2, and TAB3 (Fig. 4A). Endogenous Usp38 was constitutively associated with Il-33r in rest and Il-33–treated NIH 3T3 cells. It was noticed that the association was decreased at early time points after Il-33 stimulation and then gradually restored to the level in unstimulated cells (Fig. 4B). The decrease of association of Il-33r and Usp38 at early time points after Il-33 stimulation is correlated with activation of Il-33r–mediated signaling (Fig. 1E).
Fig. 4.
USP38 removes K27-linked polyubiquitin moieties from IL-33R. (A) USP38 associates with IL-33R. HEK293 cells (5 × 106) were transfected with the indicated plasmids. Eighteen hours after transfection, coimmunoprecipitation was performed with anti-Flag or control mouse IgG. The immunoprecipitates and lysates were analyzed by immunoblotting with the indicated antibodies. (B) Endogenous Usp38 is associated with Il-33r. NIH 3T3 cells (2 × 107) were left untreated or treated with Il-33 (40 ng/mL) for the indicated time. Coimmunoprecipitation and immunoblotting analysis were performed with the indicated antibodies. Lower shows relative levels of Usp38-associated Il-33r relative to total cellular β-actin, which was quantitated by ImageJ. n.s., nonspecific. (C) USP38 but not the C454A mutant deubiquitinates and down-regulates IL-33R. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 18 h before ubiquitination assays with the indicated antibodies. Expression levels of the transfected proteins were shown by immunoblots. (D) USP38 removes K27-linked polyubiquitin moieties from IL-33R. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 18 h before ubiquitination assays with the indicated antibodies. WT, wild type; KR, K is mutated to R; KO, K only. (E) Usp38 deficiency potentiates K27-linked polyubiquitination of Il-33r. The control (gNC) and Usp38-deficient (gUsp38) NIH 3T3 cells (2 × 107) were left untreated or treated with Il-33 (40 ng/mL) for the indicated time before ubiquitination assays with the indicated antibodies. IP, immunoprecipitation; IB, immunoblotting. All experiments were repeated at least two times with similar results.
Since USP38 is a deubiquitinating enzyme and mediates down-regulation of IL-33R, we next determined whether it deubiquitinates IL-33R. In the mammalian overexpression system, USP38 but not its enzymatic inactive mutant USP38C454A removed polyubiquitin moieties from IL-33R and down-regulated the level of IL-33R (Fig. 4C). We next determined the types of polyubiquitin chains that were removed from IL-33R by USP38. By cotransfection of IL-33R with ubiquitin mutants in which only one lysine reside is mutated to arginine (KR), we found that USP38 failed to remove polyubiquitin moieties from IL-33R only when K27 was mutated to arginine (Fig. 4D). By cotransfection of IL-33R with ubiquitin mutants that contain only a single lysine residue (KO), we found that USP38 removed K27-linked polyubiquitin moieties from IL-33R (Fig. 4D). These results suggest that USP38 deubiquitinates K27-linked polyubiquitin moieties from IL-33R. Endogenous polyubiquitination assays indicated that Il-33 stimulation increased K27-linked polyubiquitination of Il-33r in NIH 3T3 cells, and these effects were enhanced in Usp38-deficient NIH 3T3 cells (Fig. 4E). Collectively, these results suggest that USP38 is specifically associated with IL-33R and deconjugates its K27-linked polyubiquitin moieties.
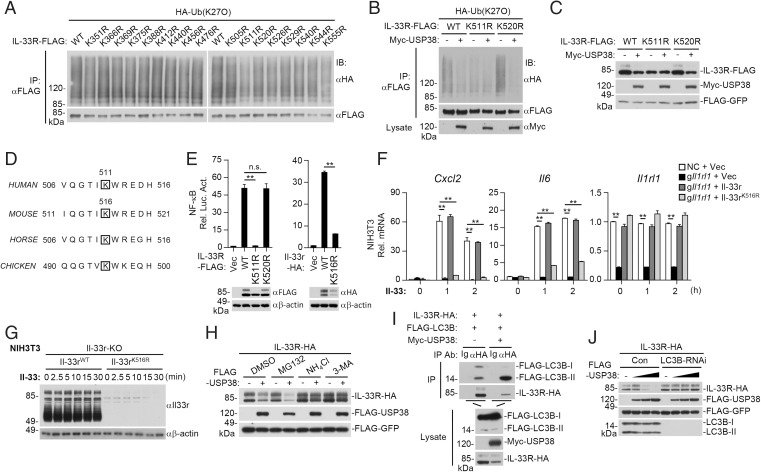
We next attempted to identify the residues of IL-33R that are modified by K27-linked polyubiquitination. We individually mutated all of the 17 lysine residues within the intracellular region of IL-33R to arginine and then examined their modifications by K27-linked polyubiquitination. As shown in Fig. 5A, K27-linked polyubiquitination of IL-33RK511R but not of the other 16 mutants was markedly decreased. Further experiments indicated that USP38 removed K27-linked polyubiquitin moieties from wild-type IL-33R and IL-33RK520R but failed to remove the low level of basal K27-linked polyubiquitination of IL-33RK511R (Fig. 5B). Consistently, USP38 also down-regulated the level of wild-type IL-33R and IL-33RK520R but not IL-33RK511R (Fig. 5C). Cycloheximide inhibition experiments indicated that wild-type IL-33R had a longer half-life than IL-33RK511R (SI Appendix, Fig. S3A). Sequence analysis showed that K511 of human IL-33R corresponds to K516 of murine Il-33r, which is conserved across various species (Fig. 5D). Confocal microscopy showed that Usp38 was colocalized with wild-type Il-33r and Il-33rK516R at the cell surfaces in untreated cells (SI Appendix, Fig. S3B). Mutation of K511 in human IL-33R or K516 in murine Il-33r impaired their ability to activate NF-κB (Fig. 5E). Reconstitution experiments indicated that wild-type Il-33r but not Il-33rK516R rescued Il-33–induced transcription of Cxcl2 and Il6 genes in Il-33r–deficient NIH 3T3 cells (Fig. 5F). In these reconstitution experiments, the mRNA levels of Il-33rK516R and wild-type Il-33r were comparable (Fig. 5F), but the protein levels of Il-33rK516R were dramatically down-regulated compared with its wild-type counterparts (Fig. 5G). These results suggest that USP38 deconjugates K27-linked polyubiquitination of IL-33R at K511 (or murine Il-33r at K516), which promotes its down-regulation and negative regulation of IL-33–triggered signaling.
Fig. 5.
USP38 deubiquitinates IL-33RK511 and causes its autophagic degradation. (A) K27-linked polyubiquitination of IL-33R and its mutants. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 18 h before ubiquitination assays. (B) USP38 mediates deubiquitination of IL-33RK511. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 18 h before ubiquitination assays. (C) Effects of USP38 on the stability of IL-33R and its mutants. HEK293 cells (5 × 105) were transfected with the indicated plasmids for 24 h before immunoblotting analysis with the indicated antibodies. (D) K511 of human IL-33R is conserved among various species. The sequences of IL-33R fragments from various species are aligned, and the conserved lysine residues are framed. (E) IL-33RK511R or Il-33rK516R loses the ability to activate NF-κB. HEK293 cells (6 × 104) were transfected with NF-κB reporter plasmid, IL-1RAP-HA, and the indicated IL-33R (or Il-33r) or its mutant plasmids for 16 h before luciferase assays and immunoblotting analysis with the indicated antibodies. (F) Reconstitution of Il-33r–deficient NIH 3T3 cells with wild-type Il-33r or Il-33rK516R. Il-33r–deficient (gIl1rl) NIH 3T3 cells were reconstituted with Il-33r-Flag, Il-33rK516R-Flag, or empty vector. Control NIH 3T3 (NC) cells were reconstituted with an empty vector. The cells (1 × 106) were then stimulated with Il-33 (5 ng/mL) for the indicated time before qPCR experiments. (G) Protein levels of wild-type Il-33r and Il-33rK516R in Il-33r–deficient NIH 3T3 cells reconstituted with wild-type Il-33r or Il-33rK516R. The cells (1 × 106) were stimulated with Il-33 (5 ng/mL) for the indicated time before immunoblotting analysis with the indicated antibodies. (H) Effects of inhibitors on USP38-mediated down-regulation of IL-33R. HEK293 cells (5 × 105) were transfected with the indicated plasmids for 6 h and then treated with NH4Cl (5 mM), MG132 (1 μM), or 3-MA (100 ng/mL) for 14 h before immunoblotting analysis with the indicated antibodies. (I) Effects of USP38 on the association of IL-33R with LC3B. HEK293 cells (5 × 106) were transfected with the indicated plasmids for 24 h before coimmunoprecipitation and immunoblotting analysis with the indicated antibodies. (J) Knockdown of LC3B inhibits USP38-mediated degradation of IL-33R. HEK293 cells (5 × 105) stably transduced with control or LC3B-RNAi were further transfected with IL-33R-HA (0.2 μg), Flag-GFP (0.1 μg), and Flag-USP38 (0, 0.4, 0.8, and 1.6 μg) for 24 h before immunoblotting analysis with the indicated antibodies. An empty vector was added to ensure that each transfection received the same amount of total DNA. Data shown in E and F are mean ± SEM. n = 3 from one representative experiment. WT, wild type; IB, immunoblotting; IP, immunoprecipitation; Vec, Vector; RNAi, RNA interference. All experiments were repeated at least two times with similar results. n.s., not significant. **P < 0.01.
We next investigated how IL-33R is down-regulated. We found that USP38-mediated degradation of IL-33R was inhibited by the lysosome inhibitor ammonium chloride (NH4Cl) and the autophagosome inhibitor 3-methyladenine (3-MA) but not by the proteasome inhibitor MG132 (Fig. 5H), suggesting that USP38 mediates degradation of IL-33R via an autophagic pathway. LC3B is an important marker of autophagy and required for fusion of the autophagosomes to the lysosomes. Confocal experiments indicated that Lc3b was colocalized with wild-type Il-33r and Il-33rK516R in rest cells (SI Appendix, Fig. S3C). During autophagy, LC3B-I is translocated from the cytoplasm to the bilayer of the autophagosomes and converts into LC3B-II (34). Coimmunoprecipitation experiments showed that IL-33R associated with LC3B-I and LC3B-II, and overexpression of USP38 dramatically enhanced the association of IL-33R with LC3B-II (Fig. 5I). Knockdown of LC3B markedly inhibited USP38-mediated degradation of IL-33R (Fig. 5J). These results suggest that USP38 promotes degradation of IL-33R through the autophagy–lysosome pathway.
TRAF6 Mediates K27-Linked Polyubiquitination of IL-33RK511.
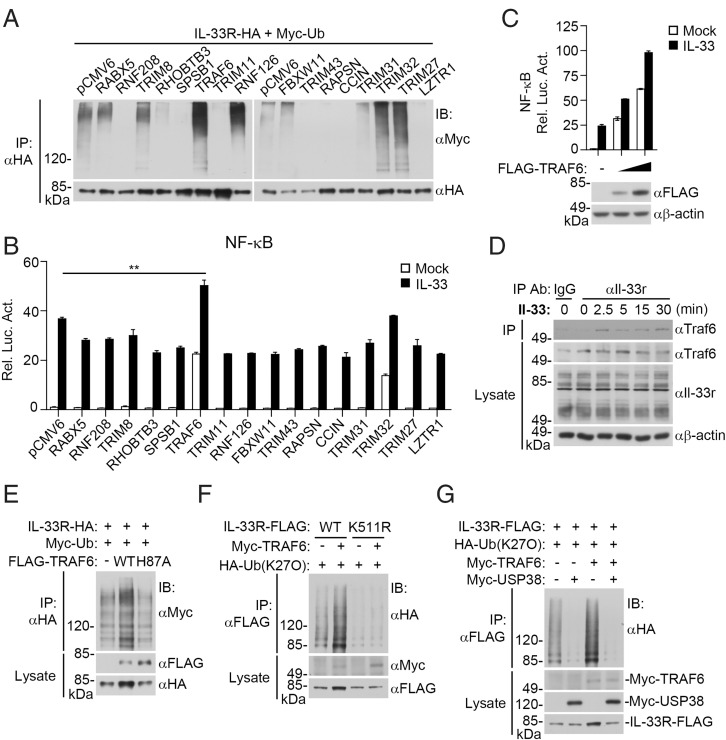
We next attempted to identify the E3 ubiquitin ligase that is responsible for mediating K27-linked polyubiquitination of IL-33RK511. In the initial cotransfection screens as described in Fig. 1A, we identified 16 ubiquitin-related enzymes that could increase the level of IL-33R. We further examined their abilities to promote IL-33R polyubiquitination. The results indicated that TRIM32, TRIM27, TRAF6, and RNF126 increased IL-33R polyubiquitination (Fig. 6A). In reporter assays, TRAF6 but not TRIM32, TRIM27, or RNF126 markedly promoted IL-33–triggered activation of NF-κB (Fig. 6B), and this effect was dose dependent (Fig. 6C). Endogenous coimmunoprecipitation experiments indicated that Traf6 was associated with Il-33r after Il-33 stimulation (Fig. 6D). In mammalian overexpression systems, TRAF6 but not its inactive H87A mutant increased K27-linked polyubiquitination of IL-33R and up-regulated its protein level (Fig. 6E). TRAF6 catalyzed K27-linked polyubiquitination of wild-type IL-33R but not of IL-33RK511R (Fig. 6F). Further experiments indicated that USP38 removed K27-linked polyubiquitination of IL-33R catalyzed by TRAF6 (Fig. 6G). These results suggest that TRAF6 mediates K27-linked polyubiquitination of IL-33RK511.
Fig. 6.
TRAF6 mediates K27-linked polyubiquitination of IL-33RK511 and its activation. (A) Screens of ubiquitin-related proteins for their effects on polyubiquitination of IL-33R. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 18 h before coimmunoprecipitation and immunoblotting analysis with the indicated antibodies. (B) Effects of ubiquitin-related proteins on IL-33–induced NF-κB activation. HEK293 IL-33R cells (6 × 104) were transfected with NF-κB reporter and the indicated ubiquitin-related protein expression plasmids for 18 h, and then, they were left untreated or treated with IL-33 (25 ng/mL) for 9 h before luciferase assays. Data shown are mean ± SEM. n = 3 from one representative experiment. **P < 0.01. (C) TRAF6 promotes IL-33–induced NF-κB activation in a dose-dependent manner. HEK293 IL-33R cells (6 × 104) were transfected with the NF-κB reporter (10 ng) and Flag-TRAF6 plasmid (0, 1, and 3 ng) for 18 h. The cells were then left untreated or treated with IL-33 (25 ng/mL) for 9 h before reporter assays and immunoblotting analysis with the indicated antibodies. (D) Endogenous Traf6 is associated with Il-33r. NIH 3T3 cells (2 × 107) were left untreated or treated with Il-33 (40 ng/mL) for the indicated time before coimmunoprecipitation and immunoblot analysis with the indicated antibodies. (E) TRAF6 but not its H87A mutant increases polyubiquitination of IL-33R. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 18 h before ubiquitination assays with the indicated antibodies. (F) TRAF6 mediates K27-linked polyubiquitination of IL-33R at K511. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 18 h before ubiquitination assays. (G) USP38 removes K27-linked polyubiquitin moieties from IL-33R conjugated by TRAF6. HEK293 cells (2 × 106) were transfected with the indicated plasmids for 18 h before coimmunoprecipitation and immunoblotting with the indicated antibodies. WT, wild type; IP, immunoprecipitation; IB, immunoblotting. All experiments were repeated at least two times with similar results.
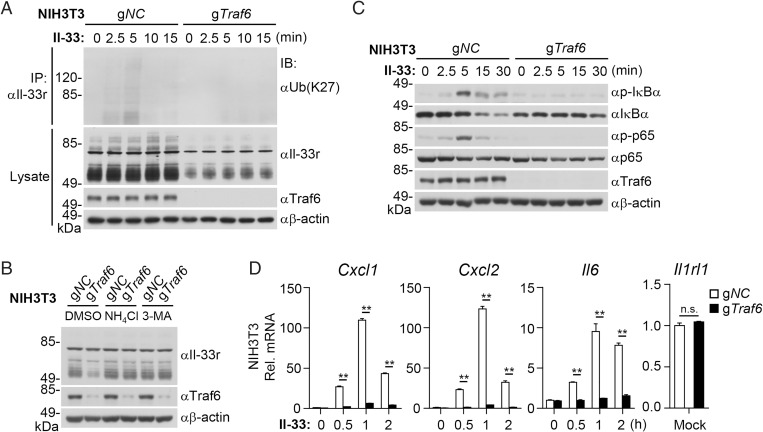
We next generated Traf6-deficient cells to investigate whether endogenous Traf6 catalyzes K27-linked polyubiquitination of Il-33r. We found that K27-linked polyubiquitination of endogenous Il-33r induced by Il-33 was inhibited in Traf6-deficient NIH 3T3 cells compared with wild-type cells (Fig. 7A). In these experiments, the protein levels of Il-33r in Traf6-deficient cells were also dramatically down-regulated compared with those in wild-type cells (Fig. 7A). In addition, treatment of 3-MA or NH4Cl reversed the down-regulation of Il-33r in Traf6-deficient cells (Fig. 7B). Consistently, Traf6 deficiency impaired Il-33–induced phosphorylation of IκBα and p65 (Fig. 7C) as well as transcription of Cxcl1, Cxcl2, and Il6 but not the Il1rl1 gene (Fig. 7D), suggesting that Traf6 is required for IL-33–triggered signaling and induction of downstream inflammatory genes.
Fig. 7.
Traf6 deficiency impairs Il-33–triggered K27-linked polyubiquitination of Il-33r and signaling. (A) Traf6 deficiency impairs Il-33–induced K27-linked polyubiquitination of endogenous Il-33r. The control (gNC) and Traf6-deficient (gTraf6) NIH 3T3 cells (2 × 107) were left untreated or treated with Il-33 (40 ng/mL) for the indicated time before ubiquitination assays with the indicated antibodies. (B) Effects of autophagy inhibitors on Il-33r levels in Traf6-deficient cells. The control (gNC) and Traf6-deficient (gTraf6) NIH 3T3 cells (2 × 107) were left untreated or treated with NH4Cl (5 mM) or 3-MA (100 ng/mL) for 14 h before immunoblotting analysis with the indicated antibodies. (C) Effects of Traf6 deficiency on Il-33–induced phosphorylation of IκBα and p65. The control and Traf6-deficient NIH 3T3 cells (1 × 106) were left untreated or treated with Il-33 (5 ng/mL) for the indicated time before immunoblot analysis with the indicated antibodies. (D) Effects of Traf6 deficiency on Il-33–induced transcription of downstream genes. The control and Traf6-deficient NIH 3T3 cells (1 × 106) were left untreated or treated with Il-33 (5 ng/mL) for the indicated time before qPCR analysis. Data shown are mean ± SEM. n = 3 from one representative experiment. DSMO, dimethyl sulfoxide; IB, immunoblotting; IP, immunoprecipitation. All experiments were repeated at least two times with similar results. n.s., not significant. **P < 0.01.
Discussion
The IL-33/IL-33R axis plays important roles in the pathogenesis of immune system–related disorders, such as pulmonary fibrosis, septic lung injury, asthma, and rheumatoid arthritis (8, 11, 13). How this axis is regulated at the receptor level remains enigmatic. It has been previously reported that the E3 ubiquitin ligase FBXL19 mediates polyubiquitination of IL-33R and negatively regulates IL-33–triggered inflammatory response (23). However, further studies are needed to elucidate the regulatory mechanisms of IL-33R. In this study, we found that the deubiquitinating enzyme USP38 was constitutively associated with IL-33R, resulting in its deubiquitination and autophagic degradation. Upon IL-33 stimulation, the E3 ubiquitin ligase TRAF6 is increasingly recruited to IL-33R and mediates K27-linked polyubiquitination of IL-33RK511, which promotes the stability and signaling of IL-33R. Our findings suggest that the level and activity of IL-33R are reciprocally regulated by polyubiquitination and deubiquitination mediated by TRAF6 and USP38, and this mechanism contributes to regulation of IL-33–triggered inflammatory response and pulmonary fibrosis.
Our results support a negative regulatory role of USP38 in IL-33–triggered signaling. Usp38 deficiency potentiated Il-33–triggered phosphorylation of IκBα and p65 as well as transcription of Cxcl1, Cxcl2, and Il6 genes, it but had no marked effects on Il-1β–induced signaling events and downstream gene transcription. Our results showed that Usp38-deficient mice displayed aggravated lung inflammatory response, such as increased eosinophilia and IL-5 levels in the lung following recombinant Il-33 treatment. It has been shown that Il-33r is critically involved in lung fibrotic pathogenesis and plays a critical role in bleomycin-triggered lung fibrosis (11). Our results indicated that Usp38-deficient mice were more susceptible to bleomycin-induced lethality and had greatly increased transcription of lung inflammatory and fibrosis-related genes in comparison with wild-type mice. Usp38-deficient mice also displayed aggravated inflammatory cell infiltration and collagen deposition as well as increased fibrotic marker α-SMA in the lung in comparison with their wild-type littermates. Treatment of Il-33r neutralizing antibody attenuated bleomycin-induced airway inflammatory damages in the lungs of both wild-type mice and Usp38−/− mice. These results suggest that Usp38 negatively regulates Il-33–triggered signaling and inflammatory response.
Our results indicated that overexpression of USP38 promoted degradation of IL-33R, whereas USP38 deficiency had opposite effects. Coimmunoprecipitation experiments indicated that USP38 was constitutively associated with IL-33R. The association was decreased at early time points after Il-33 stimulation and then gradually restored to the level in unstimulated cells, which is correlated with the activation of Il-33r. Overexpression of USP38 but not its inactive mutant USP38C454A removed K27-linked but not other types of polyubiquitin moieties from IL-33R. Consistently, USP38 deficiency increased basal as well as IL-33–induced K27-linked polyubiquitination of IL-33R and up-regulated the level of IL-33R either before or after IL-33 stimulation. Mutation of IL-33R K511 (Il-33r K516) to arginine impaired its K27-linked polyubiquitination and ability to activate NF-κB. IL-33RK511R had a shorter intracellular half-life than wild-type IL-33R. In addition, reconstitution of wild-type Il-33r but not Il-33rK516R rescued Il-33–triggered induction of downstream genes in Il-33r–deficient NIH 3T3 cells. These results suggest that USP38 mediates deubiquitination of IL-33RK511 (Il-33rK516), which promotes its down-regulation.
Our data also suggest that USP38 down-regulates IL-33R via the autophagy–lysosome pathway. The autophagy inhibitor 3-MA and lysosome inhibitor NH4Cl but not the ubiquitin–proteasome inhibitor MG132 inhibited USP38-mediated degradation of IL-33R. Association of Lc3b with Il-33r was mainly in the cytoplasm after Il-33 stimulation, and the association of Lc3b with Il-33rK516R was stronger than that of wild-type Il-33r. In addition, USP38 promoted the association of IL-33R with LC3B-II, which is a critical component in the formation of autophagosomes (34). Knockdown of LC3B inhibited USP38-mediated degradation of IL-33R. We also found that the K27-linked polyubiquitination of IL-33RK511R was impaired, and consistently, this mutant was down-regulated compared with wild-type IL-33R and could not be further down-regulated by USP38. These results suggest that deconjugation of K27-linked polyubiquitination of IL-33RK511 by USP38 leads to its autophagic degradation, thus negatively regulating IL-33–triggered signaling and inflammatory response.
Using an expression screen approach, we found that the E3 ubiquitin ligase TRAF6 was responsible for mediating K27-linked polyubiquitination of IL-33RK511. Endogenous coimmunoprecipitation experiments indicated that TRAF6 was weakly associated with IL-33R in rest cells, and this association was enhanced after IL-33 stimulation. Wild-type TRAF6 but not its inactive H87A mutant increased K27-linked polyubiquitination of IL-33RK511 as well as its stability. Deficiency of Traf6 impaired K27-linked polyubiquitination of Il-33r and caused its down-regulation in rest as well as Il-33–stimulated cells. Deficiency of Traf6 also impaired Il-33–triggered activation of downstream components as well as induction of downstream effector genes. Additionally, mutation of IL-33R K511 to arginine abolished its ability to mediate IL-33–triggered signaling. These results suggest that TRAF6 mediates K27-linked polyubiquitination of IL-33RK511, which is enhanced following IL-33 stimulation. K27-linked polyubiquitination of IL-33R promotes its stability by preventing autophagic degradation, leading to efficient onset of IL-33R–mediated signaling. Consistently, IL-33RK511R (or Il-33RK516R) was expressed at a much lower level than its wild-type counterpart, consistent with our conclusion that loss of K27-linked polyubiquitination of IL-33R promotes its autophagic degradation.
Based on our results, we propose a model on how IL-33R is reciprocally regulated by USP38 and TRAF6. In rest cells, TRAF6 is weakly associated with IL-33R, leading to a weak K27-linked polyubiquitination of IL-33RK511, whereas USP38 is strongly associated with IL-33R to deconjugate K27-linked polyubiquitination of IL-33RK511, leading to degradation of IL-33R by the autophagic pathway. Upon IL-33 stimulation, TRAF6 is increasingly recruited to IL-33R, causing increased K27-linked polyubiquitination of IL-33RK511 and promoting the stability of IL-33R by inhibiting its autophagic degradation, which leads to activation of IL-33R–mediated signaling. Our findings reveal an important mechanism on how IL-33R is precisely regulated to ensure its inactivation in rest cells and proper activation following IL-33 stimulation. Our study also suggests that USP38 and TRAF6 may serve as potential targets for the development of drugs for IL-33–triggered inflammatory diseases and fibrosis.
Materials and Methods
All animal experiments were performed in accordance with the Wuhan University Animal Care and Use Committee guidelines. The information on reagents, antibodies, cells, constructs, mice, and qPCR primers is described in SI Appendix, SI Materials and Methods. The methods for cell lines and CRISPR-Cas9 knockout, transfection, reporter assays, coimmunoprecipitation, immunoblotting analysis, ubiquitination assays, histology, bleomycin-induced pulmonary fibrosis, and statistical analysis are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Tian Xia, Heng Lin, Ru Zang, Deng Gao, Lu Feng, and other members of our laboratory for technical help and discussions. This work was supported by National Key R&D Program of China Grant 2017YFA0505800; National Natural Science Foundation of China Grants 31830024, 32188101, 31871415, and 32070775; Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences Grant 2019-I2M-5-071; and the Fundamental Research Funds for the Central Universities.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.-P.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2116279119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Liew F. Y., Girard J. P., Turnquist H. R., Interleukin-33 in health and disease. Nat. Rev. Immunol. 16, 676–689 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A., Dinarello C. A., Molgora M., Garlanda C., Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity 50, 778–795 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cayrol C., Girard J. P., Interleukin-33 (IL-33): A nuclear cytokine from the IL-1 family. Immunol. Rev. 281, 154–168 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Moussion C., Ortega N., Girard J. P., The IL-1-like cytokine IL-33 is constitutively expressed in the nucleus of endothelial cells and epithelial cells in vivo: A novel ‘alarmin’? PLoS One 3, e3331 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin N. T., Martin M. U., Interleukin 33 is a guardian of barriers and a local alarmin. Nat. Immunol. 17, 122–131 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Barnes P. J., Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 138, 16–27 (2016). [DOI] [PubMed] [Google Scholar]
- 7.McHedlidze T., et al. , Interleukin-33-dependent innate lymphoid cells mediate hepatic fibrosis. Immunity 39, 357–371 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z., Bozec A., Ramming A., Schett G., Anti-inflammatory and immune-regulatory cytokines in rheumatoid arthritis. Nat. Rev. Rheumatol. 15, 9–17 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Luzina I. G., et al. , Interleukin-33 potentiates bleomycin-induced lung injury. Am. J. Respir. Cell Mol. Biol. 49, 999–1008 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Borthwick L. A., The IL-1 cytokine family and its role in inflammation and fibrosis in the lung. Semin. Immunopathol. 38, 517–534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li D., et al. , IL-33 promotes ST2-dependent lung fibrosis by the induction of alternatively activated macrophages and innate lymphoid cells in mice. J. Allergy Clin. Immunol. 134, 1422–1432.e11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., et al. , Anti-IL-33 antibody treatment inhibits airway inflammation in a murine model of allergic asthma. Biochem. Biophys. Res. Commun. 386, 181–185 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Schmitz J., et al. , IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity 23, 479–490 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Lott J. M., Sumpter T. L., Turnquist H. R., New dog and new tricks: Evolving roles for IL-33 in type 2 immunity. J. Leukoc. Biol. 97, 1037–1048 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Conze D. B., Wu C. J., Thomas J. A., Landstrom A., Ashwell J. D., Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-kappaB activation. Mol. Cell. Biol. 28, 3538–3547 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ali S., et al. , IL-1 receptor accessory protein is essential for IL-33-induced activation of T lymphocytes and mast cells. Proc. Natl. Acad. Sci. U.S.A. 104, 18660–18665 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molofsky A. B., Savage A. K., Locksley R. M., Interleukin-33 in tissue homeostasis, injury, and inflammation. Immunity 42, 1005–1019 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayakawa H., Hayakawa M., Kume A., Tominaga S., Soluble ST2 blocks interleukin-33 signaling in allergic airway inflammation. J. Biol. Chem. 282, 26369–26380 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Bulek K., et al. , The essential role of single Ig IL-1 receptor-related molecule/Toll IL-1R8 in regulation of Th2 immune response. J. Immunol. 182, 2601–2609 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin H., et al. , MARCH3 attenuates IL-1β-triggered inflammation by mediating K48-linked polyubiquitination and degradation of IL-1RI. Proc. Natl. Acad. Sci. U.S.A. 115, 12483–12488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen R., Li M., Zhang Y., Zhou Q., Shu H. B., The E3 ubiquitin ligase MARCH8 negatively regulates IL-1β-induced NF-κB activation by targeting the IL1RAP coreceptor for ubiquitination and degradation. Proc. Natl. Acad. Sci. U.S.A. 109, 14128–14133 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin H., Li S., Shu H. B., The membrane-associated MARCH E3 ligase family: Emerging roles in immune regulation. Front. Immunol. 10, 1751 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J., et al. , F-box protein FBXL19-mediated ubiquitination and degradation of the receptor for IL-33 limits pulmonary inflammation. Nat. Immunol. 13, 651–658 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clague M. J., Urbé S., Komander D., Breaking the chains: Deubiquitylating enzyme specificity begets function. Nat. Rev. Mol. Cell Biol. 20, 338–352 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Py B. F., Kim M. S., Vakifahmetoglu-Norberg H., Yuan J., Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell 49, 331–338 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Zhang L., et al. , Upregulation of deubiquitinase PSMD14 in lung adenocarcinoma (LUAD) and its prognostic significance. J. Cancer 11, 2962–2971 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X., et al. , Regulation of TRIF-mediated innate immune response by K27-linked polyubiquitination and deubiquitination. Nat. Commun. 10, 4115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin M., et al. , USP38 inhibits type I interferon signaling by editing TBK1 ubiquitination through NLRP4 signalosome. Mol. Cell 64, 267–281 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Zhan W., et al. , USP38 regulates the stemness and chemoresistance of human colorectal cancer via regulation of HDAC3. Oncogenesis 9, 48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shalem O., et al. , Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mouratis M. A., Aidinis V., Modeling pulmonary fibrosis with bleomycin. Curr. Opin. Pulm. Med. 17, 355–361 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Wynn T. A., Ramalingam T. R., Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 18, 1028–1040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Stubbe J., Bleomycins: Towards better therapeutics. Nat. Rev. Cancer 5, 102–112 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Kuma A., Matsui M., Mizushima N., LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: Caution in the interpretation of LC3 localization. Autophagy 3, 323–328 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.