Abstract
Background: Anaplastic thyroid cancer (ATC) is a rare cancer with poor prognosis and few treatment options. The objective of this study was to investigate new immune-associated therapeutic targets by identifying ATC-derived, human leukocyte antigen (HLA) class II-presenting peptides. One protein that generated multiple peptides in ATC was chondroitin sulfate-proteoglycan-4 (CSPG4), a transmembrane proteoglycan with increased expression in multiple aggressive cancers, but not yet investigated in ATC.
Methods: We applied autologous peripheral blood T cells to ATC patient-derived xenografted mice to examine whether ATC induces a tumor-specific T cell response. We then identified peptide antigens eluted from the HLA-DQ complex in ATC patient-derived cells using mass spectrometry, detecting abundant CSPG4-derived peptides specific to the ATC sample. Next, we analyzed the surface expression level of CSPG4 in thyroid cancer cell lines and primary cell culture using flow cytometry. In addition, we used immunohistochemistry to compare the expression level and localization of the CSPG4 protein in ATC, papillary thyroid cancer, and normal thyroid tissue. We then investigated the correlation between CSPG4 expression and clinicopathological features of patients with thyroid cancer.
Results: We found that ATC tissue had a high level of HLA-DQ expression and that the patient's CD4+ T cells showed activation when exposed to ATC. By eluting the HLA-DQ complex of ATC tissue, we found that CSPG4 generated one of the most abundant and specific peptides. CSPG4 expression at the cell surface of thyroid cancer was also significantly high when determined by flow cytometry, with the majority of ATC cell lines exhibiting ∼10-fold higher mean fluorescence intensity. Furthermore, most ATC patient cases expressed CSPG4 in the cytoplasm or membrane of the tumor cells. CSPG4 expression was correlated with tumor size, extrathyroidal extension, and intercellular adhesion molecule-1 (ICAM-1) circumferential expression. CSPG4 mRNA overexpression was associated with worse overall survival in patients with ATC and poorly differentiated thyroid cancer.
Conclusions: CSPG4 expression is significantly elevated in aggressive thyroid cancers, with a strong correlation with a poor prognosis. The vast number of HLA-DQ eluted CSPG4 peptides was identified in ATC, demonstrating the potential of CSPG4 as a novel immunotherapeutic target for ATC.
Keywords: anaplastic thyroid cancer, CSPG4, thyroid cancer
Introduction
Anaplastic thyroid cancer (ATC), an aggressive subtype, makes up 1–2% of thyroid cancers and has a median survival of less than four months (1,2). Genomic mutations of thyroid cancers are well studied, with BRAFV600E mutation being the most common in papillary thyroid cancer (PTC) (3), with reported BRAF mutational rates of 40% or greater in ATC (4–6). Recently, combination therapy with a BRAFV600E inhibitor, dabrafenib, and an MEK inhibitor, trametinib, showed promising results in BRAF-mutated ATCs (7). However, a significant subset of ATCs do not have BRAF mutations, and treatment with kinase inhibitors and chemotherapeutic agents has showed limited response in most ATCs (8–10); other therapeutic targets are needed.
Recent progress in immunotherapy with checkpoint inhibitors has had a tremendous impact on cancer therapy (11); however, fewer than 20% of ATC patients had partial or complete responses to immunotherapy (12). In a recent phase II clinical trial, programmed cell death-1 (PD-1) blockade had an overall response rate of 19% and a 7% complete response rate in ATC (13). Blocking the interaction between checkpoint inhibitors and their ligands promotes T cell activities against tumor antigens. While most cancer immunotherapy efforts have focused on CD8+ T cell responses against peptide antigens presented on the major histocompatibility complex (MHC) class I, recent studies have shown that CD4+ T cells can elicit powerful tumor control by their specific interactions with tumor antigens presented on MHC II (14–16). By investigating a patient-derived ATC sample for novel MHC II-presented peptide antigens, we identified a large number of chondroitin sulfate-proteoglycan-4 (CSPG4)-derived peptides.
CSPG4, a proteoglycan first discovered in melanoma (17–19), is a transmembrane protein that can be expressed on cells either as an N-linked glycoprotein of ∼250 kDa or as a 450 kDa N-linked glycoprotein with associated proteoglycan (20). The large extracellular domain, with subdomains D1, D2, and D3, functions as a high-affinity receptor for extracellular proteins, growth factors, and integrins (20). Increased levels of CSPG4 expression have been demonstrated in many aggressive cancers, including melanomas (17), gliomas (21), mesotheliomas (22), sarcomas (23), and triple negative breast cancers (24). Moreover, CSPG4 expression has been associated with poor prognosis (23,25), hematogenous metastasis (26), and angiogenesis (27). Previous characterization of CSPG4 expression in normal and neoplastic tissues found that normal thyroid tissue did not express CSPG4 (28,29); however, CSPG4 expression has not yet been determined in thyroid cancer. Previous studies using anti-CSPG4 monoclonal antibodies (mAb) (22,30) and chimeric antigen receptor (CAR) T lymphocytes against CSPG4 (31–33) have shown promising therapeutic results in vitro and in preclinical models of other cancers.
Here we identified multiple CSPG4-derived peptides in ATC patient-derived cells after immunoprecipitation of a human leukocyte antigen (HLA) class II complex by mass spectrometry. We further analyzed CSPG4 expression in normal thyroid, PTC, and ATC patient-derived tissues and cell lines using immunohistochemistry (IHC) and flow cytometry.
Materials and Methods
Sample collection and institutional review board approval
Preoperative informed written consent was obtained from patients who underwent open neck biopsy, hemithyroidectomy, or thyroidectomy at Weill Cornell Medicine (WCM) from 2004 to 2020, and the study was approved by the WCM Institutional Review Board (IRB). Patients' demographics and tumor characteristics were collected via review of electronic medical record and pathology reports.
Cell culture
For thyroid cancer cell lines, BCPAP, 8505C, KHM-5M, and SW1736 were purchased commercially or gifted by researchers. A375-MA2 and T2 were purchased from ATCC (Manassas, VA). B lymphoblastic cell lines (B-LCL) MOU and 1331 were from Fred Hutchinson Cancer Research Center (Seattle, WA). The source of the cells, deriviation method for patient-derived ATC cells, and culture condition are described in detail in Supplementary Methods.
Isolation of peripheral blood mononuclear cells and T cell expansion
Fresh peripheral blood mononuclear cells (PBMCs) from an ATC patient were isolated using Ficoll-PaquePLUS density media (GE Healthcare) by gradient centrifugation. T cells were enriched by adding Dynabeads Human T cell Expander CD3/CD28 (ThermoFisher) in OpTmizer CTS media (ThermoFisher) supplemented with 5% human AB serum (Sigma-Aldrich), recombinant human IL7 and IL15 (both cytokines from Peprotech), and cryopreserved until usage.
Mice study
All mouse studies were approved by the WCM Institutional Animal Care and Use Committee. For xenograft experiments, we intravenously injected 0.5 million JV-ATC cells expressing fLuc per mouse into 8- to 10-week-old NOD-scid/IL2Rgnull (NSG; Jackson Laboratory) mice. The patient tumor-derived JV-ATC cells have been described previously (34). After nine days of xenograft, 1 million T cells isolated from the same donor as the tumor were injected to each xenograft. Tumor growth was tracked in vivo via whole-body luminescence imaging (In-Vivo Xtreme 4MP; Bruker). Tumor tissues from the JV-ATC xenograft were isolated and analyzed for the frequency of infiltrating T cells using an antibody cocktail against CD3, CD4, and CD8 (#319001; Biolegend). Tumor-infilitrating T cells were maintained in the CTS media, supplemented with 5% human AB serum, IL7, and IL15. These T cells isolated from the JV-ATC xenografts were cocultured with the JV-ATC tumor cells at a 1:1 ratio, and analyzed for CD69 and PD1 expression after 18 hours of coculture.
MHC-II-bound peptide isolation and liquid chromatography with tandem mass spectrometry analysis
Approximately 25 million cells of each type were used for HLA-DQ immunoprecipitation using the anti-HLA-DQ antibody (Catalog: NBP2-45041, clone SPV-L3; Novus Biologicals). Methods for peptide elution and mass spectrometry are described in Supplementary Methods.
Immunohistochemistry
Formalin-fixed, paraffin-embedded (FFPE) tissue blocks from patient tissue were stained with anti-CSPG4 (Catalog: ab139406, clone EPR9195; Abcam) on a Leica Bond system. The section was pretreated using heat-mediated antigen retrieval with Tris-EDTA buffer (pH9, epitope retrieval solution 2) for 20 minutes before antibody addition. See Supplementary Methods for detailed methods.
Flow cytometry
Key reagents and methods for live cell staining are described in Supplementary Methods. Anti-human CSPG4-APC (Catalog: ab130-117-497, clone EP-1; Miltenyi Biotec) was used to detect CSPG4 expression on live cells. The source and catalog number of the antibodies used in flow cytometry are documented in the Supplementary Methods.
Statistical analysis
Categorical variables were compared using Fisher's exact and chi-squared tests when appropriate, and continuous nonparametric variables were compared using the Wilcoxon rank-sum test. Survival curves were generated using the Kaplan–Meier method and compared using the log-rank test. Analysis was performed using Prism 8.0 (GraphPad) and Stata Version 16.1 (StataCorp). Statistical significance was set at p < 0.05.
Results
High expression of HLA-DQ in ATC
Previously, we reported establishing ATC patient-derived xenografts (PDX) and cell line, JV, which maintained the pathological and molecular features identical to the patient's original tumor (34). This JV-ATC tumor was BRAF wild type with NRAS, PTEN, and TP53 mutations. PBMCs were collected from the patient at time of surgery.
To expand tumor-reactive T cells and study their interaction with tumor cells, we administered donor-matched peripheral T cells to the JV-ATC PDX after nine days of xenograft (Supplementary Fig. S1A). Approximately one month after xenograft, we noticed that the tumor size was relatively smaller in xenografts administered with autologous T cells relative to the no treatment group. This finding suggested a possibility that T cells may be mediating blunted tumor growth.
To further examine whether those tumor-infiltrating T cells were enriched for specific T cell subsets, we isolated the regressing tumor from the JV-ATC xenograft, which was injected with the donor-matched T cells, and analyzed the tumor-infiltrating T cells by flow cytometry. Surprisingly, tumor-infiltrating T cells were predominantly CD4+ cells, skewed from the initial CD4:CD8 distribution when we first isolated from the patient's peripheral blood. The peripheral blood T cells had 70%:24% ratio of CD4:CD8 frequency, however the tumor-infiltrating T cells were greater than 97% of CD4+ T cells, with a barely detectable level of CD8+ T cells (Fig. 1A).
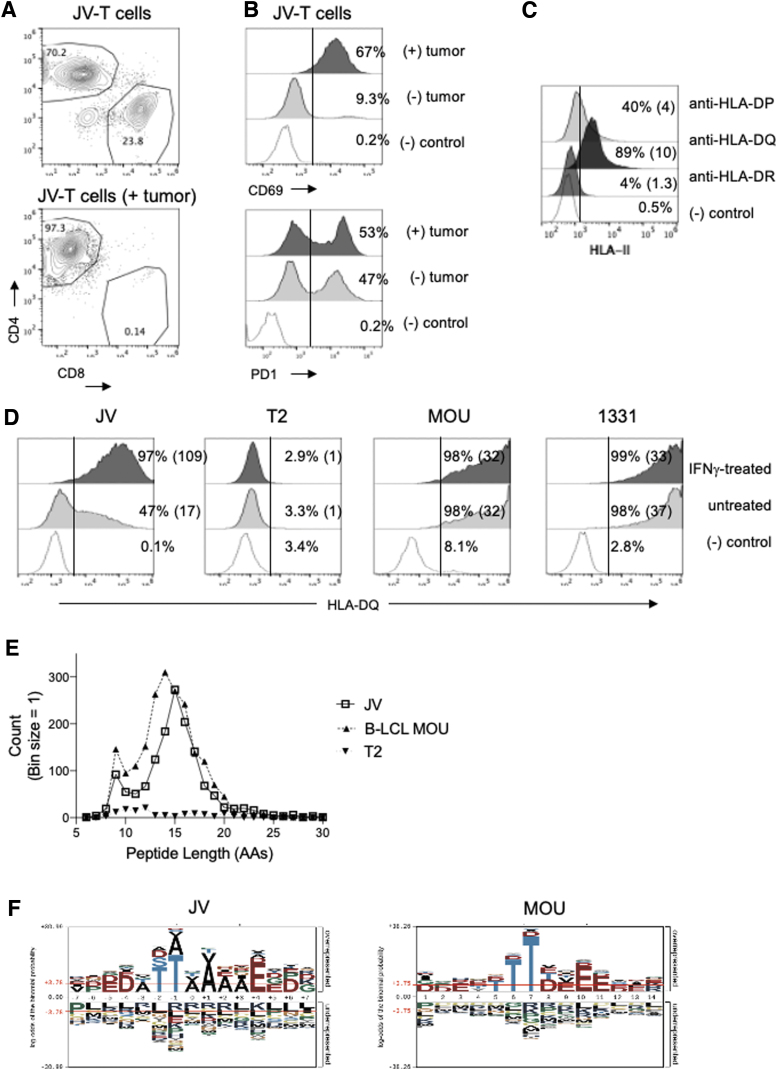
FIG. 1.
HLA-DQ-eluted peptides in JV ATC cells. (A, B) Characterization of peripheral T cells isolated from JV patient as determined by flow cytometry. (A) Frequencies of CD4:CD8 in peripheral JV-T cells (top) or in in vivo expanded JV-T cells after exposure to autologous tumor cells in vitro (bottom). (B) Expression of CD69 (top) or PD1 (bottom) in JV-T cells before or after exposure to autologous tumor cells. (C) Surface expression of MHC II molecules in JV tumor cells was determined by flow cytometry. The gates for positively stained cells were determined by staining with a secondary antibody only and were marked with vertical lines within the plots. The numbers in the parenthesis are the fold increases in mean fluorescence intensity of each MHC II expression. (D) The expression levels of HLA-DQ in uninduced and IFNγ-induced conditions (2e4 IU/mL for 48 hours) were determined in JV, T2, B-LCL MOU, and 1331 cells using SPV-L3 antibody. HLA-DQ histograms analyzed with the 1a3 antibody presented a similar profile to SPV-L3 (Supplementary Fig. S2). JV cell HLA-DQ haplotypes were HLA-DQA1*0201, HLA-DQB1*0202/HLA-DQB1*0302. MOU and 1331 B-LCL lines had DQ2.2 (HLA-DQA1*0201/HLA-DQB1*0202) and DQ8.1 (HLA-DQA1*0301/HLA-DQB1*0302), respectively. T2 cells were deficient in MHC II expression including HLA-DQ. (E) Peptide length distribution of HLA-DQ-eluted peptides from MOU, T2, and IFNγ-induced JV cells was shown as a histogram. (F) Sequence logos of eluted peptides isolated from JV-ATC (15 a.a., left) and MOU (14 a.a., right) were generated using pLogo generator (38). ATC, anaplastic thyroid cancer; B-LCL, B lymphoblastic cell line; HLA, human leukocyte antigen; MHC, major histocompatibility complex.
Next, to analyze whether those tumor-infiltrating T cells showed tumor-specific activity, we tested whether these T cells respond to the JV cells in vitro. After exposure to JV tumor cells at a 1:1 ratio, these JV tumor-isolated CD4+ T cells increased the expression of CD69, a marker of antigen-dependent T cell activation (Fig. 1B). The expression level of PD-1, an exhaustion marker, was slightly increased. These results suggested that the ATC tumor was antigenic and induced in vivo expansion of tumor-reactive CD4+ T cells, which were present in the circulating blood of the patient.
We investigated if any specific MHC II was expressed on JV-ATC cells, potentially presenting peptides to the CD4+ T cells. Surface expression level of HLA-DQ was the highest, followed by HLA-DP, whereas HLA-DR expression was negligible on the JV tumor cells (Fig. 1C). HLA-DQ expression was increased approximately eightfold when IFNγ was added to the cells for 48 hours when analyzed with the anti-HLA-DQ (1a3) antibody (35) (Supplementary Fig. S1B). HLA analysis using next-generation sequencing determined that the JV cells' HLA-DQ haplotypes were HLA-DQA1*0201 and HLA-DQB1*0202/*0302. We examined the HLA-DQ expression level with an anti-HLA-DQ (SPV-L3) antibody (Fig. 1D), which was previously used to profile HLA-DQ-restricted peptides in association with celiac disease (36). Flow cytometric analysis showed that half of JV cells expressed HLA-DQ, a slightly lower percentage of positivity compared with the 1a3 antibody (Fig. 1D). However, IFNγ treatment increased the DQ expression to approximately sixfold, almost to the same level as the result observed with the 1a3 antibody. HLA-DQ was not detectable in T2 cells, which are known to be defective for MHC II expression. B-LCL MOU and 1331 that shared DQ haplotype with JV displayed close to 100% frequency of DQ expression regardless of IFNγ treatment.
Next, we aimed to identify HLA-DQ-associated peptides enriched in JV-ATC cells and compare the number of peptides eluted in B-LCL MOU cells and T2 cells (Fig. 1E). Approximately 25 million cells of each group were used for eluting HLA-DQ-bound peptides using the SPV-L3 antibody. JV-ATC cells were stimulated with IFNγ to maximize the quantity of peptide elution. While both JV and MOU cells yielded a significant number of peptides (n = 920 for JV; n = 1239 for MOU), there were very few peptides eluted from T2 cells (n = 35) concordant with a lack of HLA-DQ expression (Supplementary Table S1). The length of the peptides varied between 6 and 30 amino acids with a peak in distribution at 14 and 15 amino acids for MOU and JV, respectively.
Peptides eluted from HLA-DQ cells were specific to ATC
We searched the immune epitope database (IEDB) for HLA-DQA1*0201/DQB1*0202 (HLA-DQ2.2) peptides (37). Similar to MOU and JV cells, the peptides eluted from the HLA-DQ2.2 complex showed the highest peaks at positions 14 and 15 (Supplementary Fig. S2). We found that specific peptides eluted in either JV or MOU cells or presented in both cells (Supplementary Table S1). Among the top 20 proteins with the most eluted peptides in each cell type, there was little overlap between MOU and JV, except for UBC, HSPA8, and HLA-B, proteins with function in homeostasis and antigen presentation (Tables 1 and 2, Supplementary Table S2). S100A8 and S100A9, Ca2+ binding proteins overexpressed in ATC cells (38), produced the most abundant peptides in JV-ATC cells, with the highest area among eluted peptides, but were not eluted in MOU cells. We isolated identical peptides, including β2-microglobulin, CD74, serglycin, and ubiquitin, from MOU cells as previously reported (36), validating our experimental procedure to identify DQ-restricted peptides presented in cells. Peptides eluted from both JV and MOU (39) exhibited a strong preference for threonine at positions 6 and 7 (Fig. 1F). After these threonine residues, the next four positions were enriched with alanine followed by glutamic acid in the peptides isolated from JV, whereas charged amino acids such as aspartic acid and glutamic acid were enriched in the peptides eluted from MOU. IEDB-registered peptides eluted from the HLA-DQ2.2 complex showed a remarkably similar sequence motif to the motif generated from MOU cells (Supplementary Fig. S2B). Overall, there were similarities in the DQ-eluted peptide length and composition, but with different types of proteins that were processed and presented between JV and MOU cells.
Table 1.
Twenty Most Abundant Proteins That Generated Peptides Eluted with HLA-DQ Specific Antibody (SPV-L3) in JV Cell Line Compared with MOU and T2
| Uniprota | Gene symbol | Description | Cell lines |
||
|---|---|---|---|---|---|
| MOUb | T2b | JVb | |||
| P05109 | S100A8 | Protein S100-A8, N-terminally processed | 0 | 0 | 239990800 |
| P06702 | S100A9 | Protein S100-A9 | 0 | 0 | 93871100 |
| P0CG48 | UBC | Polyubiquitin-C; ubiquitin | 36845093 | 82136 | 55053800 |
| P04406 | GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 11732600 | 0 | 51516553 |
| P36941 | LTBR | Tumor necrosis factor receptor superfamily member 3 | 0 | 0 | 33991000 |
| P18464 | HLA-B | HLA class I histocompatibility antigen, B-51 alpha chain | 1679600 | 0 | 31870000 |
| P01040 | CSTA | Cystatin-A, N-terminally processed | 0 | 0 | 30097800 |
| Q6UVK1 | CSPG4 | Chondroitin sulfate proteoglycan 4 | 0 | 0 | 17304600 |
| P31151 | S100A7 | Protein S100-A7 | 0 | 0 | 16113049 |
| P01903 | HLA-DRA | HLA class II histocompatibility antigen, DR alpha chain | 15392900 | 0 | 15082862 |
| P20930 | FLG | Filaggrin | 14160287 | 0 | 14160287 |
| P11142 | HSPA8 | Heat shock cognate 71 kDa protein | 65869600 | 96885 | 13812492 |
| Q9NZT1 | CALML5 | Calmodulin-like protein 5 | 0 | 0 | 13770187 |
| P07093 | SERPINE2 | Glia-derived nexin | 0 | 0 | 12985300 |
| P23381 | WARS | Tryptophan—tRNA ligase, cytoplasmic; T1-TrpRS; T2-TrpRS | 0 | 0 | 10930469 |
| P00441 | SOD1 | Superoxide dismutase [Cu-Zn] | 0 | 0 | 10864709 |
| P35527 | KRT9 | Keratin, type I cytoskeletal 9 | 9880967 | 6290028 | 10417904 |
| Q10589 | BST2 | Bone marrow stromal antigen 2 | 14997772 | 0 | 9795100 |
| P08865 | RPSA | 40S ribosomal protein SA | 189000 | 0 | 8996000 |
| P60174 | TPI1 | Triosephosphate isomerase | 0 | 0 | 8915073 |
Protein amino acid sequences were derived from the Uniprot database.
Protein area is derived from summing the intensities of all peptides that have been matched to amino acid sequences of a particular protein.
HLA, human leukocyte antigen.
Table 2.
Twenty Most Abundant Proteins That Presented Peptides with HLA-DQ Complexes in MOU Cells and Their Quantities in JV and T2 Cells
| Uniprota | Gene symbol | Description | Cell lines |
||
|---|---|---|---|---|---|
| MOUb | T2b | JVb | |||
| P61769 | B2M | Beta-2-microglobulin; beta-2-microglobulin form pI 5.3 | 8917824758 | 0 | 1302700 |
| P04233 | CD74 | HLA class II histocompatibility antigen gamma chain | 1291969600 | 1376700 | 0 |
| P0CG48 | UBC | Polyubiquitin-C; ubiquitin | 368450983 | 82136 | 55053800 |
| Q9NPF2 | CHST11 | Carbohydrate sulfotransferase 11 | 136873400 | 0 | 0 |
| P10124 | SRGN | Serglycin | 110363488 | 0 | 0 |
| P30457 | HLA-A | HLA class I histocompatibility antigen, A-66 alpha chain | 81047700 | 0 | 0 |
| P62258 | YWHAE | 14-3-3 protein epsilon | 77002235 | 0 | 0 |
| P61073 | CXCR4 | C-X-C chemokine receptor type 4 | 69844548 | 0 | 0 |
| P11142 | HSPA8 | Heat shock cognate 71 kDa protein | 65869600 | 96885 | 13812492 |
| P11836 | MS4A1 | B lymphocyte antigen CD20 | 60944700 | 0 | 0 |
| Q02818 | NUCB1 | Nucleobindin-1 | 60418500 | 0 | 0 |
| P07108 | DBI | Acyl-CoA-binding protein | 36739789 | 0 | 0 |
| P01857 | IGHG1 | Ig gamma-1 chain C region | 36209718 | 2329600 | 0 |
| P11279 | LAMP1 | Lysosome-associated membrane glycoprotein 1 | 31294400 | 0 | 7941915 |
| Q07000 | HLA-C | HLA class I histocompatibility antigen, Cw-15 alpha chain | 26873000 | 0 | 0 |
| Q15758 | SLC1A5 | Neutral amino acid transporter B (0) | 25598800 | 0 | 5772797 |
| Q9Y287 | ITM2B | Integral membrane protein 2B; BRI2, membrane form; BRI2 intracellular domain; BRI2C, soluble form; Bri23 peptide | 24537353 | 0 | 1027300 |
| P61981 | YWHAG | 14-3-3 protein gamma;14-3-3 protein gamma, N-terminally processed | 24525256 | 0 | 0 |
| Q7Z4F1 | LRP10 | Low-density lipoprotein receptor-related protein 10 | 24398500 | 0 | 277800 |
| P03989 | HLA-B | HLA class I histocompatibility antigen, B-27 alpha chain | 23919000 | 0 | 0 |
Protein amino acid sequences were derived from the Uniprot database.
Protein area is derived from summing the intensities of all peptides that have been matched to amino acid sequences of a particular protein.
CSPG4-derived peptides are presented in the HLA-DQ complex of an ATC
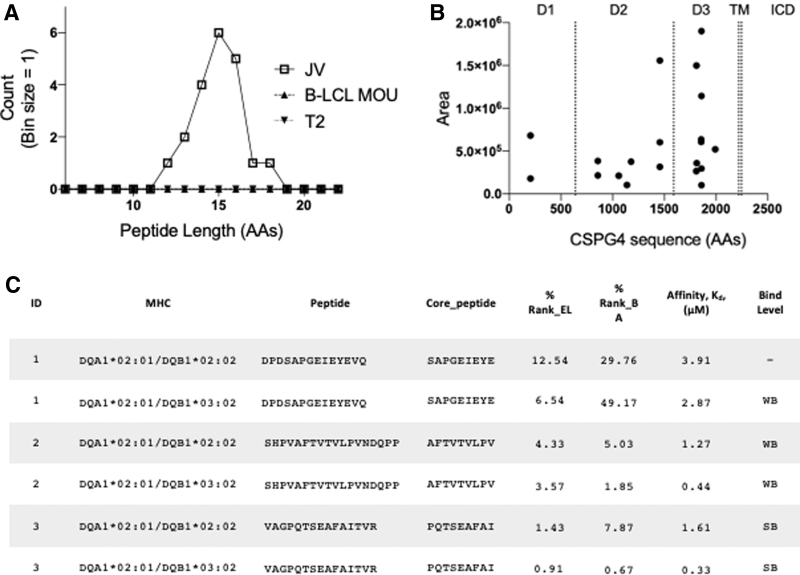
CSPG4 generated the highest peptide spectrum matches that were unique to JV-ATC cells (Supplementary Table S1). The average length of the CSPG4-derived peptides was 15 amino acids in JV cells but undetectable in MOU and T2 cells (Fig. 2A). All 20 unique peptides belonged to the three distinct extracellular domains (20) of CSPG4 (Fig. 2B). To assess the overall performance of eluted CSPG4 peptides, we estimated the affinity of eluted peptides to the respective HLA molecules by predicting binding affinities (Kd) using the NetMHCII pan-4.0 prediction program (40). We analyzed the CSPG4 peptides' binding affinity in the context of DQ2.2 and DQA1*0201/DQB1*0302, two types of possible DQ haplotypes expressed in the JV tumor cells (Table 3 and Fig. 2C). The top eluted peptide, DPDSAPGEIEYEVQ, appeared as variable lengths with multiple occurrences (Table 3), with predicted weak binding to both DQ haplotypes. The third-most abundant peptide, VAGPQTSEAFAITVR, was predicted to have a strong binding affinity in both DQ haplotypes (Fig. 2C). Overall, CSPG4 peptide binding affinities for the DQ2.2 haplotype were weaker than DQA1*0201; DQB1*0302. The predicted median binding affinities, Kd, of CSPG4 peptides were 2.39 and 0.57 μM for DQ2.2 and DQA1*0201; DQB1*0302, respectively (Table 3). Taken together, multiple HLA-DQ-restricted, CSPG4-derived peptides were isolated from ATC with strong signals, implicating high expression and antigen processing of CSPG4 in ATC cells.
FIG. 2.
HLA-DQ-eluted CSPG4 peptides in JV ATC cells. (A) Peptide length distribution of HLA-DQ-eluted CSPG4 peptides from MOU, T2, and IFNγ-induced JV cells was shown as a histogram. (B) Eluted CSPG4 peptide count areas (closed circles) were displayed along the CSPG4 sequence. Extracellular D1–3, TM, and ICD were shown on top of the plot. (C). The predicted binding affinities of the top 3 CSPG4 peptides for HLA-DQA1*02:01/DQB1*02:02 and HLA-DQA1*02:01/DQB1*03:02 were determined using the NetMHCIIpan-4.0 (38). The “Core peptide” identifies the binding core of the peptide to each “MHC” allele. “% Rank_EL” indicates the percentile rank of eluted ligand prediction score. “% Rank_BA” is calculated as a percentile rank of the predicted peptide affinity compared to a set of 100,000 random natural peptides.” Affinity is the predicted binding affinity, Kd, in μM. Threshold for SB and WB peptides was set at <2% and <10%, respectively. CSPG4, chondroitin sulfate-proteoglycan-4; D1–3, domains 1–3; ICD, intracellular domains; SB, strong binding; TM, transmembrane; WB, weak binding.
Table 3.
Unique CSPG4 Peptides Eluted from HLA-DQ Complexes in JV-Anaplastic Thyroid Cancer Tumors
| Gene symbol | Peptide | Peptide length | Area | Count | Predicted affinity, Kd (μM)a DQA1*02:01 DQB1*02:02 |
Predicted affinity, Kd (μM)a DQA1*02:01 DQB1*03:02 |
|---|---|---|---|---|---|---|
| CSPG4 | DPDSAPGEIEYEVQ | 14 | 1899000 | 1 | 3.91 | 1.72 |
| CSPG4 | SHPVAFTVTVLPVNDQPP | 18 | 1556000 | 2 | 1.27 | 0.44 |
| CSPG4 | VAGPQTSEAFAITVR | 15 | 1498000 | 2 | 1.61 | 0.33 |
| CSPG4 | DPDSAPGEIEYEV | 13 | 1143000 | 1 | 4.95 | 4.48 |
| CSPG4 | GRPTSAFSQFQIDQ | 14 | 520800 | 1 | 2.00 | 1.14 |
| CSPG4 | FSGPHSLAAFPAWGT | 15 | 680500 | 1 | 2.79 | 0.27 |
| CSPG4 | VVDPDSAPGEIEYEV | 15 | 635600 | 1 | 3.10 | 2.10 |
| CSPG4 | VVDPDSAPGEIEYEVQ | 16 | 606600 | 1 | 3.18 | 2.09 |
| CSPG4 | SHPVAFTVTVLPVNDQ | 16 | 603000 | 2 | 0.70 | 0.29 |
| CSPG4 | VTYGATARASEAVED | 15 | 383800 | 2 | 1.96 | 0.26 |
| CSPG4 | DPDSAPGEIEYE | 12 | 295400 | 2 | 7.40 | 5.94 |
| CSPG4 | GPQTSEAFAITVR | 13 | 359900 | 2 | 2.88 | 0.89 |
| CSPG4 | AGQPATAFSQQDLLD | 15 | 375100 | 1 | 0.81 | 0.69 |
| CSPG4 | SHPVAFTVTVLPVN | 14 | 315500 | 1 | 0.98 | 0.41 |
| CSPG4 | VTYGATARASEAVE | 14 | 216100 | 1 | 2.47 | 0.38 |
| CSPG4 | KDLLFGSIVAVDEPTRP | 17 | 211800 | 2 | 0.47 | 0.26 |
| CSPG4 | SVAGPQTSEAFAITVR | 16 | 266400 | 2 | 1.55 | 0.33 |
| CSPG4 | FSGPHSLAAFPAWGTQ | 16 | 178700 | 2 | 3.10 | 0.30 |
| CSPG4 | DPDSAPGEIEYEVQR | 15 | 101900 | 1 | 3.73 | 2.59 |
| CSPG4 | GGQGTIDTAVLHLDTN | 16 | 104500 | 1 | 2.31 | 1.02 |
MHC II binding affinity was predicted using the prediction program described by Reynisson et al. (40).
CSPG4, chondroitin sulfate-proteoglycan-4; MHC, major histocompatibility complex.
CSPG4 expression is correlated with ATC histology and extrathyroidal extension
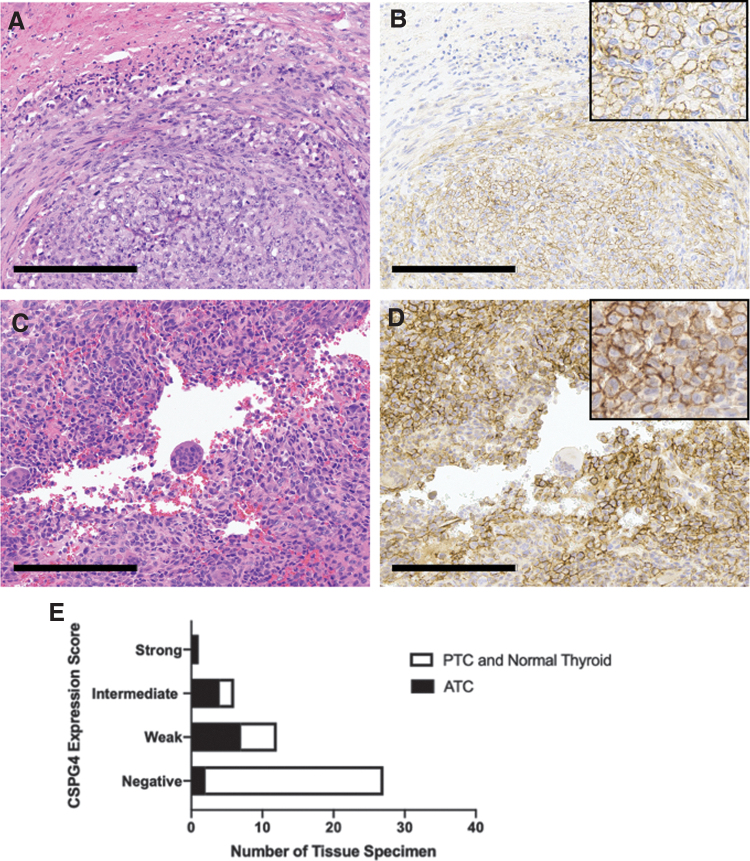
Based on the abundance of CSPG4-eluted peptides from JV-ATC cells on mass spectrometry, we evaluated CSPG4 expression histologically in surgical specimens of PTC, ATC, and adjacent normal thyroid tissue (Fig. 3). In normal thyroid tissue, staining was rare and only cytoplasmic (Table 4). Compared with normal thyroid tissue and PTC, expression was significantly increased in ATC samples (Table 4). For ATCs, 86% of tumor specimens displayed CSPG4 staining compared with 36% of PTCs. Patterns of CSPG4 staining included perinuclear, cytoplasmic, and membranous (Fig. 3A–D). Most PTCs (Supplementary Fig. S3A, B) showed negative or weak staining, while the majority of ATCs were positive for CSPG4 with weak, intermediate, and strong staining (Fig. 3E).
FIG. 3.
CSPG4 is expressed predominantly in the cytoplasm and/or membrane of ATC. (A, B) H&E (left) and membranous CSPG4 (right) staining in ATC. Scale bar = 200 μm. (C, D) H&E (left) and membranous and cytoplasmic CSPG4 (right) staining in ATC. Scale bar = 200 μm. (E) Immunohistochemistry tissue sides were scored for percentage of tumor cells stained (0 = negative, 1 = 1–29%, 2 = 30–59%, 3 = 60–100%) and intensity of staining (0 = negative, 1 = weak, 2 = intermediate, 3 = strong) by an endocrine pathologist in a blinded review. Intensity (0–3) and percentage stained (0–3) were added together for a possible total of 0–6 points (0 = negative, 1–2 = weak, 3–4 = intermediate, 5–6 = strong). Scores compared ATC with PTC and normal thyroid tissue using Fisher's exact test (p < 0.001). H&E, hematoxylin and eosin; PTC, papillary thyroid cancer.
Table 4.
CSPG4 Expression and Pattern in Thyroid Cancer Tissue Samples Including Adjacent Normal Thyroid
| n | CSPG4 0% | CSPG4 ≥ 1% | p-Valuea | |
|---|---|---|---|---|
| Histology | 0.0001 | |||
| PTC | 11 | 7 (64%) | 4 (36%) | |
| 1 nuclear 2 cytoplasmic 1 membranous |
||||
| ATC | 14 | 2 (14%) | 12 (86%) | |
| 0 nuclear 7 cytoplasmic 5 membranous |
||||
| Normal thyroid | 21 | 18 (86%) | 3 (14%) | |
| 0 nuclear 3 cytoplasmic 0 membranous |
p-Value was determined using the chi-squared test.
ATC, anaplastic thyroid cancer; PTC, papillary thyroid cancer.
Our group has previously reported the association of intercellular adhesion molecule-1 (ICAM-1) and program death-ligand-1 (PD-L1) expression with aggressive thyroid cancers (34,41,42). To assess for an association with CSPG4, all tumor samples also underwent IHC for ICAM-1 and PD-L1. CSPG4 expression was correlated with circumferential ICAM-1 expression (Table 5), a finding we previously reported as having significant correlation with poorly differentiated thyroid cancer (PDTC) and ATC tumors (41). There was no association between PD-L1 and CSPG4 expression. Clinicopathologically, CSPG4 expression was significantly associated with extrathyroidal extension (ETE), lymphovascular invasion, and tumor size (Table 5). Moreover, all four PTC samples that were positive for CSPG4 expression had ETE on pathology report, while none of the seven PTCs that were negative for CSPG4 expression had ETE. BRAFV600E mutational status on our ATC samples was limited as our institution started routine BRAFV600E testing in 2010, and there was no significant correlation with CSPG4 expression (Table 5).
Table 5.
Clinical Characteristics and Immunohistochemistry Staining of Thyroid Cancer Samples Used in This Study
| CSPG4 0% (n = 9) | CSPG4 ≥ 1% (n = 16) | p-Value | |
|---|---|---|---|
| Histology | 0.0168a | ||
| PTC | 7 (78%) | 4 (25%) | |
| ATC | 2 (22%) | 12 (75%) | |
| ICAM1 expression | >0.9999a | ||
| >1% | 9 (100%) | 16 (100%) | |
| ICAM1 pattern | 0.0168a | ||
| Apical | 7 (78%) | 4 (25%) | |
| Circumferential | 2 (22%) | 12 (75%) | |
| PDL1 expression | 0.4341a | ||
| >1% | 4 (44%) | 10 (63%) | |
| Clinical characteristics | |||
| Age | 44 (16–62) | 61 (26–86) | 0.0736b |
| Male sex | 0/9 (0%) | 5/16 (31%) | 0.0613a |
| Tumor size (cm) | 1.6 (0.5–4.8) | 3.7 (1.1–7.3) | 0.0035b |
| Lymph node metastases | 3/5 (60%) | 9/12 (75%) | 0.6000a |
| Distant metastases | 2/9 (22%) | 4/13 (31%) | >0.9999a |
| Lymphovascular invasion | 2/9 (22%) | 11/13 (85%) | 0.0073a |
| Extrathyroidal extension | 1/7 (14%) | 15/15 (100%) | <0.0001a |
| BRAFV600E mutationc | 7/8 (88%) | 6/10 (60%) | 0.3137a |
p-Values were determined using Fisher's exact test.
p-Values were determined using the Wilcoxon rank-sum test.
BRAFV600E mutational status was not assessed until 2010 at our institution.
CSPG4 is expressed on the cell surface of ATC cell lines
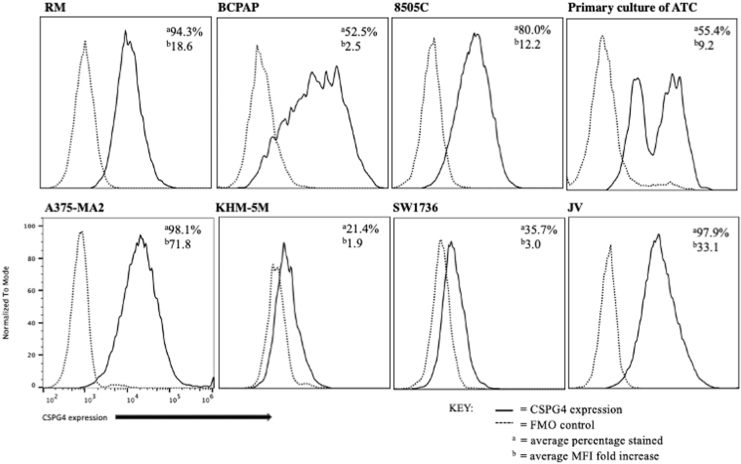
Because membranous CSPG4 expression was observed predominantly in ATC, we assessed surface expression of CSPG4 in multiple thyroid cancer cell lines and a primary culture of ATC by flow cytometry. As expected, expression in a melanoma cell line, A375-MA2, was high in the majority of cells. The JV cell line had an average of 97.9% expression with an average mean fluorescence intensity (MFI) of 33.1-fold higher. Our cell line derived from an ATC, RM, had an average of 94.3% cells expressing CSPG4 with an average MFI of 18-fold higher. Interestingly, the patient's FFPE was one of two ATCs negative for CSPG4 IHC expression (Supplementary Fig. S3C, D), while the cell line created from this tumor showed expression by flow cytometry. The percentage of 8505C cells expressing CSPG4 ranged from 67% to 90% with an average MFI of 12.2-fold increase. Other ATC cell lines, KHM-5M and SW1736, had lower CSPG4 expression. Our primary culture of a recent ATC showed an average of 55.4% of tumor-associated cells stained with an average MFI of 9.2-fold increase. The PTC line, BCPAP, had 52–53% expression of CSPG4 with an average MFI of 2.5-fold increase (Fig. 4). Overall, the majority of ATC cells tested exhibited CSPG4 at the cell surface with an ∼10-fold increase in MFI.
FIG. 4.
Some ATC cells show membranous expression of CSPG4. Representative histogram plots of CSPG4 expression and FMO controls in multiple thyroid tumor cell lines and primary tumor culture were determined by flow cytometry. These experiments were repeated at least three times independently. FMO, fluorescence minus one.
Patients with increased tumoral CSPG4 expression have worse prognosis in ATC
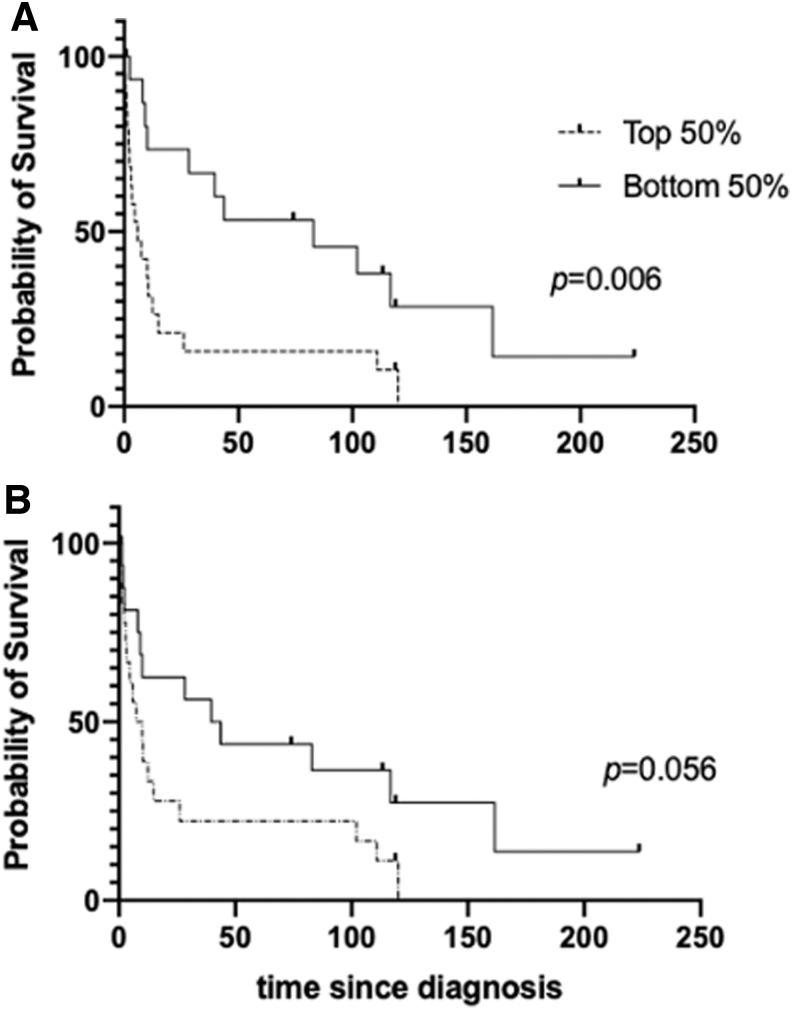
CSPG4 expression has been associated with worse prognosis in other cancers (25,43,44). To determine whether CSPG4 overexpression is correlated with survival rates in thyroid cancer, we analyzed RNA expression and clinical data in patients with PDTC and ATC from a previously published study (45). In this microarray data, there were two probes that matched CSPG4, 204736 and 214297. When we compared the overall survival (OS) for patients categorized into the top 50% and bottom 50% of CSPG4 expression for each probe, we found that patients with a higher level of CSPG4 expression had decreased OS (Fig. 5) (p-values were 0.006 and 0.056 for probes 204736 and 214297, respectively, when determined by log-rank test). Median survival was markedly decreased with elevated CSPG4 expression. These data demonstrate that PDTC and ATC patients with increased tumoral CSPG4 mRNA expression were associated with worse OS.
FIG. 5.
Advanced thyroid tumor patients with higher CSPG4 mRNA expression correlated with a shorter OS rate. (A) Kaplan–Meier survival curves were generated using the OS and mRNA expression data available from a published study on poorly differentiated thyroid cancer and ATC patients (43). Statistical difference was analyzed by comparing two cohorts with CSPG4 mRNA expression (top 50% and bottom 50%) using log-rank test with data obtained with CSPG4 probe 204736 (p = 0.006), with median survival of 5.92 months and 82.83 months for the top and bottom 50% of CSPG4 expression, respectively. (B) Another CSPG4 probe 214297 was also used to analyze OS (p = 0.056) using log-rank test with median survival of 8.78 and 41.68 months for the top and bottom 50%, respectively. OS, overall survival.
Discussion
ATC is a deadly cancer with limited treatment options in need of further therapeutic targets. A potential target, CSPG4, has not previously been studied in ATC, although it is a well-described antigen in other aggressive cancers. Given its elevated expression in malignant tissues, but restricted expression in normal tissue, CSPG4 is an attractive molecular target for cancer therapy. In this study, we demonstrated CSPG4 overexpression in ATC using our own and publicly available data from patient samples and thyroid cancer cell lines using several methods, including IHC, flow cytometry, mass spectrometry, and mRNA expression analysis.
Data regarding peptide antigens presented by the MHC II, in particular HLA-DQ, have been limited because of its highly polymorphic gene structure, with each allelic variant exhibiting different peptide binding affinities. Here we provide mass spectrometry-based characterization of HLA-DQ-restricted peptides eluted from an ATC patient-derived cell line, JV. We selected HLA-DQ to study immunopeptidomes because JV-ATC cells elicited robust activation by autologous CD4+ T cells and these cells showed the strongest expression of HLA-DQ among the complexes of MHC II. We identified 920 HLA-DQ-eluted peptides isolated uniquely from the JV cells, which need further validation to characterize their potential as tumor-associated antigens.
CSPG4 generated one of the strongest peptide signals in JV cells. The most abundant CSPG4 peptide overlapped 10 amino acids with an epitope predicted to bind HLA-DRB1*10:01 allele (epitope ID 1188394) (46), verifying its presence among immunopeptidomes. Most of the CSPG4-derived peptides we identified were novel. The addition of CD4+ T cells has been shown to potentiate T cell therapies against CSPG4 in preclinical experiments (47). Many treatments against CSPG4 have been developed using hybridoma technology for mAb targeting (48,49) or generating CAR-T cell-based therapies (31,50). The HLA-DQ peptides eluted from CSPG4 in our patient sample are prospective immunogenic agents that may help develop T cell receptor-based immune therapies (51). While we discovered significant levels of CSPG4 expression in most ATCs, it will be important to demonstrate whether CSPG4-directed adoptive T cell therapy or blockade of CSPG4 signaling will eliminate tumor growth in vivo. In addition, understanding the functional role of CSPG4 will give insight into devising the optimal strategy to target CSPG4 in ATC effectively. Given the complexity and heterogeneity of ATC tissue, CSPG4 blockade in addition to inhibitors of BRAF or MAPK/ERK signaling may achieve improved efficacy for targeting ATC (52,53).
Our IHC data show that the majority of ATCs express CSPG4, although most scored an intermediate level of staining. More ATCs stained cytoplasmic than membranous; however, flow cytometry confirmed cell surface expression in the majority of ATC samples and cell lines. Curiously, one of the two ATC specimens negative for CSPG4 using IHC on an FFPE slide had strong CSPG4 expression on flow cytometry using our cell line, RM, derived from the patient's tumor. Given this discordant data, it is possible that our IHC antibody may have technical limitations in detecting weaker or denatured CSPG4 protein expression in the FFPE tissue. Alternatively, it is possible that the tumor tissues showed heterogeneous CSPG4 expression, and the tumor cells that sustained in culture were selected for CSPG4-expressing and potentially more aggressive tumor cells. Nevertheless, given our expression data, CSPG4 is a potential target for mAb- as well as T cell-based therapies in ATC.
Limitations to our study include our small sample size, as well as our limited analysis of expression, including IHC, mass spectrometry, and mRNA data from public sources. A previous study on head and neck squamous cell carcinoma correlated CSPG4 protein expression by IHC analysis and its mRNA expression level (43). For survival analysis, our institutional data were limited, and thus, we analyzed survival using previously published RNA expression data (45). Importantly, these data showed a significantly shorter OS with increased CSPG4 expression, similar to the prognostic value of CSPG4 expression in other cancers (43,44).
In conclusion, we have identified CSPG4 as a new potential therapeutic target in ATC, an aggressive cancer with poor prognosis and limited treatment options. Our data show that ATCs have higher CSPG4 expression and should be further studied for therapeutic potential with mAb- and T cell-based therapies. Moreover, the CSPG4 peptides eluted from MHC II offer additional specific targets for designing T cell receptor-based therapies. Ultimately, developing therapeutic strategies to target CSPG4 may provide new treatment options for ATC.
Supplementary Material
Acknowledgments
The authors acknowledge the support of the Translational Research Program, the Research Animal Resource Center, and the CITI Biomedical Imaging Center, all at Weill Cornell Medicine.
Authors' Contributions
C.E.E. and M.M.J.: Acquisition, analysis, or interpretation of data for the work; article writing and revision; final approval of the version to be published; and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
D.S., A.A., V.J.R., K.J.C., T.Z., B.H., and T.S.: Acquisition, analysis, or interpretation of data for the work; article revision; final approval of the version to be published; and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
J.W.T., J.A.G., B.M.F., and R.Z.: Acquisition of data for the work; article revision; final approval of the version to be published; and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
N.D., T.F. III, and I.M.M.: Substantial contributions to the conception or design of the work; acquisition, analysis, or interpretation of data for the work; article writing and revision; final approval of the version to be published; and agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work is supported by the Emerson Collective Cancer Research Fund (ECCRF 191824-01; to I.M.M.).
Supplementary Material
References
- 1. Lin B, Ma H, Ma M, Zhang Z, Sun Z, Hsieh IY, Okenwa O, Guan H, Li J, Lv W. 2019. The incidence and survival analysis for anaplastic thyroid cancer: a SEER database analysis. Am J Transl Res 11:5888–5896. [PMC free article] [PubMed] [Google Scholar]
- 2. Mao Y, Xing M. 2016. Recent incidences and differential trends of thyroid cancer in the USA. Endocr Relat Cancer 23:313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cancer Genome Atlas Research Network 2014. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159:676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho JC, Krishnamoorthy GP, Xu B, Schultz N, Berger MF, Sander C, Taylor BS, Ghossein R, Ganly I, Fagin JA. 2016. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest 126:1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pozdeyev N, Gay LM, Sokol ES, Hartmaier R, Deaver KE, Davis S, French JD, Borre PV, LaBarbera DV, Tan AC, Schweppe RE, Fishbein L, Ross JS, Haugen BR, Bowles DW. 2018. Genetic analysis of 779 advanced differentiated and anaplastic thyroid cancers. Clin Cancer Res 24:3059–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu B, Fuchs T, Dogan S, Landa I, Katabi N, Fagin JA, Tuttle RM, Sherman E, Gill AJ, Ghossein R. 2020. Dissecting anaplastic thyroid carcinoma: a comprehensive clinical, histologic, immunophenotypic, and molecular study of 360 cases. Thyroid 30:1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Subbiah V, Kreitman RJ, Wainberg ZA, Cho JY, Schellens JHM, Soria JC, Wen PY, Zielinski C, Cabanillas ME, Urbanowitz G, Mookerjee B, Wang D, Rangwala F, Keam B. 2018. Dabrafenib and trametinib treatment in patients with locally advanced or metastatic BRAF V600-mutant anaplastic thyroid cancer. J Clin Oncol 36:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sosa JA, Elisei R, Jarzab B, Balkissoon J, Lu SP, Bal C, Marur S, Gramza A, Yosef RB, Gitlitz B, Haugen BR, Ondrey F, Lu C, Karandikar SM, Khuri F, Licitra L, Remick SC. 2014. Randomized safety and efficacy study of fosbretabulin with paclitaxel/carboplatin against anaplastic thyroid carcinoma. Thyroid 24:232–240. [DOI] [PubMed] [Google Scholar]
- 9. Iniguez-Ariza NM, Ryder MM, Hilger CR, Bible KC. 2017. Salvage lenvatinib therapy in metastatic anaplastic thyroid cancer. Thyroid 27:923–927. [DOI] [PubMed] [Google Scholar]
- 10. Savvides P, Nagaiah G, Lavertu P, Fu P, Wright JJ, Chapman R, Wasman J, Dowlati A, Remick SC. 2013. Phase II trial of sorafenib in patients with advanced anaplastic carcinoma of the thyroid. Thyroid 23:600–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hargadon KM, Johnson CE, Williams CJ. 2018. Immune checkpoint blockade therapy for cancer: an overview of FDA-approved immune checkpoint inhibitors. Int Immunopharmacol 62:29–39. [DOI] [PubMed] [Google Scholar]
- 12. Naoum GE, Morkos M, Kim B, Arafat W. 2018. Novel targeted therapies and immunotherapy for advanced thyroid cancers. Mol Cancer 17:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Capdevila J, Wirth LJ, Ernst T, Ponce Aix S, Lin CC, Ramlau R, Butler MO, Delord JP, Gelderblom H, Ascierto PA, Fasolo A, Führer D, Hütter-Krönke ML, Forde PM, Wrona A, Santoro A, Sadow PM, Szpakowski S, Wu H, Bostel G, Faris J, Cameron S, Varga A, Taylor M. 2020. PD-1 blockade in anaplastic thyroid carcinoma. J Clin Oncol 38:2620–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, Parkhurst MR, Yang JC, Rosenberg SA. 2014. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 344:641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Veatch JR, Lee SM, Fitzgibbon M, Chow IT, Jesernig B, Schmitt T, Kong YY, Kargl J, Houghton AM, Thompson JA, McIntosh M, Kwok WW, Riddell SR. 2018. Tumor-infiltrating BRAFV600E-specific CD4+ T cells correlated with complete clinical response in melanoma. J Clin Invest 128:1563–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, Blasberg R, Yagita H, Muranski P, Antony PA, Restifo NP, Allison JP. 2010. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med 207:637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ross AH, Cossu G, Herlyn M, Bell JR, Steplewski Z, Koprowski H. 1983. Isolation and chemical characterization of a melanoma-associated proteoglycan antigen. Arch Biochem Biophys 225:370–383. [DOI] [PubMed] [Google Scholar]
- 18. Harper JR, Bumol TF, Reisfeld RA. 1984. Characterization of monoclonal antibody 155.8 and partial characterization of its proteoglycan antigen on human melanoma cells. J Immunol 132:2096–2104. [PubMed] [Google Scholar]
- 19. Wilson BS, Imai K, Natali PG, Ferrone S. 1981. Distribution and molecular characterization of a cell-surface and a cytoplasmic antigen detectable in human melanoma cells with monoclonal antibodies. Int J Cancer 28:293–300. [DOI] [PubMed] [Google Scholar]
- 20. Price MA, Colvin Wanshura LE, Yang J, Carlson J, Xiang B, Li G, Ferrone S, Dudek AZ, Turley EA, McCarthy JB. 2011. CSPG4, a potential therapeutic target, facilitates malignant progression of melanoma. Pigment Cell Melanoma Res 24:1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chekenya M, Rooprai HK, Davies D, Levine JM, Butt AM, Pilkington GJ. 1999. The NG2 chondroitin sulfate proteoglycan: role in malignant progression of human brain tumours. Int J Dev Neurosci 17:421–435. [DOI] [PubMed] [Google Scholar]
- 22. Rivera Z, Ferrone S, Wang X, Jube S, Yang H, Pass HI, Kanodia S, Gaudino G, Carbone M. 2012. CSPG4 as a target of antibody-based immunotherapy for malignant mesothelioma. Clin Cancer Res 18:5352–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nicolosi PA, Dallatomasina A, Perris R. 2015. Theranostic impact of NG2/CSPG4 proteoglycan in cancer. Theranostics 5:530–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X, Osada T, Wang Y, Yu L, Sakakura K, Katayama A, McCarthy JB, Brufsky A, Chivukula M, Khoury T, Hsu DS, Barry WT, Lyerly HK, Clay TM, Ferrone S. 2010. CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. J Natl Cancer Inst 102:1496–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Svendsen A, Verhoeff JJ, Immervoll H, Brøgger JC, Kmiecik J, Poli A, Netland IA, Prestegarden L, Planagumà J, Torsvik A, Kjersem AB, Sakariassen P, Heggdal JI, Van Furth WR, Bjerkvig R, Lund-Johansen M, Enger P, Felsberg J, Brons NH, Tronstad KJ, Waha A, Chekenya M. 2011. Expression of the progenitor marker NG2/CSPG4 predicts poor survival and resistance to ionising radiation in glioblastoma. Acta Neuropathol 122:495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Vries JE, Keizer GD, te Velde AA, Voordouw A, Ruiter D, Rümke P, Spits H, Figdor CG. 1986. Characterization of melanoma-associated surface antigens involved in the adhesion and motility of human melanoma cells. Int J Cancer 38:465–473. [DOI] [PubMed] [Google Scholar]
- 27. You WK, Yotsumoto F, Sakimura K, Adams RH, Stallcup WB. 2014. NG2 proteoglycan promotes tumor vascularization via integrin-dependent effects on pericyte function. Angiogenesis 17:61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Natali P, Bigotti A, Cavalieri R, Wakabayaski S, Taniguchi M, Ferrone S. 1985. Distribution of a cross-species melanoma-associated antigen in normal and neoplastic human tissues. J Invest Dermatol 85:340–346. [DOI] [PubMed] [Google Scholar]
- 29. Ziai MR, Imberti L, Nicotra MR, Badaracco G, Segatto O, Natali PG, Ferrone S. 1987. Analysis with monoclonal antibodies of the molecular and cellular heterogeneity of human high molecular weight melanoma associated antigen. Cancer Res 47:2474–2480. [PubMed] [Google Scholar]
- 30. Hafner C, Breiteneder H, Ferrone S, Thallinger C, Wagner S, Schmidt WM, Jasinska J, Kundi M, Wolff K, Zielinski CC, Scheiner O, Wiedermann U, Pehamberger H. 2005. Suppression of human melanoma tumor growth in SCID mice by a human high molecular weight-melanoma associated antigen (HMW-MAA) specific monoclonal antibody. Int J Cancer 114:426–432. [DOI] [PubMed] [Google Scholar]
- 31. Geldres C, Savoldo B, Hoyos V, Caruana I, Zhang M, Yvon E, Del Vecchio M, Creighton CJ, Ittmann M, Ferrone S, Dotti G. 2014. T lymphocytes redirected against the chondroitin sulfate proteoglycan-4 control the growth of multiple solid tumors both in vitro and in vivo. Clin Cancer Res 20:962–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang Y, Geldres C, Ferrone S, Dotti G. 2015. Chondroitin sulfate proteoglycan 4 as a target for chimeric antigen receptor-based T-cell immunotherapy of solid tumors. Expert Opin Ther Targets 19:1339–1350. [DOI] [PubMed] [Google Scholar]
- 33. Pellegatta S, Savoldo B, Di Ianni N, Corbetta C, Chen Y, Patané M, Sun C, Pollo B, Ferrone S, DiMeco F, Finocchiaro G, Dotti G. 2018. Constitutive and TNFα-inducible expression of chondroitin sulfate proteoglycan 4 in glioblastoma and neurospheres: implications for CAR-T cell therapy. Sci Transl Med 10:eaao2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gray KD, McCloskey JE, Vedvyas Y, Kalloo OR, Eshaky SE, Yang Y, Shevlin E, Zaman M, Ullmann TM, Liang H, Stefanova D, Christos PJ, Scognamiglio T, Tassler AB, Zarnegar R, Fahey TJ, 3rd, Jin MM, Min IM. 2020. PD1 blockade enhances ICAM1-directed CAR T therapeutic efficacy in advanced thyroid cancer. Clin Cancer Res 26:6003–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shookster L, Matsuyama T, Burmester G, Winchester R. 1987. Monoclonal antibody 1a3 recognizes a monomorphic epitope unique to DQ molecules. Hum Immunol 20:59–70. [DOI] [PubMed] [Google Scholar]
- 36. Bergseng E, Dørum S, Arntzen M, Nielsen M, Nygård S, Buus S, de Souza GA, Sollid LM. 2015. Different binding motifs of the celiac disease-associated HLA molecules DQ2.5, DQ2.2, and DQ7.5 revealed by relative quantitative proteomics of endogenous peptide repertoires. Immunogenetics 67:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vita R, Mahajan S, Overton JA, Dhanda SK, Martini S, Cantrell JR, Wheeler DK, Sette A, Peters B. 2019. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res 47:D339–D343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reeb AN, Li W, Sewell W, Marlow LA, Tun HW, Smallridge RC, Copland JA, Spradling K, Chernock R, Lin RY. 2015. S100A8 is a novel therapeutic target for anaplastic thyroid carcinoma. J Clin Endocrinol Metab 100:E232–E242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Shea JP, Chou MF, Quader SA, Ryan JK, Church GM, Schwartz D. 2013. pLogo: a probabilistic approach to visualizing sequence motifs. Nat Methods 10:1211–1212. [DOI] [PubMed] [Google Scholar]
- 40. Reynisson B, Alvarez B, Paul S, Peters B, Nielsen M. 2020. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res 48:W449–W454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Min IM, Shevlin E, Vedvyas Y, Zaman M, Wyrwas B, Scognamiglio T, Moore MD, Wang W, Park S, Park S, Panjwani S, Gray KD, Tassler AB, Zarnegar R, Fahey TJ, 3rd, Jin MM. 2017. CAR T therapy targeting ICAM-1 eliminates advanced human thyroid tumors. Clin Cancer Res 23:7569–7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Buitrago D, Keutgen XM, Crowley M, Filicori F, Aldailami H, Hoda R, Liu YF, Hoda RS, Scognamiglio T, Jin M, Fahey TJ, 3rd, Zarnegar R. 2012. Intercellular adhesion molecule-1 (ICAM-1) is upregulated in aggressive papillary thyroid carcinoma. Ann Surg Oncol 19:973–980. [DOI] [PubMed] [Google Scholar]
- 43. Warta R, Herold-Mende C, Chaisaingmongkol J, Popanda O, Mock A, Mogler C, Osswald F, Herpel E, Küstner S, Eckstein V, Plass C, Plinkert P, Schmezer P, Dyckhoff G. 2014. Reduced promoter methylation and increased expression of CSPG4 negatively influences survival of HNSCC patients. Int J Cancer 135:2727–2734. [DOI] [PubMed] [Google Scholar]
- 44. Tsidulko AY, Kazanskaya GM, Kostromskaya DV, Aidagulova SV, Kiselev RS, Volkov AM, Kobozev VV, Gaitan AS, Krivoshapkin AL, Grigorieva EV. 2017. Prognostic relevance of NG2/CSPG4, CD44 and Ki-67 in patients with glioblastoma. Tumour Biol 39:1010428317724282. [DOI] [PubMed] [Google Scholar]
- 45. Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho JC, Krishnamoorthy GP, Xu B, Schultz N, Berger MF, Sander C, Taylor BS, Ghossein R, Ganly I, Fagin JA. 2016. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest 126:1052–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Marcu A, Bichmann L, Kuchenbecker L, Kowalewski DJ, Freudenmann LK, Backert L, Mühlenbruch L, Szolek A, Lübke M, Wagner P, Engler T, Matovina S, Wang J, Hauri-Hohl M, Martin R, Kapolou K, Walz JS, Velz J, Moch H, Regli L, Silginer M, Weller M, Löffler MW, Erhard F, Schlosser A, Kohlbacher O, Stevanović S, Rammensee H-G, Neidert MC 2020 The HLA Ligand Atlas—a resource of natural HLA ligands presented on benign tissues. bioRxiv:778944. [Google Scholar]
- 47. Peng L, Ko E, Luo W, Wang X, Shrikant PA, Ferrone S. 2006. CD4-dependent potentiation of a high molecular weight-melanoma-associated antigen-specific CTL response elicited in HLA-A2/Kb transgenic mice. J Immunol 176:2307–2315. [DOI] [PubMed] [Google Scholar]
- 48. Kantor RR, Ng AK, Giacomini P, Ferrone S. 1982. Analysis of the NIH workshop monoclonal antibodies to human melanoma antigens. Hybridoma 1:473–482. [DOI] [PubMed] [Google Scholar]
- 49. Mittelman A, Chen ZJ, Liu CC, Hirai S, Ferrone S. 1994. Kinetics of the immune response and regression of metastatic lesions following development of humoral anti-high molecular weight-melanoma associated antigen immunity in three patients with advanced malignant melanoma immunized with mouse antiidiotypic monoclonal antibody MK2-23. Cancer Res 54:415–421. [PubMed] [Google Scholar]
- 50. Beard RE, Zheng Z, Lagisetty KH, Burns WR, Tran E, Hewitt SM, Abate-Daga D, Rosati SF, Fine HA, Ferrone S, Rosenberg SA, Morgan RA. 2014. Multiple chimeric antigen receptors successfully target chondroitin sulfate proteoglycan 4 in several different cancer histologies and cancer stem cells. J Immunother Cancer 2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ilieva KM, Cheung A, Mele S, Chiaruttini G, Crescioli S, Griffin M, Nakamura M, Spicer JF, Tsoka S, Lacy KE, Tutt ANJ, Karagiannis SN. 2017. Chondroitin sulfate proteoglycan 4 and its potential as an antibody immunotherapy target across different tumor types. Front Immunol 8:1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu L, Favoino E, Wang Y, Ma Y, Deng X, Wang × 2011. The CSPG4-specific monoclonal antibody enhances and prolongs the effects of the BRAF inhibitor in melanoma cells. Immunol Res 50:294–302. [DOI] [PubMed] [Google Scholar]
- 53. Pucciarelli D, Lengger N, Takacova M, Csaderova L, Bartosova M, Breiteneder H, Pastorekova S, Hafner C. 2015. Anti-chondroitin sulfate proteoglycan 4-specific antibodies modify the effects of vemurafenib on melanoma cells differentially in normoxia and hypoxia. Int J Oncol 47:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.