Abstract
Background: Neurotrophic tyrosine receptor kinase (NTRK) fusion has been detected in rare types of CNS tumours, which can promote tumorigenesis. The efficacy of Trk inhibitor became a significant therapeutic interest. Our aim was to investigate whether Pan-Trk immunohistochemistry (IHC) is a reliable and efficient marker for detecting NTRK-fusion in different brain tumours.
Methods: This study included 23 patients diagnosed with different types of CNS tumours. Testing for Pan-Trk IHC with monoclonal Ab (EPR17341) has been performed on all FFPE tissues. Parallelly, NTRK-rearrangements were tested using both DNA and RNA-based next-generation sequencing (NGS) assay using TruSight Onco500 platform.
Results: The cohort included eight pilocytic astrocytomas, one oligodendroglioma, six IDHwildtype glioblastomas, four IDHmutant grade four astrocytomas, and one sample of each (astroblastoma, central neurocytoma, medulloblastoma, and liponeurocytoma). The mean age was 35 years; seven cases were in the paediatric age group, and 16 were adult. Pan-Trk expression was detected in 11 (47.8%) tumours, and 12 (52.1%) tumours showed no Pan-Trk expression. Nine Cases (82%) with different Pan-Trk expressions did not reveal NTRK-rearrangement. The other two positively expressed cases (liponeurocytoma and glioblastoma) were found to have NTRK2-fusions (SLC O 5A1-NTRK2, AGBL4-NTRK2, BEND5-NTRK2). All the 12 cases (100%) with no Pan-Trk expression have shown no NTRK-fusions. There was no statistically significant association between Pan-Trk expression and NTRK-fusion (p = 0.217). The detection of NTRK- fusions using NGS had high specificity over NTRK-fusion detection by using Pan-Trk IHC.
Conclusion: Pan-Trk IHC is not a suitable tissue-efficient biomarker to screen for NTRK-fusions in CNS tumours, however RNA-based NGS sequencing should be used as an alternative method.
Keywords: immunohistochemistry, next-generation sequencing, CNS tumours, NTRK-fusions, Pan-Trk, TruSight Oncology500
Introduction
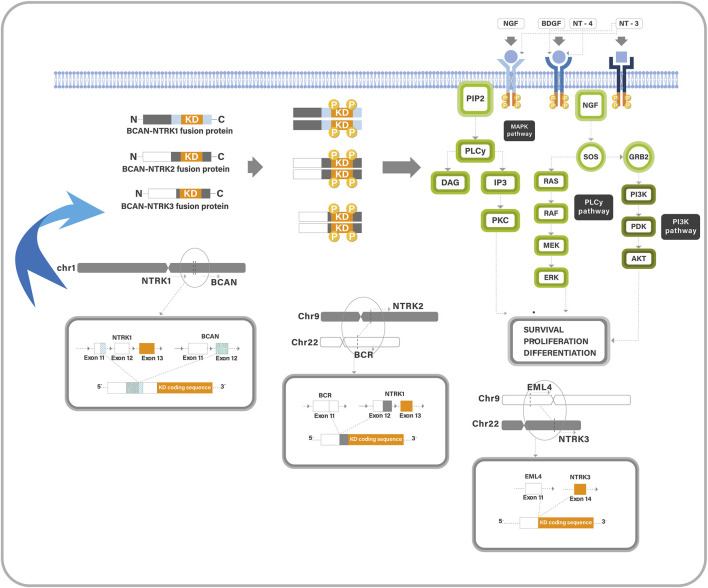
The neurotrophic tyrosine receptor kinase (NTRK) is a family member of three genes (NTRK1, NTRK2, NTRK3), which produce tyrosine kinase (TK) proteins (Trk-A, Trk-B, and Trk-C) (1). Their receptors are highly expressed in neural tissue, in which they play an important role in neuronal development, proliferation, synaptic plasticity, and cognition and memory (2). They have a similar structure; each consists of an extracellular ligand-binding domain, a transmembrane region, and an intracellular kinase domain. Each receptor has a ligand that it prefers; Trk-A has the highest affinity for neurotrophin nerve growth factor, Trk-B for brain-derived neurotrophic factor and neurotrophin-4, and Trk-C for neurotrophin-3 (1). Normally, ligands binding to the extracellular region may stimulate the kinase domain of the Trk receptor, resulting in homodimerization, phosphorylation, and activation of signaling pathways (Figure 1). Rearrangements in the NTRK gene can result in two genes fusing at the C-terminal TK-domain with N-terminal fusion partner producing altered Trk proteins. This fusion may lead to uncontrolled growth of tumour cells (1). Fusions involving the NTRK genes can be oncogenic drivers, leading to abnormal Trk receptor dimerization and constitutive activation of Trk pathways, resulting in upregulation and apoptosis resistance (2).
FIGURE 1.
A diagram describes the Trk pathway and the oncogenic mechanism of NTRK-fusions. Trk proteins contain an intracellular TK domain which promotes cell proliferation through MAPK/ERK, PLCg/PKC, and PI3K/AKT pathways. Trk-fusion proteins have a complete TK domain, and the partner gene is expressed in a homodimer, which induces ligand-independent activation of the TK domain. It also activates the cancer-associated pathways.
The prevalence of NTRK-fusion is less than 1% of all tumors. It can be found in approximately 1,500–5,000 children and adults with cancers annually (3). It has been identified in a broad range of solid tumours in adults, including salivary gland cancer, thyroid cancer, breast cancer, gastrointestinal cancers, gynecological cancers, non-small cell carcinoma of lung, and some soft tissue sarcomas (3–5). In children, these fusions have been identified in rare cases of diffuse CNS gliomas as well as in melanoma, soft-tissue sarcomas, inflammatory myofibroblastic tumours, congenital infantile fibrosarcoma, and mesoblastic nephroma (4). About 0.55–2% of all neuroepithelial tumours and gliomas contain NTRK-fusions (2).
Hechtman et al. detected four of 23 glioblastomas with NTRK-fusion (6). Torre et al. tested NTRK-fusions in 42 patients with CNS glioma of different ages. NTRK-fusion was detected in CNS gliomas of the infants (7 cases), adult cases (22 cases), and pediatric cases >1 year (13 cases). Most pediatric cases had NTRK2-fusions (69%, 9 cases), while NTRK1-fusions were found in most adult glioma cases (68%, 15 cases) (2).
Due to the efficacy of food and drug administration (FDA) approved NTRK targeted therapy in non-CNS cancers, it is important to identify patients with NTRK-fusion driven brain tumours using the accurate technique (1, 2). NTRK-fusions are generally detected at the molecular level, using fluorescent in-situ hybridization technique (FISH) or preferably a next-generation sequencing (NGS) of targeted DNA or RNA testing. However, molecular studies are still costly and time-consuming, and technical risks such as nucleic acid degradation might arise. Alternatively, using the immunohistochemistry (IHC) technique, Pan-Trk staining is typically less expensive, has a rapid turnaround time, and is more tissue-efficient for NTRK-fusion detection (6). Pan-Trk IHC expression was commonly used in non-CNS cancers because of its high specificity and sensitivity. Solomon et al. found that Pan-Trk was strongly expressed in lung, pancreatic, colorectal, thyroid, and biliary carcinomas (1). The specificity decreased in salivary gland and breast carcinomas as well as soft tissue sarcomas with neural or smooth muscle differentiation (1). Hechtman et al. has detected some glioblastomas with NTRK-fusions, which were compatible with the IHC staining and Archer RNA test (6). It was concluded that the sensitivity of IHC with Pan-Trk in their cohort was 95%, and the specificity was 100% for NTRK-fusions (6). At the same time, Solomon et al. found that the sensitivity for NTRK1 was 96% and for NTRK2 was 100%, while sensitivity for NTRK3 fusions was 79% (7). It was clear that Pan-Trk staining variability correlates with fusion partners.
Our study assessed Pan-Trk expression using IHC technique in different types of CNS tumours, and we tested its specificity, sensitivity, and accuracy with NGS technique. In addition, we explored if Pan-Trk expression can be used as a reliable biomarker immunolabeling to detect NTRK-fusions in CNS tumours.
Materials and Methods
Patients’ Stratification
This study included 23 patients, aged between 3 and 64 years and histologically diagnosed as eight cases of pilocytic astrocytomas, one case of oligodendroglioma, six cases IDHwildtype glioblastomas, four cases IDHmutant WHO grade four astrocytomas, and one case of each (astroblastoma, central neurocytoma, medulloblastoma, and liponeurocytoma) (Table 1). The study was approved by the National Biomedical Ethics Committee at King Abdulaziz University (HA-02-J-008) under a general ethical report. Patients’ clinical data were retrieved from hospital records and included patients ‘age at diagnosis, gender, tumour location and type, 2021 WHO grading, and IDH1 mutational status (Table 1). The histological diagnosis was made based on 2021 WHO classification of CNS tumours (8, 9).
TABLE 1.
Demographic data of the 23 patients with CNS tumours, including NGS findings of DNA-based and RNA-based mutations.
| Age | Gender | Location | Tumour | Grade | PanTrk | LI (%) | DNA-based mutation | RNA-based mutation | TMB | MSI | NTRK | IDH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 28 | Male | Frontal | Liponeurocytoma | II | Diffuse | 90 | None | SLCO5A1-NTRK2 | low | stable | Detected | Not done |
| 10 | Male | Posterior fossa | Pilocytic astrocytoma | I | Diffuse | 90 | BRAFV600E/TP53 | None | medium | none | Not detected | Not done |
| 54 | Male | Parietal | Astrocytoma | IV | No | 0 | IDH1/ATRX/TP53/BCOR/PTCH1 | EGFR amplification | medium | stable | Not detected | mutant |
| 33 | Male | Frontal | Oligodendroglioma | III | No | 0 | MET | None | medium | stable | Not detected | mutant |
| 40 | Male | Temporal | Astroblastoma | None | No | 0 | BRAFV600 E | None | low | stable | Not detected | wildtype |
| 8 | Male | Posterior fossa | Pilocytic astrocytoma | I | No | 0 | None | AUTS2-BRAF/PRKAR2B-BRAF | low | stable | Not detected | Not done |
| 52 | Male | Frontal | Glioblastoma | IV | Partial | 40 | TERT/PTEN/FGFR4 | AGBL4-NTRK2/BEND5-NTRK2 | low | stable | Detected | wildtype |
| 53 | Male | Frontal | Glioblastoma | IV | No | 0 | TP53 | None | high | stable | Not detected | wildtype |
| 4 | Female | Spinal | Pilocytic astrocytoma | I | No | 0 | None | None | medium | none | Not detected | Not done |
| 62 | Male | Parietal | Glioblastoma | IV | No | 0 | BRAFV600E/TP53/APC | None | low | stable | Not detected | wildtype |
| 53 | Male | Temporal | Pilocytic astrocytoma | I | Focal | 10 | TERT/ | EGFR amplification/CD4-6 gain | low | stable | Not detected | wildtype |
| 45 | Female | Parietal | Glioblastoma | IV | Diffuse | 85 | PTEN | CDK6 gain/EGFR amplification | low | stable | Not detected | wildtype |
| 40 | Male | Posterior fossa | Pilocytic astrocytoma | I | Focal | 8 | BRAFV600 E | None | medium | stable | Not detected | Not done |
| 6 | Female | Cerebellar | Medulloblastoma | IV | Diffuse | 90 | PTCH1 | CDK6 gain | low | stable | Not detected | Not done |
| 3 | Female | Frontal | Pilocytic astrocytoma | I | Focal | 10 | None | None | medium | stable | Not detected | Not done |
| 36 | Male | Lateral ventricle | Central neurocytoma | II | No | 0 | None | None | low | stable | Not detected | Not done |
| 45 | Male | Temporal | Glioblastoma | IV | No | 0 | TERT | None | medium | stable | Not detected | wildtype |
| 12 | Female | Posterior fossa | Pilocytic astrocytoma | I | No | 0 | FGFR1 | None | low | stable | Not detected | Not done |
| 14 | Male | Hypothalamic | Pilocytic astrocytoma | I | No | 0 | BARD1 | KIAA1549-BRAF | low | stable | Not detected | Not done |
| 64 | Female | Temporal | Glioblastoma | IV | Focal | 10 | ATRX/TERT | None | medium | none | Not detected | wildtype |
| 51 | Female | Frontal | Astrocytoma | IV | Partial | 45 | PTEN | None | high | stable | Not detected | mutant |
| 60 | Male | Temporal | Astrocytoma | IV | Partial | 40 | IDH1 | None | medium | none | Not detected | mutant |
| 51 | Female | Parietal | Astrocytoma | IV | Non | 0 | IDH1 | None | high | stable | Not detected | mutant |
LI, labelling index; TMB, tumour-burden; MSI, microsatellite instability.
Tumour Samples
Archival routine formalin-fixed and paraffin-embedded (FFPE) tumour tissues were collected from 23 patients with different CNS tumours. Haematoxylin and Eosin (H&E)-stained sections were re-examined by a certified neuropathologist (MK) to confirm that the histopathological diagnosis was made based on 2021 WHO classification (8, 9). One unstained positive-charged slide from each of 23 FFPE tissue blocks was prepared for Pan-Trk immunostaining.
Immunohistochemistry Technique
IHC Protocol
4-μm FFPE tissue sections were used in the process of IHC. The IHC assay was performed using anti-Pan-Trk antibody (clone EPR#17341, rabbit monoclonal antibody, Abcam, Cat# Ab181560). The procedure was performed with the ultraView DAB detection Kit (Ventana) on a BenchMark XT automated stainer from Ventana (Tucson, AZ, United States). A protocol was established so that the entire assay procedure consisted of deparaffinization with EZ Prep at 75°C, heat pre-treatment in cell conditioning medium (Ag unmasking) (CC1; Ventana) for 60 min and then primary incubation for 16 min at 37°C. The antibodies were optimized using a dilution of 1:50. The slides were counterstained with hematoxylin II and bluing reagent for 16 min. After that, the slides were removed from the slide stainer and then immersed into successive alcohol buffers at different concentrations for 3 min.
IHC Assessment
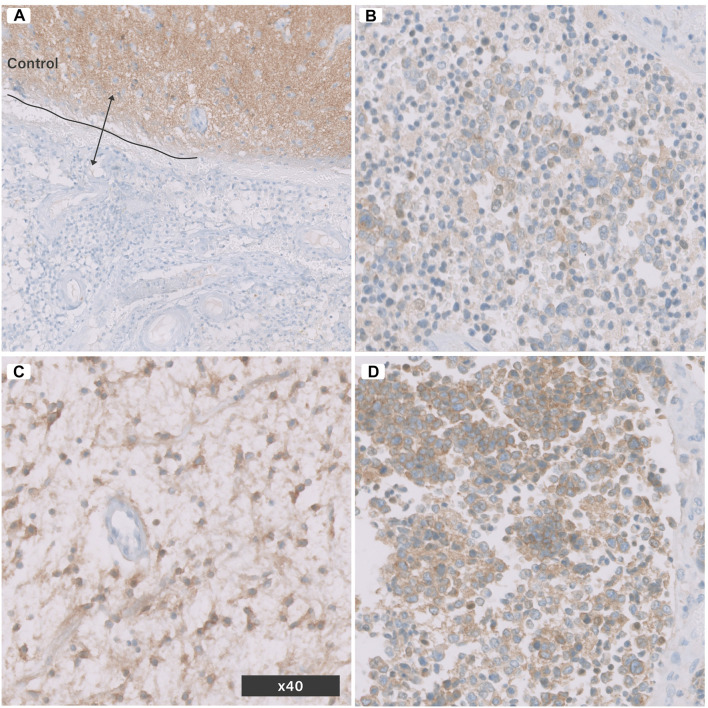
Anti Pan-Trk antibody normally stains neuropil background (Figure 2A). Each tissue section was screened at a low power field (×10) using digital microscopy, and a single hot spot of a non-necrotic area was selected to count the cells manually at a high-power field (×40). Cells expressing Pan-Trk were considered positive (stained-tumour cells), while the total cells included both stained tumour cells and non-stained tumour cells. The non-stained tumour cells included lymphocytes and other types of glial cells. The labeling index was assessed using the following equation:
The staining pattern was then categorized as i) diffusely expressed, ii) partially expressed, iii) focally expressed, and iv) non-expressed (Figures 2B–D; Table 2). The quantitative method used in the current study was typical of the method described by Kurdi et al (10).
FIGURE 2.
Pan-Trk Expression (nuclear or cytoplasmic) in brain tumours using IHC. (A) control normal brain tissue and tumour tissue negative for Pan-Trk, (B) focal expression, (C) partial expression, (D) diffuse expression. All images are in (×40) magnification.
TABLE 2.
Quantitative expression of Pan-Trk in tumour cells using a digital microscope.
| Expression | Labelling index (%) |
|---|---|
| No expression | 0 |
| Focal expression | >0–20 |
| Partial expression | >20–50 |
| Diffuse expression | >50 |
For statistical analysis, the scores were divided by 100.
Next-Generation Sequencing
NTRK rearrangement was detected by DNA-based or RNA-based NGS performed at the time of research investigation. FFPE samples were collected from 23 patients diagnosed with different histopathological diagnoses. DNA and RNA were extracted using QIAMP DNA FFPE kit and RNeasy FFPE kit, respectively. DNA and RNA quality was estimated by a Nanodrop 2000 spectrophotometer (Thermo Scientific, United States), with an OD 260/280 value between 1.8 and 2.0. Qubit (Invitrogen, United States) was used to quantify nucleic acids. The minimum accepted DNA/RNA input was 50 ng. Trusight Oncology 500 (TSO500R) high throughoutput library preparation kit (Illumina, United States) using Nextseq 550, for screening 500 gene variants including single nucleotide variants (SNVs), fusions, splice variants, copy number variants (CNVs), microsatellite instability (MSI) and tumor mutational burden (TMB). (Figures 3A,B). An enrichment-based technology was used to perform library preparation for both DNA and RNA (www.illumina.com/tso500). Genomic DNA (gDNA) sample quality was assessed using Illumina FFPE QC. Next, gDNA was sheared to 90–250 bp. The fragmentation of gDNA was optimized using ME220 Focused-ultrasonicator (Covaris, United States). End repair and A-Tailing was performed on sheared gDNA samples. RNA sample integrity was evaluated via Agilent Technologies, 2100 Bioanalyzer, using Agilent RNA 6000 Nano kit (Agilent). RNA samples were denatured and primed to synthesize complementary DNA (cDNA). UMI1 adapters containing unique indexes were ligated to DNA fragments. Short Universal Adapters 1 (SUA1) were ligated to cDNA fragments. After that, ligated fragments were purified using sample purification beads (SPB). To allow up to eight libraries to be pooled and sequenced together, unique indexing primers were added to purified gDNA and cDNA fragments to be amplified in preparation for sample multiplexing. Following TSO 500 protocol, two hybridization steps were performed. During the first step, a pool of oligos specific to 523 genes were hybridized to DNA libraries, while a pool of oligos specific to 55 genes hybridized to RNA libraries. Then, probes hybridized to the targeted regions were captured using streptavidin magnetic beads (SMB). The second hybridization step was performed to ensure specificity of captured regions. A pool of primers was used to amplify enriched libraries. The libraries were then sequenced on the illumina Nextseq 550 platform. The run data were uploaded to the Clinical Genomics Workbench (PierianDx, France). QC analysis, mapping to hg19, variant calling, and annotation were all performed (Figure 3C). The extraction, validation, hybridization, library preparation and genomic sequencing were all performed at CAP-accredited center of excellence of genomic medicine research at King Abdulaziz University.
FIGURE 3.
Next-generation sequencing (NGS) assay. (A) TruSight Oncology500 workflow from illumina integrates into lab workflows, going from nucleic acids to a variant calls in 3–4 days. (www.illumina.com/tso500), (B) TSO500 analyzes 500 cancer-relevant genes from both DNA and RNA in one integrated workflow. The assays assess multiple variant types (SNVs, indels, CNVs, splice variants, fusions, and emerging biomarkers that rely on analysis of multiple genomic loci, such TMB and MSI, (C) Clinical report generated by PierianDx after the FASTQ files and VCF being uploaded to the Clinical Genomics Workbench (PierianDx, France).
Statistical Methods
The data were described as frequencies and percentages. Fisher’s exact test was used to test the significant relationship between Pan-Trk expression and NTRK-fusions results. p-value less than 0.05 was considered significant. All statistical analyses were performed using the IBM SPSS1 ver. 24 and R-Package statistical software programs (“Circlize” version is 0.4.13).
Results
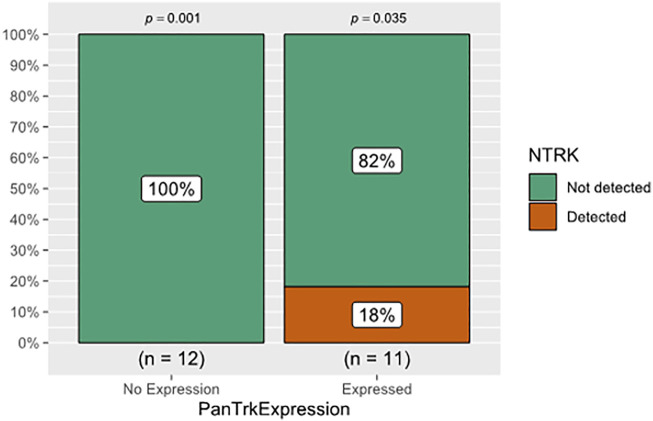
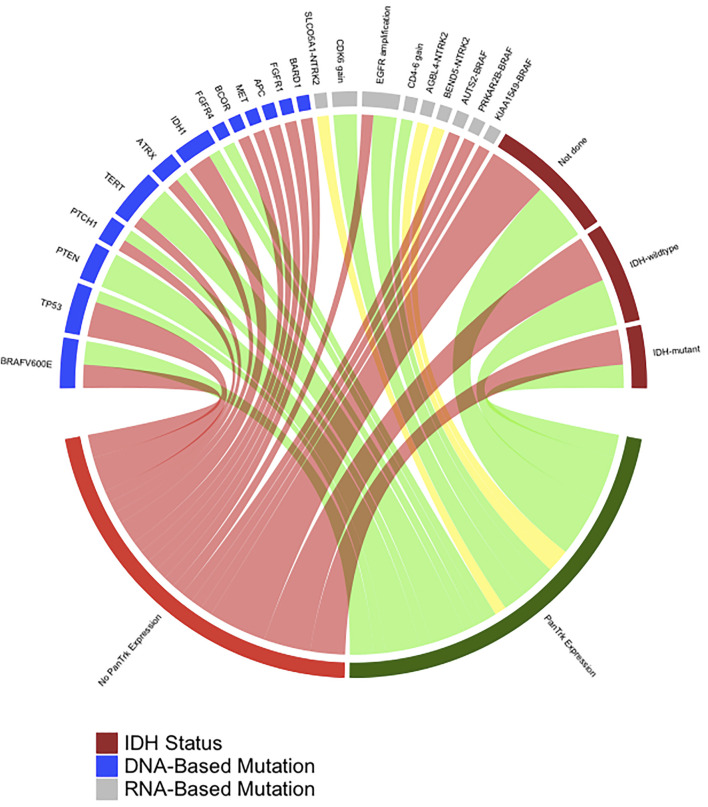
The cohort included 23 patients diagnosed with different types of CNS tumours (pilocytic astrocytomas (n = 8), oligodendroglioma (n = 1), IDHwildtype glioblastomas (n = 6), IDHmutant WHO grade 4 astrocytomas (n = 4), astroblastoma (n = 1), central neurocytoma (n = 1), medulloblastoma (n = 1) and liponeurocytoma (n = 1) (Table 1). The mean age: 35.8 years (±20.7 years); seven cases were in the paediatric-age group, and 16 cases were adult; 15 males (65.2%) and eight females (34.8%). Approximately 26.1% (n = 6) of the tumours were in the frontal lobe followed by the temporal lobe (21.7%, n = 5), parietal lobe (17.4%, n = 4), posterior fossa (21.7%, n = 5), and one case for each (lateral ventricle, hypothalamic, and spinal cord) (Table 1). Pan-Trk expression was detected in 11 tumours (47.8%) and 12 tumours (52.1%) showed no Pan-Trk expression. The Pan-Trk expressed tumours (n = 11) were clustered into diffuse expression (17.4%, n = 4), partial expression (13%, n = 3) and focal expression (17.4%, n = 4). Two of these cases (18%) (liponeurocytoma, glioblastoma) with Pan-Trk expression (diffuse, partial) were found to have NTRK2-fusions (SLC O 5A1-NTRK2, AGBL4-NTRK2, BEND5-NTRK2) and these cases were adult. Additionally, one of those cases (glioblastoma) was found to have DNA-based mutations (PTEN, TERT-promoter, and FGRF4). On the other hand, the 9 Cases (82%) with different Pan-Trk expressions did not reveal any NTRK-fusions. 100% (n = 12) of the tumours with no Pan-Trk expression have shown no NTRK-fusions (Figure 4, 5).
FIGURE 4.
The relationship between Pan-Trk expression using IHC and NTRK-fusions detection using NGS. Pan-Trk expression was detected in 11 tumours and 12 tumours showed no Pan-Trk expression. Only two of the expressed cases (18%) were found to have NTRK2-fusions, and the remaining 9 Cases (82%) did not reveal any NTRK-fusions. The 12 cases with no Pan-Trk expression showed no NTRK-fusions. There was no statistically significant association between IHC and NGS in detecting NTRK-fusion (p > 0.05).
FIGURE 5.
A diagram was generated by R-package, which shows the association between Pan-Trk expression with NTRK and non-NTRK fused tumours. In addition, it shows other detected DNA-based and RNA-based mutations by TruSight Onco500 through NGS.
There was no statistically significant association between Pan-Trk expression using IHC and NTRK-fusion using NGS (p = 0.217) (Table 3; Figure 4). This clarifies that Pan-Trk expression does not always correlate with the presence of demonstrable NTRK-fusions. The sensitivity of Pan-Trk IHC relative to NGS to detect NTRK-fusion was relatively high (100%) (two positive cases were detected by Pan-Trk IHC). However, the ability to detect negative cases by Pan-Trk IHC was observed to be 57.1%. Overall diagnostic accuracy of the PanTrk in detecting NTRK-fusions was 60.9% in the group with prevalence of 8.7% (Table 4).
TABLE 3.
The relationship between Pan-Trk expression using IHC and NTRK-fusion detection by NGS.
| Dependent: PanTrk expression | No expression | Expressed | Total | p-value | |
|---|---|---|---|---|---|
| NTRK-fusion | Detected | 0 (0.0) | 2 (18.2) | 2 (8.7) | 0.217 a |
| Not detected | 12 (100.0) | 9 (81.8) | 21 (91.3) |
Fisher’s Exact Test.
TABLE 4.
Sensitivity, specificity, and diagnostic accuracy of using NGS method over Pan-Trk IHC to detect NTRK-fusions.
| NTRK-fusion | No NTRK fusion | Total |
|---|---|---|
| Expressed-PanTrk | 2 | 11 |
| No PanTrk expression | 0 | 12 |
| Total | 2 | 23 |
| Ratios | ||
| Sensitivity | 100% | |
| Specificity | 57.1% | |
| Accuracy | 60.9% | |
| Prevalence | 8.7% | |
| Positive predictive value | 18.2% | |
| Negative predictive value | 100% | |
| Post-test disease Probability | 18.02% |
Discussion
Although NTRK-fusions have been detected in a small number of pediatric and adult tumour types, they have also been identified in other common cancers at lower frequencies. These findings suggest that a diagnostic strategy managed by NTRK-fusions biological incidence may be the best effective approach to identify patients with NTRK-fusions. Furthermore, such NTRK-fusions have now been shown to be actionable genomic signatures, predicting therapeutic responses against Trk receptors, making their detection an evolving clinical priority (11).
Screening of NTRK-fusions is usually performed at molecular level using NGS or FISH technique (12), DNA or RNA targeted testing. However, the molecular technique is costly, time-consuming, and unavailable in most centers, and sometimes associated with sampling errors due to nucleic acid degradation. Alternatively, anti-Pan-Trk (IHC) is commonly used to examine protein expression. The clone reacts with the C-terminus of Trk-A, -B, and–C and is therefore reactive with gliomas harbouring NTRK-fusions. IHC is associated with limited costs and a fast turnaround time, allowing good histological correlations and protein expression validation. The major limitation is that the antibody is restricted to Trk receptors’ wildtype epitopes, thus not specific to detect NTRK-fusions. They do not provide additional information about the fusion partner. Instead, FISH technique was found more useful than Pan-Trk IHC as a screening tool to detect NTRK-fusion prior to RNA sequencing (12).
Pan-Trk IHC can be used as an effective screening tool for most cancers. Hechtman et al. tested 23 cases of non-CNS carcinomas with NTRK-fusions, in which 16 cases showed positive fusion transcript with Archer fusion and six of them were novel rearrangements (6). The 20 cases stained positively with Pan-Trk were concordant with Archer RNA, two cases with NTRK rearrangements showed negative in both Archer fusion and IHC. A single case of a fusion-positive colorectal carcinoma with an ETV6-NTRK3 fusion was discordant with IHC, which showed no expression (6). This specificity and sensitivity were found low in CNS neoplasms due to the physiological expression of Pan-Trk receptors in normal CNS neuropil (Figures 2A– Control). Solomon et al. also reported an unsatisfactory specificity value of 20.8% in CNS gliomas (7). FISH showed better results than Pan-Trk in CNS tumours, particularly gliomas. RNA sequencing analyses are necessary in FISH positive cases with less than 30% positive nuclei, to avoid false positivity when scoring is close to the detection threshold (12).
Our results showed that Pan-Trk expression was detected in 11 tumours (47.8%) and 12 tumours (52.1%) showed no Pan-Trk expression. Out of the 11 cases, nine cases (82%) did not reveal any NTRK-rearrangement, while two cases were found to have NTRK2-fusions (SLC O 5A1-NTRK2, AGBL4-NTRK2, BEND5-NTRK2) (Figures 4, 5). The rest of the 12 cases with no Pan-Trk expression showed no NTRK-fusion. Our results also showed that NGS is the best molecular method to detect NTRK-fusions with 100% specificity compared to Pan-Trk IHC, which showed low specificity (Table 4). This is likely related to the normal physiological expression of Trk protein receptors in normal brain tissue, which may falsely predict NTRK-fusions. Moreover, using TruSightOnco500 platform replaced IHC and other molecular methods to detect a wide range of DNA-based and RNA-based mutations.
Finally, one limitation must be acknowledged in our study is, that the total number of cases analyzed for NTRK-fusions and Pan-Trk IHC is relatively low. Despite this limitation and to our best knowledge, this is the first study, globally and particularly in Saudi Arabia, that investigate the tyrosine kinases biomarkers in different CNS tumours, reflecting the accuracy of diagnostic technique on patient management.
Conclusion
Pan-Trk IHC is not a suitable tissue-efficient biomarker to screen for NTRK-fusions in CNS tumours. Its usage should be with extreme caution, and its confirmation by other techniques is warranted. RNA-based NGS sequencing should be used as an alternative method to detect NTRK-fusions. TruSightOnco500 is a wide-genomic platform that can replace IHC and other molecular techniques to screen for DNA and RNA-based mutation using FFPE tissue.
Acknowledgments
Thanks to Dr. Nadeem S. Butt (Department of Family Medicine and Community, Faculty of Medicine, Rabigh, Saudi Arabia) for statistical analysis. We also thank Dr. Nouf Alghanmi from the Bioinformatics Unit at the Center of Excellence in Genomic Medicine Research in King Abdulaziz University for R-package statistical diagram.
Data Availability Statement
The original contributions and data presented in the study are available upon request from the corresponding author.
Ethics Statement
This study was approved by the National Biomedical Ethics Committee of King Abdulaziz University (HA-02-J-008), which complies with the guidelines of the “System of ethics of research” prepared by the King Abdulaziz City for Science and Technology and approved by Royal Decree No. M/59 on August 24, 2010. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
FM, Tissue cutting, immunohistochemistry, Genetic interpretation, writing and editing; MK, Conceptualization, histological analysis, Genetic interpretation, writing and editing; SB, Clinical data provider, data analysis, writing and editing; AJS, Clinical data interpretation, writing and editing; SH, Tissue cutting, immunohistochemistry, writing and editing; YM, writing and editing; MA, Genetic testing; AD, Genetic testing, interpretation, and writing; TH, Final revision, and editing; AN, writing and editing; IF-E, Genetic interpretation, editing, writing.
Funding
Special thank to Saudi Association of Neurological Surgery (SANS), Riyadh, Saudi Arabia (Grant no. 1 R01 SANS 202050-01) and also Deanship of Scientific Research at King Abdulaziz University (Grant no. G:171-828-1440).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1. Solomon JP, Benayed R, Hechtman JF, Ladanyi M. Identifying Patients with NTRK Fusion Cancer. Ann Oncol (2019) 30:viii16–viii22. Epub 2019 Dec 24. 10.1093/annonc/mdz384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Torre M, Vasudevaraja V, Serrano J, DeLorenzo M, Malinowski S, Blandin A-F, et al. Molecular and Clinicopathologic Features of Gliomas Harboring NTRK Fusions. Acta Neuropathol Commun (2020) 8:107. 10.1186/s40478-020-00980-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amatu A, Sartore-Bianchi A, Siena S. NTRK Gene Fusions as Novel Targets of Cancer Therapy across Multiple Tumour Types. ESMO Open (2016) 1:e000023. 10.1136/esmoopen-2015-000023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lange A, Lo H-W. Inhibiting TRK Proteins in Clinical Cancer Therapy. Cancers (2018) 10:105. 10.3390/cancers10040105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vaishnavi A, Le AT, Doebele RC. TRKing Down an Old Oncogene in a new era of Targeted Therapy. Cancer Discov (2015) 5:25–34. 10.1158/2159-8290.cd-14-0765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hechtman JF, Benayed R, Hyman DM, Drilon A, Zehir A, Frosina D, et al. Pan-Trk Immunohistochemistry Is an Efficient and Reliable Screen for the Detection of NTRK Fusions. Am J Surg Pathol (2017) 41:1547–51. 10.1097/pas.0000000000000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Solomon JP, Linkov I, Rosado A, Mullaney K, Rosen EY, Frosina D, et al. NTRK Fusion Detection across Multiple Assays and 33,997 Cases: Diagnostic Implications and Pitfalls. Mod Pathol (2020) 33:38–46. 10.1038/s41379-019-0324-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a Summary. Neuro Oncol (2021) 23:1231–51. 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weller M, van den Bent M, Preusser M, Le Rhun E, Tonn JC, Minniti G, et al. EANO Guidelines on the Diagnosis and Treatment of Diffuse Gliomas of Adulthood. Nat Rev Clin Oncol (2021) 18:170–86. 10.1038/s41571-020-00447-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kurdi M, Alghamdi B, Butt NS, Baeesa S. The Relationship between CD204 M2-Polarized Tumour-Associated Macrophages (TAMs), Tumour-Infiltrating Lymphocytes (TILs), and Microglial Activation in Glioblastoma Microenvironment: a Novel Immune Checkpoint Receptor Target. Discov Onc (2021) 12:28. 10.1007/s12672-021-00423-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Drilon A, Laetsch TW, Kummar S, DuBois SG, Lassen UN, Demetri GD, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Childrenficacy of Larotrectinib in TRK Fusionepositive Cancers in Adults and Children. N Engl J Med (2018) 378:731–9. 10.1056/NEJMoa1714448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bourhis A, Caumont C, Quintin-Roué I, Magro E, Dissaux G, Remoué A, et al. Detection of NTRK Fusions in Glioblastoma: Fluorescent In Situ Hybridisation Is More Useful Than Pan-TRK Immunohistochemistry as a Screening Tool Prior to RNA Sequencing. Pathology (2021) S0031-3025(21):00429–3. 10.1016/j.pathol.2021.05.100 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions and data presented in the study are available upon request from the corresponding author.