Abstract
Rationale and Objectives:
The use of natural language processing (NLP) in radiology provides an opportunity to assist clinicians with phenotyping patients. However, the performance and generalizability of NLP across healthcare systems is uncertain. We assessed the performance within and generalizability across four healthcare systems of different NLP representational methods, coupled with elastic-net logistic regression to classify lower back pain-related findings from lumbar spine imaging reports.
Materials and Methods:
We used a dataset of 871 X-ray and magnetic resonance imaging reports sampled from a prospective study across four healthcare systems between October 2013 and September 2016. We annotated each report for 26 findings potentially related to lower back pain. Our framework applied four different NLP methods to convert text into feature sets (representations). For each representation, our framework used an elastic-net logistic regression model for each finding (i.e., 26 binary or “one-vs.-rest” classification models). For performance evaluation, we split data into training (80%, 697/871) and testing (20%, 174/871). In the training set, we used cross validation to identify the optimal hyperparameter value and then retrained on the full training set. We then assessed performance based on area under the curve (AUC) for the test set. We repeated this process 25 times with each repeat using a different random train/test split of the data, so that we could estimate 95% confidence intervals, and assess significant difference in performance between representations. For generalizability evaluation, we trained models on data from three healthcare systems with cross validation and then tested on the fourth. We repeated this process for each system, then calculated mean and standard deviation (SD) of AUC across the systems.
Results:
For individual representations, n-grams had the best average performance across all 26 findings (AUC: 0.960). For generalizability, document embeddings had the most consistent average performance across systems (SD: 0.010). Out of these 26 findings, we considered eight as potentially clinically important (any stenosis, central stenosis, lateral stenosis, foraminal stenosis, disc extrusion, nerve root displacement compression, endplate edema, and listhesis grade 2) since they have a relatively greater association with a history of lower back pain compared to the remaining 18 classes. We found a similar pattern for these eight in which n-grams and document embeddings had the best average performance (AUC: 0.954) and generalizability (SD: 0.007), respectively.
Conclusion:
Based on performance assessment, we found that n-grams is the preferred method if classifier development and deployment occur at the same system. However, for deployment at multiple systems outside of the development system, or potentially if physician behavior changes within a system, one should consider document embeddings since embeddings appear to have the most consistent performance across systems.
Keywords: Natural language processing, Lower back pain, Document embeddings, Evaluation, Lumbar spine diagnostic imaging
INTRODUCTION
Lower back pain (LBP) is a common condition, in which patients typically exhibit heterogeneous anatomic phenotypes and undergo a variety of treatments (1–3). LBP patients frequently receive spinal imaging, and findings identified in the resulting radiology reports are expected to help with phenotyping and decision-making (3). However, the association between many findings and LBP is uncertain, because findings can be present in both symptomatic and asymptomatic patients (4). As a result, patients with common aging-related findings may be recommended for LBP-related treatments that are not etiologically linked to their pain. To address this uncertainty, cohort studies and pragmatic trials have investigated patterns of care among patients with LBP and sought to explore subgroups of patients based on the presence of potentially clinically important findings (4–6). To address the interpretation of radiology findings, the Lumbar Imaging with Reporting of Epidemiology (LIRE) study assessed the effectiveness of including benchmark prevalences in the asymptomatic population for findings found in radiology reports for patients who received a diagnostic imaging test of the lumbar spine to reduce subsequent spine-related interventions at four healthcare systems: Kaiser Permanente of Washington, Kaiser Permanente of Northern California, Henry Ford Health System, and Mayo Clinic Health System (7). To further assist research investigating the relationship between findings and LBP, the accurate extraction of findings from large patient groups is needed. However, manual annotation is time-consuming. Natural Language Processing (NLP)-based classification pipelines offer an automated alternative to identify key findings in radiology reports (3).
An NLP-based classification pipeline is composed of two parts: NLP methods that extract features from free-text data and convert them to a structured format (or representation) and the machine learning (ML) model that uses these representations for classification. Text conversion or feature generation can be performed using methods that range from relatively simple domain-dependent and highly manual, to sophisticated data-driven scalable strategies (8–11). Taskspecific rule-based methods identify terms in the free-text that are typically defined by domain experts for a specific outcome of interest. Word or phrase counting methods (n-grams) convert free-text to grouped consecutive words (10). Controlled vocabulary methods convert free-text into a standardized language, using resources such as the Unified Medical Language System (UMLS)’s Metathesaurus, a large vocabulary database that contains information about biomedical and health related concepts, their various names, and the relationships among them (9,12,13). Document embedding methods use neural networks to represent the semantics of documents as vectors of continuous numerical values (11). Each of these methods produces different representations that can influence ML performance. Previous studies have investigated the classification performance of these types of NLP methods (14–16), however to the best of our knowledge only one study assessed generalizability as well (17). With this study, they investigated the performance of their embeddings on a single external dataset, however without extensive validation on multiple external datasets, there is a risk of overestimating both NLP strategies and ML models’ performance (8,17,18).
We hypothesize that NLP methods will have more heterogeneous performance characteristics on external data compared with internal data. The LIRE data provide an opportunity to conduct a systematic evaluation of the utility of different representational methods for identification of image findings in radiology reports drawn from multiple healthcare systems. To assess the reproducibility and reliability of NLP methods, we need to test our methods on multiple external datasets. The purpose of our study is to assess the (1) performance within and (2) generalizability across the four LIRE healthcare systems of different NLP-based feature extraction methods: rules, n-grams, controlled vocabulary, and document embeddings, coupled with elastic-net logistic regression (i.e., the ML model) for classifying radiology reports for LBP-related findings.
MATERIALS AND METHODS
Annotated Dataset
This was a retrospective study that utilized the same annotated dataset from a previous study that showed that ML-based models were superior to rule-based classification (3). Our work is an extension of this as we expanded our NLP methods to include controlled vocabulary and document embeddings and explicitly assessed generalizability across healthcare systems. All participating IRBs agreed that the LIRE study was minimal risk and granted waivers of both consent and Health Insurance Portability and Accountability Act authorization (IRB approval number is 476829). We used limited dataset consisting of a subsample of the LIRE cohort which consisted of approximately 250,000 patients from four healthcare systems who received a thoracic or lumbar spine plain X-ray, magnetic resonance imaging (MRI), or computed tomography (CT) between October 1, 2013 and September 30, 2016 (7). The LIRE study was a multicenter intervention study that investigated whether inserting text about finding prevalence into lumbar spine imaging reports reduced subsequent spine-related treatments (7). Once in the study, patients were followed for two years and their data was regularly collected. We randomly sampled 871 index radiology reports, the first radiology report for each patient, and stratified by system and image modality (3). The sample size was determined based on prior NLP classification tasks (19). Each report was annotated for the presence of 26 LBP-related findings (Table 1) by a team of clinical experts composed of two neuroradiologists, a physiatrist, and a physical therapist. A single report can be annotated for multiple findings. See Supplement Appendix E3 for more details on this process. These findings were based on prior research consisting of a review (20), prospective cohort study (4), and randomized control trial (21) that characterized LBP based on its causes and treatments. Out of these 26, eight were considered to be potentially clinically important: any stenosis, central stenosis, lateral stenosis, foraminal stenosis, disc extrusion, nerve root displacement compression, endplate edema, listhesis grade 2 (4,5,7). Further details of this sampling and annotation process are presented in a previous study (3).
TABLE 1.
The 26 imaging findings of our study
| Type of Finding | Imaging Finding |
|---|---|
|
| |
| Deformities | Listhesis-Grade 1 |
| Listhesis-Grade 2 or higher* | |
| Scoliosis | |
| Fracture | Fracture |
| Spondylosis | |
| Anterior Column Degeneration | Annular Fissure |
| Disc Bulge | |
| Disc Degeneration | |
| Disc Desiccation | |
| Disc Extrusion* | |
| Disc Height Loss | |
| Disc Herniation | |
| Disc Protrusion | |
| Endplate Edema or Type 1 Modic* | |
| Osteophyte-anterior column | |
| Posterior Column Degeneration | Any Stenosis* |
| Facet Degeneration | |
| Associated with Leg Pain | Central Stenosis* |
| Foraminal Stenosis* | |
| Nerve Root Contact | |
| Nerve Root Displaced/Compressed* | |
| Lateral Recess Stenosis* | |
| Nonspecific Findings and Other | Any Degeneration |
| Hemangioma | |
| Spondylolysis | |
| Any Osteophyte | |
Any stenosis refers to any of central, foraminal, lateral recess, or not otherwise specified. Any degeneration refers to any of disc degeneration, facet degeneration, or degeneration not otherwise specified. * indicates the potentially clinically important findings.
Classification Pipeline Overview
Our classification pipeline analyzed the 871 LIRE radiology reports with the goal of learning patterns that are predictive of each of the 26 findings (Fig 1). The pipeline can be separated into three steps: preprocessing, featurization, and ML.
Figure 1.
Overview of the Pipeline. (A) This visualization shows the different steps of our pipeline where we collect 871 radiology reports from our four systems, perform preprocessing to clean the text data, perform feature extraction using our four different methods. We load the n-grams, controlled vocabulary, and document embeddings feature matrices into a logistic regression model to predict the presence of these findings. For rules, we instead use a rule-based model that classifies a report as “positive” if at least one mention was non-negated and “negative” if there was no mention or all mentions of the finding were negated. We perform two types of assessments: generalizability and performance based on AUC. (B) A visual representation using the four different NLP methods to featurize the text for two example findings: fracture and any degeneration. The resulting finding-specific feature matrices are then used for the machine learning model, which uses the first column as the labels and remaining columns as features to predict the presence of these findings. AUC, Area Under the Curve; UMLS, Unified Medical Language System.
Preprocessing and Featurization
For preprocessing, we developed regular expressions to help isolate the finding and impression sections of the 871 radiology reports. For featurization, rules, n-grams, controlled vocabulary mapping, and document embedding methods were used to extract features from the finding and impression sections. Rules require domain experts to identify terms that are related to an outcome of interest. This method is timeconsuming, but since the rules were developed by clinician experts, they can be considered a proxy for clinicians’ judgement for annotations. In our implementation, we developed regular expressions based on the terms our team of clinical experts identified for each finding during the annotation process. For each report, we split the text into sentences. For each sentence, we identified the presence of a finding using the regular expression and checked for negation (22). However, the presence of findings may be uncertain as radiology reports can have terms such as “suggesting” and “not definite.” We minimized this uncertainty by coding these and other similar terms as indicating the presence of a finding. We used Java (v4.6.0), using Apache Lucene (v6.1.0), Porter Stemmer, and NegEx (23,24).
N-grams is a simple, but powerful method in NLP that converts free-text across the radiology reports to n-grams (phrases of different lengths) and indicates their presence and absence in each report (25). In our implementation, we used R (v3.6.1) package Quanteda (v2.0.1) to convert the text into un-, bi-, and trigrams.
Controlled vocabulary is a filtered version of the n-grams approach that leverages only clinically-related features from a standardized medical terminology mapped from the text (9). In our implementation, we first split the text into its constituent sentences using the maximum entropy model in the Apache OpenNLP toolkit to infer the end of a sentence (26). We then applied MetaMap Lite and an assertion classifier developed by Bejan et al., to each patient’s radiology text report to obtain standard UMLS concepts and assertions of whether they were present, absent, conditional, possible or associated with someone else (27,28). We used MetaMap Lite because previous literature demonstrated MetaMap Lite’s performance was comparable to or exceeded MetaMap and other similar methods (28). In addition, we also implemented a version of the controlled vocabulary method (controlled vocabulary filter only) that outputs recognized concepts as raw text, instead of the mapped UMLS terms to assess how a “many-to-one” mapping affects classification performance (29).
Document embeddings is a sophisticated approach that uses a neural network to convert the semantics of text into a continuous numerical vector (11). For the document embedding method, we used the Python (v3.7.3) package Gensim (v3.7.1) to implement the Distributed Bag of Words (DBOW) (11,30). We set the vector length to 600, number of epochs to 500, and allowed the model to initially learn word embeddings using the skip-gram architecture prior to learning document embeddings. We pretrained our DBOW architecture on the full text using two data sets: 522,283 radiology reports from the third version of Medical Information Mart for Intensive Care (MIMIC-III) (31), and the finding and impression sections from 255,094 unannotated reports from the LIRE study. We refer to these as document MIMIC and document LIRE, respectively. The former reflects a typical pretraining scenario and the latter assesses how pretraining on a corpus similar to our actual train/test corpus of 871 reports affects classification performance. We used these pretrained models to derive numerical vector representations of each of the 871 radiology reports. See Supplemental Appendix E1 for additional details. At the end of the featurization step, the textual data from radiology reports are represented as rules, n-grams, controlled vocabulary, controlled vocabulary filter only, document MIMIC, and document LIRE.
Rule- and Machine Learning-Based Model
For the rules, we used a rule-based model to classify a report as “positive” for a finding if at least one mention in a report was non-negated and “negative” if there was no mention or all mentions of the finding were negated. We then used the trapezoidal rule approximation to calculate the area under the curve (AUC) (32). For each non-rule representation (i.e., feature set) and the finding labels from the annotation process, we developed an elastic-net logistic regression model to predict the presence of each finding (i.e., 26 binary or “one-vs.-rest” classification models) using the R (v3.5.1) package caret (v6.0–80). Within the training set, ten-fold cross validation was used to adjust the value of our regularization parameter (lambda) to perform feature selection on our predictors by shrinking our nonimportant predictors’ coefficients towards 0. For each resulting finding-specific model, we identified the optimal threshold based on the training set’s receiver operator characteristic (ROC) plot; the threshold is the point closest to the true positive rate of 1 and false positive rate of 0 (i.e., the point closest to the top left corner of the curve) using Euclidean distance (33).
Performance and Generalizability Assessment
We used R (v3.5.1) to evaluate each representation. We used AUC of an ROC plot as the primary evaluation metric. This is because we envision our pipeline as an efficient “first-pass” screening tool intended to favor the identification of more true positives. For performance assessment, we randomly split our full dataset into 80% (697/871) for training and 20% (174/871) for evaluating our model for each finding. We assessed group-level performance by averaging the evaluation AUC across all finding-specific models and across all potentially clinically important finding-specific models, separately. We repeated this process 25 times with each independent repeat using a different random train/test split of the data, so that we could estimate 95% confidence intervals. For each finding/group, a t-test was used to assess significant performance comparing the 25 repeats of the best representation to the next best representation. We used Bonferroni correction to correct for multiple hypothesis testing; for the two groups, we considered p-value 0.025 (0.05/2 groups) to be significant, and for the 26 findings, we considered p-value 0.0019 (0.05/26 findings) to be significant. To assess generalizability across healthcare systems, we trained our model on reports from three systems and evaluated on the fourth, iteratively, for each finding. For each finding, we calculated the mean and standard deviation of the AUC across the four systems. We calculated group generalizability by averaging the AUC across all findings, and across all potentially clinically important findings for each system and then calculated the mean and standard deviation. We chose mean and standard deviation to quantify generalizability, because the former measures the quality, while the latter measures the consistency of performance across systems. For the generalizability assessment, we included all representations except for rules because they were developed using reports from all four systems, eliminating the possibility of evaluation using data unavailable at the point of algorithm development.
Additional details for these and other secondary analyses are provided in the Supplementary Materials.
RESULTS
Data Summary
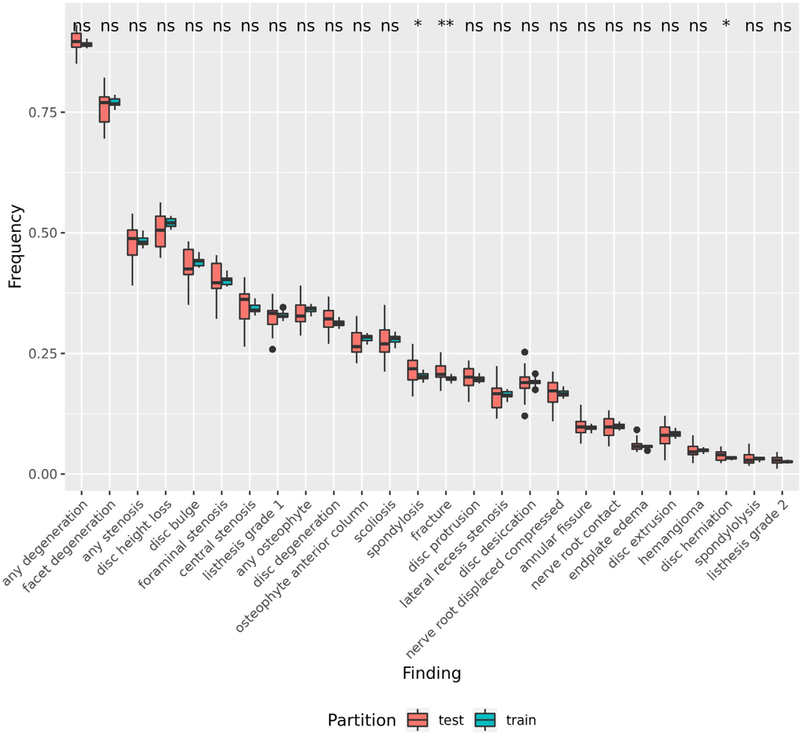
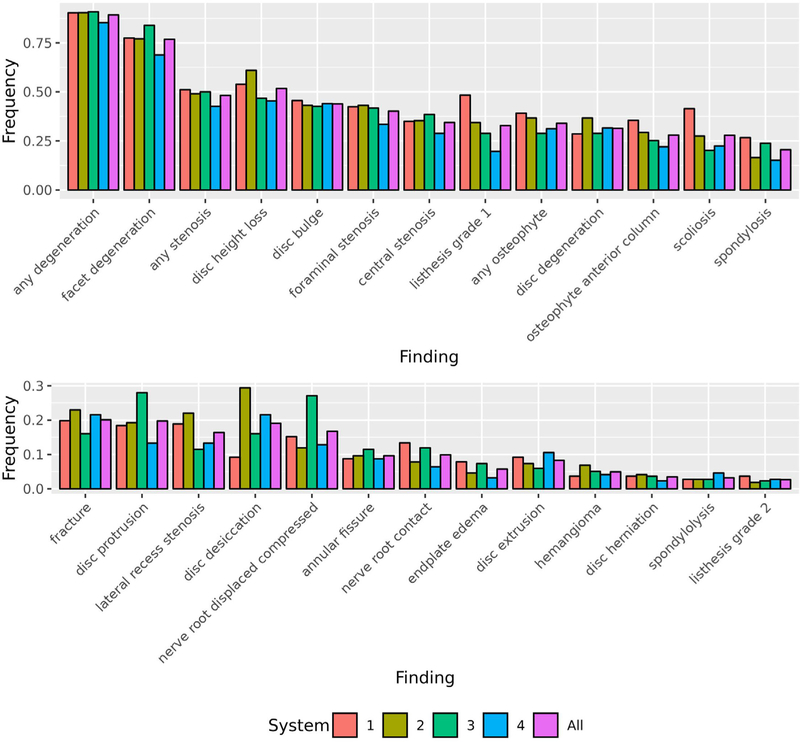
In our dataset (n = 871), we sampled reports with similar proportions of image type (i.e., X-ray and MRI) and patients’ age and gender across our healthcare systems (Table 2). For performance assessment, we’ve shown that our training set is representative of our test set for 23/26 findings by using a t-test to assess the significant difference in the prevalence between sets across the 25 repeats for each finding (Fig 2). For generalizability assessment, we found that each healthcare system was comparable since the finding label prevalence across healthcare systems is overall similar with any degeneration having the highest label prevalence (0.896) and listhesis grade 2 having the lowest label prevalence (0.028) (Fig 3).
TABLE 2.
Summary of the 871 Reports Based on Image Type, Average Text Length, and Patients’ Age and Gender Across the Four Healthcare Systems
| System | Image Type | N in Dataset | Average Text Length | Average Age | Female % |
|---|---|---|---|---|---|
|
| |||||
| Kaiser Permanente of Washington | X-Ray | 102 | 132 ± 34 | 70.35 ± 13.84 | 0.60 |
| MR | 115 | 267 ± 106 | 58.90 ± 14.35 | 0.49 | |
| Total | 217 | 203 ± 105 | 64.28 ± 15.20 | 0.54 | |
| Kaiser Permanente of Northern California | X-Ray | 104 | 143 ± 38 | 67.51 ± 16.82 | 0.61 |
| MR | 114 | 270 ± 95 | 57.1 ± 14.96 | 0.53 | |
| Total | 218 | 210 ± 97 | 62.06 ± 16.68 | 0.56 | |
| Henry Ford Health System | X-Ray | 103 | 121 ± 57 | 67.15 ± 16.04 | 0.72 |
| MR | 115 | 268 ± 152 | 58.95 ± 15.77 | 0.5 | |
| Total | 218 | 199 ± 137 | 62.96 ± 16.44 | 0.61 | |
| Mayo Clinic Health System | X-Ray | 103 | 141 ± 39 | 69.35 ± 16.15 | 0.61 |
| MR | 115 | 222 ± 104 | 55.13 ± 15.44 | 0.58 | |
| Total | 218 | 184 ± 90 | 61.85 ± 17.28 | 0.60 | |
| All | X-Ray | 413 | 134 ± 44 | 68.58 ± 15.76 | 0.63 |
| MR | 458 | 257 ± 118 | 57.52 ± 15.17 | 0.52 | |
| Total | 871 | 199 ± 109 | 62.79 ± 16.42 | 0.58 | |
We calculated the average text length for the finding and impression sections in each report, the average age of patients, and the proportion of female patients for each healthcare system and each type of report. For average text length, we calculated the average text length for both the finding and impression sections, since these sections were required for our pipeline. For both average text length and age, we included standard deviation.
Figure 2.
Comparison of the Finding Label Prevalence Between the Training and Test Set. We compared the finding label prevalence between the train and test sets across the 25 repeats. To assess a significant difference, we performed a t-test between the two sets for each finding. An asterisk indicates a significant difference, while “ns” indicates no significant difference.
Figure 3.
Comparison of the Finding Label Prevalence Across the Healthcare Systems. We compared the finding label prevalence across the four healthcare systems. 1 = Kaiser Permanente of Washington, 2 = Kaiser Permanente of Northern California, 3 = Henry Ford Health System, 4 = Mayo Clinic Health System, All = all four systems.
Comparing the Group and Finding Level Predictive Performance of Individual Representations
To assess the best performing representation, we trained and tested 26 finding-specific models for each representation and calculated finding-level and group-level AUC. On average across all findings, we found that the models generally performed well, with average AUC values above 0.87. N-grams and controlled vocabulary had the best (AUC = 0.960) and worst average (AUC = 0.879) performance, respectively (Table 3). At the finding level, n-grams had better performance than the corresponding second-best representation (which differed from finding to finding) for 22 findings, 11 of which were statistically significant. These results suggest that on average, the relatively simple methodology of n-grams is sufficient to classify our 26 findings.
TABLE 3.
Best performing individual representations based on group and finding level AUC
| Finding | Prop. | P-Value | N-Grams | Document LIRE | Rules | Document MIMIC | Controlled Vocabulary | Controlled Vocabulary Filter Only |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| All Findings | - | 1.06E-24† | 0.960 (0.949, 0.972)✦ | 0.910 (0.892, 0.929)✧ | 0.897 (0.882, 0.911) | 0.894 (0.872, 0.916) | 0.882 (0.862, 0.901) | 0.879 (0.857, 0.902) |
| Potentially Clinically Important Findings | - | 2.06E-13† | 0.954 (0.925, 0.983)✦ | 0.910 (0.878, 0.942)✧ | 0.821 (0.789, 0.852) | 0.888 (0.856, 0.920) | 0.857 (0.821, 0.894) | 0.854 (0.813, 0.895) |
| any degeneration | 0.896 | 0.03289 | 0.947 (0.906, 0.989)✦ | 0.896 (0.820, 0.972) | 0.850 (0.770, 0.931) | 0.874 (0.789, 0.958) | 0.936 (0.911, 0.961)✧ | 0.913 (0.882, 0.943) |
| facet degeneration | 0.762 | 0.13484 | 0.970 (0.940, 0.999)✦ | 0.963 (0.935, 0.991)✧ | 0.873 (0.832, 0.914) | 0.949 (0.919, 0.979) | 0.923 (0.888, 0.959) | 0.922 (0.883, 0.961) |
| disc height loss | 0.507 | 5.46E-10† | 0.931 (0.891, 0.970)✦ | 0.833 (0.791, 0.875) | 0.877 (0.829, 0.925) | 0.830 (0.774, 0.886) | 0.874 (0.812, 0.935) | 0.878 (0.826, 0.930)✧ |
| any stenosis* | 0.480 | 0.09604 | 0.972 (0.950, 0.994)✦ | 0.957 (0.930, 0.983) | 0.893 (0.856, 0.930) | 0.967 (0.945, 0.988)✧ | 0.955 (0.926, 0.984) | 0.961 (0.935, 0.987) |
| disc bulge | 0.435 | 0.00276 | 0.986 (0.972, 1.000)✦ | 0.978 (0.956, 1.000)✧ | 0.976 (0.953, 1.000) | 0.967 (0.939, 0.994) | 0.955 (0.923, 0.987) | 0.952 (0.920, 0.984) |
| foraminal stenosis* | 0.400 | 0.00153† | 0.950 (0.922, 0.978)✦ | 0.935 (0.899, 0.971) | 0.914 (0.876, 0.952) | 0.936 (0.906, 0.965)✧ | 0.932 (0.898, 0.966) | 0.930 (0.896, 0.964) |
| central stenosis* | 0.351 | 6.37E-10† | 0.950 (0.919, 0.981)✦ | 0.903 (0.876, 0.930) | 0.773 (0.731, 0.815) | 0.915 (0.884, 0.947)✧ | 0.907 (0.861, 0.953) | 0.901 (0.852, 0.950) |
| any osteophyte | 0.332 | 3.44E-07† | 0.955 (0.916, 0.994)✦ | 0.888 (0.845, 0.932) | 0.925 (0.894, 0.955)✧ | 0.886 (0.835, 0.937) | 0.875 (0.818, 0.933) | 0.880 (0.819, 0.94) |
| listhesis grade 1 | 0.324 | 0.03888 | 0.967 (0.941, 0.994)✦ | 0.927 (0.880, 0.974) | 0.958 (0.928, 0.989)✧ | 0.910 (0.858, 0.961) | 0.945 (0.903, 0.987) | 0.947 (0.904, 0.989) |
| disc degeneration | 0.322 | 0.00203 | 0.935 (0.898, 0.973)✦ | 0.830 (0.792, 0.869) | 0.904 (0.865, 0.944) | 0.784 (0.726, 0.842) | 0.909 (0.842, 0.976)✧ | 0.909 (0.863, 0.954)✧ |
| scoliosis | 0.274 | 0.03726 | 0.969 (0.943, 0.994)✦ | 0.924 (0.888, 0.961) | 0.959 (0.924, 0.994)✧ | 0.929 (0.887, 0.971) | 0.900 (0.850, 0.949) | 0.901 (0.854, 0.948) |
| osteophyte anterior column | 0.271 | 5.13E-10† | 0.953 (0.930, 0.976)✦ | 0.867 (0.820, 0.914) | 0.913 (0.872, 0.954)✧ | 0.846 (0.785, 0.907) | 0.882 (0.831, 0.933) | 0.874 (0.825, 0.923) |
| spondylosis | 0.217 | 0.39974 | 0.992 (0.977, 1.010)✦ | 0.936 (0.881, 0.99) | 0.990 (0.974, 1.010)✧ | 0.901 (0.846, 0.955) | 0.900 (0.836, 0.964) | 0.883 (0.817, 0.949) |
| fracture | 0.212 | 0.62647 | 0.949 (0.912, 0.987)✦ | 0.947 (0.921, 0.973)✧ | 0.883 (0.816, 0.95) | 0.896 (0.839, 0.953) | 0.914 (0.868, 0.960) | 0.925 (0.878, 0.972) |
| disc protrusion | 0.197 | 0.69348 | 0.977 (0.953, 1.000) | 0.940 (0.906, 0.973) | 0.927 (0.879, 0.974) | 0.910 (0.866, 0.954) | 0.982 (0.952, 1.010)✦ | 0.980 (0.948, 1.010)✧ |
| disc desiccation | 0.189 | 1.39E-05† | 0.981 (0.953, 1.010)✦ | 0.957 (0.923, 0.990) | 0.958 (0.921, 0.994)✧ | 0.923 (0.884, 0.962) | 0.817 (0.745, 0.888) | 0.822 (0.747, 0.898) |
| nerve root displaced/compressed* | 0.169 | 1.08E-06† | 0.955 (0.913, 0.996)✦ | 0.913 (0.854, 0.972)✧ | 0.785 (0.698, 0.872) | 0.907 (0.848, 0.966) | 0.870 (0.819, 0.921) | 0.864 (0.817, 0.911) |
| lateral recess stenosis* | 0.163 | 0.00235 | 0.966 (0.921, 1.010)✦ | 0.941 (0.909, 0.973) | 0.649 (0.567, 0.731) | 0.948 (0.918, 0.978)✧ | 0.843 (0.771, 0.914) | 0.844 (0.773, 0.915) |
| annular fissure | 0.099 | 0.38358 | 0.950 (0.888, 1.010)✧ | 0.944 (0.886, 1.000) | 0.957 (0.905, 1.010)✦ | 0.922 (0.848, 0.996) | 0.763 (0.667, 0.860) | 0.755 (0.650, 0.860) |
| nerve root contact | 0.097 | 1.13E-08† | 0.972 (0.949, 0.996)✦ | 0.921 (0.859, 0.982)✧ | 0.910 (0.827, 0.993) | 0.914 (0.856, 0.973) | 0.797 (0.716, 0.877) | 0.818 (0.741, 0.894) |
| disc extrusion* | 0.079 | 0.00089† | 0.994 (0.963, 1.020)✦ | 0.979 (0.954, 1.000)✧ | 0.886 (0.775, 0.997) | 0.948 (0.901, 0.995) | 0.969 (0.914, 1.020) | 0.962 (0.897, 1.030) |
| endplate edema* | 0.059 | 0.00088† | 0.916 (0.831, 1.000)✦ | 0.868 (0.765, 0.970)✧ | 0.854 (0.732, 0.976) | 0.838 (0.713, 0.963) | 0.789 (0.683, 0.896) | 0.778 (0.626, 0.930) |
| hemangioma | 0.049 | 0.0736 | 0.991 (0.950, 1.030)✧ | 0.899 (0.807, 0.991) | 0.999 (0.995, 1.000)✦ | 0.875 (0.705, 1.050) | 0.963 (0.867, 1.060) | 0.951 (0.845, 1.060) |
| disc herniation | 0.038 | 0.04629 | 0.956 (0.878, 1.003)✧ | 0.890 (0.715, 1.060) | 0.975 (0.927, 1.020)✦ | 0.912 (0.740, 1.080) | 0.760 (0.526, 0.994) | 0.786 (0.520, 1.050) |
| spondylolysis | 0.032 | 0.00567 | 0.981 (0.940, 1.020)✦ | 0.832 (0.654, 1.010) | 0.936 (0.795, 1.080)✧ | 0.866 (0.747, 0.986) | 0.754 (0.507, 1.000) | 0.785 (0.561, 1.010) |
| listhesis grade 2* | 0.028 | 0.00018† | 0.905 (0.741, 1.070)✦ | 0.799 (0.604, 0.994)✧ | 0.767 (0.580, 0.955) | 0.683 (0.486, 0.88) | 0.735 (0.559, 0.91) | 0.706 (0.510, 0.902) |
For each representation, we trained and tested 26 models (one for each finding) on 80% and 20% of the dataset, respectively. For group level, we averaged the AUC across all findings and across all potentially clinically important findings for each representation. We repeated this process 25 times with different splits of the data to calculate 95% confidence intervals. Table shows the best performing representation ordered left to right based on the All Findings row (1st row). The first column indicates the finding/group. For the findings, the second column indicates prevalence in the test set represented as a proportion. For each row, ✦ indicates best performing individual representation based on the average AUC and ✧ indicates the second-best representation. We show the 95% confidence interval in the parentheses. Finally, for each finding and group, we performed a t-test comparing the best representation’s distribution of AUC values for the 25 repeats to the second-best representation. † indicates significant comparisons with Bonferroni correction (p-value significance for the groups: 0.025 = 0.05/2 groups, p-value significance for the findings: 0.0019 = 0.05/26 findings). Finally * indicates the potentially clinically important findings. AUC = Area Under the Curve, Prop. = Proportion.
In addition to assessing the performance of n-grams, we were also interested in characterizing the performance relative to rules, a representation requiring domain-expertise, and document LIRE, an advanced data-driven representation. This comparison is of interest as each of these three representations reflect different disciplines in featurizing textual data that range from domain-expertise to advanced domain-agnostic implementations. On average across all findings, rules (AUC = 0.897) performed worse than n-grams and document LIRE. At the finding level, rules was out performed by n-grams for eight out of twelve rare findings (prevalence < 20%). Additionally, while document LIRE had better overall performance than rules, it was not the best representation for any of the findings. This may be due to the fact that document LIRE may not have been the best representation but had stable performance across findings (min AUC = 0.799, max AUC = 0.979) compared to rules (min AUC = 0.649, max AUC = 0.999). These results also suggest when considering only rare findings, n-grams still perform better than other representations.
Comparing the Group and Finding Level Generalizability Performance of Individual Representation Across Healthcare Systems
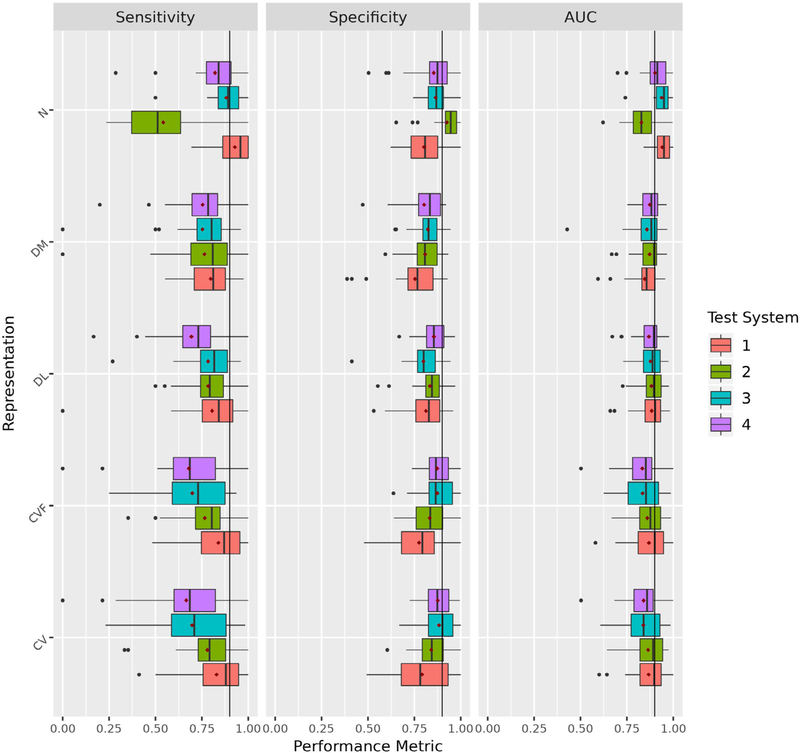
To assess the best generalizable representation, we trained 26 finding-specific models for each representation on data from three systems and tested on the fourth system, iteratively. For each finding/group, we calculated the mean and standard deviation of the test AUC across the four systems. At the group level, n-grams had the best average performance across all findings (mean AUC = 0.902) and at the finding level, it was the best performing representation for 10 findings (Table 4). The next best representation was document LIRE at the group level (mean AUC = 0.879) and it was the best method for 10 findings as well (Table 4). Interestingly, when considering standard deviation at the group level, we found that document LIRE and n-grams were the most (standard deviation = 0.010) and least consistent (standard deviation = 0.051) representations across all findings, respectively (Table 5). We found n-grams could not generalize well to system two, particularly resulting in a lower sensitivity and higher specificity compared to other representations (Fig 4); we verified this result through complementary analyses (Supplemental Appendix E3). At the finding level, document LIRE was the most consistent representation for 11 findings. These results suggest that while n-grams had relatively the best performance, it had the worst consistency across systems. Instead, document embeddings pretrained on study-specific data (document LIRE) had relatively the most consistent classification performance on average across our systems.
TABLE 4.
Best performing individual representations based on group and finding level mean of AUC across systems
| Finding | Proportion | N-Grams | Document LIRE | Document MIMIC | Controlled Vocabulary | Controlled Vocabulary Filter Only |
|---|---|---|---|---|---|---|
|
| ||||||
| All Findings | - | 0.902✦ | 0.879 | 0.868 | 0.857 | 0.853 |
| Potentially Clinically Important Findings | - | 0.898✦ | 0.890 | 0.887 | 0.834 | 0.833 |
| any degeneration | 0.896 | 0.905 | 0.881 | 0.848 | 0.927✦ | 0.874 |
| facet degeneration | 0.762 | 0.919 | 0.952✦ | 0.927 | 0.914 | 0.898 |
| disc height loss | 0.507 | 0.845✦ | 0.754 | 0.748 | 0.845✦ | 0.837 |
| any stenosis* | 0.480 | 0.907 | 0.957✦ | 0.955 | 0.946 | 0.943 |
| disc bulge | 0.435 | 0.954 | 0.963✦ | 0.953 | 0.930 | 0.929 |
| foraminal stenosis* | 0.400 | 0.885 | 0.922✦ | 0.913 | 0.904 | 0.888 |
| central stenosis* | 0.351 | 0.916✦ | 0.881 | 0.897 | 0.891 | 0.878 |
| any osteophyte | 0.332 | 0.874✦ | 0.874✦ | 0.860 | 0.831 | 0.833 |
| listhesis grade 1 | 0.324 | 0.905 | 0.899 | 0.891 | 0.930 | 0.936✦ |
| disc degeneration | 0.322 | 0.873 | 0.710 | 0.716 | 0.861 | 0.908✦ |
| scoliosis | 0.274 | 0.911✦ | 0.892 | 0.891 | 0.865 | 0.870 |
| osteophyte anterior column | 0.271 | 0.832 | 0.825 | 0.818 | 0.845✦ | 0.825 |
| spondylosis | 0.217 | 0.985✦ | 0.801 | 0.764 | 0.823 | 0.781 |
| fracture | 0.212 | 0.910 | 0.926✦ | 0.889 | 0.910 | 0.906 |
| disc protrusion | 0.197 | 0.948 | 0.935 | 0.904 | 0.977 | 0.978✦ |
| disc desiccation | 0.189 | 0.929✦ | 0.909 | 0.850 | 0.796 | 0.797 |
| nerve root displaced/compressed* | 0.169 | 0.918✦ | 0.906 | 0.894 | 0.845 | 0.855 |
| lateral recess stenosis* | 0.163 | 0.915 | 0.932 | 0.934✦ | 0.831 | 0.841 |
| annular fissure | 0.099 | 0.908 | 0.921✦ | 0.886 | 0.766 | 0.763 |
| nerve root contact | 0.097 | 0.917✦ | 0.885 | 0.854 | 0.736 | 0.746 |
| disc extrusion* | 0.079 | 0.947 | 0.972✦ | 0.925 | 0.968 | 0.959 |
| endplate edema* | 0.059 | 0.844 | 0.865✦ | 0.841 | 0.725 | 0.762 |
| hemangioma | 0.049 | 0.917 | 0.869 | 0.908 | 0.963✦ | 0.963✦ |
| disc herniation | 0.038 | 0.820 | 0.826✦ | 0.798 | 0.725 | 0.700 |
| spondylolysis | 0.032 | 0.938✦ | 0.814 | 0.849 | 0.775 | 0.804 |
| listhesis grade 2* | 0.028 | 0.838✦ | 0.740 | 0.681 | 0.657 | 0.614 |
For each representation, we trained our model on reports from three systems and evaluated on the fourth, iteratively, for each finding. For each finding, we calculated the mean of the AUC across the four systems. We calculated group-level performance by averaging the AUC across all findings, and across all potentially clinically important findings for each system and then calculated the mean across the systems. Table shows the best performing representation ordered left to right based on the All Findings row (1 st row). The first column indicates the finding/group. For the findings, the second column indicates prevalence in the test set represented as a proportion. ✦ indicates the best performing representation for that finding and group. Finally * indicates the findings that were potentially clinically important. AUC = Area Under the Curve.
TABLE 5.
Most consistent individual representations based on group and finding level standard deviation of AUC across systems
| Finding | Proportion | Document LIRE | Controlled Vocabulary | Document MIMIC | Controlled Vocabulary Filter Only | N-Grams |
|---|---|---|---|---|---|---|
|
| ||||||
| All Findings | - | 0.010✦ | 0.012 | 0.013 | 0.014 | 0.051 |
| Potentially Clinically Important Findings | - | 0.007✦ | 0.035 | 0.024 | 0.031 | 0.076 |
| any degeneration | 0.896 | 0.021 | 0.031 | 0.024 | 0.041 | 0.020✦ |
| facet degeneration | 0.762 | 0.018✦ | 0.039 | 0.022 | 0.038 | 0.064 |
| disc height loss | 0.507 | 0.033 | 0.050 | 0.010✦ | 0.057 | 0.103 |
| any stenosis* | 0.480 | 0.023 | 0.032 | 0.013✦ | 0.035 | 0.051 |
| disc bulge | 0.435 | 0.019 | 0.022 | 0.009✦ | 0.015 | 0.040 |
| foraminal stenosis* | 0.400 | 0.016✦ | 0.058 | 0.035 | 0.077 | 0.042 |
| central stenosis* | 0.351 | 0.030✦ | 0.046 | 0.038 | 0.056 | 0.043 |
| any osteophyte | 0.332 | 0.034 | 0.099 | 0.031✦ | 0.094 | 0.108 |
| listhesis grade 1 | 0.324 | 0.032 | 0.014 | 0.009✦ | 0.014 | 0.076 |
| disc degeneration | 0.322 | 0.033 | 0.052 | 0.092 | 0.049 | 0.026✦ |
| scoliosis | 0.274 | 0.017✦ | 0.051 | 0.025 | 0.043 | 0.068 |
| osteophyte anterior column | 0.271 | 0.025 | 0.084 | 0.020✦ | 0.083 | 0.088 |
| spondylosis | 0.217 | 0.040 | 0.156 | 0.036 | 0.164 | 0.015✦ |
| fracture | 0.212 | 0.019✦ | 0.028 | 0.029 | 0.039 | 0.029 |
| disc protrusion | 0.197 | 0.021 | 0.011✦ | 0.018 | 0.016 | 0.053 |
| disc desiccation | 0.189 | 0.024 | 0.052 | 0.020✦ | 0.051 | 0.090 |
| nerve root displaced/compressed* | 0.169 | 0.020 | 0.018 | 0.015 | 0.011✦ | 0.021 |
| lateral recess stenosis* | 0.163 | 0.017✦ | 0.038 | 0.019 | 0.034 | 0.063 |
| annular fissure | 0.099 | 0.015✦ | 0.064 | 0.057 | 0.065 | 0.063 |
| nerve root contact | 0.097 | 0.071 | 0.035 | 0.044 | 0.014✦ | 0.090 |
| disc extrusion* | 0.079 | 0.007✦ | 0.024 | 0.015 | 0.023 | 0.076 |
| endplate edema* | 0.059 | 0.059✦ | 0.164 | 0.059✦ | 0.091 | 0.126 |
| hemangioma | 0.049 | 0.073 | 0.048 | 0.007✦ | 0.048 | 0.105 |
| disc herniation | 0.038 | 0.046✦ | 0.079 | 0.092 | 0.065 | 0.094 |
| spondylolysis | 0.032 | 0.107 | 0.113 | 0.087 | 0.108 | 0.044✦ |
| listhesis grade 2* | 0.028 | 0.047✦ | 0.128 | 0.184 | 0.099 | 0.184 |
For each representation, we trained our model on reports from three systems and evaluated on the fourth, iteratively, for each finding. For each finding, we calculated the standard deviation of the AUC across the four systems. We calculated group-level consistency by averaging the AUC across all findings, and across all potentially clinically important findings for each system as a test set and then calculated the standard deviation across the systems. Table shows the most consistent representation ordered left to right based on the All Findings row (1st row). The first column indicates the finding/group. For the findings, the second column indicates prevalence in the test set represented as a proportion. ✦ indicates the most consistent representation for that finding and group. Finally * indicates findings that were potentially clinically important. AUC = Area Under the Curve.
Figure 4.
Assessing Generalizability of Individual Representations. We compared the generalizability of each of our representations and assessed performance using sensitivity, specificity, and AUC. For each representation, we plotted a boxplot to represent the distribution of the 26 findings for each test performance metric across healthcare systems. CV, Controlled Vocabulary; CVF, Controlled Vocabulary Filter Only; DM, Document MIMIC; DL, Document LIRE; N, N-grams. 1 = Kaiser Permanente of Washington, 2 = Kaiser Permanente of Northern California, 3 = Henry Ford Health System, and 4 = Mayo Clinic Health System.
Assessing Performance and Generalizability of Potentially Clinically Important Findings
In our previous sections, we focused on all 26 findings, however we consider eight of these findings to be potentially clinically important. As a result, we believe it’s important to present results for this important subset of findings. For performance assessment, n-grams had the best performance (AUC = 0.954), and it was significantly better than that of document LIRE, the second-best representation (AUC = 0.910) (Table 3). At the finding level, n-grams also had the best performance for all eight potentially clinically important findings, six of which were statistically significant. For generalizability assessment, n-grams had better performance (mean AUC = 0.898) than document LIRE (mean AUC = 0.890) (Table 4). At the finding level, n-grams and document LIRE had the best performance for seven of these findings. For consistency, document LIRE was the most consistent representation with standard deviation of 0.007 compared to n-grams’ 0.076 (Table 5). At the finding level, document LIRE and MIMIC had the most consistent performance for six and two potentially clinically important findings, respectively, with one tie endplate edema (Table 5). These results indicate for this subset of findings, we still observe the same trend where n-grams has the best performance, but document LIRE has the best consistency.
DISCUSSION
Manual extraction of information from radiology reports can be burdensome, making automated NLP methods attractive for such tasks. However, correctly estimating these methods’ performance across multiple healthcare systems requires an understanding of their generalizability on external datasets. In this study, we compared and contrasted the performance of different NLP methods coupled with elastic-net logistic regression to classify 26 findings related to LBP through performance and generalizability assessment. Our study suggests that if classifier development and deployment occur at the same system, then n-grams may be preferable. However, for deployment at multiple systems outside of the system of development, one should consider n-grams with the caveat that it’s consistency can vary across systems, while document embeddings pretrained on study-specific data (document LIRE) or a publicly available dataset (document MIMIC) had the most consistent performance.
Overall, based on performance assessment, n-grams, a relatively simple, data-driven, domain-agnostic method, is superior to more sophisticated methods (document embeddings and controlled vocabulary) in extracting known findings from text. These results are in line with prior studies (14,34). Additionally, for rare findings (prevalence < 20%), n-grams had the highest AUCs, which is consistent with a prior study evaluating n-grams coupled with LASSO logistic regression to classify acute LBP (prevalence of 22%) (35). However, n-grams did not generalize well across the four healthcare systems when compared to document embeddings. This performance-generalizability duality can be explained as follows: the n-grams method is dependent on the precise phrasing in the training text. When we considered performance assessment, we split the full dataset into 80% (697/871 reports) for the train set and 20% (174/871 reports) for the test set; both sets contained the four healthcare systems, and their text were representative of each other. However, when considering each system independently for the generalizability assessment, n-grams were more susceptible to overfitting, i.e., they may have contained predictors more relevant to the training systems than the test system. When comparing summary statistics of the raw text among systems, we found that system two was indeed different from the other systems (Supplemental Appendix E3) and changing classification thresholds for the models tested on this system did not affect performance. In comparison, document embeddings better captured differently worded but synonymous concepts by transforming the raw text into abstract numerical representations that reflect semantics, leading to less deviation in performance across systems but slightly worse performance overall.
Document embeddings are of particular interest because they represent a sophisticated method of featurization that are pretrained on large-scale corpora to learn more generalizable representations of text. Here, document embeddings were pretrained on two different data sources, unannotated LIRE reports (smaller but more relevant to LBP) and MIMIC-III (larger but less specific). While document LIRE overall performed better than document MIMIC, the difference was modest, suggesting that a lack of task-specific corpus is not a barrier for using document embeddings in clinical tasks. This observation is consistent with other studies (16,36,37).
Controlled vocabulary and controlled vocabulary filter only are two representations that can be considered filtered versions of the n-grams approach that leverages only clinically related features. These representations differ from each other in that the former maps the clinically relevant raw terms to standardized terms to then use as features, while the latter does not map and instead uses the clinically relevant raw terms as features. As a result, controlled vocabulary performs a “many-to-one” mapping that can affect performance. When comparing these two representations, we found that controlled vocabulary marginally outperformed controlled vocabulary filter only in both performance and generalizability. Our study indicates that this “many-to-one” mapping is not detrimental to performance, but does not provide a substantial improvement relative to using only the clinically relevant raw terms as features.
Beyond performance and generalizability, scalability and interpretability are important factors to consider when choosing a NLP-based feature extraction method. Rules are the most interpretable method, because they solely rely on domain experts to provide the synonyms to search for in text. However, this method cannot scale well as expanding the synonyms for a more complete identification of findings and larger number of findings will require more time and domain experts. In contrast, n-grams, controlled vocabulary, and document embeddings are domain-agnostic computational methods, and as a result they can scale well to a large number of radiology reports and findings. These methods differ in their interpretability. Controlled vocabulary and n-grams are the most interpretable methods as the former provides an ML model clinically relevant terms as features, while the latter provides the raw text as features. It is relatively easy for a researcher to examine a model’s features and coefficients based on either of these two methods and understand what aspects of a radiology report are predictive of the outcome of interest. Document embeddings is the least interpretable method as it uses a neural network to represent a document’s semantics as a vector of continuous values. These values are no longer interpretable as they are a result of the interactions of different word embeddings from the radiology reports in the neural network. When considering subsequent implementation, it is important to consider factors such as scalability and interpretability in addition to performance and generalizability.
There are several limitations to this study. First, our pipeline required binary annotations for findings, however the presence of findings may be uncertain as radiology reports can have terms such as “suggesting” and “not definite.” We minimized this uncertainty by coding these and other similar terms as indicating the presence of a finding. Second, while our sample size was in line with recommendations for classification tasks, larger training and testing sets could have led to less variable performances across our different NLP methods (19). Third, we evaluated the algorithms but not the entire pipeline involving the querying and transfer of data; there may be discrepancies in our performance estimates when compared to those at actual deployment. Fourth, we could not assess our rules’ generalizability, since the search terms were developed from reports from all four systems. Finally, in the case of document embeddings, because of our limited computational resources, we had to sequentially adjust hyperparameter values in the pretraining step, rather than conducting a grid search. With a more extensive hyperparameter search, we may have been able to improve performance.
Diagnostic imaging is often an early step for LBP patients that eventually leads to interventions, however the association between findings and LBP is uncertain (4). Jarvik et al. investigated this association and identified eight (potentially clinically important) findings that may be associated with a history of LBP and of these eight, nerve root contact, disc extrusion, and central stenosis may be associated with a new onset of pain (4,5,7). We’ve shown that our pipeline can automate classifying reports for these potentially clinically important findings using n-grams, and can generalize across healthcare systems using document embeddings. Our automated pipeline can assist similar studies by developing large cohorts quickly and inexpensively to investigate the association between findings and a clinical outcome within and across healthcare systems using text-based data.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by the 1) National Institutes of Health (NIH) Health Care Systems Research Collaboratory by the NIH Common Fund through cooperative agreement U24AT009676 from the Office of Strategic Coordination within the Office of the NIH Director and cooperative agreements UH2AT007766 and UH3AR066795 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), 2) University of Washington Clinical Learning, Evidence, and Research (CLEAR) Center for Musculoskeletal Disorders Administrative, Methodologic and Resource Cores and NIAMS/NIH P30AR072572, 3) National Center for Advancing Translational Sciences (NCATS) TL1 TR002318 (for CJ), and 4) National Library of Medicine (NLM) K99LM012992 (for VP). The content is solely the responsi- bility of the authors and does not necessarily represent the official views of the NIH.
We give thanks to Kathryn T James, PA, MPH for her extensive role in research conduct of the LIRE study. We give thanks to Hannu T. Huhdanpaa MD, Msc, Pradeep Suri MD, MS, and Sean D. Rundell DPT, PhD for annotating the 871 reports. We thank Eric N. Meir MS and W. Katherine Tan PhD for preprocessing the 871 reports. Finally, we thank W. Katherine Tan PhD for developing the initial pipeline that used rules and n-grams to annotate the 871 reports.
Abbreviations:
- LBP
Lower Back Pain
- NLP
Natural Language Processing
- ML
Machine Learning
- UMLS
Unified Medical Language System
- LIRE
Lumbar Imaging with Reporting of Epidemiology
- MRI
Magnetic Resonance Imaging
- CT
Computed Tomography
- DBOW
Distributed Bag of Words
- MIMIC-III
Medical Information Mart for Intensive Care
- AUC
Area Under the Curve
- ROC
Receiver Operator Characteristics
Footnotes
CODE AVAILABILITY
https://github.com/chethanjjj/LireNLPSystem
SUPPLEMENTARY MATERIALS
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.acra.2021.09.005.
Contributor Information
Chethan Jujjavarapu, Department of Biomedical Informatics and Medical Education, School of Medicine, University of Washington, Seattle, Washington.
Vikas Pejaver, Department of Biomedical Informatics and Medical Education, School of Medicine, University of Washington, Seattle, Washington.
Trevor A. Cohen, Department of Biomedical Informatics and Medical Education, School of Medicine, University of Washington, Seattle, Washington.
Sean D. Mooney, Department of Biomedical Informatics and Medical Education, School of Medicine, University of Washington, Seattle, Washington.
Patrick J. Heagerty, Department of Biostatistics, University of Washington, Seattle, Washington; Center for Biomedical Statistics, University of Washington, Seattle, Washington.
Jeffrey G. Jarvik, Department of Radiology, University of Washington, 1959 NE Pacific Street, Seattle WA 98195; Department of Neurological Surgery, University of Washington, Seattle, Washington; Department of Health Services, University of Washington, Seattle Washington; Clinical Learning, Evidence And Research Center, University of Washington, Seattle, Washington.
REFERENCES
- 1.Koes BW, Tulder MW van, Thomas S Diagnosis and treatment of low back pain. BMJ 2006; 332(7555):1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deyo RA, Rainville J, Kent DL. What can the history and physical examination tell us about low back pain? JAMA 1992; 268(6):760–765. [PubMed] [Google Scholar]
- 3.Tan WK, Hassanpour S, Heagerty PJ, et al. Comparison of natural language processing rules-based and machine-learning systems to identify lumbar spine imaging findings related to low back pain. Acad Radiol 2018; 25(11):1422–1432. Available from: https://pubmed.ncbi.nlm.nih.gov/29605561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarvik JJ, Hollingworth W, Heagerty P, et al. The Longitudinal Assessment of Imaging and Disability of the Back (LAIDBack) study: baseline data. Spine 2001; 26(10):1158–1166. [DOI] [PubMed] [Google Scholar]
- 5.Jarvik JG, Hollingworth W, Heagerty PJ, et al. Three-year incidence of low back pain in an initially asymptomatic cohort: clinical and imaging risk factors. Spine 2005; 30(13):1541–1548. [DOI] [PubMed] [Google Scholar]
- 6.Kim S-Y, Lee I-S, Kim B-R, et al. Magnetic resonance findings of acute severe lower back pain. Ann Rehab Med 2012; 36(1):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarvik JG, Comstock BA, James KT, et al. Lumbar Imaging With Reporting Of Epidemiology (LIRE)—protocol for a pragmatic cluster randomized trial. Contemp Clin Trials 2015; 45(Pt B):157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pons E, Braun LMM, Hunink MGM, et al. Natural language processing in radiology: a systematic review. Radiology 2016; 279(2):329–343. [DOI] [PubMed] [Google Scholar]
- 9.Aronson A Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp 2001: 17–21. [PMC free article] [PubMed] [Google Scholar]
- 10.Brown PF, deSouza PV, Mercer RL, et al. Class-based n-gram models of natural language. Computat Linguist 1992; 18:467–479. [Google Scholar]
- 11.Le QV, Mikolov T. Distributed representations of sentences and documents. Arxiv [Internet] 2014. abs/1405.4053. Available from: http://arxiv.org/abs/1405.4053.
- 12.Bodenreider O The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res 2004; 32(suppl_1):D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Metathesaurus. In: UMLS referen®ce manual [Internet]. n.d. Available from: https://www.ncbi.nlm.nih.gov/books/NBK9684/
- 14.Zech J, Pain M, Titano J, et al. Natural language based machine learning models for the annotation of clinical radiology reports. Radiology. 2018; 287(2):171093. [DOI] [PubMed] [Google Scholar]
- 15.Brown AD, Kachura JR. Natural language processing of radiology reports in patients with hepatocellular carcinoma to predict radiology resource utilization. J Am Coll Radiol 2019; 16(6):840–844. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Liu S, Afzal N, et al. A comparison of word embeddings for the biomedical natural language processing. J Biomed Inform 2018; 87:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee I, Chen MC, Lungren MP, et al. Radiology report annotation using intelligent word embeddings: applied to multi-institutional chest CT cohort. J Biomed Inform 2018; 77:11–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai X, Xie D, Madsen KH, et al. Generalizability of machine learning for classification of schizophrenia based on resting-state functional MRI data. Hum Brain Mapp 2019; 41(1):172–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dreyer KJ, Kalra MK, Maher MM, et al. Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: validation study. Radiology. 2005; 234(2):323–329. [DOI] [PubMed] [Google Scholar]
- 20.Atlas SJ, Deyo RA. Evaluating and managing acute low back pain in the primary care setting. J Gen Intern Med 2001; 16(2):120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birkmeyer NJO, Weinstein JN, Tosteson ANA, et al. Design of the Spine Patient Outcomes Research Trial (SPORT). Spine. 2002; 27(12):1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson K Programming techniques: regular expression search algorithm. Commun Acm 1968; 11(6):419–422. [Google Scholar]
- 23.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001; 34(5):301–310. [DOI] [PubMed] [Google Scholar]
- 24.P MF. An algorithm for suffix stripping. Program [Internet] 1980; 14 (3):130–137. doi: 10.1108/eb046814. Available from: https://doi.org/. [DOI] [Google Scholar]
- 25.Harris ZS. Distributional structure. Word. 2015; 10(2 3):146–162. [Google Scholar]
- 26.Severance C The apache software foundation: Brian Behlendorf. Computer 2012; 45(10):8–9. [Google Scholar]
- 27.Bejan CA, Vanderwende L, Xia F, et al. Assertion modeling and its role in clinical phenotype identification. J Biomed Inform 2013; 46(1):68–74. [DOI] [PubMed] [Google Scholar]
- 28.Dina D-F, Willie J R, Alan R A. MetaMap Lite: an evaluation of a new Java implementation of MetaMap. J Am Med Inform Assn [Internet] 2017; 24 (4):841–844. doi:10.1093/jamia/ocw177. Available from: https://doi.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yetisgen-Yildiz M, Pratt W. The effect of feature representation on MEDLINE document classification. AMIA Annu Symp Proc 2005: 849–853. [PMC free article] [PubMed] [Google Scholar]
- 30.REHCJREK R, SOJKA P Software framework for topic modelling with large corpora. In: In: Proceedings of LREC 2010 workshop New Challenges for NLP Frameworks; 2010. p. 45–50. [Google Scholar]
- 31.Alistair EWJ, Tom JP, Lu S, et al. MIMIC-III, a freely accessible critical care database. Sci Data [Internet] 2016; 3(1):160035.doi:10.1038/sdata.2016.35. Available from: https://doi.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bamber D The area above the ordinal dominance graph and the area below the receiver operating characteristic graph. J Math Psychol 1975; 12(4):387–415. [Google Scholar]
- 33.Perkins NJ, Schisterman EF. The inconsistency of “optimal” cutpoints obtained using two criteria based on the receiver operating characteristic curve. Am J Epidemiol [Internet] 2006; 163(7):670–675. Available from: https://europepmc.org/articles/PMC1444894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen P-H, Zafar H, Galperin-Aizenberg M, et al. Integrating natural language processing and machine learning algorithms to categorize oncologic response in radiology reports. J Digit Imaging 2018; 31(2):178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miotto R, Percha BL, Glicksberg BS, et al. Identifying acute low back pain episodes in primary care practice from clinical notes: observational study. JMIR Med Inform 2020; 8(2):e16878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Li H-J, Wang J, et al. Adapting word embeddings from multiple domains to symptom recognition from psychiatric notes. Amia Jt Summits Transl Sci Proc Amia Jt Summits Transl Sci 2018; 2017:281–289. [PMC free article] [PubMed] [Google Scholar]
- 37.Serguei VSP, Greg F, Reed M, et al. Corpus domain effects on distributional semantic modeling of medical terms. Bioinformatics [Internet] 2016; 32(23):3635–3644. doi:10.1093/bioinformatics/btw529. Available from: https://doi.org/. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.