Abstract
The clue behind the SARS-CoV-2 origin is still a matter of debate. Here, we report that SARS-CoV-2 has gained a novel spike protein S1–N-terminal domain (S1-NTD). In our CLuster ANalysis of Sequences (CLANS) analysis, SARS-CoV/SARS-CoV-2 S1-NTDs displayed a close relationship with OC43 and HKU1. However, in the complete and S1-NTD-free spike protein, SARS-CoV/SARS-CoV-2 revealed closeness with MERS-CoV. Further, we have divided the S1-NTD of SARS-CoV-2 related viruses into three distinct types (Type-I to III S1-NTD) and the S1-NTD of viruses associated with SARS-CoVs into another three classes (Type-A to C S1-NTD) using CLANS and phylogenetic analyses. In particular, the results of our study indicate that SARS-CoV-2, RaTG13, and BANAL-20-52 viruses carry Type-I-S1-NTD and other SARS-CoV-2-related-bat viruses have Type-II and III. In addition, it was revealed that the Pangolin-GX and Pangolin-Guangdong lineages inherited Type-I-like and Type–II–like S1-NTD, respectively. Then our CLANS study shows the potential for evolution of Type-I and Type-III S1-NTD from SARS-CoV-related viruses Type-A and Type-B S1-NTDs, respectively. Furthermore, our analysis clarifies the possibility that Type-II S1-NTDs may have evolved from Type-A-S1-NTD of SARS-CoV-related viruses through Type-I S1-NTDs. We also observed that BANAL-20-103, BANAL-20-236, and Pangolin-Guangdong-lineage viruses containing Type–II–like S1-NTD are very close to SARS-CoV-2 in spike genetic areas other than S1-NTD. Possibly, it suggests that the common ancestor spike gene of SARS-CoV-2/RaTG13/BANAL-20-52-like virus may have evolved by recombining the Pangolin-Guangdong/BANAL-20-103/BANAL-20-236-like spike gene to Pangolin-GX-like Type-I-like-S1-NTD in the unsampled bat or undiscovered intermediate host or possibly pangolin. These may then have evolved into SARS-CoV-2, RaTG13, and BANAL-20-52 virus spike genes by host jump mediated evolution. The potential function of the novel Type-I-S1-NTD and other types of S1-NTDs needs to be studied further to understand better its importance in the ongoing COVID-19 outbreak and for future pandemic preparedness.
Keywords: SARS-CoV-2, The origin of SARS-CoV-2, Spike protein, S1–N-Terminal domain, Pandemic outbreak
1. Introduction
The origin of the SARS-CoV-2 virus, which suddenly emerged in December 2019 in Wuhan, China (Wu et al., 2020; Zhou et al., 2020a) and spread rapidly worldwide, remains unanswered. Subsequently, SARS-CoV-2 related viruses' complete genomes were sequenced from bat samples collected before and after the SARS-CoV-2 outbreak in humans and previously collected pangolins samples (Zhou et al., 2020a, 2020b, 2021a; Wacharapluesadee et al., 2021; Lam et al., 2020; Xiao et al., 2020). On the other hand, all SARS-CoV-2-related bat and pangolin viruses, except RaTG13&BANAL-20-52, have the high genetic diversity with the SARS-CoV-2 spike gene (Zhou et al., 2020b, 2021a; Wacharapluesadee et al., 2021; York, 2021; Sarah Temmam et al., 2021), which is vital in determining virus entry, tissue tropism, host-range, human-human infection, and host immune responses (Walls et al., 2020; Chi et al., 2020; Greaney et al., 2021; Suryadevara et al., 2021; Li, 2015, 2016; Hulswit et al., 2016). The SARS-CoV-2 spike protein is divided into a signal peptide, S1, and S2 subunits. The S1 subunit further divides into two independent receptor binding domains such as sugar receptor binding S1-NTD (14–305 amino acid residues) and protein receptor binding S1-RBD (319–541 amino acid residues) (Chi et al., 2020; Li, 2015; Wang et al., 2020; Huang et al., 2020). Further, it is widely accepted that the coronaviruses (CoVs) obtained the S1-NTD from host galectins through gene capture and then gained the S1-RBD through gene duplication from the S1-NTD (Li, 2015, 2016). Due to the presence of S1-RBD in the tip of the spike head, S1-NTD is perhaps positioned underneath the S1-RBD (Li, 2015; Li et al., 2006; Beniac et al., 2006). S1-RBD is more possibly exposed to the host immune system and evolves faster than S1-NTD (Li, 2015). Further, extensive N-linked glycan shielding in the S1-NTD is expected to play a role in immune evasion (Walls et al., 2020; Watanabe et al., 2020; Piccoli et al., 2020; Rogers et al., 2020). Therefore, the less diversified S1-NTDs are considered a more reliable domain for the coronaviruses to bind the sugar receptors, thereby facilitating the S1-RBDs to search their high-affinity protein receptors (Li, 2015). Remarkably, the level of diversity and interaction of these domains with the host receptor is a critical determinant of the coronavirus host range, cross-species infection, tissue tropism, and host immune responses (Li, 2015, 2016; Hulswit et al., 2016). However, over-the-counter studies have shown that genetic variation and evolution in SARS-CoV-2 S1-RBD are of more significant concern (Zhou et al., 2020b, 2021b; Wacharapluesadee et al., 2021; Wang et al., 2020; Wrobel et al., 2020; Ou et al., 2020). On the other hand, the genetic diversity and evolutionary origin of the SARS-CoV-2 S1-NTDs are not yet fully established.
In this study, in particular, we present evidence that the SARS-CoV-2 virus originated with novel Type-I-S1-NTD in its spike protein. Furthermore, we report the presence of different S1-NTDs in SARS-CoV-2-related-bat-CoVs (except RaTG13 & BANAL-20-52, which displays Type-I-S1-NTD). Next, we report the potential for various genetic recombinations in SARS-CoV-2-related-bat-CoVs, especially in the S1-NTD regions of the spike gene. Finally, we suggest the possibility that the spike gene of the common precursor SARS-CoV-2/RaTG13/BANAL-20-52-like viruses may have evolved by recombination.
2. Materials and method
2.1. Genetic diversity analysis
2.1.1. Phylogenetic analysis
We used the NCBI and GISAID databases to retrieve the SARS-CoV-2 and related bat coronavirus sequences. For the phylogenetic study of SARS-CoV-2 complete genome, entire spike gene, and phylogenetic tree to classify the S1-NTD at nucleotide and protein levels, PhyML 3.3_1 was utilized, with the Evolutionary model GTR for nucleotide/LG for amino acids, Equilibrium frequencies Empirical for nucleotide/ML- Model for amino acids, number of categories (n = 4) for the discrete gamma model, SPR (Subtree Pruning and Regraphing) was used for tree topology search with optimizing parameters such as tree topology, branch length, and model parameter, and Likelihood aLRT statistics was used to test the branch support. Further, the comparison phylogenetic analysis between the nucleotide sequences of S1-NTD and the spike gene without S1-NTDs and phylogenetic tree for the S1-RBD region (22561-23161 nt) was performed in MEGA7, with Maximum Likelihood method based on the General Time Reversible model, BioNJ algorithms, and Neighbor-Join to a matrix of pairwise distances estimated using the Maximum Composite Likelihood methodology, the topology was selected with a superior log-likelihood value, bootstrap tests (1000 replicates), gamma distribution (G) (categories = 5) with the invariant (I) site (G + I).
2.1.2. Net between group mean distance (NBGMD) analysis
The NBGMD was calculated using the MEGA7 through the Kimura 2-parameter model. The gamma distribution (shape parameter = 5) was used as a model to measure rate variation among sites, a bootstrap test (1000 replicates) was used. To estimate the Standard error and ambiguous sites were removed from the analysis for each sequence pair. Standard error estimates were displayed above the diagonal.
2.2. Recombination analysis
2.2.1. SimPlot analysis
The percent identity between the query and reference sequences was determined using the SimPlot 3.5.1 program (Paraskevis et al., 2020). The nucleotide sequences were first aligned in MEGA7 before being exported to SimPlot 3.5.1 for additional analysis. Using the Kimura two-parameter model, we employed the 500 base pair of the window at a 50 base pair step to measure the identity between the query and reference sequences.
2.2.2. Recombination detection program (RDP) analysis
We used RDP4 (Martin et al., 2015) to detect potential recombination events in SARS-CoV-2 and its related viruses. MEGA7 was used to align the nucleotides, which were then exported to RDP4 for additional processing. Then we analyzed with default parameter values for the BOOTSCAN, GENECONV, Chimaera, RDP, MaxChi, SISCAN, and 3seq methods, and a minimum of four or more approaches was assessed for probable recombination using a Bonferroni corrected p-value cut-off (0.05). The BOOTSCAN method-based images are displayed in the figures.
2.3. CLANS (CLuster ANalysis of sequences) analysis
The CLANS analysis was performed in the online Toolkit software (https://toolkit.tuebingen.mpg.de/tools/clans). The protein sequences retrieved from the NCBI database were subjected to the pairwise sequence similarity calculation using the online CLANS analysis in the MPI Bioinformatics Toolkit with a scoring matrix of BLOSUM80 and BLAST HSP's (High Scoring Pair) up to a p-value of 1e−4. Next, the CLANS files obtained from the Toolkit were visualized in a Java application (clans.jar) 21. Minimum 1,00,000 rounds were used to show the sequences connection and clusters in the clans. jar application.
2.4. Determining the selection and mutation pressure
2.4.1. Effective number of codons (ENc)
One of the approaches for determining the codon usage bias is the effective number of codons used from 61 codons for the 20 amino acids, ranging from 20 to 61. ENc values less than 35 suggest strong codon bias, while ENc values greater than 50 imply broad random codon usage(29,30). ENc values were calculated using an online server in this investigation (http://ppuigbo.me/programs/CAIcal/)(Puigbo et al., 2008).
2.4.2. ENc-GC3s plot
The ENc values are plotted against the third position of GC3s of codon values in this study to establish the key variables affecting the codon usage bias, such as selection or mutation pressure(32). The expected curve was determined by estimating the expected ENc values for each GC3S as recommended in previous publications (Wang et al., 2018; Tian et al., 2020). The ENc and GC3s for each gene were obtained from an online CAI analysis server (http://ppuigbo.me/programs/CAIcal/)(Puigbo et al., 2008). When codon bias is just driven by mutation pressure, the genes will lie on or near the expected curve. However, when codon bias is influenced by selection and other variables, the genes will fall well below the expected curve (Wang et al., 2018; Tian et al., 2020).
2.4.3. Neutrality plot analysis
The GC12 values of codons are plotted against the GC3 values in a neutrality plot to determine the degree of influence of mutation pressure and natural selection on codon usage patterns. The GC12 and GC3 values were retrieved for S1-NTD (region 6) nucleotide sequences from the online CAI analysis server (http://ppuigbo.me/programs/CAIcal/)(Puigbo et al., 2008).
2.4.4. Parity Rule 2 (PR2)-bias plot
To measure the mutation pressure and natural selection affecting the codon usage bias, we plotted the AT bias [A3/(A3+T3)] against the GC bias [G3/(G3+C3)](32). Using the ACUA Software (Vetrivel et al., 2007), the A3, T3, G3, and C3 values of nucleotide sequences of S1-NTD (region 6) were calculated.
3. Results
3.1. Genetic diversity in SARS-CoV-2 related bat coronaviruses at the complete genome level
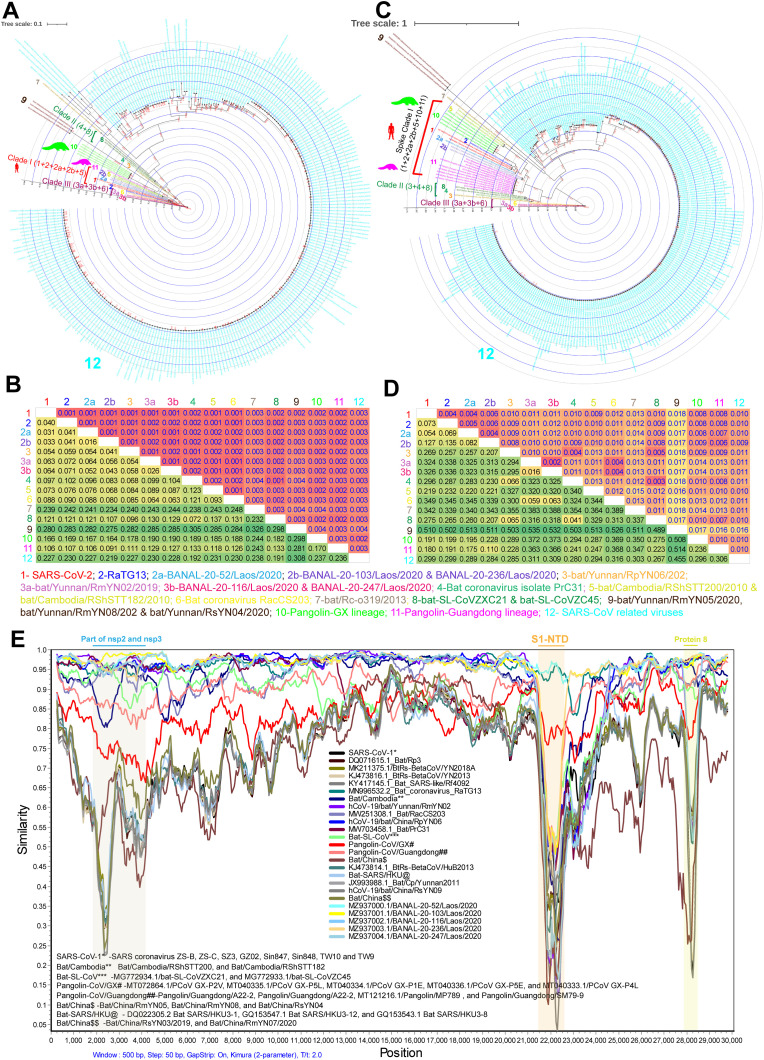
To understand the genetic diversity between the SARS-CoV-2 and SARS-CoV-related-bat-CoVs at the whole genome level, we performed the phylogenetic analysis using a total of 282 complete genome sequences. The details of different sequences are presented in supplementary data 1. In this complete genome phylogenetic analysis, the topology of phylogenetic trees is consistent with previous studies (Zhou et al., 2021a) (Fig. 1 A). Our study revealed that SARS-CoV-2-related-bat-CoVs have three distinct clades at the whole genome level (Fig. 1A). Of these, Clade-I viruses (RaTG13; BANAL-20-52, BANAL-20-103, BANAL-20-236, RShSTT182h, and RShSTT200) showed only 1.6–7.6% nucleotide diversity between themselves and the SARS-CoV-2. However, clade II viruses (bat/PrC31, bat-SL-CoVZC45, and bat-SL-CoVZXC21) expressed around 10–13% nucleotide diversity with SARS-CoV-2, Clade-I, and Clade III viruses (Fig. 1B). Further, clade III viruses (BANAL-20-116, BANAL-20-247, RmYN02, and RacCS203) expressed about 5.4–8.8%, 4.3–9.3%, and 9.9–13.1% nucleotide diversity with SARS-CoV-2, Clade-I, and Clade II viruses, respectively (Fig. 1B). Similarly, Pangolin- Guangdong lineage viruses showed association with the clade I virus (Fig. 1B).
Fig. 1.
The genetic diversity in SARS-CoV-2. (A&B) The Phylogenetic tree (A) and the net between-group mean distance (B), respectively, use the complete genome's nucleotide sequences of the SARS-CoV-2 and its related bat/pangolin viruses and SARS-CoV related viruses. SARS-CoV-2 related bat viruses n = 18; SARS-CoV-2 related pangolin viruses n = 6; SARS-CoV related viruses n = 258. The different group-specific viruses sequence details are listed in supplementary data 1. (C&D) The Phylogenetic tree (C) and the net between-group mean distance (D) used the complete spike gene nucleotide sequences of the SARS-CoV-2 and its related bat/pangolin viruses and SARS-CoV related viruses. SARS-CoV-2 related bat viruses n = 18; SARS-CoV-2 related pangolin viruses n = 16; SARS-CoV related viruses n = 256. The different group-specific viruses sequence details are listed in supplementary data 1. (E) The representative Similarity plot is based on the complete genomes nucleotide sequences of the SARS-CoV-2, SARS-CoV-2-related bat/pangolin viruses, and SARS-CoV related viruses. SARS-CoV-2 is used as a reference sequence query. First, we performed the similarity plot analysis with a total of 284 complete genome sequences of SARS-CoV-2 related bat/pangolin viruses and SARS-CoV-related viruses. Next, representative sequences are selected presented here. Details are listed in supplementary data 1.
3.2. Genetic diversity and evolution of SARS-CoV-2 spike gene
Subsequently, we were interested in understanding the spike gene's genetic diversity, a critical determinant of the coronavirus host range, tissue tropism, and host immune responses (Li, 2015, 2016; Hulswit et al., 2016). To understand the spike gene genetic diversity, first, we performed phylogenetic and NBGMD analysis using 290 sequences. The details of different sequences are declared in supplementary data 1. In this complete spike gene phylogenetic analysis, the topology of phylogenetic trees is consistent with previous studies (Zhou et al., 2021a) (Fig. 1C). In this analysis, we noticed that the spike genes of SARS-CoV-2-related-bat-CoVs viruses contain three distinct clades of spike-clade I, spike-clade II, and spike-clade III (Fig. 1C & D). The spike-clade I is formed by SARS-CoV-2, RaTG13, BANAL-20-52, BANAL-20-103, BANAL-20-236, RShSTT200, RShSTT182, pangolin-Guangdong, and pangolin-GX lineages. SARS-CoV-2, RaTG13, and BANAL-20-52 are closely related to each other. We observed similar results in similarity plot analysis; the RaTG13 & BANAL-20-52 viruses showed the closest genetic relationship to SARS-CoV-2 (Fig. 1E). At the same time, spike-clade I viruses BANAL-20-103 & BANAL-20-236 revealed 8.2–13.5% nucleotide diversity with SARS-CoV-2/RaTG13/BANAL-20-52 (Fig. 1C & D). Significantly, the pangolin-Guangdong and pangolin-GX lineages have 18% and 19% nucleotide diversity, respectively, with the spike gene of the SARS-CoV-2 (Fig. 1C & D). Next, RpYN06, Bat/PrC31, bat-SL-CoVZXC21, and bat-SL-CoVZC45 viruses formed spike-clade II. Similarly, RmYN02, BANAL-20-116, BANAL-20-247, and Bat/RacCS203 viruses formed spike-clade III. Further, we observed that spike-clade II and spike-clade III have 20–35% nucleotide diversity with spike-clade I (Fig. 1C & D).
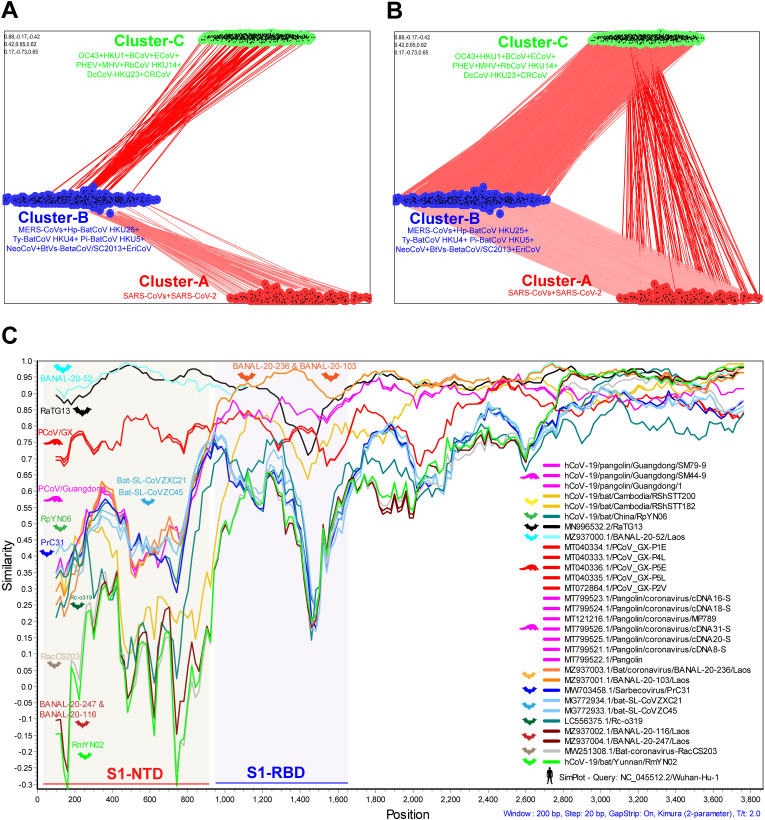
Next, to understand the evolution of SARS-CoV-2 spike protein, we used 1160 complete spike proteins from beta coronaviruses OC43, HKU1, MERS-CoVs, SARS-CoV, SARS-CoV-2, and related viruses for CLANS analysis. The complete spike proteins of SARS-CoV and SARS-CoV-2 viruses have a very high p-value and cannot be separated in CLANS analysis (Fig. 2 A). Furthermore, we observed that the spike proteins of beta coronavirus viruses split into three distinct clusters at the p-value threshold of 1e−200 in CLANS (Fig. 2A). Cluster-A is made up of spike proteins of SARS-CoV and SARS-CoV-2 viruses. Cluster-B was then formed by spike proteins associated with MERS-CoVs. Finally, cluster-C is produced by the spike proteins of OC43, HKU1, Bovine coronavirus (BCoV), Porcine hemagglutinating encephalomyelitis virus (PHEV), Equine coronavirus (ECoV), and Rodent coronaviruses. The details about the cluster-specific sequences are provided in supplementary data 2.
Fig. 2.
Evolution and genetic diversity in the SARS-CoV-2 spike gene. (A&B) CLANS (pairwise sequences similarity network) analysis was performed using the spike protein of beta coronaviruses that can infect humans and their related spike protein of viruses that can infect animals. A total of 1160 amino acid sequences of complete spike proteins were used in this analysis. The details of strains that belong to different clusters are presented in supplementary data 2. (A) The p-value threshold of 1e−200 in CLANS is used to show the lines connecting the sequences. (B) The p-value threshold of 1e−165 in CLANS is used to indicate the lines connecting the sequences. (C) The representative Similarity plot is based on the complete spike gene nucleotide sequences of the SARS-CoV-2 and SARS-CoV-2-related bat/pangolin viruses. SARS-CoV-2 is used as a reference sequence query.
Furthermore, Cluster-B (MERS-CoVs) at the p-value threshold of 1e−200 formed a direct relationship with cluster-A (SARS-CoV and SARS-CoV-2) and cluster-C (OC43, HKU1 & others) viruses. But at this threshold level, there is no direct link between cluster-A (SARS-CoV and SARS-CoV-2) and cluster-C (OC43, HKU1 & others) viruses (Fig. 2A). Then, at the p-value threshold of 1e−165, these three clusters showed a sequence similarity network with each other (Fig. 2B). However, cluster-A (SARS-CoV and SARS-CoV-2) is more closely related to cluster-B (MERS-CoVs) than cluster-C (OC43, HKU1 & others) (Fig. 2B). These results indicate that cluster-A (SARS-CoV and SARS-CoV-2 spike protein) may have originated from cluster-B (MERS-CoVs) related viruses.
Further, spike gene region S1-NTD (21578-22560 nt) and region S1-RBD (22561-23161 nt) are highly diversified regions in the spike gene (Fig. 2C). Region S1-RBD revealed two distinct RBD-clusters in the SARS-CoV-2 and SARS-CoV-2-related-bat-CoVs (Supplementary Fig. 1A and 1B). The SARS-CoV-2, RaTG13, BANAL-20-52, BANAL-20-103, BANAL-20-236, RShSTT182, RShSTT200, Pangolin-GX, and Pangolin-Guangdong formed an RBD-cluster-I (Supplementary Fig. 1A and 1B). Surprisingly, the SARS-CoV-2-related-bat-CoVs of bat-SL-CoVZXC21, bat-SL-CoVZC45, RpYN06, bat/PrC31, bat/RacCS203, RmYN02, BANAL-20-116, and BANAL-20-247 were formed RBD-cluster-II with SARS-CoV-related-bat-CoVs (Supplementary Fig. 1A and 1B). These results indicate that there is a high potential for widespread recombination in the RBD region.
3.3. Gain of novel S1-NTD in SARS-CoV-2
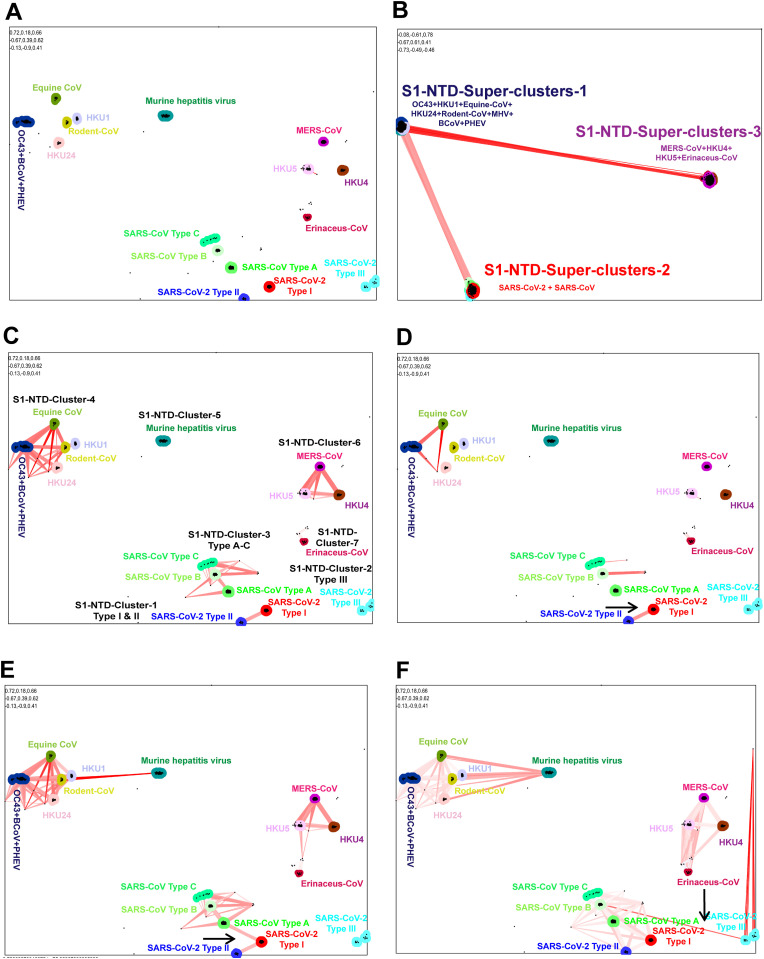
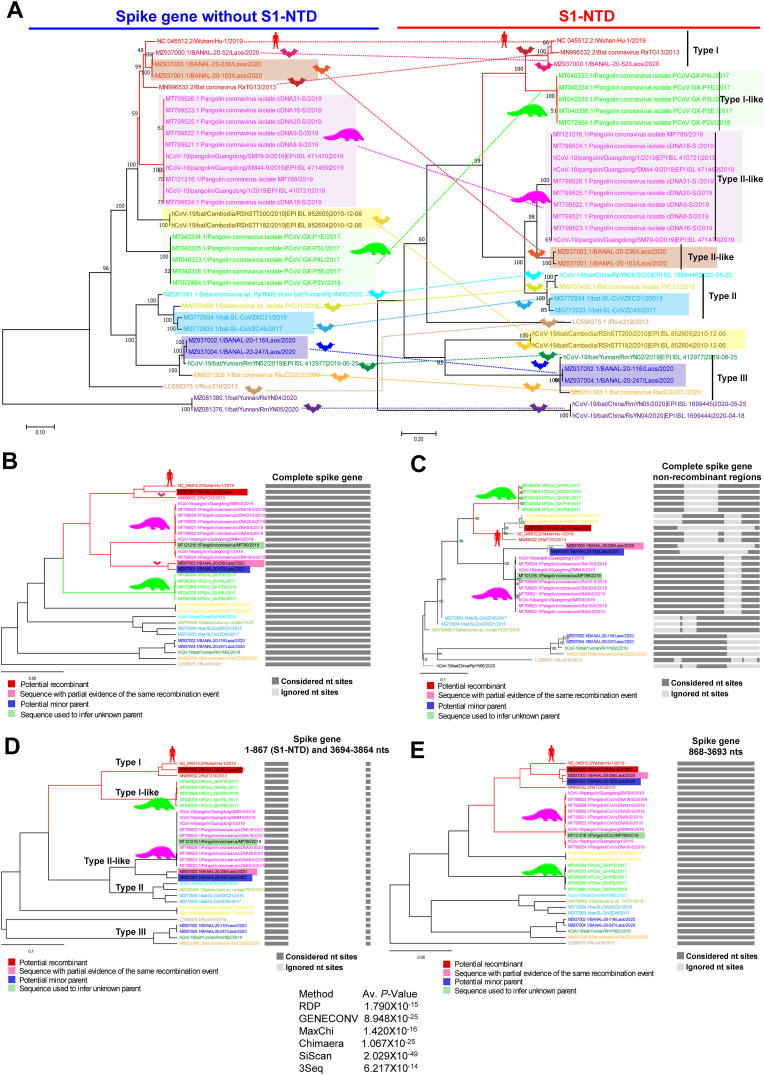
Significantly, using SimPlot and NCBI BLAST analysis, we learned that SARS-CoV-2 has a much higher genetic diversity compared to SARS-CoV-2-related-bat-CoVs (except RaTG13&BANAL-20-52) in regions containing S1-NTDs (Fig. 1E). First, we used pairwise sequence similarity network/CLANS analysis to determine the evolutionary origin of the amino acid scale SARS-CoV-2 S1-NTD. To perform this analysis, we retrieved the amino acid sequences of S1-NTDs of 1379 SARS-CoV, SARS-CoV-2, MERS-CoVs, OC43, HKU1, and BCoVs-related viruses from the NCBI public database. We used the p-value threshold of 1e−168 in CLANS to classify S1-NTDs of Betacoronavirus (Fig. 3 A). Accordingly, we ranked the S1-NTDs of SARS-CoV-2 and its related viruses into three distinct types (Type-I to III). The SARS-CoV-2, RaTG13, BANAL-20-52, and Pangolin-GX S1-NTDs formed the Type-I-S1-NTD (Fig. 3A). Type–II–S1-NTD was created by the S1-NTDs of SL-CoVZXC21, bat-SL-CoVZC45, RpYN06, Bat/PrC31, BANAL-20-103, BANAL-20-236, and Pangolin-Guangdong (Fig. 3A). Type–III–S1-NTD group was formed by RmYN02, RacCS203, BANAL-20-116, and BANAL-20-247. Further, we observed three different types of S1-NTDs (Type A to C S1-NTDs) in the SARS-CoV and its related viruses. Details of strains corresponding to different types provided in Supplementary Data 3).
Fig. 3.
Origin and evolution of the SARS-CoV-2 S1-NTD. CLANS (pairwise sequences similarity network) analysis was performed using the S1-NTDs of beta coronaviruses that can infect humans and their related spike protein of viruses that can infect animals. A total of 1379 amino acid sequences of S1-NTDs were used in this analysis. The details of strains that belong to different clusters are presented in supplementary data 3. (A) We used the p-value threshold of 1e−168 in CLANS to classify the S1-NTDs of beta coronavirus. (B) At the p-value threshold of 1e−10 in CLANS, the S1-NTDs of beta coronavirus formed three super-clusters. (C) With a p-value threshold of 1e−130, the S1-NTDs of beta coronavirus formed seven significant orphan clusters that showed no connections with each other. (D) At the p-value threshold of 1e−160, the Type-I-S1-NTD and Type–II–S1-NTD of SARS-CoV-2 related S1-NTDs revealed evolutionary links. The black arrow denotes the link. (E) At the p-value threshold of 1e−120, the Type-I-S1-NTD of SARS-CoV-2 showed a direct connection with Type-A-S1-NTDs SARS-CoV. The black arrow indicates the link. (F) At the p-value threshold of 1e−103, Type–III–S1-NTD revealed a direct correlation with type-B-S1-NTDs of SARS-CoV-related bat-CoVs. The black arrow represents the link.
At the p-value threshold of 1e−10 in CLANS, we observed three super-clusters (Fig. 3B). The OC43, HKU1, BCoVs, PHEV, ECoV, Rodent coronavirus S1-NTDs created the S1-NTD-superclusters-1 (Fig. 3B). The SARS-CoVs and SARS-CoV-2 related S1-NTDs formed the S1-NTD-superclusters-2 (Fig. 3B). MERS-CoVs related S1-NTDs generated the S1-NTD-superclusters-3. We also observed that the super-clusters-2 (SARS-CoVs and SARS-CoV-2 related S1-NTDs) were directly linked with the super-clusters-1 (OC43, HKU1, and other associated viruses) (Fig. 3B). And super-clusters-2 (SARS-CoVs and SARS-CoV-2 related S1-NTDs) displayed the evolutionary link with super-clusters-3 (MERS-CoV related S1-NTDs) through the super-clusters-1 (OC43, HKU1, and other associated viruses) (Fig. 3B). These results indicate that the S1-NTDs of SARS-CoV, SARS-CoV-2, and their related viruses are more closely related to OC43/HKU1/BCoV/PHEV/ECoV/MHV S1-NTDs than to S1-NTDs associated with MERS-CoVs.
In particular, at the p-value threshold of 1e−130, we observed seven significant orphan clusters without a pairwise sequence similarity network relationship to each other (Fig. 3C). S1-NTD-Cluster-1 was developed by Type-I-S1-NTD and Type–II–S1-NTD of SARS-CoV-2 related-CoVs S1-NTDs (Fig. 3C). S1-NTD-Cluster-2 contains Type–III–S1-NTD of SARS-CoV-2-related-CoVs S1-NTDs (Fig. 3C). S1-NTD-Cluster-3 was generated by the SARS-CoV-related S1-NTDs (Fig. 3C). The OC43, HKU1, BCoV, PHEV, and ECoV viruses S1-NTD-Cluster-4, the murine hepatitis virus S1-NTD-Cluster-5, MERS-CoVs S1-NTD-Cluster-6, and finally Erinaceus-CoVs S1-NTDs formed S1-NTD-cluster-7 (Fig. 3C). These results show that the viruses that cause significant outbreaks in humans or animals have their unique S1-NTD. It is also clear that SARS-CoV-2 has acquired its own novel S1-NTD.
Interestingly, at the p-value threshold of 1e−160, the Type-I-S1-NTD and Type–II–S1-NTD of SARS-CoV-2 related S1-NTDs displayed the pairwise sequence similarity network relationship (Fig. 3D). Then, at the p-value threshold of 1e−120, SARS-CoV-2's Type-I-S1-NTD showed a direct evolutionary relationship with SARS-CoV's Type-A-S1-NTDs (Fig. 3E). And the Type–II–S1-NTD displayed the ties with the SARS-CoV Type-A-S1-NTDs through Type-I-S1-NTD of SARS-CoV-2 (Fig. 3E). Similarly, at the p-value threshold of 1e−103, Type–III–S1-NTDs of SARS-CoV-2-related viruses showed the direct link with Type-B-S1-NTDs of SARS-CoV-related-bat-CoVs (Fig. 3F). However, the Type–III–S1-NTDs did not form a direct relationship with the Type-I-S1-NTD and Type–II–S1-NTD (Fig. 3F). These results suggest that Type-I-S1-NTD of SARS-CoV-2 may have evolved from SARS-CoV S1-NTD, and then Type–II–S1-NTD must have emerged from this Type-I-S1-NTD. Mainly Type–III–S1-NTD seems to have evolved from SARS-CoV-related-bat-CoVs independent to Type-I and Type–II–S1-NTDs.
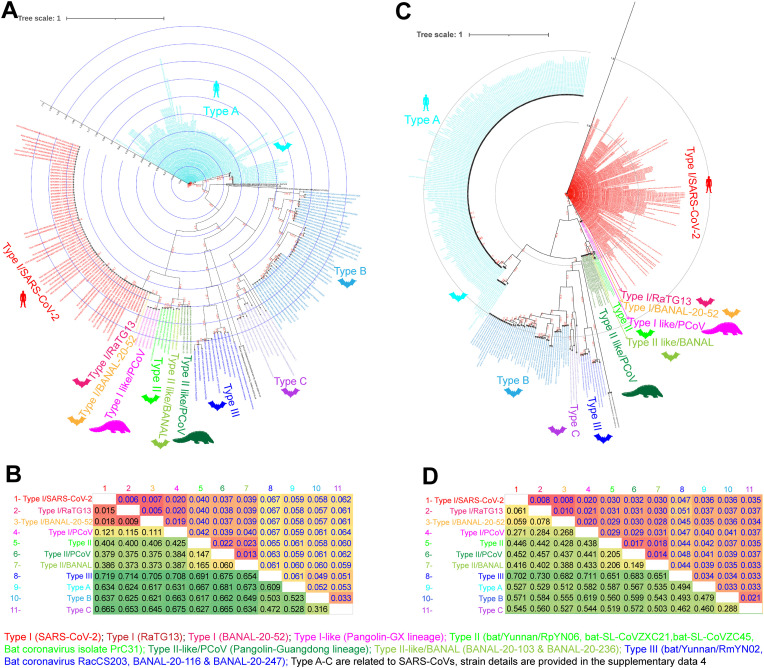
Subsequently, we validated the CLANS results using phylogenetic, and net between-group mean distance (NBGMD) analysis. We observed that in the phylogenetic tree, the S1-NTDs of SARS-CoV-2 viruses split into three groups (Type I to III S1-NTDs) and the S1-NTDs of SARS-CoV viruses into another three groups (Fig. 4 A), as seen in the CLANS classification. We then followed the following three parameters to classify S1-NTDs properly. (i) clustering networks at a p-value threshold of 1e−168 in the CLANS analysis used by BLOSUM80; (ii) gamma distribution (shape parameter = 5) and pairwise deletion of gaps/missing data with Equal Input model showing NBGMD over 30% in MEGA7; and finally (iii) we considered individual types with phylogenetic topology along with CLANS and NBGMD results. This phylogenetic tree was built using the PhyML 3.3_1, with the evolutionary model LG, equilibrium frequencies ML/Model, number of categories (n = 4) for the discrete gamma model, SPR was used for tree topology search with optimizing parameters such as tree topology, branch length, and model parameter, and Likelihood aLRT statistics was used to test the branch support. Considering the above three parameters, we classified the S1-NTDs of SARS-CoV-2-related viruses into three types (Type I to Type III) at amino acid sequence levels (Fig. 3A; Fig. 4A; Fig. 4B). We have similarly classified the S1-NTDs of SARS-CoV-related viruses into three other types (Type A to Type C) (Fig. 3A; Fig. 4A; Fig. 4B) (sequence details are provided in the supplementary Data 4).
Fig. 4.
The genetic diversity in spike gene of SARS-CoV-2 related viruses. (A&B) The Phylogenetic tree (A) and the net between-group mean distance (B), respectively, use the amino acid sequences of S1-NTDs of the SARS-CoV-2 and its related bat/pangolin viruses and SARS-CoV related viruses. A total of 236 amino acid sequences of S1-NTDs were used in this analysis. The different Type-specific virus strains details are presented in supplementary data 4. (C&D) The Phylogenetic tree (C) and the net between-group mean distance (D), respectively, use the nucleotide sequences of S1-NTDs of the SARS-CoV-2 and its related bat/pangolin viruses and SARS-CoV related viruses. A total of 462 nucleotide sequences of S1-NTDs were used in this analysis. The different Type-specific virus strains details are presented in supplementary data 4.
We then asked if these could be divided into distinct types at the nucleotide levels, as seen at the amino acid sequence levels. For this, we used two parameters as follows (i) gamma distribution (shape parameter = 5) with Kimura 2-parameter model and pairwise deletion of gaps/missing data showing NBGMD above 30% in MEGA7; and (ii) those with phylogenetic topology along with the support of NBGMD results considered as distinct types. This phylogenetic tree was built in PhyML 3.3_1, with the evolutionary model GTR, Equilibrium frequencies empirical, number of categories (n = 4) for the discrete gamma model, SPR was used for tree topology search with optimizing parameters such as tree topology, branch length, and model parameter, and Likelihood aLRT statistics was used to test the branch support. We observed that the S1-NTDs of SARS-CoV-2-related viruses were divided into three types (Type I to Type III) and the S1-NTDs of SARS-CoV-related viruses into three different types (Type A to Type C) at nucleotide levels also, as seen in the amino acid sequence levels (Fig. 4C; Fig. 4D). Sequence details are provided in the supplementary Data 4.
SARS-CoV-2, RaTG13, BANAL-20-52, and Pangolin-GX S1-NTDs formed a separate cluster at the p-value threshold of 1e−168 in CLANS (Fig. 3A). As well as showed 30% diversity with other Types of S1-NTDs in NBGMD at the amino acid (Fig. 4B) and nucleotide sequence levels (Fig. 4D), and grouped in phylogenetic topology (Fig. 4A; Fig. 4C), so we classified these as Type-I-S1-NTD. Remarkably, diversity between SARS-CoV-2, RaTG13, and BANAL-20-52 S1-NTDs was found to be less than 1.8% and 7.8% at the amino acid (Fig. 4B) and nucleotide sequence (Fig. 4D) levels, respectively. However, Pangolin-GX S1-NTDs expressed 11–12% and 26.8–28% diversity compared to SARS-CoV-2/RaTG13/BANAL-20-52 S1-NTDs at the amino acid (Fig. 4B) and nucleotide sequence (Fig. 4D) levels, respectively. We considered Pangolin-GX S1-NTDs as Type-I-like S1-NTDs for the above reasons.
Similarly, the S1-NTDs of SL-CoVZXC21, bat-SL-CoVZC45, RpYN06, Bat/PrC31, BANAL-20-103, BANAL-20-236, and Pangolin-Guangdong formed a separate cluster at the p-value threshold of 1e−168 in CLANS (Fig. 3A). And these viruses showed NBGMD of more than 30% with other Types S1-NTDs, at amino acid (Fig. 4B) and nucleotide sequence (Fig. 4D) levels, and in phylogenetic topology (Fig. 4A; Fig. 4C); therefore, we classified these as Type–II–S1-NTD. We further noted that there are three distinct groups in the Type–II–S1-NTD in phylogenetic topology formed by the amino acid and nucleotide sequence (Fig. 4A; Fig. 4C). These include SL-CoVZXC21, bat-SL-CoVZC45, RpYN06, and Bat/PrC31 S1-NTDs split into one group and the Pangolin-Guangdong lineage and BANAL-20-103/BANAL-20-236 into another two separate groups (Fig. 4A; Fig. 4C). Pangolin-Guangdong or BANAL-20-103/BANAL-20-236 S1-NTDs have revealed a diversity of 14.7–16.5% and 20.6% at the amino acid (Fig. 4B) and nucleotide (Fig. 4D) levels, respectively, compared to SL-CoVZXC21/bat-SL-CoVZC45/RpYN06/PrC31 S1-NTDs. Moreover, Pangolin-Guangdong lineage and BANAL-20-103/BANAL-20-236 viruses showed 6% and 14.9% genetic diversity between S1-NTDs at the amino acid (Fig. 4B) and nucleotide sequence (Fig. 4D) levels, respectively. Hence, we classified SL-CoVZXC21/bat-SL-CoVZC45/RpYN06/PrC31 S1-NTDs as Type–II–S1-NTDs. Further, the Pangolin-Guangdong lineage and BANAL-20-103/BANAL-20-236 viruses S1-NTDs as Type–II–like-PCoV and Type–II–like-BANAL S1-NTDs, respectively. Next, the Type–III–S1-NTD group was created by RmYN02, RacCS203, BANAL-20-116, and BANAL-20-247 virus S1-NTD with p-value threshold of 1e−168 in CLANS (Fig. 3A), and this was also reflected in NBGMD (Fig. 4B; Fig. 4D) and phylogenetic topology (Fig. 4A; Fig. 4C).
Just as we classified the S1-NTDs of SARS-CoV-2-related-CoV viruses, we used the same parameters in CLANS (Fig. 3A), NBGMD (Fig. 4B; Fig. 4D), and phylogenetic topology (Fig. 4A; Fig. 4C) to classify the S1-NTDs of SARS-CoV-related-CoV viruses into another three types (Type A to C). We noted that these Type-A-S1-NTDs are derived from S1-NTDs of SARS-CoV viruses isolated from humans, civets, and bats. However, Type-B-S1-NTDs and Type-C-S1-NTDs were almost entirely made up of S1-NTDs of SARS-CoV viruses isolated from the bat. Sequence details are provided in the supplementary Data 4.
Collectively, these findings indicate that Type-I-S1-NTD (region 6) is present only in SARS-CoV-2/RaTG13/BANAL-20-52. Further, the Pangolin-GX lineage, next to RaTG13/BANAL-20-52, contains 12% and 27% amino acid and nucleotide diversity, respectively, close to SARS-CoV-2 by Type-I-like-S1-NTD (Fig. 4B Fig. 4D). At the same time, another pangolin lineage, the pangolin-Guangdong lineage, showed a close relationship with Type–II–S1-NTD, but this lineage showed a somewhat similar relationship to RaTG13/BANAL-20-52/SARS-CoV-2 in RBD (Supplementary Fig. 1A and 1B). Similarly, BANAL-20-103/BANAL-20-236 with Type–II–S1-NTDs showed close association with SARS-CoV-2 in RBD as well as RaTG13 and BANAL-20-52 (Supplementary Fig. 1A and 1B). From here, it may be predicted that the spike gene of viruses such as SARS-CoV-2/RaTG13/BANAL-20–52 may have evolved from the BANAL-20-103/BANAL-20-236/Pangolin-Guangdong spike gene by recombining with the Type-I-like-S1-NTD of pangolin-GX.
3.4. Recombination event in the SARS-CoV-2 spike gene
SARS-CoV-2, RaTG13, and BANAL-20-52 viruses revealed 3.1–4.1% genetic diversity at the whole genome (Figs. 1B), 5.4–7.3% at the entire spike gene (Figs. 1D), 5.2–7.7% at the spike gene without S1-NTD (Supplementary Figure 1C), and 5.9–7.8% at S1-NTD (Fig. 4D), which indicate that all of these three viruses are likely to be originated from a common ancestor. BANAL-20-103/BANAL-20-236 viruses compared with SARS-CoV-2/RaTG13/BANAL-20-52 viruses expressed the genetic diversity of 1.6–4.1% at the whole genome level (Fig. 1B); 8.2–13.5% at spike gene (Fig. 1D); 0.6–6.6% at the spike gene without S1-NTD (Supplementary Figure 1C); and 38.8–41.6% at the S1-NTD (Fig. 4D). Given such a large genetic diversity, especially in the S1-NTD alone, we can assume that these viruses may have inherited the S1-NTD elsewhere, possibly through recombination. Furthermore, since similarity plot analysis observed significant genetic variation in S1-NTD than in other parts of the spike gene of SARS-CoV-2-related viruses (Fig. 2C), we compared the phylogenetic tree with the spike gene without-S1-NTD and with just S1-NTD. Correspondingly, BANAL-20-103/BANAL-20-236 viruses grouped with SARS-CoV-2/RaTG13/BANAL-20-52 viruses at spike gene without S1-NTD in phylogenetic analysis (Fig. 5 A). In a phylogenetic analysis of S1-NTD, BANAL-20-103/BANAL-20-236 viruses are grouped with Type–II–S1-NTDs (Fig. 5A). Similarly, the Pangolin-Guangdong lineage is associated with SARS-CoV-2/RaTG13/BANAL-20-52 at the spike gene without S1-NTD and with Type–II–S1-NTDs in S1-NTD (Fig. 5A). In contrast, the pangolin-GX lineage is the closest to SARS-CoV-2/RaTG13/BANAL-20-52 in S1-NTDs (Fig. 5A). Moreover, this pangolin-GX lineage has expressed more genetic diversity than the BANAL-20-103/BANAL-20-236/pangolin-Guangdong viruses against SARS-CoV-2/RaTG13/BANAL-20-52 viruses at the complete genome-scale (Fig. 1B), spike (Fig. 1D), and spike without S1-NTD scales (Supplementary Figure 1C). From these results, it can be inferred that the ancestral spike gene of SARS-CoV-2/RaTG13/BANAL-20–52 may have originated from recombination of the pangolin-GX Type-I-like-S1-NTD with the BANAL-20-103/BANAL-20-236/pangolin-Guangdong viruses spike gene.
Fig. 5.
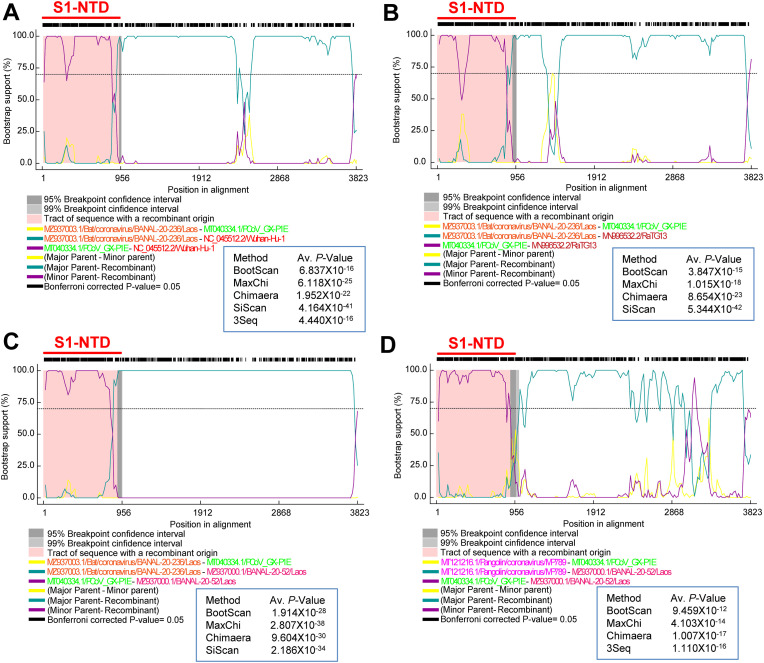
Recombination in the spike gene of SARS-CoV-2. (A) Compared the phylogenetic tree using the spike gene without S1-NTD of SARS-CoV-2 related bat/Pangolin viruses with the S1-NTD-based phylogenetic group. The first tree was constructed using SARS-CoV-2 related bat/pangolin viruses spike gene without S1-NTD. The second tree utilized only S1-NTD nucleotide sequences of SARS-CoV-2 related bat/pangolin viruses. (B to E) Phylogenetic tree generated from the RDP using the complete spike sequences of the SARS-CoV-2 related bat/pangolin viruses. The recombination events in the spike gene S1-NTD of SARS-CoV-2 and its related bat/pangolin viruses are supported by RDP, GENECONV, MaxChi, Chimaera, SiScan, and 3Seq. The RDP-based phylogenetic tree using (B) complete spike gene with UPGMA phylogenetic analysis; (C) non-recombinant regions of the entire spike gene with FastNJ phylogenetic analysis; (D) recombinant region of spike gene 1–867 nts (S1-NTD) with UPGMA phylogenetic analysis and 3694–3864 nts; and (E) recombinant region of the spike gene 868–3693 nts with UPGMA phylogenetic analysis.
To explore the possible recombination events in the SARS-CoV-2 spike gene, we performed the RDP analysis. In RDP analysis, BANAL-20-103/BANAL-20-236/pangolin-Guangdong viruses showed more excellent proximity to SARS-CoV-2/RaTG13/BANAL-20-52 in the UPGMA phylogenetic analysis, which ignored recombinant areas using the entire spike gene (Fig. 5B). But in the FastNJ phylogenetic analysis that utilized parts of the non-recombination spike gene, we found that the pangolin-GX lineages revealed greater closeness with SARS-CoV-2/RaTG13/BANAL-20-52 (Fig. 5C). Interestingly, we also noted that the pangolin-GX lineages were more closely related to SARS-CoV-2/RaTG13/BANAL-20-5 in recombined regions 1–867 (S1-NTD) and regions 3694-3864 nts spike gene (Fig. 5D) in RDP analysis. However, in the recombination region of spike gene 868-3693 nts, BANAL-20-103/BANAL-20-236/pangolin-Guangdong viruses showed the most closeness with SARS-CoV-2/RaTG13/BANAL-20-52 (Fig. 5E). Furthermore, more than four recombination analysis methods (BOOTSCAN, GENECONV, Chimaera, RDP, MaxChi, SISCAN, and 3seq) suggest the possible origin of the SARS-CoV-2/RaTG13/BANAL-20-52 viruses spike gene through recombination of BANAL-20-103/BANAL-20-236/pangolin-Guangdong spike gene with pangolin-GX S1-NTD (the representative BOOTSCAN images presented in the Fig. 6 A–D). Altogether, our results suggest that the common progenitor SARS-CoV-2/RaTG13/BANAL-20-52-like virus spike gene might have evolved in unsampled bat or pangolin or undiscovered intermediate host through recombination of pangolin-Guangdong/BANAL-20-103/BANAL-20-236-like spike gene with pangolin-GX-like S1-NTD. Finally, SARS-CoV-2, RaTG13, and BANAL-20-52 virus spike genes perhaps evolved from the common progenitor of the SARS-CoV-2/RaTG13/BANAL-20-52-like virus spike gene through host jump mediated evolution.
Fig. 6.
Recombination in the Pangolin-Guangdong/BANAL-20-103/BANAL-20-236 spike gene with Pangolin-GX-like Type-I-like-S1-NTD. (A) Representative RDP-based BOOTSCAN depicts the recombination between the BANAL-20-236/Laos and PCoV_GX-P1E in the spike gene. The recombination event is supported by BootScan, MaxChi, Chimaera, SiScan, and 3Seq. It can be inferred that the SARS-CoV-2 spike gene may have been formed by recombining the spike gene of BANAL-20-103/BANAL-20-236 with the S1-NTD of the Pangolin-GX lineage. Because strains BANAL-20-103 and BANAL-20-236 have 98.82% nucleotide sequence identity in the spike gene. Similarly, viruses in the pangolin-GX lineages share 99.5% nucleotide sequence identity in the spike gene. (B) Representative BOOTSCAN depicts the possible origin of the spike gene of RaTG13 by recombining the spike gene of BANAL-20-103/BANAL-20-236 to the S1-NTD of the Pangolin-GX lineage. The recombination event is supported by BootScan, MaxChi, Chimaera, and 3Seq. (C) Representative BOOTSCAN depicts the possible origin of the spike gene of BANAL-20-52 by recombining the spike gene of BANAL-20-103/BANAL-20-236 to the S1-NTD of the Pangolin-GX lineage. The recombination event is supported by BootScan, MaxChi, Chimaera, and 3Seq. (D) Representative BOOTSCAN depicts the possible origin of the spike gene of BANAL-20-52 by recombining the spike gene of Pangolin-Guangdong to the S1-NTD of the Pangolin-GX lineage. The recombination event is supported by BootScan, MaxChi, Chimaera, and 3Seq. Viruses in the pangolin-GX lineages hold 99.5% nucleotide sequence identity in the spike gene. Viruses in the pangolin- Guangdong lineages also share 99.9% nucleotide sequence identity in the spike gene.
3.5. Common and single-origin of SARS-CoV-2 S1-NTDs in humans
In this study, we found that SARS-CoV-2-related viruses contain three distinct S1-NTDs. Then we wanted to find out which of these three S1-NTDs are associated with the ongoing SARS-CoV-2 outbreak. For this, we retrieved highly matching S1-NTDs by subjecting S1-NTDs to each of the viruses in the Type-I-like, Type-II, and Type–III–S1-NTD group to NCBI and GISAID BLSAT analysis. Next, we subjected these retrieved S1-NTDs to phylogenetic analysis. None of the retrieved S1-NTDs in this phylogenetic analysis developed an evolutionary intermediate between SARS-CoV-2 and RaTG13 or SARS-CoV-2-related-bat-CoVs viruses (Fig. 4A; Fig. 4C). Instead, they grouped with SARS-CoV-2 S1-NTD (Fig. 4A; Fig. 4C), which refers to the common and single-origin of SARS-CoV-2 S1-NTDs in humans.
3.6. Recombination event in the SARS-CoV-2 related bat virus with Type-III S1-NTDs
SARS-CoV-2-related viruses containing these Type-III S1-NTDs (RmYN02, RacCS203, BANAL-20-116, and BANAL-20-247) in CLANS showed a direct pairwise similarity relationship with the S1-NTDs of SARS-CoV-related viruses (Fig. 3F). These Type-III S1-NTDs revealed a close relationship with S1-NTDs of SARS-CoV-related viruses in phylogenetic (Fig. 4A; Fig. 4C) and NBGMD (Fig. 4B; Fig. 4D) analysis, as seen in CLANS. However, SARS-CoV-2-related viruses containing these Type-III S1-NTDs displayed genetic diversity with SARS-CoV-2 at 6.3–8.8%, 32.4–34.9%, 21–23.8%, and 70.2%, respectively, at the whole genome level (Fig. 1B), spike (Fig. 1D), spike except S1-NTD (Supplementary Fig. 1C) and S1-NTD level (Fig. 4D). Further similarity plot analysis revealed that SARS-CoV-2-related viruses with these Type-III S1-NTDs showed significant diversity in S1-NTD with SARS-CoV-2 than other genetic regions (Fig. 1E). From these results, it can be speculated that SARS-CoV-2-related viruses containing these Type-III S1-NTDs may have inherited S1-NTD from SARS-CoV-related viruses and other genetic regions (except S1-NTD) from SARS-CoV-2-related viruses.
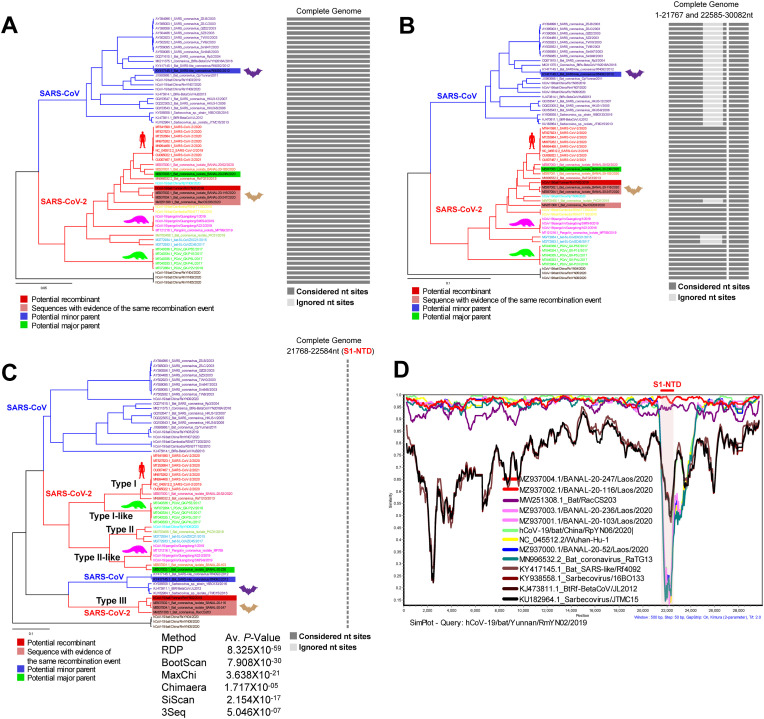
To explore the possible recombination events in the SARS-CoV-2-related viruses containing these Type-III S1-NTDs, we performed the RDP analysis. SARS-CoV-2-related viruses containing the Type-III S1-NTDs (RmYN02, RacCS203, BANAL-20-116, and BANAL-20-247) in phylogenetic trees created using complete genome (Fig. 7 A), and entire genome sequence except for S1-NTDs (Fig. 7B) in RDP analysis showed association with SARS-CoV-2-related viruses. As expected, SARS-CoV-2-related viruses containing these Type-III S1-NTDs revealed a close relationship with S1-NTDs of SARS-CoV-related viruses in a phylogenetic tree created using only S1-NTDs (Fig. 7C). After this, we can see that this result is also reflected in the similarity plot analysis (Fig. 7D). From all of these results, an unsampled virus might have evolved from the recombination of SARS-CoV-2-related viruses genome with the S1-NTD of SARS-CoV-related viruses. It then appears that SARS-CoV-2-related viruses containing Type-III S1-NTDs might have emerged from this unsampled virus.
Fig. 7.
The possible recombination-mediated evolution of Type-III S1-NTD in the SARS-CoV-2 related viruses. (A to C) Phylogenetic tree generated from the RDP using the complete genome sequences of the representative SARS-CoV-2 and SARS-CoV. The recombination events in the spike gene Type-III S1-NTD of SARS-CoV-2-related bat viruses are supported by RDP, BootScan, MaxChi, Chimaera, SiScan, and 3Seq. The RDP-based phylogenetic tree using (A) complete genome with UPGMA phylogenetic analysis; (B) recombinant area of the 1–21767 and 22585-30082 nts with UPGMA phylogenetic analysis; and (C) recombinant region of 21768–22584 nt (S1-NTD) nts with UPGMA phylogenetic analysis. (D) Similarity plot based on the complete genomes nucleotide sequences of the representative SARS-CoV-2 and SARS-CoV. The RmYN02 is used as a reference sequence query. It indicates the possible recombination-mediated evolution of Type-III S1-NTDs in the SARS-COV-2 related viruses from the SARS-CoVs S1-NTD.
3.7. Recombination event in the spike gene of SARS-CoV-2 related cambodian bat viruses
Cambodian bat coronaviruses (RShSTT200 & RShSTT182) clustered with S1-NTDs of SARS-CoV related viruses in CLANS (Fig. 3A). These Cambodian bats CoVs (RShSTT200 & RShSTT182) revealed a close relationship with S1-NTDs of SARS-CoV-related viruses in phylogenetic (Fig. 4A; Fig. 4C) analysis, as seen in CLANS. However, Cambodian bat CoVs displayed genetic diversity with SARS-CoV-2 at 7.3%, 21.9%, and 12.5%, respectively, at the whole genome level (Fig. 1B), spike (Fig. 1D), and spike except S1-NTD (Supplementary Figure 1C). Further similarity plot analysis revealed that Cambodian bat CoVs showed significant diversity in S1-NTD with SARS-CoV-2 than other genetic regions (Fig. 1E). These results indicate that Cambodian bat CoVs may have obtained S1-NTD from SARS-CoV-related viruses and other genetic regions (except S1-NTD) from SARS-CoV-2-related viruses.
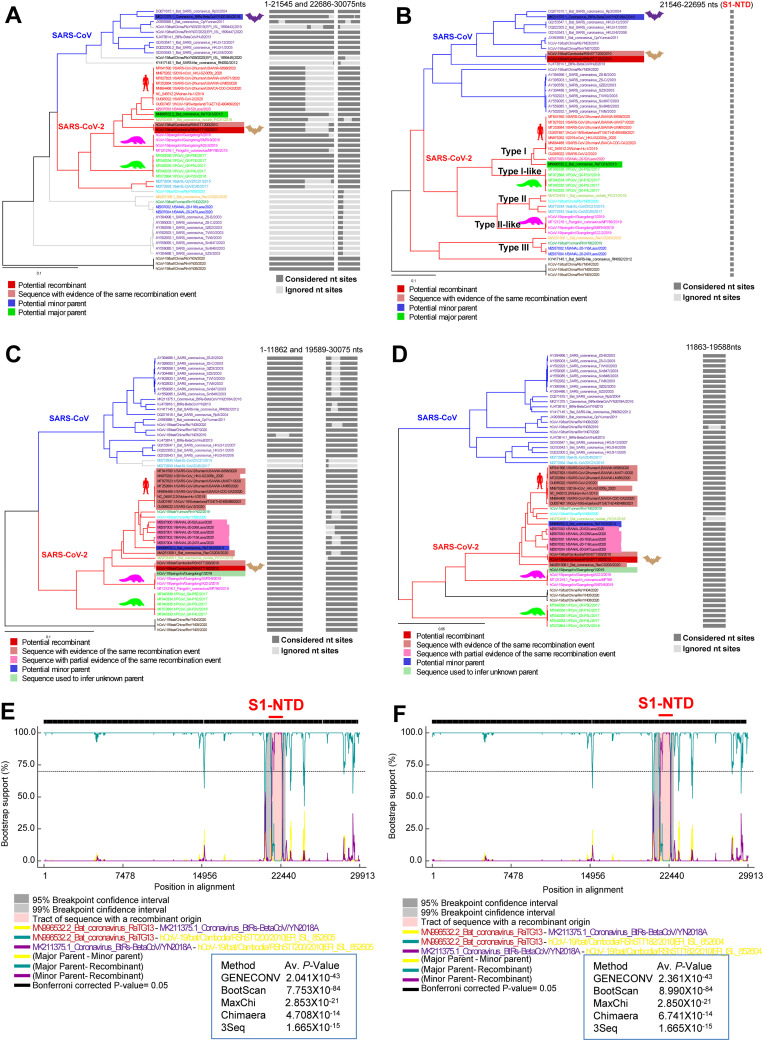
Then, we did an RDP analysis to explore the feasibility of this recombination. In this RDP (Chimaera) analysis, RShSTT200/RShSTT182 viruses were grouped with RaTG13 in the UPGMA phylogenetic tree using 1–21545 and 22686-30075 nts (Fig. 8 A). However, RShSTT200/RShSTT182 viruses in the UPGMA phylogenetic tree used by 21546–22695 nts (S1-NTD) were grouped with S1-NTD of SARS-CoV-related bat coronaviruses (Fig. 8B). In UPGMA phylogenetic tree using 1–21545 and 22686-30075 nts, some parts of RShSTT200/RShSTT182 viruses were ignored by recombination, and we then examined the origin of this ignored area. The RShSTT200/RShSTT182 viruses in the UPGMA phylogenetic tree using 1–11862 and 19589-30075 nts were somewhat associated with RaTG13 (Fig. 8C). However, tree derived using 11863-19588 nts RShSTT200/RShSTT182 viruses were more closely related to RaTG13 (Fig. 8D). Furthermore, more than four recombination analysis methods (BOOTSCAN, GENECONV, Chimaera, RDP, MaxChi, SISCAN, and 3seq) suggest the possible origin of the RShSTT200/RShSTT182 viruses through recombination of RaTG13 virus with SARS-CoV-related bat coronavirus S1-NTD (MK211375.1_Coronavirus_BtRs-BetaCoV/YN2018A) (the representative BOOTSCAN images presented in the Fig. 8E–F).
Fig. 8.
The SARS-CoV-2 related Cambodian bat viruses perhaps obtained S1-NTD from SARS-CoV. (A to D) RDP program was used to generate the phylogenetic tree. (A) RShSTT200/RShSTT182 viruses in the UPGMA tree of 1–21545 and 22686-30075 nts are grouped with SARS-Cov-2 related viruses. (B) RShSTT200/RShSTT182 viruses in the UPGMA tree of 21546–22695 nts (S1-NTD) are grouped with SARS-CoV-related bat coronaviruses. (C) RShSTT200/RShSTT182 viruses in the UPGMA tree of 1–11862 and 19589-30075 nts are grouped with SARS-Cov-2 related viruses. (D) RShSTT200/RShSTT182 viruses in the UPGMA tree of 11863–19588 nts are grouped with SARS-Cov-2-related viruses. It is ignored regions in Fig. 8A. (E–F) Representative RDP-based BOOTSCAN depicts the Cambodia bat virus RShSTT200 (E) and RShSTT182 (F) as a recombinant SARS-CoV-2 virus spike gene with S1-NTD of SARS-CoV-related viruses. The recombination event is supported by GENECONV, BootScan, MaxChi, Chimaera, and 3Seq.
3.8. Selection pressure in the S1-NTDs of SARS-CoV-2/SARS-CoVs
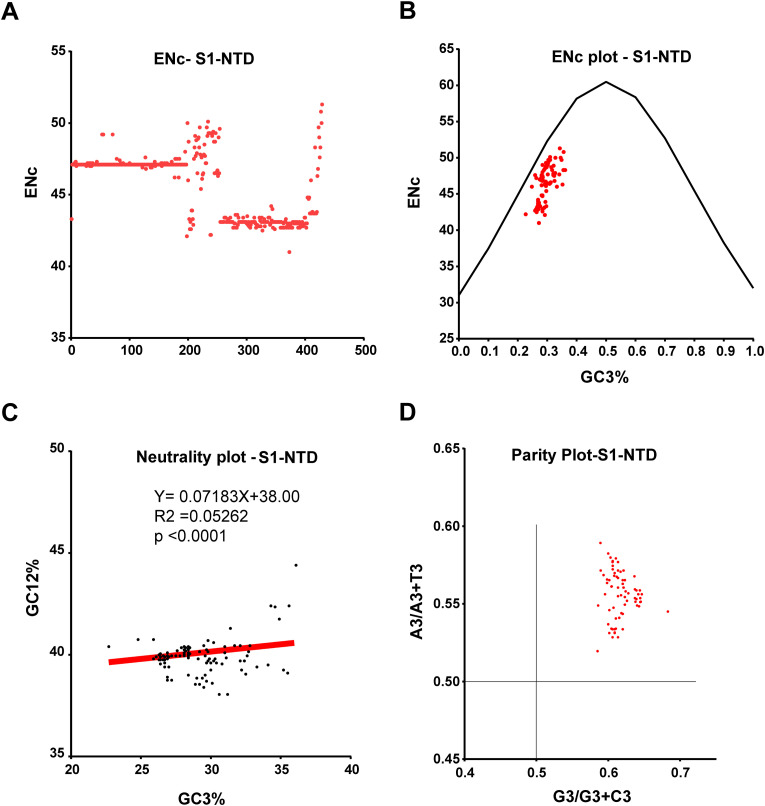
ENc values < 35 indicate high codon bias, and values > 50 show general random codon usage (Zhao et al., 2016; Wang et al., 2018). Here S1-NTD nucleotide sequence of SARS-CoV-2/SARS-CoVs viruses displayed the average ENc 45.62 ± SD2.21 (n = 429), suggesting the moderate codon use bias (Fig. 9 A). We further performed ENc-GC3s plot analysis where the ENc values are plotted against the GC3s values (GC at the 3rd position in the codon) to determine the significant factors such as selection or mutation pressure affecting the codon usage bias (Tian et al., 2020). In this analysis, genes whose codon bias is affected by mutations will lie on or around the expected curve, whereas genes whose codon bias is affected by selection and other factors will lie beneath the expected curve (Wang et al., 2018; Tian et al., 2020). Interestingly, we observed that a vast majority of the points fall below the expected curve for the S1-NTD (region 6) of SARS-CoV-2/SARS-CoVs (Fig. 9B), indicating the substantial influence of selection pressure than mutation pressure. Similarly, neutrality plot analysis was done where GC12 values (average GC content percentage at the first and second position in the codon) are plotted against GC3 values to evaluate the degree of impact of mutation and natural selection pressure on the codon usage. Our neutrality plot analysis revealed that the S1-NTD (region 6) of SARS-CoV-2/SARS-CoVs had a slope of 0.07183X (Y = 0.07183X+38.00, R2 = 0.05262; p < 0.0001) (Fig. 9C), indicating that the mutation pressure and natural selections were 7.1% and 92.9%, respectively. Finally, we performed Parity rule 2 bias analysis, where the AT bias [A3/(A3+ T3)] is plotted against GC-bias [G3/(G3 + C3)] to determine whether mutation pressure and natural selection affect the codon usage bias (Tian et al., 2020). If A = T and G = C, it indicates no mutation pressure and natural selection, while any discrepancies indicate mutation pressure and natural selection. In our analysis, we found uneven A3>T3 and G3>C3 numbers in the S1-NTD (region 6) of SARS-CoV-2/SARS-CoVs sequences (Fig. 9D), suggesting the existence of mutation and selection pressure. Collectively, these results indicate the presence of intense selection pressure in this region.
Fig. 9.
Selection pressure in the S1-NTDs. (A) ENc values indicate the effective number of codons usage of S1-NTD of SARS-CoV-2 and SARS-CoV. The ENc values are plotted in the Y-axis, and the sequence number is plotted in the X-axis. A total of 434 S1-NTD nucleotide sequences are used, and the sequence details are presented in supplementary data 5. (B) ENc plotted, the ENc values of the S1-NTD of SARS-CoV-2 and SARS-CoV plotted against GC3s (n = 434). The black curved line is the expected ENc value. (C) Neutrality plot analysis of the GC12 and the GC3 for S1-NTD of SARS-CoV-2 and SARS-CoV (n = 434). (D) Parity Rule 2 (PR2)-bias plot analysis for S1-NTD of SARS-CoV-2 and SARS-CoV (n = 434). A total of 434 S1-NTD nucleotide sequences are used, and the sequence details are presented in supplementary data 5.
4. Discussions
In this work, we report that the SARS-CoV-2 gained the novel S1-NTD in its genome. Most studies focused on the rapid spread of the SARS-CoV-2 due to its S1-RBD diversity to bind with the human ACE2 receptor and the presence of a specific furin cleavage site (FCS) (Zhou et al., 2020b; Wang et al., 2013; Coutard et al., 2020; Ramanathan et al., 2021; Andersen et al., 2020; Zhang et al., 2020; Zhang and Holmes, 2020; Segreto and Deigin, 2021). However, the furin cleavage site is present not only in SARS-CoV-2 but also present in MERS-CoVs, HKU1, and HCV-OC43 (Zhou et al., 2020b; Andersen et al., 2020; Zhang and Holmes, 2020; Wu and Zhao, 2020) and the SARS-CoV-2 without FCS also can infect target cells efficiently in the presence of trypsin or human airway trypsin-like protease (HAT) (Xia et al., 2020). Further, the S1-RBD for the human ACE2 is also present in SARS-CoV; however, SARS-CoV-2 has improved binding to ACE2 receptor than SARS-CoV (Lan et al., 2020; Shang et al., 2020). Moreover, SARS-CoV-2 has been shown to infect efficiently 293T and Vero cells without human ACE2 transfection (Suryadevara et al., 2021; Nie et al., 2021). Therefore, it could be inferred that the SARS-CoV-2-S1-RBD and FCS might cause the severity of the infection. In addition, some other factors also contribute to the increased transmissibility of disease in humans, which is the hallmark of SARS-CoV-2 compared to SARS-CoV and MERS-CoV.
Compared to other coronaviruses, SARS-CoV-2 has a large flat, non-sunken sialic acid-binding domain with extended and divergent loop sections of the SARS-CoV-2 S1-NTD. The above modification may improve viral transmission and infectivity by facilitating sialic acid-binding ability (Lundstrom et al., 2020; Vandelli et al., 2020; Baker et al., 2020; Seyran et al., 2020; Sun, 2021). Furthermore, glycosylated cell surface proteins and lipids are present in most types of cells and play a role in immunity and cell-cell communication (Li, 2015, 2016; Dove, 2001; Ghazarian et al., 2011; Schwegmann-Wessels and Herrler, 2006). Perhaps, SARS-CoV-2 can first attach to any cells with the S1-NTD to the cell surface sugar receptor. Further, it possibly allows the S1-RBD to search for the specific receptor for the virus entry into the cell, dramatically increasing infection's transmissibility. The SARS-CoV-2 S1-NTD specific monoclonal antibody (mAb) treatment showed the role of S1-NTD in the receptor post-attachment virus entry (without affecting the binding of the S1-RBD with the ACE2) and cell-cell fusion, thereby altogether abolishing the clinical onset of the infection (Suryadevara et al., 2021; McCallum et al., 2021a). Further, most of the highly transmissible SARS-CoV-2 variants of Mink Cluster 5, B.1.1.7, B.1.351, P.1, B.1.617.1, B.1.617.2, B.1.526, B.1.525, B.1.177, B.1.427 and B.1.429 were linked to the mutations/deletions in the S1-NTDs in natural infection (Suryadevara et al., 2021; McCallum et al., 2021a, 2021b; McCarthy et al., 2021; Annavajhala et al., 2021; Hodcroft et al., 2021; Kemp et al., 2021; Meng et al., 2021; Avanzato et al., 2020) and also in the experimental condition (McCallum et al., 2021a). Furthermore, the Cambodian bats' viruses (RShSTT200 and RShSTT182) are recombinant of SARS-CoV-2/RaTG13/BANAL-20-52-like genome with S1-NTD of SARS-CoV-related bat coronavirus, that was not detected in the ongoing pandemics. In contrast, bat/RaTG13/BANAL-20-52 related viruses (Type-1-S1-NTD) established pandemic, indicating the novel Type-I-S1-NTD in the SARS-CoV-2 genome might be the reason for its rapid pandemic spread. However, the functional importance of novel Type-1-S1-NTD is needed to be studied extensively in experimental conditions.
Importantly, SARS-CoV-2/SARS-CoVs complete spike proteins showed the evolutionary closer relationship with the MERS-CoVs-related spike proteins than OC43/HKU1/BCoV/ECoV/PHEV/MHV in CLANS analysis. However, the evolutionary proximity of S1-NTDs of SARS-CoV-2/SARS-CoV viruses exposes to S1-NTDs of OC43/HKU1/BCoV/ECoV/PHEV/MHV viruses rather than MERS-CoV viruses. Next, SARS-CoV-2/SARS-CoVs spike protein without S1-NTD showed an evolutionary closer relationship with the MERS-CoVs-related spike proteins than OC43/HKU1/BCoV/ECoV/PHEV/MHV (Supplementary Fig. 1D and 1E). From these results, it can be inferred that SARS-CoV-2/SARS-CoV viruses may have acquired S1-NTD from one virus (OC43/HKU1/BCoV/ECoV/PHEV/MHV) and other parts of the non-S1-NTD spike protein from another (MERS-CoVs-related spike proteins). These events may have taken place through genetic recombination. Though the OC43, HKU1, BCoV, ECoV, PHEV, and MHV S1-NTDs displayed the close-fitting pairwise similarity network at the p-value threshold of 1e−130, each specific viruses formed independent orphan clusters at the p-value threshold of 1e−168. Similarly, MERS-CoV, HKU4, HKU5, and Erinaceus-CoV created separate orphan clusters at the p-value threshold of 1e−168. Next, at the p-value threshold of 1e−168, the S1-NTDs of SARS-CoV-2 viruses revealed three distinct clusters (Type I-III), and the S1-NTDs of SARS-CoV viruses clustered to another three different groups (Type A-C). The SARS-CoV virus that caused the outbreak in 2003 contained Type-A S1-NTD, and it had 52.3% and 47.9% amino acid and nucleotide sequence diversity against Type-B S1-NTD. Likewise, SARS-CoV-2 holds the Type-I S1-NTD. The SARS-CoV-2 has the Type-I S1-NTD displayed 38–71% amino acid/nucleotide sequence diversity against the Type-II/Type-III S1-NTDs. These results indicate that beta coronaviruses that cause various major outbreaks have their own unique S1-NTDs.
Furthermore, in most genetic areas other than S1-NTD, the Cambodian bat coronaviruses RShSTT200 & RShSTT182 are closely related to SARS-CoV-2. But reveals a close relationship with SARS-CoV-related viruses in the S1-NTD region. Similarly, SARS-CoV-2-related BANAL-20-103, BANAL-20-236, BANAL-20-116, BANAL-20-247, RShSTT182h, RShSTT200, RmYN02, RacCS203, and RpYN06 viruses displayed substantial diversity against SARS-CoV-2 in S1-NTD. In conjunction with this, S1-NTDs of RmYN02, RacCS203, BANAL-20-116, and BANAL-20-247 viruses containing Type-III S1-NTD are more closely related to SARS-CoV-related bat viruses S1-NTD than SARS-CoV-2 in CLANS, Phylogenetic, NBGMD, similarity plot, and RDP analysis. These suggest that the RmYN02, RacCS203, BANAL-20-116, and BANAL-20-247 viruses containing Type-III S1-NTD may have evolved through recombination SARS-CoV-2 genome with SARS-CoV-related bat viruses S1-NTD. Altogether, it brings to light that the genetic recombination/exchange of S1-NTD between SARS-CoV and SARS-CoV-2 viruses is generally possible. When analyzed by Boni et al., 2020) after removing recombinant regions by three independent methods, it has been reported that SARS-CoV-2 is not originated by recombination of any of the sarbecovirus detected to date (Boni et al., 2020). It is noteworthy that only SARS-CoV-2 related viruses such as RaTG13, Pangolin-Guangdong CoV, Pangolin-GX CoV, bat-SL-CoVZC45, and bat-SL-CoVZXC21 were used in the 2020 study by Boni et al., 2020) (Boni et al., 2020). Other SARS-CoV-2-related viruses such as BANAL-20-52, BANAL-20-103, BANAL-20-236, BANAL-20-116, BANAL-20-247, RShSTT182h, and RShSTT200, bat/PrC31, RpYN06, RmYN02, and RacCS203 were not used because the sequences of these viruses were not available at the time. Furthermore, Makarenkov et al., 2021 (Makarenkov et al., 2021), Xia et al., 2020 (Xiao et al., 2020), and Lam et al., 2020 (Li et al., 2020) studies reported that SARS-CoV-2 may have originated from the recombination of S1-RBD of pangolin-Guangdong CoVs with RaTG13. It is noteworthy that these studies also used only the sequences of SARS-CoV-2 related viruses used by Boni et al., 2020) (Xiao et al., 2020; Boni et al., 2020; Makarenkov et al., 2021; Li et al., 2020).
In addition, after RaTG13&BANAL-20-52 viruses, pangolin-GX viruses form the most genetic collaboration in the S1-NTD region of the SARS-CoV-2 virus. Similarly, BANAL-20-103&BANAL-20-236 viruses containing Type-II S1-NTD exhibit a genetic relationship parallel to BANAL-20-52 in the RBD region of the SARS-CoV-2. In addition, pangolin-Guangdong viruses with Type-II S1-NTD form genetic interactions somewhat similar to RaTG13 in the RBD region of the SARS-CoV-2 virus. But pangolin-Guangdong viruses have a more significant relationship than RaTG13 in the RBD region of the SARS-CoV-2 virus at the level of the critical amino acids that bind to the human ACE2 receptor. We believe from all of this analysis that many years ago, a virus spike gene such SARS-CoV2/RaTG13/BANAL-20-52-like may have been formed by recombination of pangolin-Guangdong/BANAL-20-103/BANAL-20-236-like spike genes with pangolin-GX-like Type-I-like-S1-NTD. The spike gene of SARS-CoV2, RaTG13, and BANAL-20-52 viruses might then have evolved from these SARS-CoV2/RaTG13/BANAL-20-52-like viruses with somewhat similar S1-NTDs and slightly different RBD regions. The spike gene of BANAL-20-52 and SARS-CoV-2 viruses retain the critical amino acids that bind to the human ACE2 receptor, but some of these amino acids are mutated in RaTG13. Finally, the spike gene of SARS-CoV-2 viruses might have been evolved with the novel S1-NTD, the critical amino acids that bind to the human ACE2 receptor and furin cleavage sites. However, we emphasize the importance of the novel S1-NTD and its essential functions to be discovered in the future.
In conclusion, SARS-CoV-2 gained the novel S1-NTD and probably originated through recombination and host jump mediated evolution. Identifying the potent host receptor for the different types of S1-NTDs to determine its role in viral spread, host range, and severity of infection is needed to be investigated. Further, structure-based studies on the importance of mutations in the Type-I-S1-NTD and monitoring of mutations in this domain are warranted to predict the future direction of the current pandemic; to design novel antiviral molecules to develop vaccines, and neutralize antibodies to prevent and control the ongoing and prospective COVID-19 outbreaks.
Data availability
We retrieved the SARS coronavirus's related nucleotide sequences from publicly available NCBI and GISAID databases. The accession numbers and sequence names are indicated in respective Figures and supplementary data.
Funding
P.A.D is a DST-INSPIRE faculty is supported by research funding from the Government of India (DST/INSPIRE/04/2016/001067). P.A.D is supported by research funding from the Science and Engineering Research Board, Department of Science and Technology, Government of India (CRG/2018/002192).
Author contributions
PAD performed most of the bioinformatics experiments. KN assisted PAD for most of the bioinformatics works. PAD and KN wrote the first draft of the manuscript. KD edited and proofread the manuscript. PAD conceived the study, designed experiments, and wrote the final version of the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Director, IISc, Bangalore, India, The Chair, Department of Microbiology and Cell Biology, Indian Institute of Science, Bengaluru, India, and Prof. Nagalingam R. Sundaresan, Department of Microbiology and Cell Biology, Indian Institute of Science, Bengaluru, India for the providing working place at IISc, Bangalore.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envres.2022.113047.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
(A&B) The RBD region (22561-23161 nt) specific (A) phylogenetic tree and (B) net between-group mean distance for SARS-CoV-2 related viruses. (C) The spike gene without S1-NTD specific net between-group mean distance for SARS-CoV-2 related viruses. A total of 292 nucleotide sequences of spike gene without S1-NTDs were used in this analysis. The different group-specific viruses sequence details are listed in supplementary data 6. (D&E) CLANS analysis was performed using the spike protein without S1-NTD of beta coronaviruses that can infect humans and their related spike protein of viruses that can infect animals. A total of 2014 amino acid sequences of spike protein without S1-NTD were used in this analysis. The details of strains that belong to different clusters are presented in supplementary data 7. (D) The p-value threshold of 1e−170 in CLANS is used to show the lines connecting the sequences. (E) The p-value threshold of 1e−155 in CLANS is used to indicate the lines connecting the sequences.
References
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annavajhala M.K., Mohri H., Zucker J.E., Sheng Z., Wang P., Gomez-Simmonds A., Ho D.D., Uhlemann A.C. 2021. A Novel SARS-CoV-2 Variant of Concern, B.1.526, Identified in New York. medRxiv : the Preprint Server for Health Sciences. [Google Scholar]
- Avanzato V.A., Matson M.J., Seifert S.N., Pryce R., Williamson B.N., Anzick S.L., Barbian K., Judson S.D., Fischer E.R., Martens C., et al. Case study: prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised individual with cancer. Cell. 2020;183 doi: 10.1016/j.cell.2020.10.049. 1901-1912 e1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker A.N., Richards S.J., Guy C.S., Congdon T.R., Hasan M., Zwetsloot A.J., Gallo A., Lewandowski J.R., Stansfeld P.J., Straube A., et al. The SARS-COV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 2020;6:2046–2052. doi: 10.1021/acscentsci.0c00855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beniac D.R., Andonov A., Grudeski E., Booth T.F. Architecture of the SARS coronavirus prefusion spike. Nat. Struct. Mol. Biol. 2006;13:751–752. doi: 10.1038/nsmb1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni M.F., Lemey P., Jiang X., Lam T.T., Perry B.W., Castoe T.A., Rambaut A., Robertson D.L. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nature microbiology. 2020;5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir. Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove A. The bittersweet promise of glycobiology. Nat. Biotechnol. 2001;19:913–917. doi: 10.1038/nbt1001-913. [DOI] [PubMed] [Google Scholar]
- Ghazarian H., Idoni B., Oppenheimer S.B. A glycobiology review: carbohydrates, lectins and implications in cancer therapeutics. Acta Histochem. 2011;113:236–247. doi: 10.1016/j.acthis.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., et al. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29 doi: 10.1016/j.chom.2020.11.007. 44-57 e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft E.B., Zuber M., Nadeau S., Vaughan T.G., Crawford K.H.D., Althaus C.L., Reichmuth M.L., Bowen J.E., Walls A.C., Corti D., et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature. 2021 doi: 10.1038/s41586-021-03677-y. [DOI] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulswit R.J., de Haan C.A., Bosch B.J. Coronavirus spike protein and tropism changes. Adv. Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R.P., Ferreira I., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S., et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020;583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annual review of virology. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Berardi M., Li W., Farzan M., Dormitzer P.R., Harrison S.C. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 2006;80:6794–6800. doi: 10.1128/JVI.02744-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Giorgi E.E., Marichannegowda M.H., Foley B., Xiao C., Kong X.P., Chen Y., Gnanakaran S., Korber B., Gao F. Emergence of SARS-CoV-2 through recombination and strong purifying selection. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom K., Seyran M., Pizzol D., Adadi P., Mohamed Abd El-Aziz T., Hassan S.S., Soares A., Kandimalla R., Tambuwala M.M., Aljabali A.A.A., et al. 2020. Viewpoint: Origin of SARS-CoV-2. Viruses; p. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarenkov V., Mazoure B., Rabusseau G., Legendre P. Horizontal gene transfer and recombination analysis of SARS-CoV-2 genes helps discover its close relatives and shed light on its origin. BMC ecology and evolution. 2021;21:5. doi: 10.1186/s12862-020-01732-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus evolution. 2015;1 doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184 doi: 10.1016/j.cell.2021.03.028. 2332-2347 e2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., Bassi J., Marco A., Chen A., Walls A.C., Iulio J.D., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., et al. 2021. SARS-CoV-2 Immune Evasion by Variant B.1.427/B.1.429. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng B., Kemp S.A., Papa G., Datir R., Ferreira I., Marelli S., Harvey W.T., Lytras S., Mohamed A., Gallo G., et al. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the Alpha variant B.1.1.7. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie J., Li Q., Zhang L., Cao Y., Zhang Y., Li T., Wu J., Liu S., Zhang M., Zhao C., et al. Functional comparison of SARS-CoV-2 with closely related pangolin and bat coronaviruses. Cell discovery. 2021;7:21. doi: 10.1038/s41421-021-00256-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. : journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2020;79 doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccoli L., Park Y.J., Tortorici M.A., Czudnochowski N., Walls A.C., Beltramello M., Silacci-Fregni C., Pinto D., Rosen L.E., Bowen J.E., et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183 doi: 10.1016/j.cell.2020.09.037. 1024-1042 e1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbo P., Bravo I.G., Garcia-Vallve S. CAIcal: a combined set of tools to assess codon usage adaptation. Biol. Direct. 2008;3:38. doi: 10.1186/1745-6150-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M., Ferguson I.D., Miao W., Khavari P.A. SARS-CoV-2 B.1.1.7 and B.1.351 spike variants bind human ACE2 with increased affinity. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarah Temmam K.V., Eduard Baquero Salazar, Sandie Munier, Max Bonomi, Béatrice Régnault, Bounsavane Douangboubpha, Yasaman Karami, Delphine Chretien, Daosavanh Sanamxay, Vilakhan Xayaphet, Phetphoumin Paphaphanh, Vincent Lacoste, Somphavanh Somlor, Khaithong Lakeomany, Nothasin Phommavanh, Philippe Pérot, Flora Donati, Thomas Bigot, Michael Nilges, Félix Rey, Sylvie van der Werf, Paul Brey, Marc Eloit. 2021. Coronaviruses with a SARS-CoV-2-like Receptor-Binding Domain Allowing ACE2-Mediated Entry into Human Cells Isolated from Bats of Indochinese Peninsula. [Google Scholar]
- Schwegmann-Wessels C., Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconj. J. 2006;23:51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segreto R., Deigin Y. The genetic structure of SARS-CoV-2 does not rule out a laboratory origin: SARS-COV-2 chimeric structure and furin cleavage site might be the result of genetic manipulation. Bioessays : news and reviews in molecular, cellular and developmental biology. 2021;43 doi: 10.1002/bies.202000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyran M., Takayama K., Uversky V.N., Lundstrom K., Palu G., Sherchan S.P., Attrish D., Rezaei N., Aljabali A.A.A., Ghosh S., et al. The structural basis of accelerated host cell entry by SARS-CoV-2dagger. FEBS J. 2020 doi: 10.1111/febs.15651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.L. The role of cell surface sialic acids for SARS-CoV-2 infection. Glycobiology. 2021 doi: 10.1093/glycob/cwab032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., et al. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. Cell. 2021;184 doi: 10.1016/j.cell.2021.03.029. 2316-2331 e2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H.F., Hu Q.M., Xiao H.B., Zeng L.B., Meng Y., Li Z. Genetic and codon usage bias analyses of major capsid protein gene in Ranavirus. Infect. Genet. Evol. 2020;84 doi: 10.1016/j.meegid.2020.104379. [DOI] [PubMed] [Google Scholar]
- Vandelli A., Monti M., Milanetti E., Armaos A., Rupert J., Zacco E., Bechara E., Delli Ponti R., Tartaglia G.G. Structural analysis of SARS-CoV-2 genome and predictions of the human interactome. Nucleic Acids Res. 2020;48:11270–11283. doi: 10.1093/nar/gkaa864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetrivel U., Arunkumar V., Dorairaj S. ACUA: a software tool for automated codon usage analysis. Bioinformation. 2007;2:62–63. doi: 10.6026/97320630002062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S., Tan C.W., Maneeorn P., Duengkae P., Zhu F., Joyjinda Y., Kaewpom T., Chia W.N., Ampoot W., Lim B.L., et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 2021;12:972. doi: 10.1038/s41467-021-21240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. 281-292 e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Shi X., Jiang L., Zhang S., Wang D., Tong P., Guo D., Fu L., Cui Y., Liu X., et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Xing H., Yuan Y., Wang X., Saeed M., Tao J., Feng W., Zhang G., Song X., Sun X. Genome-wide analysis of codon usage bias in four sequenced cotton species. PLoS One. 2018;13 doi: 10.1371/journal.pone.0194372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181 doi: 10.1016/j.cell.2020.03.045. 894-904 e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhao S. Furin cleavage sites naturally occur in coronaviruses. Stem Cell Res. 2020;50 doi: 10.1016/j.scr.2020.102115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal transduction and targeted therapy. 2020;5:92. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J.J., Li N., Guo Y., Li X., Shen X., et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. 2020;583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- York A. Searching for relatives of SARS-CoV-2 in bats. Nat. Rev. Microbiol. 2021 doi: 10.1038/s41579-021-00595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.Z., Holmes E.C. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. 2020;181:223–227. doi: 10.1016/j.cell.2020.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. : CB. 2020;30 doi: 10.1016/j.cub.2020.03.063. 1346-1351 e1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Zheng H., Xu A., Yan D., Jiang Z., Qi Q., Sun J. Analysis of codon usage bias of envelope glycoprotein genes in nuclear polyhedrosis virus (NPV) and its relation to evolution. BMC Genom. 2016;17:677. doi: 10.1186/s12864-016-3021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Chen X., Hu T., Li J., Song H., Liu Y., Wang P., Liu D., Yang J., Holmes E.C., et al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr. Biol. : CB. 2020;30 doi: 10.1016/j.cub.2020.09.030. 2196-2203 e2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Ji J., Chen X., Bi Y., Li J., Wang Q., Hu T., Song H., Zhao R., Chen Y., et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021;184 doi: 10.1016/j.cell.2021.06.008. 4380-4391 e4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Ji J., Chen X., Bi Y., Li J., Wang Q., Hu T., Song H., Zhao R., Chen Y., et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021 doi: 10.1016/j.cell.2021.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A&B) The RBD region (22561-23161 nt) specific (A) phylogenetic tree and (B) net between-group mean distance for SARS-CoV-2 related viruses. (C) The spike gene without S1-NTD specific net between-group mean distance for SARS-CoV-2 related viruses. A total of 292 nucleotide sequences of spike gene without S1-NTDs were used in this analysis. The different group-specific viruses sequence details are listed in supplementary data 6. (D&E) CLANS analysis was performed using the spike protein without S1-NTD of beta coronaviruses that can infect humans and their related spike protein of viruses that can infect animals. A total of 2014 amino acid sequences of spike protein without S1-NTD were used in this analysis. The details of strains that belong to different clusters are presented in supplementary data 7. (D) The p-value threshold of 1e−170 in CLANS is used to show the lines connecting the sequences. (E) The p-value threshold of 1e−155 in CLANS is used to indicate the lines connecting the sequences.
Data Availability Statement
We retrieved the SARS coronavirus's related nucleotide sequences from publicly available NCBI and GISAID databases. The accession numbers and sequence names are indicated in respective Figures and supplementary data.