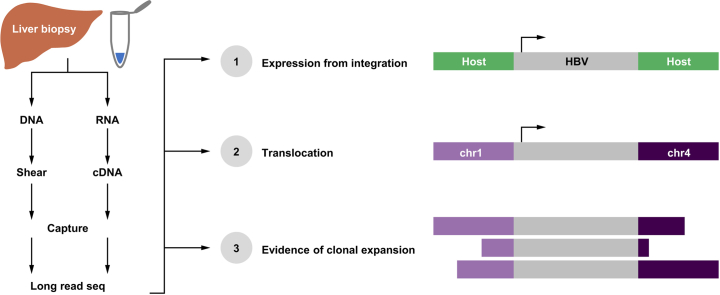
Abstract
Background & Aims
HBV infects over 257 million people worldwide and is associated with the development of hepatocellular carcinoma (HCC). Integration of HBV DNA into the host genome is likely a key driver of HCC oncogenesis. Here, we utilise targeted long-read sequencing to determine the structure of HBV DNA integrations as well as full isoform information of HBV mRNA with more accurate quantification than traditional next generation sequencing platforms.
Methods
DNA and RNA were isolated from fresh frozen liver biopsies collected within the GS-US-174-0149 clinical trial. A pan-genotypic panel of biotinylated oligos was developed to enrich for HBV sequences from sheared genomic DNA (∼7 kb) and full-length cDNA libraries from poly-adenylated RNA. Samples were sequenced on the PacBio long-read platform and analysed using a custom bioinformatic pipeline.
Results
HBV-targeted long-read DNA sequencing generated high coverage data spanning entire integrations. Strikingly, in 13 of 42 samples (31%) we were able to detect HBV sequences flanked by 2 different chromosomes, indicating a chromosomal translocation associated with HBV integration. Chromosomal translocations were unique to each biopsy sample, suggesting that each originated randomly, and in some cases had evidence of clonal expansion. Using targeted long-read RNA sequencing, we determined that upwards of 95% of all HBV transcripts in patients who are HBeAg-positive originate from cccDNA. In contrast, patients who are HBeAg-negative expressed mostly HBsAg from integrations.
Conclusions
Targeted lso-Seq allowed for accurate quantitation of the HBV transcriptome and assignment of transcripts to either cccDNA or integration origins. The existence of multiple unique HBV-associated inter-chromosomal translocations in non-HCC CHB patient liver biopsies suggests a novel mechanism with mutagenic potential that may contribute to progression to HCC.
Lay summary
Fresh frozen liver biopsies from patients infected with HBV were subjected to targeted long-read RNA and DNA sequencing. Long-read RNA sequencing captures entire HBV transcripts in a single read, allowing for resolution of overlapping transcripts from the HBV genome. This resolution allowed us to quantify the burden of transcription from integrations vs. cccDNA origin in individual patients. Patients who were HBeAg-positive had a significantly larger fraction of the HBV transcriptome originating from cccDNA compared with those who were HBeAg-negative. Long-read DNA sequencing captured entire integrated HBV sequences including multiple kilobases of flanking host sequence within single reads. This resolution allowed us to describe integration events flanked by 2 different host chromosomes, indicating that integrated HBV DNA are associated with inter-chromosomal translocations. This may lead to significant transcriptional dysregulation and drive progression to HCC.
Keywords: Integrated HBV DNA, Chronic HBV, Chromosomal translocations, Clonal expansion, Long-read sequencing, Targeted sequencing
Abbreviations: cccDNA, covalently closed circular DNA; CCS, circular consensus sequence; CHB, chronic hepatitis B; contig, contiguous sequence; DNA-Seq, DNA sequencing; DR1, direct repeat 1; dslDNA, double-stranded linear DNA; FFPE, formalin-fixed paraffin-embedded; gDNA, genomic DNA; HCC, hepatocellular carcinoma; N/A, nucleos(t)ide analogue; NHEJ, non-homologous end-joining; PEG-IFNα, pegylated interferon α; pgRNA, pre-genomic RNA; rcDNA, relaxed circular DNA; RIN, RNA integrity number; RNA-Seq, RNA sequencing; targeted Iso-Seq, targeted long-read RNA-sequencing; TDF, tenofovir disoproxil fumarate; TERT, telomerase reverse transcriptase; WGS, whole genome sequencing
Graphical abstract
Highlights
-
•
Fresh frozen liver biopsies from patients with CHB were subjected to targeted long-read RNA and DNA sequencing.
-
•
Inter-chromosomal translocations associated with HBV integration events detected in one-third of patients.
-
•
Chromosomal translocations were unique to each biopsy sample, suggesting that each originated randomly.
-
•
A larger fraction of the HBV transcriptome originates from cccDNA in patients who are HBeAg-positive.
Introduction
HBV infects over 296 million people worldwide and is a leading cause of hepatocellular carcinoma (HCC).1 Chronic infection with HBV and HCV is thought to lead to HCC through chronic inflammation that eventually leads to fibrosis and cirrhosis. However, unlike HCV, HBV also integrates into the host genome.2 In HCC patients, HBV integrations are often found in or near oncogenes such as telomerase reverse transcriptase (TERT) leading to dysregulated transcription.3,4 In addition, some integrations generate chimeric transcripts and proteins that have demonstrated oncogenic properties.[5], [6], [7] Although integrated HBV DNA is thought to be a dead end in terms of viral replication, integrated HBV can produce viral proteins, creating an obstacle to complete viral antigen clearance. In addition, integration may lead to chromosomal instability, which could be a key contributor for progression to HCC although the mechanism is not clearly defined.8
HBV is a small DNA virus with a 3.2 kbp covalently closed circular DNA (cccDNA) genome that replicates through an RNA intermediate known as pre-genomic RNA (pgRNA).9 pgRNA is a 3.5 kb transcript that is polyadenylated and packaged into virions along with the HBV reverse transcriptase. Reverse transcription of pgRNA typically results in the formation of a relaxed circular DNA (rcDNA) species that is partially single stranded. However, a minor species of HBV DNA is also generated in about 10% of capsids known as double-stranded linear DNA (dslDNA). It is thought that dslDNA is the substrate for integration.10 Evidence supports a mechanism that includes the host non-homologous end-joining (NHEJ) machinery that inserts HBV dslDNA into double-stranded breaks in the host chromosome upon infection.11 Integrations occur early post-infection and have been detected 3 days post-infection in tissue culture, 4 weeks post-infection in the woodchuck model and in paediatric patient samples.[12], [13], [14], [15]
Accumulation of integrated HBV sequences occurs throughout the natural progression of HBV disease and has been shown to randomly occur throughout the human genome.16 Integrations have been mapped to all human chromosomes without any detectable hotspots. The structure of dslDNA separates the core open reading frame (ORF) from its promoter, however, PreS1, PreS2/S, and X transcripts can still theoretically be generated from integrated HBV DNA.11,17 For this reason, it is believed that a large amount of HBsAg in the liver is expressed from integrated HBV DNA. Over time, some hepatocytes harbouring HBV integrations may be subject to clonal expansions.18,19 In particular, HCC samples demonstrate high clonality for integrations, often associated with integrations located in or near oncogenes.3 Periods of chronic hepatitis that correlate with hepatocyte death are thought to contribute to clonal expansion of hepatocytes with integrated HBV.20 Perhaps some integration events, primarily those in or near oncogenes, are preferentially expanded during this process leading to pre-neoplastic collections of cells in the infected liver.
We developed a novel method that combines target enrichment of DNA or RNA libraries for long-read sequencing with a custom analytical pipeline to characterise integrations in chronic hepatitis B (CHB) patient liver biopsies. This allows for the resolution of the entire architecture of each integration event, not only short reads mapping to virus–host junctions. Target enrichment for HBV DNA increases our sequencing coverage by >2,000-fold resulting in high coverage long-read DNA sequencing datasets. We demonstrate the existence of HBV-associated inter-chromosomal translocations in non-HCC CHB liver biopsies. We provide evidence for clonal expansion of these chromosomal translocation events as well as transcriptional activity associated with the production of HBsAg. In addition, we differentiate and quantify the transcriptional burden from cccDNA vs. integrated HBV DNA using targeted long-read RNA-sequencing (targeted Iso-Seq). We demonstrate that most of the transcription in patients who are HBeAg-negative comes from integrated HBV DNA.
Materials and methods
Liver biopsy collection
Liver biopsies were obtained from treatment-naïve patients who were HBeAg-positive and -negative enrolled in a phase IV clinical trial of tenofovir disoproxil fumarate (TDF) ± pegylated-interferon-α (PEG-IFNα) (GS-US-174-0149). Sixty-three patients participated in voluntary liver biopsy donation including 56 formalin-fixed paraffin-embedded and 67 fresh frozen samples. Ten patients were biopsied longitudinally at baseline and Week 96. Patient samples analysed were collected from 8 countries (USA, Korea, Turkey, Hong Kong, Poland, The Netherlands, Greece, and Germany). All patients signed an informed consent form before screening and in accordance with local regulatory and ethics committee requirements. The experimental protocol in these trials was approved by Gilead Sciences and all local regulatory agencies (see ClinicalTrials.gov: NCT01940471). This is the same sample set with matching random patient numbers as our companion manuscript describing the liver immune microenvironment in the same patient population.21
RNA-Seq
Total DNA and RNA was isolated from all fresh frozen biopsies at Expression Analysis – Q2 Solutions using the Allprep kit (Qiagen). Fifty-three of 67 samples met the quantity and quality standards for sequencing. Of these 53 samples, 41 were from baseline, 12 were from Week 96 and included were 7 longitudinal pairs (Table S1). Sequencing libraries were prepared for Illumina sequencing using TruSeq Stranded Total Gold with RiboZero. Sequencing was performed at Expression Analysis – Q2 Solutions. FASTQ reads were delivered to Gilead Sciences for analysis.
Targeted PacBio and targeted Iso-Seq
Genomic DNA (gDNA) was sheared to ∼7.5 kbp using a G-tube (Covaris) and purified using the AMPure PB DNA beads (Pacific Biosciences). Sheared DNA was barcoded using pre-annealed sample indexes (IDT) using the Kapa Hyper Prep library kit (Kapa Biosystems). Barcoded DNA was amplified using the Takara LA polymerase (Clontech) and analysed using a DNA high-sensitivity chip on the Bioanalyzer 2100 (Agilent). A total of 42 DNA samples were analysed by targeted long-read DNA-Seq (Table S1).
Liver biopsy RNA was analysed for quality on the Bioanalyzer 2100 (Agilent). A total of 43 samples with an RNA integrity number (RIN) higher than 8.5 were analysed by targeted Iso-Seq (Table S1). RNA was converted to full-length cDNA using the SMARTer cDNA synthesis kit (Takara Bio). A custom 3′ SMART CDS primer IIA containing a unique molecular index upstream of the polyT was generated for cDNA amplification, AAGCAGTGGTATCAACGCAGAGTACNNNNNNNNNNNNTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTN∗V. cDNA was amplified using the HiFi PCR kit (Kapa biosystems) and customised amplification primers that added PacBio sample indexes (IDT). cDNA was checked for quality and quantity using a Nanodrop and a Bioanalyzer 2100 with a DNA high-sensitivity chip.
A custom panel of 120-bp biotinylated oligos was designed to be compatible with the xGen Lockdown platform (IDT). The panel consists of 144 HBV targeting probes and 16 probes targeting host genes (Fig. 1C). Barcoded DNA and cDNA libraries were enriched for HBV sequences by incubating with our probe pool at 65°C for 4 h, capturing with streptavidin Dynabeads at 65°C for 45 min and washing using the xGen Hybridization and Wash kit (IDT). Captured DNA sequences were amplified using the Takara LA polymerase.
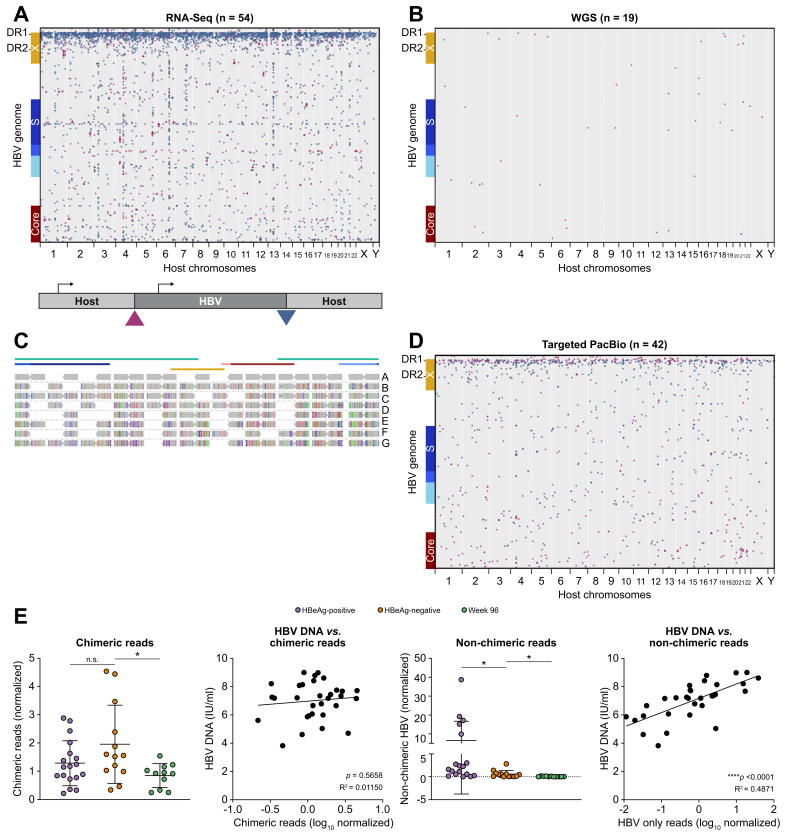
Fig. 1.
A combination of genomics approaches were developed and utilised to characterise HBV integrations in CHB patient liver biopsies.
(A) Liver biopsies obtained through the Gilead 174-0149 clinical trial were processed to extract both DNA and RNA. Bulk short-read RNA-Seq was performed and multiple reads that were chimeric for host and HBV sequences were identified and mapped across the human genome (x-axis) and HBV genome (y-axis). Triangle orientation reflects the directionality of each chimeric junction. (B) WGS was performed on 19 patients with corresponding RNA-Seq and chimeric reads were mapped using the same algorithm as shown in part A. (C) We developed a custom target enrichment panel that was coupled with PacBio sequencing to generate long reads containing HBV integrations. (D) Chimeric reads were obtained from all 42 samples. Chimeric reads were mapped across the HBV genome (y-axis) and human genome (x-axis). Triangle orientation reflects the directionality of each chimeric junction. (E) Targeted PacBio recovered HBV reads that were chimeric with host sequences and HBV reads that were not. These 2 read types were quantified and correlated to HBeAg status as well as peripheral HBV DNA levels. Statistical analyses were performed using unpaired t tests and linear regression analysis. CHB, chronic hepatitis B; RNA-Seq, RNA sequencing; WGS, whole genome sequencing.
Enriched DNA libraries were sequenced on the Sequel II (Pacific Biosciences) at the Arizona Genomics Institute at the University of Arizona (Tuscon, AZ, USA). Enriched cDNA libraries were sequenced on the Sequel II (Pacific Biosciences) at the McDonnell Genomics Institute at Washington University (St. Louis, MO, USA). Individual subreads were converted to circular consensus sequence (CCS) reads. CCS reads were used for analysis.
Integration analyses
For genotypes A through D, a reference sequence was made by consensus sequence calling respective HBV sequences downloaded from the National Center for Biotechnology Information. As the HBV genome is not circular, a 2× length sequence for each genotype was created by concatenating two copies of the consensus sequence, starting with the ATG of the core ORF. Reads were aligned to their respective sample’s 2× consensus sequence combined with human genome reference hg38 using BWA,22 STAR,23 or minimap224 for whole genome sequencing (WGS), RNA-Seq, or PacBio data, respectively. Paired-end reads were merged with BWA pemerge. Reads aligning at least partially to HBV were converted from .bam to .bed format. Using custom R scripts (R Foundation for Statistical Computing, Vienna, Austria), sequences with split alignment were used to infer chimerism between HBV and host. Junctions between HBV and host were inspected for barcode and adapter sequences to remove potential concatemer artefacts. Nearest gene analysis was performed using bedtools25 and Gencode26 gene annotation version 32.
For junctions with at least 25 supporting reads from PacBio sequencing, reads from each junction were used for de novo assembly of integration sites using multiple sequence alignment followed by indel removal using R (R Foundation for Statistical Computing). If any reads encompassed two junctions, reads from either junction were combined to create a single contiguous sequence (contig). Contigs were aligned to hg38 + HBV reference sequences, converted to .bed format using bedtools, then annotated for HBV ORFs and visualised in R (R Foundation for Statistical Computing). For select contigs, reads aligning to the respective junction were realigned using minimap2 or BWA then visualised in IGV.27
Results
Short-read sequencing reveals location and transcriptional activity of integrated HBV
DNA and RNA were collected from 53 core needle liver biopsy samples from the Gilead Sciences clinical trial (GS-174-0149): 41 samples taken at baseline and 12 taken at Week 96 (Table S1). Using RNA-Seq, we identified HBV-to-host genome junctions using chimeric reads with sequence fragments aligning to HBV and human genomes. In all samples examined, we detected chimeric RNAs indicating that all samples contained transcriptionally active HBV integrations (Fig. 1A). Consistent with previous studies, we observed no hotspots across the human genome and chimeric reads were detected in each of the 23 human chromosomes. However, we did identify 2 hotspots in the HBV genome: 1 major hotspot near direct repeat 1 (DR1) and a minor hotspot within the HBsAg ORF, consistent with previous studies in HCC (Fig. 1A).16 As RNA-Seq can only capture transcriptionally active integrations, we applied WGS to 19 samples to observe the corresponding DNA sequences within liver biopsies and compare those with the transcriptional signatures described above (Fig. 1B). Although all samples contained transcriptionally active integrations, we only detected chimeric junctions in 11 of 19 samples by WGS (Fig. 1B), suggesting standard WGS is not sensitive enough to study HBV DNA integrations in non-tumour tissues. In addition to low coverage, WGS failed to resolve the internal structure of integrated HBV DNA as only short chimeric fragments flanking each integration could be mapped.
Development of targeted long-read sequencing for HBV integrations
We developed a targeted long-read sequencing method that could increase the sensitivity for HBV sequences and resolve entire integrations (Fig. 1C).28 A custom panel of biotinylated oligos was developed to enrich for integrated HBV DNA from long-read sequencing libraries and the subsequent enriched libraries were subjected to PacBio long-read sequencing (Fig. 1C and D). The resulting dataset demonstrated enrichment for HBV sequences typically >2,000-fold compared with non-enriched samples. We detected integrated HBV DNA in all 42 samples tested (Fig. 1D, Table S2). We confirmed the integration hotspot on the HBV genome at DR1 and that integrations were detected in all human chromosomes without significant hotspots. Interestingly, we did not observe a hotspot in the middle of the HBsAg ORF as we did by RNA-Seq, suggesting mid-S chimeric transcripts may be generated via splicing. We captured both chimeric and non-chimeric HBV sequences. Although chimeric sequences must originate from integrated HBV DNA, the origin of non-chimeric sequences is ambiguous and could originate from any HBV DNA isoform or a less than full-length fragment of an integrated HBV DNA. However, as non-chimeric DNAs correlated strongly with peripheral HBV DNA, it suggests that most originated from a replicative species (Fig. 1E).
Targeted long-read sequencing reveals the comprehensive architecture of integrated HBV DNA
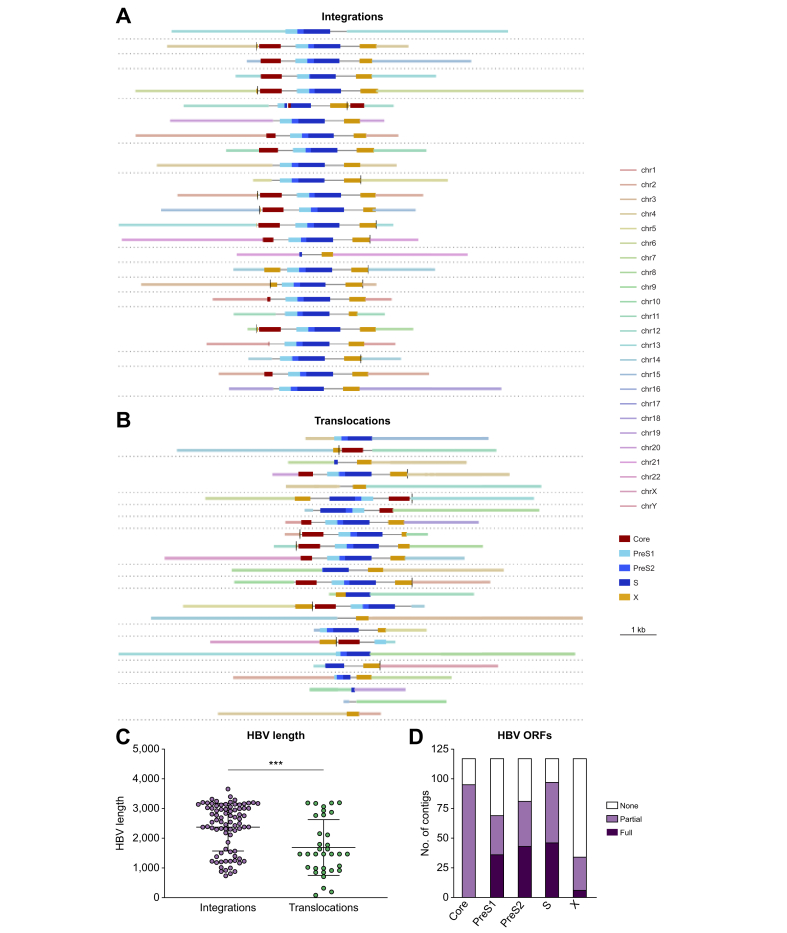
We observed a wide variety of integration patterns, architecture and burden amongst the patients examined. Shown in Fig. 2A is a selection of unique integration events, all supported by >25 reads that contained host sequence flanking both ends of the inserted HBV sequence. Integrations were detected that were comprised of the entire HBV genome beginning and ending with DR1, fitting the model suggesting that dslDNA is the substrate for integration (Table S2).10 However, most integrations, including many of those associated with DR1, resulted in truncated Core and X ORFs (Fig. 2D). The HBV sequences varied in length, sometimes comprising the entire dslDNA and other times comprising of subgenomes, with a mean length of 2,372 bp (Fig. 2C). In addition, there were several integrations that consisted of HBV sequences longer than the 3.2 kbp HBV genome (Fig. 2C). Although we did not find integrations that could produce pgRNA including the canonical polyadenylation site, the existence of longer than full-length integrations suggest that it is theoretically possible.
Fig. 2.
Heterogeneity observed amongst HBV integration patterns within and between patients.
(A) Schematics for a collection of contigs generated from individual integration events assembled using targeted PacBio. The contigs are those with the highest level of read support. (B) HBV-associated chromosomal translocations where HBV sequences are flanked by sequences from 2 different human chromosomes. The contigs highlighted were those with the highest level of read support. (C) Length distributions of the HBV sequences from integrated HBV DNA vs. those that were associated with inter-chromosomal translocations. Statistical comparisons were performed using an unpaired t test. (D) Assessment of the ORFs associated with integrated HBV DNA. Data were only generated from contigs where both chimeric ends were validated by sequencing. Plotted are the number of contigs with full, partial, or no coverage for a given HBV ORF. contig, contiguous sequence; ORF, open reading frame.
We also detected several HBV-associated inter-chromosomal translocations (denoted as translocations), that is integrated HBV sequences flanked by 2 distinct human chromosomes (Fig. 2B, chromosomes represented by different coloured bars). Shown in Fig. 2B is a selection of unique translocation events, all supported by >25 reads that contained host sequences from 2 unique chromosomes flanking the inserted HBV sequence. HBV sequences associated with these translocations were typically shorter than those associated with intra-chromosomal integrations (2,372 bp vs. 1,689 bp mean length), suggesting that the HBV DNA was shortened during the chromosomal rearrangement. However, we observed multiple translocations that consisted of nearly full-length HBV insertions. In total, chromosomal translocations were found in 13 of 42 samples.
Not only was heterogeneity observed at the structural level, integration patterns were also completely unique between and within patients. No two patients had identical integration events or patterns shared. Also, longitudinal biopsies collected from several patients did not exhibit the same integration events observed at baseline and at Week 96.
Quantification of the HBV transcriptional burden in patient liver biopsies
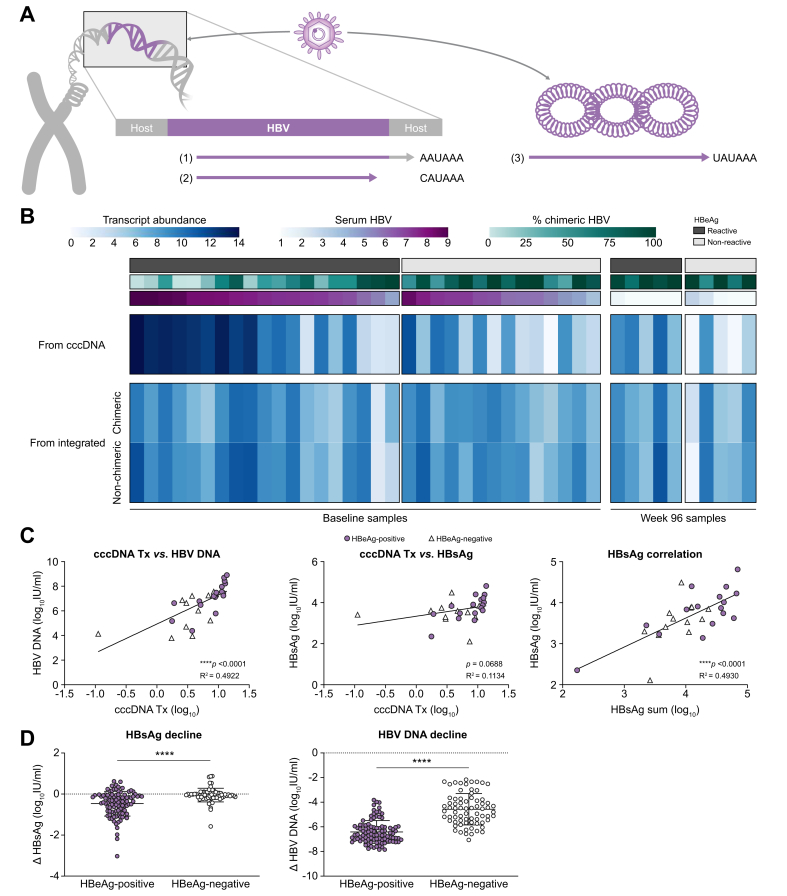
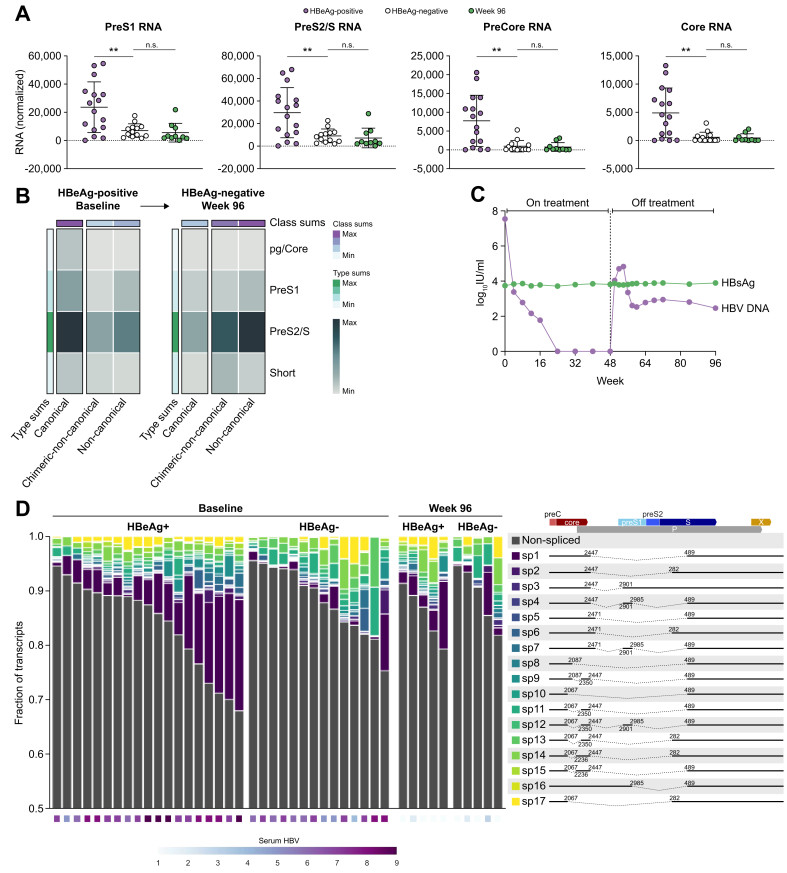
We have previously demonstrated that transcripts from cccDNA vs. from integrated HBV DNA have unique 3′ ends (Fig. 3A).28 All transcripts from cccDNA terminate at the canonical HBV poly(A) sequence downstream of the HBV X ORF. In contrast, 2 transcript types are associated with integrated HBV DNA. The first are chimeric transcripts that read through from HBV into the host genome and terminate at the nearest host poly(A) site. These transcripts often contain over 1 kb of host sequence appended to the 3′ end of each HBV RNA. The second integrated transcript type is non-chimeric and terminates within the integrated HBV DNA at a non-canonical poly(A) site located within the X ORF.29 For each sample, we quantified these 3 transcript types using targeted Iso-Seq and correlated these findings with both clinical and targeted DNA sequencing data (Fig. 3B). Notably, patients who are HBeAg-positive contain a much higher level of cccDNA transcription compared with patients who are HBeAg-negative (∗∗p = 0.0097). This cccDNA transcription correlates strongly with peripheral HBV DNA levels (Fig. 3C). In contrast, the amount of HBV transcription from integrations was not significantly different between patients who were HBeAg-positive and HBeAg-negative. However, because of the change in transcription from cccDNA, the ratio of integrated HBV transcripts to cccDNA transcripts was much higher in patients who were HBeAg-negative. These data indicate that the majority of transcription in patients who were HBeAg-negative originates from integrated HBV DNA and not cccDNA, consistent with previous studies.17 Although transcription from cccDNA did demonstrate a reasonable correlation to peripheral HBsAg, when we calculated a sum of all HBsAg transcription for each sample, the correlation to peripheral HBsAg increased dramatically (Fig. 3C).
Fig. 3.
Differentiation and quantification of the transcriptional burden from cccDNA vs. integrated HBV DNA.
(A) Differentiation of transcripts from cccDNA vs. integrated HBV DNA by characterising unique 3′ tails. (B) Quantification of each transcript type as cccDNA, chimeric integrated, or non-chimeric integrated in each of the 31 baseline samples. Transcriptional signatures are correlated to serum HBV DNA as well as the ratio of chimeric to non-chimeric HBV sequences obtained from targeted DNA-Seq. (C) Correlation of cccDNA transcription to corresponding peripheral HBV DNA and HBsAg levels. Additionally, we summed all HBsAg transcripts from cccDNA and integrated origins and demonstrate a strong correlation to the corresponding peripheral HBsAg levels. Patients who are HBeAg-positive are represented by purple circles and those who are HBeAg-negative by white triangles. Correlations were calculated using a linear regression. (D) HBsAg and HBV DNA decline in the periphery at Week 48 in all patients from 174-0149 treated with TDF alone for 120 weeks (Arm C). Statistical comparison was performed using an unpaired t test. cccDNA, covalently closed circular DNA; DNA-Seq, DNA sequencing.
As nucleos(t)ide analogue (N/A) treatment targets viral replication and not integrated HBV, these data imply that patients who are HBeAg-negative should experience little to no HBsAg decline following N/A treatment. As these samples originated from a clinical trial investigating the effects of TDF ± PEG-IFNα, we were able to examine HBsAg decline following TDF-only treatment in patients who were HBeAg-positive and HBeAg-negative (Fig. 3D). We demonstrate that at Week 48 in patients treated only with TDF, the mean change in serum HBsAg in patients who were HBeAg-negative (N = 71) was -0.05 log10 IU/ml compared with -0.47 log10 IU/ml in patients who were HBeAg-positive (N = 99). Although biopsies were not taken for most of these patients, these results are consistent with our sequencing data in patients who were HBeAg-positive indicating that they have more target available leading to a larger TDF-induced HBsAg decline.
HBV RNA isoforms differentiated in CHB liver biopsies
Targeted Iso-Seq also resolved overlapping HBV isoforms such as Core, PreCore, PreS1, and PreS2/S RNAs. Consistent with higher viral load and cccDNA transcription in patients who were HBeAg-positive, all isoforms were expressed highest in these patients (Fig. 4A). However, patients who were HBeAg-negative still expressed a significant amount of PreS1 and PreS2/S transcripts, mostly from integrated HBV DNA.
Fig. 4.
Differentiation and quantification of all HBV RNA isoforms in CHB liver biopsies.
(A) All HBV isoforms including PreCore, Core, PreS1, and PreS2/S RNAs are expressed at the highest levels in biopsies from patients who were HBeAg-positive. Statistical comparison was performed using an unpaired t test. (B) HBV transcript isoform analysis from an individual patient sampled longitudinally at baseline and Week 96. This patient experienced HBeAg loss before Week 96. (C) Serum HBsAg and HBV DNA levels from the same patient from baseline through Week 96. (D) Identification of all known HBV splice isoforms in CHB liver biopsies. The ratio of spliced to non-spliced RNAs as well as the types of splice isoforms expressed were variable between patients. CHB, chronic hepatitis B.
We applied isoform analysis on longitudinal samples from 1 patient who was HBeAg-positive at baseline, treated with 48 weeks of TDF and 16 weeks of PEG-IFNα and experienced HBeAg-loss (but remained anti-HBe negative) before Week 96. The patient was off treatment at the time of biopsy at Week 96 with a peripheral viral load of 2.46 log10 IU/ml HBV DNA and 3.89 log10 IU/ml HBsAg (Fig. 4C). Isoform analysis of this patient demonstrates a large decline in transcription from cccDNA between baseline and Week 96 (Fig. 4B). The majority of HBsAg produced at Week 96 appears to come from integrated HBV DNA. This patient had 3.74 log10 IU/ml HBsAg at baseline, indicating that despite a dramatic reduction in transcription from cccDNA, no decline in HBsAg was observed.
Finally, we performed a splice isoform analysis using the known HBV splice isoforms, SP1–SP17.30,31 As previously reported, SP1 was the most common splice isoform identified in our patient cohort (Fig. 4D).32 The ratio of spliced to non-spliced HBV RNAs ranged from 2% to over 20% and was highest in patients who were HBeAg-positive. Using a 3ʹ poly(A) analysis, the majority of spliced HBV RNAs originated from cccDNA, and accordingly, splicing was more prevalent in HBeAg-positive patients with higher cccDNA transcriptional burden. In addition to characterising the known splice isoforms, Iso-Seq revealed many novel spliced transcripts that contained up to 3 splice junctions in a single HBV transcript. Although most spliced transcripts were longer preCore or Core transcripts, we noted a variety of spliced PreS1 isoforms.
Clonal expansion and expression from integrated HBV DNA
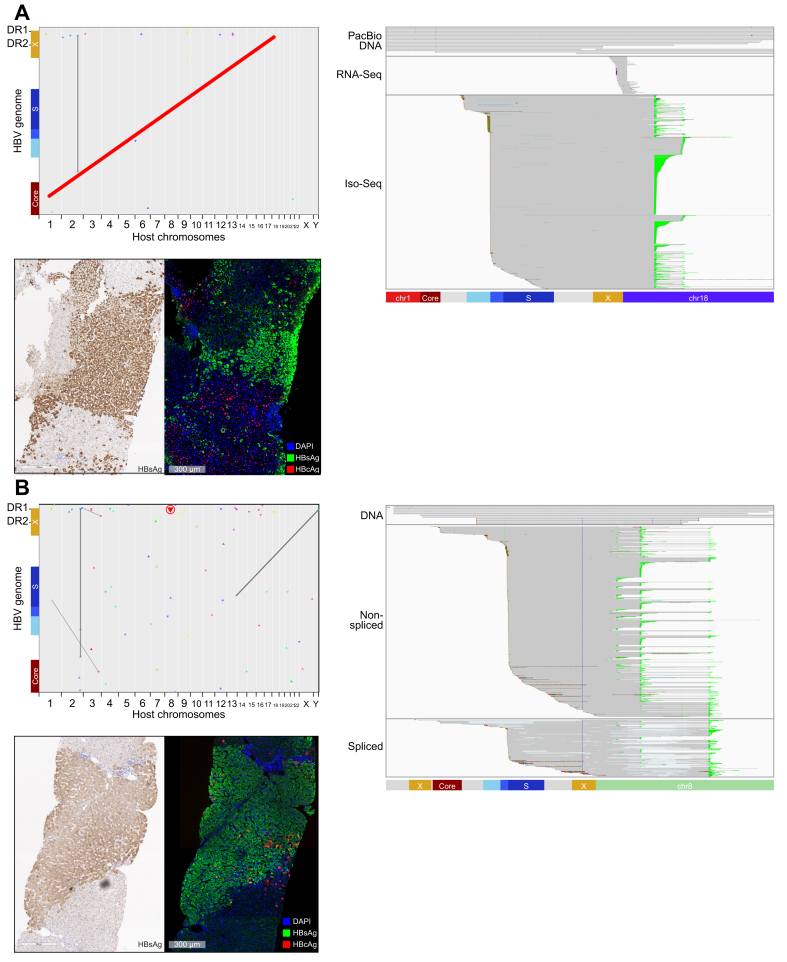
Several of the integrations and translocations observed demonstrate unique architecture and provide insight into clonal expansion. Long reads with variable lengths must have originated from unique DNA molecules. Therefore, if multiple reads align to an integration or translocation without evidence of being PCR duplicates, they should have originated from different cells. As examples, we highlight 2 such events and their associated transcriptional signatures. The first patient had multiple unique reads mapping to a chromosome 1 to chromosome 18 translocation event (Fig. 5A, highlighted in red). This patient was HBeAg-negative and corresponding liver antigen staining indicated high HBsAg burden but low HBcAg staining. The HBsAg staining revealed patches of similar HBsAg staining patterns, suggesting clonal expansions (Fig. 5A). Short-read RNA-Seq data was mapped to this chromosomal translocation indicating that it is transcriptionally active. Targeted Iso-Seq confirmed that this integration generated chimeric PreS1 and PreS2/S transcripts and utilised 2 different host poly(A) sites within chromosome 18.
Fig. 5.
Identification of a clonally expanded and transcriptionally active HBV-associated chromosomal translocation.
(A) Matching FFPE and fresh frozen baseline biopsies were analysed from a 36-year-old White male who was HBeAg-negative. This patient had HBV DNA load of 7.22 log10 IU/ml and HBsAg load of 3.90 log10 IU/ml. We obtained dozens of unique reads confirming an HBV-associated chromosomal translocation between chromosome 1 and chromosome 18. We also obtained targeted Iso-Seq and bulk RNA-Seq reads indicating transcriptional activity from the same integration. Two FFPE slides were stained. As part of a multiplex panel, HBcAg, HBsAg, and NaKATPase were overlaid. In addition, a chromogenic HBsAg stain was performed. (B) Matching FFPE and fresh frozen baseline biopsies were analysed from a 24-year-old Asian male who was HBeAg-positive. This patient had a HBV DNA load of 7.54 log10 IU/ml and a HBsAg load of 3.74 log10 IU/ml. We obtained a number of reads confirming an integration consisting of 4,119 bp of continuous HBV sequence. We also obtained targeted Iso-Seq reads indicating transcriptional activity from the same integration. Two FFPE slides were stained. As part of a multiplex panel, HBcAg, HBsAg, and NaKATPase were overlaid. In addition, a chromogenic HBsAg stain was performed. FFPE, formalin-fixed paraffin-embedded; RNA-Seq, RNA sequencing; targeted Iso-Seq, targeted long-read RNA-sequencing.
The second clonal expansion featured includes an unusually long HBV sequence (Fig. 5B). Although we only captured 1 chimeric end, the resolved HBV sequence was 4,119 bp long. Unlike most of the integrations characterised, this integration appeared capable of producing preCore and Core transcripts. This particular integration event generates an abundance of unique transcripts, all chimeric with chromosome 8 at the 3′ end. These transcripts express all of Core, PreS1, and PreS2/S and utilise 3 different host poly(A) sequences (Fig. 5B highlighted in green). Additionally, there are many splice isoforms associated with this integration. Splicing was associated with all 3 viral transcript types and was also observed within the host sequence. This unique integration event highlights the complexity of integrations within HBV patients as this single integration event has the potential to generate dozens of unique proteins.
Integrations upregulate nearby host transcription
Short-read RNA-Seq was used for host transcriptomic analysis. For each chimeric junction that was observed at the RNA or DNA level, we calculated transcriptional changes for the nearest genes (Fig. S2). Previous studies in HCC liver biopsies have demonstrated that many genes near integrations are transcriptionally upregulated owing to the presence of Core enhancer regions within the X ORF found at the 3′ end of integrations.3 These types of analyses are obscured in CHB patients as the level of clonal expansion is much lower and the diversity of integration events far higher than the HCC samples. However, we could detect gene expression that was modestly upregulated for genes near HBV integrations (Fig. S2). Although the fold increase was low overall, we would only expect increased gene expression in those cells harbouring each integration event. The fact that any changes in gene expression were detected using bulk RNA-Seq indicates that the transcriptional changes at the single cell level may be large.
Discussion
We developed a custom HBV target enrichment strategy coupled with long-read RNA-Seq and DNA-Seq and applied it to patient liver biopsies to provide the most comprehensive description to date of the burden, architecture, and transcriptional activity of integrated HBV DNA.28 Several recent studies that utilise patient HCC tissue have described integrated HBV using either long-read sequencing or target enrichment strategies, but have not coupled these 2 powerful platforms.[33], [34], [35] Analysis of integrated HBV DNA demonstrated that only a small fraction of integrations appeared to consist of full-length HBV sequences (Fig. 2A). However, most integrations still utilise DR1/2 junction points at the 3′ end, but significant heterogeneity was observed at the 5′ end of integrations, leading to a range of HBV insertion lengths. In addition, these methods revealed HBV-associated chromosomal translocations in CHB patient liver biopsies. Nearly one-third of the samples analysed (13/42) contained HBV-associated chromosomal translocations (Fig. 2B and Table S2). HBV-associated inter-chromosomal translocations typically included shorter HBV sequences (1,689 bp) compared with HBV integrations (2,372 bp) (Fig. 2C). We utilised targeted Iso-Seq to differentiate and quantify transcriptional burden from integrations vs. cccDNA. Patients who were HBeAg-positive had a much higher transcriptional burden from cccDNA compared with patients who were HBeAg-negative (Fig. 3B). The transcriptional burden from integrations was not statistically different between HBeAg-positive and HBeAg-negative samples, however, the ratio of transcriptional activity between integrated HBV DNA and cccDNA was much higher in patients who were HBeAg-negative. This indicates that the majority of HBsAg in serum of patients who were HBeAg negative originates from integrated HBV.
Significant heterogeneity of integration events was observed among and within patients. No 2 integration events were the same, and no 2 samples had similar integration patterns. Additionally, in the small number of longitudinal samples that we analysed, no integrations that were present at baseline were also present at Week 96. This result supports a highly random mechanism for HBV integration, likely driven by random breakage of the host chromosome and insertion of full-length, or partial-length dslDNA that usually contains DR1/2 at the 3′ end.10 Despite not seeing similar integrations or translocation between samples, we did see multiple integrations and translocations that had matched chimeric transcripts. This indicates that in samples with high sequencing depth we are capturing the majority of HBV reads from each sample. We have previously applied this technique to several HCC cell lines and recovered reads from each integration event.28 As HCC samples appear to be enriched for integrations in specific genes such as TERT and MLL4, progression to HCC likely involves selective clonal expansion of hepatocytes that happen to contain an oncogenic integration event.19
For the first time, HBV-associated chromosomal translocations have been detected in HBV-infected, non-HCC patients. As liver biopsies represent a small portion of the whole liver, we hypothesise that translocations may be even more prominent than detected. We previously characterised HBV-associated chromosomal translocations in HCC cell lines and validated most of them using spectral karyotyping.28 At the time, it was unclear whether HBV-associated chromosomal translocations would be translatable to patient samples, or whether these translocations were an artefact of long-term culturing.36 In addition, a recent study in HCC samples, also demonstrated the existence of HBV-associated chromosomal translocations in patients.37 These data imply that chromosomal translocations may be a natural consequence of HBV infection. As most translocations consisted of less than full-length HBV sequences, a potential mechanism involves random breakage of an initial longer integration that undergoes incorrect repair. However, we did observe several longer HBV sequences intersecting chromosomal translocations, suggesting that they may occur during the initial integration event as well. Chromosomal translocations can be clonally expanded and as a result may significantly contribute to the HBsAg burden in patients (Fig. 5A). Given that translocations can lead to dysregulation of host gene expression, we hypothesise that this may play a role in progression to HCC. Characterisation of the translocation burden in HCC samples will be critical for defining how the translocations contribute to liver cancer.
Targeted Iso-Seq allowed for the resolution of overlapping transcripts from cccDNA and integrated HBV DNA. This method also allowed for the quantification of each of the HBV transcript types such as pgRNA, PreS1 RNA, and PreS2/S RNA etc. By integrating clinical parameters with the sequencing analysis, it was revealed that patients who were HBeAg-positive had a large amount of transcription from cccDNA whereas patients who were HBeAg-negative mostly transcribed PreS1 and PreS2/S from integrations. Notably, the amount of transcription from integrations was not significantly different between patients who were HBeAg-positive and HBeAg-negative. The major difference between these 2 groups is that patients who are HBeAg-negative have far less active viral replication. As TDF impacts reverse transcription, it should only impact transcription from cccDNA and not integrated HBV DNA. We observed that patients who are HBeAg-negative only realised minor declines in HBsAg from baseline to Week 48 on TDF. The patient population what was HBeAg-positive experienced nearly 10-fold more HBsAg decline (0.05 vs. 0.47 log10 IU/ml HBsAg decline) (Fig. 3D). Finally, we compared baseline vs. Week 96 samples longitudinally, and found that integrated HBV DNA transcription is not significantly impacted by TDF treatment whereas transcription from cccDNA is markedly reduced in DNA-suppressed patients. This has broad impact on HBV cure strategies and implies that immune modulators are likely required to eliminate cells bearing integrated HBV DNA to achieve HBsAg-loss.
Although HBV and HCV both lead to the progression to fibrosis, cirrhosis, and HCC, there are some key differences between these 2 diseases. There is evidence for progression to HCC in the absence of cirrhosis in HBV-infected patients, mostly in southern Africa.38 Chronic inflammation is thought to lead to the fibrosis and cirrhosis that drives HCC in patients infected with HBV and HCV. However, the unique feature of non-cirrhotic HCC development in patients who are infected with HBV may be driven by integrations or translocations. We have found chromosomal translocations in all HCC cell lines that we have examined to date and others have now found chromosomal translocations in HCC liver biopsies.28,37 Application of this technology to additional HCC samples will be critical to establishing the contributions of translocations to disease progression. As integrated HBV DNA has been identified in paediatric patients,15 the development of chromosomal translocations may occur earlier in natural infection. As HBV treatments are now generic, safe, and do not lead to the development of drug resistance39 the benefits of early treatment may dramatically outweigh the benefits of waiting for liver disease onset.20
Financial support
The study was funded by Gilead Sciences Inc.
Authors’ contributions
Experimental design: RR, NvB, BF. Library preparation and enrichment: NvB, DH, LM. Bioinformatics: RR, CS, PP, RM. Sample collection: HC, PM, MB. Sample handling: VS, NB, NB. Pathology and image analysis: ST, NvB. Manuscript writing: NvB, RR, BF. Manuscript revisions: LL, HM, AG.
Data availability statement
Data are available upon request.
Conflicts of interest
All authors are employed by Gilead Sciences Inc.
Please refer to the accompanying ICMJE disclosure forms for further details.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2022.100449.
Supplementary data
The following are the supplementary data to this article:
References
- 1.World Health Organization . 2017. Hepatitis B.https://www.who.int/news-room/fact-sheets/detail/hepatitis-b [Google Scholar]
- 2.Tu T., Budzinska M.A., Shackel N.A., Urban S. HBV DNA integration: molecular mechanisms and clinical implications. Viruses. 2017;9:75. doi: 10.3390/v9040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toh S.T., Jin Y., Liu L., Wang J., Babrzadeh F., Gharizadeh B., et al. Deep sequencing of the hepatitis B virus in hepatocellular carcinoma patients reveals enriched integration events, structural alterations and sequence variations. Carcinogenesis. 2013;34:787–798. doi: 10.1093/carcin/bgs406. [DOI] [PubMed] [Google Scholar]
- 4.Jiang Z., Jhunjhunwala S., Liu J., Haverty P.M., Kennemer M.I., Guan Y., et al. The effects of hepatitis B virus integration into the genomes of hepatocellular carcinoma patients. Genome Res. 2012;22:593–601. doi: 10.1101/gr.133926.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lau C.C., Sun T., Ching A.K., He M., Li J.W., Wong A.M., et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25:335–349. doi: 10.1016/j.ccr.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 6.Graef E., Caselmann W.H., Hofschneider P.H., Koshy R. Enzymatic properties of overexpressed HBV-mevalonate kinase fusion proteins and mevalonate kinase proteins in the human hepatoma cell line PLC/PRF/5. Virology. 1995;208:696–703. doi: 10.1006/viro.1995.1201. [DOI] [PubMed] [Google Scholar]
- 7.Takada S., Koike K. Trans-activation function of a 3′ truncated X gene-cell fusion product from integrated hepatitis B virus DNA in chronic hepatitis tissues. Proc Natl Acad Sci U S A. 1990;87:5628–5632. doi: 10.1073/pnas.87.15.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishii T., Tamura A., Shibata T., Kuroda K., Kanda T., Sugiyama M., et al. Analysis of HBV genomes integrated into the genomes of human hepatoma PLC/PRF/5 cells by HBV sequence capture-based next-generation sequencing. Genes (Basel) 2020;11:661. doi: 10.3390/genes11060661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeger C.Z.F., Mason W.S. 6th edn. Wolters Kluwer/Lippincott Williams & Wilkins Health; Philadelphia, PA: 2014. Hepadnaviruses. Field's Virology; pp. 3376–3436. [Google Scholar]
- 10.Yang W., Summers J. Integration of hepadnavirus DNA in infected liver: evidence for a linear precursor. J Virol. 1999;73:9710–9717. doi: 10.1128/jvi.73.12.9710-9717.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bill C.A., Summers J. Genomic DNA double-strand breaks are targets for hepadnaviral DNA integration. Proc Natl Acad Sci U S A. 2004;101:11135–11140. doi: 10.1073/pnas.0403925101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Summers J., Jilbert A.R., Yang W., Aldrich C.E., Saputelli J., Litwin S., et al. Hepatocyte turnover during resolution of a transient hepadnaviral infection. Proc Natl Acad Sci U S A. 2003;100:11652–11659. doi: 10.1073/pnas.1635109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu T., Budzinska M.A., Vondran F.W.R., Shackel N.A., Urban S. Hepatitis B virus DNA integration occurs early in the viral life cycle in an in vitro infection model via sodium taurocholate cotransporting polypeptide-dependent uptake of enveloped virus particles. J Virol. 2018;92:e02007–e02017. doi: 10.1128/JVI.02007-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimbi G.C., Kramvis A., Kew M.C. Integration of hepatitis B virus DNA into chromosomal DNA during acute hepatitis B. World J Gastroenterol. 2005;11:6416–6421. doi: 10.3748/wjg.v11.i41.6416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yaginuma K., Kobayashi H., Kobayashi M., Morishima T., Matsuyama K., Koike K. Multiple integration site of hepatitis B virus DNA in hepatocellular carcinoma and chronic active hepatitis tissues from children. J Virol. 1987;61:1808–1813. doi: 10.1128/jvi.61.6.1808-1813.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Podlaha O., Wu G., Downie B., Ramamurthy R., Gaggar A., Subramanian M., et al. Genomic modeling of hepatitis B virus integration frequency in the human genome. PLoS One. 2019;14 doi: 10.1371/journal.pone.0220376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wooddell C.I., Yuen M.F., Chan H.L., Gish R.G., Locarnini S.A., Chavez D., et al. RNAi-based treatment of chronically infected patients and chimpanzees reveals that integrated hepatitis B virus DNA is a source of HBsAg. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aan0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tu T., Mason W.S., Clouston A.D., Shackel N.A., McCaughan G.W., Yeh M.M., et al. Clonal expansion of hepatocytes with a selective advantage occurs during all stages of chronic hepatitis B virus infection. J Viral Hepat. 2015;22:737–753. doi: 10.1111/jvh.12380. [DOI] [PubMed] [Google Scholar]
- 19.Mason W.S., Jilbert A.R., Litwin S. Hepatitis B virus DNA integration and clonal expansion of hepatocytes in the chronically infected liver. Viruses. 2021;13:210. doi: 10.3390/v13020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mason W.S., Gill U.S., Litwin S., Zhou Y., Peri S., Pop O., et al. HBV DNA integration and clonal hepatocyte expansion in chronic hepatitis B patients considered immune tolerant. Gastroenterology. 2016;151:986–998.e984. doi: 10.1053/j.gastro.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Buuren N., Ramirez R., Turner S., Chen S., Suri V., Aggarwal A., et al. Characterization of the liver immune microenvironment in chronic HBV infected patient liver biopsies. J Hep Rep. 2022;4:10038. doi: 10.1016/j.jhepr.2021.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramirez R., van Buuren N., Gamelin L., Soulette C., May L., Han D., et al. Targeted long-read sequencing reveals comprehensive architecture, burden and transcriptional signatures from HBV-associated integrations and translocations in HCC cell lines. J Virol. 2021;95 doi: 10.1128/JVI.00299-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilger C., Velhagen I., Zentgraf H., Schroder C.H. Diversity of hepatitis B virus X gene-related transcripts in hepatocellular carcinoma: a novel polyadenylation site on viral DNA. J Virol. 1991;65:4284–4291. doi: 10.1128/jvi.65.8.4284-4291.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong S., Revill P. Overview of hepatitis B viral replication and genetic variability. J Hepatol. 2016;64:S4–S16. doi: 10.1016/j.jhep.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen J., Wu M., Wang F., Zhang W., Wang W., Zhang X., et al. Hepatitis B virus spliced variants are associated with an impaired response to interferon therapy. Sci Rep. 2015;5:16459. doi: 10.1038/srep16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim C.S., Sozzi V., Littlejohn M., Yuen L.K.W., Warner N., Betz-Stablein B., et al. Quantitative analysis of the splice variants expressed by the major hepatitis B virus genotypes. Microb Genom. 2021;7 doi: 10.1099/mgen.0.000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W., Wei W., Hou F., Xu H., Cui X. The integration model of hepatitis B virus genome in hepatocellular carcinoma cells based on high-throughput long-read sequencing. Genomics. 2022;11:23–30. doi: 10.1016/j.ygeno.2021.11.025. [DOI] [PubMed] [Google Scholar]
- 34.Zhuo Z., Rong W., Li H., Li Y., Luo X., Liu Y., et al. Long-read sequencing reveals the structural complexity of genomic integration of HBV DNA in hepatocellular carcinoma. NPJ Genom Med. 2021;6:84. doi: 10.1038/s41525-021-00245-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peneau C., Imbeaud S., La Bella T., Hirsch T.Z., Caruso S., Calderaro J., et al. Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut. 2021 doi: 10.1136/gutjnl-2020-323153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander J.J. In: Neoplasms of the Liver. Okuda K., Ishak K.G., editors. Springer; Tokyo: 1987. Human hepatoma cell lines; pp. 47–56. [Google Scholar]
- 37.Alvarez E.G., Demeulemeester J., Otero P., Jolly C., Garcia-Souto D., Pequeno-Valtierra A., et al. Aberrant integration of hepatitis B virus DNA promotes major restructuring of human hepatocellular carcinoma genome architecture. Nat Commun. 2021;12:6910. doi: 10.1038/s41467-021-26805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brechot C., Gozuacik D., Murakami Y., Paterlini-Brechot P. Molecular bases for the development of hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) Semin Cancer Biol. 2000;10:211–231. doi: 10.1006/scbi.2000.0321. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y., Corsa A.C., Buti M., Cathcart A.L., Flaherty J.F., Miller M.D., et al. No detectable resistance to tenofovir disoproxil fumarate in HBeAg+ and HBeAg- patients with chronic hepatitis B after 8 years of treatment. J Viral Hepat. 2017;24:68–74. doi: 10.1111/jvh.12613. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon request.