Abstract
Clarithromycin resistance in Helicobacter pylori is due to point mutation within the 23S rRNA. We examined the growth rates of different types of site-directed mutants and demonstrated quantitatively the competitive growth advantage of A-to-G mutants over other types of mutants by a multiplex sequencing assay. The results provide a rational explanation of why A-to-G mutants are predominantly observed among clarithromycin-resistant clinical isolates.
Clarithromycin is commonly used in triple therapy regimens for the treatment of Helicobacter pylori infections (8, 12). Development of H. pylori strains resistant to clarithromycin accounts for the majority of the treatment failures (1, 4). Previous studies demonstrated that clarithromycin resistance of H. pylori is due to point mutations in the 23S rRNA (3, 5, 9, 10, 11, 13, 14). A-to-G mutations at position 2142 or 2143 were predominantly observed in clarithromycin-resistant H. pylori clinical isolates. Very few cases of A-to-C mutation and no A-to-T mutation were reported. Recently, we have constructed in vitro site-directed H. pylori mutants using the wild-type strain UA802 for which the MIC of clarithromycin is 0.004 μg/ml and demonstrated that five types of point mutation within 23S rRNA gene may confer clarithromycin resistance (15). A-to-G and A-to-C mutations at the same position mediate identical MICs (MIC = 16 μg/ml for mutation of A2142 to G or C; MIC = 4 μg/ml for mutation of A2143 to G or C). Note that the A2142C mutation confers a higher MIC than the A2143G mutation, and the A2142T mutation is also able to provide an intermediate-level MIC (4 μg/ml). In the meantime, a similar study was published by Debets-Ossenkopp et al. (2). The overall patterns of MICs for the different types of mutants are similar between the two studies, although the MICs for all of the strains studied by Debets-Ossenkopp et al. (2) are much higher than those noted in our study (15). This could be due to the difference in the susceptibilities of the wild-type strains used and/or to the methods used for MIC testing. The lower MIC for A-to-T mutants may partly account for their absence in clinical isolates. Apparently, however, the rare occurrence of A-to-C mutants cannot be explained by the MIC levels.
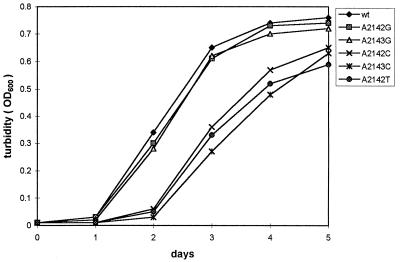
What then is the main factor that gives the A-to-G mutants a selective advantage? Since the mutations associated with clarithromycin resistance are within the 23S rRNA, it is possible that A-to-C or A-to-T mutations impair ribosome function in protein synthesis but that A-to-G mutations do not, no matter whether clarithromycin is bound or not. Thus we hypothesize that A-to-G mutations have a growth advantage over other types of mutation. Indeed, while constructing in vitro site-directed mutants, we noticed that the growth of the A-to-C or A-to-T mutants was significantly slower than that of wild type or A-to-G mutants. This result was similar to that observed by Debets-Ossenkopp et al. (2). In this study, we examined the growth curves of the five types of mutants. H. pylori Clar cells were incubated in 37% brain heart infusion–0.3% yeast extract broth plus 5% fetal bovine serum and 1-μg/ml clarithromycin at 37°C under microaerobic conditions. As a control, wild-type cells were incubated under the same condition without clarithromycin. Cell growth was monitored by determining optical density at 600 nm (OD600) of the culture each day after inoculation (Fig. 1). The growth of the two A-to-G mutants was similar to that of the wild-type strain. In contrast, the A-to-C or A-to-T mutants grew more slowly, having an extra 1-day lag compared to the wild type and the A-to-G mutants.
FIG. 1.
Growth of H. pylori Clar mutants compared with that of the wild-type, strain UA802. A representative experiment (N = 3) is shown.
To demonstrate the competitive growth advantage of the A-to-G mutants over other types of mutants, the same amounts (judged by OD600) of different Clar mutant cells at late-log phase were inoculated together in the above broth medium and allowed to grow in the presence of 1-μg/ml clarithromycin for 4 days. At the end of the growth experiment, we determined the fractions of particular mutant strains in the mixed culture by using a multiplex sequence analysis. This technology was pioneered by Palejwala et al. (6) to determine mutation frequency and specificity at mutational hot spots and has been well validated and extensively used in recent years by us (for example, see references 7 and 16). For the particular purpose used here, however, appropriate modifications were made. First, total chromosomal DNA was isolated from the mixed culture, and a 1,093-bp fragment carrying the mutation site at the center was PCR amplified with the primer pair HP1 (5′TTGGAGGGAAGGCAAATCCA3′) and HP2 (5′ACGTTCTGAACCCAGCTCGC3′), which were designed based on the 23S rRNA gene sequence (11). The PCRs were performed with Vent DNA polymerase (NEB) in the presence of 1.5 mM MgCl2 and cycled at 94°C for 1 min, 60°C for 1 min, and 72°C for 3 min for 28 cycles. The PCR products were gel purified with the QiaxII kit (Qiagen) and were used for subsequent multiplex sequencing.
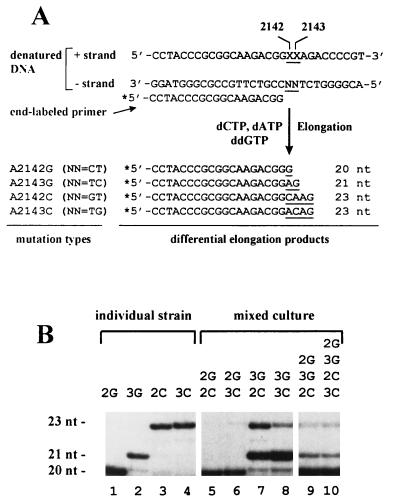
The principle of the multiplex sequence analysis is described in Fig. 2A. A 0.01-pmol sample of DNA fragment (the 1,093-bp PCR product) was heat denatured and annealed with 0.33 pmol of a 5′ 32P-end-labeled 19-mer primer (HP3, 5′CCTACCCGCGGCAAGACGG3′). The annealed primer was elongated by Taq DNA polymerase in Taq buffer (Boehringer Mannheim) in the presence of 20 μM (each) dATP and dCTP and 200 μM ddGTP, and cycled at 94°C for 20 s, 60°C for 20 s, and 75°C for 20 s for 20 cycles. Under these conditions, limited primer extension occurs such that elongation on each type of template DNA results in a product of unique length. The elongation products were fractionated by high-resolution gel (16% polyacrylamide–8 M urea) electrophoresis, and the proportion of each product was determined from densitometric analyses of autoradiographs.
FIG. 2.
Multiplex sequence analysis for detecting the fractions of different Clar mutant cells in a mixed culture. (A) Experimental strategy. Chromosomal DNAs were isolated from the mixed cultures, and a 1,093-bp fragment, carrying the mutation site in the center, was PCR amplified. The DNA plus strand is equivalent to the sequence of the 23S rRNA, within which the XX (underlined) represent mutation sites. In wild-type cells, XX = AA, whereas in the mutant cells they are changed, respectively, to GA (A2142G), AG (A2143G), CA (A2142C), and AC (A2143C). The identity of NN in the DNA minus strand are indicated in parentheses for each mutant. The minus strand serves as template for elongation of the 5′ end 32P-labelled 19-mer primer. In the presence of dCTP, dATP, and ddGTP, the elongation will result in products of unique length depending on mutation types. (B) Autoradiograph of a multiplex sequencing gel. Primer elongation on DNAs from an individual strain (control, lanes 1 to 4) gave rise to products of unique length marked on the left (nt, nucleotide). As indicated on the top, lanes 5 to 8 are elongation on DNA templates obtained from mixed cultures of two mutant strains, whereas lanes 9 and 10 are those from mixed cultures of three or four strains, respectively. Abbreviations: 2G, A2142G; 3G, A2143G; 2C, A2142C; 3C, A2143C.
Figure 2B is a representative autoradiograph of the multiplex sequencing gel. As the control, the primer elongation on DNA from each individual mutant strain produced a unique band with expected length (lanes 1 to 4). Lanes 5 to 10 are analyses for DNAs obtained from mixed cultures with various combinations of the Clar mutants (mixtures of two, three, and four strains of interest). The quantitative data obtained from densitometric analyses of the autoradiographs are shown in Table 1. In the 4-day-grown mixed culture of A2142G and A2142C mutants, which were initially inoculated in the same amount, 99% were A2142G mutants. The same predominance was found for A2142G over A2143C mutants. The ratios of competitive accumulation for A2143G to A2142C or to A2143C mutants were 68:30 and 89:7, respectively. Note that in these two mixtures a small percentage of A2142G mutant, which was not included in the initial inoculation, was detected. Actually, 1 to 2% of this type of mutation was also observed in the 4-day cultures of a single strain of A2143G, A2142C, or A2143C (Fig. 2B, lanes 2 to 4). Although this mutation may occur in the 4-day growing stage from other types of mutation originally included in the mixture, such high frequency could not be valid. According to our preliminary observation, the mutation rate (from wild type to Clar) is below 10−7. Most probably, the small percentage of A-to-G mutation detected by multiplex sequencing arose during the assay process, namely, due to DNA polymerase error at the PCR step for the amplification of the template DNA fragments and/or due to incomplete elongation at the subsequent sequencing step. It may not represent the real percentage of A-to-G mutants in the mixed culture. Thus, this assay system may have up to a 4% overestimation for the A2142G mutation.
TABLE 1.
Competitive growth patterns of H. pylori Clar mutantsa
| Strain mixtureb | Fractions (%) in 4-day-grown culture
|
|||
|---|---|---|---|---|
| 2G | 3G | 2C | 3C | |
| 2G + 2C | 99 | <1 | ||
| 2G + 3C | 99 | <1 | ||
| 3G + 2C | 2c | 68 | 30 | |
| 3G + 3C | 4c | 89 | 7 | |
| 2G + 3G + 2C | 84 | 14 | <2 | |
| 2G + 3G + 2C + 3C | 86 | 12 | 2d | 2d |
Data are deduced from densitometric analyses of autoradiographs shown in Fig. 2B. The percentage of a particular strain in the mixed population is averaged from three determinations with numbers rounded to the nearest integer.
Strain mixtures were equal inoculations. Abbreviations 2G, A2142G; 3G, A2143G; 2C, A2142C; 3C, A2143C.
As explained in the text, this percentage may not necessarily represent that of real A2142G mutants in the mixed culture. Accordingly, the fraction of A2142G mutant in all the mixed cultures may need to be adjusted by subtracting 2% to 4%. This adjustment will have no significant effect on the overall pattern.
A total of 2% for both types of A-to-C mutants, which are indistinguishable in the assay.
Examination of the mixtures of three or four strains indicated that the mixed population consists of about 85% A2142G and 13% A2143G, and two A-to-C mutants together account for 2% of the whole population. We did not include the A2142T mutant for multiplex sequence analysis in this study, because a different nucleotide combination in the assay is required to detect its elongation product. From its growth curve, however, it appears likely that the A2142T mutant is similar to the A2143C mutant in competitive growth.
Based on the patterns of competitive growth as well as the individual growth of different Clar mutant strains, we conclude that the order of preference of competitive accumulation is A2142G > A2143G >>> A2142C > A2143C (A2142T). If the same is true in vivo, once an A-to-G mutation occurs (spontaneously or drug induced), the other types of mutation that exist in the same environment, if any, are likely to be overgrown after a period of time. A-to-C or A-to-T mutants could be isolated only when an A-to-G mutant has not appeared at that particular gastric niche. Our results provide a rational explanation for the mutation pattern observed in clinical isolates. As we discussed previously (15), an additional possible mechanism yet to be identified, by which the A-to-G mutations are preferentially produced in H. pylori, may also contribute to the observed predominance of A-to-G mutations. Further genetic studies are needed to test this hypothesis.
Acknowledgments
This work was supported in part by funding from the Canadian Bacterial Diseases Network (Centers of Excellence Program) to D.E.T., who is a Medical Scientist with the Alberta Heritage Foundation for Medical Research (AHFMR), and by PHS grants to M. Z. H. G.W. is a recipient of a postdoctoral fellowship from the Canadian Association of Gastroenterology and Astra Canada in association with an MRC-PMAC award as well as a fellowship from AHFMR.
REFERENCES
- 1.Cayla R, Zerbib F, Talbi P, Megraud F, Lamouliatte H. Pre- and posttreatment clarithromycin resistance of Helicobacter pylori strains: a key factor of treatment failure. Gut. 1995;37(Suppl. 1):A55. [Google Scholar]
- 2.Debets-Ossenkopp Y J, Brinkman A B, Kuipers E J, Vandenbroucke-Grauls C M J E, Kusters J G. Explaining the bias in the 23S rRNA gene mutations associated with clarithromycin resistance in clinical isolates of Helicobacter pylori. Antimicrob Agents Chemother. 1998;42:2749–2751. doi: 10.1128/aac.42.10.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Debets-Ossenkopp Y J, Sparrius M, Kusters J G, Kolkman J J, Vandenbroucke-Grauls C M J E. Mechanism of clarithromycin resistance in clinical isolates of Helicobacter pylori. FEMS Microbiol Lett. 1996;142:37–42. doi: 10.1111/j.1574-6968.1996.tb08404.x. [DOI] [PubMed] [Google Scholar]
- 4.Goddard A F, Logan R P H. Antimicrobiol resistance and Helicobacter pylori. J Antimicrob Chemother. 1996;37:639–643. doi: 10.1093/jac/37.4.639. [DOI] [PubMed] [Google Scholar]
- 5.Occhialini A, Urdaci M, Doucet-Populaire F, Bebear C M, Lamouliatte H, Megraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palejwala V A, Rzepka R W, Simha D, Humayun M Z. Quantitative multiplex sequence analysis of mutational hot spots. Frequency and specificity of mutations induced by a site-specific ethenocytosine in M13 viral DNA. Biochemistry. 1993;32:4105–4111. doi: 10.1021/bi00066a036. [DOI] [PubMed] [Google Scholar]
- 7.Rahman M S, Dunman P M, Wang G, Murphy H S, Humayun M Z. Effect of UVM induction on mutation fixation at non-pairing and mispairing DNA lesions. Mol Microbiol. 1996;22:747–755. doi: 10.1046/j.1365-2958.1996.d01-1723.x. [DOI] [PubMed] [Google Scholar]
- 8.Rene W M, Hulst V D, Keller J J, Rauws E A J, Tytgat G N J. Treatment of Helicobacter pylori infection: a review of the world literature. Helicobacter. 1996;1:6–19. doi: 10.1111/j.1523-5378.1996.tb00003.x. [DOI] [PubMed] [Google Scholar]
- 9.Stone G G, Shortridge D, Versalovic J, Beyer J, Flamm R K, Ghoneim A T, Tanaka K. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone G G, Shortridge D, Flamm R K, Versalovic J, Beyer J, Idler K, Zulawinski L, Tanaka K. Identification of a 23S rRNA gene mutation in clarithromycin-resistant Helicobacter pylori. Helicobacter. 1996;1:227–228. doi: 10.1111/j.1523-5378.1996.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 11.Taylor D E, Ge Z, Purych D, Lo T, Hiratsuka K. Cloning and sequence analysis of the two copies of 23S rRNA genes from Helicobacter pylori and association of clarithromycin resistance with 23S rRNA mutations. Antimicrob Agents Chemother. 1997;41:2621–2628. doi: 10.1128/aac.41.12.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vaira D, Holton J, Miglioli M, Menegatti M, Mule P, Barbara L. Peptic ulcer disease and Helicobacter pylori infection. Curr Opin Gastroenterol. 1994;10:98–104. [Google Scholar]
- 13.Versalovic J, Shortridge D, Kibler K, Griffy M V, Bryer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Versalovic J, Osato M S, Spakovsky K, Dore M P, Reddy R, Stone G G, Shortridge D, Flamm R K, Tanaka S K, Graham D Y. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997;40:283–286. doi: 10.1093/jac/40.2.283. [DOI] [PubMed] [Google Scholar]
- 15.Wang G, Taylor D E. Site-specific mutations in the 23S rRNA gene of Helicobacter pylori confer two types of resistance to macrolide-lincosamide-streptogramin B antibiotics. Antimicrob Agents Chemother. 1998;42:1952–1958. doi: 10.1128/aac.42.8.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, Palejwala V A, Dunman P M, Aviv D H, Murphy H S, Rahman M S, Humayun M Z. Alkylating agents induce UVM, a recA-independent inducible mutagenic phenomenon in Escherichia coli. Genetics. 1995;141:813–823. doi: 10.1093/genetics/141.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]