Abstract
When the lethal action of a C-8 methoxyl fluoroquinolone against clinical isolates of Mycobacterium tuberculosis in liquid medium was measured, the compound was found to be three to four times more effective (as determined by measuring the 90% lethal dose) than a C-8-H control fluoroquinolone or ciprofloxacin against cells having a wild-type gyrA (gyrase) gene. Against ciprofloxacin-resistant strains, the C-8 methoxyl group enhanced lethality when alanine was replaced by valine at position 90 of the GyrA protein or when aspartic acid 94 was replaced by glycine, histidine, or tyrosine. During infection of a human macrophage model by wild-type Mycobacterium bovis BCG, the C-8 methoxyl group lowered survival 20- to 100-fold compared with the same concentration of a C-8-H fluoroquinolone. The C-8 methoxyl fluoroquinolone was also more effective than ciprofloxacin against a gyrA Asn94 mutant of M. bovis BCG. In an M. tuberculosis-macrophage system the C-8 methoxyl group improved fluoroquinolone action against both quinolone-susceptible and quinolone-resistant clinical isolates. Thus, a C-8 methoxyl group enhances the bactericidal activity of quinolones with N1-cyclopropyl substitutions; these data encourage further refinement of fluoroquinolones as antituberculosis agents.
The resurgence of tuberculosis in New York City during the early 1990s was accompanied by an alarming incidence of antibiotic resistance in the causative organism, Mycobacterium tuberculosis: isolates that were resistant to as many as seven different agents emerged (10). Among the resistant strains were the so-called W isolates, a group of clonal isolates in which the genomic distribution of IS6110 is the same (2). Disease caused by these resistant strains was particularly difficult to treat when patients were coinfected with human immunodeficiency virus type 1 (12). After a few years, the New York City outbreak was suppressed, largely by improved institutional infection control and by the use of directly observed therapy that ensured patient compliance with antibiotic regimens (9). However, the cost of the outbreak was considerable (3, 9). Moreover, W-type isolates appear to have spread to other cities (1, 2) where treatment failures still occur (1). This experience left us with the conviction that the arsenal of antituberculosis agents is too small.
We have focused our effort on the fluoroquinolones. In bacteria other than mycobacteria, these compounds attack DNA gyrase and the related enzyme DNA topoisomerase IV (reviewed in reference 8); it is likely that gyrase is also the target in mycobacteria, since first-step mutations obtained with laboratory strains of M. tuberculosis map in gyrA and gyrB, the two genes encoding gyrase (17, 22). Moreover, the general features of quinolone action are similar in mycobacteria and Escherichia coli (7), most clinical strains that are resistant to fluoroquinolones carry a mutation in gyrA (6, 21), and gyrase isolated from a resistant strain of Mycobacterium smegmatis is resistant to fluoroquinolones in vitro (4). Thus, knowledge gained from studies with other bacteria can be used to seek more effective antimycobacterial fluoroquinolones.
Two relevant observations were made recently. First, intracellular action of the quinolones occurs as two distinct steps: bacteriostatic drug-gyrase-DNA complexes form, and then lethal double-strand DNA breaks are released from the complexes (5). This distinction between blocking growth and killing cells is potentially important because quinolone treatment induces the SOS response (8), which is modestly mutagenic (19a). If induced mutants are not killed before the resistant form of the target topoisomerase is fully expressed, the quinolone treatment might contribute to the formation of resistant cells. Thus, quinolone potency assessment should include direct measurement of lethal activity, which is not always predictable from standard bacteriostasis assays (27). The second observation is that substituents at the C-8 position increase fluoroquinolone potency, especially against first-step gyrase and topoisomerase IV resistance mutants (6, 13, 15, 16, 27, 28). This observation led to the finding that fewer resistant mutants are selected when bacteria are challenged with C-8 methoxyl (C8-OMe) fluoroquinolones than with C-8-hydrogen (C8-H) derivatives (6, 27). The next step is to determine whether C8-OMe fluoroquinolones are also particularly lethal for M. tuberculosis and its gyrA mutants.
In the present study we first tested the expectation that a C8-OMe fluoroquinolone is more bactericidal than its C8-H control or the clinical standard ciprofloxacin against gyrA+ and gyrA resistance mutants of M. tuberculosis growing in liquid medium. This was the case. We then examined fluoroquinolone susceptibility of Mycobacterium bovis bacillus Calmette-Guérin (BCG) and M. tuberculosis during infection of the human monocytic cell line THP1 following induced differentiation into macrophage-like cells (23). Susceptibility paralleled results obtained with cultures grown in liquid medium. Thus, C8-OMe fluoroquinolones are likely to be much more effective antituberculosis agents than currently used compounds, such as ciprofloxacin, particularly because their activity against gyrA mutants will decrease the outgrowth of resistant strains that arise during treatment.
MATERIALS AND METHODS
Bacterial strains and culture methods.
Bacterial strains used in this study are listed in Table 1. Strains of M. tuberculosis were clinical isolates obtained from the Public Health Research Institute (PHRI) Tuberculosis Center. M. bovis BCG (substrain Pasteur), isolate KD1295, was obtained from Mounsef Tiza, PHRI. Seed lots of M. tuberculosis and M. bovis BCG were prepared in the following way. Aliquots (1 ml) were removed from frozen cultures (−70°C), thawed at room temperature, and inoculated into plastic bottles containing 50 ml of 7H9 liquid medium (Middlebrook 7H9; Difco Laboratories, Detroit, Mich.) with 10% albumin-dextrose complex and 0.05% Tween 80. These cultures were incubated at 37°C with rolling (Low Profile Roller; Stovall Life Sciences, Greensboro, N.C.) until stationary phase was reached, as determined by measuring culture turbidity (A600). Dimethyl sulfoxide or bovine calf serum was added to a final concentration of 10%, and 0.5- or 1-ml aliquots were frozen at −70°C. For each experiment a frozen aliquot was thawed, diluted at least 100-fold, and grown as described above to exponential phase (determined by culture turbidity and confirmed by counting colonies that grew on 7H10 agar). All experiments with M. tuberculosis were carried out in a biosafety level 3 containment facility.
TABLE 1.
Bacterial strains
| Strain | IS6110 typea | Mutation in GyrAb | Drug(s) to which the strain is resistantc |
|---|---|---|---|
| M. tuberculosis | |||
| TN913 | C | None (WT) | None |
| TN1626 | W | None (WT) | I, S, R, EM, KN |
| TN1625 | W | A90V | I, S, R, EM, KN, C, ET, CM |
| TN1627 | W | D94Y | I, S, R, EM, KN, C |
| TN565 | W | D94H | I, S, R, EM, KN, C, ET |
| TN606 | W | D94G | I, S, R, EM, C |
| M. bovis BCG | |||
| KD1295 | None (WT) | ||
| CX1d | D94N | C |
RFLP DNA pattern as described in reference 24.
The genetic mutations specifying the amino acid changes occur in the quinolone resistance-determining region of gyrA (26). WT, wild type.
Abbreviations: I, isoniazid; S, streptomycin; R, rifampin; EM, ethambutol; C, ciprofloxacin; KN, kanamycin; ET, ethionamide; CM, capreomycin.
First-step spontaneous gyrA mutant of KD1295 (7).
Fluoroquinolone action against M. tuberculosis grown in liquid media.
Ciprofloxacin was a product of Miles Laboratories, and compounds PD161148 (C8-OMe) and PD160793 (C8-H) were obtained from Parke-Davis Pharmaceutical Co. The fluoroquinolones were prepared as solutions of 10 mg/ml in 0.1 N NaOH and were stored at −80°C. Dilutions were prepared in 7H9 or RPMI 1640 medium immediately before use.
M. tuberculosis isolates were grown to exponential phase by rolling-culture incubation (about 106 to 107 CFU/ml) and distributed as 3-ml aliquots in glass screw-cap tubes. Fluoroquinolones were added to various concentrations, and bacterial cultures were incubated for 5 days with rolling (the incubation time, which from earlier work [7] was expected to allow moderate survival at a moderate fluoroquinolone concentration, was chosen to parallel the macrophage studies described below). Serial dilutions were plated on 7H10 agar lacking drugs; colonies were counted after incubation of agar plates for 4 to 5 weeks at 37°C.
Fluoroquinolone action against mycobacteria in human macrophages.
Cells of human monocytic cell line THP1 were cultured in RPMI 1640 medium supplemented with 10% bovine calf serum (HyClone Laboratories, Logan, Utah) lacking antibiotics. THP1 cells (2.5 × 105 cells in 0.5 ml) were seeded into the wells of 24-well tissue culture plates. Each well contained a 12-mm-diameter glass coverslip. After incubation at 37°C for 24 h, phorbol 12-myristate 13-acetate (PMA; Sigma Chemical Co., St. Louis, Mo.) was added to each well at a final concentration of 100 nM. This treatment caused the cells to differentiate into adherent, macrophage-like cells (23). Cells were incubated overnight, and the growth medium was replaced by 0.5 ml of fresh RPMI 1640 medium containing 10% bovine calf serum or 20% human serum (Sigma Chemical Co.) lacking PMA. After one additional hour of incubation, mycobacteria were added as described below.
Prior to infection of macrophages, mycobacteria (5 ml) growing exponentially were harvested by centrifugation at 5,000 × g for 15 min, washed once with 5 ml of phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, 1 mM MgCl2), and resuspended in 5 ml of PBS. One milliliter of bacterial suspension was transferred to a 15-ml plastic tube that was then immersed in water and sonicated at room temperature for 15 s (model 2210 ultrasonic cleaner; Bransonic, Danbury, Conn.) to disperse bacterial clumps. Microscopic examination for acid-fast-stained bacteria revealed that sonication broke most of the bacterial clumps into single cells (a few small clumps of 2 to 5 bacteria were detected; clumps containing 10 or more cells were rare). Mycobacteria (1 × 105 to 5 × 105 CFU) were added to 2.5 × 105 to 5 × 105 differentiated THP1 cells per well present on coverslips in tissue culture plates. Plates were incubated for 3 to 4 h, and extracellular bacteria were removed by three washes with warm (37°C) PBS (1 ml was added to each well and then removed by aspiration). Fresh growth medium (0.5 ml) was then added to the infected monolayers, and they were cultured for 2 or 5 days with various concentrations of fluoroquinolone (from earlier work with M. bovis BCG, 2 days was expected to be an early kinetic point and 5 days was expected to be a moderately long one but short enough to avoid problems associated with macrophage death, which arose at about 7 to 8 days). Then medium was removed, monolayers were lysed by treatment with 0.5% sodium dodecyl sulfate, serial dilutions were prepared, and 50-μl aliquots were plated onto 7H10 agar. The numbers of CFU were determined after 4 to 5 weeks of incubation at 37°C. In preliminary control experiments, removal of fluoroquinolones by washing cells prior to plating had no effect on the number of colonies detected, probably because dilution was sufficient to eliminate carryover effects. Thus, in the experiments reported, aliquots were plated directly after dilution.
The extent of infection and numbers of intracellular bacilli per macrophage were determined by light microscopy of control cultures that were not treated with fluoroquinolone. Glass coverslips to which infected THP1 cells were attached were treated with 10% formaldehyde for 20 min to kill mycobacteria, and then cells were stained with acid-fast stain (Difco). About 10 to 30% of adhered THP1 cells were found to be infected, and the number of bacteria per cell ranged from 1 to 10. At the end of the 5-day incubation period 85% of the macrophages remained intact and attached to coverslips; only 10% of the bacteria present in cells had been released into the medium. Thus, cell lysis occurring during the incubation period had little effect on the interpretation of the data.
RESULTS
Lethal action of fluoroquinolones against M. tuberculosis grown in liquid medium.
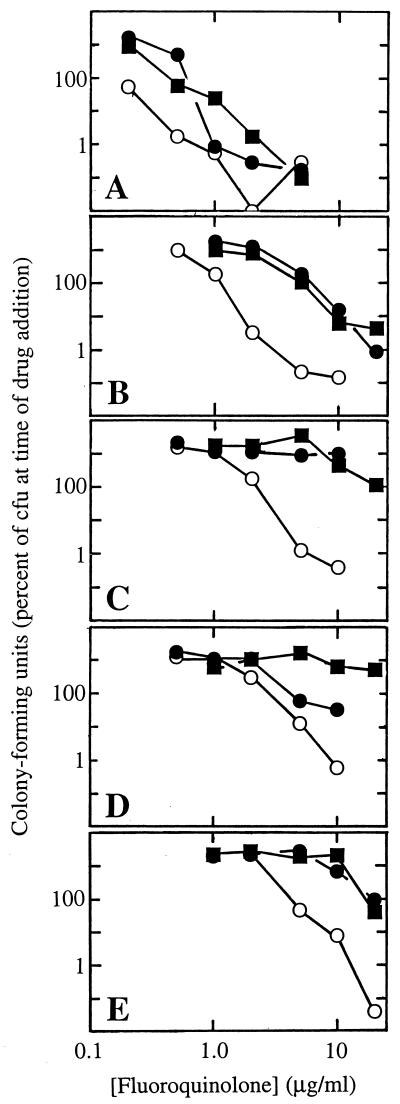
Work with E. coli (27), Staphylococcus aureus (13, 28), and M. bovis BCG (6) showed that a C8-OMe group makes fluoroquinolones more lethal, especially to moderately resistant gyrA mutants or, in the case of S. aureus, to parC mutants. To determine whether this is also true for clinical isolates of M. tuberculosis, we compared a C8-OMe compound (PD161148) with its C8-H control (PD160793) and ciprofloxacin (structures are shown in Fig. 1). Cultures were grown to mid-log phase and then incubated for 5 days with various concentrations of fluoroquinolone. The CFU of surviving bacteria were then counted. At low fluoroquinolone concentrations the cultures continued to grow, and so differences in cell number between effective and ineffective concentrations or compounds were sometimes many orders of magnitude (Fig. 2). The C8-OMe fluoroquinolone was more lethal than the other two compounds to the gyrA+ isolate TN1626 (Fig. 2A). This result was quantified by comparing the concentrations required to kill a given fraction of the population. For example, the doses required to kill 90% of the cells (LD90s of TN1626 were 0.38, 0.8, and 1.3 μg/ml for PD161148 (C8-OMe), PD160793 (C8-H), and ciprofloxacin, respectively. The ciprofloxacin-resistant isolate TN1625, which contained a valine substitution for alanine at position 90 of GyrA, was also more susceptible to the C8-OMe compound (LD90s were 1.5, 9, and 11 μg/ml for PD161148, PD160793, and ciprofloxacin, respectively [Fig. 2B]). Three other resistant strains, which had substitutions of glycine, histidine, or tyrosine for aspartic acid at position 94 of GyrA, were also killed effectively by the C8-OMe compound, although higher concentrations were required (LD90s were 3.2, 5.6, and 10 μg/ml, respectively [Fig. 2C to E]). PD160793 (C8-H) and ciprofloxacin showed less activity against these strains. Thus, while a C8-OMe group increased fluoroquinolone lethality to wild-type and ciprofloxacin-resistant M. tuberculosis in general, the mutations at position 94 conferred resistance to higher doses than a mutation at position 90. These amino acid differences are consistent with an earlier report in which ciprofloxacin displayed more bacteriostatic activity against the mutant with a substitution at position 90 than against the mutants with substitutions at position 94 (25).
FIG. 1.
Fluoroquinolone structures. The arrow indicates the C-8 position.
FIG. 2.
Lethal action of fluoroquinolones against clinical isolates of M. tuberculosis. Isolates obtained from the PHRI collection were grown to exponential phase as described in Materials and Methods. Fluoroquinolones were then added to the indicated concentrations, and incubation was continued for 5 days. Then cultures were diluted and plated for counting of CFU, which are expressed as percentages of the numbers of CFU at the time of drug addition. (A) Strain TN1626 (gyrA+); (B) strain TN1625 (containing A90V resistant gyrase); (C) strain TN606 (containing D94G resistant gyrase); (D) strain TN565 (containing D94H resistant gyrase); (E) strain TN1627 (containing D94Y resistant gyrase). Symbols: open circles, PD161148 (C8-OMe); solid circles, PD160793 (C8-H); solid squares, ciprofloxacin. Similar results were obtained in a replicate experiment.
Fluoroquinolone activity against M. bovis BCG in human macrophages.
We next examined the activities of fluoroquinolones in a human macrophage model infected with M. bovis BCG. For the wild-type bacterial strain, survival in the presence of the C8-OMe compound was 20 to 100 times lower than in the presence of the C8-H control or ciprofloxacin at 2 μg/ml (Table 2). With the gyrA Cipr mutant, the C8-OMe compound saturated at the lowest concentration tested while the two C8-H compounds allowed four to six times more survival at the highest concentration (Table 2). Similar results were obtained in two replicate experiments (data not shown). Thus, a C8-OMe group improves fluoroquinolone action against M. bovis BCG in macrophages.
TABLE 2.
Results for M. bovis BCG treated with fluoroquinolones while inside human macrophages
| Strain | Fluoroquinolone | Concn (μg/ml) | No. of CFU ata:
|
Survival (% of that on day 0) | |
|---|---|---|---|---|---|
| Day 0 | Day 2 postinfection | ||||
| KD1295 (WT) | None | 0 | 3.7 × 104 | 2.2 × 104 | 59.4 |
| PD161148 (C8-OMe) | 2 | 4.0 × 101 | 0.11 | ||
| 6 | 0 | 0 | |||
| 10 | 0 | 0 | |||
| PD160793 (C8-H) | 2 | 7.6 × 102 | 2.0 | ||
| 6 | 3.3 × 102 | 0.89 | |||
| 10 | 0 | 0 | |||
| Ciprofloxacin | 2 | 6.0 × 103 | 16 | ||
| 6 | 1.5 × 102 | 0.41 | |||
| 10 | 0 | 0 | |||
| CX1 (Cipr) | None | 0 | 6.3 × 104 | 2.0 × 104 | 32 |
| PD161148 (C8-OMe) | 2 | 2.6 × 103 | 4 | ||
| 6 | 2.5 × 103 | 4 | |||
| 10 | 2.8 × 103 | 4 | |||
| PD160793 (C8-H) | 2 | 8.1 × 104 | 130 | ||
| 6 | 4.2 × 104 | 67 | |||
| 10 | 1.5 × 104 | 24 | |||
| Ciprofloxacin | 2 | 3.5 × 104 | 55 | ||
| 6 | 3.7 × 104 | 59 | |||
| 10 | 1.0 × 104 | 16 | |||
Values are means for two wells. Similar results were obtained in two replicate experiments.
In each of three experiments with M. bovis BCG there was little or no net bacterial growth during the 2 days of infection; consequently, the fluoroquinolone experiments described above addressed only lethal activity. Control experiments with longer incubation times showed that from day 2 to 5 the number of bacteria increased 10- to 20-fold (data not shown). Thus, these experiments were performed during lag phase (a lag in growth during infection of macrophages has also been observed with M. avium [14]). We conclude that exponential growth is not required to observe increased lethality arising from a C8-OMe group.
Fluoroquinolone activity against M. tuberculosis in human macrophages.
For examination of M. tuberculosis, bacteria were recovered after 5 days of incubation, which was enough time to allow untreated controls to increase at least 20-fold. The data were normalized in two ways. Compared to the data for a no-drug control measured at day 0, the data represent a minimum measure of bacterial killing. Compared to the data for the control measured at day 5, the data reflect relative growth. As expected, the gyrA mutations reduced susceptibility to all compounds (Table 3). Against the two gyrA+ strains TN913 and TN1626, the effect of the C8-OMe fluoroquinolone differed little from that of its C8-H control, but the C8-OMe fluoroquinolone was much more effective than the control compound against the Asp94-Gly and Asp94-His ciprofloxacin-resistant mutants (Table 3). Action by the C8-OMe compound against the Asp94-Tyr mutant TN1627 (Table 3) was seen only when culture growth was considered. With this strain the C8-H compound PD160793 showed little activity. In general, these results were consistent with the ability of the two compounds to kill cells grown in liquid medium (Fig. 2), although against wild-type cells (strain TN1626) the C8-H compound was more active than expected.
TABLE 3.
Survival of M. tuberculosis treated with fluoroquinolones while inside human macrophages
| Strain | Fluoroquinolone | Concn (μg/ml) | No. of CFU recovereda
|
Survival (% of that on day 0) | Growth (% of that on day 5)b | |
|---|---|---|---|---|---|---|
| Day 0 | Day 5 | |||||
| TN913 (gyrA+) | None | 0 | 8.3 × 104 | 1.6 × 106 | ++c | 100 |
| PD161148 (C8-OMe) | 2 | 3.5 × 103 | 4 | 0.2 | ||
| 4 | 5.4 × 103 | 6 | 0.3 | |||
| PD16073 (C8-H) | 2 | 2.9 × 103 | 3 | 0.2 | ||
| 4 | 5.2 × 103 | 6 | 0.3 | |||
| Ciprofloxacin | 2 | 1.4 × 105 | ++ | 8.8 | ||
| 4 | 4.9 × 104 | 59 | 3.1 | |||
| TN1626 (gyrA+) | None | 0 | 8.2 × 103 | 4.6 × 105 | ++ | 100 |
| PD161148 (C8-OMe) | 2 | 1.9 × 103 | 23 | 0.4 | ||
| 4 | 1.1 × 103 | 13 | 0.2 | |||
| 8 | 3.0 × 103 | 37 | 0.7 | |||
| PD160793 (C8-H) | 2 | 4.0 × 104 | ++ | 8.7 | ||
| 4 | 1.0 × 103 | 12 | 0.2 | |||
| 8 | 1.6 × 103 | 20 | 0.3 | |||
| Ciprofloxacin | 2 | 6.7 × 104 | ++ | 15 | ||
| 4 | 2.0 × 104 | ++ | 4.3 | |||
| 8 | 6.0 × 103 | 73 | 1.3 | |||
| TN606 (containing D94G-resistant gyrase) | None | 7 × 104 | 4.1 × 106 | ++ | 100 | |
| PD161148 (C8-OMe) | 2 | 5.3 × 105 | ++ | 13 | ||
| 4 | 5.3 × 104 | 76 | 1.3 | |||
| 8 | 8.7 × 103 | 12 | 0.2 | |||
| PD160793 (C8-H) | 2 | 4.0 × 106 | ++ | 98 | ||
| 4 | 1.9 × 106 | ++ | 46 | |||
| 8 | 8.0 × 105 | ++ | 20 | |||
| Ciprofloxacin | 2 | 2.0 × 106 | ++ | 49 | ||
| 4 | 1.6 × 106 | ++ | 39 | |||
| 8 | 1.0 × 106 | ++ | 24 | |||
| TN565 (containing D94H-resistant gyrase) | None | 0 | 1.3 × 105 | 4.4 × 106 | ++ | 100 |
| PD161148 (C8-OMe) | 2 | 4.1 × 105 | ++ | 9.3 | ||
| 4 | 4.8 × 105 | ++ | 11 | |||
| 8 | 2.2 × 104 | 17 | 0.5 | |||
| PD160793 (C8-H) | 2 | 1.5 × 106 | ++ | 34 | ||
| 4 | 1.5 × 106 | ++ | 34 | |||
| 8 | 7.5 × 105 | ++ | 17 | |||
| Ciprofloxacin | 2 | 1.2 × 106 | ++ | 27 | ||
| 4 | 1.57 × 106 | ++ | 36 | |||
| 8 | 1.84 × 106 | ++ | 42 | |||
| TN1627 (containing D94Y-resistant gyrase) | None | 0 | 2.3 × 104 | 1.3 × 106 | ++ | 100 |
| PD161148 (C8-OMe) | 2 | 5.3 × 105 | ++ | 41 | ||
| 4 | 1.6 × 105 | ++ | 12 | |||
| 8 | 8.9 × 104 | ++ | 6.8 | |||
| PD160793 (C8-H) | 2 | 7.6 × 105 | ++ | 58 | ||
| 4 | 6.2 × 105 | ++ | 48 | |||
| 8 | 5.0 × 105 | ++ | 38 | |||
| Ciprofloxacin | 2 | 4.0 × 105 | ++ | 31 | ||
| 4 | 9.6 × 105 | ++ | 74 | |||
| 8 | 5.5 × 105 | ++ | 42 | |||
Values are averages of two determinations. Similar results were obtained in two additional experiments.
Values below 100% indicate that growth was inhibited.
++, growth occurred but was not quantified, and so survival was not calculated.
Ciprofloxacin was clearly less potent. When the gyrA+ isolates were treated with this fluoroquinolone, 2 to 30 times more CFU were recovered than following treatment with the C8-OMe compound at the same dose, and ciprofloxacin had little bactericidal effect (Table 3). Differences could not be quantified with the ciprofloxacin-resistant isolates because ciprofloxacin showed no lethal action. In each case where growth was considered, PD161148 (C8-OMe) was found to be more active (Table 3). These were the results expected from comparisons made with cells grown in liquid medium (Fig. 2).
Comparison of gyrA+ strains TN913 and TN1626 was also of interest. TN913 is a member of a group exhibiting IS6110 restriction fragment length polymorphism (RFLP) pattern C. It represents the major type found in the New York City outbreak of the early 1990s. Members of this type are susceptible to all antituberculosis agents. TN1626 is a member of the group exhibiting RFLP type W, the major multidrug-resistant group in the outbreak. As shown in Table 3, strain TN913 exhibited a slightly lower level of survival than TN1626 when challenged with the new fluoroquinolones.
DISCUSSION
In the experiments described above, a C8-OMe group increased the bactericidal action of fluoroquinolones against clinical isolates of M. tuberculosis grown in culture or after infection of THP1-derived macrophages. This was true when gyrase was wild type (Fig. 2A) and when it contained mutations that were associated with clinical resistance to ciprofloxacin (Fig. 2B to E). These data help explain why the C8-OMe derivative allows fewer resistant mutants to arise in mycobacterial populations challenged with fluoroquinolones (6). Greater lethality to wild-type cells probably restricts SOS-dependent mutagenesis, greater lethality to preexisting and induced mutants makes their survival less likely, and greater bacteriostatic activity against mutants (6) lowers the fluoroquinolone concentration required to prevent outgrowth of surviving mutants. It is likely that increased lethality due to the C8-OMe group is also exerted against M. tuberculosis growing in the human monocytic cell line THP1 after PMA-induced differentiation (Table 3). However, our data for macrophages do not distinguish between direct killing of M. tuberculosis by the fluoroquinolones and enhancement of sensitivity to macrophage-based lethality.
The increased effectiveness of C8-OMe fluoroquinolones could arise in two ways. First, the C8-OMe moiety could affect factors such as uptake, efflux, and detoxification. Second, the C8-OMe group might confer greater specific activity against DNA gyrase. The two possibilities, which are not mutually exclusive, can be distinguished by comparing lethal action against a gyrA mutant after normalization to lethal action against wild-type cells, assuming that the two strains differ mainly at the gyrA locus. For the C8-OMe compound PD161148, the ratio of the LD90 for strain TN1625 (GyrAr) to that for strain TN1626 (gyrA+) was about 4 (1.5/0.38). For the C8-H compounds PD160793 and ciprofloxacin, the ratios were 11 (9/0.8) and 8 (11/1.3), respectively. The lower ratio for PD161148 suggests that the C8-OMe group increases specific activity against gyrase, a conclusion that should now be confirmed in vitro (15).
Some gyrA+ clinical isolates of M. tuberculosis appear to be more susceptible to fluoroquinolones than others. For example, isolate TN913, a representative of the strain most frequently recovered during the recent tuberculosis outbreak in New York City, is more susceptible to PD161148 (C8-OMe) than is TN1626, the gyrA+ representative of the multidrug-resistant W strains (Table 3). The group of isolates represented by TN913 (IS6110 RFLP pattern C [18]) has not been associated with antibiotic resistance (18). In contrast, isolates with the W IS6110 RFLP pattern are resistant to four first-line antituberculosis agents, and many members of the group have acquired additional markers, probably from failed therapy (Table 1). The reasons for these differences between gyrA+ strains are not known.
Fluoroquinolone concentrations used to kill M. tuberculosis inside human macrophages were similar to levels used previously with mouse macrophages (20), but they were higher than needed when the bacteria were grown in liquid culture. Since quinolones are not very effective against nongrowing cells (reviewed in reference 8), a requirement for high concentration would be expected if the bacterial population in macrophages did not grow when exposed to fluoroquinolone. Quinolones differ in their abilities to kill nongrowing cells, so it may be possible to find derivatives that are especially effective in macrophage culture. It is less likely that the fluoroquinolones were taken up poorly by THP1 cells, since mammalian cells tend to concentrate fluoroquinolones (11, 19). Nor is there reason to believe that the greater potency of the C8-OMe compound arose from increased accumulation by macrophages, since parallel results were obtained in broth culture. Regardless of the reasons for the differences in fluoroquinolone susceptibility between mycobacteria grown in liquid culture and those grown in macrophages, it is clear that PD161148 (C8-OMe) is considerably more effective than ciprofloxacin, a compound already in clinical use. Thus, C8-OMe fluoroquinolones may be useful therapeutically against M. tuberculosis.
ACKNOWLEDGMENTS
We thank M. Gennaro, S. Kayman, B. Kreiswirth, and X. Zhao for critical comments on the manuscript.
This work was supported by grants AI35257 and AI37877 from the National Institutes of Health.
REFERENCES
- 1.Agerton T, Valway S, Gore B, Pozsik C, Plikaytis B, Woodley C, Onorato I. Transmission of a highly drug-resistant strain (strain W1) of Mycobacterium tuberculosis. JAMA. 1997;278:1073–1077. [PubMed] [Google Scholar]
- 2.Bifani P, Plikaytis B B, Kapur V, Stockbauer K, Pan X, Lusfty M, Moghazeh S, Eisner W, Daniel T, Kaplan M, Crawford J T, Musser J M, Kreiswirth B N. Origin and interstate spread of a New York City multidrug resistant Mycobacterium tuberculosis clone family: adverse implications for tuberculosis control in the 21st century. JAMA. 1996;275:452–457. [PubMed] [Google Scholar]
- 3.Bloom B, Murray C. Tuberculosis: commentary on a reemergent killer. Science. 1992;251:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 4.Cambau E, Jarlier V. Resistance to quinolones in mycobacteria. Res Microbiol. 1995;147:52–59. doi: 10.1016/0923-2508(96)80204-x. [DOI] [PubMed] [Google Scholar]
- 5.Chen C-R, Malik M, Snyder M, Drlica K. DNA gyrase and topoisomerase IV on the bacterial chromosome: quinolone-induced DNA cleavage. J Mol Biol. 1996;258:627–637. doi: 10.1006/jmbi.1996.0274. [DOI] [PubMed] [Google Scholar]
- 6.Dong Y, Xu C, Zhao X, Domagala J, Drlica K. Fluoroquinolone action against mycobacteria: effects of C-8 substituents on growth, survival, and resistance. Antimicrob Agents Chemother. 1998;42:2978–2984. doi: 10.1128/aac.42.11.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drlica K, Xu C, Wang J-Y, Burger R M, Malik M. Fluoroquinolone action in mycobacteria: similarity with effects in Escherichia coli and detection by cell lysate viscosity. Antimicrob Agents Chemother. 1996;40:1594–1599. doi: 10.1128/aac.40.7.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drlica K, Zhao X. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frieden T, Fujiwara P, Washko R, Hamburg M. Tuberculosis in New York City—turning the tide. N Engl J Med. 1995;333:229–233. doi: 10.1056/NEJM199507273330406. [DOI] [PubMed] [Google Scholar]
- 10.Frieden T, Sterling T, Pablos-Mendez A, Kilburn J, Cauthen G, Dooley S. The emergence of drug-resistant tuberculosis in New York City. N Engl J Med. 1993;328:521–526. doi: 10.1056/NEJM199302253280801. [DOI] [PubMed] [Google Scholar]
- 11.Garcia I, Pascual A, Guzman M C, Perea E. Uptake and intracellular activity of sparfloxacin in human polymorphonuclear leukocytes and tissue culture cells. Antimicrob Agents Chemother. 1992;36:1053–1056. doi: 10.1128/aac.36.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horn D, Hewlett D, Alfalla C, Patel A, Brudney K, Crawford J, Alland D, Kreiswirth B, Opal S, Peterson S. Clinical experience with rifampin-isoniazid-streptomycin-ethambutol (RISE)-resistant tuberculosis. Infect Dis Clin Pract. 1996;5:68–72. [Google Scholar]
- 13.Ito T, Matsumoto M, Nishino T. Improved bactericidal activity of Q-35 against quinolone-resistant staphylococci. Antimicrob Agents Chemother. 1995;39:1522–1525. doi: 10.1128/aac.39.7.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson J L, Ellner J J, Shiratsuchi H. Monocyte-Mycobacterium avium complex interactions: studies of potential virulence factors for humans. Immunol Ser. 1994;60:263–279. [PubMed] [Google Scholar]
- 15.Kitamura A, Hoshino K, Kimura Y, Hayakawa I, Sato K. Contribution of the C-8 substituent of DU-6859a, a new potent fluoroquinolone, to its activity against DNA gyrase mutants of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1467–1471. doi: 10.1128/aac.39.7.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klopman G, Fercu D, Li J-Y, Rosenkranz H S, Jacobs M R. Antimycobacterial quinolones: a comparative analysis of structure-activity and structure-cytotoxicity relationships. Res Microbiol. 1996;147:86–96. doi: 10.1016/0923-2508(96)80209-9. [DOI] [PubMed] [Google Scholar]
- 17.Kocagoz T, Hackbarth C J, Unsal I, Rosenberg E, Nikaido H, Chambers H F. Gyrase mutations in laboratory-selected fluoroquinolone-resistant mutants of Mycobacterium tuberculosis H37Ra. Antimicrob Agents Chemother. 1996;40:1768–1774. doi: 10.1128/aac.40.8.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreiswirth B, Moss A. Genotyping multidrug-resistant M. tuberculosis in New York City. In: Rom W, Garay S, editors. Tuberculosis. Boston, Mass: Little, Brown and Co.; 1995. pp. 199–209. [Google Scholar]
- 19.Pascual A, Garcia I, Perea E. Fluorometric measurements of ofloxacin uptake by human polymorphonuclear leukocytes. Antimicrob Agents Chemother. 1989;33:653–656. doi: 10.1128/aac.33.5.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19a.Power E, Phillips I. Correlation between umuC induction and Salmonella mutagenicity assay for quinolone antimicrobial agents. FEMS Microbiol Lett. 1993;112:251–254. doi: 10.1111/j.1574-6968.1993.tb06458.x. [DOI] [PubMed] [Google Scholar]
- 20.Rastogi N, Blom-Potar M. Intracellular bactericidal activity of ciprofloxacin and ofloxacin against Mycobacterium tuberculosis H37Rv multiplying in the J-774 macrophage cell line. Zentbl Bakteriol. 1990;273:195–199. doi: 10.1016/s0934-8840(11)80249-5. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan E A, Kreiswirth B N, Palumbo L, Kapur V, Musser J M, Ebrahimzadeh A, Frieden T R. Emergence of fluoroquinolone-resistant tuberculosis in New York City. Lancet. 1995;345:1148–1150. doi: 10.1016/s0140-6736(95)90980-x. [DOI] [PubMed] [Google Scholar]
- 22.Takiff H E, Salazar L, Guerrero C, Philipp W, Huang W M, Kreiswirth B, Cole S T, Jacobs W R, Jr, Telenti A. Cloning and nucleotide sequence of the Mycobacterium tuberculosis gyrA and gyrB genes and detection of quinolone resistance mutations. Antimicrob Agents Chemother. 1994;38:773–780. doi: 10.1128/aac.38.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, Tada K. Induction of maturation in cultured human monocytic leukemia cells by phorbol ester. Cancer Res. 1982;42:1530–1536. [PubMed] [Google Scholar]
- 24.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu C, Kreiswirth B N, Sreevatsan S, Musser J M, Drlica K. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug resistant Mycobacterium tuberculosis. J Infect Dis. 1996;174:1127–1130. doi: 10.1093/infdis/174.5.1127. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao X, Xu C, Domagala J, Drlica K. DNA topoisomerase targets of the fluoroquinolones: a strategy for avoiding bacterial resistance. Proc Natl Acad Sci USA. 1997;94:13991–13996. doi: 10.1073/pnas.94.25.13991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao X, Wang J-Y, Xu C, Dong Y, Zhou J, Domagala J, Drlica K. Killing of Staphylococcus aureus by C-8-methoxy fluoroquinolones. Antimicrob Agents Chemother. 1998;42:956–958. doi: 10.1128/aac.42.4.956. [DOI] [PMC free article] [PubMed] [Google Scholar]