Abstract
Multidrug-resistant Vibrio cholerae O1 El Tor strains isolated during the 1994 outbreak of cholera in Albania and Italy were characterized for the molecular basis of antibiotic resistance. All strains were found to be resistant to tetracycline, streptomycin, spectinomycin, trimethoprim, sulfathiazole, and the vibriostatic compound O/129 (2,4-diamino-6,7-diisopropylteridine). Resistance genes were self-transferable by a conjugative plasmid of about 60 MDa, with the exception of spectinomycin resistance, which was conferred by the aadA1 gene cassette located in the bacterial chromosome within a class 1 integron. The resistance to trimethoprim and O/129 was conferred by the dfrA1 gene, which was present on the plasmid. Although the dfrA1 gene is known to be borne on an integron cassette, class 1, 2, or 3 intI genes were not detected as part of the plasmid DNA from the strains studied.
Since 1977, outbreaks of cholera caused by resistant Vibrio cholerae strains have been reported in Africa, Asia, and America (5, 16, 17, 19, 22, 23, 34). The therapeutic and prophylactic treatment of cholera with antibiotics like tetracycline has probably contributed to the sporadic appearance of drug-resistant strains in different geographical areas. Multiple-antibiotic resistance in V. cholerae has been described, frequently upon the acquisition of R plasmids belonging to the conjugative group C (9, 10, 32, 33).
In September 1994 an epidemic of cholera due to V. cholerae O1 El Tor occurred in Albania (11, 35), and somewhat later in autumn, 12 cases of cholera were reported on the southeast coast of Italy, in the province of Bari (36). Twenty clinical V. cholerae O1 strains from Albania and 15 (12 clinical and 3 environmental) strains from Italy were ribotyped (26); results confirmed the clonal origin of all the Albanian and Italian isolates and showed a different ribotype for a V. cholerae strain isolated in Italy in the same year, from an imported case of cholera (Fig. 1).
FIG. 1.
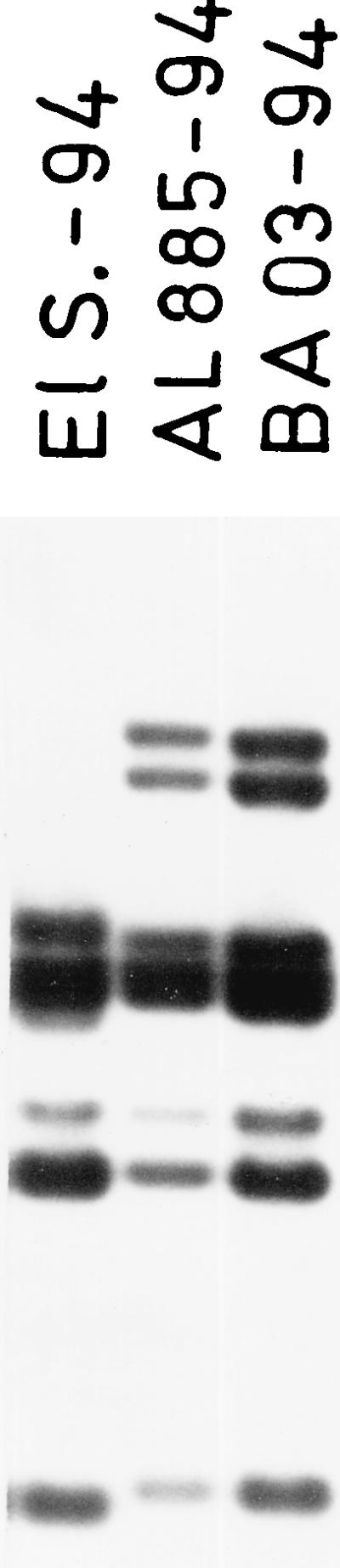
16 S and 23 S BglI ribotypes of representative V. cholerae O1 isolates from the 1994 outbreak in Albania (AL 885-94) and Italy (BA 03-94) are compared with a V. cholerae O1 strain isolated from an imported case in Italy from the same year (El S -94).
All the Albanian and Italian V. cholerae epidemic strains showed an identical R type, being resistant to streptomycin, spectinomycin, tetracycline, trimethoprim, and sulfathiazole, as assessed according to the National Committee for Clinical Laboratory Standards guidelines (24). They were also resistant to 150 μg of O/129 (2,4-diamino-6,7-diisopropylpteridine). All isolates were susceptible to ampicillin, nalidixic acid, chloramphenicol, kanamycin, and ciprofloxacin.
The 15 Albanian and 13 Italian isolates were further characterized with regard to their genetic basis of resistance. Conjugation experiments with Escherichia coli K-12 14R525 as the recipient strain showed that the resistance to streptomycin, tetracycline, trimethoprim, sulfathiazole, and O/129 was self-transferable. MICs of trimethoprim, spectinomycin, streptomycin, and tetracycline were measured for V. cholerae O1 strains and for their transconjugants, according to NCCLS guidelines (25), and demonstrated that transconjugants have acquired the multiple-resistance phenotype of the V. cholerae donor strains, except for spectinomycin resistance, which was not transferred.
A single plasmid of ∼60 MDa was identified in V. cholerae isolates and in transconjugants and tested for incompatibility group (Inc). We found that the V. cholerae R plasmid did not belong to the IncC group, as frequently reported for other plasmids previously identified in V. cholerae isolates (32, 33). The following standard plasmids were tested by conjugation experiments: R40a for IncC=A, RHH72 for IncB=O, R1drd for IncFII, TP154 for IncH1, TP245 for IncH2, TP110 for IncI1, RJ66a for IncIw, pIP69 for IncM, RPC3 for IncN, RP4 for IncP, Rst1 for IncT, Rsa for IncW, and R6K for IncX (1, 7, 12). The IncQ group was tested by Southern blot hybridization with the inc-rep Q probe from the plasmid bank (generously provided by W. M. Maas) as described by Couturier et al. (6). The V. cholerae plasmid was compatible with all the plasmids used and did not hybridize with the IncQ specific probe.
The presence of a previously unknown type of plasmid conferring resistance to drugs of choice in the treatment of cholera represents a novel finding in the description of the antibiotic resistance mechanisms of V. cholerae O1 human isolates.
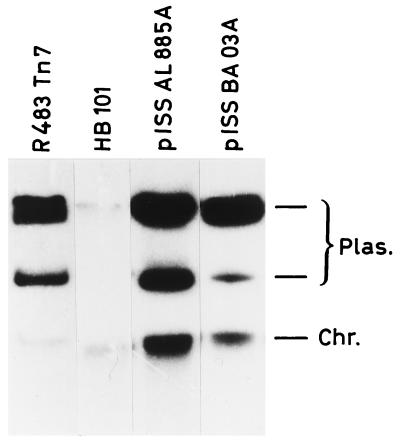
On the V. cholerae R plasmid we identified the gene conferring trimethoprim and O/129 resistance. Specific primers for the known dfr gene types were used in PCR experiments (Table 1). A 268-bp positive amplicon was obtained by using the DH1 and DH2 primers, which were designed to amplify the dfrA1 gene (8). The DNA sequence (28) of this DH1-DH2 PCR product demonstrated that trimethoprim resistance was conferred by the dfrA1 gene. This gene was localized on the V. cholerae R plasmid by Southern blot hybridization using the DH1-DH2 PCR product as a probe. Figure 2 shows hybridization results obtained with plasmid DNA extracted from AL885 and BA03 V. cholerae strains. The R483::Tn7 plasmid coding for the dfrA1 gene and the genomic DNA extracted from the HB101 E. coli strain were used as positive and negative controls, respectively.
TABLE 1.
Primers used in PCR amplification experiments
| Gene or inte-gron primer | Primer sequence | Primer site | EMBL accession no. | Reference |
|---|---|---|---|---|
| Gene | ||||
| dfrA1 DH1 | 5′ CAAGTTTACATCTGACAATGAGAACGTAT 3′ | 429–459 | X00926 | 8 |
| dfrA1 DH2 | 5′ ACCCTTTTGCCAGATTTGGTA 3′ | 706–686 | X00926 | This study |
| dfrA1 DH9 | 5′ AGCTGTTCACCTTTGGC 3′ | 1059–1043 | K00052 | 29 |
| dfrB1 DH3 | 5′ GTTGGACTCAAATGATGACAATGTAGTTG 3′ | 430–460 | S76821 | 36 |
| dfrB1 DH4 | 5′ CCAAATTTGATAGCAATAGTT 3′ | 696–676 | S76821 | This study |
| dfrV DH5 | 5′ CCTGGACGGCCGATAATGACAACGTAATAG 3′ | 1501–1531 | X12868 | 30 |
| dfrV DH6 | 5′ CCAAATTTGATAGCAATAGTT 3′ | 1767–1747 | X12868 | This study |
| dfrVIII DH7 | 5′ AAGCGCTGGAGCTTCCGGGTGTTCGTGACG 3′ | 906–935 | U09273 | 3 |
| dfrVIII DH8 | 5′ ATTCTGTAAGCTCCTTTTTAC 3′ | 1211–1190 | U09273 | This study |
| Integron | ||||
| 5′-CS | 5′ GGCATCCAAGCAGCAAG 3′ | 1190–1206 | M73819 | 18 |
| 3′-CS | 5′ AAGCAGACTTGACCTGA 3′ | 1342–1326 | M73819 | 18 |
| aadA1a | 5′ TCGATGACGCCAACTAC 3′ | 464–448 | X02340 | 15 |
| Int1A | 5′ AAAACCGCCACTGCGCCGTTA 3′ | 1300–1278 | M95287 | This study |
| Int1B | 5′ GAAGACGGCTGCACTGAACG 3′ | 100–115 | M95287 | This study |
| Int2A | 5′ ATGTCTAACAGTCCATTTTTAAATTCTA 3′ | 1474–1495 | AJ002782 | This study |
| Int2B | 5′ AAATCTTTAACCCGCAAACGC 3′ | 1917–1887 | AJ002782 | This study |
| Int3A | 5′ GTGGCGCAGGGTGTGGAC 3′ | 194–211 | D50438 | 2 |
| Int3B | 5′ ACAGACCGAGAAGGCTTATG 3′ | 959–939 | D50438 | 2 |
| sul1 | 5′ CTTCGATGAGAGCCGGCGGC | 924–943 | X12869 | 30 |
| sul1R | 5′ GCAAGGCGGAAACCCGCGCC | 1360–1341 | X12869 | 30 |
FIG. 2.
Localization of the dfrA1 gene within the V. cholerae R plasmid. Plasmid DNA was extracted from AL885 (pISS AL 885A) and BA03 (pISS BA 03A) V. cholerae isolates and hybridized with the DH1-DH2 PCR product as a specific probe for the dfrA1 gene. E. coli containing the R483 plasmid that includes Tn7 harboring the dfrA1 gene cassette and genomic DNA from E. coli HB101 were used as positive and negative controls, respectively. Chromosomal DNA lit up because of a weak trapping of the plasmid. Plasmid molecular weight was estimated by comparing the plasmid’s mobility during agarose gel electrophoresis with those of R40a, R483, RP4, RSa, and S6K plasmids (22).
It has been suggested that the high and still increasing prevalence of trimethoprim resistance could be due to the frequent association of dfr genes to integrons of both class 1 and 2, particularly in Tn21 and in Tn7 derivative transposons (14, 30, 31).
The dfrA1 gene we found in V. cholerae isolates was not associated with Tn21 or Tn7 integrons, since the V. cholerae R plasmid did not react with specific probes for class 1 (intI1) and class 2 (intI2) integrase genes during Southern blot hybridization. The intI1 and intI2 probes were obtained by PCR amplification of the pACYC184::Tn21 (4) and R483::Tn7 (29) plasmids (data not shown), using the Int1A-Int1B and Int2A-Int2B primers shown in Table 1, respectively. The presence of class 3 integrons was excluded by PCR amplification with the primers Int3A and Int3B as described by Arakawa et al. (2). Type 1 sulfonamide resistance (encoded by sul1), which is exclusively associated with the class 1 integron (27), was not detected on the 60-MDa plasmid (data not shown), suggesting that the dfrA1 gene is not a cassette which has been segregated from a class 1 integron originally responsible for its insertion. Although our data on the V. cholerae plasmid suggest that the ubiquitous dfrA1 was not identified as an integron-borne gene cassette, we cannot exclude the possibility that this gene is inserted in a new type of integron.
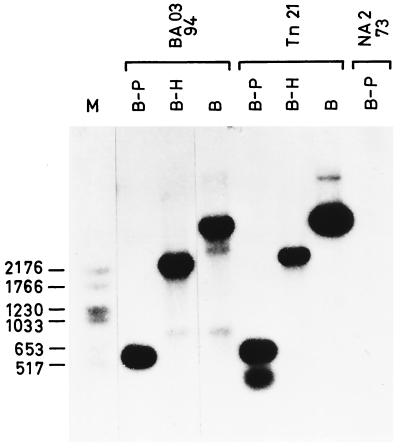
Interestingly, a class 1 integron was detected in the V. cholerae isolates when genomic DNA, instead of plasmid DNA, was probed with intI1. Genomic DNA was digested with BamHI-PvuII, BamHI-HindIII, and BamHI enzymes, which characteristically recognized specific sites in 5′ and 3′ conserved regions of class 1 integrons (13), and it was then compared with DNA from the integron carried by the Tn21 transposon (Fig. 3). The susceptible V. cholerae NA2 strain isolated in Italy during an outbreak of cholera in 1973 was used as a negative control. Hybridization results (shown in Fig. 3 for the BA03 isolate) indicated the presence in the V. cholerae genome of an integron with a restriction pattern identical to that carried by the Tn21 transposon (In2). This integron is known to carry the aadA1 gene cassette, which confers spectinomycin resistance (27). The presence of the aadA1 gene cassette within a class 1 integron on the chromosome of V. cholerae strains was confirmed by PCR amplifications using the IntIA-IntIB, 5′-CS–3′-CS, 5′-CS–aadA1, and sul1-sul1R (18, 30) primer pairs listed in Table 1 (data not shown). This class 1 integron accounts for the nontransferable spectinomycin resistance phenotype exhibited by the V. cholerae isolates.
FIG. 3.
Southern blot analysis of V. cholerae class 1 integron. Genomic DNA was extracted from a V. cholerae BA03 isolate, an E. coli strain harboring the pACYC184::Tn21 plasmid, and a V. cholerae NA2 strain; digested with BamHI-PvuII (B-P), BamHI-HindIII (B-H), and BamHI (B); and hybridized with the intI1 probe specific for the class 1 integrase gene. Molecular size standards (in base pairs) are indicated to the left of the DNA ladder (lane M).
The presence of class 1, 2, and 3 integrons has never been reported before in Vibrio isolates, since a distinctive class of integrase (intI4) has been recently described in Vibrio spp. The intI4 integrase was found to be associated with the V. cholerae repeats and probably represents the class 1, 2, and 3 integrase gene ancestor (20). The observation that a class 1 integron has been acquired by the V. cholerae genome represents an interesting finding, implying that the antibiotic resistance in this species may evolve through integron-mediated acquisition of further gene cassettes.
Acknowledgments
We thank Alexander Sallabanda and Asmi Dibra (Public Health Institute, Tirana, Albania), Giovanni Rizzo (University of Bari, Bari, Italy), and Eugenio Bordi (Spallanzani Hospital, Rome, Italy) for providing isolates and Sergio Arena, Ildo Benedetti, and Susanna Mariotti for technical assistance. We thank Werner K. Maas for generously providing the inc/rep plasmid bank.
This work was partially supported by the UNICEF Program Control of acute diarrheal diseases including cholera in Albania.
REFERENCES
- 1.Anderson E S, Threlfall E J. The characterization of plasmids in the enterobacteria. J Hyg. 1974;72:471–487. doi: 10.1017/s0022172400023718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barg N L, Register S, Thomson C, Amyes S. Sequence identity with type VIII and association with IS176 of type IIIc dihydrofolate reductase from Shigella sonnei. Antimicrob Agents Chemother. 1995;39:112–116. doi: 10.1128/aac.39.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colonna B, Bernardini M, Micheli G, Maimone F, Nicoletti M, Casalino M. The Salmonella wien virulence plasmid pZM3 carries Tn1935, and multiresistant trasposon containing a composite IS1936-kanamycin resistant element. Plasmid. 1988;20:221–231. doi: 10.1016/0147-619x(88)90028-5. [DOI] [PubMed] [Google Scholar]
- 5.Coppo A, Colombo M, Pazzani C, Bruni R, Mohamud K A, Omar K H, Mastrandrea S, Salvia A M, Rotigliano G, Maimone F. Vibrio cholerae in the horn of Africa: epidemiology, plasmids, tetracycline resistance gene amplification and comparison between O1 and non O1 strains. Am J Trop Med Hyg. 1995;53:351–359. doi: 10.4269/ajtmh.1995.53.351. [DOI] [PubMed] [Google Scholar]
- 6.Couturier M, Bex F, Berquist P L, Maas W K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988;52:375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta N. Plasmid classification: incompatibility grouping. In: Timmis K N, Puhler A, editors. Proceedings of the Symposium on Plasmids of Medical Environmental and Commercial Importance—1979. Amsterdam, The Netherlands: Elsevier/North-Holland Biomedical Press; 1979. pp. 3–12. [Google Scholar]
- 8.Fling M E, Richards C. Nucleotide sequence of the trimethoprim resistant dihydrofolate reductase gene harboured by Tn7. Nucleic Acids Res. 1983;11:5147–5158. doi: 10.1093/nar/11.15.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Glass R I, Huq M I, Lee J V, Threlfall E J, Khan M R, Alim A R, Rowe B, Gross R J. Plasmid-borne multiple drug resistance in Vibrio cholerae serogroup O1 biotype El Tor: evidence for a point-source outbreak in Bangladesh. J Infect Dis. 1983;147:204–209. doi: 10.1093/infdis/147.2.204. [DOI] [PubMed] [Google Scholar]
- 10.Goldstein F, Gerbaud G, Courvalin P. Transposable resistance to trimethoprim and O/129 in Vibrio cholerae. J Antimicrob Chemother. 1986;17:559–569. doi: 10.1093/jac/17.5.559. [DOI] [PubMed] [Google Scholar]
- 11.Greco D, Luzzi I, Sallabanda A, Dibra A, Kakarricy E, Shapo L. Cholera in the Mediterranean: outbreak in Albania. Eurosurveillance. 1995;1:1–2. [PubMed] [Google Scholar]
- 12.Grindley N D, Grindley J N, Anderson E S. R factors compatibility groups. Mol Gen Genet. 1972;119:287–297. doi: 10.1007/BF00272087. [DOI] [PubMed] [Google Scholar]
- 13.Hall R M, Brown H J, Brookes D E, Stokes H W. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J Bacteriol. 1994;176:6286–6294. doi: 10.1128/jb.176.20.6286-6294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall R M, Collis C M. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. doi: 10.1111/j.1365-2958.1995.tb02368.x. [DOI] [PubMed] [Google Scholar]
- 15.Hollingshead S, Vapnek D. Nucleotide sequence analysis of gene encoding a streptomycin/spectinomycin adenyltransferase. Plasmid. 1985;13:17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- 16.Huq A, Alam M, Parveen S, Colwell R R. Occurrence of resistance to vibriostatic compound O/129 in Vibrio cholerae O1 isolated from clinical and environmental samples in Bangladesh. J Clin Microbiol. 1992;30:219–221. doi: 10.1128/jcm.30.1.219-221.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jesudason M V, John T J. Transferable trimethoprim resistance of Vibrio cholerae O1 encountered in southern India. Trans R Soc Trop Med Hyg. 1990;84:136–137. doi: 10.1016/0035-9203(90)90407-6. [DOI] [PubMed] [Google Scholar]
- 18.Levesque C, Piché L, Larose C, Roy P H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maimone F, Coppo A, Pazzani C, Ismail S O, Guerra R, Procacci P, Rotigliano G, Omar K H. Clonal spread of multiply resistant strains of Vibrio cholerae O1 in Somalia. J Infect Dis. 1986;153:802–803. doi: 10.1093/infdis/153.4.802. [DOI] [PubMed] [Google Scholar]
- 20.Mazel D, Dychinco B, Webb V A, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 21.Myers J A, Sanchez D, Elwell L P, Falkow S. Simple agarose gel electrophoretic method for the identification and characterization of plasmid deoxyribonucleic acid. J Bacteriol. 1976;127:1529–1537. doi: 10.1128/jb.127.3.1529-1537.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mhalu F S, Mmari P W, Ijumba J. Rapid emergence of El Tor Vibrio cholerae resistant to antimicrobial agents during first six months of fourth cholera epidemic in Tanzania. Lancet. 1979;i:345–347. doi: 10.1016/s0140-6736(79)92889-7. [DOI] [PubMed] [Google Scholar]
- 23.Nath G, Sanyal S C. Emergence of Vibrio cholerae O1 resistant to vibriostatic agent O/129. Lancet. 1992;340:366–367. doi: 10.1016/0140-6736(92)91438-e. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standards M2-A4. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. Standard methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A2. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1990. [Google Scholar]
- 26.Popovic T, Bopp C A, Olsvik O, Kiehlbauch J A. Ribotyping in molecular epidemiology. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology. Washington, D.C: American Society for Microbiology; 1993. pp. 573–583. [Google Scholar]
- 27.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Simonsen C C, Chen E Y, Levinson A D. Identification of the type I trimethoprim-resistant dihydrofolate reductase specified by Escherichia coli R-plasmid R483: comparison with procaryotic and eucaryotic dihydrofolate reductases. J Bacteriol. 1983;155:1001–1008. doi: 10.1128/jb.155.3.1001-1008.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundström L, Radström P, Swedberg G, Sköld O. Site-specific recombination promotes linkage between trimethoprim and sulfonamide resistance genes. Sequence characterization of dhfrV and sulI and a recombination active locus of Tn21. Mol Gen Genet. 1988;213:191–201. doi: 10.1007/BF00339581. [DOI] [PubMed] [Google Scholar]
- 31.Sundström L, Roy P H, Sköld O. Site-specific insertion of three structural gene cassettes in transposon Tn7. J Bacteriol. 1991;173:3025–3028. doi: 10.1128/jb.173.9.3025-3028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabtieng R, Wattanasri S, Echeverria P, Seriwatana J, Bodhidatta L, Chatkaeomorakot A, Rowe B. An epidemic of Vibrio cholerae El Tor Inaba resistant to several antibiotics with a conjugative group C plasmid coding for type II dihydrofolate reductase in Thailand. Am J Trop Med Hyg. 1989;41:680–686. doi: 10.4269/ajtmh.1989.41.680. [DOI] [PubMed] [Google Scholar]
- 33.Threlfall E J, Rowe B, Huq I. Plasmid-encoded multiple antibiotic resistance in Vibrio cholerae El Tor from Bangladesh. Lancet. 1980;i:1247–1248. doi: 10.1016/s0140-6736(80)91701-8. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 34.Threlfall E J, Said B, Rowe B. Emergence of multiple drug resistance in Vibrio cholerae O1 El Tor from Ecuador. Lancet. 1993;342:1173. doi: 10.1016/0140-6736(93)92156-n. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. Cholera, Albania. Weekly Epidemiol Rec. 1994;69:280. [Google Scholar]
- 36.World Health Organization. Cholera, Italy. Weekly Epidemiol Rec. 1994;69:325. [Google Scholar]
- 37.Young H K, Qumsieh M J, McIntosh M L. Nucleotide sequence and genetic analysis of the type Ib trimethoprim-resistant, Tn4132-encoded dihydrofolate reductase. J Antimicrob Chemother. 1994;34:715–725. doi: 10.1093/jac/34.5.715. [DOI] [PubMed] [Google Scholar]