Abstract
Cysteine desulfurases are pyridoxal-5'-phosphate (PLP)-dependent enzymes that mobilize sulfur derived from the l-cysteine substrate to the partner sulfur acceptor proteins. Three cysteine desulfurases, IscS, NifS, and SufS, have been identified in ISC, NIF, and SUF/SUF-like systems for iron-sulfur (Fe-S) cluster biosynthesis, respectively. These cysteine desulfurases have been investigated over decades, providing insights into shared/distinct catalytic processes based on two types of enzymes (type I: IscS and NifS, type II: SufS). This review summarizes the insights into the structural/functional varieties of bacterial and eukaryotic cysteine desulfurases involved in Fe-S cluster biosynthetic systems. In addition, an inactive cysteine desulfurase IscS paralog, which contains pyridoxamine-5'-phosphate (PMP), instead of PLP, is also described to account for its hypothetical function in Fe-S cluster biosynthesis involving this paralog. The structural basis for cysteine desulfurase functions will be a stepping stone towards understanding the diversity and evolution of Fe-S cluster biosynthesis.
Keywords: cofactors, PLP-dependent enzymes, reaction intermediates, structure-function relationship, sulfur metabolism
Significance
Recent X-ray crystallography, cryo-EM, NMR and SAXS analyses unveiled common and distinct features of two different types of PLP-dependent cysteine desulfurases involved in Fe-S cluster biosynthetic systems. The significance of the type-dependent local structural difference is highlighted in this review, which will give clues for understanding not only the details of the cysteine desulfurase mechanisms, but also the diversity of the Fe-S cluster biosynthetic systems.
Introduction
Iron-sulfur (Fe-S) clusters are inorganic cofactors composed of Fe ions and sulfides, and are found in most organisms (Fig. 1) [1–8]. Fe-S clusters are utilized by Fe-S proteins that play critical roles in many biological processes, such as the respiratory chain, the photosystem, and regulation of gene expression. For example, ferredoxins, which are well-known electron transfer proteins, utilize Fe-S clusters as redox centers [9]. As another example, aconitase utilizes a [4Fe-4S] cluster as a Lewis acid at its active site [10]. Radical S-adenosyl-l-methionine (SAM) enzymes are also important Fe-S enzymes that catalyze a variety of reactions via radical formation using SAM [11]. In addition to Fe-S proteins with canonical Fe-S clusters (i.e., [2Fe-2S], [3Fe-4S] and [4Fe-4S] clusters) (Fig. 1A), some unique Fe-S enzymes having unusual Fe-S clusters are also of interest in terms of their cluster derivative chemistry (e.g., nitrogenases [12], [FeFe]-hydrogenase [13], and Ni,Fe-carbon monoxide dehydrogenase [14]).
Figure 1 .
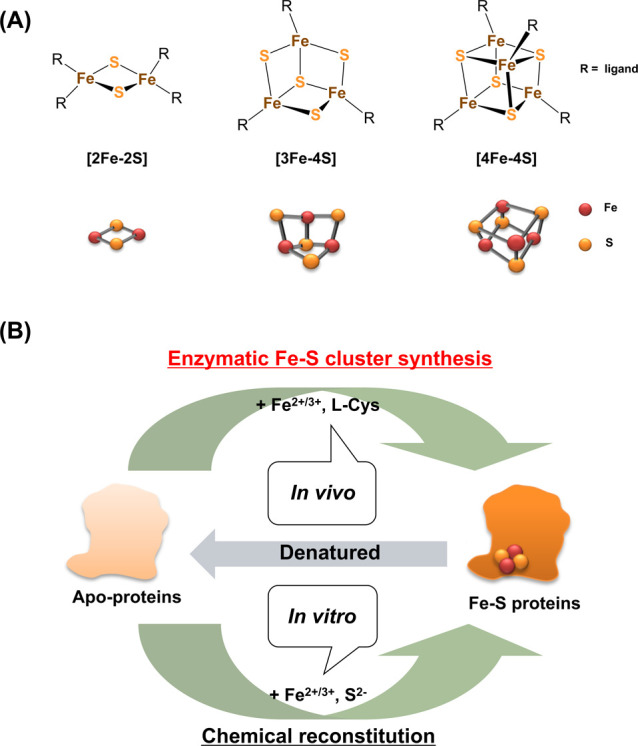
(A) Chemical structures, and ball-and-stick models of the three types of Fe-S clusters: [2Fe-2S], [3Fe-4S], and [4Fe-4S]. (B) Schematic representation of Fe-S cluster synthesis, and the maturation of Fe-S proteins in vivo and in vitro.
The Fe-S clusters in proteins are coordinated by amino acid ligands, which are typically Cys residues, and occasionally His, Asp/Glu with metal-coordinating ability (Fig. 1A) [15,16]. So far, various types of Fe-S clusters with non-amino acid ligands have been synthesized by chemists [15,16]. The studies on chemical synthesis of Fe-S clusters greatly contributed to the understanding of their chemical properties. In contrast, it has been difficult to clarify the biological systems involved in the de novo synthesis of Fe-S clusters in cells. In the past, Fe-S clusters in cells were believed to be synthesized non-enzymatically (spontaneously) from Fe ions and sulfides (Fig. 1B), because they could be chemically reconstituted in vitro to certain apo-Fe-S proteins (e.g. ferredoxin) using Fe ions and sulfides [17]. However, in the 1980s, the enzymatic Fe-S cluster synthesis systems were identified through biochemical and genetic analyses, using several bacteria such as Escherichia coli and Azotobacter vinerandii [18,19].
Currently, three distinct Fe-S cluster biosynthetic systems are identified in the case of bacteria. They are called ISC, SUF, and NIF systems, based on the names of the iron-sulfur cluster (ISC), sulfur mobilization (SUF), and the nitrogen fixation (NIF) (Fig. 2) [2,20]. The ISC system consists of seven components encoded by iscSUA-hscBA-fdx-iscX in the isc operon [21–40]. Among the members, cysteine desulfurase IscS and Fe-S cluster assembling scaffold IscU are essential for the biosynthesis. The E. coli SUF system consists of six components encoded by sufABCDSE in the suf operon [41–57] In the SUF system, cysteine desulfurase SufS functions as IscS. However, the other components in the SUF system are rather different from the members in the ISC system. SufB, SufC, and SufD form a protein complex SufBCD serving as an Fe-S cluster-assembling scaffold. SufE is a sulfur-transfer protein to deliver the inorganic sulfur from SufS to the SufBCD complex. These features are unique for the SUF system compared to the ISC system, which means two systems are distinct from each other (Fig. 3). The NIF system is also different from the ISC and SUF systems. The NIF system is simply composed of two components, cysteine desulfurase NifS and Fe-S cluster-assembling scaffold NifU, which are encoded by nifSU [19,58–67]. Actually, the NIF system was first discovered as components of the nitrogenase maturation system in nitrogen-fixing bacteria [68,69]. However, nifSU can also be found in non-nitrogen-fixing bacteria (e.g. Helicobacter pylori), where the ISC and SUF systems are absent. Therefore, the NIF system in these bacteria is used as a general Fe-S cluster biosynthetic system, rather than a nitrogenase-specific Fe-S-type cluster biosynthetic system.
Figure 2 .
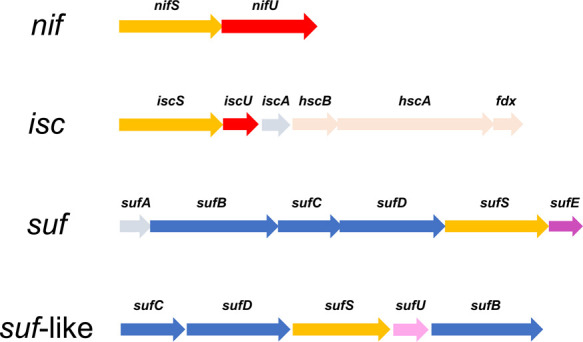
Gene clusters encoding bacterial Fe-S cluster biosynthetic systems: NIF, ISC, SUF, and SUF-like systems. All systems contain cysteine desulfurases, encoded by nifS, iscS, and sufS (yellow arrows). The nifU and iscU genes (red arrows) encode NifU and IscU, which are Fe-S cluster biosynthesis scaffold proteins in the NIF and ISC systems, respectively. In the SUF and SUF-like systems, a SufBCD protein complex encoded by sufB, sufC, and sufD (blue arrow) are regarded as a Fe-S cluster biosynthetic scaffold. The sufE gene (magenta arrow) and sufU gene (pink arrow) encode the sulfur-transfer proteins SufE and SufU, respectively. The other genes are not strictly essential under normal conditions, but become critical under certain growth conditions.
Figure 3 .
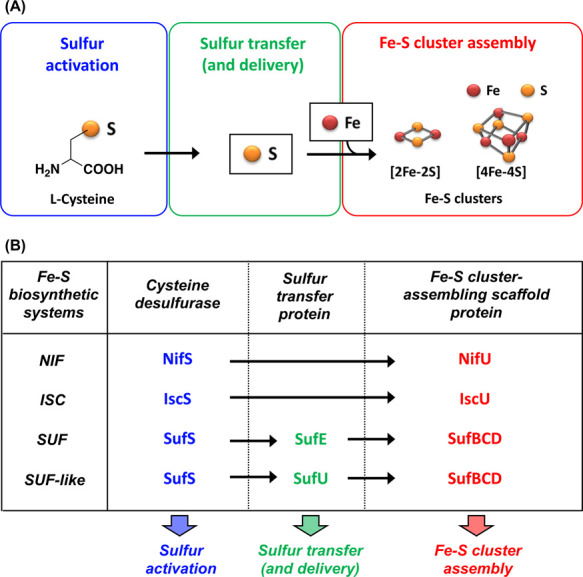
(A) General scheme for the biosynthesis of Fe-S clusters. (B) Components involved in the bacterial Fe-S cluster biosynthetic systems: ISC, NIF, SUF, and SUF-like systems. The first step involves sulfur activation of the substrate l-cysteine by cysteine desulfurases in all the systems. From here, there are two pathways for sulfur-mobilization. In the NIF and ISC systems, sulfur is delivered to the Fe-S cluster biosynthetic scaffold in the IscS-IscU or NifS-NifU complexes and used for Fe-S cluster assembly. In the SUF and SUF-like systems, sulfur is delivered from SufS to an Fe-S cluster biosynthetic scaffold in the SufBCD complex via sulfur delivery proteins SufE and SufU, respectively.
More recently, the SUF-like system (Fig. 2) was identified and characterized in some gram-positive bacteria (e.g. Bacillus subtilis) [70–75]. The SUF-like system contains five components encoded by the sufCDSUB operon. Hence, SufS and SufBCD exist in both the SUF-like system and in the SUF system. SufU, however, is unique to the SUF-like system [76–79], and is structurally homologous to IscU in the ISC system. Considering this, the SUF-like system is regarded as a chimeric system of the ISC and SUF systems. Biochemical, genetic and structural analyses of SufU have recently been conducted, confirming that SufU is a sulfur-transfer protein from SufS to SufBCD as SufE in the SUF system (Fig. 3). This finding is very curious because SufU and IscU belong to a “U-type protein family”, in which the members designated as U-type proteins (e.g., SufU, IscU, NifU) typically have IscU-type folds (an α+β tertiary fold, with three antiparallel β-strands and four α-helices in a compact globular structure) and are found in Fe-S cluster biosynthetic machineries [20]. However, SufU and IscU have different functions: SufU for a sulfur transfer and IscU for an Fe-S cluster assembly. A possible structural evolution of U-type proteins has been discussed elsewhere [75].
In contrast to Fe-S cluster assembling scaffolds, the cysteine desulfurases do not differ significantly [21,58,59,80–84], because they share some structural and functional features [85–87]. However, recent intensive studies have demonstrated that the cysteine desulfurases are also important for understanding the distinct mechanisms of Fe-S cluster biosynthetic systems. In particular, type difference of the cysteine desulfurases (type I: IscS and NifS and type II: SufS) [85] is highlighted in this review. Not only the cysteine desulfurases, but also cysteine desulfurase-partner complexes are also discussed by focusing on the type difference. Furthermore, cysteine desulfurase NFS1 in the mitochondrial ISC system [88–91], and an inactive cysteine desulfurase paralog, IscS from an archaeon Archaeoglobus fulgidus, [92,93] are introduced, which will provide a clue to consider the diversity and evolution of cysteine desulfurases and Fe-S cluster biosynthetic systems.
Two Distinct Types of Cysteine Desulfurases
Cysteine desulfurases are pyridoxal-5'-phosphate (PLP)-dependent enzymes that catalyze sulfur mobilization from the l-cysteine substrate to the partner sulfur-accepting proteins (Figs. 3, 4). The Fe-S cluster biosynthetic machineries NIF, ISC, and SUF/SUF-like systems contain NifS, IscS, and SufS as cysteine desulfurases, respectively. The overall structures of these cysteine desulfurases are almost identical (Figs. 4A, 4B). These enzymes exhibit a very similar homodimeric architecture. The N-terminal region consists of the core region, and the C-terminal region comprising α-helices is extended from the central core region to the ends of the homodimeric architecture. The active sites of cysteine desulfurases contain PLP with some strictly conserved residues, Lys, Arg, His, and Cys [85]. This PLP is covalently attached to the conserved Lys via a Schiff base, forming the PLP-Lys internal aldimine (Fig. 4C).
Figure 4 .
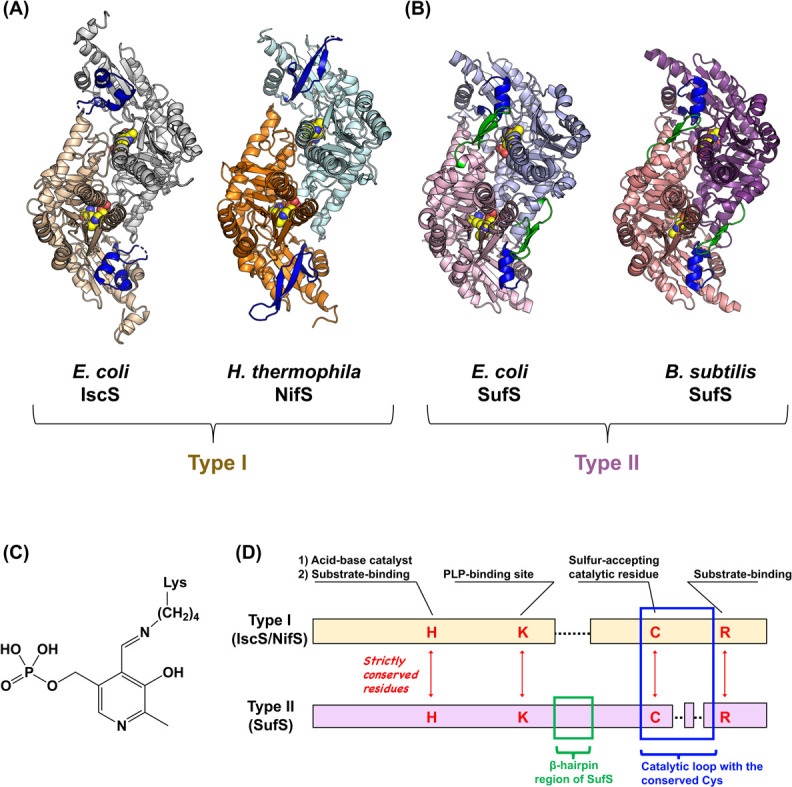
(A) Structures of type I cysteine desulfurases, Escherichia coli IscS [PDB ID: 3lvm] and Hydrogenimonas thermophila NifS [PDB ID: 5zsp]. (B) Structures of type II cysteine desulfurases, E. coli SufS [PDB ID: 6o10] and Bacillus subtilis SufS [PDB ID: 5zs9]. Both types of cysteine desulfurases show homodimeric architectures, and have the PLP-Lys internal aldimine (shown as spheres) at their active sites. Catalytic loops are colored in blue. The β-hairpin region specific for type II SufS enzymes are colored in green. (C) Chemical structure of the PLP-Lys internal aldimine. (D) Primary structure comparisons of type I and type II cysteine desulfurases. H, K, C, and R indicate the conserved His, Lys, Cys, and Arg residues, respectively. The type I enzyme has a longer catalytic loop than the type II (shown in a blue box). Moreover, the type II enzyme has a β-hairpin region (shown in a green box) near the conserved Lys, whereas the type I does not have it.
Notably, the cysteine desulfurases can be categorized into two types: NifS and IscS (type I) and SufS (type II). [83,94]. There are several local structural differences between two types (Fig. 4D). For example, a β-hairpin region of SufS [95–97] is distinguished from the corresponding part of IscS/NifS, which lacks such a region (Fig. 5). This β-hairpin region is close to the PLP-Lys and the conserved Cys residue in the catalytic loop in SufS. Crystal structure analysis of E. coli SufS [95] and its variants revealed that this hairpin is movable, which may mediate the interaction of SufS with its partner SufE (Fig. 3B) [98,99].
Figure 5 .
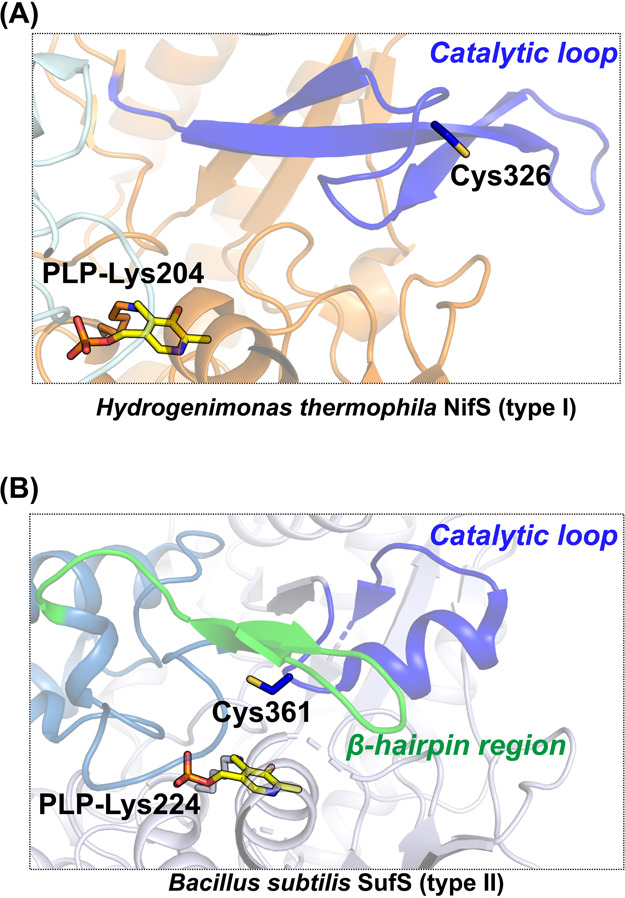
(A) Active site structure of Hydrogenimonas thermophila NifS [PDB ID: 5zsp]. (B) Active site structure of Bacillus subtilis SufS [PDB ID: 5zs9]. Catalytic loops in both NifS and SufS are colored in blue. The β-hairpin region of SufS is colored in green. The PLP-Lys moieties and catalytic cysteine residues are shown in stick models.
Catalytic loops are also different between two types (Figs. 4, 5) [84,85]. The type I loop is usually approximately 10 amino acids longer than the type II loop. Crystal structure analysis of types I and II enzymes demonstrated that the structural folds of the loops are different (Figs. 4, 5). The loops of type I cysteine desulfurases, IscS and NifS (e.g., H. pylori NifS) are mostly or partially disordered (Fig. 4), which may indicate their flexibility. The exception here exists in Hydrogenimonas thermophila NifS, which showed that the loop region includes a β-sheet (Fig. 5). In contrast, loops of type II enzymes, (e.g., SufS from E. coli and B. subtilis), are folded as α-helices (Fig. 5). The conserved Cys residue of SufS is positioned in the turn between two α-helices of the catalytic loop. X-ray crystallographic snapshot analysis was carried out for trapping the catalytic intermediates of the two types of cysteine desulfurases, NifS and SufS, and the intermediates were compared to these resting states. [84] (Fig. 6) This analysis provided insights into the common and distinct features of the two types of cysteine desulfurases. In both types, the structures of PLP-l-Cys external aldimines showed polar interactions at equivalent positions. For example, the SH group of PLP-l-Cys interacted with the imidazole group of the side chain of the conserved His. The carboxy group of PLP-l-Cys was also tightly bound to the guanidium of the side chain of the conserved Arg. The PLP-l-Ala external aldimine in both types also showed an almost identical conformation. Structural analysis of inactivated SufS variants was also performed, resulting in the structural characterization of some PLP-l-amino acid adducts, including a ketimine-type intermediate [82].
Figure 6 .
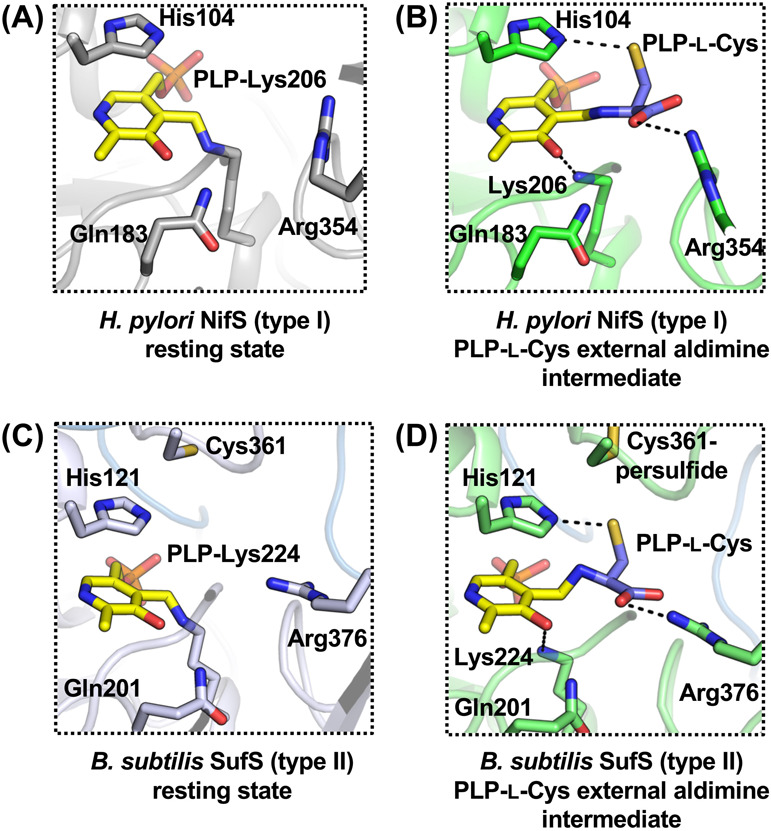
(A) Active site structure of Helicobacter pylori NifS in the resting state [PDB ID: 5wt2]. (B) Active site structure of H. pylori NifS in an intermediate state of PLP-l-Cys external aldimine [PDB ID: 6kg0]. (C) Active site structure of Bacillus subtilis SufS in the resting state [PDB ID: 5zs9]. (D) Active site structure of B. subtilis SufS in an intermediate state of PLP-l-Cys external aldimine with Cys361-persulfide [PDB ID: 6kfz]. Dashed lines indicate key polar interactions for binding of the substrate l-cysteine into the active sites.
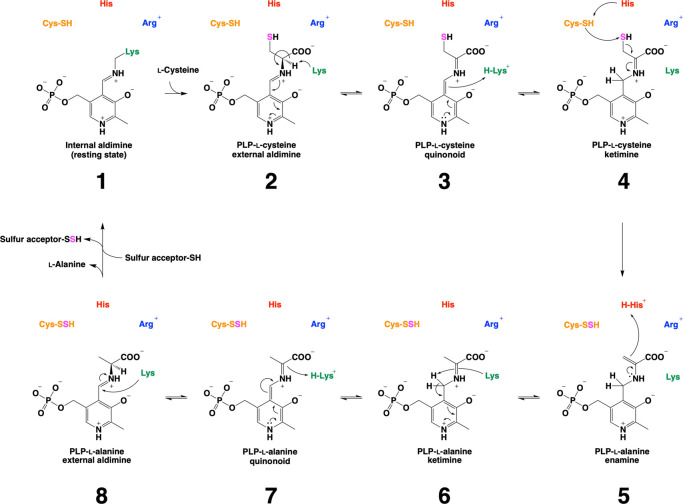
Based on recent studies, the catalytic cycle of cysteine desulfurases has been revisited (Fig. 7) [82,84,85]. The substrate l-cysteine initially reacts with the PLP-Lys internal aldimine (1), resulting in the formation of the corresponding external PLP-l-Cys aldimine (2) via a Schiff-base-exchange reaction. The conserved His works to stabilize the PLP-l-Cys external aldimine (2) via polar interaction. After that, the conserved His serves as an acid-base catalyst, to proceed conversion of the PLP-l-Cys external aldimine (2) to the corresponding PLP-l-Cys quinonoid (3), and then to the PLP-l-Cys ketimine (4) [82]. At this stage, the SH group of the PLP-l-Cys ketimine (4) is attacked by a nucleophilic cysteine residue on the catalytic loop, resulting in cysteine persulfide (Cys-SSH) and PLP-l-Ala enamine (5), which is further converted to PLP-l-Ala ketimine (6). The PLP-l-Ala ketimine (6) is converted to PLP-l-Ala quinonoid (7), and then to PLP-l-Ala aldimine (8) via acid-base catalysis with the conserved Lys. Finally, the PLP-l-Ala aldimine (8) was converted to the resting state of the PLP-Lys internal aldimine (1) via the release of l-Ala. The Cys-SSH donates the sulfur to the partner sulfur acceptor protein.
Figure 7 .
Catalytic mechanism of cysteine desulfurases. The reaction initiates from the resting state where the PLP-Lys internal aldimine exists. From here, l-cysteine (l-Cys) is covalently bound to the PLP moiety (1) via a Schiff-base exchange reaction, yielding the PLP-l-Cys external aldimine (2). This aldimine is stabilized via polar interactions. For example, the conserved Arg interacts with the carboxy group of the cysteine moiety of the PLP-l-Cys external aldimine (2). Also, the conserved His interacts with the SH group of the PLP-l-Cys, although it also functions as an acid-base catalyst during cysteine desulfurase catalysis. Further, the PLP-l-Cys external aldimine (2) is converted to the corresponding ketimine (4) via the quinonoid (3). Then, the catalytic Cys residue attacks the SH group of the PLP-l-Cys ketimine (4), yielding persulfurated Cys (Cys-SSH) and PLP-l-alanine (PLP-l-Ala) enamine (5). This enamine (5) is subsequently converted to PLP-l-Ala ketimine (6), the quinonoid (7), and the external aldimine (8) in a stepwise manner via acid-base catalysis. Finally, the Cys-SSH transfers the sulfur to the sulfur-accepting Cys residue of the partner proteins. Together with, l-Ala is released from the PLP-l-Ala external aldimine (8), which is to be the resting state, PLP-Lys internal aldimine (1).
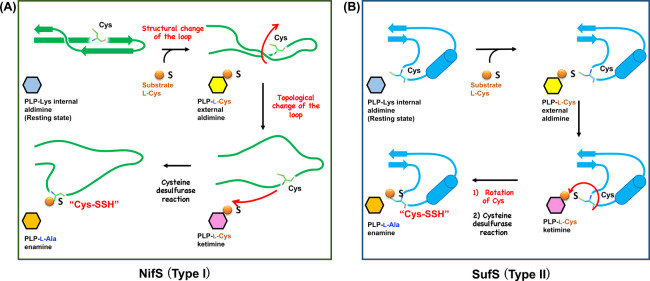
Despite the shared intermediates, behaviors of the catalytic loops of NifS and SufS are distinct, which has been proposed by the X-ray crystallographic snapshot analysis (Fig. 8) [84]. In the conversion of the resting state of the PLP-Lys internal aldimine to the PLP-l-Cys intermediate of H. thermophila NifS, the catalytic loop becomes partially unstructured. It has been proposed that this structural transition was caused by the binding of substrate l-cysteine to the conserved Arg at the active site on the basis of the observation of a slight change of the orientation of Arg [84]. However, the topology of the loop in the PLP-l-Cys intermediate state is still not ready for a nucleophilic reaction by the conserved Cys residue (Fig. 8A, top). In this state, the Cys residue is sterically hindered by the partially-folded loop. Thus, it can be hypothesized that the topology of the loop may be necessarily changed for making the Cys residue close to the PLP site. Then, the Cys residue could finally attack the PLP-l-Cys ketimine, resulting in cysteine desulfurase reaction (Fig. 8A, bottom). After the cysteine desulfurase reaction, the catalytic loop could be structured again with the Cys-SSH in the catalytic cycle of NifS. Indeed, the structure of NifS after the cysteine desulfurase reaction exhibited the folded catalytic loop with the Cys-SSH [84]. When NifS and a sulfur accepting partner NifU co-exist, the catalytic loop having the Cys-SSH should be moving toward NifU, for the sulfur transfer (Fig. 3).
Figure 8 .
Proposed conformational and topological changes of the catalytic loops of the two types of the cysteine desulfurases. (A) NifS (type I). (B) SufS (type II). The schematic representation for type I is based on Hydrogenimonas thermophila NifS, whose loop forms a β-sheet in the resting state, though many of type I enzymes show mostly or partially disordered loops. The Cys residue on the type I loop was far from the PLP site in the resting state. Thus, the structural change of the catalytic loop must occur, to make the conserved Cys serve as a nucleophile toward the PLP-l-Cys ketimine for the cysteine desulfurase reaction. In contrast, the type II catalytic loop (e.g., the catalytic loop of Bacillus subtilis SufS) forms an α-helix with a turn including the catalytic Cys residues. This Cys residue is already in proximity to the PLP-site. Thus, only a slight conformational change of the Cys residue of the type II is needed for the cysteine desulfurase reaction. Notably, it is not completely clarified whether and how the loop conformational change is related to the conversion of the PLP-l-Cys external aldimine to the PLP-l-Cys ketimine because of no structures of the ketimine forms of type I and II wild-type enzymes, although this conversion is supposed to proceed via an acid-base catalysis.
Unlike type I, the catalytic loop of type II B. subtilis SufS maintained the α-helix fold in the forms with PLP-l-Cys and PLP-l-Ala species (Fig. 8B). The conformational change in SufS catalysis was only the change in orientation of Cα-Cβ-Sγ of the conserved Cys residue in the catalytic loop. This small conformational change, however, is sufficient for SufS catalysis, since the SH group of the catalytic Cys residue could already be in proximity to the SH group of the PLP-l-Cys moiety for the nucleophilic attack and subsequent formation of the Cys-SSH in the catalytic loop.
Structures of Cysteine Desulfurase-Partner Complexes in the ISC, NIF and SUF Systems
Cysteine desulfurases interact with their sulfur-accepting partner proteins to form protein complexes (Fig. 9) [100]. Within these complexes, the sulfane sulfur of Cys-SSH of cysteine desulfurases is transferred to sulfur acceptor proteins. In the case of the ISC system, IscS-IscU serves not only as a sulfur-transferring complex, but also as an Fe-S cluster-assembling complex (Fig. 3). There are several crystal structures available for IscS-IscU complexes, such as E. coli IscS-IscU (Fig. 9A) [39] and A. fulgidus IscS-IscU D35A (Fig. 9B) [40], where the conserved Asp was substituted by Ala in IscU (Fig. 9B, bottom) [33,34,101–104]. Based on these structures and available biochemical data, it has been proposed that the assembly of the [2Fe-2S] cluster takes place at the interface of IscS and IscU. However, it should be noted that the actual ligands for the biosynthesis of the [2Fe-2S] cluster in IscS-IscU wild-type have not been fully resolved yet, since only the structure of the [2Fe-2S]-bound form of IscS-IscU D35A variant (Fig. 9B) is available as an Fe-S cluster-bound state. In other words, there is no available structure for wild-type IscS-IscU with an Fe-S cluster. Also, it is known that the substitution of Asp by Ala is known to render IscU inactive in vivo [35,105]. To further understand the mechanisms that underlie the assembly of the [2Fe-2S] cluster in IscS-IscU, it is necessary to characterize the structures of both an intermediate and a [2Fe-2S]-bound wild-type IscS-IscU.
Figure 9 .
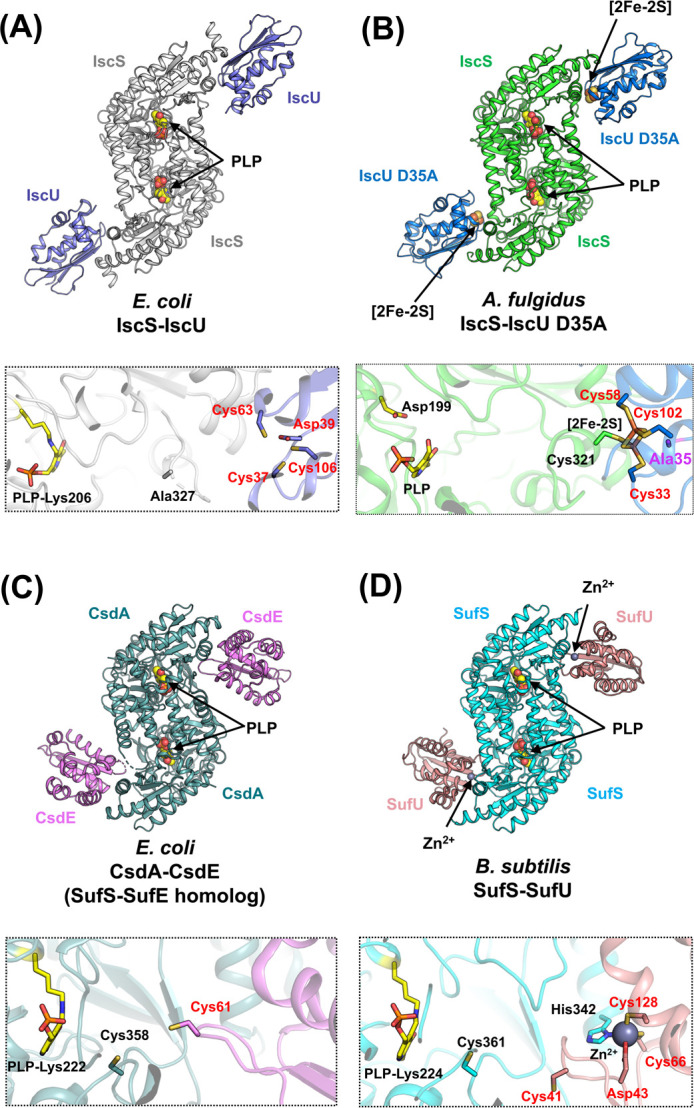
Overall structures and the close-up views of the interfaces of cysteine desulfurase-partner protein complexes. (A) E. coli IscS-IscU [PDB ID: 3lvl]. (B) A. fulgidus IscS-IscU D35A [PDB ID: 4eb5]. (C) E. coli CsdA-CsdE [PDB ID: 4lw4] (a SufS-SufE homolog). (D) B. subtilis SufS-SufU [PDB ID: 5xt5]. In the close-up views, key amino acid residues (except for Ala327 in IscS-IscU) and PLPs are shown as sticks. The loop region including Cys328 (the nucleophilic Cys in catalysis) in E. coli IscS is disordered. The black- and red-colored labels in the close-up views of the complex interfaces indicate the residues from the cysteine desulfurases and their partner proteins, respectively. The residue of Ala35 of A. fulgidus IscU D35A was shown in a stick model in magenta. It should be noted that the structure of A. fulgidus IscS-IscU D35A was modeled as a PLP-bound form, whereas the structure of A. fulgidus IscS alone harbored pyridoxamine-5'-phosphate (PMP) rather than PLP. The reason for the cofactors being different between A. fulgidus IscS-IscU D35A and A. fulgidus IscS is still elusive, but will be elaborated on in the section titled “An inactive cysteine desulfurase paralog with PMP rather than PLP”.
Compared to the IscS-IscU, the NifS-NifU complex is not fully characterized yet because structures of NifU and NifS-NifU are not available. However, it has been known that NifU is uniquely composed of three domains (N-terminal, middle, and C-terminal domains) [2] for functioning as Fe-S cluster-assembling scaffold in NIF system, which is distinct from IscU in ISC system. The N-terminal domain of NifU is structurally homologous to IscU, which suggests that the NifS-NifU complex may function as the IscS-IscU complex [62,63,67]. The middle domain is a [2Fe-2S] cluster-bound domain, which probably functions as an electron donor [60,62]. The C-terminal domain is a type of Nfu-like protein with two conserved Cys residues [64,106–108]. The functions of the middle- and C-terminal domains have not yet been fully resolved; however, some insights into their possible roles have been revealed. To gain further insights into the functions of NifS-NifU, the structure of NifU and NifS-NifU needs to be resolved.
Conversely, the SufS-SufE complex in the SUF system specifically functions in sulfur-mobilization (Fig. 3) [109–111]. The X-ray crystal structure of CsdA-CsdE [112,113] (a SufS-SufE homolog) has been determined (Fig. 9C) [114,115], from which mechanism of SufS-SufE function can also be deduced [116]. Notably, the location of CsdE, which is bound next to the PLP site of CsdA is rather different from that of IscU to IscS. This CsdE-binding location in CsdA-CsdE is suitable for facile sulfur transfer [115]. This type of facile sulfur-transfer process could also occur in SufS-SufE system because of the high degree of homology of SufS-SufE to CsdA-CsdE.
Structure of the SufS-SufU Complex in the SUF-like System
The SUF-like system utilizes SufS-SufU, a cysteine desulfurase-partner complex (Fig. 9D). Uniquely, complex formation and sulfur-transfer mechanisms of the SufS-SufU complex are dependent on the Zn site of SufU (Fig. 10A). In B. subtilis SufU, the Zn-coordination site has four conserved amino acid ligands, i.e. Cys41, Asp43, Cys66, and Cys128. However, in the B. subtilis SufS-SufU complex, the Zn site contains Asp43, Cys66, and Cys128 of SufU, and His342 of SufS, which resulted from a ligand swapping [116]. This His residue is unique to SufS associated with SufU in the SUF-like system [75,116,117], but not to SufS associated with SufE in the SUF system. The importance of His342 was confirmed by studying the B. subtilis SufS H342Y variant, which showed no interaction to SufS and very low activity. Gene complementation analysis also demonstrated the specific interaction between SufS and SufU [75].
Figure 10 .
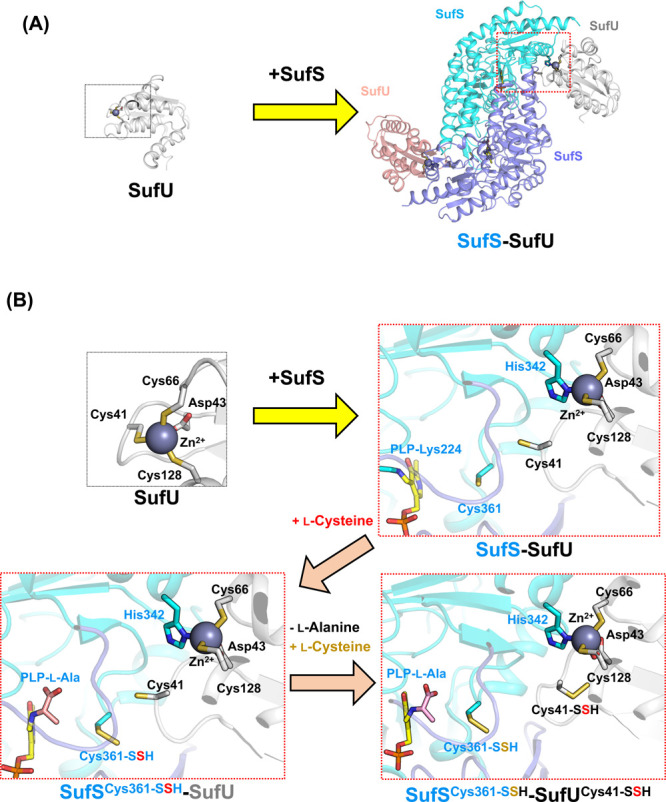
(A) Schematic representation of SufS-SufU complex formation. Two SufU [PDB ID: 2azh] molecules are bound to SufS, resulting in the SufS-SufU complex [PDB ID: 5xt5]. The Zn-binding sites were indicated by dotted line boxes. (B) Structural changes in Zn-binding site of SufU in the formation of the SufS-SufU complex and during cysteine desulfurase reaction and sulfur-transfer [PDB ID: 2azh, 5xt5, 5xt6]. Cys41 of SufU is exchanged with His342 of SufS via ligand-swapping. The His ligand for binding to Zn is conserved only in SufS in SUF-like systems having SufU, but not in SufS in SUF systems having SufE. After the SufS-SufU complex formation, substrate l-cysteine reacts with the PLP-Lys of SufS for cysteine desulfurase reaction, yielding Cys361-persulfide (Cys361-SSH) of SufS and PLP-l-Ala aldimine (PLP-l-Ala). Then, l-alanine is released and Cys361-persulfide transfers its sulfur to Cys41 of SufU. Afterwards, the second l-cysteine substrate comes to the PLP-Lys, which resulted from the release of l-alanine from PLP-l-Ala. The second catalytic turnover cycle occurs, which yield Cys361-SSH. As a result, SufS-SufU complex with two persulfides, Cys361-SSH of SufS and Cys41-SSH of SufU, was formed.
When B. subtilis SufS-SufU complex forms, Cys41 of B. subtilis SufU moves toward the PLP site. The position of Cys41 is suitable for accepting sulfur from persulfurated Cys361 (Cys361-SSH) of SufS. X-ray crystallographic snapshot analysis of SufS-SufU revealed two intermediates, which proposed the sulfur transfer route: sulfur is transferred from the PLP site (PLP-l-Cys) to Cys361 of SufS, then to Cys41 of SufU (Fig. 10B) [116].
Distinct Features between the Two Types of Cysteine Desulfurases-Partner Complexes
Difference in catalytic loops between type I and II enzymes may also be important for discussion of distinct features of the cysteine desulfurase-partner complexes. The type I IscS-IscU complex should utilize a “longer” catalytic loop for dual functions: the sulfur transfer and Fe-S cluster assembly. During these processes by IscS-IscU, this loop should change its conformation largely and flexibly, because there is a long distance between IscS and IscU. More importantly, IscS-IscU can interact with other partners (e.g., Fdx, IscX, and CyaY) [118–123]. Based on studies of IscS-IscU with other partners using X-ray crystallography, NMR and SAXS, it is hypothesized that the motions of the catalytic loop may be affected by the association/dissociation of these partners to IscS-IscU, contributing to a precise Fe-S cluster assembly. This kind of discussion will also be shown in the section concerning human type I cysteine desulfurase (see below).
Conversely, type II SufS-SufU and SufS-SufE (CsdA-CsdE) may have evolved to specifically function in sulfur transfer, with a “shorter” catalytic loop. Indeed, CsdA-CsdE and SufS-SufU sulfur-transfer intermediates have demonstrated that sulfur-transfer is possibly achieved by small conformational changes of the catalytic loops. Also, no additional partners to SufS-SufE is reported, which is different from the case of IscS-IscU. Interestingly, it is known that SufS-SufE is relatively resistant to oxidative species, whereas IscS-IscU not [109–111]. This feature of SufS-SufE may be considered based on the structure of the shorter loop enabling the facile sulfur-transfer.
For further understanding type-dependent features of the complexes, it is necessary to determine the structures of other type I and II cysteine desulfurase-partner complexes, e.g, NifS-NifU. Also, it is worthwhile to examine whether cysteine desulfurases can interact with not only sulfur acceptor proteins, but also the additional partners. For example, type II SufS-SufU from B. subtilis is supposed to bind to a frataxin-type protein [83]. However, this possible association of B. subtilis frataxin-like protein with SufS-SufU was reported before unveiling the X-ray crystal structure of the SufS-SufU complex [116], and there are no additional reports of three-dimensional structures of B. subtilis frataxin-like-protein-bound SufS-SufU. To gain further idea, structural analysis of the frataxin-like-protein-bound SufS-SufU complex might be necessarily conducted, followed by comparison of it to the other ternary complexes such as IscS-IscU-IscX.
Structures of Eukaryotic Cysteine Desulfurase NFS1 in Complex with its Partners in the Mitochondrial ISC System
The mitochondrial ISC system [124] has recently been of great interest in research related to not only the diversity and evolution of Fe-S cluster biosynthesis, but also the relationship of the ISC system to human diseases [125]. The mitochondrial ISC system is a more complicated system than that of bacteria, containing 18 known proteins. However, the basic reaction steps (i.e., sulfur activation and Fe-S cluster assembly) are well conserved between the bacterial and mitochondrial ISC systems. The key components of the mitochondrial ISC system are type I cysteine desulfurase NFS1 and the Fe-S cluster biosynthetic scaffold ISCU [126], which are homologous to IscS and IscU, respectively. In addition, NFS1 and ISCU interact with each other, similar to IscS and IscU [39,40].
However, there are several unique features of the NFS1 and NFS1-ISCU systems [127] compared to the IscS and IscS-IscU systems [39,40]. For example, NFS1 must form a complex with ISD11 [128–130], a member of the LYRM protein family [131]. Moreover, ISD11 must be in complex with the acyl carrier protein (ACP) [132], which plays a role in mitochondrial fatty acid synthesis and lipoic acid formation. The NFS1-ISD11-ACP complex, in which E. coli ACP was used instead of mitochondrial ACP, has been isolated as a functional complex to study its characteristic properties [133]. X-ray crystal structure analysis of NFS1-ISD11-ACP (Fig. 11A) [127] demonstrated that the overall folds and PLP-binding sites of NFS1 and IscS are very similar. However, NFS1 shows disordered segments that are not observed in IscS. For example, the N-terminal region (e.g., the residues from Tyr85 to Gly96) are disordered. The unusually disordered region of the dimer interface of NFS1 are in contact with the PLP moiety. Related to the disordered region, the dimeric architecture of NFS1 appears to be flexible compared to that of IscS. Considering this, it has been proposed that an NFS1 domain orientation may be related to the functions of NFS1-involved protein complexes. This hypothesis is supported by the fact that the disordered regions of NFS1-ISD11-ACP are more structured in NFS1-ISD11-ACP-ISCU in both Zn2+-bound and metal-free states (Figs. 11B, 11C).
Figure 11 .
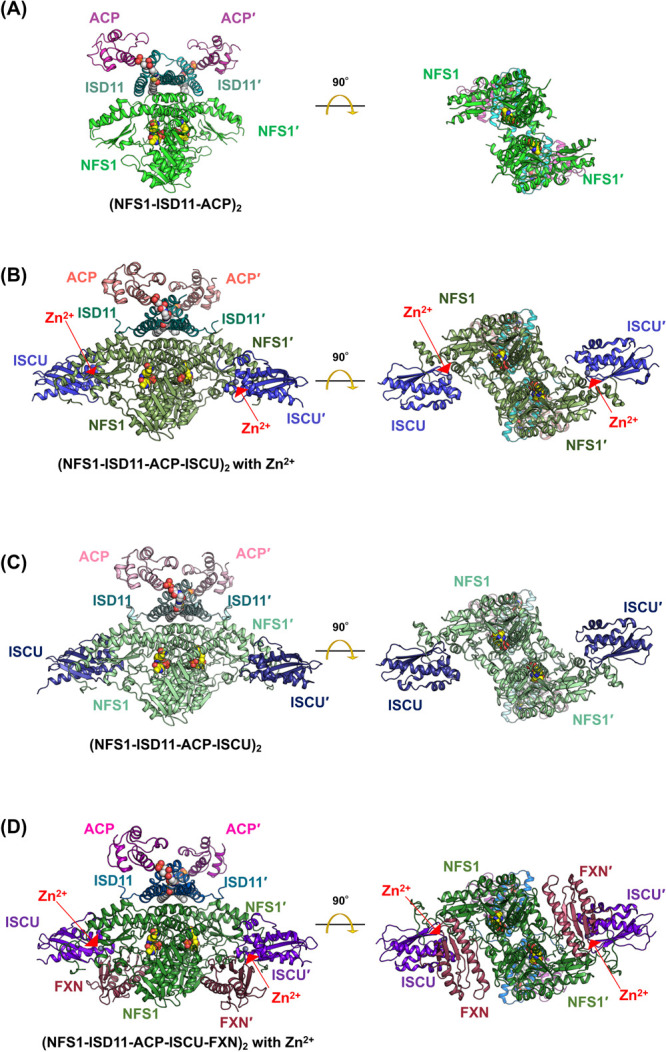
Structures of cysteine desulfurase NFS1 in complex with its partners in the mitochondrial ISC system. (A) NFS1-ISD11-ACP [PDB ID: 5wgb]. (B) NFS1-ISD11-ACP-ISCU with Zn2+ [PDB ID: 5wlw]. (C) NFS1-ISD11-ACP-ISCU [PDB ID: 5wkp]. (D) NFS1-ISD11-ACP-ISCU-FXN [PDB ID: 6nzu]. PLP moieties are shown in sphere models (colored in yellow for carbon, red for oxygen, and blue for nitrogen). The fatty acid-linked (S)-dodecanoyl-4'-phosphopantetheine (Ligand ID: 8Q1) molecules in ISD11-ACP interfaces are shown in sphere models (colored in white for carbon, red for oxygen, and blue for nitrogen). Zn2+ ions are shown in grey sphere models.
Unlike the N-terminal regions, the catalytic Cys-containing loops in both NFS1 and IscS are commonly disordered, as found in other type I enzymes [127]. However, visualizing an intermediate state of the NFS1 catalytic loop (Fig. 11D) has recently been reported [134] using cryo-electron microscopy (cryo-EM) analysis of NFS1-ISD11-ACP-ISCU in complex with frataxin (FTX) [135–139]. In this case, FTX can be likely to contribute to the stabilization of the intermediate state of the loop, which may also give a hint for understanding the system of IscS-IscU with other partners.
The comparison of NFS1 with not only IscS, but also SufS will give further insights into structural evolution of the cysteine desulfurases. Actually, type II E. coli SufS also utilizes a structural change of the β-hairpin region close to the PLP [95–97]. This structural change of SufS upon the binding of the partner is similar to that of NFS1, although SufS and NFS1 are different cysteine desulfurase types. Perhaps, structural evolution of the cysteine desulfurases may be rather complicated as expected. In other words, the evolution may not be just from type I IscS to type I NFS1. Clearly, structural studies of a variety of the type I and II cysteine desulfurases including eukaryotic ones are necessary for understanding the diversity and evolution of the cysteine desulfurases.
An Inactive Cysteine Desulfurase Paralog with PMP Rather than PLP
An unusual example of IscS from A. fulgidus (Af IscS) has recently been reported using biochemical and X-ray crystallographic analyses (Fig. 12) [92,93]. Interestingly, Af IscS wild-type (WT) has Asp199 and pyridoxamine-5'-phosphate (PMP) at positions equivalent to the conserved Lys and PLP of type I cysteine desulfurases, respectively. Because of the lack of PLP-Lys, Af IscS WT has no l-cysteine desulfurase activity. Further studies on the Af IscS D199K variant showed that it also has PMP. Thus, it was proposed that Af IscS may function solely as a scaffold protein, providing one Cys ligand to bind to the [2Fe-2S] cluster onto the Af IscS-IscU complex [40].
Figure 12 .
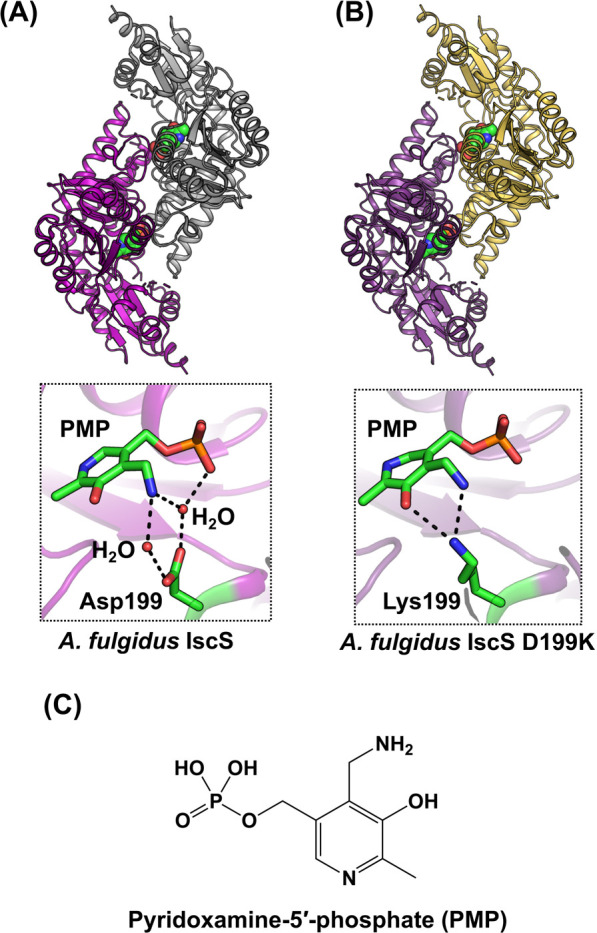
Structures of A. fulgidus IscS, an inactive cysteine desulfurase paralog, containing pyridoxamine-5'-phosphate (PMP). (A) A. fulgidus IscS wild-type [PDB ID: 4hvk]. (B) D199K variant [PDB ID: 4r5f]. (C) Chemical structure of PMP.
However, certain open questions remain, for example, how does [2Fe-2S] cluster assembly proceed without a cysteine desulfurase function in IscS-IscU from A. fulgidus? In a previous report [93], authors hypothesized that sulfide ion (S2–) might be a sulfur source for de novo synthesized Fe-S cluster onto Af IscS-IscU, since A. fulgidus is a sulfate-reducing archaeon that can produce S2–. Still, it is uncertain why “PMP” was bound in Af IscS wild-type (WT) and D199K [92,93], and why “PLP” was bound in [2Fe-2S]-cluster-bound Af IscS-IscU D35A [40]. Further studies are needed for clarifying their PLP or PMP-binding property and roles of these paralogs in Fe-S cluster biosynthesis. Here, it should be noted that not all archaea have such an unusual IscS without PLP. For example, Methanosarcina acetivorans, which is one of the methanogenic archaea, has a functional (but not essential) ISC system [140]. This ISC system is composed of iscSU and M. acetivorans IscS was characterized as a typical PLP-dependent cysteine desulfurase, not the PMP-bound type.
Concluding Remarks and Outlook
This review summarizes the findings of recent studies conducted on cysteine desulfurases involved in Fe-S cluster biosynthesis. These findings provide insights into their catalytic mechanisms involving common intermediates and the utilization of distinct features between the two types, including human cysteine desulfurase NFS1 and the inactive cysteine paralog Af IscS. Notably, the multidisciplinary approaches using state-of-the-art techniques (e.g., X-ray crystallography, cryo-EM, NMR and SAXS) in structural biology is of great importance for understanding not only details of cysteine desulfurases mechanisms, but also the diversity and evolution of Fe-S cluster biosynthesis. Also, it is necessary to develop anaerobic sample-handling techniques in structure-based studies of the Fe-S cluster biosynthetic reactions involving the cysteine desulfurases and other partners because of instability of the Fe-S clusters against oxidative species.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
T.F., R.N., K.K., and Y.T. wrote the manuscript and prepared the figures.
Acknowledgments
This work was financially supported by JSPS KAKENHI Grants 17K14510 (to T.F.) and 20H03204 (to Y.T.), and by a Sasakawa Scientific Research Grant from The Japan Science Society (to R.N.).
References
- [1].Beinert, H., Holm, R. H., Munck, E.. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 277, 653–659 (1997). https://doi.org/10.1126/science.277.5326.653 [DOI] [PubMed] [Google Scholar]
- [2].Johnson, D. C., Dean, D. R., Smith, A. D., Johnson, M. K.. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74, 247–281 (2005). https://doi.org/10.1146/annurev.biochem.74.082803.133518 [DOI] [PubMed] [Google Scholar]
- [3].Lill, R. Function and biogenesis of iron-sulphur proteins. Nature 460, 831–838 (2009). https://doi.org/10.1038/nature08301 [DOI] [PubMed] [Google Scholar]
- [4].Py, B., Barras, F.. Building Fe-S proteins: Bacterial strategies. Nat. Rev. Microbiol. 8, 436–446 (2010). https://doi.org/10.1038/nrmicro2356 [DOI] [PubMed] [Google Scholar]
- [5].Mettert, E. L., Kiley, P. J.. Fe-S proteins that regulate gene expression. Biochim. Biophys. Acta 1853, 1284–1293 (2015). https://doi.org/10.1016/j.bbamcr.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pain, D., Dancis, A.. Roles of Fe-S proteins: From cofactor synthesis to iron homeostasis to protein synthesis. Curr. Opin. Genet. Dev. 38, 45–51 (2016). https://doi.org/10.1016/j.gde.2016.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Roche, B., Aussel, L., Ezraty, B., Mandin, P., Py, B., Barras, F.. Reprint of: Iron/sulfur proteins biogenesis in prokaryotes: Formation, regulation and diversity. Biochim. Biophys. Acta 1827, 923–937 (2013). https://doi.org/10.1016/j.bbabio.2013.05.001 [DOI] [PubMed] [Google Scholar]
- [8].Rees, D. C., Howard, J. B.. The interface between the biological and inorganic worlds: iron-sulfur metalloclusters. Science 300, 929–931 (2003). https://doi.org/10.1126/science.1083075 [DOI] [PubMed] [Google Scholar]
- [9].Liu, J., Chakraborty, S., Hosseinzadeh, P., Yu, Y., Tian, S., Petrik, I., et al. Metalloproteins containing cytochrome, iron-sulfur, or copper redox centers. Chem. Rev. 114, 4366–4469 (2014). https://doi.org/10.1021/cr400479b [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Beinert, H., Kennedy, M. C., Stout, C. D.. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 96, 2335–2374 (1996). https://doi.org/10.1021/cr950040z [DOI] [PubMed] [Google Scholar]
- [11].Broderick, J. B., Duffus, B. R., Duschene, K. S., Shepard, E. M.. Radical S-adenosylmethionine enzymes. Chem. Rev. 114, 4229–4317 (2014). https://doi.org/10.1021/cr4004709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Seefeldt, L. C., Yang, Z. Y., Lukoyanov, D. A., Harris, D. F., Dean, D. R., Raugei, S., et al. Reduction of substrates by nitrogenases. Chem. Rev. 120, 5082–5106 (2020). https://doi.org/10.1021/acs.chemrev.9b00556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lubitz, W., Ogata, H., Rüdiger, O., Reijerse, E.. Hydrogenases. Chem. Rev. 114, 4081–4148 (2014). https://doi.org/10.1021/cr4005814 [DOI] [PubMed] [Google Scholar]
- [14].Can, M., Armstrong, F. A., Ragsdale, S. W.. Structure, function, and mechanism of the nickel metalloenzymes, CO dehydrogenase, and acetyl-CoA synthase. Chem. Rev. 114, 4149–4174 (2014). https://doi.org/10.1021/cr400461p [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Holm, R. H., Lo, W.. Structural conversions of synthetic and protein-bound iron-sulfur clusters. Chem. Rev. 116, 13685–13713 (2016). https://doi.org/10.1021/acs.chemrev.6b00276 [DOI] [PubMed] [Google Scholar]
- [16].Ohta, S., Ohki, Y.. Impact of ligands and media on the structure and properties of biological and biomimetic iron-sulfur clusters. Coord. Chem. Rev. 338, 207–225 (2017). https://doi.org/10.1016/j.ccr.2017.02.018 [Google Scholar]
- [17].Malkin, R., Rabinowitz, J. C.. The reconstitution of clostridial ferredoxin. Biochem. Biophys. Res. Commun. 23, 822–827 (1966). https://doi.org/10.1016/0006-291x(66)90561-4 [DOI] [PubMed] [Google Scholar]
- [18].Takahashi, Y., Mitsui, A., Hase, T., Matsubara, H.. Formation of the iron-sulfur cluster of ferredoxin in isolated chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 83, 2434–2437 (1986). https://doi.org/10.1073/pnas.83.8.2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Jacobson, M. R., Cash, V. L., Weiss, M. C., Laird, N. F., Newton, W. E., Dean, D. R.. Biochemical and genetic-analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol. Gen. Genet. 219, 49–57 (1989). https://doi.org/10.1007/BF00261156 [DOI] [PubMed] [Google Scholar]
- [20].Ayala-Castro, C., Saini, A., Outten, F. W.. Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 72, 110–125 (2008). https://doi.org/10.1128/mmbr.00034-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zheng, L., Cash, V. L., Flint, D. H., Dean, D. R.. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273, 13264–13272 (1998). https://doi.org/10.1074/jbc.273.21.13264 [DOI] [PubMed] [Google Scholar]
- [22].Takahashi, Y., Nakamura, M.. Functional assignment of the ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster involved in the assembly of Fe-S clusters in Escherichia coli. J. Biochem. 126, 917–926 (1999). https://doi.org/10.1093/oxfordjournals.jbchem.a022535 [DOI] [PubMed] [Google Scholar]
- [23].Nakamura, M., Saeki, K., Takahashi, Y.. Hyperproduction of recombinant ferredoxins in Escherichia coli by coexpression of the ORF1-ORF2-iscS-iscU-iscA-hscB-hscA-fdx-ORF3 gene cluster. J. Biochem. 126, 10–18 (1999). https://doi.org/10.1093/oxfordjournals.jbchem.a022409 [DOI] [PubMed] [Google Scholar]
- [24].Tokumoto, U., Takahashi, Y.. Genetic analysis of the isc operon in Escherichia coli involved in the biogenesis of cellular iron-sulfur proteins. J. Biochem. 130, 63–71 (2001). https://doi.org/10.1093/oxfordjournals.jbchem.a002963 [DOI] [PubMed] [Google Scholar]
- [25].Tokumoto, U., Nomura, S., Minami, Y., Mihara, H., Kato, S., Kurihara, T., et al. Network of protein-protein interactions among iron-sulfur cluster assembly proteins in Escherichia coli. J. Biochem. 131, 713–719 (2002). https://doi.org/10.1093/oxfordjournals.jbchem.a003156 [DOI] [PubMed] [Google Scholar]
- [26].Tanaka, N., Kanazawa, M., Tonosaki, K., Yokoyama, N., Kuzuyama, T., Takahashi, Y.. Novel features of the ISC machinery revealed by characterization of Escherichia coli mutants that survive without iron-sulfur clusters. Mol. Microbiol. 99, 835–848 (2016). https://doi.org/10.1111/mmi.13271 [DOI] [PubMed] [Google Scholar]
- [27].Agar, J. N., Krebs, C., Frazzon, J., Huynh, B. H., Dean, D. R., Johnson, M. K.. IscU as a scaffold for iron-sulfur cluster biosynthesis: Sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry 39, 7856–7862. (2000). https://doi.org/10.1021/bi000931n [DOI] [PubMed] [Google Scholar]
- [28].Smith, A. D., Agar, J. N., Johnson, K. A., Frazzon, J., Amster, I. J., Dean, D. R., et al. Sulfur transfer from IscS to IscU: The first step in iron-sulfur cluster biosynthesis. J. Am. Chem. Soc. 123, 11103–11104 (2001). https://doi.org/10.1021/ja016757n [DOI] [PubMed] [Google Scholar]
- [29].Urbina, H. D., Silberg, J. J., Hoff, K. G., Vickery, L. E.. Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J. Biol. Chem. 276, 44521–44526 (2001). https://doi.org/10.1074/jbc.M106907200 [DOI] [PubMed] [Google Scholar]
- [30].Kato, S., Mihara, H., Kurihara, T., Takahashi, Y., Tokumoto, U., Yoshimura, T., et al. Cys-328 of IscS and Cys-63 of IscU are the sites of disulfide bridge formation in a covalently bound IscS/IscU complex: Implications for the mechanism of iron-sulfur cluster assembly. Proc. Natl. Acad. Sci. U.S.A. 99, 5948–5952 (2002). https://doi.org/10.1073/pnas.082123599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ramelot, T. A., Cort, J. R., Goldsmith-Fischman, S., Kornhaber, G. J., Xiao, R., Shastry, R., et al. A. Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site. J. Mol. Biol. 344, 567–583 (2004). https://doi.org/10.1016/j.jmb.2004.08.038 [DOI] [PubMed] [Google Scholar]
- [32].Liu, J. Y., Oganesyan, N., Shin, D. H., Jancarik, J., Yokota, H., Kim, R., et al. Structural characterization of an iron-sulfur cluster assembly protein IscU in a zinc-bound form. Proteins 59, 875–881 (2005). https://doi.org/10.1002/prot.20421 [DOI] [PubMed] [Google Scholar]
- [33].Shimomura, Y., Kamikubo, H., Nishi, Y., Masako, T., Kataoka, M., Kobayashi, Y., et al. Characterization and crystallization of an IscU-type scaffold protein with bound [2Fe-2S] cluster from the hyperthermophile, Aquifex aeolicus. J. Biochem. 142, 577–586 (2007). https://doi.org/10.1093/jb/mvm163 [DOI] [PubMed] [Google Scholar]
- [34].Shimomura, Y., Wada, K., Fukuyama, K., Takahashi, Y.. The asymmetric trimeric architecture of [2Fe-2S] IscU: Implications for its scaffolding during iron-sulfur cluster biosynthesis. J. Mol. Biol. 383, 133–143 (2008). https://doi.org/10.1016/j.jmb.2008.08.015 [DOI] [PubMed] [Google Scholar]
- [35].Tanaka, N., Yuda, E., Fujishiro, T., Hirabayashi, K., Wada, K., Takahashi, Y.. Identification of IscU residues critical for de novo iron-sulfur cluster assembly. Mol. Microbiol. 112, 1769–1783 (2019). https://doi.org/10.1111/mmi.14392 [DOI] [PubMed] [Google Scholar]
- [36].Sato, S., Matsushima, Y., Kanazawa, M., Tanaka, N., Fujishiro, T., Kunichika, K., et al. Evidence for dynamic in vivo interconversion of the conformational states of IscU during iron-sulfur cluster biosynthesis. Mol. Microbiol. 115, 807–818 (2021). https://doi.org/10.1111/mmi.14646 [DOI] [PubMed] [Google Scholar]
- [37].Kunichika, K., Nakamura, R., Fujishiro, T., Takahashi, Y.. The structure of the dimeric state of IscU harboring two adjacent [2Fe-2S] clusters provides mechanistic insights into cluster conversion to [4Fe-4S]. Biochemistry 60, 1569–1572 (2021). https://doi.org/10.1021/acs.biochem.1c00112 [DOI] [PubMed] [Google Scholar]
- [38].Chandramouli, K., Unciuleac, M. C., Naik, S., Dean, D. R., Huynh, B. H., Johnson, M. K.. Formation and properties of [4Fe-4S] clusters on the IscU scaffold protein. Biochemistry 46, 6804–6811 (2007). https://doi.org/10.1021/bi6026659 [DOI] [PubMed] [Google Scholar]
- [39].Shi, R., Proteau, A., Villarroya, M., Moukadiri, I., Zhang, L. H., Trempe, J. F., et al. Structural basis for Fe-S cluster assembly and tRNA thiolation mediated by IscS protein-protein interactions. PLoS Biol. 8, e1000354 (2010). https://doi.org/10.1371/journal.pbio.1000354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Marinoni, E. N., de Oliveira, J. S., Nicolet, Y., Raulfs, E. C., Amara, P., Dean, D. R., et al. (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer. Angew. Chem. Int. Ed. Engl. 51, 5439–5442 (2012). https://doi.org/10.1002/anie.201201708 [DOI] [PubMed] [Google Scholar]
- [41].Takahashi, Y., Tokumoto, U.. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277, 28380–28383 (2002). https://doi.org/10.1074/jbc.C200365200 [DOI] [PubMed] [Google Scholar]
- [42].Rangachari, K., Davis, C. T., Eccleston, J. F., Hirst, E. M. A., Saldanha, J. W., Strath, M., et al. SufC hydrolyzes ATP and interacts with SufB from Thermotoga maritima. FEBS Lett. 514, 225–228 (2002). https://doi.org/10.1016/S0014-5793(02)02369-4 [DOI] [PubMed] [Google Scholar]
- [43].Outten, F. W., Wood, M. J., Munoz, F. M., Storz, G.. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J. Biol. Chem. 278, 45713–45719 (2003). https://doi.org/10.1074/jbc.M308004200 [DOI] [PubMed] [Google Scholar]
- [44].Ollagnier-de-Choudens, S., Lascoux, D., Loiseau, L., Barras, F., Forest, E., Fontecave, M.. Mechanistic studies of the SufS-SufE cysteine desulfurase: Evidence for sulfur transfer from SufS to SufE. FEBS Lett. 555, 263–267 (2003). https://doi.org/10.1016/S0014-5793(03)01244-4 [DOI] [PubMed] [Google Scholar]
- [45].Outten, F. W., Djaman, O., Storz, G.. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52, 861–872 (2004). https://doi.org/10.1111/j.1365-2958.2004.04025.x [DOI] [PubMed] [Google Scholar]
- [46].Goldsmith-Fischman, S., Kuzin, A., Edstrom, W. C., Benach, J., Shastry, R., Xiao, R., et al. The SufE sulfur-acceptor protein contains a conserved core structure that mediates interdomain interactions in a variety of redox protein complexes. J. Mol. Biol. 344, 549–565 (2004). https://doi.org/10.1016/j.jmb.2004.08.074 [DOI] [PubMed] [Google Scholar]
- [47].Eccleston, J. F., Petrovic, A., Davis, C. T., Rangachari, K., Wilson, R. J. M. I.. The kinetic mechanism of the SufC ATPase: The cleavage step is accelerated by SufB. J. Biol. Chem. 281, 8371–8378 (2006). https://doi.org/10.1074/jbc.M513455200 [DOI] [PubMed] [Google Scholar]
- [48].Kitaoka, S., Wada, K., Hasegawa, Y., Minami, Y., Fukuyama, K., Takahashi, Y.. Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery. FEBS Lett. 580, 137–143 (2006). https://doi.org/10.1016/j.febslet.2005.11.058 [DOI] [PubMed] [Google Scholar]
- [49].Layer, G., Gaddam, S. A., Ayala-Castro, C. N., Choudens, S. O., Lascoux, D., Fontecave, M., et al. SufE transfers sulfur from SufS to SufB for iron-sulfur cluster assembly. J. Biol. Chem. 282, 13342–13350 (2007). https://doi.org/10.1074/jbc.M608555200 [DOI] [PubMed] [Google Scholar]
- [50].Wada, K., Sumi, N., Nagai, R., Iwasaki, K., Sato, T., Suzuki, K., et al. Molecular dynamism of Fe-S cluster biosynthesis implicated by the structure of the SufC2-SufD2 complex. J. Mol. Biol. 387, 245–258 (2009). https://doi.org/10.1016/j.jmb.2009.01.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Saini, A., Mapolelo, D. T., Chahal, H. K., Johnson, M. K., Outten, F. W.. SufD and SufC ATPase activity are required for iron acquisition during in vivo Fe-S cluster formation on SufB. Biochemistry 49, 9402–9412 (2010). https://doi.org/10.1021/bi1011546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wollers, S., Layer, G., Garcia-Serres, R., Signor, L., Clemancey, M., Latour, J. M., et al. Iron-sulfur (Fe-S) cluster assembly: The SufBCD complex is a new type of Fe-S scaffold with a flavin redox cofactor. J. Biol. Chem. 285, 23331–23341 (2010). https://doi.org/10.1074/jbc.M110.127449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Boyd, E. S., Thomas, K. M., Dai, Y. Y., Boyd, J. M., Outten, F. W.. Interplay between oxygen and Fe-S cluster biogenesis: Insights from the Suf pathway. Biochemistry 53, 5834–5847 (2014). https://doi.org/10.1021/bi500488r [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Pérard, J., Ollagnier de Choudens, S.. Iron-sulfur clusters biogenesis by the SUF machinery: Close to the molecular mechanism understanding. J. Biol. Inorg. Chem. 23, 581–596 (2018). https://doi.org/10.1007/s00775-017-1527-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Hirabayashi, K., Yuda, E., Tanaka, N., Katayama, S., Iwasaki, K., Matsumoto, T., et al. Functional dynamics revealed by the structure of the SufBCD complex, a novel ATP-binding cassette (ABC) protein that serves as a scaffold for iron-sulfur cluster biogenesis. J. Biol. Chem. 290, 29717–29731 (2015). https://doi.org/10.1074/jbc.M115.680934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Yuda, E., Tanaka, N., Fujishiro, T., Yokoyama, N., Hirabayashi, K., Fukuyama, K., et al. Mapping the key residues of SufB and SufD essential for biosynthesis of iron-sulfur clusters. Sci. Rep. 7, 9387 (2017). https://doi.org/10.1038/s41598-017-09846-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Blahut, M., Sanchez, E., Fisher, C. E., Outten, F. W.. Fe-S cluster biogenesis by the bacterial Suf pathway. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118829 (2020). https://doi.org/10.1016/j.bbamcr.2020.118829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zheng, L., White, R. H., Cash, V. L., Jack, R. F., Dean, D. R.. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 90, 2754–2758 (1993). https://doi.org/10.1073/pnas.90.7.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zheng, L., White, R. H., Cash, V. L., Dean, D. R.. Mechanism for the desulfurization of l-cysteine catalyzed by the nifS gene product. Biochemistry 33, 4714–4720 (1994). https://doi.org/10.1021/bi00181a031 [DOI] [PubMed] [Google Scholar]
- [60].Fu, W. G., Jack, R. F., Morgan, T. V., Dean, D. R., Johnson, M. K.. nifU gene-product from Azotobacter vinelandii is a homodimer that contains two identical [2Fe-2S] clusters. Biochemistry 33, 13455–13463 (1994). https://doi.org/10.1021/bi00249a034 [DOI] [PubMed] [Google Scholar]
- [61].Olson, J. W., Agar, J. N., Johnson, M. K., Maier, R. J.. Characterization of the NifU and NifS Fe-S cluster formation proteins essential for viability in Helicobacter pylori. Biochemistry 39, 16213–16219 (2000). https://doi.org/10.1021/bi001744s [DOI] [PubMed] [Google Scholar]
- [62].Agar, J. N., Yuvaniyama, P., Jack, R. F., Cash, V. L., Smith, A. D., Dean, D. R., et al. Modular organization and identification of a mononuclear iron-binding site within the NifU protein. J. Biol. Inorg. Chem. 5, 167–177 (2000). https://doi.org/10.1007/s007750050361 [DOI] [PubMed] [Google Scholar]
- [63].Dos Santos, P. C., Smith, A. D., Frazzon, J., Cash, V. L., Johnson, M. K., Dean, D. R.. Iron-sulfur cluster assembly: NifU-directed activation of the nitrogenase Fe protein. J. Biol. Chem. 279, 19705–19711 (2004). https://doi.org/10.1074/jbc.M400278200 [DOI] [PubMed] [Google Scholar]
- [64].Tokumoto, U., Kitamura, S., Fukuyama, K., Takahashi, Y.. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: Functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J. Biochem. 136, 199–209 (2004). https://doi.org/10.1093/jb/mvh104 [DOI] [PubMed] [Google Scholar]
- [65].Ali, V., Shigeta, Y., Tokumoto, U., Takahashi, Y., Nozaki, T.. An intestinal parasitic protist, Entamoeba histolytica, possesses a non-redundant nitrogen fixation-like system for iron-sulfur cluster assembly under anaerobic conditions. J. Biol. Chem. 279, 16863–16874 (2004). https://doi.org/10.1074/jbc.M313314200 [DOI] [PubMed] [Google Scholar]
- [66].Yuvaniyama, P., Agar, J. N., Cash, V. L., Jothnson, M. K., Dean, D. R.. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc. Natl. Acad. Sci. U.S.A. 97, 599–604 (2000). https://doi.org/10.1073/pnas.97.2.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Smith, A. D., Jameson, G. N. L., Dos Santos, P. C., Agar, J. N., Naik, S., Krebs, C., et al. NifS-mediated assembly of [4Fe-4S] clusters in the N- and C-terminal domains of the NifU scaffold protein. Biochemistry 44, 12955–12969 (2005). https://doi.org/10.1021/bi051257i [DOI] [PubMed] [Google Scholar]
- [68].Burén, S., Jiménez-Vicente, E., Echavarri-Erasun, C., Rubio, L. M.. Biosynthesis of nitrogenase cofactors. Chem. Rev. 120, 4921–4968 (2020). https://doi.org/10.1021/acs.chemrev.9b00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Jasniewski, A. J., Lee, C. C., Ribbe, M. W., Hu, Y.. Reactivity, mechanism, and assembly of the alternative nitrogenases. Chem. Rev. 120, 5107–5157 (2020). https://doi.org/10.1021/acs.chemrev.9b00704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kobayashi, K., Ehrlich, S. D., Albertini, A., Amati, G., Andersen, K. K., Arnaud, M., et al. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 100, 4678–4683 (2003). https://doi.org/10.1073/pnas.0730515100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Huet, G., Daffe, M., Saves, I.. Identification of the Mycobacterium tuberculosis SUF machinery as the exclusive mycobacterial system of [Fe-S] cluster assembly: Evidence for its implication in the pathogen’s survival. J. Bacteriol. 187, 6137–6146 (2005). https://doi.org/10.1128/Jb.187.17.6137-6146.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Huet, G., Castaing, J. P., Fournier, D., Daffé, M., Saves, I.. Protein splicing of SufB is crucial for the functionality of the Mycobacterium tuberculosis SUF machinery. J. Bacteriol. 188, 3412–3414 (2006). https://doi.org/10.1128/jb.188.9.3412-3414.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Riboldi, G. P., Larson, T. J., Frazzon, J.. Enterococcus faecalis sufCDSUB complements Escherichia coli sufABCDSE. FEMS Microbiol. Lett. 320, 15–24 (2011). https://doi.org/10.1111/j.1574-6968.2011.02284.x [DOI] [PubMed] [Google Scholar]
- [74].Roberts, C. A., Al-Tameemi, H. M., Mashruwala, A. A., Rosario-Cruz, Z., Chauhan, U., Sause, W. E., et al. The Suf iron-sulfur cluster biosynthetic system is essential in Staphylococcus aureus, and decreased Suf function results in global metabolic defects and reduced survival in human neutrophils. Infect Immun. 85, e00100-17 (2017). https://doi.org/10.1128/iai.00100-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Yokoyama, N., Nonaka, C., Ohashi, Y., Shioda, M., Terahata, T., Chen, W., et al. Distinct roles for U-type proteins in iron-sulfur cluster biosynthesis revealed by genetic analysis of the Bacillus subtilis sufCDSUB operon. Mol. Microbiol. 107, 688–703 (2018). https://doi.org/10.1111/mmi.13907 [DOI] [PubMed] [Google Scholar]
- [76].Riboldi, G. P., Verli, H., Frazzon, J.. Structural studies of the Enterococcus faecalis SufU [Fe-S] cluster protein. BMC Biochem. 10, 3 (2009). https://doi.org/10.1186/1471-2091-10-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Albrecht, A. G., Netz, D. J. A., Miethke, M., Pierik, A. J., Burghaus, O., Peuckert, F., et al. SufU is an essential iron-sulfur cluster scaffold protein in Bacillus subtilis. J. Bacteriol. 192, 1643–1651 (2010). https://doi.org/10.1128/Jb.01536-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Albrecht, A. G., Peuckert, F., Landmann, H., Miethke, M., Seubert, A., Marahiel, M. A.. Mechanistic characterization of sulfur transfer from cysteine desulfurase SufS to the iron-sulfur scaffold SufU in Bacillus subtilis. FEBS Lett. 585, 465–470 (2011). https://doi.org/10.1016/j.febslet.2011.01.005 [DOI] [PubMed] [Google Scholar]
- [79].Riboldi, G. P., de Oliveira, J. S., Frazzon, J.. Enterococcus faecalis SufU scaffold protein enhances SufS desulfurase activity by acquiring sulfur from its cysteine-153. Biochim. Biophys. Acta 1814, 1910–1918 (2011). https://doi.org/10.1016/j.bbapap.2011.06.016 [DOI] [PubMed] [Google Scholar]
- [80].Kaiser, J. T., Clausen, T., Bourenkow, G. P., Bartunik, H. D., Steinbacher, S., Huber, R.. Crystal structure of a NifS-like protein from Thermotoga maritima: Implications for iron sulphur cluster assembly. J. Mol. Biol. 297, 451–464 (2000). https://doi.org/10.1006/jmbi.2000.3581 [DOI] [PubMed] [Google Scholar]
- [81].Cupp-Vickery, J. R., Urbina, H., Vickery, L. E.. Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J. Mol. Biol. 330, 1049–1059 (2003). https://doi.org/10.1016/s0022-2836(03)00690-9 [DOI] [PubMed] [Google Scholar]
- [82].Blahut, M., Wise, C. E., Bruno, M. R., Dong, G., Makris, T. M., Frantom, P. A., et al. Direct observation of intermediates in the SufS cysteine desulfurase reaction reveals functional roles of conserved active-site residues. J. Biol. Chem. 294, 12444–12458 (2019). https://doi.org/10.1074/jbc.RA119.009471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Blauenburg, B., Mielcarek, A., Altegoer, F., Fage, C. D., Linne, U., Bange, G., et al. Crystal structure of Bacillus subtilis cysteine desulfurase SufS and its dynamic interaction with frataxin and scaffold protein SufU. PLoS One 11, e0158749 (2016). https://doi.org/10.1371/journal.pone.0158749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Nakamura, R., Hikita, M., Ogawa, S., Takahashi, Y., Fujishiro, T.. Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes. FEBS J. 287, 1138–1154 (2020). https://doi.org/10.1111/febs.15081 [DOI] [PubMed] [Google Scholar]
- [85].Black, K. A., Dos Santos, P. C.. Shared-intermediates in the biosynthesis of thio-cofactors: Mechanism and functions of cysteine desulfurases and sulfur acceptors. Biochim. Biophys. Acta 1853, 1470–1480 (2015). https://doi.org/10.1016/j.bbamcr.2014.10.018 [DOI] [PubMed] [Google Scholar]
- [86].Mihara, H., Esaki, N.. Bacterial cysteine desulfurases: Their function and mechanisms. Appl. Microbiol. Biotechnol. 60, 12–23 (2002). https://doi.org/10.1007/s00253-002-1107-4 [DOI] [PubMed] [Google Scholar]
- [87].Mueller, E. G. Trafficking in persulfides: Delivering sulfur in biosynthetic pathways. Nat. Chem. Biol. 2, 185–194 (2006). https://doi.org/10.1038/nchembio779 [DOI] [PubMed] [Google Scholar]
- [88].Braymer, J. J., Freibert, S. A., Rakwalska-Bange, M., Lill, R.. Mechanistic concepts of iron-sulfur protein biogenesis in biology. Biochim. Biophys. Acta Mol. Cell Res. 1868, 118863 (2021). https://doi.org/10.1016/j.bbamcr.2020.118863 [DOI] [PubMed] [Google Scholar]
- [89].Kispal, G., Csere, P., Prohl, C., Lill, R.. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18, 3981–3989 (1999). https://doi.org/10.1093/emboj/18.14.3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Schilke, B., Voisine, C., Beinert, H., Craig, E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 96, 10206–10211 (1999) https://doi.org/10.1073/pnas.96.18.10206 [DOI] [PMC free article] [PubMed]
- [91].Garland, S. A., Hoff, K., Vickery, L. E., Culotta, V. C.. Saccharomyces cerevisiae ISU1 and ISU2: members of a well-conserved gene family for iron-sulfur cluster assembly. J. Mol. Biol. 294, 897–907 (1999). https://doi.org/10.1006/jmbi.1999.3294 [DOI] [PubMed] [Google Scholar]
- [92].Yamanaka, Y., Zeppieri, L., Nicolet, Y., Marinoni, E. N., de Oliveira, J. S., Odaka, M., et al. Crystal structure and functional studies of an unusual l-cysteine desulfurase from Archaeoglobus fulgidus. Dalton Trans. 42, 3092–3099 (2013). https://doi.org/10.1039/C2DT32101G [DOI] [PubMed] [Google Scholar]
- [93].Pagnier, A., Nicolet, Y., Fontecilla-Camps, J. C.. IscS from Archaeoglobus fulgidus has no desulfurase activity but may provide a cysteine ligand for [Fe2S2] cluster assembly. Biochim. Biophys. Acta 1853, 1457–1463 (2015). https://doi.org/10.1016/j.bbamcr.2014.10.015 [DOI] [PubMed] [Google Scholar]
- [94].Selbach, B., Earles, E., Dos Santos, P. C.. Kinetic analysis of the bisubstrate cysteine desulfurase SufS from Bacillus subtilis. Biochemistry 49, 8794–8802 (2010). https://doi.org/10.1021/bi101358k [DOI] [PubMed] [Google Scholar]
- [95].Dunkle, J. A., Bruno, M. R., Outten, F. W., Frantom, P. A.. Structural evidence for dimer-interface-driven regulation of the type II cysteine desulfurase, SufS. Biochemistry 58, 687–696 (2019). https://doi.org/10.1021/acs.biochem.8b01122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kim, D., Singh, H., Dai, Y., Dong, G., Busenlehner, L. S., Outten, F. W., et al. Changes in protein dynamics in Escherichia coli SufS reveal a possible conserved regulatory mechanism in type II cysteine desulfurase systems. Biochemistry 57, 5210–5217 (2018). https://doi.org/10.1021/acs.biochem.7b01275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Dunkle, J. A., Bruno, M. R., Frantom, P. A.. Structural evidence for a latch mechanism regulating access to the active site of SufS-family cysteine desulfurases. Acta Crystallogr. D Struct. Biol. 76, 291–301 (2020). https://doi.org/10.1107/s2059798320000790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Dai, Y., Kim, D., Dong, G., Busenlehner, L. S., Frantom, P. A., Outten, F. W.. SufE D74R substitution alters active site loop dynamics to further enhance SufE interaction with the SufS cysteine desulfurase. Biochemistry 54, 4824–4833 (2015). https://doi.org/10.1021/acs.biochem.5b00663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Singh, H., Dai, Y., Outten, F. W., Busenlehner, L. S.. Escherichia coli SufE sulfur transfer protein modulates the SufS cysteine desulfurase through allosteric conformational dynamics. J. Biol. Chem. 288, 36189–36200 (2013). https://doi.org/10.1074/jbc.M113.525709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zheng, C., Dos Santos, P. C.. Metallocluster transactions: Dynamic protein interactions guide the biosynthesis of Fe-S clusters in bacteria. Biochem. Soc. Trans. 46, 1593–1603 (2018). https://doi.org/10.1042/bst20180365 [DOI] [PubMed] [Google Scholar]
- [101].Foster, M. W., Mansy, S. S., Hwang, J., Penner-Hahn, J. E., Surerus, K. K., Cowan, J. A.. A mutant human IscU protein contains a stable [2Fe−2S]2+ center of possible functional significance. J. Am. Chem. Soc. 122, 6805–6806 (2000). https://doi.org/10.1021/ja000800+ [Google Scholar]
- [102].Wu, S. P., Wu, G., Surerus, K. K., Cowan, J. A.. Iron-sulfur cluster biosynthesis. Kinetic analysis of [2Fe-2S] cluster transfer from holo ISU to apo Fd: Role of redox chemistry and a conserved aspartate. Biochemistry 41, 8876–8885 (2002). https://doi.org/10.1021/bi0256781 [DOI] [PubMed] [Google Scholar]
- [103].Unciuleac, M. C., Chandramouli, K., Naik, S., Mayer, S., Huynh, B. H., Johnson, M. K., et al. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry 46, 6812–6821 (2007). https://doi.org/10.1021/bi6026665 [DOI] [PubMed] [Google Scholar]
- [104].Raulfs, E. C., O’Carroll, I. P., Dos Santos, P. C., Unciuleac, M. C., Dean, D. R.. In vivo iron-sulfur cluster formation. Proc. Natl. Acad. Sci. U.S.A. 105, 8591–8596 (2008). https://doi.org/10.1073/pnas.0803173105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Johnson, D. C., Unciuleac, M. C., Dean, D. R.. Controlled expression and functional analysis of iron-sulfur cluster biosynthetic components within Azotobacter vinelandii. J. Bacteriol. 188, 7551–7561 (2006). https://doi.org/10.1128/jb.00596-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Benoit, S. L., Holland, A. A., Johnson, M. K., Maier, R. J.. Iron-sulfur protein maturation in Helicobacter pylori: Identifying a Nfu-type cluster carrier protein and its iron-sulfur protein targets. Mol. Microbiol. 108, 379–396 (2018). https://doi.org/10.1111/mmi.13942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Nishio, K., Nakai, M.. Transfer of iron-sulfur cluster from NifU to apoferredoxin. J. Biol. Chem. 275, 22615–22618 (2000). https://doi.org/10.1074/jbc.C000279200 [DOI] [PubMed] [Google Scholar]
- [108].Yabe, T., Yamashita, E., Kikuchi, A., Morimoto, K., Nakagawa, A., Tsukihara, T., et al. Structural analysis of Arabidopsis CnfU protein: An iron-sulfur cluster biosynthetic scaffold in chloroplasts. J. Mol. Biol. 381, 160–173 (2008). https://doi.org/10.1016/j.jmb.2008.05.072 [DOI] [PubMed] [Google Scholar]
- [109].Sendra, M., de Choudens, S. O., Lascoux, D., Sanakis, Y., Fontecave, M.. The SUF iron-sulfur cluster biosynthetic machinery: Sulfur transfer from the SUFS-SUFE complex to SUFA. FEBS Lett. 581, 1362–1368 (2007). https://doi.org/10.1016/j.febslet.2007.02.058 [DOI] [PubMed] [Google Scholar]
- [110].Dai, Y. Y., Outten, F. W.. The E. coli SufS-SufE sulfur transfer system is more resistant to oxidative stress than IscS-IscU. FEBS Lett. 586, 4016–4022 (2012). https://doi.org/10.1016/j.febslet.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Selbach, B. P., Pradhan, P. K., Dos Santos, P. C.. Protected sulfur transfer reactions by the Escherichia coli Suf system. Biochemistry 52, 4089–4096 (2013). https://doi.org/10.1021/bi4001479 [DOI] [PubMed] [Google Scholar]
- [112].Loiseau, L., Ollagnier-de Choudens, S., Lascoux, D., Forest, E., Fontecave, M., Barras, F.. Analysis of the heteromeric CsdA-CsdE cysteine desulfurase, assisting Fe-S cluster biogenesis in Escherichia coli. J. Biol. Chem. 280, 26760–26769 (2005). https://doi.org/10.1074/jbc.M504067200 [DOI] [PubMed] [Google Scholar]
- [113].Trotter, V., Vinella, D., Loiseau, L., de Choudens, S. O., Fontecave, M., Barras, F.. The CsdA cysteine desulphurase promotes Fe/S biogenesis by recruiting Suf components and participates to a new sulphur transfer pathway by recruiting CsdL (ex-YgdL), a ubiquitin-modifying-like protein. Mol. Microbiol. 74, 1527–1542 (2009). https://doi.org/10.1111/j.1365-2958.2009.06954.x [DOI] [PubMed] [Google Scholar]
- [114].Kim, S., Park, S.. Structural changes during cysteine desulfurase CsdA and sulfur acceptor CsdE interactions provide insight into the trans-persulfuration. J. Biol. Chem. 288, 27172–27180 (2013). https://doi.org/10.1074/jbc.M113.480277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Fernández, F. J., Ardá, A., López-Estepa, M., Aranda, J., Peña-Soler, E., Garces, F., et al. Mechanism of sulfur transfer across protein-protein interfaces: The cysteine desulfurase model system. ACS Catal. 6, 3975–3984 (2016). https://doi.org/10.1021/acscatal.6b00360 [Google Scholar]
- [116].Fujishiro, T., Terahata, T., Kunichika, K., Yokoyama, N., Maruyama, C., Asai, K., et al. Zinc-ligand swapping mediated complex formation and sulfur transfer between SufS and SufU for iron-sulfur cluster biogenesis in Bacillus subtilis. J. Am. Chem. Soc. 139, 18464–18467 (2017). https://doi.org/10.1021/jacs.7b11307 [DOI] [PubMed] [Google Scholar]
- [117].Jin, H. S., Dhanasingh, I., Sung, J. Y., La, J. W., Lee, Y., Lee, E. M., et al. The sulfur formation system mediating extracellular cysteine-cystine recycling in Fervidobacterium islandicum AW-1 is associated with keratin degradation. Microb. Biotechnol. 14, 938–952 (2021). https://doi.org/10.1111/1751-7915.13717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Kakuta, Y., Horio, T., Takahashi, Y., Fukuyama, K.. Crystal structure of Escherichia coli Fdx, an adrenodoxin-type ferredoxin involved in the assembly of iron-sulfur clusters. Biochemistry 40, 11007–11012 (2001). https://doi.org/10.1021/bi010544t [DOI] [PubMed] [Google Scholar]
- [119].Shimomura, Y., Takahashi, Y., Kakuta, Y., Fukuyama, K.. Crystal structure of Escherichia coli YfhJ protein, a member of the ISC machinery involved in assembly of iron-sulfur clusters. Proteins 60, 566–569 (2005). https://doi.org/10.1002/prot.20481 [DOI] [PubMed] [Google Scholar]
- [120].Kim, J. H., Bothe, J. R., Frederick, R. O., Holder, J. C., Markley, J. L.. Role of IscX in iron-sulfur cluster biogenesis in Escherichia coli. J. Am. Chem. Soc. 136, 7933–7942 (2014). https://doi.org/10.1021/ja501260h [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Kim, J. H., Frederick, R. O., Reinen, N. M., Troupis, A. T., Markley, J. L.. [2Fe-2S]-ferredoxin binds directly to cysteine desulfurase and supplies an electron for iron-sulfur cluster assembly but is displaced by the scaffold protein or bacterial frataxin. J. Am. Chem. Soc. 135, 8117–8120 (2013). https://doi.org/10.1021/ja401950a [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Kim, J. H., Bothe, J. R., Alderson, T. R., Markley, J. L.. Tangled web of interactions among proteins involved in iron–sulfur cluster assembly as unraveled by NMR, SAXS, chemical crosslinking, and functional studies. Biochim. Biophys. Acta 1853, 1416–1428 (2015). https://doi.org/10.1016/j.bbamcr.2014.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Adinolfi, S., Iannuzzi, C., Prischi, F., Pastore, C., Iametti, S., Martin, S. R., et al. Bacterial frataxin CyaY is the gatekeeper of iron-sulfur cluster formation catalyzed by IscS. Nat. Struct. Mol. Biol. 16, 390–396 (2009). https://doi.org/10.1038/nsmb.1579 [DOI] [PubMed] [Google Scholar]
- [124].Lill, R., Freibert, S. A.. Mechanisms of mitochondrial iron-sulfur protein biogenesis. Annu. Rev. Biochem. 89, 471–499 (2020). https://doi.org/10.1146/annurev-biochem-013118-111540 [DOI] [PubMed] [Google Scholar]
- [125].Stehling, O., Wilbrecht, C., Lill, R.. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie 100, 61–77 (2014). https://doi.org/10.1016/j.biochi.2014.01.010 [DOI] [PubMed] [Google Scholar]
- [126].Mühlenhoff, U., Gerber, J., Richhardt, N., Lill, R.. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 22, 4815–4825 (2003). https://doi.org/10.1093/emboj/cdg446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Boniecki, M. T., Freibert, S. A., Mühlenhoff, U., Lill, R., Cygler, M.. Structure and functional dynamics of the mitochondrial Fe/S cluster synthesis complex. Nat. Commun. 8, 1287 (2017). https://doi.org/10.1038/s41467-017-01497-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Adam, A. C., Bornhövd, C., Prokisch, H., Neupert, W., Hell, K.. The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 25, 174–183 (2006). https://doi.org/10.1038/sj.emboj.7600905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wiedemann, N., Urzica, E., Guiard, B., Müller, H., Lohaus, C., Meyer, H. E., et al. Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 25, 184–195 (2006). https://doi.org/10.1038/sj.emboj.7600906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Shi, Y., Ghosh, M. C., Tong, W. H., Rouault, T. A.. Human ISD11 is essential for both iron-sulfur cluster assembly and maintenance of normal cellular iron homeostasis. Hum. Mol. Genet. 18, 3014–3025 (2009). https://doi.org/10.1093/hmg/ddp239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Angerer, H. The superfamily of mitochondrial Complex1_LYR motif-containing (LYRM) proteins. Biochem. Soc. Trans. 41, 1335–1341 (2013). https://doi.org/10.1042/bst20130116 [DOI] [PubMed] [Google Scholar]
- [132].Van Vranken, J. G., Jeong, M. Y., Wei, P., Chen, Y. C., Gygi, S. P., Winge, D. R., et al. The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis. eLife 5, e17828 (2016). https://doi.org/10.7554/eLife.17828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Cai, K., Frederick, R. O., Tonelli, M., Markley, J. L.. Mitochondrial cysteine desulfurase and ISD11 coexpressed in Escherichia coli yield complex containing acyl carrier protein. ACS Chem. Biol. 12, 918–921 (2017). https://doi.org/10.1021/acschembio.6b01005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Fox, N. G., Yu, X., Feng, X., Bailey, H. J., Martelli, A., Nabhan, J. F., et al. Structure of the human frataxin-bound iron-sulfur cluster assembly complex provides insight into its activation mechanism. Nat. Commun. 10, 2210 (2019). https://doi.org/10.1038/s41467-019-09989-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Yoon, T., Cowan, J. A.. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. J. Am. Chem. Soc. 125, 6078–6084 (2003). https://doi.org/10.1021/ja027967i [DOI] [PubMed] [Google Scholar]
- [136].Schmucker, S., Martelli, A., Colin, F., Page, A., Wattenhofer-Donzé, M., Reutenauer, L., et al. Mammalian frataxin: An essential function for cellular viability through an interaction with a preformed ISCU/NFS1/ISD11 iron-sulfur assembly complex. PLoS One 6, e16199 (2011). https://doi.org/10.1371/journal.pone.0016199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Colin, F., Martelli, A., Clémancey, M., Latour, J. M., Gambarelli, S., Zeppieri, L., et al. Mammalian frataxin controls sulfur production and iron entry during de novo Fe4S4 cluster assembly. J. Am. Chem. Soc. 135, 733–740 (2013). https://doi.org/10.1021/ja308736e [DOI] [PubMed] [Google Scholar]
- [138].Tsai, C. L., Barondeau, D. P.. Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry 49, 9132–9139 (2010). https://doi.org/10.1021/bi1013062 [DOI] [PubMed] [Google Scholar]
- [139].Fox, N. G., Martelli, A., Nabhan, J. F., Janz, J., Borkowska, O., Bulawa, C., et al. Zinc(II) binding on human wild-type ISCU and Met140 variants modulates NFS1 desulfurase activity. Biochimie 152, 211–218 (2018). https://doi.org/10.1016/j.biochi.2018.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Deere, T. M., Prakash, D., Lessner, F. H., Duin, E. C., Lessner, D. J.. Methanosarcina acetivorans contains a functional ISC system for iron-sulfur cluster biogenesis. BMC Microbiol. 20, 323 (2020). https://doi.org/10.1186/s12866-020-02014-z [DOI] [PMC free article] [PubMed] [Google Scholar]