Abstract
Background:
This study explores whether genomic profiles of colorectal liver metastasis (CRLM) patients with early-onset (EO, <50-years-old) and screening age (SA) primary diagnosis are associated with overall survival (OS).
Methods:
All patients undergoing hepatectomy between 2002–2017 were identified and tumor specimens with next-generation sequencing data were catalogued. Gene and signaling-level alterations were checked for association with OS from primary diagnosis accommodating for left-truncated survival.
Results:
Of 1822 patients, 333 were sequenced–127 (38%) EO-CRLM and 206 (62%) SA-CRLM patients. More aggressive features presented in EO-CRLM patients–synchronous metastatic presentation (83% vs 75%, p<0.001) and primary node-positive disease (71% vs 61%, p<0.001). The median OS from primary diagnosis was 11.8 years (95%CI=7.94-NA). Five-year OS did not differ by age (p=0.702). On multivariable analysis, altered APC[EO-CRLM:(HR=0.37, p=0.018) vs SA-CRLM:(HR=0.61, p=0.260)], BRAF[EO-CRLM:(HR=4.38, p=0.007) vs SA-CRLM:(HR=4.78, p=0.032)], and RAS-TP53[EO-CRLM:(HR=2.82, p=0.011) vs SA-CRLM:(HR=2.35, p=0.003)] associated with OS.
Conclusions:
Despite bearing more aggressive features, EO-CRLM patients had similar genomic profiles and survival as SA-CRLM patients. Better performance status in younger patients leading to increased treatment tolerance may partly explain this. As screening and treatment strategies from older patients are applied to younger patients, genomic predictors of biology identified historically in older cohorts could apply to early-onset patients.
Keywords: Colorectal neoplasms, hepatectomy, cancer genes, precision medicine, cancer biomarker
INTRODUCTION
The incidence and mortality from colorectal cancer are rising among patients diagnosed before the screening age of 50-years-old.1 At presentation, younger patients more commonly have colorectal liver metastases (CRLM) due to delayed detection from the lack of screening.2 In response, the American Cancer Society recently updated their guidelines to recommend beginning colonoscopy screening at 45-years-old for average risk adults.3 In support of this change are multiple reports indicating that younger patients with colorectal cancer have more aggressive features on presentation relative to older patients such as advanced primary tumor T-stage, more frequent node-positive specimens, and higher grade disease.2,4,5
Despite bearing more aggressive features at diagnosis, younger patients do not have significantly different overall survival compared to their older counterparts.2,6 Comparable outcomes in younger patients despite delayed diagnosis could reflect more indolent biology or increased feasibility for aggressive interventions in this cohort. Expanding beyond traditional clinical profiles of colorectal cancer tumor biology, growing interest in novel genomic profiles are beginning to guide selection of therapies. For example, patients with tumors bearing KRAS alteration resist epidermal growth factor receptor inhibitors and CRLM patients with BRAF-altered tumors have poor prognosis with some exceptions.7–9 A recent report on genomic differences between colorectal cancer patients below 40-years-old and above 50-years-old found few genomic differences but did not explore overall survival based on detected alterations.10
Little is known regarding the genomic profiles of CRLM tumors of younger patients and their clinical correlates. The aim of this study is to determine whether unique genomic profiles predict survival for early-onset (EO) compared to screening-age (SA) CRLM patients.
METHODS
Patient Selection
All patients with CRLM undergoing resection by the Hepatopancreatobiliary service with a primary colorectal tumor diagnosis between January 2002 and October 2017 were identified from a prospectively maintained hepatectomy database. Any patients that died or were lost to follow up within 30 days of their operation were excluded. Race and ethnicity data were not recorded into this database during the study period. Recorded data regarding clinicopathologic characteristics, surgical history, and follow up were retrieved. The clinical risk score (CRS) was defined as a composite of points added for a node-positive primary colorectal tumor, preoperative carcinoembryonic antigen (CEA) > 200 ng/mL, largest CRLM > 5 cm, multifocal disease, and disease-free interval from primary colon tumor diagnosis to liver metastasis detection < 12 months.11 High CRS was defined as a score of 3 or higher. Primary tumors originating in the right colon were detected from the proximal cecum to the distal transverse colon and left colon tumors occurred anywhere from the splenic flexure to the distal sigmoid colon. Tumors arising in the rectosigmoid junction or distally were considered rectal. Preoperative chemotherapy was considered treatment with any local or systemic agent within 6 months before planned hepatectomy. Adjuvant chemotherapy was considered any local or systemic agent started within 6 months postoperatively without evidence for recurrent disease. This study was approved by the Institutional Review Board. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Genomic Analysis
A subset of sequenced patients was identified from the larger clinical cohort excluding those with microsatellite instability, POLE mutations, or low tumor purity. When sequencing data from both the primary colorectal and CRLM tumor were available, the former was used given its proximity to the date of primary diagnosis. Genomic profiles of CRLM tumors were considered eligible for inclusion based on prior work noting high genomic concordance between matched primary and CRLM lesions, but not extrahepatic tumors.12,13 Patients with only extrahepatic specimens sequenced were thus excluded. Paired DNA from tumor specimens and matching normal tissue were reviewed by pathologists experienced in colorectal tumor and CRLM diagnosis, grading, and staging. These specimens underwent targeted next-generation sequencing using the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay - a platform designed to identify point mutations, copy number alterations, and select gene fusions in 341 to 468 cancer-associated genes.14 Mutational burden was adjusted by megabase (mB) per IMPACT panel size based on precedent from prior work.15 Sequenced genomic data was stored for analysis on a secure server for large-scale cancer genomics data (cBioPortal for Cancer Genomics).16 Actionable genetic alterations were defined by the OncoKb database as somatic alterations conferring some heightened response or resistance to therapy relative to the wild-type configuration.17 In addition to signaling-level pathways, select genomic targets previously reported to be associated with oncologic outcomes such as amplification of 20q genes (BCL2L1, DNMT3B, SRC) and co-altered RAS-TP53 were investigated.18–22
Statistical Analysis
All patients with a primary colorectal tumor diagnosis before 50-years-old were considered EO; those diagnosed at 50-years-old or later were considered SA. Clinicopathologic characteristics of all patients were summarized using descriptive statistics and compared using Chi-square tests for categorical and Wilcoxon Rank-sum tests for the continuous variables between EO-CRLM and SA-CRLM patients. Genes altered in at least 3% of the IMPACT sequencing data and known to be associated with oncologic outcomes were included for analysis.
Given recent changes to colorectal cancer screening guidelines3 and the homology of mutations identified from analysis of primary and liver metastasis specimens12,13, overall survival (OS) was estimated from the date of primary diagnosis (as opposed to CRLM diagnosis) until the date of death or last follow up using Kaplan-Meier (KM) methods. Those alive at last follow up were censored at the date of last follow-up. Given that the risk of death began following hepatectomy and no deaths were observed during the interval between primary tumor and CRLM diagnoses, the numbers at risk from the time of primary tumor diagnosis under the survival curves were not provided. In addition, survival methodologies accommodating for the left-truncation data and different entry times into the risk set were applied.23,24 Cox proportional hazards models accounting for the left-truncated entry time were employed to study the association of clinical and genomic characteristics on OS. A Cox model was constructed for each gene with alterations associated with OS by including gene indicator, age-group, and an interaction term for both genomic alteration and age-group. Each of the models were further adjusted for CRS since this has been previously established as a potential confounder in this disease group.11 Multiplicity testing correction was used to adjust the p-values. Two-sided p-values less than 0.05 were considered statistically significant. All analyses were performed using R version 3.4.1 (R Foundation for Statistical Computing, Vienna, Austria) or SAS 9.3 (The SAS Institute, Cary, NC).
RESULTS
Clinical Characteristics of All Resected and Sequenced CRLM Patients
Of 1822 CRLM patients, 1252 (68.7%) had a primary diagnosis at or after the SA and 570 (31.3%) were EO. In the EO-CRLM subgroup, 32 (5.6%) patients were 30-years-old or younger at diagnosis, 146 (25.6%) between 40 to 45-years-old, and 244 (42.8%) were 45 to 49-years-old. The EO-CRLM subgroup had notably more aggressive disease with more node-positive primary specimens (71% vs 61%, p<0.001), synchronous metastatic presentation (83% vs 75%, p<0.001), high CRS disease (51% vs 38%, p<0.001), preoperative systemic (75% vs 65%, p<0.001) and hepatic artery infusion (HAI) chemotherapy exposure (5% vs 2%, p<0.001), as well as shorter median time from primary tumor diagnosis to hepatectomy (35 vs 43 weeks, p<0.001), as noted in Table 1. Similar, but fewer significant differences were noted between the EO-CRLM and SA-CRLM subgroups in the subset of 333 sequenced CRLM patients as shown in Table 2.
Table 1:
Clinicopathologic characteristics of all resected colorectal liver metastasis (CRLM) patients, stratified by age at primary colorectal tumor diagnosis.
| All patients | Early-Onset | Screening Age | p-value | ||
|---|---|---|---|---|---|
|
| |||||
| Number of patients | 1822 | 570 (31.3) | 1252 (68.7) | ||
|
| |||||
| Age at Diagnosis, years | Median (range) | 57 (18, 90) | 44 (18, 49) | 62 (50, 90) | |
|
| |||||
| Gender | Male | 1008 (55.3) | 306 (53.7) | 702 (56.1) | 0.400 |
| Female | 814 (44.7) | 264 (46.3) | 550 (43.9) | ||
|
| |||||
| Primary Tumor Location | Right Colon | 517 (28.4) | 118 (20.7) | 399 (31.9) | <0.001 |
| Left Colon | 700 (38.4) | 228 (40.0) | 472 (37.7) | ||
| Rectum | 597 (32.8) | 223 (39.1) | 374 (29.9) | ||
| Multifocal | 6 (0.3) | 1 (0.2) | 5 (0.4) | ||
| Colon, NOS | 2 (0.1) | 0 (0.0) | 2 (0.2) | ||
|
| |||||
| Primary Pathologic Nodal Status | N0 | 636 (34.9) | 161 (28.2) | 475 (37.9) | <0.001 |
| N+ | 1170 (64.2) | 405 (71.1) | 765 (61.1) | ||
| Unknown | 16 (0.9) | 4 (0.7) | 12 (1.0) | ||
|
| |||||
| Disease Free Interval | < 12 months | 1413 (77.6) | 474 (83.2) | 939 (75.0) | <0.001 |
| ≥ 12 months | 331 (18.2) | 72 (12.6) | 259 (20.7) | ||
| Unknown | 78 (4.3) | 24 (4.2) | 54 (4.3) | ||
|
| |||||
| Preoperative CEA | < 200 ng/mL | 1535 (84.2) | 476 (83.5) | 1059 (84.6) | 0.058 |
| ≥ 200 ng/mL | 101 (5.5) | 41 (7.2) | 60 (4.8) | ||
| Unknown | 186 (10.2) | 53 (9.3) | 133 (10.6) | ||
|
| |||||
| Largest CRLM Size | < 5 cm | 1398 (76.6) | 443 (77.5) | 955 (76.1) | 0.300 |
| ≥ 5 cm | 357 (19.8) | 103 (18.2) | 254 (20.4) | ||
| Unknown | 67 (3.7) | 24 (4.2) | 43 (3.4) | ||
|
| |||||
| Number of CRLM | Solitary | 606 (33.2) | 156 (27.2) | 450 (35.9) | <0.001 |
| Multifocal | 1144 (62.8) | 390 (68.6) | 754 (60.2) | ||
| Unknown | 72 (4.0) | 24 (4.2) | 48 (3.8) | ||
|
| |||||
| Clinical Risk Score (CRS) | Low Risk (0–2) | 811 (44.5) | 205 (36.0) | 606 (48.4) | <0.001 |
| High Risk (3–5) | 763 (41.9) | 290 (50.9) | 473 (37.8) | ||
| Unknown | 248 (13.6) | 75 (13.2) | 173 (13.8) | ||
|
| |||||
| Preoperative Chemotherapy | Yes | 1243 (68.2) | 425 (74.6) | 818 (65.3) | <0.001 |
| No | 579 (31.8) | 145 (25.4) | 434 (34.7) | ||
|
| |||||
| Hepatic Artery Infusion Chemotherapy | None | 995 (54.6) | 246 (43.2) | 749 (59.8) | <0.001 |
| Preoperative | 61 (3.3) | 31 (5.4) | 30 (2.4) | ||
| Adjuvant | 678 (37.2) | 258 (45.3) | 420 (33.5) | ||
| Salvage | 88 (4.8) | 35 (6.1) | 53 (4.2) | ||
|
| |||||
| Time from Primary Diagnosis to Resection, weeks | Median (range) | 40 (23, 82) | 35 (20, 69) | 43 (24, 90) | <0.001 |
|
| |||||
| Extent of Hepatectomy | Minor | 1139 (62.5) | 356 (62.5) | 783 (62.5) | >0.900 |
| Major | 683 (37.5) | 214 (37.5) | 469 (37.5) | ||
|
| |||||
| Ablation at Hepatectomy | Yes | 337 (18.5) | 107 (18.8) | 230 (18.4) | 0.900 |
| No | 1485 (81.5) | 463 (81.2) | 1022 (81.6) | ||
Table 2:
Clinicopathologic characteristics of sequenced resected colorectal liver metastasis (CRLM) patients, stratified by age at primary colorectal tumor diagnosis.
| All patients | Early-Onset | Screening Age | p-value | ||
|---|---|---|---|---|---|
|
| |||||
| Number of patients | 333 | 127 (38.1) | 206 (61.9) | ||
|
| |||||
| Age at Diagnosis, years | Median (range) | 54 (23, 84) | 45 (23, 49) | 60 (50, 84) | |
|
| |||||
| Gender | Male | 176 (52.9) | 65 (51.2) | 111 (53.9) | 0.700 |
| Female | 157 (47.1) | 62 (48.8) | 95 (46.1) | ||
|
| |||||
| Primary Tumor Location | Right Colon | 80 (24.0) | 28 (22.0) | 52 (25.2) | 0.700 |
| Left Colon | 142 (42.6) | 56 (44.1) | 86 (41.7) | ||
| Rectum | 104 (31.2) | 42 (33.1) | 62 (30.1) | ||
| Multifocal | 6 (1.8) | 1 (0.8) | 5 (2.4) | ||
| Colon, NOS | 1 (0.3) | 0 (0) | 1 (0.5) | ||
|
| |||||
| Primary Pathologic Nodal Status | N0 | 100 (30.0) | 27 (21.3) | 73 (35.4) | 0.008 |
| N+ | 232 (69.7) | 100 (78.7) | 132 (64.1) | ||
| Unknown | 1 (0.3) | 0 (0) | 1 (0.5) | ||
|
| |||||
| Disease Free Interval | < 12 months | 266 (79.9) | 106 (83.5) | 160 (77.7) | 0.300 |
| ≥ 12 months | 67 (20.1) | 21 (16.5) | 46 (22.3) | ||
|
| |||||
| Preoperative CEA | < 200 ng/mL | 290 (87.1) | 109 (85.8) | 183 (88.8) | 0.200 |
| ≥ 200 ng/mL | 28 (8.4) | 14 (11.0) | 14 (6.8) | ||
| Unknown | 15 (4.5) | 6 (4.2) | 9 (4.4) | ||
|
| |||||
| Largest CRLM Size | < 5 cm | 269 (80.8) | 104 (81.9) | 165 (80.1) | 0.800 |
| ≥ 5 cm | 64 (19.2) | 23 (18.1) | 41 (19.9) | ||
|
| |||||
| Number of CRLM | Solitary | 85 (25.5) | 30 (23.6) | 55 (26.7) | >0.900 |
| Multifocal | 248 (74.5) | 97 (76.4) | 151 (73.3) | ||
|
| |||||
| Clinical Risk Score (CRS) | Low Risk (0–2) | 146 (43.8) | 43 (33.9) | 103 (50.0) | 0.005 |
| High Risk (3–5) | 171 (51.4) | 78 (61.4) | 93 (45.1) | ||
| Unknown | 16 (4.8) | 6 (4.7) | 10 (4.9) | ||
|
| |||||
| Preoperative Chemotherapy | Yes | 216 (64.9) | 85 (66.9) | 131 (63.6) | 0.600 |
| No | 117 (35.1) | 42 (33.1) | 75 (36.4) | ||
|
| |||||
| Hepatic Artery Infusion Chemotherapy | None | 76 (22.8) | 16 (12.6) | 60 (29.1) | <0.001 |
| Preoperative | 0 (0) | 0 (0) | 0 (0) | ||
| Adjuvant | 220 (66.1) | 100 (78.7) | 120 (58.3) | ||
| Salvage | 37 (11.1) | 11 (8.7) | 26 (12.6) | ||
|
| |||||
| Time from Primary Diagnosis to Resection, weeks | Median (range) | 32 (17, 69) | 26 (16, 57) | 35 (18, 75) | 0.024 |
|
| |||||
| Extent of Hepatectomy | Minor | 237 (71.2) | 88 (69.3) | 149 (72.3) | 0.600 |
| Major | 96 (28.8) | 39 (30.7) | 57 (27.7) | ||
|
| |||||
| Ablation at Hepatectomy | Yes | 75 (22.5) | 28 (22.0) | 47 (22.8) | >0.900 |
| No | 258 (77.5) | 99 (78.0) | 159 (77.2) | ||
Survival of All Resected and Sequenced CRLM Patients
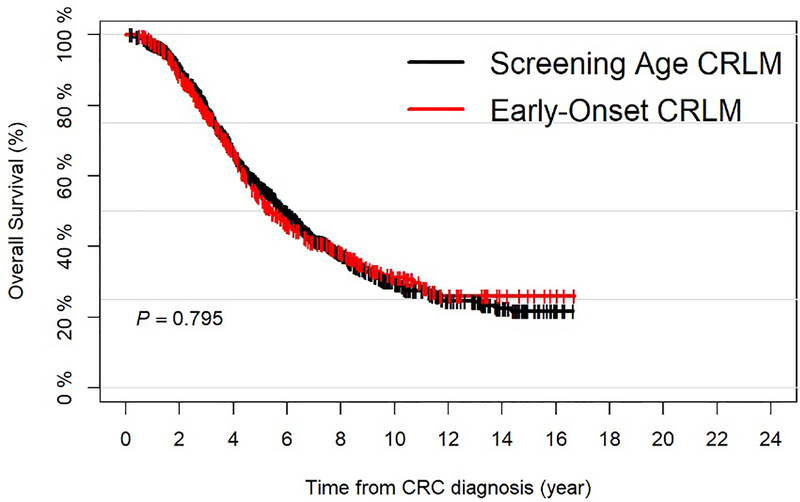
The median follow up for all surviving patients (n=928) was 5.1 years (range=0.2–16.7). The median OS was 5.8 years (95%CI=5.5–6.2) and the 5-year OS was 55.9% (95%CI=53.3–58.7). No significant difference in OS was noted between the EO-CRLM and SA-CRLM subgroups (Figure 1A).
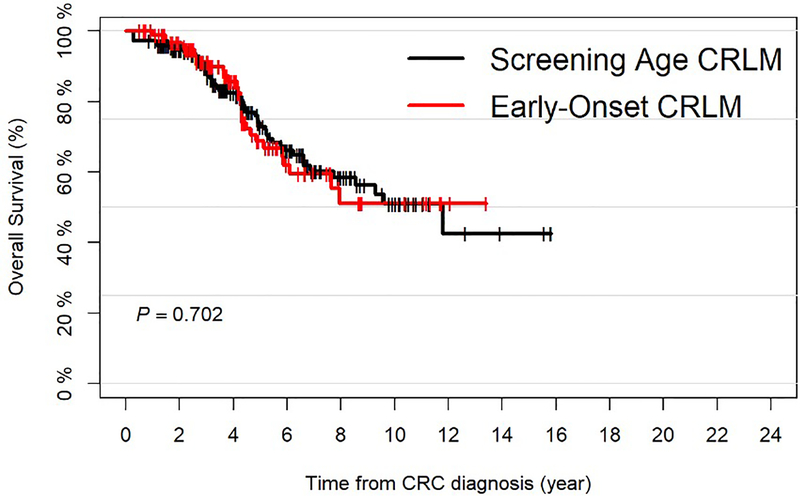
Figure 1.
A: Overall Survival for all Resected Colorectal Liver Metastasis (CRLM) Patients, Stratified by Age at Primary Colorectal Tumor Diagnosis; B: Overall Survival for all Sequenced and Resected Colorectal Liver Metastasis (CRLM) Patients, Stratified by Age at Primary Colorectal Tumor Diagnosis.
The median follow up after primary diagnosis among sequenced survivors was 3.8 years (range=0.5–15.8) and the median OS was 11.8 years (95%CI=7.94-NA). The 5-year OS was 71.6% (95%CI=65.2–78.6%). Again, no significant difference in OS was found when stratifying patients by age at primary diagnosis (Figure 1B).
Genomic Profiles and Associations with Survival in Resected CRLM Patients
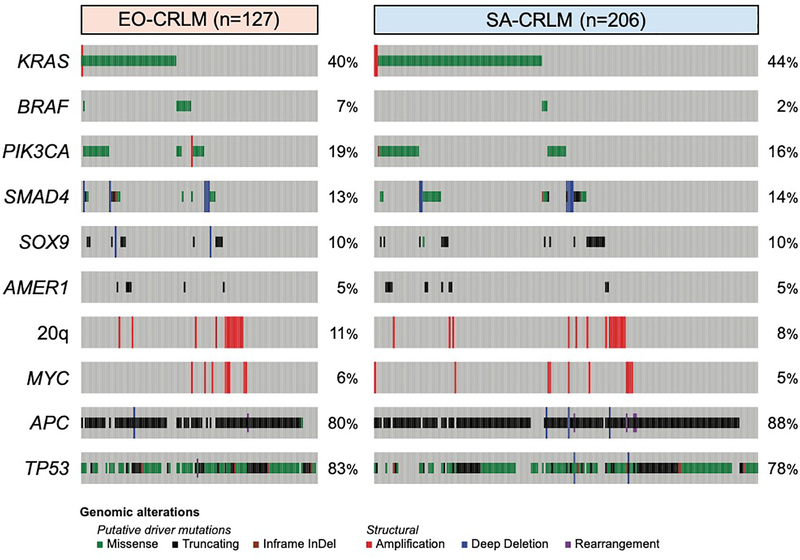
The most commonly altered genes were APC (n=283/333, 85%), TP53 (n=267/333, 80%), KRAS (n=141/333, 42%), and PIK3CA (n=56/333, 17%, Figure 2). The most commonly altered signaling-level pathways were Wnt (n=291/333, 87%), p53 (n=273/333, 82%), RTK/RAS (n=211/333, 63%), RAS (n=154/333, 46%), and PI3K (n=86/333, 26%). No single gene or pathway alteration was enriched in either age group. Alterations in APC (p=0.029), KRAS (p=0.033), BRAF (p<0.001), SMAD4 (p=0.026), BCL2L1 (p=0.039), DNMT3B (p=0.049), and RAS-TP53 co-alteration (p<0.001) were significantly associated with OS in the sequenced subset (Supplemental Figure 1).
Figure 2:
Heatmap of Sequenced and Resected Colorectal Liver Metastasis Patients Stratified by Age at Primary Colorectal Tumor Diagnosis. 20q alterations include mutations in BCL2L1, DNMT3B, and SRC.
Risk Factors Associated with Survival After Resection in CRLM Patients
On univariate analysis, the association between clinical and genetic factors with survival was significant for high CRS (HR=1.62, 95%CI=1.01–2.62, p=0.047), as well as alterations in APC (HR=0.51, 95%CI=0.29–0.88, p=0.015), BRAF (HR=4.21 95%CI=1.80–9.88, p=0.001), KRAS (HR=1.62, 95%CI=1.03–2.54, p=0.037), SMAD4 (HR=1.87, 95%CI=1.07–3.25, p=0.027), and RAS-TP53 co-alteration (HR=2.24, 95%CI=1.42–3.53, p<0.001). A significant association between age at primary diagnosis and OS was not detected.
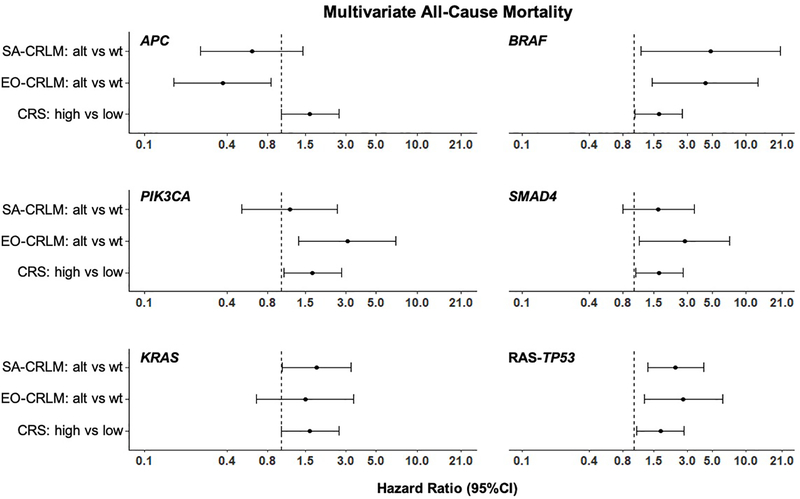
Interaction models for mortality were constructed between altered genes and age at primary diagnosis adjusting for CRS (Figure 3). Similar magnitudes and directions for mortality were detected for EO-CRLM and SA-CRLM patients with altered BRAF (HR=4.32, 95%CI=1.46–12.77, p=0.008 and HR=4.80, 95%CI=1.14–20.17, p=0.032, respectively) and co-altered RAS-TP53 (HR=2.76, 95%CI=1.24–6.16, p=0.013 and HR=2.36, 95%CI=1.32–4.21, p=0.004, respectively). Similar directions but different magnitudes for mortality were detected when EO-CRLM patients were altered compared to SA-CRLM patients at APC (HR=0.37, 95%CI=0.16–0.85, p=0.018 vs HR=0.61, 95%CI=0.26–1.45, p=0.264), PIK3CA (HR=3.06, 95%CI=1.34–6.98, p=0.008 vs HR=1.15, 95%CI=0.51–2.58, p=0.731), and SMAD4 (HR=2.82, 95%CI=1.12–7.10, p=0.028 vs HR=1.64, 95%CI=0.79–3.42, p=0.184). No significant interaction was detected between age subgroup and the alterations. High CRS was independently associated with increased mortality in the presence of altered KRAS (HR=1.63, 95%CI=1.00–2.64, p=0.048), BRAF (HR=1.66, 95%CI=1.03–2.70, p=0.039), PIK3CA (HR=1.70, 95%CI=1.05–2.76, p=0.031), SMAD4 (HR=1.68, 95%CI=1.04–2.74, p=0.035), and co-altered RAS-TP53 (HR=1.72, 95%CI=1.06–2.80, p=0.029).
Figure 3:
Forest Plots of Multivariable Survival Models. Each panel represents a multivariable regression model with genetic alteration, age (SA-CRLM vs EO-CRLM), interaction between genetic alteration/age, and CRS. SA=Screening Age, EO=Early-Onset, CRLM=Colorectal liver metastasis, CRS=Clinical risk score, alt=alteration, wt=wild-type.
DISCUSSION
With the incidence and mortality from colorectal cancer rising among younger patients,1 new discriminatory biomarkers for aggressive disease are needed to further investigate this phenomenon and guide therapeutic interventions. Despite the growing understanding of genomic correlates for tumor biology, it remains unclear if findings from sequencing studies conducted in historically older cohorts can be applied to younger patients. This report explored differences in the clinical and genomic profiles of resectable CRLM patients with primary diagnosis at EO or SA as well as their associations with OS.
Although significant differences in clinical features were noted between EO-CRLM and SA-CRLM patients, no significant difference in survival was detected. Additionally, tumor alterations did not vary between these age groups possibly suggesting that presentation of EO-CRLM patients with more aggressive disease could be due to delayed diagnosis rather than a predilection for more hostile genomic profiles. Interestingly, the magnitude of risk for mortality with altered APC, PIK3CA, and SMAD4 differed between the EO-CRLM and SA-CRLM subgroups.
The presentation of EO-CRLM patients with more aggressive clinical features recapitulates earlier work noting more aggressive clinical characteristics in CRLM patients diagnosed at a younger age.2 Therapeutic factors may also have contributed to more aggressive malignant degeneration. Preoperative chemotherapy exposure was more common in the EO-CRLM subgroup and has been associated with more aggressive genomic profiles.25 Additionally, HAI chemotherapy given at all sequences (preoperative, adjuvant, salvage) was more common in the EO-CRLM subgroup likely reflecting the pursuit of more aggressive interventions in younger patients with higher volume disease.
Despite having different clinical features, a significant difference in OS was not observed between EO-CRLM and SA-CRLM patients. Most earlier reports found no significant difference in OS between younger and older colorectal cancer patients.2,6,26 The lack of a survival difference despite more aggressive disease presenting in EO-CRLM patients could be related to better performance status in this cohort or better tolerance for more aggressive systemic regimens and surgical interventions.4,27 Studies identifying survival differences by age dichotomized their cohorts to compare SA-CRLM patients with younger groups than reported herein. Lieu et al noted a parabolic relationship with risk for mortality where the youngest (under 20-years-old) and oldest (over 80-years-old) metastatic colorectal cancer patients had the worst outcomes.26 Of note, age lost its association with OS in that study when the cohort was adjusted for metastatic site. Sultan et al reported outcomes for a 30-year cohort from the Surveillance, Epidemiology, and End Results (SEER) database noting diminished 5-year survival for colorectal cancer patients diagnosed under 20-years-old relative to older patients.28 Khan et al similarly reported that colorectal cancer patients under 30-years-old undergoing resection had lower 5-year disease-specific survival compared to a cohort undergoing resection over 50-years-old.29 While it is possible that the tumor biology of colorectal cancer specimens obtained from very young adults differs from those obtained from older EO-CRLM patients, this report was underpowered to detect this as only 32 patients of the overall clinical cohort were less than 30-years-old.
The genomic profile of the sequenced cohort resembles that of prior study cohorts of resectable CRLM patients reporting, for example, TP53 and KRAS altered in 55–100%, and 34–62%, respectively.30–34 This predictable pattern of genomic alterations highlights the feasibility of using novel non-invasive methods developed in older cohorts, such as circulating tumor cells or circulating DNA, in screening or surveillance strategies for high-risk EO-CRLM patients.35,36
Despite EO-CRLM and SA-CRLM cohorts bearing similar genomic profiles, the impact of altered genes on OS varied. Notably, altered SMAD4 and PIK3CA were associated with significantly greater risk for all-cause mortality on multivariate analyses in the EO-CRLM subgroup only. Earlier work concurs that SMAD4 alteration is associated with worse OS in resectable CRLM patients.37,38 Altered PIK3CA was previously reported to have no association with survival,39,40 however, Yamashita et al noted an association between co-altered APC-PIK3CA and worse OS in resectable CRLM patients after chemotherapy exposure.41 In this study, altered APC alone was associated with increased risk for improved OS in the EO-CRLM group after accounting for CRS. A report from Jorissen et al revealed similar results, with wild-type APC associated with worse OS in patients with microsatellite stable primary tumors.42 An association between APC alteration and OS in CRLM patients, however, has not been previously reported.
Altered BRAF and co-altered RAS-TP53 were associated with worse OS with a higher mortality hazard magnitude in both subgroups, recapitulating earlier work in CRLM patients.8,33,43 Co-altered RAS-TP53 has been noted to be enriched in unresectable CRLM cohorts,44 however, data describing outcomes for patients with co-alteration in otherwise low-risk, resectable disease is lacking. Published preclinical work supports a mechanism for interplay between loss-of-function p53 alteration and RAS activation in colorectal cancer.18 Additionally, altered TP53 correlates negatively with cytolytic immune cell activity contributing to worse OS.19 Furthermore, altered KRAS was associated with significantly greater risk for worse OS in the SA-CRLM group but not the EO-CRLM group. Altered KRAS has been associated with worse outcomes in earlier work,45,46 however, select strategies (e.g. HAI chemotherapy) have been shown to result in better survival regardless of KRAS status.47 Although multiple gene alterations were associated with OS in one age group and not the other, lack of a significant interaction between these genes and age as a covariate suggests that the impact on OS may not depend on age at primary diagnosis. Instead, other characteristics of patients in these age groups may be related to how tumor alterations impact survival. Future work investigating the interaction between tumor alterations and age-specific factors is necessary to clarify this.
Limitations
Several limitations to this study merit mention. First, the retrospective nature of this study introduces limitations inherent to this design including the selection bias of analyzing genomic data from patients chosen by their oncologists for next generation sequencing. For instance, although no significant difference in OS was detected between the EO-CRLM and SA-CRLM subgroups in the clinical and sequenced cohorts, several clinicopathologic differences were noted between these subgroups in the clinical cohort and fewer differences in the sequenced subset. The EO-CRLM subgroup had more aggressive features than the SA-CRLM patients in the clinical cohort and the sequenced EO-CRLM subgroup had relatively fewer differences compared to the sequenced SA-CRLM patients, suggesting that SA-CRLM patients with higher risk disease underwent sequencing. Conversely, this change could represent removal of the fraction of EO-CRLM patients with microsatellite instability or POLE mutations that are associated with more aggressive tumor biology. Although multivariable analyses were performed to adjust for clinical and genomic selection differences, the presence of a significant relationship with a genomic profile and OS in one age subgroup and not the other should not be interpreted as an absence of association in the latter. This study was not powered to determine this. Second, use of next generation sequencing of tumor specimens, as utilized in this study, may not offer the optimal representation of genomic aberrations for metastatic disease. New interest in using circulating tumor cells or circulating DNA as a liquid biopsy is quickly becoming a competitive option to detect mutations.48 Although there is strong concordance between alterations found in resected primary colon and CRLM tumors,12,13 some alterations (e.g. TP5349) are more commonly found in metastatic CRLM tumors.
CONCLUSIONS
Although the incidence and mortality of colorectal cancer are rising in younger patients that present with more aggressive clinical characteristics, differences in genomic profiles or survival were not detected. Notably, differences in the impact of tumor alterations on survival were found to vary between age groups, however, the mechanism for this difference needs further clarification. As screening and treatment strategies from older patients are applied to younger patients, genomic predictors of biology identified historically in older cohorts could apply to early-onset patients as well.
Supplementary Material
Supplemental Figure 1: Overall Survival by Genomic Alteration in Resected Colorectal Liver Metastasis Patients. A: APC, B: PIK3CA, C: KRAS, D: BRAF, E: BCL2L1, F: DNMT3B, G: SRC, H: 20q amplification, I: SMAD4, J: RAS-TP53 co-alteration.
SYNOPSIS.
The incidence and mortality from colorectal cancer are rising among patients younger than the screening age of 50-years-old. Novel age-related genomic signatures among resected colorectal liver metastasis patients may have implications toward therapeutic decision making and are explored herein.
FUNDING SOURCES:
This work was supported in part by the NIH/NCI P30 CA008748 Cancer Center Support Grant.
Footnotes
CONFLICTS OF INTEREST: The authors have no conflicts of interest to report.
REFERENCES
- 1.Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020. [DOI] [PubMed] [Google Scholar]
- 2.de Haas RJ, Wicherts DA, Salloum C, et al. Long-term outcomes after hepatic resection for colorectal metastases in young patients. Cancer. 2010;116(3):647–658. [DOI] [PubMed] [Google Scholar]
- 3.Wolf AMD, Fontham ETH, Church TR, et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin. 2018;68(4):250–281. [DOI] [PubMed] [Google Scholar]
- 4.O’Connell JB, Maggard MA, Liu JH, Etzioni DA, Livingston EH, Ko CY. Do young colon cancer patients have worse outcomes? World J Surg. 2004;28(6):558–562. [DOI] [PubMed] [Google Scholar]
- 5.Yeo H, Betel D, Abelson JS, Zheng XE, Yantiss R, Shah MA. Early-onset Colorectal Cancer is Distinct From Traditional Colorectal Cancer. Clin Colorectal Cancer. 2017;16(4):293–299 e296. [DOI] [PubMed] [Google Scholar]
- 6.Liang H, Wang XN, Wang BG, et al. Prognostic factors of young patients with colon cancer after surgery. World J Gastroenterol. 2006;12(9):1458–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Cutsem E, Kohne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360(14):1408–1417. [DOI] [PubMed] [Google Scholar]
- 8.Margonis GA, Buettner S, Andreatos N, et al. Association of BRAF Mutations With Survival and Recurrence in Surgically Treated Patients With Metastatic Colorectal Liver Cancer. JAMA Surg. 2018;153(7):e180996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagniere J, Dupre A, Gholami SS, et al. Is Hepatectomy Justified for BRAF Mutant Colorectal Liver Metastases?: A Multi-institutional Analysis of 1497 Patients. Ann Surg. 2018. [DOI] [PubMed] [Google Scholar]
- 10.Lieu CH, Golemis EA, Serebriiskii IG, et al. Comprehensive Genomic Landscapes in Early and Later Onset Colorectal Cancer. Clin Cancer Res. 2019;25(19):5852–5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230(3):309–318; discussion 318–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vakiani E, Janakiraman M, Shen R, et al. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol. 2012;30(24):2956–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brannon AR, Vakiani E, Sylvester BE, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15(8):454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17(3):251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stadler ZK, Battaglin F, Middha S, et al. Reliable Detection of Mismatch Repair Deficiency in Colorectal Cancers Using Mutational Load in Next-Generation Sequencing Panels. J Clin Oncol. 2016;34(18):2141–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parada LF, Land H, Weinberg RA, Wolf D, Rotter V. Cooperation between gene encoding p53 tumour antigen and ras in cellular transformation. Nature. 1984;312(5995):649–651. [DOI] [PubMed] [Google Scholar]
- 19.McMurray HR, Sampson ER, Compitello G, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453(7198):1112–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruin SC, Klijn C, Liefers GJ, et al. Specific genomic aberrations in primary colorectal cancer are associated with liver metastases. BMC Cancer. 2010;10:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ptashkin RN, Pagan C, Yaeger R, et al. Chromosome 20q Amplification Defines a Subtype of Microsatellite Stable, Left-Sided Colon Cancers with Wild-type RAS/RAF and Better Overall Survival. Mol Cancer Res. 2017;15(6):708–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chun YS, Passot G, Yamashita S, et al. Deleterious Effect of RAS and Evolutionary High-risk TP53 Double Mutation in Colorectal Liver Metastases. Ann Surg. 2019;269(5):917–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein JP, Moeschberger ML. Survival Analysis - Techniques for Censored and Truncated Data. 2nd ed. New York, NY: Springer-Verlag; 2003. [Google Scholar]
- 24.Foreman AJ, Lai GP, Miller DP. Surviving Left Truncation Using PROC PHREG. ICON Clinical Research; 2008; San Francisco, CA. [Google Scholar]
- 25.Andreou A, Kopetz S, Maru DM, et al. Adjuvant chemotherapy with FOLFOX for primary colorectal cancer is associated with increased somatic gene mutations and inferior survival in patients undergoing hepatectomy for metachronous liver metastases. Ann Surg. 2012;256(4):642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieu CH, Renfro LA, de Gramont A, et al. Association of age with survival in patients with metastatic colorectal cancer: analysis from the ARCAD Clinical Trials Program. J Clin Oncol. 2014;32(27):2975–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kneuertz PJ, Chang GJ, Hu CY, et al. Overtreatment of young adults with colon cancer: more intense treatments with unmatched survival gains. JAMA Surg. 2015;150(5):402–409. [DOI] [PubMed] [Google Scholar]
- 28.Sultan I, Rodriguez-Galindo C, El-Taani H, et al. Distinct features of colorectal cancer in children and adolescents: a population-based study of 159 cases. Cancer. 2010;116(3):758–765. [DOI] [PubMed] [Google Scholar]
- 29.Khan SA, Morris M, Idrees K, et al. Colorectal cancer in the very young: a comparative study of tumor markers, pathology and survival in early onset and adult onset patients. J Pediatr Surg. 2016;51(11):1812–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilat N, Grunberger T, Langle F, et al. Assessing the TP53 marker type in patients treated with or without neoadjuvant chemotherapy for resectable colorectal liver metastases: a p53 Research Group study. Eur J Surg Oncol. 2015;41(5):683–689. [DOI] [PubMed] [Google Scholar]
- 31.Loes IM, Immervoll H, Sorbye H, et al. Impact of KRAS, BRAF, PIK3CA, TP53 status and intraindividual mutation heterogeneity on outcome after liver resection for colorectal cancer metastases. Int J Cancer. 2016;139(3):647–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng L, Hong S, Gao J, Li J. Whole-Exome Sequencing Characterized the Landscape of Somatic Mutations and Pathways in Colorectal Cancer Liver Metastasis. J Oncol. 2019;2019:2684075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Datta J, Smith JJ, Chatila WK, et al. Coaltered Ras/B-raf and TP53 Is Associated with Extremes of Survivorship and Distinct Patterns of Metastasis in Patients with Metastatic Colorectal Cancer. Clin Cancer Res. 2020;26(5):1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cavallaro P, Bordeianou L, Stafford C, et al. Impact of Single-organ Metastasis to the Liver or Lung and Genetic Mutation Status on Prognosis in Stage IV Colorectal Cancer. Clin Colorectal Cancer. 2020;19(1):e8–e17. [DOI] [PubMed] [Google Scholar]
- 35.Connor AA, McNamara K, Al-Sukhni E, et al. Central, But Not Peripheral, Circulating Tumor Cells are Prognostic in Patients Undergoing Resection of Colorectal Cancer Liver Metastases. Ann Surg Oncol. 2016;23(7):2168–2175. [DOI] [PubMed] [Google Scholar]
- 36.Narayan RR, Goldman DA, Gonen M, et al. Peripheral Circulating Tumor DNA Detection Predicts Poor Outcomes After Liver Resection for Metastatic Colorectal Cancer. Ann Surg Oncol. 2019;26(6):1824–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mizuno T, Cloyd JM, Vicente D, et al. SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur J Surg Oncol. 2018;44(5):684–692. [DOI] [PubMed] [Google Scholar]
- 38.Kawaguchi Y, Kopetz S, Newhook TE, et al. Mutation Status of RAS, TP53, and SMAD4 is Superior to Mutation Status of RAS Alone for Predicting Prognosis after Resection of Colorectal Liver Metastases. Clin Cancer Res. 2019;25(19):5843–5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Isella C, Mellano A, Galimi F, et al. MACC1 mRNA levels predict cancer recurrence after resection of colorectal cancer liver metastases. Ann Surg. 2013;257(6):1089–1095. [DOI] [PubMed] [Google Scholar]
- 40.Frankel TL, Vakiani E, Nathan H, et al. Mutation location on the RAS oncogene affects pathologic features and survival after resection of colorectal liver metastases. Cancer. 2017;123(4):568–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamashita S, Chun YS, Kopetz SE, et al. APC and PIK3CA Mutational Cooperativity Predicts Pathologic Response and Survival in Patients Undergoing Resection for Colorectal Liver Metastases. Ann Surg. 2017. [DOI] [PubMed] [Google Scholar]
- 42.Jorissen RN, Christie M, Mouradov D, et al. Wild-type APC predicts poor prognosis in microsatellite-stable proximal colon cancer. Br J Cancer. 2015;113(6):979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tran B, Kopetz S, Tie J, et al. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117(20):4623–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith JJ, Chatila WK, Sanchez-Vega F, et al. Genomic stratification beyond Ras/B-Raf in colorectal liver metastasis patients treated with hepatic arterial infusion. Cancer Med. 2019;8(15):6538–6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nash GM, Gimbel M, Shia J, et al. KRAS mutation correlates with accelerated metastatic progression in patients with colorectal liver metastases. Ann Surg Oncol. 2010;17(2):572–578. [DOI] [PubMed] [Google Scholar]
- 46.Goffredo P, Utria AF, Beck AC, et al. The Prognostic Impact of KRAS Mutation in Patients Having Curative Resection of Synchronous Colorectal Liver Metastases. J Gastrointest Surg. 2018. [DOI] [PubMed] [Google Scholar]
- 47.Gholami S, Kemeny NE, Boucher TM, et al. Adjuvant Hepatic Artery Infusion Chemotherapy is Associated With Improved Survival Regardless of KRAS Mutation Status in Patients With Resected Colorectal Liver Metastases: A Retrospective Analysis of 674 Patients. Ann Surg. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narayan RR, Babicky ML, Goldman DA, et al. Change in Circulating Tumor DNA After Hepatic Resection for Metastatic Colorectal Cancer. Paper presented at: Americas Hepato-Pancreato-Biliary Association; 2018; Miami, FL. [Google Scholar]
- 49.Yaeger R, Chatila WK, Lipsyc MD, et al. Clinical Sequencing Defines the Genomic Landscape of Metastatic Colorectal Cancer. Cancer Cell. 2018;33(1):125–136 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Overall Survival by Genomic Alteration in Resected Colorectal Liver Metastasis Patients. A: APC, B: PIK3CA, C: KRAS, D: BRAF, E: BCL2L1, F: DNMT3B, G: SRC, H: 20q amplification, I: SMAD4, J: RAS-TP53 co-alteration.