Abstract
Cx3cr1+ monocyte-derived macrophages (moMacs) are recruited to tissues after injury and are known to have profibrotic effects, but the cell-cell interactions and specific pathways that regulate this polarization and function are incompletely understood. Here we investigate the role of moMac-derived Pdgfa in bleomycin-induced lung fibrosis in mice. Deletion of Pdgfa with Cx3cr1-CreERT2 decreased bleomycin-induced lung fibrosis. Among a panel of in vitro macrophage polarizing stimuli, robust induction of Pdgfa was noted with IL10 in both mouse and human moMacs. Likewise, analysis of single-cell data revealed high expression of the receptor IL10RA in moMacs from human fibrotic lungs. Studies with IL10-GFP mice revealed that IL10-expressing cells were increased after injury in mice and colocalized with moMacs. Notably, deletion of IL10ra with Csf1r-Cre: IL10ra fl/fl mice decreased both Pdgfa expression in moMacs and lung fibrosis. Taken together, these findings reveal a novel, IL10-dependent mechanism of macrophage polarization leading to fibroblast activation after injury.
Keywords: IL10, lung injury, macrophages, Pdgfa, tissue fibrosis
INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is characterized by progressive scarring of the lung and has a survival rate of only 2–3 years without lung transplant (1). Currently, only two antifibrotic therapies are US Food and Drug Administration (FDA)-approved, and neither achieves a durable reversal of the fibrotic process. Hence, novel targets are urgently needed. Studies in the recent literature suggest that macrophages can have profibrotic effects and thus could represent a novel therapeutic target in the disease (2–5). In multiple tissues, the macrophage lineage with the most profibrotic profile is the monocyte-derived macrophage (moMac); moMacs are recruited to the site of injury, directly interact with fibroblasts, and secrete mediators that induce fibroblast activation (2, 4–10).
One such mediator is thought to be platelet-derived growth factor (PDGF), which was found to have a paracrine trophic function for fibroblasts in vitro (11). Furthermore, PDGF was increased in macrophages derived from the bronchial lavage in patients with IPF (12). We recently found that the specific isoform Pdgfa is upregulated in murine Cx3cr1-expressing moMacs found in regions of fibrotic scar as well as in scar-associated macrophages in lung samples from patients with IPF (4). However, the dependence of fibrosis on this moMac Pdgfa expression has not been demonstrated. Here, we found that Cx3cr1-CreERT2-mediated deletion of moMac Pdgfa resulted in decreased lung fibrosis in the bleomycin model. We then screened macrophage-polarizing cytokines and found that moMac IL10ra signaling was necessary for Pdgfa expression by moMacs and lung fibrosis. Taken together, these data elucidate the regulation and mechanism of profibrotic function of macrophages after injury.
METHODS
Mice
Cx3cr1-CreERT2 (Cx3cr1tm2.1(cre/ERT2) Jung), Ai14 (Rosa26-LSL-tdTomato), IL10-GFP, IL10ra floxed, Csf1r-Cre, and Pdgfra floxed mice were obtained from the Jackson Laboratory. Col1a2-CreERT2 mice were obtained from Bin Zhou (13). Pdgfa floxed mice were obtained from Johanna Andrae (14). Mice were housed in specific-pathogen-free conditions in the animal barrier facility of the University of California, San Francisco, and genotyping was performed by PCR. Mice were matched for sex and age across groups and were between the ages of 6 and 10 wk at the time of the study. Group allocation was randomized in cases where identical genotypes were used. All experiments were performed in compliance with ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines (15).
Bleomycin Lung Injury
Mice were intratracheally instilled with 3 U/kg clinical-grade bleomycin (Fresenius Kabi) on day 0 and lungs were harvested 7- or 21-days post injury, after perfusing the lung with 1X PBS. For the inducible Cre models (Cx3cr1-CreERT2 and Col1a2-CreERT2), 2 mg of tamoxifen (No. T5648, Sigma) was injected into the mice intraperitoneally every alternate day.
Hydroxyproline Assay
Hydroxyproline assay was performed as described previously (4). Briefly, snap-frozen lung samples were homogenized and mixed with 50% Trichloroacetic acid (No. T6399, Sigma) before incubating them overnight in 12 N HCl (No. 144, Fisher) at 110°C. The following day, the samples were reconstituted with water for 2 h. After reconstitution, they were mixed with 1.4% chloramine T (No. 85739, Sigma) in 10% isopropanol (No. A416, Fisher) and 0.5 M sodium acetate (No. 241245, Sigma). Finally, they were mixed with Ehrlich’s solution (No. 03891, Sigma) and incubated at 65°C for 15 min. Absorbance was measured at 550 nm, and the values were computed on the generated standard curve.
Sirius Red/Fast Green
Staining with Sirius Red (No. 26357-02, Electron Microscopy Science) and Fast Green FCF (No. F99-10, Fisher Scientific) was performed on sections prepared from mouse lungs frozen in OCT. Sections were stained with Sirius Red/Fast Green solution (0.125% Fast Green FCF and 0.1% Sirius Red in saturated picric acid) for 60 min followed by a 2-min 0.01 N HCl wash, rinsed in 70% ethanol, and air-dried overnight before imaging. Images were acquired using a bright-field microscope (Zeiss Axio scan.Z1). The images were quantified using ImageJ with at least three different regions of interest from each sample.
Primary Human Monocyte Isolation
Primary monocytes were isolated from healthy donor blood (Vitalant, San Francisco, CA). 15 mL of blood from TRIMA (Terumo) apheresis residual was diluted 1:1 with a 3 mM EDTA PBS 2% FBS buffer. The mixture was layered on top of a Ficoll-Paque density gradient medium and centrifuged at 1,200 g for 30 min at room temperature. The PBMC layer was collected and resuspended in 500 µL of ACK (ammonium-chloride-potassium) lysis buffer (No. A1049201, Gibco) and incubated at room temperature for 3 min. 40 mL of 3 mM EDTA in PBS with 2% FBS buffer was added, and cells were centrifuged for 2 min at 1,200 g. PBMC were then washed twice with 50 mL 3 mM EDTA in PBS with 2% FBS and centrifuged for 2 min at 1,200 g. Cells were counted, and 5 × 107 PBMC were resuspended in 8.5 mL of 3 mM EDTA in PBS with 2% FBS, and monocytes were isolated using EasySep Human Monocyte Isolation Kit (No. 19359, StemCell Technologies) following the manufacturer’s protocol.
In Vitro Differentiation and Polarization of Primary Human Monocyte-Derived Macrophages
Six-well plates were seeded with 4 × 106 freshly isolated cells per well with 4 mL of differentiation medium [RPMI 1640 (No. 11875119, Gibco)], supplemented with 10% FBS, 1% Penicillin-Streptomycin, and 50 µg/mL human M-CSF (No. 300-25, PeproTech) for macrophage differentiation. After 7 days, culture medium was removed and replaced with maintenance medium [RPMI 1640 (No. 11875119, Gibco) supplemented with 10% FBS, 1% Penicillin-Streptomycin] for M0 MDMs or maintenance medium supplemented with 100 ng/mL LPS (List Biological Laboratories) for M1 polarization; 20 ng/mL IL4 (No. 200-04, PeproTech) and 20 ng/mL IL13 (No. 200-13, PeproTech) for M2a polarization; 1 µg/mL LPS (List Biological Laboratories) and 10 ng/mL IL1β (No. 200-01B, PeproTech) for M2b polarization; 10 ng/mL IL10 (No. 200-10, PeproTech) for M2c polarization; or 50 ng/mL IL6 (No. 200-06, PeproTech) for M2d polarization. After 72 h of culture, supernatants were collected for ELISA and cells were harvested for RNA isolation.
Isolation of Monocyte-Derived Macrophages and Treatment of Cells
Total cells were flushed out from femur of mice and red blood cells were lysed using ACK lysis buffer (No. A1049201, Gibco). Monocytes were isolated using Bone Marrow Monocyte Isolation Kit (No. 130-100-629, Miltenyi Biotech) following the manufacturer’s protocol. To generate moMacs, isolated cells were cultured in Iscove’s Modified Dulbecco’s Medium (No. 122440053, Thermo Fisher) supplemented with 10% FCS (No. A31605, Gibco) and M-CSF (No. 315-02, Peprotech) for 5 days. moMacs were treated with murine recombinant IL10 (No. 210-10, Peprotech) for 24 h, and cells were lysed for RNA isolation.
qRT-PCR
For human monocyte-derived macrophages, total RNA was isolated using AllPrep DNA/RNA Mini Kit (No. 80204, Qiagen) following the manufacturer’s protocol. Sixty nanograms of RNA were used for cDNA synthesis using QuantiTect Reverse Transcription Kit (No. 205311, Qiagen) following the manufacturer’s recommendation. TaqMan reaction was performed using the following cycle: incubation at 50°C for 2 min, incubation at 95°C for 10 min, then 40 cycles of 95°C for 15 s then 70°C for 1 min, on an Applied Biosystems ViiA 7 Real-Time PCR System. The reaction mixture consisted of 2 µL of 1:10 diluted cDNA, 16 µL of Applied Biosystems TaqMan Universal PCR Master Mix with UNG (uracil-N-glycoslyase; No. 4440038, Thermo Fisher Scientific), and 2 µL of PrimeTime qPCR Probe Assays (PDGFA:Hs.PT.5839166652, RPL37A: Hs.PT.58.20168410). Each sample was performed in three technical replicates. Data were analyzed using the Applied Biosystems ViiA 7 software using RPL37A as a housekeeping gene.
For mouse cells, RNA was isolated using RNAeasy Mini Kit (No. 74106, Qiagen) following the manufacturer’s protocol. RNA quality was assessed using a spectrophotometer (Nanodrop, Thermo Fisher). cDNA from these RNA was synthesized using iScript Reverse Transcription Supermix (No. 1708841, Bio-Rad). SYBR Green PCR Master Mix (No. 4309155, Applied Biosystems) was used for running qPCR on ViiA 7 Real-time PCR system (Applied Biosystems), with normalization to Gapdh or 18 s. The following primer pairs were used for qRT-PCR (all 5′ to 3′):
Pdgfa Forward: GACCAGATGTGAGGTGAGATG
Pdgfa Reverse: GTGCTACAGTACTTGCTTTGATG
Gapdh Forward: AGTATGACTCCACTCACGGCAA
Gapdh Reverse: TCTCGCTCCTGGAAGATGGT
18s Forward: GCAATTATTCCCCATGAACG
18s Reverse: GGCCTCACTAAACCATCCAA
Immunofluorescence
IL10-GFP mice were euthanized at steady state or 7 days after bleomycin injury. Lungs were perfused with 1× PBS and inflated with 1:1 of optimal cutting temperature (OCT, No. 4585, Fisher Scientific) compound and 30% sucrose in PBS. The samples were then fixed in 4% PFA (15710, Electron Microscopy Sciences) overnight. The next day, the samples were washed with 1X PBS and embedded in OCT. Sections (15-µm) were cut from OCT-embedded tissue and blocked with 3% goat serum (No. 31872, Thermo Fisher), 1% BSA (No. BP1600-100, Fisher), and 0.3% Triton X-100 (No. BP151, Fisher) in PBS. The samples were then stained with anti-GFP primary antibody (1:100, No. ab13970, Abcam) followed by goat anti-chicken Alexa Fluor 488 secondary antibody (1:200, No. ab150173, Abcam). Images were acquired on a custom-built wide-field epifluorescence microscope built around a Zeiss Axiovert 200 M microscope body and controlled with Micro-Manager 2.0 software.
Human PDGF-AA ELISA
Supernatants from cultured human macrophages were used for quantification of human PDGFA following the manufacturer’s protocol (No. ELH-PDGFAA-1, RayBiotech).
Single-Cell Dissociation for FACS
Lungs were perfused with 1X PBS and suspended in RPMI 1640 medium containing 10 mM HEPES, 0.2% collagenase (No. 10103586001, Roche), 0.1 mg/mL Dispase II (No. 4942078001, Sigma), and 2,000 U/mL DNase I (No. 4716728001, Roche). After passing the cells through a 70-µm strainer, cells were stained with following antibodies at 4°C for 30 min: CD45-FITC (1:200, No. 11–0451082, clone 30-F11, Invitrogen), CD11b-Pe/Cy7 (1:200, No. 25–0112-81, clone:M1/70, Invitrogen), CD64-APC (1:400, No. 139305, clone:X54-5/7.1, BioLegend), and IL10ra-PE (1:100, No. 112705, clone:1B1.3a, BioLegend). Gating for FACS in Fig. 1B was as per Aran et al. (4) Cells were sorted on a BD FACSAria2 or on Sony SH800, and data were analyzed using FlowJo 10.8 software.
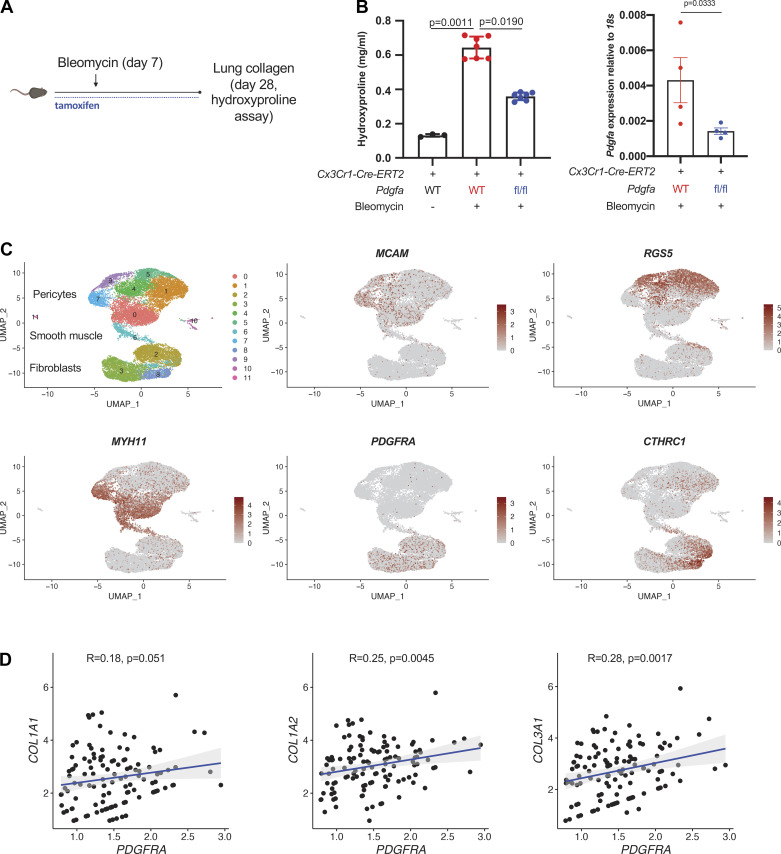
Figure 1.
moMac Pdgfa drives lung fibrosis after bleomycin injury. A: experimental design for inducible, cell type-specific deletion in lung moMacs. B, left: lung hydroxyproline assay in Cx3cr1-CreERT2: Pdgfa fl/fl mice and Cx3cr1-CreERT2 controls (n = 3, 7, and 6 mice/condition as shown). P value is for one-way ANOVA with Dunn’s multiple-comparisons testing; right: qPCR for Pdgfa in macrophages (TdTomato+ cells) sorted from Cx3cr1-CreERT2: Pdgfa fl/fl: R26-loxP-STOP-loxp-TdTomato mice or Cx3cr1-CreERT2: R26-loxP-STOP-loxp-TdTomato controls (n = 4 mice/condition). P value is for one-sided Student’s t test. C: UMAP of scRNA-Seq data for mesenchymal cells from patients with IPF (n = 3 separate individuals) (16). Marker genes shown in feature plots correspond to specific mesenchymal cell types. D: scatter plots show the correlation between PDGFRA and collagen gene expression within the fibroblast compartment; Spearman correlation coefficients and associated P values are shown. IPF, idiopathic pulmonary fibrosis; moMacs, monocyte-derived macrophages; scRNA-Seq, single-cell RNA-Seq. [A was created with BioRender.com.]
Analysis of Published scRNA-Seq Data
Single-cell data from Tsukui et al. [(16); GSE132771] and Haberman et al. [(17); GSE135893] were used for the analysis of gene expression and clustering using Seurat 4.0.2 (18). For the Haberman et al. data set, macrophages were identified using SingleR (4). Computational QC protocols were followed to exclude genes that were found in less than three cells and to remove cells with less than 200 genes. Patient samples were analyzed in aggregate. Gene expression was normalized using the default parameter “LogNormalize” and calculated for the top 2,000 most variable genes for principle component analysis after doublet removal with DoubletFinder. Analysis of the number of principle components was computed using the ElbowPlot function to prevent overfitting on the UMAP plot.
Statistics and Sample Size
Statistical analysis was performed using GraphPad Prism 8. Student’s t test was used for comparison between two groups and one-way ANOVA followed by Dunn’s multiple-comparison test was used for comparison of statistical significance between more than two groups. Wilcoxon rank-sum test was used for comparison of gene expression in single-cell RNA-Seq data. P values are mentioned in individual graphs, and a value less than 0.05 was considered statistically significant. Group sizes for in vivo experiments were determined based on calculation of statistical power based on previous effect sizes and variability in the bleomycin model, and investigators were blinded to group assignment during the study.
Study Approval
Animal experiments were approved by the Institutional Animal Care and Use Committee at the University of California, San Francisco. Experiments with human donor cells were approved by the Institutional Review Board of the University of California, San Francisco.
RESULTS
moMac Pdgfa in Lung Fibrosis
Our previous work identified Pdgfa as a potential profibrotic mediator expressed uniquely in Cx3cr1-expressing monocyte-derived macrophages in both mouse and human fibrotic tissues (4). We now tested whether moMac Pdgfa was necessary for bleomycin-induced lung fibrosis using Cx3cr1-CreERT2: Pdgfa fl/fl mice. moMac-specific Pdgfa deletion decreased lung fibrosis in the bleomycin murine model (Figs. 1, A and B). Testing the relevance of Pdgfa to the fibroblast response, we crossed a fibroblast-specific Cre line to a floxed allele of the receptor for Pdgfa, Pdgfra. Col1a2-CreERT: Pdgfra fl/fl mice had decreased fibrosis compared with controls (Supplemental Fig. S1; see https://doi.org/10.6084/m9.figshare.18708140.v1).
To determine the clinical significance of PDGFRA expression in IPF, we analyzed a recent data set of IPF lung mesenchymal cells sequenced by scRNA-Seq (16). Marker analysis identified cell types by their characteristic markers—RGS5 and MCAM (pericytes), MYH11 (smooth muscle cells), and PDGFRA and CTHRC1 (fibroblasts and pathological fibroblasts, respectively) (Fig. 1C). Importantly, in fibroblasts, PDGFRA expression positively correlated with expression of collagens essential for fibrosis, COL1A1, COL1A2, and COL3A1 (Fig. 1D), consistent with our murine data implicating the Pdgfa-Pdgfra axis in lung fibrosis.
IL10 Polarizes Macrophages for Pdgfa Expression
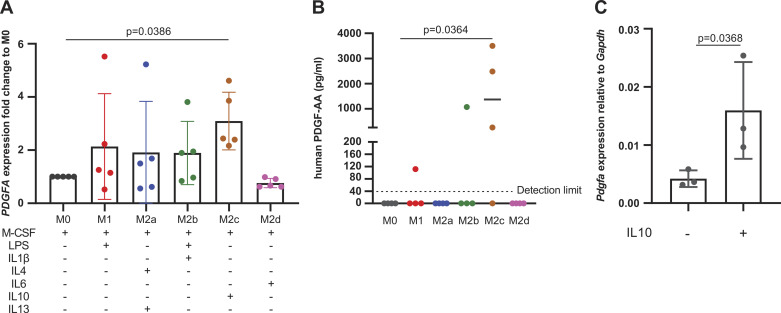
We wished to determine niche-specific factors driving the observed profibrotic state of moMacs. Although M2 polarization has long been proposed to be necessary for the profibrotic function of macrophages, specific candidate mediators have not been extensively tested by conditional deletion in vivo. Therefore, we first performed a screen of the effects of M1- and M2-polarizing factors on the expression of Pdgfa in monocyte-derived macrophages. Interestingly, polarizing human peripheral blood-derived monocytes with M2c conditions (M-CSF1 + IL10) resulted in marked induction of PDGFA at the mRNA and protein levels (Fig. 2, A and B). We then confirmed that IL10 induced Pdgfa expression in murine bone marrow monocyte-derived macrophages (Fig. 2C).
Figure 2.
IL10 induces Pdgfa in moMacs. A: qPCR for PDGFA in human peripheral blood moMacs polarized with factors shown in the table (n = 5 individual donors/condition). B: ELISA for PDGFA in supernatants from cultured moMacs treated in A (n = 4 individual donors/condition). C: qPCR for Pdgfa in murine bone marrow-derived macrophages treated with recombinant IL10. P values for (A) and (B) are for one-way ANOVA with Dunn’s multiple-comparison testing. P value in (C) is for one-tailed Student’s t test. moMacs, monocyte-derived macrophages.
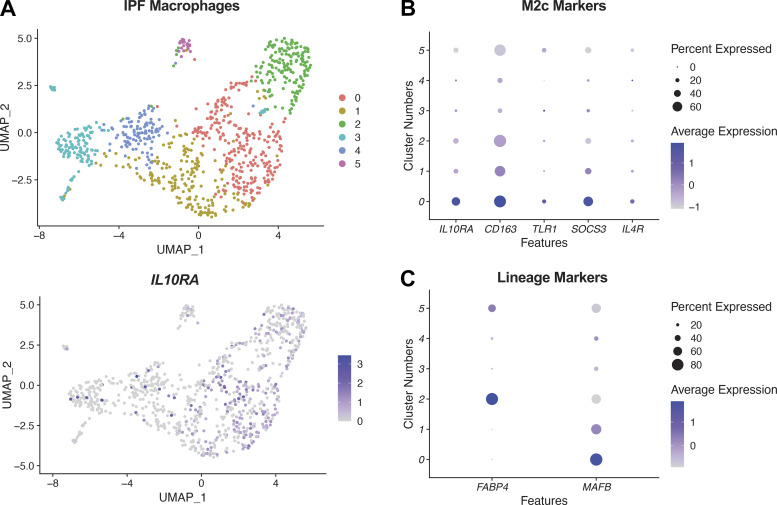
We next examined macrophage heterogeneity at the single-cell level in IPF, focusing on expression of the IL10RA, the gene encoding the receptor for IL10. Interestingly, analysis of macrophages from 17 IPF samples revealed expression of IL10RA to be focused if not exclusive to a subset of macrophages, cluster 0, whose gene expression was consistent with M2c polarization based on marker expression [Fig. 3, A and B; (19, 20)]. These macrophages expressed the moMac marker MAFB, which is not expressed highly in resident alveolar macrophages (21–23), and did not express the alveolar macrophage lineage marker FABP4 [Fig. 3C; (24)]. These data are consistent with the potential of moMac polarization by IL10 as a feature of fibrotic disease and motivated further functional studies in vivo.
Figure 3.
IL10RA expression in human profibrotic macrophages. A, top: scRNA-Seq analysis of macrophages identified by SingleR (4) from IPF lungs (17) (n = 17 separate individuals); bottom: feature plot for IL10RA. B: dot plot of expression of M2c markers for clusters identified in A. C: expression of lineage specific markers (alveolar macrophages, FABP4, and moMacs, MAFB) for clusters identified in A. IPF, idiopathic pulmonary fibrosis; moMacs, monocyte-derived macrophages; scRNA-Seq, single-cell RNA-Seq.
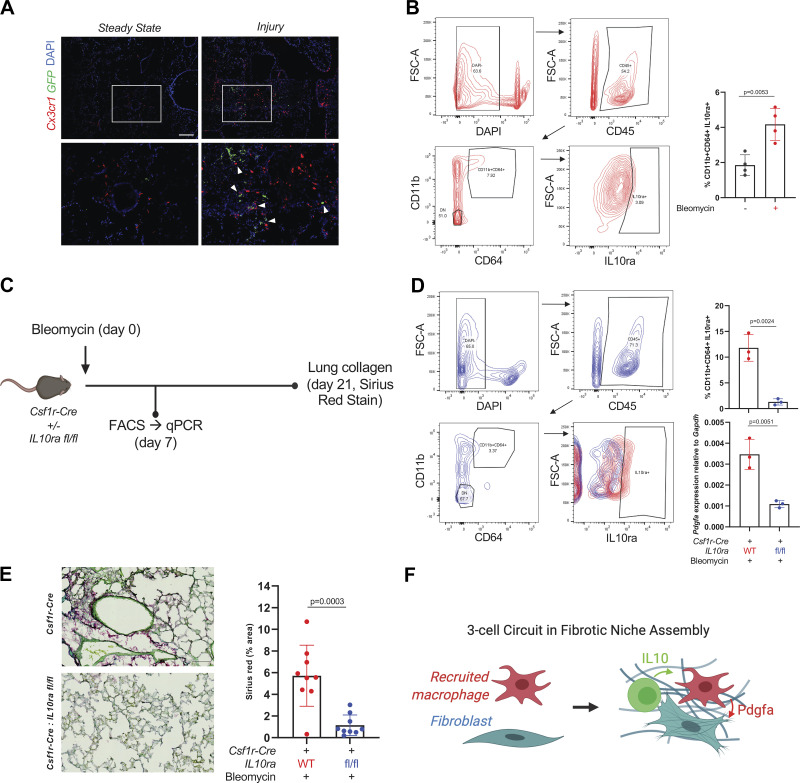
IL10-Expressing Cells Localizing to moMacs Drive Pdgfa Expression after Injury
At the 7-day time point after bleomycin injury, IL10-expressing cells including dendritic cells and Tregs have been detected by flow cytometry using IL10-GFP reporter mice (7), but their localization relative to moMacs by cross-sectional imaging has not been reported. Therefore, we crossed the IL10-GFP reporter mice to Cx3cr1-CreERT2: R26-LSL-tdTomato mice to determine whether IL10-expressing cells localize to moMacs by immunofluorescence. Consistent with a role for IL10-expressing cells in polarizing moMacs in the early fibrotic period, IL10-expressing cells in some cases were found in close proximity to macrophages in the early fibrotic period after lung injury (Fig. 4A). Moreover, IL10ra expression by flow cytometry in moMacs was found to increase after injury compared with steady state (Fig. 4B). To test the effect of IL10 on moMac expression of Pdgfa, we used Csf1r-Cre: IL10ra fl/fl mice (Fig. 4C). Remarkably, Pdgfa expression in CD11b+CD64+ moMacs sorted after injury from Csf1r-Cre: IL10ra fl/fl mice was markedly reduced compared with Csf1-Cre controls without floxed IL10ra (Fig. 4D). Importantly, lung collagen measured by Sirius Red stain at 21 days after bleomycin was also reduced (Fig. 4E). Taken together, these studies reveal a profibrotic, IL10-Pdgfa-Pdgfra circuit involving IL10-expressing cells [likely Tregs or dendritic cells based on previous results at the same time point after bleomycin (7) but whose identity remains to be further characterized across the fibrotic period], macrophages, and fibroblasts (Fig. 4F).
Figure 4.
IL10-expressing cells localize to moMacs after lung injury and induce Pdgfa expression. A: immunofluorescence of lung sections from Cx3cr1-CreERT2: R26-LSL-TdTomato: IL10-GFP mice at steady state and 7 days after injury with bleomycin. Magnified regions of interest are shown in the lower panels, with arrowheads marking sites of colocalization of Cx3cr1+ and IL10-expessing cells. Scale bar = 50 µm. Data are representative of n = 4 mice for injured and n = 2 mice for steady state. B: representative flow cytometry for IL10ra in CD11b+CD64+ moMacs from WT mice 7 days after bleomycin injury. Quantitation is for percentage of IL10ra+ moMacs at steady state and 7 days after bleomycin (n = 4 mice/group). P value is for two-sided Student’s t test. C: experimental design for IL10ra deletion with Csf1r-Cre used for D and E. D: representative flow cytometry for IL10ra in CD11b+CD64+ moMacs after bleomycin injury in Csf1r-Cre: IL10ra fl/fl mice (blue) or Csf1r-Cre controls (red). Quantitation is for percentage of IL10ra+ moMacs (top) and for qPCR for Pdgfa expression in sorted CD11b+CD64+ moMacs from WT and KO mice (bottom; n = 3 mice/group). P values are for two-sided Student’s t tests. E: representative image from Sirius Red staining of lungs at 21 days after bleomycin in Csf1r-Cre: IL10ra fl/fl mice (bottom) or Csf1r-Cre controls (top). Quantitation is for three separate images for each mouse (n = 3 mice/condition). P value shown is for two-sided Student’s test. Scale bar = 100 µm. F: schematic of cell-cell interaction in the fibrotic niche. KO, knockout; moMacs, monocyte-derived macrophages; scRNA-Seq, single-cell RNA-Seq; WT, wild-type. [C and F were created with BioRender.com.]
DISCUSSION
Our study reveals a novel role for IL10ra signaling in moMacs as a determinant of their profibrotic function during the response to lung injury. IL10 was identified among a panel of M1 and M2 polarizing factors as a Pdgfa-inducing cytokine in vitro; in vivo, IL10ra signaling in moMacs drove their expression of Pdgfa, which, in turn, was necessary for lung fibrosis. It is worth considering previous data in which IL10 KO mice were in fact found to have increased fibrosis (7). This result was attributed to IL10’s well-characterized anti-inflammatory role, given that the magnitude of inflammation in the first few days after bleomycin injury period determines fibrotic outcomes; for this reason, studies focused on fibrosis are recommended by working groups to focus interventions after this early injury period (25). In other data, when IL10 was overexpressed, in the absence of inflammation or injury, lung fibrosis was likewise increased (26). Resolving these discrepant results, we hypothesized that during the fibrotic period—that is, subsequent to the early inflammatory period after injury—IL10 may have an M2-polarizing, profibrotic effect on macrophages. To test this hypothesis, we used moMac-specific IL10ra deletion in the bleomycin lung injury model. This experiment demonstrated the dependence of both moMac expression of Pdgfa and lung fibrosis on moMac IL10ra.
Our study has important caveats. First, targeting monocyte-derived macrophages was accomplished with Cre drivers that narrow recombination significantly but imperfectly to moMacs. For example, natural killer (NK) cells are to some extent targeted by Cx3cr1-CreERT2, and both myeloid and lymphoid lineages evidence expression of Csf1r-Cre (27). Second, we have not characterized the kinetics of recruitment or the lineage of IL10-expressing cells observed. Previous results indicate a contribution of both regulatory T cells and dendritic cells (7), and the time course of recruitment of IL10-expressing cells in the bleomycin model, and indeed in other lung injury models, should be worked out.
To our knowledge, this study reveals for the first time the role of IL10 signaling in the functional polarization of macrophages in lung fibrosis. Furthermore, we find that this macrophage phenotype is clinically relevant, as IL10RA+ macrophages represented a specific cluster with monocyte-derived lineage identity. Importantly, the results indicate that moMac IL10RA could be pursued as a potential therapeutic target for IPF. Taken together, our findings enhance the understanding of cell-cell interactions across immune and stromal lineages and highlight the role of multicellular circuits in driving tissue responses to injury.
SUPPLEMENTAL DATA
Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.18708140.v1.
GRANTS
This work was supported by institutional discretionary funds (to M.Bh.), a grant from the National Institutes of Health (NIH), University of California, San Francisco-Gladstone Institute of Virology & Immunology Center for AIDS Research P30-AI027763 (to S.K.P.), and NIH Grant R01 AI150449 (to S.K.P.).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
M.Bh. and A.B. conceived and designed research; A.B., K.B., M.Bo., T.Y.C., R.W., and P.T. performed experiments; A.B., K.B., L.M., and M.Bo. analyzed data; S.K.P. and M.Bh. interpreted results of experiments; A.B. and M.Bh. prepared figures; M.Bh. drafted manuscript; A.B., S.K.P., and M.Bh. edited and revised manuscript; A.B., K.B., M.Bo., L.M., T.Y.C., R.W., P.T., S.K.P., and M.Bh. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors acknowledge the assistance provided by the University of California, San Francisco (UCSF) Parnassus flow core for analysis of the flow cytometry data and by the UCSF Parnassus Biological Imaging Development CoLab (BIDC) for microscopy training and support.
REFERENCES
- 1.Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med 378: 1811–1823, 2018. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 2.Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med 214: 2387–2404, 2017. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, Wu S, Lang R, Iredale JP. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest 115: 56–65, 2005. doi: 10.1172/JCI22675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, Chak S, Naikawadi RP, Wolters PJ, Abate AR, Butte AJ, Bhattacharya M. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 20: 163–172, 2019. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Satoh T, Nakagawa K, Sugihara F, Kuwahara R, Ashihara M, Yamane F, Minowa Y, Fukushima K, Ebina I, Yoshioka Y, Kumanogoh A, Akira S. Identification of an atypical monocyte and committed progenitor involved in fibrosis. Nature 541: 96–101, 2017. doi: 10.1038/nature20611. [DOI] [PubMed] [Google Scholar]
- 6.Larson-Casey JL, Deshane JS, Ryan AJ, Thannickal VJ, Carter AB. Macrophage Akt1 kinase-mediated mitophagy modulates apoptosis resistance and pulmonary fibrosis. Immunity 44: 582–596, 2016. doi: 10.1016/j.immuni.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson MS, Madala SK, Ramalingam TR, Gochuico BR, Rosas IO, Cheever AW, Wynn TA. Bleomycin and IL-1β-mediated pulmonary fibrosis is IL-17A dependent. J Exp Med 207: 535–552, 2010. doi: 10.1084/jem.20092121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hulsmans M, Sam F, Nahrendorf M. Monocyte and macrophage contributions to cardiac remodeling. J Mol Cell Cardiol 93: 149–155, 2016. doi: 10.1016/j.yjmcc.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Heide D, Weiskirchen R, Bansal R. Therapeutic targeting of hepatic macrophages for the treatment of liver diseases. Front Immunol 10: 2852, 2019. doi: 10.3389/fimmu.2019.02852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wen Y, Yan HR, Wang B, Liu BC. Macrophage heterogeneity in kidney injury and fibrosis. Front Immunol 12: 681748, 2021. doi: 10.3389/fimmu.2021.681748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou X, Franklin RA, Adler M, Jacox JB, Bailis W, Shyer JA, Flavell RA, Mayo A, Alon U, Medzhitov R. Circuit design features of a stable two-cell system. Cell 172: 744–757.e17, 2018. doi: 10.1016/j.cell.2018.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martinet Y, Rom WN, Grotendorst GR, Martin GR, Crystal RG. Exaggerated spontaneous release of platelet-derived growth factor by alveolar macrophages from patients with idiopathic pulmonary fibrosis. N Engl J Med 317: 202–209, 1987. doi: 10.1056/NEJM198707233170404. [DOI] [PubMed] [Google Scholar]
- 13.He L, Huang X, Kanisicak O, Li Y, Wang Y, Li Y, Pu W, Liu Q, Zhang H, Tian X, Zhao H, Liu X, Zhang S, Nie Y, Hu S, Miao X, Wang QD, Wang F, Chen T, Xu Q, Lui KO, Molkentin JD, Zhou B. Preexisting endothelial cells mediate cardiac neovascularization after injury. J Clin Invest 127: 2968–2981, 2017. doi: 10.1172/JCI93868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouveia L, Betsholtz C, Andrae J. PDGF-A signaling is required for secondary alveolar septation and controls epithelial proliferation in the developing lung. Development 145: dev161976, 2018. doi: 10.1242/dev.161976. [DOI] [PubMed] [Google Scholar]
- 15.Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol 8: e1000412, 2010. doi: 10.1371/journal.pbio.1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsukui TS, Wetter JB, Wilson-Kanamori JR, Hazelwood LA, Henderson NC, Adams TS, Schupp JC, Poli SD, Rosas IO, Kaminski N, Matthay MA, Wolters PJ, Sheppard D. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Commun 11: 1920, 2020. doi: 10.1038/s41467-020-15647-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, Peter L, Chung MI, Taylor CJ, Jetter C, Raju L, Roberson J, Ding G, Wood L, Sucre JMS, Richmond BW, Serezani AP, McDonnell WJ, Mallal SB, Bacchetta MJ, Loyd JE, Shaver CM, Ware LB, Bremner R, Walia R, Blackwell TS, Banovich NE, Kropski JA. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv 6: eaba1972, 2020. doi: 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36: 411–420, 2018. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao Y, Xu XH, Jin L. Macrophage polarization in physiological and pathological pregnancy. Front Immunol 10: 792, 2019. doi: 10.3389/fimmu.2019.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lurier EB, Dalton D, Dampier W, Raman P, Nassiri S, Ferraro NM, Rajagopalan R, Sarmady M, Spiller KL. Transcriptome analysis of IL-10-stimulated (M2c) macrophages by next-generation sequencing. Immunobiology 222: 847–856, 2017. doi: 10.1016/j.imbio.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma'ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ; Immunological Genome Consortium. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 13: 1118–1128, 2012. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soucie EL, Weng Z, Geirsdóttir L, Molawi K, Maurizio J, Fenouil R, Mossadegh-Keller N, Gimenez G, VanHille L, Beniazza M, Favret J, Berruyer C, Perrin P, Hacohen N, Andrau JC, Ferrier P, Dubreuil P, Sidow A, Sieweke MH. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science 351: aad5510, 2016. doi: 10.1126/science.aad5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu X, Briseño CG, Durai V, Albring JC, Haldar M, Bagadia P, Kim KW, Randolph GJ, Murphy TL, Murphy KM. Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J Exp Med 213: 2553–2565, 2016. doi: 10.1084/jem.20160600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McQuattie-Pimentel AC, Ren Z, Joshi N, Watanabe S, Stoeger T, Chi M, Lu Z, Sichizya L, Aillon RP, Chen CI, Soberanes S, Chen Z, Reyfman PA, Walter JM, Anekalla KR, Davis JM, Helmin KA, Runyan CE, Abdala-Valencia H, Nam K, Meliton AY, Winter DR, Morimoto RI, Mutlu GM, Bharat A, Perlman H, Gottardi CJ, Ridge KM, Chandel NS, Sznajder JI, Balch WE, Singer BD, Misharin AV, Budinger GRS. The lung microenvironment shapes a dysfunctional response of alveolar macrophages in aging. J Clin Invest 131: e140299, 2021. doi: 10.1172/JCI140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenkins RG, Moore BB, Chambers RC, Eickelberg O, Konigshoff M, Kolb M, Laurent GJ, Nanthakumar CB, Olman MA, Pardo A, Selman M, Sheppard D, Sime PJ, Tager AM, Tatler AL, Thannickal VJ, White ES; ATS Assembly on Respiratory Cell and Molecular Biology. An Official American Thoracic Society Workshop Report: use of animal models for the preclinical assessment of potential therapies for pulmonary fibrosis. Am J Respir Cell Mol Biol 56: 667–679, 2017. doi: 10.1165/rcmb.2017-0096ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun L, Louie MC, Vannella KM, Wilke CA, LeVine AM, Moore BB, Shanley TP. New concepts of IL-10-induced lung fibrosis: fibrocyte recruitment and M2 activation in a CCL2/CCR2 axis. Am J Physiol Lung Cell Mol Physiol 300: L341–L353, 2011. doi: 10.1152/ajplung.00122.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCubbrey AL, Allison KC, Lee-Sherick AB, Jakubzick CV, Janssen WJ. Promoter specificity and efficacy in conditional and inducible transgenic targeting of lung macrophages. Front Immunol 8: 1618, 2017. doi: 10.3389/fimmu.2017.01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. S1: https://doi.org/10.6084/m9.figshare.18708140.v1.