Abstract
OBJECTIVE
Maternal glycemic dysregulation during pregnancy increases the risk of adverse health outcomes in her offspring, a risk thought to be linearly related to maternal hyperglycemia. It is hypothesized that changes in offspring DNA methylation (DNAm) underline these associations.
RESEARCH DESIGN AND METHODS
To address this hypothesis, we conducted fixed-effects meta-analyses of epigenome-wide association study (EWAS) results from eight birth cohorts investigating relationships between cord blood DNAm and fetal exposure to maternal glucose (Nmaximum = 3,503), insulin (Nmaximum = 2,062), and area under the curve of glucose (AUCgluc) following oral glucose tolerance tests (Nmaximum = 1,505). We performed lookup analyses for identified cytosine-guanine dinucleotides (CpGs) in independent observational cohorts to examine associations between DNAm and cardiometabolic traits as well as tissue-specific gene expression.
RESULTS
Greater maternal AUCgluc was associated with lower cord blood DNAm at neighboring CpGs cg26974062 (β [SE] −0.013 [2.1 × 10−3], P value corrected for false discovery rate [PFDR] = 5.1 × 10−3) and cg02988288 (β [SE]−0.013 [2.3 × 10−3], PFDR = 0.031) in TXNIP. These associations were attenuated in women with GDM. Lower blood DNAm at these two CpGs near TXNIP was associated with multiple metabolic traits later in life, including type 2 diabetes. TXNIP DNAm in liver biopsies was associated with hepatic expression of TXNIP. We observed little evidence of associations between either maternal glucose or insulin and cord blood DNAm.
CONCLUSIONS
Maternal hyperglycemia, as reflected by AUCgluc, was associated with lower cord blood DNAm at TXNIP. Associations between DNAm at these CpGs and metabolic traits in subsequent lookup analyses suggest that these may be candidate loci to investigate in future causal and mediation analyses.
Introduction
Gestational diabetes mellitus (GDM) has major health consequences for both mother and child (1–3). Even among women without GDM, maternal hyperglycemia and hyperinsulinemia have been associated with increased risk for pregnancy complications (1) and offspring cardiometabolic disease (3). The latter relationships are hypothesized to be mediated by alterations in epigenetic factors, including DNA methylation (DNAm), laid down during prenatal development (4). Single cohort studies have reported associations between GDM or maternal glycemic measures and offspring DNAm (5–8). The most comprehensive study to date has been a Pregnancy and Childhood Epigenetics (PACE) consortium meta-analysis of epigenome-wide association studies (EWAS) with assessment of the association between GDM diagnosis and cord blood DNAm (9). This study did not find evidence for robust associations between mother’s GDM status and offspring DNAm at the single cytosine-guanine dinucleotide (CpG) level, suggesting that GDM may not influence changes in the fetal epigenome. However, this may also be partly explained by methodological limitations such as the heterogeneous definitions of GDM, differences in GDM treatment across cohorts, or limited statistical power to identify changes across the DNA methylome (N = 317 cases of GDM). In addition, GDM diagnosis is a clinical threshold dichotomizing glucose levels, yet linear associations have been reported between various measures of glucose metabolism and offspring outcomes (3). We therefore opted to evaluate continuous measures of maternal glycemic dysregulation in relation to offspring DNAm.
In the current study, we conducted fixed-effects meta-analyses of EWAS investigating associations between continuous maternal glucose, insulin, and area under the curve of glucose (AUCgluc) measures from an oral glucose tolerance tests (OGTT) conducted during pregnancy and cord blood DNAm. We used AUCgluc as one of our exposures of interest, as glucose measures at different OGTT time points show similar linear associations with health outcomes (1) and capture both fasting and nonfasting maternal glycemic regulation (10). The findings from the meta-analyses were subsequently looked up in complementary observational studies for assessment of whether the variation of DNAm at identified CpGs also potentially associated with cardiometabolic traits in children (11) and adults (12). Additionally, we performed lookup analyses investigating relationships between DNAm at these CpGs and gene expression in two relevant human tissues (13).
Research Design and Methods
Participating Cohorts
Seven cohorts with cord blood DNAm and fasting glycemic data in midpregnancy participated in the meta-analyses (Table 1 and Supplementary Material). These cohorts were from Southeast Asia (Singapore: Growing Up in Singapore Towards healthy Outcomes [GUSTO] [14]), North America (Canada: Genetics of Glucose regulation in Gestation and Growth [Gen3G] [15], U.S.: Healthy Start [16]), and Europe (Finland: the Finnish Gestational Diabetes [FinnGeDi] Study [7,17] and Prediction and Prevention of Preeclampsia and Intrauterine Growth Restriction [PREDO] study [18], France: EDEN [19], and Belgium: the ENVIRonmental influence ON early AGEing [ENVIRONAGE] [20]). One cohort, in the Generation R Study (21) (the Netherlands), had nonfasting glycemic data (included in a secondary analysis). Apart from FinnGeDi (17), all studies included general population–based birth cohorts. Recruitment for the FinnGeDi control subjects (FinnGeDi-control) was similar to that for the other cohorts, while the FinnGeDi case subjects (FinnGeDi-GDM) were recruited and glycemic markers were measured much earlier in pregnancy (12–16 weeks). Therefore, data for FinnGeDi-GDM and FinnGeDi-control were analyzed separately. Ethics approval and informed consent were obtained following national and international standards.
Table 1.
Cohort characteristics
| Cohort | Ancestry | Array | Sample size n (% female) | Mat. age, years | Mat. prepregnancy BMI, kg/m2 | Multiparous (%) | GA at glycemic measure, days | GA at birth, days | FG, mmol/L | FI, pmol/L | AUCgluc, mmol/L*min |
|---|---|---|---|---|---|---|---|---|---|---|---|
| EDEN | French European | 450k | 53 (41.5) | 31.1 (5.7) | 24.4 (5.7) | 72 | 172 (19) | 264 (11) | 4.38 (0.45) | — | 882 (111) |
| FinnGeDi-control | Finnish European | EPIC | 236 (45.3) | 31.5 (5.2) | 25.6 (4.8) | 50 | 191 (18) | 282 (8) | 4.66 (0.29) | — | 759 (100) |
| FinnGeDi-GDM | Finnish European | EPIC | 266 (50.0) | 32.5 (5.4) | 27.8 (6.1) | 56 | 165 (46) | 278 (9) | 5.27 (0.49) | — | 982 (132) |
| Gen3G | European | EPIC | 451 (47.5) | 28.2 (4.3) | 28.0 (5.5) | 67 | 185 (7) | 276 (7) | 4.19 (0.38) | 64 (73) | 725 (129) |
| GUSTO | Chinese, Malay, Indian | 450k | 264 (49.4) | 30.1 (5.4) | 23.5 (5.1) | 54 | 186 (19) | 274 (7) | 4.40 (0.49) | — | — |
| Healthy Start | Caucasian, Hispanic, African American | 450k | 532 (48) | 27.6 (6.2) | 26.0 (6.8) | 42 | 125 (23) | 277 (8) | 4.27 (0.39) | 92 (61) | 867 (144) |
| PREDO | Finnish European | 450k | 552 (47.5) | 33.5 (5.8) | 28.8 (6.4) | 67 | 185 (24) | 280 (9) | 4.89 (0.46) | — | 822 (142) |
| ENVIRONAGE | European | EPIC | 103 (45.6) | 30.5 (4.5) | 23.9 (4.1) | 43 | 181 (24) | 279 (9) | 4.55 (0.71) | — | 892 (146) |
| Generation R Study† | Dutch European | 450k | 1,101 (49) | 31.5 (4.1) | 24.0 (3.9) | 39 | 92 (12) | 282 (9) | 4.33 (0.78 | 141 (130) | — |
Data are means (SD) unless otherwise indicated. Mat., maternal.
Nonfasting glucose and insulin data.
Meta-analysis: Participants and Exclusion Criteria
We provided the analysis plan with R scripts for running the EWAS to all interested cohorts (Supplementary Material). Investigators for cohorts measured DNAm in cord blood using either the Illumina Infinium HumanMethylation450 (450k) or Illumina MethylationEPIC (EPIC) BeadChip array, which was normalized as investigators deemed appropriate (Supplementary Material, including Supplementary Table 1). Only term singletons (gestational age [GA] >37 weeks) were included in the analyses. We excluded siblings and offspring from mothers with type 1 or type 2 diabetes prior to the pregnancy.
Glycemia-Related Traits (Exposure)
We investigated three glycemia-related traits as continuous exposures: fasting glucose (FG) (in millimoles per liter), fasting insulin (FI) (in picomoles per liter, log2 transformed), and AUCgluc (mmol/L*min). For each cohort, maternal blood samples were collected by trained professionals. If multiple measurements were available during pregnancy, the earliest measurement was used. If samples were collected during an OGTT, the glucose and/or insulin concentration at the start of the OGTT was used as the “fasting” measure. The Generation R Study had standardized, but nonfasting, glucose and insulin measurements available (N ≈ 1,100) (6). The OGTT were performed with a bolus of 50 g (ENVIRONAGE), 75 g (FinnGeDi, PREDO, and Gen3G), or 100 g pure glucose (EDEN, Healthy Start) in accordance with respective national guidelines. The AUCgluc was calculated from glucose concentrations (in millimoles per liter) measured at time 0, 60, and 120 min with the method of Matthew et al. BMJ 1990, appendix II.
Cohort-Specific Analyses
For all analyses, DNAm was analyzed as normalized untransformed β values. β values denote DNAm levels, where 0 approximates 0% and 1 approximates 100%. Effect estimates were converted to percentages throughout the manuscript with multiplication of the β values by 100. Investigators for each cohort performed EWAS on glucose/insulin/AUCgluc using robust linear regression (rlm) from the R MASS package with the White estimator for robust SEs, as implemented in the R package sandwich (22), which leads to a model robust for outlying β values and heteroscedasticity. We used the β values of each CpG as the outcome and each of the glycemic variables as the predictor in separate models. Directed acyclic graphs (Supplementary Material) were used to investigate and determine the necessary minimal set of covariates to include in the model. Each EWAS was adjusted for the sex of the child (female/male), GA at maternal glycemic samplings (days), maternal age (years), GA at birth (days), parity (nulliparous yes/no), and imputed cord blood cell proportions (23) from the estimateCellCounts function in the minfi R package (24) with use of the “Bakulski reference” data set for cord blood. In addition, investigators of the cohorts were instructed to adjust for cohort-specific variables as needed (Supplementary Material). EWAS results from each cohort were evaluated with the R QCEWAS package (25).
Meta-analysis
After quality control, we filtered out all probes that 1) did not map to unique genomic locations, 2) overlapped single nucleotide polymorphisms (minor allele frequency >5% in 1,000 genomes), or 3) had >0.2 mean β value differences between the 450k and EPIC array (26). EWAS often suffer from deflation/inflation (λ) and bias (μ) (as apparent in quantile-quantile plots [QQ-plots]) in the test statistic distribution, which may lead to spurious findings (27). We therefore used the R Bioconductor package bacon to estimate and mitigate the λ and μ for each EWAS (27) (Supplementary Tables 2, 3, and 4). A fixed-effects meta-analysis with inverse variance weighting was then run for the cohort-specific bacon adjusted results for FG, FI, and AUCgluc with the R package metafor (28). We also ran leave-one-out analyses for all probes using metafor. Heterogeneity was assessed with the Cochran Q test. In the meta-analysis with FG as an exposure, we observed genome-wide heterogeneity (Supplementary Fig. 1A), and the EDEN cohort was identified as the source of heterogeneity (Supplementary Fig. 1B), so in the final FG meta-analysis we excluded EDEN (N = 2,404). The addition of nonfasting data from the Generation R Study did not introduce heterogeneity (Supplementary Figs. 2 and 3). Among the six cohorts for which values were provided for AUCgluc, in EDEN (N = 32), ENVIRONAGE (N = 86), and Healthy Start (N = 48) only measurements of women at high risk of developing GDM were included. There was heterogeneity in the meta-analysis (Supplementary Fig. 4A), which was mitigated with omission of these three cohorts (Supplementary Fig. 4B–D). The removal of the FinnGeDi-GDM sample had no effect on heterogeneity (Supplementary Fig. 4E). Therefore, in this meta-analysis we excluded EDEN, ENVIRONAGE, and Healthy Start but included the FinnGeDi-GDM sample. The meta-analyses were performed by two independent analysts to reduce the possibility of human error. All reported P values are two sided, and multiple testing corrections were performed with use of Benjamini-Hochberg (i.e., false discovery rate [FDR]). P values corrected by FDR are designated as PFDR. P values that were not corrected by FDR (for instance, from lookup analyses) are designated as Pnominal. In EWAS meta-analyses, raw Pnominal values <1 × 10−6 were deemed suggestive and PFDR values <0.05 were considered statistically significant. All probes were annotated to the human reference genome version 19, build 37. Meta-analysis results are deposited to the EWAS catalog (29), Zenodo DOI https://doi.org/10.5281/zenodo.5886997. The presence of differentially met-hylated regions (DMR) in relation to the glycemic exposures was evaluated with the R packages ipDMR (30) and DMRcate (31), with use of each respective meta-analysis test statistic file. A DMR was considered robust if identified with both methods.
Cross-sectional Lookup Analyses
For the Study in TEENs of the natural course of type 1 DIABetes (TEENDIAB) cohort (11) (Germany) and the Northern Finland birth cohort of 1966 (NFBC1966) (12), DNAm data were provided for blood of children and adults, respectively, for conducing cross-sectional lookup analyses for loci of interest with cardiometabolic phenotypes. In addition, investigators for the Biological Atlas of Severe Obesity (ABOS) study (France) (13) provided DNAm and RNA-sequencing data for liver and muscle tissue from adult women with obesity who had undergone gastric bypass surgery (Supplementary Material). In all three cohorts, we used rlm to determine the association between DNAm at specific probes and each phenotype of interest. In the TEENDIAB cohort analyses, we adjusted for the child’s sex, the age of the child (years), maternal type 1 diabetes status (binary), six imputed blood cell types (32), parental socioeconomic status (low, medium, and high), and batch (sentrix position). In the NFBC1966 (adults), we adjusted for sex, the imputed blood cell types (32), socioeconomic status (low, medium, or high), and batch. For the ABOS cohort, we adjusted for age (years), BMI, and type 2 diabetes status (binary).
Results
Cohort Summaries
Characteristics of each cohort are described in Table 1. Mean maternal age ranged from 27.6 to 33.5 years and mean BMI from 23.9 to 28.8 kg/m2. The French EDEN cohort had the lowest mean FG (4.3 mmol/L), while the Finnish cohorts had the highest mean FG (FinnGeDi-control 4.6 mmol/L, FinnGeDi-GDM 5.3 mmol/L, PREDO 4.9 mmol/L). Mean FI differed between Gen3G (64 pmol/L) and Healthy Start (92 pmol/L), likely due to a lack of standardization of this measurement.
Glucose and Insulin
The maternal FG meta-analysis (N = 2,404, λ = 1.047, µ = 0.056) yielded evidence for an association between FG and DNAm at CpG cg26104143 (β [SE] −0.26 [0.04], PFDR = 6.6 × 10−3, N = 2,404) (Table 2). This CpG (chromosome [chr]4: 41874579–41874580) is located upstream of TMEM33. The heterogeneity for association at this specific CpG was considerable (I2 = 42%) and driven by the ENVIRONAGE cohort (Supplementary Fig. 5), as the association was attenuated and no longer significant after exclusion of ENVIRONAGE (β [SE] −0.09 [0.07], Pnominal = 0.19). Adding nonfasting glucose data from the Generation R Study did not reveal CpGs reaching statistical significance (PFDR > 0.073, N = 3,503, λ = 1.042, µ = 0.059) (Table 2). No robust DMRs were identified for FG.
Table 2.
Cord blood DNAm associations with maternal glycemic traits (P value <1.0 × 10−6)
| Glycemic trait | Probe identifier | Position (hg19) | Nearest gene | Restricting to fasting participants | Including nonfasting participants | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | β (SE) | P † | I 2 | N | β (SE) | P † | I 2 | ||||
| Glucose | cg26104143 | chr4: 41869579 | TMEM33 | 2,404 | −0.26 (0.04) | 7.9 × 10−9 | 42.7 | 3,503 | −0.18 (0.033) | 1.1 × 10−7 | 62.2 |
| Glucose | cg26974062 | chr1: 145440734 | TXNIP | 1,056 | −3.0 (0.56) | 3.0 × 10−7 | 0 | 1,056 | −3.0 (0.56) | 2.6 × 10−7 | 0 |
| Glucose | cg21686486 | chr2: 172377802 | CYBRD1 | 1,056 | 1.2 (0.22) | 3.2 × 10−7 | 57.4 | 1,056 | 1.2 (0.22) | 2.8 × 10−7 | 57.4 |
| Insulin | cg21139325 | chr6: 32729470 | HLA-DQB2 | 961 | 0.55 (0.11) | 2.8 × 10−7 | 0 | 2,062 | 0.16 (0.029) | 3.1 × 10−7 | 15.2 |
| AUCgluc | cg26974062 | chr1: 145440734 | TXNIP | 953 | −0.013 (2.1 × 10−3) | 6.3 × 10−9 | 52.1 | ||||
| AUCgluc | cg02988288 | chr1: 145440445 | TXNIP | 953 | −0.013 (2.3 × 10−3) | 7.9 × 10−8 | 60.4 | ||||
| AUCgluc | cg09049566 | chr5: 132165605 | SHROOM1 | 1,505 | −2.0 × 10−3 (3.9 × 10−4) | 9.2 × 10−7 | 1.9 | ||||
Overview of the meta-analysis results with a P value <1.0 × 10−6 after correction for inflation/bias with the bacon R package. The used rlm with robust SEs was as follows: β value ˜ glycemic trait + GA at maternal sampling + sex of the child + imputed cord blood cell proportions + maternal age + GA at birth + parity and cohort-specific (technical) variables.
P value after correction for inflation and bias with the bacon R package. Correction is based on the entire distribution of test statistics of each meta-analysis and may therefore (slightly) differ between the fasted and combined meta-analyses as the sample size is increased for many CpGs.
Next, we investigated FI, which was measured in Gen3G (N = 438) and Healthy Start (N = 523). We did not find evidence of a statistically significant association between maternal FI and DNAm in offspring cord blood (PFDR > 0.11, N = 961, λ = 1.027, µ = −0.078). Adding nonfasting insulin data from the Generation R Study did not reveal CpGs reaching statistical significance (PFDR > 0.14, N = 2,062, λ = 1.036, µ = 0.004). The CpGs at which DNAm was nominally associated with FI or FG (Pnominal < 1 × 10−6) are presented in Table 2. No robust DMRs were identified for FI.
Glycemic Excursion During the OGTT
The AUCgluc meta-analysis that included data from FinnGeDi, Gen3G, and PREDO (N = 1,505, λ = 1.027, µ = −0.004) identified significant associations between a higher AUCgluc and lower DNAm at cg26974062 (β [SE] −0.013 [2.1 × 10−3], PFDR = 5.1 × 10−3, N = 953) and cg02988288 (β SE−0.013 [2.3 × 10−3], PFDR = 0.031, N = 953). These two CpGs are located in thioredoxin interacting protein (TXNIP) (cg26974062 at chr1, 145440734, and cg02988288 at chr1, 145440445) (Fig. 1A). The meta-analysis on FG identified suggestive associations with lower DNAm at both TXNIP CpGs (Table 2) (cg26974062, β [SE] −3.0 [0.56], Pnominal = 3.0 × 10−7, N = 1,056; cg02988288, −3.2 [0.64], Pnominal = 1.8 × 10−6, N = 1,056), consistent with the direction of effect observed in our EWAS for AUCgluc.
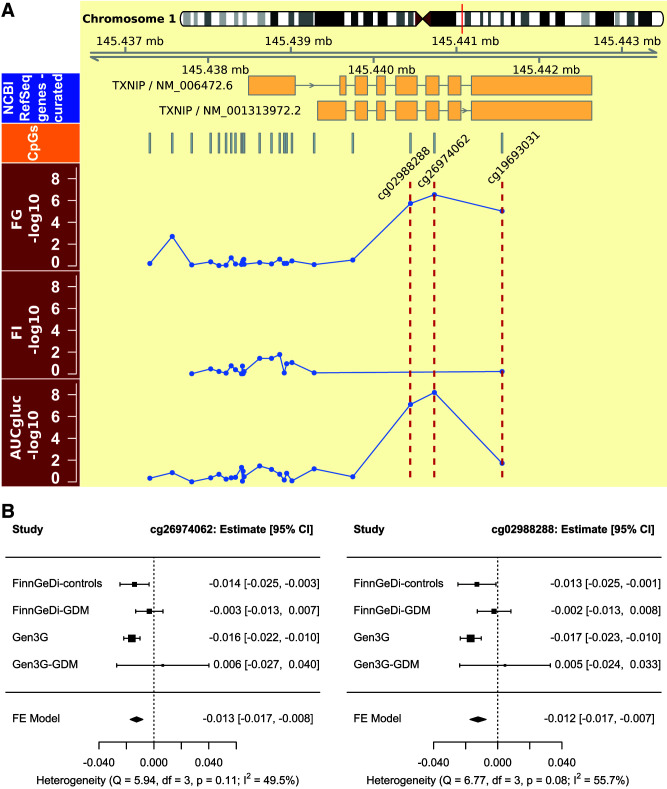
Figure 1.
Overview of findings at TXNIP. A: Chromosomal and gene map for the TXNIP locus (top), followed with the locations of the CpGs incorporated in the meta-analysis. Highlighted with red dotted lines are CpGs cg02988288, cg26974062, and cg19693031 in the panels with −log10 nominal P values for the meta-analyses on FG, FI, and AUCgluc for the measured CpGs in TXNIP. B: Forest plot for the AUC of an OGTT meta-analysis stratified by GDM status for the two CpGs that were genome-wide significant. Gen3G-GDM, GDM case subjects from the Gen3G cohort; NCBI, National Center for Biotechnology Information; FE, fixed-effect.
DNAm at the probes located upstream (+5 kb) of these CpGs were not associated with AUCgluc (Pnominal > 0.29). Directly downstream of the newly identified CpGs, DNAm at cg19693031 (chromosome 1: 145441552) has been associated previously with multiple adult metabolic traits and the risk of type 2 diabetes development (33). In our data set, cord blood DNAm at cg19693031 was nominally associated with a greater AUCgluc (β [SE] −1.0 × 10−5 [4.4 × 10−6], Pnominal = 0.019, N = 1,505) and with higher maternal FG (−0.4 [0.1], Pnominal = 9.4 × 10−6, N = 2,404) but not with FI (Pnominal = 0.60) (Fig. 1A). However, this region was not designated as a DMR and we did not identify any robust DMRs for AUCgluc.
Despite mitigation of genome-wide heterogeneity, heterogeneity was high for associations between AUCgluc and cord blood DNAm at cg26974062 (I2 = 52.1%) and cg02988288 (I2 = 60.3%). Both CpGs are represented on the EPIC array but not the 450k array (unlike cg19693031, which is present on both). Therefore, both probes were only available for the two FinnGeDi groups and Gen3G. The heterogeneity for these probes originated from a lack of association among offspring born to FinnGeDi-GDM mothers (Supplementary Fig. 6). A similar observation was made in stratification of Gen3G participants by GDM status (Fig. 1B). However, there was no statistical evidence of interaction to support a moderating effect of GDM in either the FinnGeDi or Gen3G cohorts (PAUCgluc × GDM > 0.10). Excluding GDM pregnancies from the AUCgluc meta-analysis did not reveal any additional CpGs, apart from cg26974062 and cg02988288, reaching statistical significance thresholds (data not shown).
Cross-sectional Lookups
To investigate whether DNAm at the two newly identified CpG sites in TXNIP may play a role in offspring metabolic health, we investigated associations between blood DNAm at these two CpGs and metabolic phenotypes at various time points across the life span. First, we did an in silico lookup analysis using data from TEENDIAB (11), a prospective study where DNAm (EPIC array) was measured in the blood of children (4–19 years of age) born to mothers with (N = 162) or without (N = 221) type 1 diabetes, a condition characterized by relative maternal hyperglycemia during pregnancy in the majority of women, despite tight glycemic targets. Exposure to maternal type 1 diabetes in utero was associated with lower child blood DNAm at both cg26974062 (β [SE] −0.76 [0.34], Pnominal = 0.024) and cg02988288 (−0.89 [0.29], Pnominal = 2.4 × 10−3), and the directions of effect were consistent with our analyses of AUCgluc and FG. In contrast, child blood DNAm at the four CpGs with suggestive associations with FG and FI (see Table 2) did not show associations with in utero exposure to maternal type 1 diabetes (P > 0.05).
Next, we investigated cross-sectional associations between blood DNAm at these loci and metabolic phenotypes in childhood and adulthood. At both TXNIP CpGs, lower DNAm in childhood blood was associated with higher child HOMA of insulin resistance and, for cg02988288, higher FI (Table 3 and Supplementary Table 5). Similarly, using metabolic traits in adults at 46 years of age in NFBC1966, we observed consistent negative cross-sectional associations between blood DNAm at cg26974062 and cg02988288 and all of the metabolic traits tested (serum glucose, insulin, AUCgluc, HbA1c, and BMI) (Table 3 and Supplementary Table 4). In contrast, of the CpGs that showed suggestive associations with FG and FI in our meta-analysis (Table 2), we only found cg21139325 to be nominally associated with adult BMI (Supplementary Table 6).
Table 3.
Cross-sectional associations of blood DNAm at cg02988288 and metabolic phenotypes in childhood and adulthood
| TEENDIAB participants (German Europeans ages 4–19 years [49.6% female])§ | NFBC1966 participants (Finnish Europeans ages 46 years [56% female])† | |||||
|---|---|---|---|---|---|---|
| β (SE) | P | N | β (SE) | P | N | |
| Fasting plasma glucose (mmol/L) | −0.37 (0.30) | 0.22 | 366 | −0.71 (0.16) | 1.20 × 10−5 | 680 |
| Fasting plasma insulin (pmol/L) | −0.41 (0.17) | 0.014 | 369 | −0.044 (0.013) | 9.9 × 10−4 | 685 |
| AUCgluc (mmol/L*min) | −1.4 × 10−3 (1.5 × 10−3) | 0.33 | 232 | −2.1 × 10−3 (6.5 × 10−4) | 1.3 × 10−3 | 589 |
| BMI (kg/m2) | −7.3 × 10−2 (5.3 × 10−2) | 0.17 | 383 | −0.077 (0.022) | 5.0 × 10−4 | 693 |
| Body fat (bio-impedence) | NA | NA | NA | −0.039 (0.014) | 4.3 × 10−3 | 671 |
| Waist-to-hip ratio | −0.14 (0.17) | 0.42 | 365 | NA | NA | NA |
| HOMA-IR | −0.29 (8.4 × 10−2) | 5.0 × 10−4 | 365 | NA | NA | NA |
| HbA1c (mmol/L) | −8.5 × 10−3 (4.7 × 10−2) | 0.85 | 361 | −0.090 (0.024) | 2.6 × 10−4 | 693 |
| Type 2 diabetes | NA | NA | NA | −1.46 (0.52) | 4.7 × 10−3 | 507 |
HOMA-IR, HOMA of insulin resistance. NA, not available for assessment.
Outcome of analyses in the TEENDIAB cohort. Columns denote the results from an rlm with robust SEs adjusting for sex, age at blood draw, batch, imputed cell heterogeneity, maternal type 1 diabetes status, and parental socioeconomic status.
Outcome of analyses in NFBC1966. The results from an rlm with robust SEs adjusting for sex, batch, imputed cell proportions, and socioeconomic status.
Finally, we investigated DNAm levels at cg26974062 and cg02988288 and TXNIP expression measured in muscle and liver biopsies of women with obesity in the ABOS cohort (13). Lower DNAm at cg26974062 (β [SE] −1.1 × 10−2 [5.2 × 10−3], Pnominal = 0.031, N = 319) and cg02988288 (−4.5 × 10−2 [1.2 × 10−2], Pnominal = 3.2 × 10−4, N = 319) was associated with higher TXNIP gene expression in liver but not in muscle (N = 71). In contrast, the CpGs with suggestive associations with FG and FI (Table 2) were not associated with gene expression (Supplementary Table 7).
Lookups in Literature
We checked the CpGs that we identified (Table 2) in the EWAS catalog (29), which documents (suggestive) associations (P < 10−4). Cord blood DNAm at cg26974062 had a nominal association with maternal 1-h glucose in the UK Pregnancies Better Eating and Activity Trial (UPBEAT) (5). Next, we checked recently published data on maternal HbA1c levels and cord blood DNAm (Gen3G [N = 412]) (8), and both TXNIP probes showed nominal associations with maternal HbA1c (cg02988288, β [SE] −4.5 [0.16], Pnominal = 3.9 × 10−3; cg26974062, −3.8 [1.5], Pnominal= 0.012) in a direction consistent with that of our AUCgluc and FG meta-analyses. None of the other CpGs with suggestive associations in Table 2 were associated with maternal HbA1c. Finally, the CpGs that showed (suggestive) associations with FG, FI, and AUCgluc were not associated with GDM (or probes were not available) in the prior PACE meta-analysis (Pnominal > 0.48) (9).
In “reverse lookups,” we found little evidence for the reported associations with maternal FG and 1-h or 2-h glucose from UPBEAT participants cord blood analyses: only 5 of 609 reported CpGs for 1-h or 2-h glucose were nominally associated with AUCgluc with the same direction of effect (namely, cg24914185, cg13874780, cg04322572, cg03795071, and cg23913963) (5). Cord blood DNAm at a CpG near URGCP reportedly associated with maternal HbA1c (8) was not associated with any glycemic trait in our meta-analyses (Pnominal > 0.074), and none of the CpGs located in DMRs identified for GDM in a prior PACE report were associated either (9) (Pnominal > 0.18).
Conclusions
We did not find evidence for robust associations between maternal prenatal glucose and insulin levels and offspring DNAm in cord blood (9). Collectively, these findings might argue against the hypothesis that maternal hyperglycemia during pregnancy and later childhood health phenotypes can be mediated by changes in DNAm (4). However, our meta-analysis of AUCgluc did reveal inverse associations with cord blood DNAm at two CpG sites located within an exon of TXNIP (cg26974062 and cg02988288). In analyses stratified by GDM status, these associations were only observed among participants without GDM. Consistent with an interpretation that this association reflects an association with maternal hyperglycemia, we found that exposure to higher maternal FG, HbA1c and maternal type 1 diabetes was also nominally associated with a lower DNAm in TXNIP in (cord) blood. In addition, we found suggestive associations with liver gene expression and multiple metabolic traits.
TXNIP encodes for a thioredoxin-interacting protein involved in the regulation of glucose-sensing and redox processes. Several studies meta-analyzed by Walaszczyk et al. (33) have reported associations between blood DNAm at cg19693031 (also located in TXNIP) and lipid traits, type 2 diabetes, and prediabetes. Upon lookup in the results of our present meta-analysis, we observed evidence for associations between maternal AUCgluc and FG and cord blood DNAm levels for cg19693031, which is located downstream of cg26974062 and cg02988288. Furthermore, we found that the methylation at TXNIP was negatively associated with TXNIP gene expression in the liver, but not in skeletal muscle, further supporting the role of liver TXNIP as a future therapeutic target. In fact, a TXNIP inhibitor (SRI-37330) is currently under investigation as a therapeutic target for diabetes (34). We found both cg26974062 and cg02988288 to be associated with multiple cardiometabolic traits. To date, only one other study has reported an association for both probes, namely, with type 2 diabetes (35). Both probes are unique to the EPIC array; it is therefore possible that these associations were missed in previous studies, which have largely used the 450k array.
Interestingly, we observed a high level of heterogeneity for the associations between DNAm at cg26974062 and cg02988288 and maternal AUCgluc potentially due to a lack of association among participants with GDM. In included studies, the women with GDM were instructed to self-monitor their blood glucose, modify their diet and physical activity, and, if necessary, use pharmacologic agents aiming to normalize their blood glucose levels. Adequate glucose control can prevent GDM-associated pregnancy complications, but the effect of GDM treatment on long-term offspring health remains an unresolved question (36). We speculate that by moderating maternal hyperglycemia during the last trimester of pregnancy, GDM treatment may also influence the association between maternal AUCgluc and cord blood DNAm at TXNIP. Consistent with this hypothesis, it was reported that the associations between maternal glycemia during pregnancy and cord blood DNAm were attenuated as a result of UPBEAT where mothers were randomized to a lifestyle intervention during pregnancy (5). In this latter study, lower cord blood DNAm at cg26974062 was nominally associated with higher maternal 1-h glucose (with a direction of effect consistent with our AUCgluc meta-analysis) and cord blood DNAm at cg02988288 was associated with GDM; however, the association between GDM and cord blood DNAm at cg02988288 did not seem attenuated by the UPBEAT lifestyle intervention.
Our study has several limitations. First, while this collaborative effort is, to our knowledge, the largest inquiry on this topic to date, our sample size remains modest and may have been underpowered to detect some smaller associations against the null hypothesis (37). Second, our meta-analysis covered a small fraction of the known 28 million CpGs of the human epigenome. This limitation is somewhat remedied by the EPIC array, as it covers most known enhancers (26), which may be particularly sensitive to prenatal exposures (38). However, this array was used for only half of the cohorts. Another known limitation (37) is that we measured DNAm in (cord)blood and our results may be influenced by tissue heterogeneity and may not extrapolate to other tissues. Similarly, genetic variation may likewise influence DNAm. With the exception of cg21139325 (HLA-DQB2) (39), no genetic variation was reported to be associated with blood DNAm among the identified CpGs (in Table 2). Another important consideration is that nutritional status and maternal glucose levels as early as gestational weeks 4–12 have been associated with postnatal growth (40), and studies of prenatal famine exposure have indicated that early gestation is an especially sensitive window for remethylation, which happens during this period (37). We are unable to test the influence of gestational timing of maternal glycemic exposures. However, in a recent study with comparison of the association between early and late measures of maternal HbA1c during pregnancy and cord blood DNAm, no evidence was found for robust associations related to early pregnancy exposure (8).
In conclusion, our meta-analyses of maternal glycemic traits identified one sole exon of TXNIP at which higher maternal hyperglycemia, as reflected by higher AUCgluc (and to a lesser extend FG, type 1 diabetes, and HbA1c), was robustly associated with lower cord blood DNAm, and we found that these associations were attenuated in treated GDM pregnancies. We found little evidence for additional associations between maternal glucose and insulin levels during midpregnancy/late pregnancy and the cord blood methylome. In suggestive lookup analyses, TXNIP blood DNAm in childhood was similarly associated with prenatal exposure to maternal type 1 diabetes. TXNIP blood DNAm later in life was cross-sectionally associated with glycemic and anthropometric variables. Thus, future investigations of the links of in utero hyperglycemia exposure, DNAm at TXNIP, and cardiometabolic health across the life course are warranted.
Article Information
Acknowledgments. The authors thank the participants of the precision nutri-Epigenetic approach to tackle the PRECISion nutri-Epigenetic approach to tackle the mother-to-child transmission of impaired glucose metabolism (PREcisE) project for interesting discussion and feedback.
Duality of Interest. N.K. and Y.S.C. are part of an academic consortium that has received research funding from Abbott Nutrition, Nestec, and Danone. Y.S.C. has received reimbursement for speaking at conferences sponsored by companies selling nutritional products. No other potential conflicts of interest relevant to this article were reported.
These parties did not have any role in this research, the content of the manuscript, or the decision to publish.
Funding. E.W.T. was supported by a VENI grant from the Netherlands Organization for Scientific Research (91617128). This work was funded by the Joint Programming Initiative – A Healthy Diet for a Healthy Life (JPI HDHL) (proposal number 655). In the U.K. it is jointly funded by the Medical Research Council and the Biotechnology and Biological Sciences Research Council (grant MR/S03658X/1), in Spain by Instituto de Salud Carlos III (PCI2018-093147), in Germany by the German Federal Ministry of Education and Research (FKZ 01EA1905), in the Netherlands by ZonMw (529051023), and in France by French National Research Agency (ANR18-HDHL-0003-05). J.R. and S.S. received funding from the Healthy Diet for a Healthy Life (JPI HDHL) (PREcisE proposal no. 655) and the European Union's Horizon 2020 research and innovation program under grant agreement nos. 733206 (LifeCycle), 824989 (EUCAN-Connect), 874739 (Longitools), and 848158 (EarlyCause). Information regarding funding for the contributing cohorts can be found in Supplementary Material.
Author Contributions. The contributor roles taxonomy (CRediT) was as follows: E.W.T., C.G.H., M.-F.H., and S.S. contributed to study conceptualization. E.W.T. developed study methodology. E.W.T., D.L.J.-Q., J.R., and R.O. undertook the investigation. E.W.T., D.L.J.-Q., J.R., R.O., R.A., M.L.G., L.K.K., I.Y.L., G.P., J.T., A.P.S., and S.H. contributed to formal analysis. E.W.T. and D.L.J.-Q. performated the validation. I.Y.L., E.B., R.C., R.G., P.D.G., E.Ke., N.K., S.M., T.S.N., F.P., M.P., V.R., K.H.T., C.W., A.G.Z., I.A.-M., Y.S.C., D.D., J.F.F., B.H., V.W.V.J., J.L., M.V., A.B., P.F., S.H., E.Ka., M.-R.J., M.-F.H., M.C., A.K., J.K.Y.C., B.R., and S.S. provided (cohort) resources. E.W.T. and D.L.J.-Q. contributed to meta-analysis data curation. E.W.T. and S.S. contributed to writing the original draft of the manuscript. All authors contributed to review and editing of the manuscript. E.W.T. undertook the visualization. M.-R.J., C.G.H., M.-F.H., and S.S. contributed to study supervision. E.W.T., E.T., M.-R.J., C.G.H., M.-F.H., R.P.M.S.-T., and S.S. contributed to project administration. E.W.T., P.P., R.G., P.D.G., N.K., T.S.N., K.R., A.G.Z., I.A.-M., L.B., Y.S.C., D.D., J.F.F., B.H., V.W.V.J., J.L., M.V., S.H., E.Ka., M.-R.J., M.-F.H., R.P.M.S.-T. and S.S. contributed to funding acquisition. E.W.T. and S.S. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
C.G.H., M.-F.H., and S.S. made equal contributions.
This article contains supplementary material online at https://doi.org/10.2337/figshare.17209262.
References
- 1. Metzger BE, Lowe LP, Dyer AR, et al.; HAPO Study Cooperative Research Group . Hyperglycemia and adverse pregnancy outcomes. N Engl J Med 2008;358:1991–2002 [DOI] [PubMed] [Google Scholar]
- 2. Metzger BE, Lowe LP, Dyer AR, et al.; HAPO Study Cooperative Research Group . Hyperglycemia and Adverse Pregnancy Outcome (HAPO) Study: associations with neonatal anthropometrics. Diabetes 2009;58:453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vääräsmäki M, Pouta A, Elliot P, et al. Adolescent manifestations of metabolic syndrome among children born to women with gestational diabetes in a general-population birth cohort. Am J Epidemiol 2009;169:1209–1215 [DOI] [PubMed] [Google Scholar]
- 4. Hjort L, Novakovic B, Grunnet LG, et al. Diabetes in pregnancy and epigenetic mechanisms-how the first 9 months from conception might affect the child’s epigenome and later risk of disease. Lancet Diabetes Endocrinol 2019;7:796–806 [DOI] [PubMed] [Google Scholar]
- 5. Antoun E, Kitaba NT, Titcombe P, et al.; UPBEAT Consortium . Maternal dysglycaemia, changes in the infant’s epigenome modified with a diet and physical activity intervention in pregnancy: secondary analysis of a randomised control trial. PLoS Med 2020;17:e1003229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geurtsen ML, Jaddoe VWV, Gaillard R, Felix JF. Associations of maternal early-pregnancy blood glucose and insulin concentrations with DNA methylation in newborns. Clin Epigenetics 2020;12:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Canouil M, Khamis A, Keikkala E, et al. Epigenome-wide association study reveals methylation loci associated with offspring gestational diabetes mellitus exposure and maternal methylome. Diabetes Care 2021;44:1992–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Juvinao-Quintero DL, Starling AP, Cardenas A, et al. Epigenome-wide association study of maternal hemoglobin A1c in pregnancy and cord blood DNA methylation. Epigenomics 2021;13:203–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Howe CG, Cox B, Fore R, et al. Maternal gestational diabetes mellitus and newborn DNA methylation: findings from the Pregnancy and Childhood Epigenetics consortium. Diabetes Care 2020;43:98–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang C, Wei Y, Sun W, Yang H. The area under the curve (AUC) of oral glucose tolerance test (OGTT) could be a measure method of hyperglycemia in all pregnant women. Open J Obstet Gynecol 2019;09:186–195 https://www.scirp.org/journal/paperinformation.aspx?paperid=90450 [Google Scholar]
- 11. Ziegler AG, Meier-Stiegen F, Winkler C; TEENDIAB Study Group . Prospective evaluation of risk factors for the development of islet autoimmunity and type 1 diabetes during puberty--TEENDIAB: study design. Pediatr Diabetes 2012;13:419–424 [DOI] [PubMed] [Google Scholar]
- 12. Rantakallio P. The longitudinal study of the Northern Finland birth cohort of 1966. Paediatr Perinat Epidemiol 1988;2:59–88 [DOI] [PubMed] [Google Scholar]
- 13. Margerie D, Lefebvre P, Raverdy V, et al. Hepatic transcriptomic signatures of statin treatment are associated with impaired glucose homeostasis in severely obese patients. BMC Med Genomics 2019;12:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soh S-E, Tint MT, Gluckman PD, et al.; GUSTO Study Group . Cohort profile: Growing Up in Singapore Towards healthy Outcomes (GUSTO) birth cohort study. Int J Epidemiol 2014;43:1401–1409 [DOI] [PubMed] [Google Scholar]
- 15. Guillemette L, Allard C, Lacroix M, et al. Genetics of Glucose regulation in Gestation and Growth (Gen3G): a prospective prebirth cohort of mother-child pairs in Sherbrooke, Canada. BMJ Open 2016;6:e010031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Starling AP, Liu C, Shen G, et al. Prenatal exposure to per- and polyfluoroalkyl substances, umbilical cord blood DNA methylation, and cardio-metabolic indicators in newborns: the Healthy Start study. Environ Health Perspect 2020;128:127014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Keikkala E, Mustaniemi S, Koivunen S, et al. Cohort profile: the Finnish Gestational Diabetes (FinnGeDi) study. Int J Epidemiol 2020;49:762–763g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Girchenko P, Lahti M, Tuovinen S, et al. Cohort profile: Prediction and Prevention of Preeclampsia and Intrauterine Growth Restriction (PREDO) study. Int J Epidemiol 2017;46:1380–1381g [DOI] [PubMed] [Google Scholar]
- 19. Heude B, Forhan A, Slama R, et al.; EDEN mother-child cohort study group . Cohort profile: the EDEN mother-child cohort on the prenatal and early postnatal determinants of child health and development. Int J Epidemiol 2016;45:353–363 [DOI] [PubMed] [Google Scholar]
- 20. Janssen BG, Madhloum N, Gyselaers W, et al. Cohort profile: the ENVIRonmental influence ON early AGEing (ENVIRONAGE): a birth cohort study. Int J Epidemiol 2017;46:1386–1387m [DOI] [PubMed] [Google Scholar]
- 21. Kooijman MN, Kruithof CJ, van Duijn CM, et al. The Generation R Study: design and cohort update 2017. Eur J Epidemiol 2016;31:1243–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeileis A. Object-oriented computation of sandwich estimators. J Stat Softw 2006;16:1–16 [Google Scholar]
- 23. Salas LA, Koestler DC, Butler RA, et al. An optimized library for reference-based deconvolution of whole-blood biospecimens assayed using the Illumina HumanMethylationEPIC BeadArray. Genome Biol 2018;19:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fortin JP, Triche TJ Jr, Hansen KD. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2017;33:558–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van der Most PJ, Küpers LK, Snieder H, Nolte I. QCEWAS: automated quality control of results of epigenome-wide association studies. Bioinformatics 2017;33:1243–1245 [DOI] [PubMed] [Google Scholar]
- 26. Pidsley R, Zotenko E, Peters TJ, et al. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 2016;17:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Iterson M, van Zwet EW; BIOS Consortium . Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol 2017;18:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Viechtbauer W. Conducting meta-analyses in R with the metafor. J Stat Softw 2010;36:1–48 [Google Scholar]
- 29. Battram T, Yousefi P, Crawford G, et al. The EWAS Catalog: a database of epigenome-wide association studies. 2 February 2021 [preprint]. OSF Preprints. Available from https://osf.io/837wn/ [DOI] [PMC free article] [PubMed]
- 30. Xu Z, Xie C, Taylor JA, Niu L. ipDMR: identification of differentially methylated regions with interval P-values. Bioinformatics 2021;37:711–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Peters TJ, Buckley MJ, Statham AL, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 2015;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reinius LE, Acevedo N, Joerink M, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One 2012;7:e41361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walaszczyk E, Luijten M, Spijkerman AMW, et al. DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case-control sample of the Lifelines study. Diabetologia 2018;61:354–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thielen LA, Chen J, Jing G, et al. Identification of an anti-diabetic, orally available small molecule that regulates TXNIP expression and glucagon action. Cell Metab 2020;32:353–365.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Albao DS, Cutiongco-de la Paz EM, Mercado ME, et al. Methylation changes in the peripheral blood of Filipinos with type 2 diabetes suggest spurious transcription initiation at TXNIP. Hum Mol Genet 2019;28:4208–4218 [DOI] [PubMed] [Google Scholar]
- 36. Hartling L, Dryden DM, Guthrie A, Muise M, Vandermeer B, Donovan L. Benefits and harms of treating gestational diabetes mellitus: a systematic review and meta-analysis for the U.S. Preventive Services Task Force and the National Institutes of Health Office of Medical Applications of Research. Ann Intern Med 2013;159:123–129 [DOI] [PubMed] [Google Scholar]
- 37. Heijmans BT, Mill J. Commentary: the seven plagues of epigenetic epidemiology. Int J Epidemiol 2012;41:74–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tobi EW, Goeman JJ, Monajemi R, et al. DNA methylation signatures link prenatal famine exposure to growth and metabolism. Nat Commun 2014;5:5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Min JL, Hemani G, Hannon E, et al.; BIOS Consortium . Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet 2021;53:1311–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dong L, Liu E, Guo J, et al. Relationship between maternal fasting glucose levels at 4-12 gestational weeks and offspring growth and development in early infancy. Diabetes Res Clin Pract 2013;102:210–217 [DOI] [PubMed] [Google Scholar]