Abstract
It has become increasingly clear that cytochromes P450 can cycle back and forth between two extreme conformational states termed the closed and open states. In the well-studied cytochrome P450cam, the binding of its redox partner, putidaredoxin (Pdx), shifts P450cam toward the open state. Shifting to the open state is thought to be important in the formation of a proton relay network essential for O–O bond cleavage and formation of the active Fe(IV)=O intermediate. Another important intermediate is the oxy–P450cam complex when bound to Pdx. Trapping this intermediate in crystallo is challenging owing to its instability, but the CN− complex is both stable and an excellent mimic of the O2 complex. Here we present the P450cam–Pdx structure complexed with CN−. CN− results in large conformational changes including cis/trans isomerization of proline residues. Changes include large rearrangements of active-site residues and the formation of new active-site access channel that we have termed channel 2. The formation of channel 2 has also been observed in our previous molecular dynamics simulations wherein substrate binding to an allosteric site remote from the active site opens up channel 2. This new structure supports an extensive amount of previous work showing that distant regions of the structure are dynamically coupled and underscores the potentially important role that large conformational changes and dynamics play in P450 catalysis.
Graphical Abstract

INTRODUCTION
Cytochrome P450cam (CYP101A1) is a bacterial heme monooxygenase that catalyzes the regio and stereoselective hydroxylation of d-camphor to form 5-exo-hydroxycamphor. This is the first step in the oxidative assimilation of camphor as an energy source for its host organism Pseudomonas putida. Since its discovery, P450cam has served as an important model for structure and function relationships in P450 enzymes.1–5
Figure 1 shows the P450 catalytic cycle. In P450cam, NADH reducing equivalents first are transferred from the FAD of putidaredoxin reductase (PdR) to the Fe2S2 ferredoxin, putidaredoxin (Pdx), and then, Pdx delivers electrons to P450cam. A hallmark of the P450cam system is that the second electron transfer step can be supported by only Pdx, so it has long been thought that Pdx plays an effector/allosteric role.6 This effector role has been demonstrated by numerous spectroscopic methods,7–13 but the underlying conformational dynamics that give rise to its specificity have only recently begun to emerge. Crystal structures of P450–redox partner complexes are challenging and rare with only three structures to date.14–16 The most recent are the covalent and noncovalent structures of P450cam–Pdx determined by both X-ray crystallography and NMR.16,17 These structures provided the first direct structural evidence of how allosteric effects of Pdx may be achieved. The binding of Pdx to substrate-bound P450 pushes P450cam toward the open conformation, consistent with earlier spectroscopic evidence that demonstrated Pdx binding shifted substrate-bound P450cam back toward the low-spin state.13 This was hypothesized to allow for the formation of a water-mediated proton relay network to enter the active-site channel and free active-site residue Asp251 from a strong ion pair with the F helix residue Arg186 to then participate in a proton delivery relay to the distal heme-bound oxygen for O–O bond scission.16,18
Figure 1.
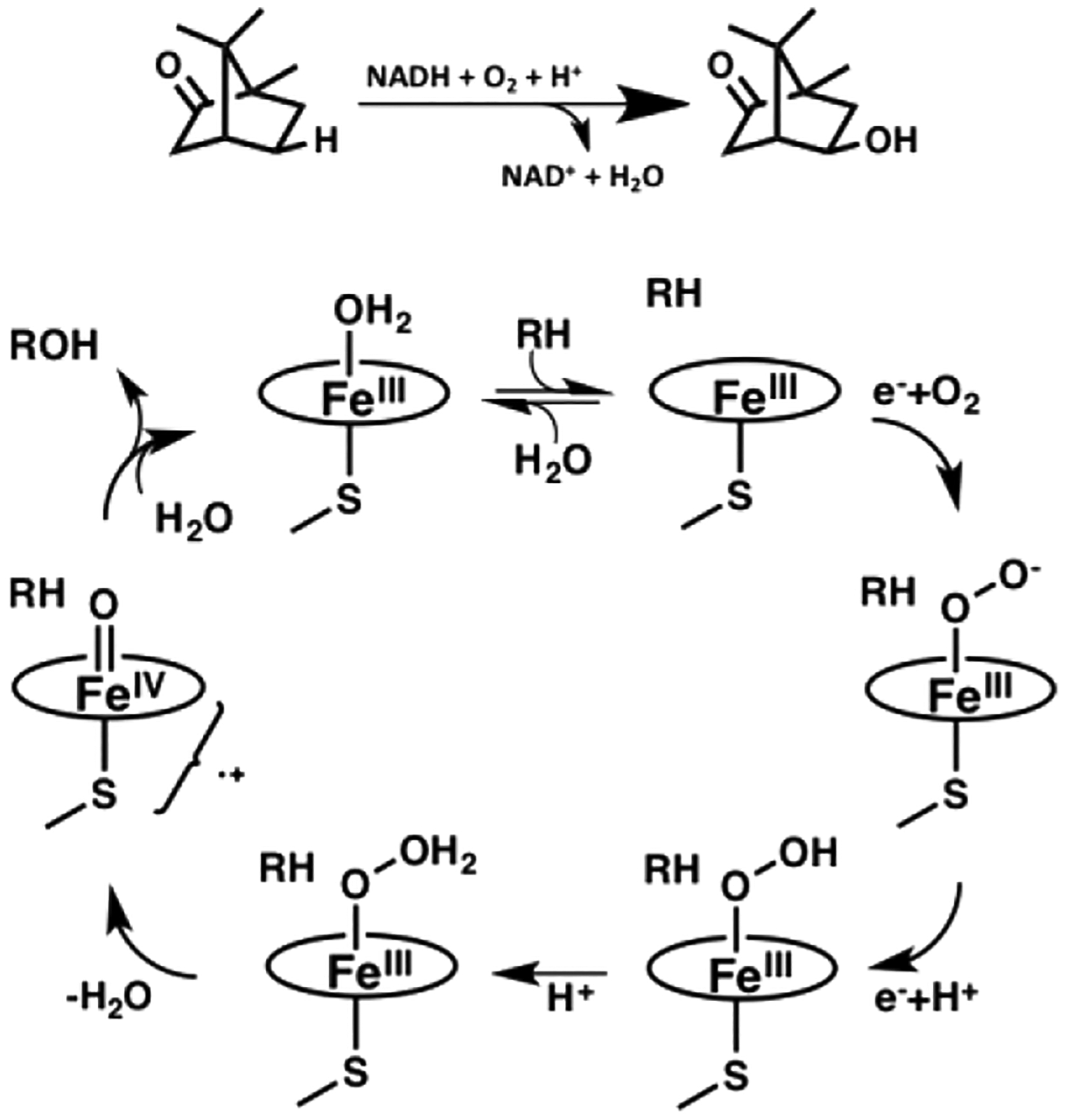
P450cam reaction cycle.
Related to both substrate and Pdx binding is perhaps the most important step of the P450 catalytic cycle, the binding and activation of molecular dioxygen. The synergistic timing of O2 binding, activation, and turnover is critical to ensure efficient coupling and prevent unproductive turnover and release of O2 as reactive oxygen species (ROS) such as superoxide or peroxide. While these interactions have been extensively investigated within the context of P450cam–camphor interactions,6,19 Pdx’s involvement with substrate and O2 dynamics is not well-understood. While the structure of substrate-bound P450cam and dioxygen has been solved, determination of the analogous structure of the oxy-complex bound to Pdx is not as straightforward. It has been previously demonstrated that in the presence of substrate and Pdx, X-ray radiation alone can reduce either P450cam and/or Pdx to initiate substrate turnover and form product within the crystal.16 Beyond the challenges related to the instability of the oxy-complex, formation of the oxygenated intermediate within the crystallized complex would surely bring a similar result upon X-ray exposure.
In order to study the effects of oxygenated intermediates of P450, both cyanide and carbon monoxide have been utilized as stable mimics of O2.20,21 However, binding of CO to P450 does not confer many of the conformational changes that are known to occur upon dioxygen binding.8,21 This may be due to electronic changes that occur upon CO binding and a lack of charge on the distal oxygen. Oxy-complexes of heme proteins are best described as ferric-superoxide rather than ferrous-oxy so the distal O2 oxygen atom carries a negative charge.22,23. The ferric-superoxide complex is electronically similar to the ferriccyanide complex, as CN− has a negative charge on the distal nitrogen. Indeed, the P450cam–CN− complex results in the same changes in local protein and solvent structure, while CO and NO complexes do not.20,24–26
In this paper, we present a substrate-bound Pdx–P450cam complexed with cyanide as an axial ligand at a resolution of 2.15 Å. Quite unexpectedly, we found that cyanide induces large structural changes that result in the formation of a new opening to the active site we have termed channel 2. Our recent molecular dynamics (MD) simulations showed that binding of the substrate, camphor, to a site on the protein well-removed from the active site results in the formation of this same channel 2.27 Thus, the present work provides experimental verification of structural change predicted by MD simulations as well as defines a novel conformational state of P450cam that may have relevance to enzyme function.
MATERIALS AND METHODS
Crystallization.
P450cam–Pdx crystals were prepared by hanging drop method as previously described.16 To prepare the CN−-bound complex, crystals were soaked in mother liquor containing 50 mM KCN for 15 min. Mother liquor supplemented with 15% glycerol was used as a cryoprotectant, and crystals were flash frozen in liquid nitrogen before data collection.
Data Collection and Refinement.
Data were collected from single crystals at the Stanford Synchrotron Radiation Lightsource (SSRL). Diffraction images were indexed, integrated, and scaled using Mosflm and Scala in the CCP4 package.28 The P450cam–Pdx complex crystal structure (PDB ID: 4JWS)16 without cofactor was used as a search model in molecular replacement using Phaser. The final structure contains two molecules of the P450cam–Pdx complex per asymmetric unit. The Phenix suite29 was used for structure refinement. All reflections were used for refinement except for 5% excluded for Rfree calculations.29 The structural model was revised in real space with the program COOT30,31 based on sigma-A-weighted 2Fo-Fc and Fo-Fc electron density maps. The final refinement statistics are given in Table S1. The final structure was refined to 2.15 Å resolution with an Rfree of 25.6% and Rwork of 19.8%.
Structural Analysis.
The final refined model consists of two molecules of complex (P450cam A-chain + Pdx C-chain, P450cam B-chain + Pdx D-chain) in the asymmetric unit. All four chains are highly ordered, and more than 97.0% of the residues were located in the core region of Ramachandran plots as determined by MolProbity.32 We did not observe any significant change in Pdx, although during data collection, the Fe2S2 center in Pdx very likely gets reduced. When Pdx is reduced, the 45–46 peptide flips, allowing the peptide NH group to donate a hydrogen bond to the Fe2S2 center in the reduced state. In our structure, this peptide is in the reduced conformation.
RESULTS AND DISCUSSION
In recent work, we used MD simulations that demonstrated how allosteric control of P450cam by a second molecule of camphor may provide a mechanism of activation by opening a primary and secondary channel.27 To date, the formation of the second channel has never been verified experimentally. Owing to the limits of classical MD, the role of O2 binding, product formation, and Pdx interaction were not demonstrated in our proposed model, but these processes are intricately governed by protein–substrate interactions. P450cam and Pdx were covalently cross-linked using a bismaleimide cross-linker between two non-native cysteines far from the protein–protein interface. The protein complex was crystallized and soaked with excess cyanide. To confirm the binding of CN−, a UV–vis spectrum of the crystal was taken using the microfocus beamline (Figure S1), and the Soret peak is significantly red-shifted, indicating the presence of bound CN−. The most significant changes between the complexes with and without CN− bound are the widening of the F/G loop and opening of the B′ channel (channel 2, Figure 2). This indicates that the binding of CN− to the heme induces large, ordered conformational rearrangements within the crystal.
Figure 2.

(A) P450cam (white) with the B–C loop (red) forming channel 2 in complex with Pdx (orange). (B) Rotated by 90° counterclockwise to show the allosteric site (green). (C) A cross section of the complex (PDB ID: 6NBL)
In our previous work, we suggested how substrate binding to a second site may prime the opening of a second channel and how Pdx may preferentially bind to this open structure to contribute to product egress via mechanical coupling. Overlaying the CN−-bound complex structure with our prior simulation egress event reveals significant similarities (Figure S2). Unlike the other P450cam–Pdx complexes, the F/G helices have moved to a completely open conformation. This allows for breakage of the Arg186–Asp251 ion pair, which, in our structure, is present in two rotameric conformations revealing how binding of dioxygen and Pdx may break this pair, allowing for Asp251 to participate in proton delivery as suggested by Tripathi et al.16 Polder maps demonstrate the bifurcation of the Asp251 residue (Figure 3). The rotamer where Asp251 is oriented in toward the active site is in an ideal position to mediate proton transfer to dioxygen.
Figure 3.
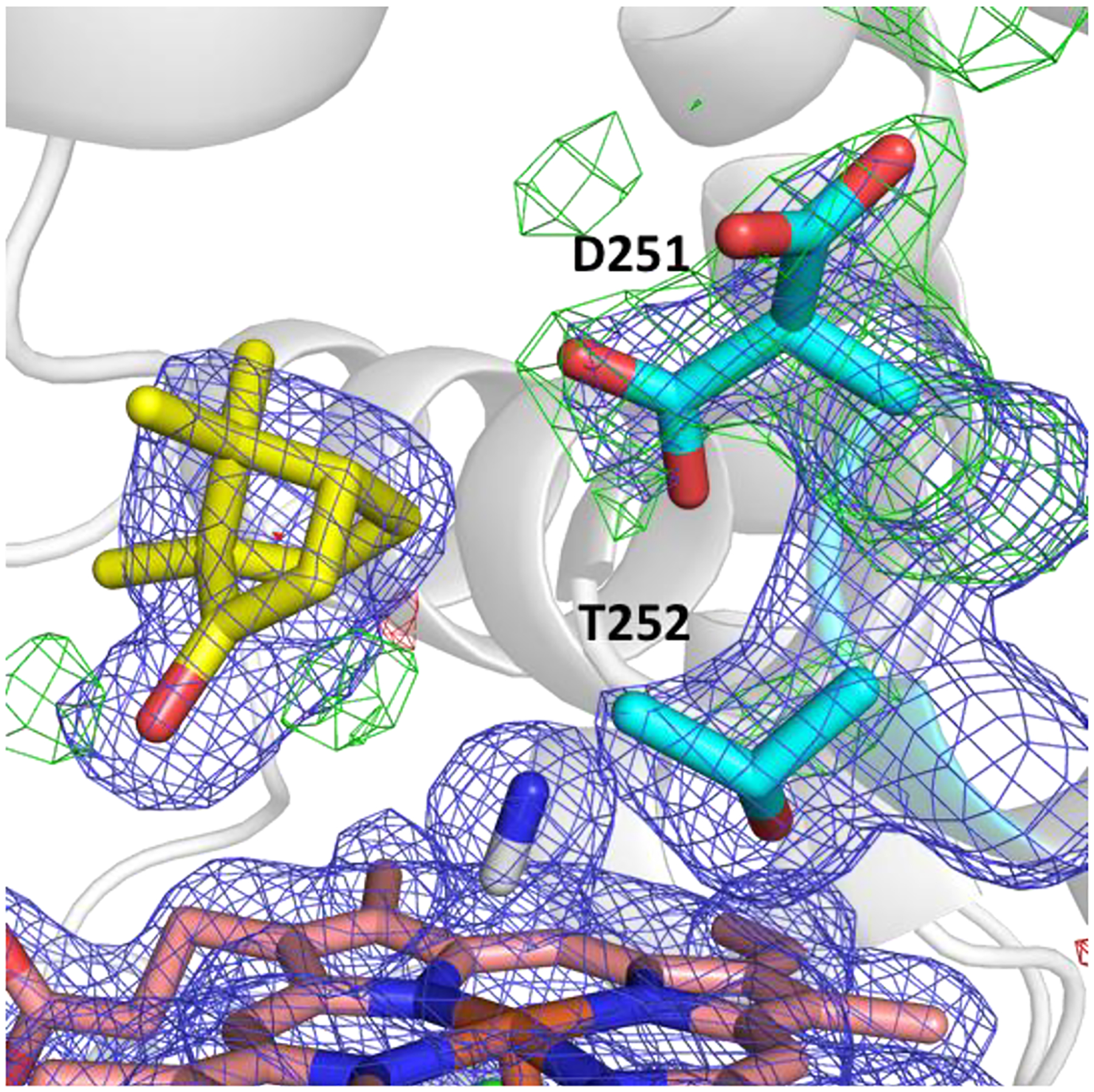
Polder map of Asp251 in two rotameric conformations. 2Fo-Fc at 1 σ (blue) and polder map at 4 σ (green).
However, the substrate access channel (channel 1) is not completely exposed in either of the molecules, as the B′ helix has lost nearly all secondary structure but remains hydrogen-bonded to the F/G loop. The movement of the F/G loop and the unfolding of the B′ helix leads to channel 2 formation, while channel 1 remains closed. This new channel provides an egress path for product and is associated with dynamic movement of Tyr96. This is important, since in the closed state, Tyr96 provides an H-bond to the camphor carbonyl oxygen. However, when CN− binds to the P450cam–Pdx complex, Tyr96 flips out of the active site (Figure 4C). To demonstrate the dynamics of this channel, we previously utilized the Ser83 to Ser102 Cα distance as a measure of channel 2 formation, which in all crystal structures where the B loop is ordered, is ~5 Å.27 In the CN− complex structure, however, the S83 to S102 distance is ~9.5 Å, which is in good agreement with our simulations that in order for substrate egress to occur channel 2 opens to beyond 7 Å.
Figure 4.
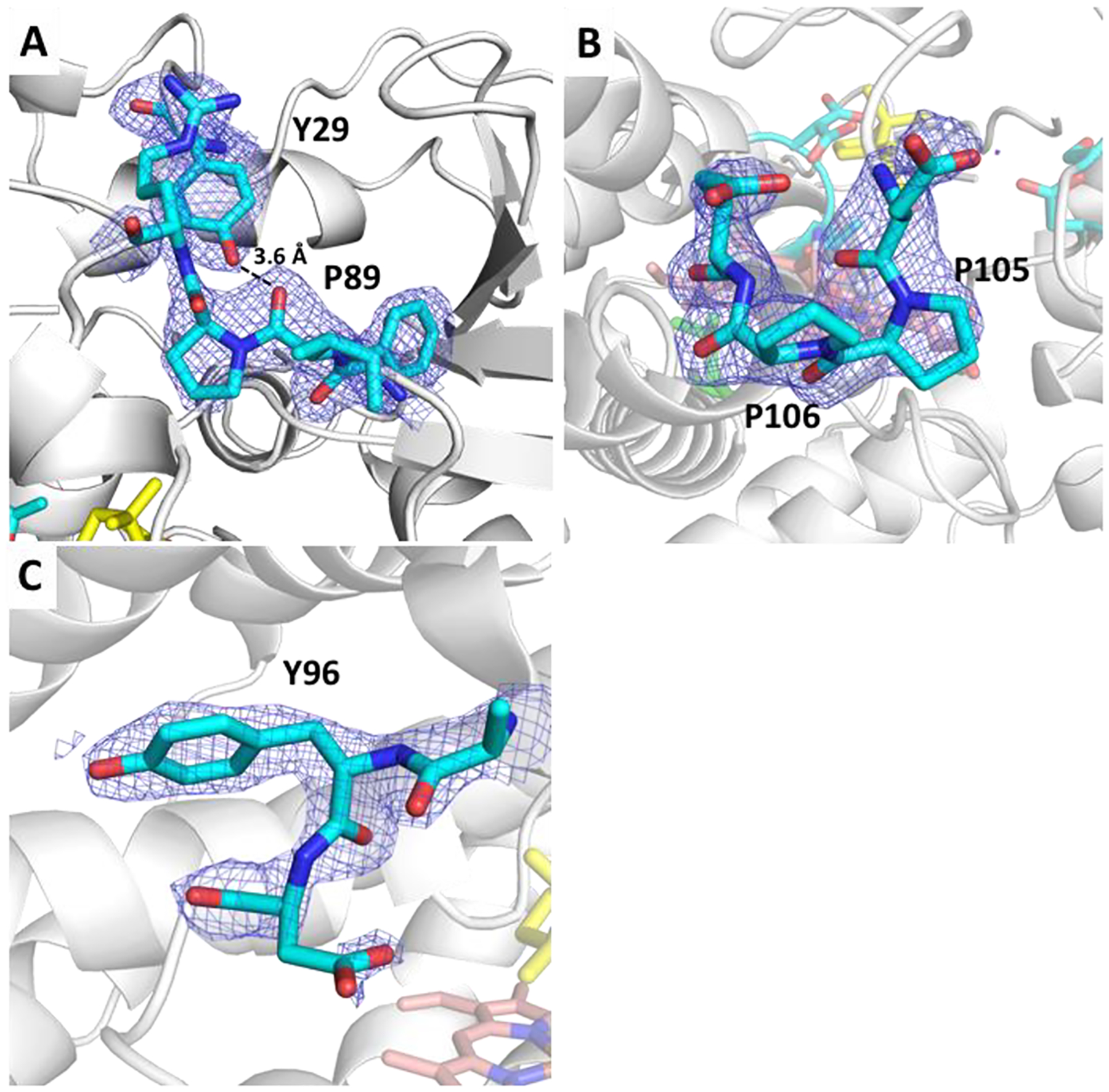
Several changes occur in the B–C loop of P450cam. In the structure, two prolines undergo cis–trans isomerization including (A) Pro89, which also moves to a distance of 3.6 Å from Tyr29, as well as (B) Pro105. (C) Tyr96 also rotates out of the active site where it H-bonds with the camphor carbonyl oxygen. Camphor (yellow); heme (red). 2Fo-Fc maps at 1 σ (blue).
NMR studies hypothesized that conformational switching in P450cam is dictated by an X-Pro cis–trans isomerization, but this has never been observed in a crystal structure. Specifically, OuYang et al. identified Pro89 as the most likely candidate for controlling this process.33 They observed two distinct camphor orientations when substrate-bound P450cam was reduced and bound to carbon monoxide (CYP-S-CO) and was then titrated with reduced Pdx (Pdxr). The time scales of the observed chemical shifts were in good agreement with reported time scales of catalyzed proline cis–trans isomerization. The binding of CN− to P450cam–Pdx results in the cis to trans isomerization of the Ile88–Pro89 bond, which breaks a bifurcated hydrogen bond from the carbonyl of Pro89 to the NH groups of Ala92 and Gly93 that allows the B′ helix to lose secondary structure but remain hydrogen-bonded to the F/G loop. NMR-directed molecular dynamics suggested that the barrier of this isomerization is lowered by a distortion of the ideally planar Ile88–Pro89 O–C–N–C∂ (ω) dihedral from 180 to ~166° in the trans form. The conclusions of those simulations are supported by the Ile88–Pro89 ω dihedral angles in our structure, which are distorted to 167 and 169°. Tyr29 was also hypothesized to be important in the controlling the isomerization process and, when compared to the closed structure, has moved from a distance of ~2.7 to 3.6 Å, nearly out of hydrogen bonding range (Figure 4A). Pro106 also undergoes cis to trans isomerization assisting in the opening of channel 2 and forces greater alignment of the backbone carbonyls effectively increasing the stability of the C-helix (Figure 4B). The Pro–Pro motif at the end of the B–C loop is conserved in a number of bacterial P450s and suggests that this mechanism may not be unique to P450cam.34
One surprising observation from the structure is the orientation of the 7-propionate group. In every crystallographic structure of P450cam deposited in the Protein Data Bank, the propionates take on identical conformations, with a 7-propionate C1A–C2A–CAA–CBA dihedral between −100 and −110°. In the cyanide structure presented here, the 7-propionate dihedral has rotated by ~180° to 89.9 and 85.3°, respectively (Figure 5). Hayashi et al. reconstituted P450cam with a “one-legged” heme to demonstrate how the 7-propionate along with Asp297, Arg299, and Gln322 act to protect the active site from solvent entry.35 Asp297 is believed to be protonated (H++ server: http://biophysics.cs.vt.edu/) and involved in hydrogen bonding with heme 7-propionate. In the CN− complex, Asp297 breaks this hydrogen bond with the propionate, revealing how Asp297 may participate in hydrogen bonding to camphor after binding O2 allowing for retained regio-stereoselectivity. In our unrestrained MD simulations, this same H-bond breakage of 7-propionate is associated with substrate egress. (One NMR structure exhibits a similar geometry, but the heme structure was determined by molecular dynamics.)
Figure 5.
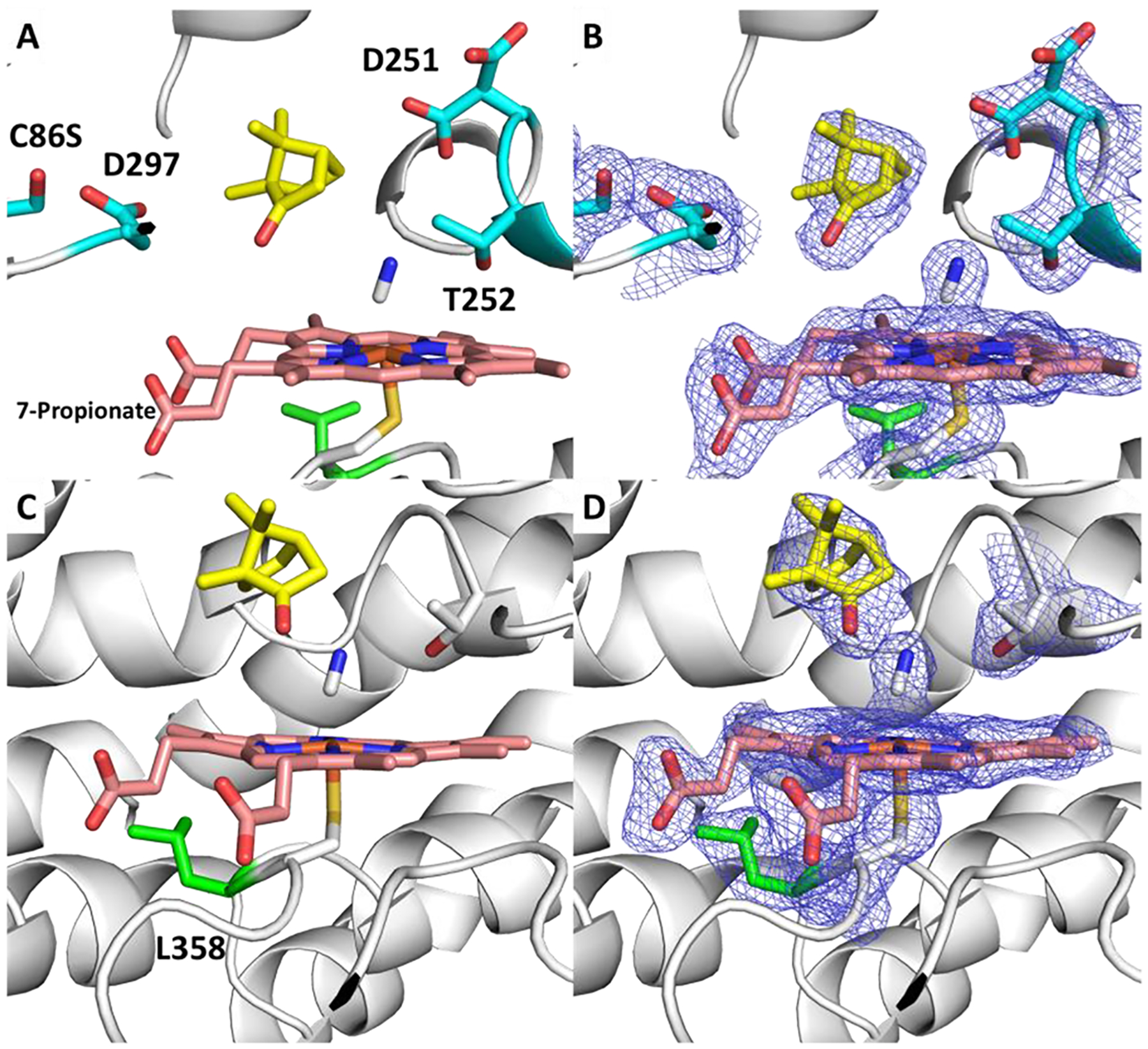
Two different views of the active site of P450cam with camphor (yellow) and CN (gray) bound to heme (red). Leu358 (green) has rotated to accommodate Pdx and pushes on the heme, where the 7-propionate has rotated and broken its interactions with Asp297. 2Fo-Fc contoured at 1 σ (blue, right).
We previously postulated that the dihedral rotation of the Leu358 side chain had relevance in Pdx binding and product formation.27 Once again, Leu358 N–Cα–Cβ–Cγ rotates ~130° from the open–closed structure (2CPP) and ~−70° to a dihedral angle of 59.3° and 56.9° in each monomer, respectively. This rotation allows for Pdx to create a tighter interface with P450cam and has been associated with the “push” effect coupled to changes on the distal side of the heme that favors the open form of P450cam.8,9,36 Here, we can see how Leu358 rotates upon CN− and Pdx binding and induces changes on the distal side of the heme as well as the heme itself that favor an open state.
Why CN− binding induces such large changes illustrates the intricate coupling of distant regions of P450cam with one another, which have been well-documented by NMR studies.33,36–43 To make room for CN−, the I helix must move. Since the F and G helices contact the I helix and the F/G loop region contacts the B′ helix, all of these regions move in concert. These large changes are possible because Pdx is holding P450cam in the partially open state. In the closed state, O2 and CN− binding, where P450cam is locked down, these large changes cannot take place. Even so, O2 and CN− binding to closed P450cam results in local change in the I helix, which is in the direction toward the open state.20,25 Pdx binding releases these restraints, thereby enabling the protein to undergo these large conformational rearrangements.
CONCLUSIONS
The P450cam–Pdx–CN− structure illustrates the magnitude of conformational changes that P450s can undergo and defines a new conformational state of P450cam. We were quite surprised that these large changes including cis–trans proline isomerization can occur within the confines of a crystal lattice. Although CN− is a good mimic of O2, the current structure cannot represent an active complex owing to the location of the camphor in channel 1. In our original structure of the P450cam–Pdx complex, the product, 5-exo-hydroxycamphor, is positioned in the substrate binding site, but a second camphor molecule is bound just above the product in channel 1 (PDB ID: 4JX1). In the CN−-complex, camphor is positioned about 2 Å up the channel away from the productive binding site and thus cannot represent the structure just prior to O–O bond cleavage and substrate hydroxylation. We postulate that opening of channel 2 has allowed productively bound substrate to escape, while rearrangements of the F/G and B′ regions trap the second camphor molecule in channel 1. It is unlikely that O2 alone can result in the same changes we observe with CN−, primarily because the oxy–P450cam complex is very unstable when bound to oxidized Pdx, while clearly, the CN− complex is quite stable. This is probably because Pdx shifting P450cam to the open state in the absence of electron transfer promotes rapid autoxidation of the oxy-complex. It thus seems more likely that the stability of the CN− complex has enabled trapping of P450cam in this new open conformational state. One could argue that this is simply an artifact of crystallization/soaking, although it can also be argued that proteins cannot adopt conformational states in crystallo that are not also accessible in solution. What provides an additional level of confidence that the CN−-induced structural changes are functionally relevant is the consistency with our previous MD simulations that show an allosterically regulated change in structure resulting in the formation of channel 2, very similar to what happens when CN− binds. This is reasonable, since the regions in the immediate vicinity of CN− binding in the I helix and those of the proposed allosteric substrate binding site ~16 Å away are mechanically coupled. Perturbing one perturbs the other. Therefore, the CN− complex provides a snapshot along the reaction coordinate after substrate hydroxylation and product egress through channel 2, while the substrate molecule in channel 1 is poised for movement to the productive binding site once Pdx dissociates.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant GM57353 (T.L.P.). The authors would like to acknowledge the San Diego Supercomputing Center (SDSC) for the use of the Triton Shared Computing Cluster (TSCC). We also thank the SSRL beamline staff for their support during remote X-ray diffraction data collection.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b13079.
Figure S1: Single-crystal UV–vis of P450cam–Pdx–CN complex; Figure S2: Alignment of P450cam crystal structures and molecular dynamics snapshots; Table S1: X-ray crystallography data collection and refinement statistics (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Poulos TL Heme enzyme structure and function. Chem. Rev 2014, 114 (7), 3919–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Poulos TL; Finzel BC; Gunsalus IC; Wagner GC; Kraut J The 2.6-A crystal structure of Pseudomonas putida cytochrome P-450. J. Biol. Chem 1985, 260 (30), 16122–16130. [PubMed] [Google Scholar]
- (3).Katagiri M; Ganguli BN; Gunsalus IC A soluble cytochrome P-450 functional in methylene hydroxylation. J. Biol. Chem 1968, 243 (12), 3543–3546. [PubMed] [Google Scholar]
- (4).Dus K; Katagiri M; Yu CA; Erbes DL; Gunsalus IC Chemical characterization of cytochrome P-450cam. Biochem. Biophys. Res. Commun 1970, 40 (6), 1423–30. [DOI] [PubMed] [Google Scholar]
- (5).Gunsalus IC; Sligar SG Equilibrium states and dynamic reactions of iron in the camphor monoxygenase system. Adv. Exp. Med. Biol 1976, 74, 254–62. [DOI] [PubMed] [Google Scholar]
- (6).Lipscomb JD; Sligar SG; Namtvedt MJ; Gunsalus IC Autooxidation and hydroxylation reactions of oxygenated cytochrome P-450cam. J. Biol. Chem 1976, 251 (4), 1116–1124. [PubMed] [Google Scholar]
- (7).Gunsalus IC; Sligar SG Redox regulation of cytochrome P450cam mixed function oxidation by putidaredoxin and camphor ligation. Biochimie 1976, 58 (1–2), 143–7. [DOI] [PubMed] [Google Scholar]
- (8).Nagano S; Tosha T; Ishimori K; Morishima I; Poulos TL Crystal structure of the cytochrome p450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding. J. Biol. Chem 2004, 279 (41), 42844–9. [DOI] [PubMed] [Google Scholar]
- (9).Tosha T; Yoshioka S; Ishimori K; Morishima I L358P mutation on cytochrome P450cam simulates structural changes upon putidaredoxin binding: the structural changes trigger electron transfer to oxy-P450cam from electron donors. J. Biol. Chem 2004, 279 (41), 42836–43. [DOI] [PubMed] [Google Scholar]
- (10).Myers WK; Lee YT; Britt RD; Goodin DB The conformation of P450cam in complex with putidaredoxin is dependent on oxidation state. J. Am. Chem. Soc 2013, 135 (32), 11732–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Liou SH; Mahomed M; Lee YT; Goodin DB Effector Roles of Putidaredoxin on Cytochrome P450cam Conformational States. J. Am. Chem. Soc 2016, 138 (32), 10163–72. [DOI] [PubMed] [Google Scholar]
- (12).Liou SH; Myers WK; Oswald JD; Britt RD; Goodin DB Putidaredoxin Binds to the Same Site on Cytochrome P450cam in the Open and Closed Conformation. Biochemistry 2017, 56 (33), 4371–4378. [DOI] [PubMed] [Google Scholar]
- (13).Unno M; Christian JF; Benson DE; Gerber NC; Sligar SG; Champion PM Resonance Raman Investigations of Cytochrome P450cam Complexed with Putidaredoxin. J. Am. Chem. Soc 1997, 119 (28), 6614–6620. [Google Scholar]
- (14).Sevrioukova IF; Li H; Zhang H; Peterson JA; Poulos TL Structure of a cytochrome P450-redox partner electron-transfer complex. Proc. Natl. Acad. Sci. U. S. A 1999, 96 (5), 1863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Strushkevich N; MacKenzie F; Cherkesova T; Grabovec I; Usanov S; Park HW Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc. Natl. Acad. Sci. U. S. A 2011, 108 (25), 10139–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Tripathi S; Li H; Poulos TL Structural basis for effector control and redox partner recognition in cytochrome P450. Science 2013, 340 (6137), 1227–30. [DOI] [PubMed] [Google Scholar]
- (17).Hiruma Y; Hass MA; Kikui Y; Liu WM; Olmez B; Skinner SP; Blok A; Kloosterman A; Koteishi H; Lohr F; Schwalbe H; Nojiri M; Ubbink M The structure of the cytochrome p450cam-putidaredoxin complex determined by paramagnetic NMR spectroscopy and crystallography. J. Mol. Biol 2013, 425 (22), 4353–65. [DOI] [PubMed] [Google Scholar]
- (18).Gerber NC; Sligar SG A role for Asp-251 in cytochrome P-450cam oxygen activation. J. Biol. Chem 1994, 269 (6), 4260–4266. [PubMed] [Google Scholar]
- (19).Sjodin T; Christian JF; Macdonald ID; Davydov R; Unno M; Sligar SG; Hoffman BM; Champion PM Resonance Raman and EPR investigations of the D251N oxycytochrome P450cam/putidaredoxin complex. Biochemistry 2001, 40 (23), 6852–9. [DOI] [PubMed] [Google Scholar]
- (20).Fedorov R; Ghosh DK; Schlichting I Crystal structures of cyanide complexes of P450cam and the oxygenase domain of inducible nitric oxide synthase-structural models of the short-lived oxygen complexes. Arch. Biochem. Biophys 2003, 409 (1), 25–31. [DOI] [PubMed] [Google Scholar]
- (21).Raag R; Poulos TL Crystal structure of the carbon monoxide-substrate-cytochrome P-450CAM ternary complex. Biochemistry 1989, 28 (19), 7586–92. [DOI] [PubMed] [Google Scholar]
- (22).Luthra A; Denisov IG; Sligar SG Spectroscopic features of cytochrome P450 reaction intermediates. Arch. Biochem. Biophys 2011, 507 (1), 26–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Huang X; Groves JT Oxygen Activation and Radical Transformations in Heme Proteins and Metalloporphyrins. Chem. Rev 2018, 118 (5), 2491–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Conner KP; Woods CM; Atkins WM Interactions of cytochrome P450s with their ligands. Arch. Biochem. Biophys 2011, 507 (1), 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Schlichting I; Berendzen J; Chu K; Stock AM; Maves SA; Benson DE; Sweet RM; Ringe D; Petsko GA; Sligar SG The catalytic pathway of cytochrome p450cam at atomic resolution. Science 2000, 287 (5458), 1615–22. [DOI] [PubMed] [Google Scholar]
- (26).Kuhnel K; Blankenfeldt W; Terner J; Schlichting I Crystal structures of chloroperoxidase with its bound substrates and complexed with formate, acetate, and nitrate. J. Biol. Chem 2006, 281 (33), 23990–8. [DOI] [PubMed] [Google Scholar]
- (27).Follmer AH; Mahomed M; Goodin DB; Poulos TL Substrate-Dependent Allosteric Regulation in Cytochrome P450cam (CYP101A1). J. Am. Chem. Soc 2018, 140 (47), 16222–16228. [DOI] [PubMed] [Google Scholar]
- (28).Battye TG; Kontogiannis L; Johnson O; Powell HR; Leslie AG iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr., Sect. D: Biol. Crystallogr 2011, 67 (4), 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Adams PD; Afonine PV; Bunkoczi G; Chen VB; Davis IW; Echols N; Headd JJ; Hung LW; Kapral GJ; Grosse-Kunstleve RW; McCoy AJ; Moriarty NW; Oeffner R; Read RJ; Richardson DC; Richardson JS; Terwilliger TC; Zwart PH PHENIX: a comprehensive Python-based system for macro-molecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66 (2), 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Emsley P; Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr 2004, 60 (12), 2126–2132. [DOI] [PubMed] [Google Scholar]
- (31).Emsley P; Lohkamp B; Scott WG; Cowtan K Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66 (4), 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Williams CJ; Headd JJ; Moriarty NW; Prisant MG; Videau LL; Deis LN; Verma V; Keedy DA; Hintze BJ; Chen VB; Jain S; Lewis SM; Arendall WB 3rd; Snoeyink J; Adams PD; Lovell SC; Richardson JS; Richardson DC MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27 (1), 293–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).OuYang B; Pochapsky SS; Dang M; Pochapsky TC A functional proline switch in cytochrome P450cam. Structure 2008, 16 (6), 916–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Nelson DR The cytochrome p450 homepage. Hum Genomics 2009, 4 (1), 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Hayashi T; Harada K; Sakurai K; Shimada H; Hirota S A role of the heme-7-propionate side chain in cytochrome P450cam as a gate for regulating the access of water molecules to the substrate-binding site. J. Am. Chem. Soc 2009, 131 (4), 1398–400. [DOI] [PubMed] [Google Scholar]
- (36).OuYang B; Pochapsky SS; Pagani GM; Pochapsky TC Specific effects of potassium ion binding on wild-type and L358P cytochrome P450cam. Biochemistry 2006, 45 (48), 14379–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Pochapsky TC; Lyons TA; Kazanis S; Arakaki T; Ratnaswamy G A structure-based model for cytochrome P450cam-putidaredoxin interactions. Biochimie 1996, 78 (8–9), 723–33. [DOI] [PubMed] [Google Scholar]
- (38).Wei JY; Pochapsky TC; Pochapsky SS Detection of a high-barrier conformational change in the active site of cytochrome P450cam upon binding of putidaredoxin. J. Am. Chem. Soc 2005, 127 (19), 6974–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Asciutto EK; Madura JD; Pochapsky SS; OuYang B; Pochapsky TC Structural and dynamic implications of an effector-induced backbone amide cis-trans isomerization in cytochrome P450cam. J. Mol. Biol 2009, 388 (4), 801–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Dang M; Pochapsky SS; Pochapsky TC Spring-loading the active site of cytochrome P450cam. Metallomics 2011, 3 (4), 339–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Asciutto EK; Young MJ; Madura J; Pochapsky SS; Pochapsky TC Solution structural ensembles of substrate-free cytochrome P450(cam). Biochemistry 2012, 51 (16), 3383–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Colthart AM; Tietz DR; Ni Y; Friedman JL; Dang M; Pochapsky TC Detection of substrate-dependent conformational changes in the P450 fold by nuclear magnetic resonance. Sci. Rep 2016, 6, 22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Asciutto EK; Pochapsky TC Some Surprising Implications of NMR-directed Simulations of Substrate Recognition and Binding by Cytochrome P450cam (CYP101A1). J. Mol. Biol 2018, 430 (9), 1295–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


