Abstract
Background
The heterogeneity of osteoarthritis (OA) significantly limits the effectiveness of pharmacological treatments in an unselected patient population. In this context, the identification of OA subtypes is meaningful for the development of therapies that target specific types of OA pathogenesis.
Methods
Expression array profiles of 70 OA and 36 control synovial samples were extracted from the GEO database. Unsupervised consensus clustering was performed based on the most variable genes to identify OA subclusters. Next, Joint samples from OA patients were obtained. We divided the OA patient into two subpopulations according to synovial ADCY7 levels. Synovium and cartilage samples from different OA subpopulations were evaluated. In addition, we established a high-fat diet (HFD)-induced rat OA model. We evaluated OA progression, lipid metabolism, synovitis and fibroblast-like synoviocytes (FLS) function in this HFD-induced OA model.
Results
70 OA patients were categorized into three distinct subclusters. We noted that one subcluster was characterized by synovial lipid metabolism disorder GO terms. We further identified the most noticeable KEGG pathway “Regulation of lipolysis in adipocytes” in this subcluster as well as the most significantly differentially expressed gene, ADCY7. We found that the ADCY7 high expressing group (32.6%) exhibited features of synovial inflammatory lipolysis epithelial-mesenchymal transition (EMT) tendency, as well as faster join space narrowing. The HFD induced OA-like degeneration in rat joints. We observed similar synovial inflammatory lipolysis and EMT in FLS, characterized by higher proliferative and invasive activity and elevated proinflammatory and procatabolic properties. ADCY7 was highly expressed in the synovium of the HFD-OA model rats and the inhibition of ADCY7 effectively attenuated these HFD-induced degenerative changes as well as synovial inflammatory lipolysis and FLS dysfunction. In HFD-FLSs, ADCY7 promoted the phosphorylation of PKA as well as its downstream lipid droplet-associated protein PLIN1 and hormone-sensitive lipase (HSL). The inhibition of PKA largely alleviated ADCY7-mediated HFD-FLS dysfunction.
Conclusions
We described a synovial EMT and lipid metabolism disorder in the pathogenesis of OA. This novel mechanism may represent a currently undefined OA subtype. ADCY7 is a potential molecular marker of this pathomechanism.
The Translational potential of this article
Utilizing synovial samples from OA patients, we identified a subpopulation with high ADCY7 expression. This may represent a currently undefined OA subtype and explain the clinical phenomenon of more severe synovial inflammation in obese OA patients. In addition, we established an HFD-induced OA rat model and found an upregulation of ADCY7 in the synovium. We confirmed that the inhibition of ADCY7 could effectively attenuate HFD-induced degenerative changes as well as the inflammatory lipolysis and FLS dysfunction observed in the rat model. This suggests that ADCY7 and its downstream pathways are potential pharmacological targets for treating this lipid-metabolism-disorder-related OA mechanism.
Keywords: Adenylyl cyclase 7, Osteoarthritis, Disease type, Fibroblast-like synoviocytes, Epithelial-mesenchymal transition
1. Introduction
Osteoarthritis (OA) is a degenerative disease involving the whole joint. In North America and Europe, the incidence of structural OA of the hands and knee among adults ≥65 years has been reported to be 60% and 33%, respectively [1]. Currently, pharmacological treatments for OA are mainly limited to temporary relief of symptoms, whereas no disease-modifying OA drugs have gained clinical acceptance [2]. In 2018, OA was labeled a “serious disease with an unmet medical need” by the FDA [3]. This has been partly attributed to the heterogeneity of OA, which makes it challenging to develop a “one-size-fits-all” therapy for an unselected patient population [4].
OA affects the whole joint and involves multiple tissues and cell types. These tissues and cells in different patients or at different stages in the same patients can exhibit differential pathological features. Cases of OA resulting from different risk factors (“mechanical”, “inflammatory” and “metabolic”) may have completely different pathogeneses and differential responses to treatments. In this context, defining OA subtypes appears meaningful for the development of therapies that target specific OA pathogenesis (e.g., matrix degradation, bone remodeling, low-grade inflammation and Wnt signaling) [5]. There have been a considerable number of strategies applied to identify OA subtypes. Clinical characteristics, imaging observations and biochemical markers were the most frequently used variables [4,6]. These strategies have certain limitations in revealing the underlying pathomechanisms and can provide only very limited information for the discovery of novel drug targets. Using “big data” from “omics” to identify OA subtypes has been a novel strategy in the last decade. In particular, high-throughput omics techniques are quite helpful for discovering unknown OA pathomechanisms [6]. Recently, using omics data obtained from cartilage and synovia, researchers have defined OA subtypes associated with inflammation and extracellular matrix metabolism [7,8]. In this study, using transcriptome data from 7 GEO database series, we identified a novel OA subtype associated with synovial lipid metabolism disorder. This subtype, which accounts for approximately 32–36% of case of OA, is characterized by inflammatory lipolysis of the synovia and the dysfunction of fibroblast-like synoviocytes (FLSs). This may represent a previously undiscovered pathomechanism of OA. Here, we investigated this subtype in detail and explored the contribution of a key molecule, adenylyl cyclase 7 (ADCY7), to OA progression.
2. Materials and methods
This study protocol was approved by the institutional review board (IRB) of the Third Xiangya Hospital, Central South University (No: 2020-S221), and a signed written consent form was obtained from all study subjects. All experiments involving human tissues and animals were performed in accordance with guidelines approved by the IRB. Each sample was processed only after the receipt of a signed informed consent form.
2.1. Data collection
The expression array profiles and partial clinical information of 70 OA and 36 control synovium samples were extracted from GSE12021, GSE32317, GSE55235, GSE55457, GSE55584, GSE82107 and GSE93698 in the GEO database (Supplementary Table 1) (https://www.ncbi.nlm.nih.gov/geo/), among which 70 OA synovium samples were regarded as the training cohort for subsequent analysis. All raw data were preprocessed with RMA algorithm normalization by the “affy” R package. The batch effect was removed by the “removeBatchEffect” function in the “limma” R package. The expression matrix of the validation cohort containing 85 OA synovium samples [7] was extracted from GitHub (https://github.com/Ellen1101/OA-subtype), 21 lipid metabolism-related pathways and corresponding gene sets were extracted from the KEGG database (http://www.kegg.jp/blastkoala/) [9], and 5 lipid metabolism-related gene sets were extracted from MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp) (Supplementary Table 2) [10,11].
2.2. Identification of OA subclusters and functional annotation
The 872 most variable genes across the training cohort were identified with the expression MAD >0.5. Unsupervised consensus clustering was performed based on the 872 genes with the “ConsensusClusterPlus” R package to identify OA subclusters. Partitioning around medoids (pam) algorithm and Spearman distance were applied, and 1000 repetitions were used to guarantee the stability of the classification. The clearest boundary and most appropriate consistency were observed when consensus matrix k = 3 (Supplementary Fig. 1a and b). Heatmaps were produced by “pheatmap” R package. GO term analysis and KEGG pathway analysis were conducted with the “clusterProfiler” and “org.Hs.e.g.,.db” R packages, while visualization of the enrichment results was performed using the R package “ggplot2”.
2.3. Human tissue collection and cell isolation
Human synovium and cartilage samples were obtained during total knee arthroplasty (advanced OA, n = 11, females, 59.80 ± 4.47 years), arthroscopic debridement (early-stage OA, only synovium was obtained, n = 22, females, 54.95 ± 7.59 years) and amputation (non-OA, n = 10, 45.91 ± 8.90 years) (Supplementary Table 3). The cartilage was obtained from the tibial plateau and distal femur of the knee. The harvested synovium and cartilage were digested with 0.3% collagenase type I (Sigma, USA) and 1 mg/ml collagenase type II (Sigma, USA), respectively, in DMEM (Gibco, USA) with penicillin (100 U/mL)/streptomycin (100 mg/ml, Gibco, USA) and 10% FBS (Gibco, USA) for 3 h at 37 °C. The cells were passed through a 70-mm nylon cell strainer and collected by centrifugation at 250g for 5 min. The cells were then resuspended and plated in a 60-mm diameter culture dish, with the medium replaced every 3 days. Synovial fluid was collected for the measurement of glycerol (glycerol assay, Sigma, USA) and FFAs (NEFA assay, Wako Diagnostics, USA).
2.4. High-fat diet (HFD) induced OA model
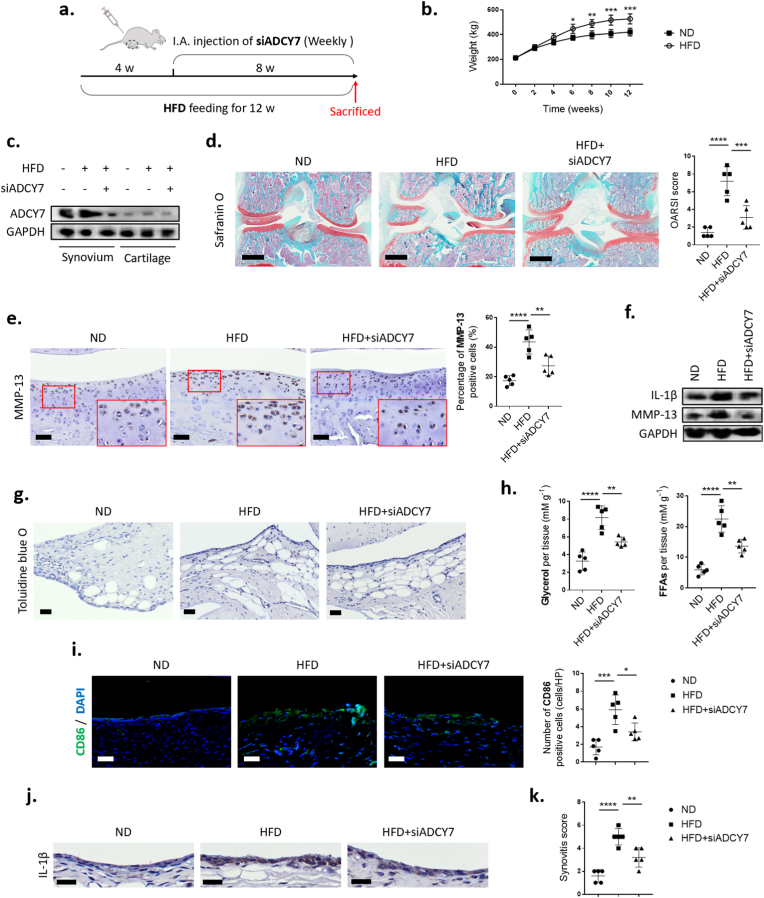
Eight-month-old SD rats were provided by the Department of Laboratory Animals of Central South University (Changsha, China). The high-fat diet (HFD) consisted of 17.5% fructose, 39.5% sweetened condensed milk, 20% beef tallow (H) or SFA (20% LA (HLA), 20% MA (HMA), 20% PA (HPA) or 20% SA (HSA)), 15.5% powdered standard rat chow, 2.5% HMW salt mixture and 5% water [12]. The HFD rats were also given drinking water supplemented with 25% fructose. The rats had ad libitum access to food and water during the 12-week protocol. After 4 weeks of HFD feeding, the rats were injected intra-articularly with 30 μl siRNA targeting ADCY7 (2′OME + 5′chol + 5-FAM-modified, GenePharma, China) once a week for 8 weeks. The body weight of the rats were measured every two weeks. After HFD feeding for 12 weeks, the rats were sacrificed for evaluation (Fig. 4a).
Fig. 4.
ADCY7 plays a key role in an HFD-induced OA model. (a). Illustration of a HFD induced rat OA model. siRNA targeting ADCY7 was injected intra-articularly (I.A.) beginning at week 4 and continuing for 8 weeks. (b) Body weight curves of the rats (n = 3). (c). Protein levels of ADCY7 in rat synovium and cartilage. (d). Safranin O staining and OARSI scoring of articular cartilage (n = 5, bar = 800 μm) (e–f). MMP-13 and IL-1β expression in cartilage assessed by immunohistochemistry (e, n = 5, bar = 100 μm) and western blot (f) (g–h). Lipid accumulation and lipolysis in synovium assessed by toluidine blue staining (g, bar = 100 μm) and levels of lipolytic products (h, n = 5) (i–k). Synovial inflammation assessed by M1 macrophage infiltration (I), IL-1β immunohistochemistry (j) and synovitis scoring (k) (n = 5, bar = 100 μm). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
2.5. Histological evaluation
Joint samples were collected and fixed in 4% paraformaldehyde (G-Clone, China) for 48 h, decalcified in 10% EDTA (G-Clone, China) for 4 weeks, embedded in paraffin and cut into sections (5 μm). Immunohistochemistry was performed using the DAB staining method. Briefly, the slides were deparaffinized and rehydrated, followed by antigen retrieval performed using sodium citrate at 95 °C. The slides were then blocked with 3% bovine serum albumin for 30min at room temperature and incubated with primary antibodies against ADCY7 (Thermo Fisher, USA), MMP13 (Affinity, China), IL-1β (Affinity, China), CD86 (a surface marker of M1 macrophages, Abcam, USA), CD55 (surface marker of FLS, Abcam, USA) overnight at 4 °C and a secondary antibody (Affinity, China), or a fluorescent secondary antibody (Affinity, China), for 30 min at 37 °C. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 10 min at room temperature before stained with 3, 30-diaminobenzidine tetra hydrochloride (DAB) and counterstained with hematoxylin. Slices of rat knee joints were also stained with safranin O/fast green (G-Clone, China) and toluidine blue (G-Clone, China). The Osteoarthritis Research Society International (OARSI) scoring system was used to evaluate the OA cartilage pathology [13]. A synovitis scoring system was used to evaluate the degree of synovitis [14].
2.6. Functional assessment of FLS
The siRNA targeting ADCY7 and the full-length rat ADCY7 cDNA (NCBI: NM_053396, ligated into the pCDNA 3.1 vector by the HindIII and NotI restriction enzymes (Takara Bio Inc., China)) was transfected into FLSs at passage 2 by Lipofectamine 2000 (Invitrogen, USA) when cells reached 70–80% confluence. FLSs at passage 3 were used for subsequent functional assessments. For the MTT assay, 5 × 104 cells were plated into 96-well plates in DMEM (10% FBS). Cell viability was estimated on day 2 after incubation with MTT for 4 hours and was read at 550 nm with a spectrophotometer. For the CFU-F assay, 1 × 103 cells were plated into a 60-mm culture dish in DMEM (10% FBS). The medium was changed every 3 days. After 14 days, the cells were fixed with 4% paraformaldehyde (G-Clone, China) and stained with 0.1% crystal violet (G-Clone, China). For the EdU assay, 2 × 105 cells were plated into 6-well plates in DMEM (10% FBS). A Click-iT Plus EdU Alexa Fluor 488 Imaging Kit (Invitrogen, USA) was used when cells reached 50–60% confluence according to the manufacturer's protocol. 24-well transwells were used for cell migration and invasion assay. For the migration assay, 5 × 104 cells in serum-free medium were seeded into the upper chamber. For the invasion assay, Matrigel was evenly inoculated into the upper chamber and allowed to form a gel at 37 °C. A total of 1 × 105 cells in serum-free medium were seeded into the upper chamber, and 20% FBS in DMEM was added to the lower chamber. After incubation, cells on the lower surface of the membrane were stained with 10% crystal violet (G-Clone, China). For cell morphology observation, the F-actin cytoskeleton was stained with rhodamine-phalloidin (diluted to 50 mg/ml, Sigma, USA) for 40 min at room temperature shielded from light when cells reached 40–50% confluence and visualized by confocal microscopy (FV1000; Olympus, Tokyo, Japan). Cells that were 80–90% confluent in 6-well plates were collected and used for RT-qPCR and Western blot analysis (See the following sections for details).
2.7. Western blot
Total proteins obtained from cells and tissues were subjected to SDS–PAGE (Biosharp, China) and then transferred to PVDF (Millipore, USA) membranes and blocked in 5% skimmed milk for 1 h. The membranes were incubated overnight at 4 °C with primary antibodies against ADCY7 (Thermo Fisher, USA), IL-1β (Affinity, China), MMP-13 (Affinity, China), N-cadherin (Affinity, China), E-cadherin (Affinity, China), vimentin (Affinity, China), α-SMA (CST, USA), p-PKA substrate (CST, USA), p-PLIN1(Abcam, USA), PLIN1 (Abcam, USA), p-HSL (CST, USA) and GAPDH. The following day, the membranes were incubated with fluorophore-conjugated secondary antibody (LI-COR Corp., NE). Protein bands were imaged with an enhanced LI-COR Odyssey infrared imaging system (LI-COR Corp., NE). The protein levels were normalized to the GAPDH levels.
2.8. RT-qPCR
ChamQ Universal SYBR qPCR Master Mix (Vazyme, China) was used for RT-qPCR according to the manufacturer's protocol. An initial denaturation step was carried out at 95 °C for 15 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 56 °C for 30 s, and extension at 72 °C for 30 s. Gene transcription levels were normalized to those of GAPDH. The primer design is shown in Supplementary Table 4.
2.9. Statistical analysis
Values in the text and figures are expressed as the mean ± SD unless otherwise noted. All statistical analysis was performed with SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). For calculation of significant differences in experiments with two groups, unpaired two-tailed Student's t test was applied when the data were normal distribution. The Mann–Whitney U test was employed when the data were not normal distribution. For experiments with more than two groups, ANOVA with Tukey or Sidak test was applied. R version 4.0.3 was used for omics data analyses. Differentially expressed genes (DEGs) between two subclusters were calculated with the R package “limma” (|log2 FC| ≥ 0.8 and adjusted P < 0.05). The statistical significance of the gene expression distribution among the three clusters was estimated by Kruskal–Wallis tests. P values in GO and KEGG enrichment analyses were adjusted using “Benjamini & Hochberg” method. Differences with p < 0.05 were considered statistically significant.
3. Results
3.1. Identification of three OA subclusters based on the synovial mRNA expression array
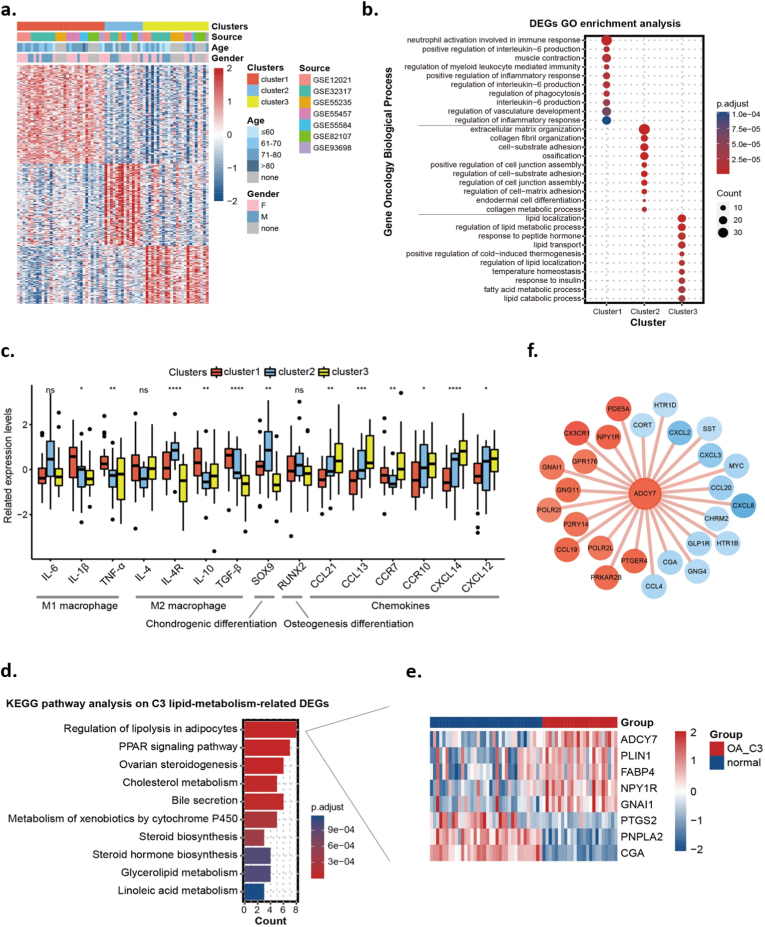
To explore the biological heterogeneity and potential subtypes of OA, we extracted the mRNA expression array profiles of 70 OA synovium samples from the GEO database. Based on the top 872 variable genes (MAD >0.5), 70 OA patients were categorized into three distinct subclusters via unsupervised clustering: 32 (45.7%) in Cluster 1 (C1), 14 (20%) in Cluster 2 (C2) and 24 (34.3%) in Cluster 3 (C3) (Fig. 1a). Next, we utilized GO enrichment analysis to identify functional annotations for each cluster based on the featured DEGs. The results indicated that C1_OA was characterized by immunity and inflammatory GO terms, including neutrophil activation involved in the immune response, regulation of IL-6 production and regulation of the inflammatory response; C2_OA was characterized by extracellular matrix-related GO terms, including collagen fibril organization and cell-substrate adhesion; C3_OA was characterized by lipid metabolism-related GO terms, including lipid localization, regulation of lipid metabolic process and lipid catabolic process (Fig. 1b). Fig. 1c compares the expression of several osteoarthritic synovitis-related genes across the three clusters.
Fig. 1.
Identification of three OA subclusters based on a synovial mRNA expression array. (a). Unsupervised consensus clustering based on the most variable genes of 70 OA synovium samples. (b). GO enrichment analysis of DEGs in the three OA subclusters. (c). The expression of osteoarthritic synovitis genes across the three subclusters. (d). KEGG pathway analysis of lipid metabolism-related DEGs of the C3_OA subcluster. (e). Eight specific DEGs were enriched in the pathway “Regulation of lipolysis in adipocytes”. (f). PPI network of ADCY7-related DEGs between C3_OA subcluster and control group. Orange and blue circles represent genes with high and low expression in the C3_OA subcluster, respectively. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns, no significance.
To verify our classification, a validation cohort of 85 OA synovium samples was extracted from GitHub. Based on the 540 feature DEGs previously acquired among the training cohort subclusters (Supplementary Figure 1c), 85 OA patients were also categorized into three similar subclusters: 37.6% in validation Cluster 1 (VC1), which was characterized by immune and inflammatory GO terms; 25.9% in validation Cluster 2 (VC2), which was characterized by cell membrane function-related GO terms; and 36.5% in validation Cluster 3 (VC3), which was characterized by lipid- metabolism-related GO terms (Supplementary Figure 1d and e).
3.2. An OA subcluster associated with ADCY7-related synovial lipolysis
Next, we performed further analyses aimed at the lipid metabolism-related C3_OA subcluster that has not been previously described. A total of 743 DEGs in the C3_OA subcluster were identified (with stricter screening criteria P < 0.05, |LogFC| > 0.8), among which 67 DEGs were regarded as correlated with lipid metabolism according to the summarized lipid metabolism-related pathways and genes from the KEGG and MSigDB databases (Supplementary Figure 1f). KEGG enrichment analysis based on these 67 DEGs identified the most noticeable pathway “Regulation of lipolysis in adipocytes” (Fig. 1d). Eight specific DEGs were enriched in this pathway, including ADCY7, PLIN1, FABP4, NPY1R and GNAI1, which were upregulated, and PTGS2, PNPLA2 and CGA, which were downregulated (Fig. 1e). Notably, ADCY7 had the highest statistical significance among these DEGs. Fig. 1f shows the PPI network between ADCY7 and the other encoded proteins among the 743 screened DEGs.
3.3. The OA subpopulation with high ADCY7 expression is characterized by synovial inflammatory lipolysis and FLS dysfunction
We examined the synovium of 10 non-OA, 22 early-stage OA, and 11 advanced OA patients and found that ADCY7 was upregulated in 32.6% of synovium from OA patients, including 11 (50%) from early-stage OA and 3 (27%) from advanced OA. According to their ADCY7 levels, we divided these patients into two subpopulations (Fig. 2a). The mean BMI of the ADCY7_High group was slightly higher than that of the ADCY7_Low group (p = 0.0677). However, the narrowing of the joint space in the ADCY7_High group was significantly faster than that in the ADCY7_Low group (p = 0.0006) (Supplementary Table 3). ADCY7 was mainly expressed in the synovial lining layer and surrounding adipose tissue, especially in areas with marked synovial hyperplasia and lipid accumulation (Fig. 2b). We observed lipolytic features (adipocyte shrinkage and increased stromal cell septum) in the perisynovial adipose tissue of the ADCY7_High group, accompanied by increased levels of the lipolytic products glycerol and free fatty acids (FFAs) in synovial fluid (Fig. 2c and d). In parallel, we detected higher expression of inflammatory cytokines (IL-1β, IL-6 and TNF-α) and chemokines (CCL21, CCL13 and CXCL14) in the synovium with high ADCY7 expression (Fig. 2e), accompanied by increased MMP-13 production and M1 macrophage (CD86+) infiltration (Fig. 2f and g). In addition, we observed some clusters of cells with high MMP-13 expression on the cartilage surface in the ADCY7_High group, and surface marker identification suggested that these cells might be derived from synovial FLSs (CD55+) (Fig. 2h).
Fig. 2.
Evaluation of the ADCY7 high-expressing OA subpopulation (a,b). ADCY7 expression in OA synovium. Red dashed lines divide the patients into high and low subpopulations according to their ADCY7 levels (10 non-OA, 22 early-stage OA and 11 advanced OA synovium were used) (c,d). Lipolysis of perisynovial adipose tissue as measured by toluidine blue staining (c) and levels of lipolytic products in synovial fluid (d, n = 8, bar = 100 μm) (e–g). Synovial inflammation assessed by inflammatory cytokines and chemokines expression (e, n = 8), MMP-13 production (f) and M1 macrophage infiltration (g, n = 8, bar = 100 μm). (h). The cluster of MMP-13 high-expressing cells (marked by white arrowheads) on the cartilage surface (bar = 100 μm). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
FLSs derived from the ADCY7_High group exhibited significantly enhanced proliferative and invasive activity (Fig. 3a–c), whereas the chondrocytes showed no differences in the expression of genes associated with phenotype and cellular function (Fig. 3d).
Fig. 3.
Functional assessment of FLSs and chondrocytes (a–c). Assessment of proliferation (a), Colony-forming units (b), and Transwell invasive activity (c) of FLSs derived from patients with differential ADCY7 levels (n = 6, bar = 100 μm). (d). Expression of chondrocyte phenotypic and functional genes (n = 6). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
3.4. ADCY7 plays a key role in a lipid metabolism disorder-related OA model
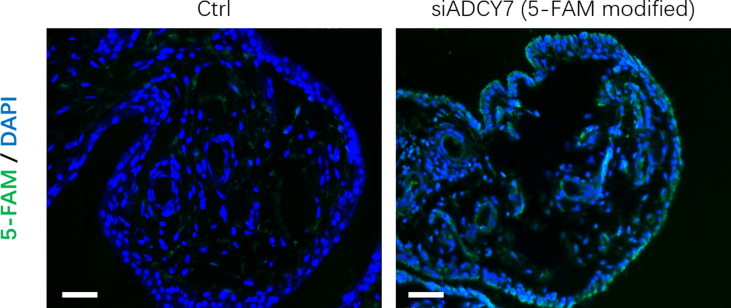
The rat knee OA model was induced by a 12-week high-fat diet (HFD). Compared with the normal diet (ND) group, the HFD for 12 weeks increased the body weight of the rats by an average of 25.4% (Fig. 4b). HFD increased the expression of ADCY7 in rat synovium, which was inhibited by siADCY7 (Fig. 4c). Fluorescent labeled siADCY7 was observed in approximately 61% of synovial cells (Supplementary Figure 2). Histologically, the HFD resulted in typical features of early OA (cartilage thinning and proteoglycan loss) (Fig. 4d). The proinflammatory factor IL-1β and the cartilage matrix-degrading enzyme MMP-13 were highly expressed in chondrocytes from the HFD group (Fig. 4e and f).
Notably, the HFD induced elevated expression of ADCY7 in the rat synovium (Fig. 4c). We found that the inhibition of ADCY7 by intra-articular injections of siADCY7 significantly alleviated HFD-induced cartilage degeneration as well as upregulation of IL-1β and MMP-13 in chondrocytes (Fig. 4d–f). Compared with the ND group, the HFD group exhibited marked lipid accumulation in the synovium. These accumulated lipids showed obvious lipolytic features, accompanied by elevated levels of lipolytic products (glycerol and FFAs) in the synovial fluid. siADCY7 further reinforced this lipid accumulation to a small extent but significantly inhibited lipolysis and reduced the levels of lipolytic products (Fig. 4g and h). In parallel, we observed infiltration of M1 macrophages, as well as elevated expression of IL-1β by the lining layer cells, in the synovium of the HFD group. These features, which were associated with synovial inflammatory lipolysis, were also alleviated with the inhibition of ADCY7 (Fig. 4i–k).
3.5. ADCY7 regulates FLS dysfunction in the HFD-induced OA model
We subsequently assessed the role of ADCY7 in the cellular function of FLSs, the major cell types of the synovial lining layer. Compared with FLSs derived from the ND group, FLSs derived from the HFD group (HFD-FLSs) exhibited elevated expression of ADCY7. In this part of the study, we transfected these HFD-FLSs with siRNA and pCDNA 3.1-ADCY7 to explore the role of ADCY7 in HFD-induced FLS dysfunction (Fig. 5a). We found that the HFD induced an epithelial-mesenchymal transition (EMT) tendency in HFD-FLSs. HFD-FLSs exhibited significantly higher proliferative activity (as measured by EdU assays) and colony-forming ability (as measured by CFU-F counts) than ND-FLSs (Fig. 5c and d). Transwell assays suggested that HFD feeding promoted the migratory and invasive activities of FLSs (Fig. 5e), which was confirmed by the increased pseudopodia in the morphological observation of cytoskeletal F-actin (Fig. 5f). The inflammatory cytokines TNF-α and IL-1β, proinflammatory macrophage chemoattractant protein-1 (MCP-1) and matrix-degrading MMP-1 and MMP-13 were also elevated in the HFD-FLSs (Fig. 5g).
Fig. 5.
ADCY7 regulates FLS dysfunction in the HFD-OA model. (a). ADCY7 expression in FLSs derived from the HFD-OA model. Cells were transfected with siRNA-ADCY7 and pCDNA-ADCY7 in vitro (n = 3). (b). Expression of epithelial-mesenchymal transition (EMT)-related markers in FLSs (n=3).(c–e). Assessment of EdU proliferation (c), Colony-forming units (d) and Transwell migration/invasion (e) of FLSs (n = 6, bar = 100 μm). (f). Morphology of cytoskeletal F-actin in FLSs. Pseudopodia are marked by white arrowheads (bar = 10 μm). (g). Expression of pro-inflammatory factors in FLSs (n = 6). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
We found that these HFD-induced EMT were, to varying degrees, dependent on the presence of ADCY7. In particular, the inhibition of ADCY7 significantly suppressed the invasive activity and the expression of IL-1β and MMPs in HFD-FLSs (Fig. 5c–g). The changes of EMT markers N-cadherin and E-cadherin induced by HFD also depend largely on the expression of ADCY7 (Fig. 5b).
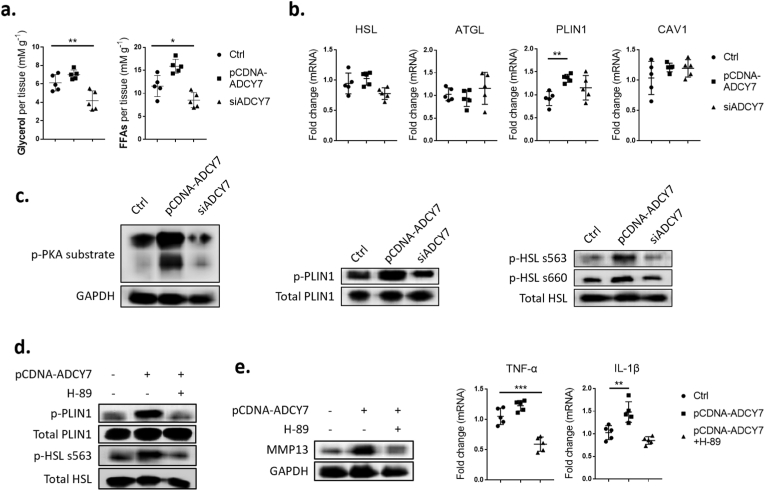
3.6. ADCY7 promotes PKA-mediated lipolysis pathway in HFD-FLS
By regulating ADCY7 expression, we found that ADCY7 plays a critical role in promoting the intracellular lipolysis of HFD-FLSs (Fig. 6a). ADCY7 elevated the phosphorylation level of the phosphokinase PKA, as well as its downstream lipid droplet-associated protein PLIN1 and hormone-sensitive lipase (HSL) (Fig. 6c). However, with the exception of PLIN1, we failed to detect changes in the expression of lipolysis-related proteins (Fig. 6b). Next, we blocked PKA activity using its inhibitor H-89. We found that inhibition of PKA activity blocked the ADCY7-mediated phosphorylation of PLIN1 and HSL, and largely alleviated the dysfunction of HFD-FLS, including increased MMP-13 and the proinflammatory cytokines IL-1β (Fig. 6d and e).
Fig. 6.
ADCY7 promotes PKA-mediated lipolysis pathway in HFD-FLSs (a–c). Assessment of lipolytic product levels (a), lipolysis-related gene expression (b) and phosphorylation of PKA and its downstream PLIN1 and HSL (c) in HFD-FLSs treated with siRNA-ADCY7 and pCDNA-ADCY7 in vitro(d–e). Assessment of PLIN1/HSL phosphorylation (d) and proinflammatory factor (MMP-13/TNF-α/IL-β) expression (e) in HFD-FLSs treated with pCDNA-ADCY7 and PKA inhibitor H-89 (n = 5). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
4. Discussion
Research is currently ongoing to identify OA subtypes for guiding treatment decisions. Multiple strategies have been proposed to distinguish OA subtypes [4,6,15]. Imaging and molecular markers can objectively reflect the characteristics of different patients and provide reliable evidences for the identification of subtypes. In particular, molecular markers are directly related to the pathological mechanism of OA. Molecular markers can sensitively reflect the characteristics of different microenvironment changes and metabolic disorders and are often used for the early diagnosis and subtype identification of OA [16]. For example, several studies identified OA subtypes characterized by higher levels of inflammatory markers [17,18]. Lv Z. et al. reviewed the characteristics of molecular markers in different stages of knee OA from before OA to late-stage OA and proposed four subtypes of progressive OA [16]. These strategies are helpful for patients with different OA subtypes to choose the most effective disease-modifying drugs according to their pathological characteristics.
Using “big data” from “omics” to group closely related items into clusters through analytical methods is a novel strategy [19]. The advantage of this strategy is that it can reveal the undiscovered OA subtypes and underlying pathological mechanisms through bioinformatics tools. By analyzing cartilage transcriptome data, Yuan C et al. identified four OA subclusters with different metabolic activities: an inflammation subcluster, two extracellular-matrix (ECM) metabolic disorder subclusters and an activated sensory neuron subcluster [7]. Considering the inherent homogeneity of cells and ECM in cartilage, the metabolic differences of cartilage between different subclusters are more likely to be affected by the synovium-synovial fluid microenvironment. Yuan C et al. analyzed tissue crosstalk in the knee joint and found that the synovium had the most dominant regulatory effect on different metabolic subclusters of cartilage [7]. Therefore, the synovium may be the driving factor of these OA subclusters. Utilizing the GEO database, we acquired transcriptome data of 70 OA synovium samples from 7 series. By unsupervised clustering, we identified three OA subclusters, including two subclusters related to inflammation and extracellular matrix metabolism and, notably, a novel lipid-metabolism-disorder-related subcluster that has not been reported before (Fig. 1a and b). This novel subcluster, which accounts for more than 30% of OA cases, is characterized by the KEGG pathway “regulation of lipolysis” as well as the differentially expressed gene ADCY7 (Fig. 1d and e). This may represent a currently undefined OA subtype and explain the clinical phenomenon of more severe synovial inflammation in obese OA patients [18].
Close associations between OA and lipid metabolism have been observed [20,21]. Substantial stores of lipid deposits have been observed in osteoarthritic chondrocytes [22]. Recently, as an association of several adipokines with OA has been discovered, the importance of adipokines in the pathogenesis of OA has begun to gain attention [23]. These adipokines, produced by periarticular adipose tissue or by the joint tissue itself, can directly affect joint health and regulate inflammatory responses to promote OA progression [24]. Lipolysis is defined as the catabolism of triacylglycerol stored in lipid droplets into glycerol and free fatty acids (FFAs) [25]. The latter, as common adipokines, exert important influences on the intra-articular microenvironment and cellular function [26]. Lipolysis also activates inflammatory responses. Through excessive release of adipokines such as FFAs, lipolysis induces the infiltration of M1 macrophages (proinflammatory) and activates inflammation within local tissues [27]. In turn, inflammatory cytokines (e.g., TNF-α, IL-6, etc.) released by inflammatory tissues further promoted lipolysis, thus forming a local vicious cycle of inflammatory lipolysis. Straub RH et al. observed features of lipolysis (increased density of sympathetic nerve fibers and more fragmented lipid droplets in adipocytes) surrounding the inflammatory synovium [28]. We divided the OA patient into two subpopulations according to synovial ADCY7 levels. In the ADCY7_High group, we observed features of synovial inflammatory lipolysis, including adipocyte shrinkage, increased stromal cell septum and lipolytic products, the infiltration of M1 macrophages and upregulation of inflammatory cytokines (Fig. 2c–g). It is possible that this is a critical mechanism of the chronic low-grade synovitis observed in some obese OA patients. However, the cause of the synovial inflammatory lipolysis is currently unclear. A high fat diet (HFD) is a common method to establish an animal model of systemic lipid metabolism disorder [29]. Several studies found that HFD induced synovial inflammation, macrophage infiltration and cartilage destruction in OA animal models [[30], [31], [32]]. We observed similar features in our rat HFD-induced OA model, accompanied by the high expression of ADCY7 (Fig. 4). Although it is not certain that these rats have exactly the same pathogenic mechanism, the method provides an available animal model for us to study this novel OA subtype.
The lipolytic microenvironment may also contribute to joint degeneration by directly affecting cellular functions within the joint. FFAs, the main product of lipolysis, are not only energy sources but also potent signaling molecules [25]. Studies have focused on the effects of FFAs on multiple joint cells, including chondrocytes, osteoblasts, osteoclasts and synoviocytes [[33], [34], [35], [36], [37], [38]]. In the HFD-induced rat OA model, with the enhancement of synovial lipolysis, FLSs transformed into a destructive phenotype characterized by high proliferative activity, high invasive and migratory activity and enhanced pro-inflammatory metabolism (Fig. 5), while chondrocytes exhibited high matrix-degrading activity (Fig. 4e and f). The inhibition of ADCY7 alleviated the phenotypic changes of these cells and the cartilage degeneration to a considerable extent. Considering that ADCY7 is mainly expressed in synovium and has a low level in cartilage (Fig. 4c), this lipolysis is likely to mainly affect the function of synovial cells and then indirectly affect chondrocytes through enhanced proinflammatory and procatabolic metabolism of synovia. Interestingly, however, we did not detect differences in the expression of phenotypic genes in chondrocytes from ADCY7_high OA patients (Fig. 3d). The main reason for this contradiction may be the different severity of OA in the samples. In animal studies, cartilage samples were derived from early-stage OA induced by 12 weeks of an HFD. In contrast, human chondrocytes were obtained from patients with advanced OA during total knee arthroplasty. The severe destruction and degeneration of cartilage are likely to mask the original phenotypic differences.
The mechanism by which the lipolytic microenvironment regulates FLS function is currently unknown. Studies have found that some specific FFAs can induce the release of inflammatory mediators from FLSs [34,39]. However, we found that intracellular pro-lipolytic signaling mediated through ADCY7 can directly affect the functions of FLSs (Fig. 5). This indicated that the dysfunction of FLSs may be caused not only by FFAs released from adipocyte lipolysis but also, more likely, by the disturbed intracellular lipid metabolism in FLSs. Ahn JK et al. analyzed the metabolomic profile of FLSs with an aggressive phenotype and found a significant increase in multiple metabolites, including those represented by FFAs [40]. They speculated that this aggressive phenotype might be related to alterations in lipid metabolism within FLSs. This may represent a currently unrecognized pathomechanism for OA. However, how intracellular lipid metabolism causes FLS dysfunction is currently unknown. Additional studies are necessary to elucidate its exact mechanism.
Multiple mechanisms may be responsible for synovial lipolysis in OA [41]. KEGG analysis revealed that ADCY7 had the highest statistical significance among the characteristic DEGs enriched in the “lipolysis” pathway in C3_OA subcluster (Fig. 1e). Adenylyl cyclases (ADCYs) are responsible for catalyzing the formation of cyclic adenosine monophosphate (cAMP) from adenosine triphosphate (ATP). ADCY7 is a membrane-bound isoform that is activated by a variety of extracellular signals [42]. Notably, ADCYs are key proteins in the pathway by which catecholamines stimulate lipolysis [41]. However, to our knowledge, the role of ADCYs in OA pathogenesis has not been previously studied, and only a few studies have addressed its role in inflammatory or aging-related diseases. Duan B et al. found that macrophages derived from ADCY7-deficient mice produced more of the proinflammatory cytokine TNF-α in response to stimulation [43]. Vatner SF et al. found that ADCY5 promotes age-related oxidative stress and that ADCY5 knockdown significantly increases longevity and healthful aging [44]. Utilizing synovial samples from OA patients, we identified a subpopulation with high ADCY7 expressing (Fig. 2a). In addition, we established an HFD-induced OA rat model and found an upregulation of ADCY7 in the synovium (Fig. 4c). In both cases, upregulated ADCY7 was strongly associated with features of synovial inflammatory lipolysis and FLS dysfunction. Although at present we do not have sufficient evidence that these two conditions, together with the C3_OA subcluster identified by synovial mRNA expression arrays, represent the same OA subtype, we confirmed that the inhibition of ADCY7 could effectively attenuate HFD-induced degenerative changes as well as the inflammatory lipolysis and FLS dysfunction observed in the rat model. This suggests that ADCY7 and its downstream pathways are potential pharmacological targets for treating this lipid-metabolism-disorder-related OA mechanism. Further study is required to clarify the more detailed role of ADCYs in the pathomechanisms of joint degeneration.
Funding
This work was supported by the National Natural Science Foundation of China (82,072,501), Science and Technology Innovation Leading Plan of High Tech Industry in Hunan Province (2020SK2011), Youth Fund Project of Natural Science Foundation of Hunan Province (2020JJ5848), and Medical Research Development Fund Project (WS865C).
Declaration of competing interest
No competing financial interests exist.
Acknowledgments
Junjie Huang: Formulation and evolution of overarching research goals and aims and writing the initial draft; Xinxing Wang: Application of statistical, mathematical, computational, or other formal techniques to analyze or synthesize study data; Song Wu: Acquisition of the financial support for the project leading to this publication; Xu Cao: Complete the main experiment and acquisition of the financial support for the project leading to this publication; Zhi Cui: Management and coordination responsibility for the research activity planning and execution; Zhiyu Ding and Yong Chen: Data Curation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jot.2022.02.007.
Contributor Information
Song Wu, Email: 600240@csu.edu.cn.
Xinxing Wang, Email: 2204140419@csu.edu.cn.
Junjie Huang, Email: hjjhuang@hotmail.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Fig. S2.
References
- 1.van Saase J.L., van Romunde L.K., Cats A., Vandenbroucke J.P., Valkenburg H.A. Epidemiology of osteoarthritis: zoetermeer survey. Comparison of radiological osteoarthritis in a Dutch population with that in 10 other populations. Ann Rheum Dis. 1989;48:271–280. doi: 10.1136/ard.48.4.271. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martel-Pelletier J., Barr A.J., Cicuttini F.M., Conaghan P.G., Cooper C., Goldring M.B., et al. Osteoarthritis. Nat Rev Dis Prim. 2016;2:16072. doi: 10.1038/nrdp.2016.72. l. [DOI] [PubMed] [Google Scholar]
- 3.Osteoarthritis: Structural endpoints for the development of drugs, devices, and biological products for treatment.
- 4.Deveza L.A., Melo L., Yamato T.P., Mills K., Ravi V., Hunter D.J. Knee osteoarthritis phenotypes and their relevance for outcomes: a systematic review. Osteoarthritis Cartilage. 2017;25:1926–1941. doi: 10.1016/j.joca.2017.08.009. l. [DOI] [PubMed] [Google Scholar]
- 5.Latourte A., Kloppenburg M., Richette P. Emerging pharmaceutical therapies for osteoarthritis. Nat Rev Rheumatol. 2020;16:673–688. doi: 10.1038/s41584-020-00518-6. l. [DOI] [PubMed] [Google Scholar]
- 6.Van Spil W.E., Kubassova O., Boesen M., Bay-Jensen A.C., Mobasheri A. Osteoarthritis phenotypes and novel therapeutic targets. Biochem Pharmacol. 2019;165:41–48. doi: 10.1016/j.bcp.2019.02.037. l. [DOI] [PubMed] [Google Scholar]
- 7.Yuan C., Pan Z., Zhao K., Li J., Sheng Z., Yao X., et al. Classification of four distinct osteoarthritis subtypes with a knee joint tissue transcriptome atlas. Bone Res. 2020;8:38. doi: 10.1038/s41413-020-00109-x. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinberg J., Southam L., Fontalis A., Clark M.J., Jayasuriya R.L., Swift D., et al. Linking chondrocyte and synovial transcriptional profile to clinical phenotype in osteoarthritis. Ann Rheum Dis. 2021;80:1070–1074. doi: 10.1136/annrheumdis-2020-219760. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J., Li Q., Su Z., Sun Q., Zhao Y., Feng T., et al. Lipid metabolism gene-wide profile and survival signature of lung adenocarcinoma. Lipids Health Dis. 2020;19:222. doi: 10.1186/s12944-020-01390-9. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panchal S.K., Poudyal H., Iyer A., Nazer R., Alam M.A., Diwan V., et al. High-carbohydrate, high-fat diet-induced metabolic syndrome and cardiovascular remodeling in rats. J Cardiovasc Pharmacol. 2011;57:611–624. doi: 10.1097/FJC.0b013e31821b1379. l. [DOI] [PubMed] [Google Scholar]
- 13.Pritzker K.P., Gay S., Jimenez S.A., Ostergaard K., Pelletier J.P., Revell P.A., et al. Osteoarthritis cartilage histopathology: grading and staging. Osteoarthritis Cartilage. 2006;14:13–29. doi: 10.1016/j.joca.2005.07.014. l. [DOI] [PubMed] [Google Scholar]
- 14.Takebe K., Rai M.F., Schmidt E.J., Sandell L.J. The chemokine receptor CCR5 plays a role in post-traumatic cartilage loss in mice, but does not affect synovium and bone. Osteoarthritis Cartilage. 2015;23:454–461. doi: 10.1016/j.joca.2014.12.002. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deveza L.A., Nelson A.E., Loeser R.F. Phenotypes of osteoarthritis: current state and future implications. Clin Exp Rheumatol. 2019;37(Suppl 120):64–72. l. [PMC free article] [PubMed] [Google Scholar]
- 16.Lv Z., Yang Y.X., Li J., Fei Y., Guo H., Sun Z., et al. Molecular classification of knee osteoarthritis. Front Cell Dev Biol. 2021;9:725568. doi: 10.3389/fcell.2021.725568. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siebuhr A.S., Petersen K.K., Arendt-Nielsen L., Egsgaard L.L., Eskehave T., Christiansen C., et al. Identification and characterisation of osteoarthritis patients with inflammation derived tissue turnover. Osteoarthritis Cartilage. 2014;22:44–50. doi: 10.1016/j.joca.2013.10.020. l. [DOI] [PubMed] [Google Scholar]
- 18.Meulenbelt I., Kloppenburg M., Kroon H.M., Houwing-Duistermaat J.J., Garnero P., Hellio-Le G.M., et al. Clusters of biochemical markers are associated with radiographic subtypes of osteoarthritis (OA) in subject with familial OA at multiple sites. The GARP study. Osteoarthritis Cartilage. 2007;15:379–385. doi: 10.1016/j.joca.2006.09.007. l. [DOI] [PubMed] [Google Scholar]
- 19.Ruiz-Romero C., Rego-Perez I., Blanco F.J. What did we learn from 'omics' studies in osteoarthritis. Curr Opin Rheumatol. 2018;30:114–120. doi: 10.1097/BOR.0000000000000460. l. [DOI] [PubMed] [Google Scholar]
- 20.Aspden R.M., Scheven B.A., Hutchison J.D. Osteoarthritis as a systemic disorder including stromal cell differentiation and lipid metabolism. Lancet. 2001;357:1118–1120. doi: 10.1016/S0140-6736(00)04264-1. l. [DOI] [PubMed] [Google Scholar]
- 21.Gkretsi V., Simopoulou T., Tsezou A. Lipid metabolism and osteoarthritis: lessons from atherosclerosis. Prog Lipid Res. 2011;50:133–140. doi: 10.1016/j.plipres.2010.11.001. l. [DOI] [PubMed] [Google Scholar]
- 22.Lippiello L., Walsh T., Fienhold M. The association of lipid abnormalities with tissue pathology in human osteoarthritic articular cartilage. Metabolism. 1991;40:571–576. doi: 10.1016/0026-0495(91)90046-y. l. [DOI] [PubMed] [Google Scholar]
- 23.Xie C., Chen Q. Adipokines: new therapeutic target for osteoarthritis? Curr Rheumatol Rep. 2019;21:71. doi: 10.1007/s11926-019-0868-z. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang T., He C. Pro-inflammatory cytokines: the link between obesity and osteoarthritis. Cytokine Growth Factor Rev. 2018;44:38–50. doi: 10.1016/j.cytogfr.2018.10.002. l. [DOI] [PubMed] [Google Scholar]
- 25.Zechner R., Zimmermann R., Eichmann T.O., Kohlwein S.D., Haemmerle G., Lass A., et al. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metabol. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brouwers H., von Hegedus J., Toes R., Kloppenburg M., Ioan-Facsinay A. Lipid mediators of inflammation in rheumatoid arthritis and osteoarthritis. Best Pract Res Clin Rheumatol. 2015;29:741–755. doi: 10.1016/j.berh.2016.02.003. l. [DOI] [PubMed] [Google Scholar]
- 27.Contreras G.A., Strieder-Barboza C., De Koster J. Symposium review: modulating adipose tissue lipolysis and remodeling to improve immune function during the transition period and early lactation of dairy cows. J Dairy Sci. 2018;101:2737–2752. doi: 10.3168/jds.2017-13340. l. [DOI] [PubMed] [Google Scholar]
- 28.Straub R.H., Lowin T., Klatt S., Wolff C., Rauch L. Increased density of sympathetic nerve fibers in metabolically activated fat tissue surrounding human synovium and mouse lymph nodes in arthritis. Arthritis Rheum. 2011;63:3234–3242. doi: 10.1002/art.30516. l. [DOI] [PubMed] [Google Scholar]
- 29.Perry R.J., Kim T., Zhang X.M., Lee H.Y., Pesta D., Popov V.B., et al. Reversal of hypertriglyceridemia, fatty liver disease, and insulin resistance by a liver-targeted mitochondrial uncoupler. Cell Metabol. 2013;18:740–748. doi: 10.1016/j.cmet.2013.10.004. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larranaga-Vera A., Lamuedra A., Perez-Baos S., Prieto-Potin I., Pena L., Herrero-Beaumont G., et al. Increased synovial lipodystrophy induced by high fat diet aggravates synovitis in experimental osteoarthritis. Arthritis Res Ther. 2017;19:264. doi: 10.1186/s13075-017-1473-z. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Munter W., van den Bosch M.H., Sloetjes A.W., Croce K.J., Vogl T., Roth J., et al. High LDL levels lead to increased synovial inflammation and accelerated ectopic bone formation during experimental osteoarthritis. Osteoarthritis Cartilage. 2016;24:844–855. doi: 10.1016/j.joca.2015.11.016. l. [DOI] [PubMed] [Google Scholar]
- 32.Brunner A.M., Henn C.M., Drewniak E.I., Lesieur-Brooks A., Machan J., Crisco J.J., et al. High dietary fat and the development of osteoarthritis in a rabbit model. Osteoarthritis Cartilage. 2012;20:584–592. doi: 10.1016/j.joca.2012.02.007. l. [DOI] [PubMed] [Google Scholar]
- 33.Harasymowicz N.S., Dicks A., Wu C.L., Guilak F. Physiologic and pathologic effects of dietary free fatty acids on cells of the joint. Ann N Y Acad Sci. 2019;1440:36–53. doi: 10.1111/nyas.13999. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez-Garcia O., Rogers N.H., Smith R.G., Lotz M.K. Palmitate has proapoptotic and proinflammatory effects on articular cartilage and synergizes with interleukin-1. Arthritis Rheumatol. 2014;66:1779–1788. doi: 10.1002/art.38399. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haywood J., Yammani R.R. Free fatty acid palmitate activates unfolded protein response pathway and promotes apoptosis in meniscus cells. Osteoarthritis Cartilage. 2016;24:942–945. doi: 10.1016/j.joca.2015.11.020. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sekar S., Wu X., Friis T., Crawford R., Prasadam I., Xiao Y. Saturated fatty acids promote chondrocyte matrix remodeling through reprogramming of autophagy pathways. Nutrition. 2018;54:144–152. doi: 10.1016/j.nut.2018.02.018. l. [DOI] [PubMed] [Google Scholar]
- 37.Loef M., Schoones J.W., Kloppenburg M., Ioan-Facsinay A. Fatty acids and osteoarthritis: different types, different effects. Joint Bone Spine. 2019;86:451–458. doi: 10.1016/j.jbspin.2018.07.005. l. [DOI] [PubMed] [Google Scholar]
- 38.Sekar S., Shafie S.R., Prasadam I., Crawford R., Panchal S.K., Brown L., et al. Saturated fatty acids induce development of both metabolic syndrome and osteoarthritis in rats. Sci Rep. 2017;7:46457. doi: 10.1038/srep46457. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frommer K.W., Schaffler A., Rehart S., Lehr A., Muller-Ladner U., Neumann E. Free fatty acids: potential proinflammatory mediators in rheumatic diseases. Ann Rheum Dis. 2015;74:303–310. doi: 10.1136/annrheumdis-2013-203755. l. [DOI] [PubMed] [Google Scholar]
- 40.Ahn J.K., Kim S., Hwang J., Kim J., Kim K.H., Cha H.S. GC/TOF-MS-based metabolomic profiling in cultured fibroblast-like synoviocytes from rheumatoid arthritis. Joint Bone Spine. 2016;83:707–713. doi: 10.1016/j.jbspin.2015.11.009. l. [DOI] [PubMed] [Google Scholar]
- 41.Duncan R.E., Ahmadian M., Jaworski K., Sarkadi-Nagy E., Sul H.S. Regulation of lipolysis in adipocytes. Annu Rev Nutr. 2007;27:79–101. doi: 10.1146/annurev.nutr.27.061406.093734. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price T., Brust T.F. Adenylyl cyclase 7 and neuropsychiatric disorders: a new target for depression? Pharmacol Res. 2019;143:106–112. doi: 10.1016/j.phrs.2019.03.015. l. [DOI] [PubMed] [Google Scholar]
- 43.Duan B., Davis R., Sadat E.L., Collins J., Sternweis P.C., Yuan D., et al. Distinct roles of adenylyl cyclase VII in regulating the immune responses in mice. J Immunol. 2010;185:335–344. doi: 10.4049/jimmunol.0903474. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vatner S.F., Pachon R.E., Vatner D.E. Inhibition of adenylyl cyclase type 5 increases longevity and healthful aging through oxidative stress protection. Oxid Med Cell Longev. 2015;2015:250310. doi: 10.1155/2015/250310. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.