Abstract
Our objective was to review and exemplify how selected applications of artificial intelligence (AI) might facilitate and improve inflammatory bowel disease (IBD) care and to identify gaps for future work in this field. IBD is highly complex and associated with significant variation in care and outcomes. The application of AI to IBD has the potential to reduce variation in healthcare delivery and improve quality of care. AI refers to the ability of machines to mimic human intelligence. The range of AI’s ability to perform tasks that would normally require human intelligence varies from prediction to complex decision-making that more closely resembles human thought. Clinical applications of AI have been applied to study pathogenesis, diagnosis, and patient prognosis in IBD. Despite these advancements, AI in IBD is in its early development and has tremendous potential to transform future care.
Keywords: Gastroenterology, IBD: clinical trials, Gastroenterology, IBD: genetics, Gastroenterology, IBD: pre-clinical treatment and novel therapies, Gastroenterology, screening and diagnosis, Gastroenterology
Background
Artificial intelligence (AI) refers to a branch of science focused on innovation in computer applications requiring the replication or simulation of human intelligence. The quintessential example is automated classification of images to recognize whether they contain certain elements, such as automated facial recognition in photos. AI has transformed other industries in powerful ways. Every day examples include the use of spam filters on email, personalized Netflix recommendations, and smart assistants like iPhone’s Siri. There are many terms which relate to AI including machine learning (e.g. random forests and boosting) and deep learning (e.g. recurrent neural networks and convolutional neural networks)—all of which refer to ways in which algorithms can learn to recognize patterns to solve complex problems that mimics human decision making. The practice of medicine is fraught with inefficiencies and therefore has tremendous potential to benefit from AI innovation. This is facilitated by the large amount of information available for data analytics, including clinical, imaging, genomic, proteomic, microbial, and metabolomics data. AI in healthcare is in its infancy but is rapidly coming into use due to the availability of large, well-described datasets and more powerful computing capability to study these datasets.
Inflammatory bowel disease (IBD) is a chronic relapsing condition that affects approximately 3 million Americans, and 7 million people throughout the world, with costs of approximately $23 000 per patient per year.1,2 IBD care is complex, and variation in care and outcomes is common. For example, numerous studies show significant variation in IBD management among academic centers and between academic and private practice gastroenterologists.3 The application of AI to IBD has the potential to reduce variation in healthcare delivery and improve quality of care. Our objective was to review and exemplify how selected applications of AI might facilitate and improve IBD care and to identify gaps for future work in this field.
What is AI?
AI refers to the ability of machines to perform tasks that would normally require human intelligence. This includes deduction and reasoning, knowledge representation, planning, natural language processing, learning, perception, and the ability to manipulate and move objects. Long-term goals of AI research include achieving creativity, social intelligence, and general (human level) intelligence.4 Examples include machine learning, neural networks, and deep learning.
Developing AI algorithms typically requires input data to train the prediction model. For example, teaching a computer to recognize faces requires first showing the computer a dataset of various images where human faces have been pre-labeled. The more diverse the training data, the better the prediction model will work in the real world. For the example of face recognition, training data that includes images with various lighting, skin tones, and backgrounds will enable a more accurate model when these variations occur in predicting faces in the real world. Determining the appropriate training data, or input data, is a critical component of this technology and has significant ethical and practical implications. For example, if an AI model is trained to predict the diagnosis of IBD from genetic markers using input data from Caucasians, that model will likely be inaccurate in the diagnosis of non-Caucasian patients. Also, if input data are misclassified (e.g. if stool is erroneously labeled as a polyp), this misclassification will propagate further into the algorithm.
Machine learning is a form of AI where data analysis focuses on pattern recognition. Machine learning, in its simplest form, uses algorithms to learn from training data and then makes predictions about real world objects based on knowledge gained from the training set. Machine learning requires human input to label the training data, so the algorithm can learn and then apply these labels to unlabeled data.5 For example, algorithms derived to identify patients at risk for healthcare overuse can enable identification of at risk patients for intervention.6 Machine learning approaches in IBD have included the prediction of treatment response to thiopurines as an alternative to thiopurine metabolite measurement, disease prognosis in newly diagnosed patients with IBD using biomarker patterns, the development of extra intestinal manifestations using combinations of genetic markers, and IBD diagnosis based on genome wide association studies (GWAS) variants.7–9
Deep learning algorithms have increasingly been used to analyze data achieving high prediction performance, especially with data that is large-scale in volume and complexity. As compared to traditional machine learning methods, deep learning requires less time-consuming preprocessing and feature engineering. In particular, it can be applied to unstructured and unlabeled input data such as images and texts, rather than requiring manual extraction of features from raw data to be used as inputs, such as in traditional machine learning. For example, deep learning methods based on convolutional neural networks have been used in IBD to predict disease severity using endoscopic images and obtained superior prediction accuracy.10
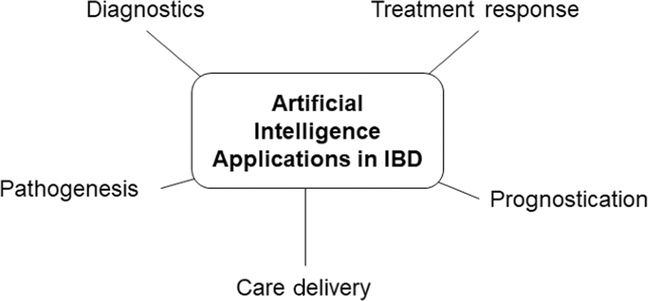
There are many opportunities for AI to benefit the practice of IBD (Fig. 1). Clinical applications of AI have been applied to study pathogenesis, diagnosis, and patient prognosis in IBD. There are also a wide variety of applications for AI in translational research as it relates to genomic, proteomic, metabolomics, and microbiome data. However, for the purposes of this review, we will focus on selected clinical applications of AI in IBD.
Figure 1.
Applications of AI in IBD: There are many opportunities for AI to benefit the practice of IBD. Clinical applications of AI have been applied to study pathogenesis, diagnosis, treatment response, prognostication, and care delivery in IBD.
IBD pathogenesis
IBD is a complex disease with multiple underlying influences including genomic, proteomic, metabolomic, environmental, and lifestyle mediators.11,12 The amount of data we have accumulated on each of these levels to study IBD pathogenesis is growing exponentially. However, robust tools are key to sorting and analyzing this large quantity of data. Artificial neural network analysis is one approach that could be leveraged to understand the complexity of IBD pathogenesis and identify potential features of importance. The feasibility of this approach has been demonstrated in a study that classified 18 227 Crohn’s disease patients and healthy controls by immunochip single nucleotide polymorphism genetic information and demonstrated the neural network approach as a powerful method to identify genetic variants.12 There is a lot of potential for similar methods of classifying patients with IBD into subtypes, including an understanding of the regulators that may drive IBD pathogenesis in each subtype, though more work needs to be done. There are numerous potential targets for applying an AI model to study the pathogenesis of IBD. For example, current research is applying these models to not only genetics but also the microbiome and lifestyle factors such as antibiotic exposure and diet. Although the clinical application of this research may be limited, they may lead to a better understanding of novel treatment targets and risk stratification.
IBD diagnostics and classification
The diagnosis of an IBD exacerbation requires an understanding of a complete clinical picture, including a patient’s history of present illness, physical examination, laboratory testing and imaging, endoscopic examination, and histology. Endoscopy is a cornerstone of IBD management. Endoscopy can be utilized to identify inflammation including its pattern, (segmental or continuous, mild or severe), location, and to differentiate inflammation from noninflammatory pathology, such as dysplasia. AI-based diagnostic systems can provide real-time automated detection, such as what has been described with computer-aided diagnosis systems to recognize gastric cancer.13 Image recognition is an important application of AI in medicine. Computer-aided automation of image interpretation can play an important role in endoscopy for IBD patients as it relates to characterizing lesions, classifying the severity of inflammation, and defining mucosal healing.14,15 Endoscopic assessments are naturally operator-dependent with high inter-observer variability. A study comparing the reliability of experienced gastroenterologists to a deep learning-based convolutional neural network for the identification of ulcerative colitis endoscopic activity showed comparable performance for scoring still images.10 In addition, automated full motion video analysis shows promise for generating Mayo endoscopic scores in clinical trial settings, offering the potential for improved standardization and reproducibility of disease assessments.16
AI also has the potential to reduce the need for histologic confirmation and reduce the cost of colonoscopy. Takenaka et al. constructed a deep neural network using colonoscopy images of ulcerative colitis patients to identify endoscopic disease severity. In a prospective validation of their algorithm, they were able to identify endoscopic remission with 90.1% (95% confidence interval [CI], 89.2–90.9) accuracy and a kappa of 0.798 (95% CI, 0.780–0.814) as compared to endoscopists’ assessment.14 This neural network based algorithm predicted histologic remission with 92.9% (95% CI, 92.1–93.7) accuracy and a kappa of 0.859 (95% CI, 0.841–0.875), as compared to pathologists’ interpretation.14
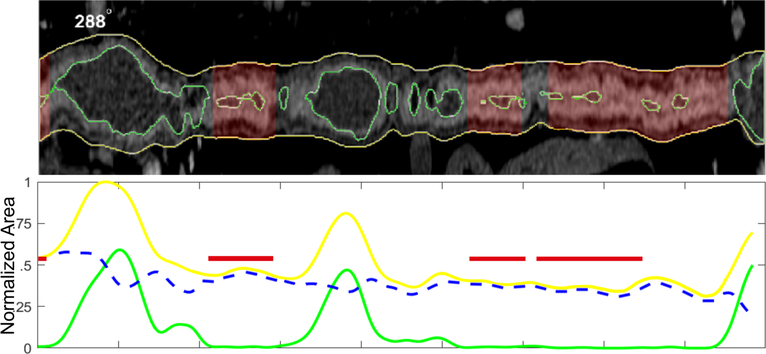
The application of computer-based automation to imaging interpretation would have similar benefits in reducing variation among radiologists’ interpretations. Computer vision image analysis performed similarly to experienced radiologists in the assessment of CT enterography evidence of structural bowel damage in Crohn’s disease.17 Such AI image interpretation systems offer not only the potential for replicating expert assessment but also can provide detailed iterative measurements which may improve the personalization of therapeutic decision making and prognosis (Fig. 2). Similar algorithms have been applied to abdominal MR data.18 The application of machine learning in medical imaging is in its infancy with many potential novel applications, including wireless capsule endoscopy and histology. This may also potentially play a role in helping to better classify patients with IBD unclassified.
Figure 2.
Conceptual application of AI for cross sectional imaging in IBD: Machine learning methods offer the potential for more than replicating expert interpretation, but enhanced disease quantification. CT or MR-enterography can be segmented into anatomic regions relevant for IBD or other disease using artificial intelligence techniques. In this conceptual example, regions of diseased bowel can be predicted using extracted measures of bowel wall thickness, lumen diameter, and total bowel dilation. Of more value is the opportunity to better quantify intestinal disease using direct area and volume measurements, which are expected to aid personalization of care in IBD.17 Outerwall;,Lumen;,Thickness;, Disease.
IBD treatment and prognostication
Prediction of treatment response has been a valuable application of AI in IBD. Our group previously derived several machine learning algorithms with the objective of predicting treatment response among patients with IBD. Thiopurines have a narrow therapeutic window. A dose that is too low will lead to inadequate treatment response, whereas doses that are too high may result in substantial immunosuppression and toxicity. Current therapeutic drug monitoring relies on thiopurine metabolite testing, which is not personalized as the pharmacokinetics of these drugs vary widely between individuals. In addition, the sensitivity and specificity of these tests are 62% and 72%, respectively.19 Furthermore, thiopurine monitoring is expensive, with costs as high as $268 per test, and can take up to 5 days to result.19 These challenges provide an opportunity for machine learning applications. To address this gap, a machine learning algorithm was created to predict treatment response and risk of adverse effects, and performed at least as well if not better than standard of care with 6-thioguanine nucleotide testing alone with an area under the receiver operator curve (AuROC) of 0.856 (95% CI, 0.793–0.919).19 In comparison, traditional thiopurine metabolite testing predicts thiopurine responders with an AuROC of 0.594 (95% CI, 0.546–0.642).19 This machine learning algorithm carries no added cost as it relies on standard blood counts and chemistries. This algorithm was externally validated in a clinical trial cohort using SONIC data.20 While this prediction model has been implemented in our institutional electronic health record to facilitate thiopurine optimization, barriers to widespread use have included information technology infrastructure challenges, changes in electronic medical record software requiring remapping of variable fields, and slow adoption of this predictive tool as part of routine clinical workflow. Other challenges include a reduction in thiopurine use in our institution.
To examine the application of machine learning models to predict treatment response to biologics, we used clinical trial data from GEMINI I and II to predict the likelihood of corticosteroid-free endoscopic remission in response to vedolizumab among patients with IBD. We used data collected during the first 6 weeks of therapy to accurately distinguish patients who were likely to achieve remission at week 52 of vedolizumab therapy. At baseline, we found that patients with a very high fecal calprotectin prior to initiation of vedolizumab were more likely to fail the therapy.21 After 6 weeks of therapy, fecal calprotectin and vedolizumab level trends predicted successful response with an AuROC of 0.73 (95% CI, 0.65–0.82).21 In a similar design, we were able to examine treatment response to ustekinumab in Crohn’s disease with an AuROC of 0.78 (95% CI, 0.69–0.87) using clinical trial data.22 These studies have the potential to direct the personalization of treatment with the right medication for the right patient at the right time.
With the wide availability of electronic medical record data, imaging data, and more, AI-based algorithms are also primed for treatment prognostication. In a prior machine learning application, our group used longitudinal data from the electronic medical record to predict IBD-related hospitalizations and steroid use.23 Other studies have focused on the predictive value of genome wide association studies and microbiome data in understanding treatment response. Cushing et al., for example, identified unique expression profiles in Crohn’s disease patients that may predict post-operative disease recurrence.24 They looked at gene transcript expression from ileal tissue obtained from operative specimens of Crohn’s disease patients to identify unique expression profiles. This was used to train a machine learning model which was able to accurately classify which patients would have an indolent postoperative course. There were two distinct profiles—depending whether patients had been exposed to anti-tumor necrosis factor therapies.
Limitations
Applications of AI in IBD need to be considered in the context of its limitations and challenges. AI has the potential to transform the role of physicians in caring for patients. While there have been general concerns of AI-based applications replacing physicians, it is much more likely that AI will facilitate care by physicians. Many AI-based applications rely on pattern recognition for identification of key features. This may not be generalizable across settings, clinics, or physicians given variation in practice patterns. The applicability of AI-based algorithms in less representative contexts, such as in the developing world, needs to be further studied and validated. In addition, it is important to recognize that while AI-based algorithms may help us to understand IBD pathogenesis and treatment responses, and to facilitate the care of patients with IBD, they do not speak to causal inference. AI-based algorithms are only as useful as the available data, and missing information may limit its applicability. Further, many machine learning (e.g., Lasso regression, random forest, etc.) and deep learning (e.g., convolutional neural networks, recurrent neural networks, etc.) approaches exist, and therefore, examination of the benefits and rationale for the use of one method over another is an important step in AI-based applications.
To many, AI-based approaches are a black box, and therefore, education on the derivation of AI models, their applicability, and their limitations in understanding causality or functionality is important to its widespread use. Model interpretation remains a challenge for many popular and powerful AI/machine learning-based models, such as random forest, neural network, and the support vector machine that are often referred to as black-box methods. To gain insights into some of the supervised machine learning models, some potential approaches have been developed to increase model interpretation including tools for interpreting/visualizing the impact of a single predictor variable on the predicted outcome. Though these methods are for a single predictor variable, they can be applied iteratively to each predictor variable, thus obtaining an entire picture of conditional impacts of the predictor variables on the predicted outcome. Some traditional visualization approaches include partial dependence plots (PDP)25,26; variable effect characteristic (VEC) curves27; and individual conditional expectation (ICE) plots.28 In addition, there have been newer methods of model interpretation including the SHapley Additive exPlanation (SHAP) values. SHAP values measure how each feature contributes to the model in either a positive or negative way, similar to feature importance.29–31 The SHAP values have the benefit that each individual would have their own set of SHAP values. This individual-level SHAP value provides a more personalized approach to understanding which variables are important.
It is also important to note that AI and machine learning based models provide probabilities of a prediction, not an absolute answer. It is important to understand performance characteristics of these models, such as positive predictive value, negative predictive value, sensitivity, and specificity; as well as discrimination, calibration, and misclassification.32 We also need to consider the possibility of false positives, and its unintended consequences, including further testing. More work needs to be done to clarify the ethical implications of AI models which have risk of significant selection bias if not trained on appropriate data. Furthermore, the legal implications of who is responsible for any harm done due to AI application need to be further explored.
Future directions
Ultimately AI in IBD is in its early development with tremendous potential to transform care. While we discuss selected clinical applications of AI in IBD (Table 1), there are many other opportunities for its application. Thinking toward delivering better care, imagine if rather than wait for an appointment to see a gastroenterologist, a patient could have an initial conversation with a chatbot that is immediate and autonomous. Chatbots utilize natural language processing to extract useful information and follow-up recommendations. This can improve accessibility and timeliness of care and has the potential to also standardize care. A recent study demonstrated that natural language processing was accurate in identifying patients with ulcerative colitis or Crohn’s disease from the electronic medical record more accurately than historic models relying on billing codes.33 However, the use of chatbots in a consultative dialog is in its infancy. A more recent study was able to categorize large amounts of electronic messages between patients and providers to demonstrate the potential to develop subsequent natural language processing based algorithms to support a chatbot that could handle most IBD patient questions and concerns.34 Current efforts in this space are focused on automating responses to questions using machine learning methods rather than hand constructed responses which reduces versatility.35–38
Table 1.
Applications of AI in IBD and examples in the literature
| AI application in IBD | Example | Sample size: total no. of samples (no. with outcome) | Type of validation | Performance characteristic of best model |
|---|---|---|---|---|
|
| ||||
| Pathogenesis | Isakov et al.11 used known IBD genes from GWAS studies to train a machine learning model which was then able to prioritize & identify novel IBD-risk genes from a comprehensive list of 16 390 genes. The model successfully differentiated IBD-risk vs non-IBD genes. | 513 intestinal biopsies (329 IBD biopsies) | 75% Training 25% Testing |
AuROC of 0.829. (95% CI not reported) |
| Diagnostics | Stidham et al.10 used colonoscopy images from patients with ulcerative colitis to build a deep learning model which categorized images as remission (Mayo 0–1) or disease (Mayo 2–3). The model successfully distinguished between these two categories. It was also able to identify exact Mayo subscores similar to agreements between three experienced human-reviewers and had similar accuracy for identifying moderate to severe disease in colonoscopy videos. | 2778 patients (Remission 1922; Diseased 856) | 80% Training 10% Tuning 10% Testing |
AuROC of 0.970 (95% CI, 0.967–0.972) |
| Treatment | Waljee et al.19 used age and standard laboratory data from individuals on thiopurine therapy to train a machine learning model to differentiate clinical responders from non-responders. This model was able to distinguish between these two categories more accurately than traditional 6-TGN metabolite testing. It was also able to identify nonadherence and thiopurine shunters. | 240 patients (131 clinical responders) | 70% Training 30% Testing |
AuROC of 0.856 (95% CI, 0.793–0.919) |
| Prognostics | Cushing et al.24 used gene transcript expression from ileal tissue obtained from operative specimens from Crohn’s patients to identify unique expression profiles. This was used to train a machine learning model which was able to accurately classify which patients would have an indolent postoperative course. | 24 anti-TNF naïve patients (6 with Rutgeerts i0) | Out-of-bag validation | Estimate of error rate 8.33% in correctly predicting i0 Rutgeerts score |
| Care delivery | Zand et al.34 used 8324 messages from IBD patients to develop a NLP model which was able to categorize conversational dialogs into distinct categories (symptoms, medications, appointments, labs, payments, communications, procedures, miscellaneous). This algorithm accurately categorized these conversations. | 424 patients | Human reviewer | In 95% of cases, there were minor or no differences between the NLP model and three physician reviewers |
NLP, natural language processing.
Further, since many AI-based algorithms are derived in a particular context, it is important to think about generalizability of algorithms across context. To address the question of generalizability, our group and others have begun to apply treatment-based algorithms to other settings and data sources. Additionally, prospective studies of AI-based algorithms are necessary to demonstrate efficacy, conforming to the new CONSORT-AI and SPIRIT guidelines for clinical trials.39–41 Finally, while many AI-based algorithms have been described, few if any have successfully been implemented into practice. How best to implement and integrate these algorithms with electronic medical record interfaces is unclear, but likely requires an understanding of barriers and facilitators to adoption. In conclusion, it is important to recognize that AI models cannot replace the patient-provider relationship and will be important tools to augment our ability to deliver care. The increasing availability of clinical data, the inefficiencies in IBD care, and challenges to diagnosis, treatment, and prognostication provide an opportunity for AI to improve IBD care. Therefore, a basic understanding of AI, its applicability, and associated limitations is important as we begin to incorporate more AI applications in patient care.
References
- 1.Alatab S, Sepanlou SG, Ikuta K et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020; 5: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehta F Report: economic implications of inflammatory bowel disease and its management. Am. J. Manag. Care 2016; 22: s51–60. [PubMed] [Google Scholar]
- 3.Berry SK, Siegel CA, Melmed GY. Quality improvement initiatives in inflammatory bowel disease. Curr. Gastroenterol. Rep 2017. Aug; 19: 1–5. [DOI] [PubMed] [Google Scholar]
- 4.Fjelland R Why general artificial intelligence will not be realized. Humanit Soc. Sci. Commun. 2020; 7: 1–9. [Google Scholar]
- 5.Rajkomar A, Dean J, Kohane I. Machine learning in medicine. N. Engl. J. Med 2019; 380: 1347–58. [DOI] [PubMed] [Google Scholar]
- 6.Ganguli I, Gordon WJ, Lupo C et al. Machine Learning and the pursuit of high-value health care. NEJM Catalyst Innovations in Care Delivery 2020; 1. [Google Scholar]
- 7.Waljee AK, Sauder K, Patel A et al. Machine learning algorithms for objective remission and clinical outcomes with thiopurines. J. Crohns Colitis 2017; 11: 801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biasci D, Lee JC, Noor NM et al. A blood-based prognostic biomarker in IBD. Gut 2019; 68: 1386–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bottigliengo D, Berchialla P, Lanera C et al. The role of genetic factors in characterizing extra-intestinal manifestations in Crohn’s disease patients: are Bayesian machine learning methods improving outcome predictions? J. Clin. Med 2019; 8: 865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stidham RW, Liu W, Bishu S et al. Performance of a deep learning model vs human reviewers in grading endoscopic disease severity of patients with ulcerative colitis. JAMA Netw. Open 2019; 2: e193963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isakov O, Dotan I, Ben-Shachar S. Machine learning-based gene prioritization identifies novel candidate risk genes for inflammatory bowel disease. Inflamm. Bowel Dis. 2017; 23: 1516–23. [DOI] [PubMed] [Google Scholar]
- 12.Romagnoni A, Jégou S, Van Steen K et al. Comparative performances of machine learning methods for classifying Crohn’s disease patients using genome-wide genotyping data. Sci. Rep. 2019; 9: 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirasawa T, Aoyama K, Tanimoto T et al. Application of artificial intelligence using a convolutional neural network for detecting gastric cancer in endoscopic images. Gastric Cancer 2018; 21: 653–60. [DOI] [PubMed] [Google Scholar]
- 14.Takenaka K, Ohtsuka K, Fujii T et al. Development and validation of a deep neural network for accurate evaluation of endoscopic images from patients With ulcerative colitis. Gastroenterology 2020; 158: 2150–7. [DOI] [PubMed] [Google Scholar]
- 15.Bossuyt P, Vermeire S, Bisschops R. Scoring endoscopic disease activity in IBD: artificial intelligence sees more and better than we do. Gut 2020; 69: 788–9. [DOI] [PubMed] [Google Scholar]
- 16.Yao H, Najarian K, Gryak J et al. Fully automated endoscopic disease activity assessment in ulcerative colitis. Gastrointest. Endosc 2020; 0. [DOI] [PubMed] [Google Scholar]
- 17.Stidham RW, Enchakalody B, Waljee AK et al. Assessing small bowel stricturing and morphology in crohn’s disease using semi-automated image analysis. Inflamm. Bowel Dis. 2020; 26: 734–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R, Cai Y, Lee IK et al. Evaluation of a convolutional neural network for ovarian tumor differentiation based on magnetic resonance imaging. Eur. Radiol. 2020: 1–12. 10.1007/s00330-020-07266-x [DOI] [PubMed] [Google Scholar]
- 19.Waljee AK, Joyce JC, Wang S et al. Algorithms outperform metabolite tests in predicting response of patients with inflammatory bowel disease to thiopurines. Clin. Gastroenterol. Hepatol 2010; 8: 143–50. [DOI] [PubMed] [Google Scholar]
- 20.Waljee AK, Sauder K, Zhang Y, Zhu J, Higgins PDR. External validation of a thiopurine monitoring algorithm on the SONIC clinical trial dataset. Clin. Gastroenterol. Hepatol. 2018; 16: 449–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waljee AK, Liu B, Sauder K et al. Predicting corticosteroid-free endoscopic remission with vedolizumab in ulcerative colitis. Aliment. Pharmacol. Ther. 2018; 47: 763–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waljee AK, Wallace BI, Cohen-Mekelburg S et al. Development and validation of machine learning models in prediction of remission in patients with moderate to severe Crohn disease. JAMA Netw. Open 2019; 2: e193721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waljee AK, Lipson R, Wiitala WL et al. Predicting hospitalization and outpatient corticosteroid use in inflammatory bowel disease patients using machine learning. Inflamm. Bowel Dis. 2018; 24: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cushing KC, McLean R, McDonald KG et al. Predicting risk of postoperative disease recurrence in Crohn’s disease: patients with indolent Crohn’s disease have distinct whole transcriptome profiles at the time of first surgery. Inflamm. Bowel Dis. 2019; 25: 180–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hastie T, Tibshirani R, Friedman J. Random forests. In 2009. p. 587–604. [Google Scholar]
- 26.Friedman JH. Greedy function approximation: a gradient boosting machine. Ann Stat. 2001; 29: 1189–232. [Google Scholar]
- 27.Cortez P, Embrechts MJ. Using sensitivity analysis and visualization techniques to open black box data mining models. Inf. Sci. (Ny) 2013; 225: 1–17. [Google Scholar]
- 28.Goldstein A, Kapelner A, Bleich J, Pitkin E. Peeking inside the black box: visualizing statistical learning with plots of individual conditional expectation. J. Comput. Graph. Stat. 2015; 24: 44–65. [Google Scholar]
- 29.Lundberg SM, Erion G, Chen H et al. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell 2020; 2: 56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundberg SM, Nair B, Vavilala MS et al. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2018; 2: 749–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundberg SM, Lee SI. A unified approach to interpreting model predictions. In: Advances in neural information processing systems. Neural information processing systems foundation; 2017. p. 4766–75. [Google Scholar]
- 32.Singal AG, Higgins PDR, Waljee AK. A primer on effectiveness and efficacy trials. Clin. Transl. Gastroenterol. 2014; 5: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ananthakrishnan AN, Cai T, Savova G et al. Improving case definition of Crohn’s disease and ulcerative colitis in electronic medical records using natural language processing: a novel informatics approach. Inflamm. Bowel Dis. 2013; 19: 1411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zand A, Sharma A, Stokes Z et al. An exploration into the use of a chatbot for patients with inflammatory bowel diseases: retrospective cohort study. J. Med. Internet Res. 2020; 22: e15589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou L, Gao J, Li D, Shum H-Y. The design and implementation of XiaoIce, an empathetic social chatbot. Comput. Linguist. 2018; 46: 53–93. [Google Scholar]
- 36.Unsupervised Deep Learning for Vertical Conversational Chatbots|by Ramesh VC|Chatbots Magazine [Internet]. [Google Scholar]
- 37.The 7 Steps of Machine Learning. From detecting skin cancer, to sorting …|by Yufeng G|Towards Data Science [Internet]. [Google Scholar]
- 38.Huang J, Zhou M, Yang D. Extracting chatbot knowledge from online discussion forums [Internet]. [Google Scholar]
- 39.Liu X, Rivera SC, Moher D, Calvert MJ, Denniston AK. Reporting guidelines for clinical trial reports for interventions involving artificial intelligence: the CONSORT-AI extension. The BMJ 2020; 370: m3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cruz Rivera S, Liu X, Chan AW et al. Guidelines for clinical trial protocols for interventions involving artificial intelligence: the SPIRIT-AI extension. Nat. Med. 2020; 26: 1351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olivera P, Danese S, Jay N, Natoli G, Peyrin-Biroulet L. Big data in IBD: a look into the future. Nat. Rev. Gastroenterol. Hepatol 2019; 16: 312–21. [DOI] [PubMed] [Google Scholar]