Summary
The success of small molecule therapeutics that promotes degradation of critical cancer targets has fueled an intense effort to mimic this activity with bispecific molecules called PROTACs (proteolysis targeting chimeras). The simultaneous binding of PROTACs to a ligase and target can induce proximity-driven ubiquitination and degradation. VHL and CRBN are the two best characterized PROTAC ligases, but the rules governing their cellular activities remain unclear. To establish these requirements and extend them to new ligases, we screened a panel of 56 cell lines with two potent PROTACs that utilized VHL, MZ1, or CRBN, dBET1 to induce degradation of BRD4. With notable exceptions, MZ1 was broadly active in the panel whereas dBET1 was frequently inactive. A search for predictive biomarkers of PROTAC activity found that expression and mutation of VHL and CRBN were themselves predictors of PROTAC activity in the cell line panel.
Subject areas: Bioengineering, Cancer, Molecular biology
Graphical Abstract

Highlights
-
•
Profiling of diverse tumor types establishes the broad utility of VHL-based PROTACs
-
•
CRBN-dependent dBET1 activity is frequently suppressed in cancer cell lines
-
•
Copy loss and mRNA expression predict the activity of VHL and CRBN-dependent PROTACs
Bioengineering; Cancer; Molecular biology
Introduction
Intense interest in targeted protein degradation is in part driven by the clinical success of the IMiDs, small molecule therapeutics including thalidomide, lenalidomide, and pomalidomide, which are used to treat multiple myeloma, MDS, and mantle cell lymphoma (Zhu et al., 2011; Lopez-Girona et al., 2012; Lu et al., 2014; Robertson, 2001; Patel and Bihani, 2018). IMiDs have myriad activities including teratogenic effects, immunosuppression, and anticancer properties. More than fifty years after their discovery, the direct target of IMIDs was revealed to be CRBN, a substrate receptor for the CRL4CRBN ubiquitin ligase. IMiD binding to CRBN promotes the ubiquitination and degradation of novel substrate proteins including Ikaros, Aiolos, and CK1α (Kronke et al., 2015; Gandhi et al., 2014; Lu et al., 2014). The IMiDs powerfully demonstrate the advantages of induced protein degradation as a therapeutic strategy. These advantages include inhibition through elimination of the target rather than blocking a single activity, prolonged inhibition of the target even after drug is eliminated, and the potential to reach targets that were once thought undruggable by binding to protein domains distant from active domains (Kronke et al., 2014, 2015).
Structural studies indicate that IMiDs act as small molecular glues akin to the plant hormone auxin, which binds to a ubiquitin ligase to facilitate novel substrate interactions. The binding of lenalidomide to CRBN in the “tri-TRP” pocket generates a surface for neo-substrates like CK1α, which bind through a zinc finger conserved in all IMiD-CRBN neo-substrates. These studies further demonstrate that the IMiD is buried on the surface of CRBN leaving just its glutarimide ring solvent exposed. The glutarimide ring makes necessary contacts with residues of the zinc finger of CK1α but additional critical contacts between the surfaces of CK1α and CRBN are also observed (Petzold et al., 2016). Thus, the ternary complex of CRBN-IMiD and substrate is driven by both protein-protein and protein-IMiD interactions that lead to cooperative binding. Through this mechanism, lenalidomide induces binding of CRBN ligase with neo-substrates such as Ikaros, Aiolos, and CK1α, catalyzing their ubiquitination and eventual destruction, and driving potent anticancer effects.
An effort to derivatize and re-engineer IMiDs to make more potent effective molecules and to induce degradation of new substrate proteins is underway and there are some early signs of success, but a bigger prize would be the discovery of new glues that replicate the mechanism of action of the IMiDs with entirely new ligases (Hansen et al., 2018; Ishoey et al., 2018; Matyskiela et al., 2016). Some hope that this will be possible, comes from the discovery that SPLAMs (splicing inhibitor sulfonamides) promote binding of RBM39 to DCAF15, also a DDB1/CUL4-associated ubiquitin substrate receptor protein. While it is encouraging that another glue-like adaptor to the ubiquitin proteasome system has been discovered, it is hard to imagine how to prospectively engineer molecules with similar glue-like activities to direct ubiquitin ligases unrelated to CRBN or DCAF15 to destroy neo-substrate targets (Han et al., 2017; Uehara et al., 2017).
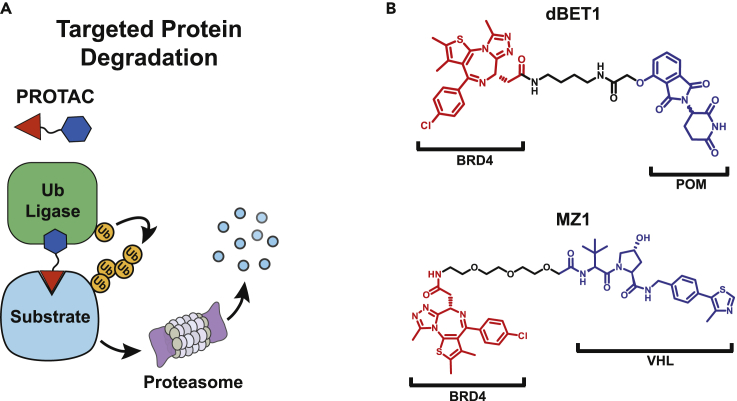
PROTACs (protein targeting chimeras) represent a rapidly emerging alternative strategy that employs ligase and substrate ligands to drive ligase and neo-substrate ternary complex formation through truly bifunctional small molecules (Figures 1A and 1B). The first examples of PROTACs contained tethered peptide degron sequences that are recognized by a ubiquitin ligase complex and were linked to small molecule ligands of a substrate protein (Sakamoto et al., 2001). PROTAC1 exemplified this strategy by linking the IKKβ degron recognized by β-TrCP to ovalbumin, a ligand for MetAP-2. PROTAC1 promoted BTRCP/SKP1-dependent ubiquitination of MetAP-2 in partially purified biochemical systems; however, cellular activity was only achieved through microinjection (Sakamoto et al., 2001). Though these first PROTACs were far short of pharmaceutical agents, they powerfully demonstrated the flexibility of the BTRCP/SKPI ligase and the potential of PROTACs as chemical tools. In the years following PROTAC1, relatively small improvements to the strategy were made culminating in an all-small-molecule PROTAC derived from Nutlin3a, an inhibitor of MDM2 (Schneekloth et al., 2004, 2008; Sakamoto et al., 2003). This jump to all-small-molecule PROTACs triggered efforts to improve ligase binders in order to achieve nanomolar cell potency and in vivo activity (Zhang et al., 2018). The discovery of a potent VHL ligand, VHL1, inhibitors of IAP proteins, and IMiDs as CRBN binders, led to a revolution in the PROTAC field, which has rapidly progressed in the last few years (Paiva and Crews, 2019).
Figure 1.
PROTACs are bispecific molecules that tether a substrate protein to a Ub-ligase complex to promote ubiquitination and subsequent degradation of the substrate protein in cells
(A) PROTACs are formed from a substrate binder and a ligand for a Ub-ligase receptor protein (R). The VHL and CRBN receptor proteins can be brought into complex with E2 proteins by associating with adapter proteins, a cullin, and a RING domain protein (Ri). Formation of a substrate-PROTAC-ligase complex promotes transfer of ubiquitin from the E2 subunit to substrate protein (S). Substrate proteins marked with Ub-chains are destined for proteasome-dependent degradation.
(B) BET ligand JQ1 is linked to either pomalidomide or VHL1 to form dBET1 and MZ1, two potent PROTACs that target BRD4 for degradation.
PROTACs have been employed to degrade a diverse set of target proteins that include receptor tyrosine kinases, hormone receptors, signaling kinases, chromatin modifying enzymes, and transcription factors (Chopra et al., 2019; Kang et al., 2018; Burslem et al., 2018). In contrast, only a handful of ubiquitin ligases have been used for these PROTACs and most published work has focused on proven PROTAC ligases, CRBN and VHL. This focus has largely been due to practical considerations; a great deal of effort is required to identify new ligase ligands as these proteins are difficult to “drug” and biochemical characterization is likely a pre-requisite of a successful chemistry campaign.
Despite these challenges, expansion of the repertoire of ligases offers the promise of building PROTAC molecules for any intracellular protein with ligases that might enhance disease specificity or target selectivity. However, little is known about many Ub-ligases, including their expression, intracellular distribution, requisite partner proteins, and post-translational regulation. Prior to purification and biochemical characterization of the ligase complex and final screening of the protein to identify novel binders, these questions need to be addressed to reduce time and resources spent. To inform choices for novel ligases, we sought to understand the limitations of two of the most well-studied PROTAC ligases, VHL and CRBN, through two potent BRD4-PROTACs, MZ1 and dBET1 (Figure 1B).
The impressive in vitro activity of both molecules is well documented, but we hypothesized that this activity might vary among cell lines representing many different cancer types (Winter et al., 2015; Zengerle et al., 2015). To this end, we generated activity data for MZ1 and dBET1 in a panel of 56 cell lines with publicly available genomic expression and mutation data and determined how changes in expression and mutation of VHL and CRBN and associated factors impacted PROTAC activity. We envisioned that similar genomic data could be used to predict the activity of other PROTAC ligases and provide a comparison to the two archetypes, VHL and CRBN.
Results
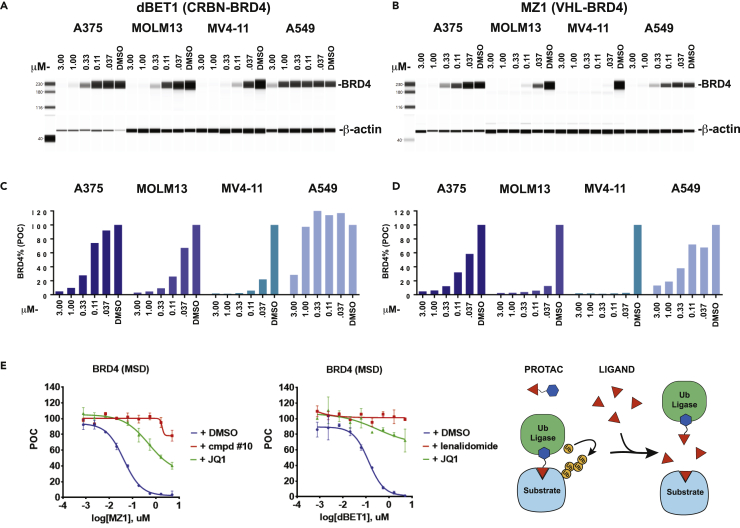
Much of the initial evaluation of the first VHL and CRBN engaging PROTACs, MZ1 and dBET1, was performed in vitro with cell lines originating from hematological cancers (Zengerle et al., 2015, Winter et al., 2015; Alqahtani et al., 2019). To better understand the utility of these PROTACs to degrade BRD4 in other cancer types, we screened four cell lines to reproduce the activity previously described. We used capillary electrophoresis to track the disappearance of BRD4 with increasing concentrations of either MZ1, a VHL engaging BRD4 degrader, or dBET1, a CRBN-dependent BRD4 degrader. As described by their inventors, each of these PROTACs was potent and efficient, eliminating nearly all BRD4 protein in 4 h in both MV4-11 and MOLM13 cells with nano-molar concentrations of compound (Figures 2A and 2B). Surprisingly, the activity of both PROTACs was not equivalent in every cell line and the non-small cell lung cancer cell line, A549, appeared to demonstrate little degradation of BRD4 with dBET1 (Figure 2A). Because the two PROTACs remain constant and the cell line is the only variable in this exploration, we surmised that the activity of the PROTACs was largely dependent on the activity of the ubiquitin ligase in each of the cell lines. To test this possibility in a larger number of cell lines, we developed a sandwich electrochemical ELISA (MSD) assay (Wurz et al., 2018). This assay yielded results comparable to capillary gel electrophoresis (R2 >0 .98) (Figures 2C, 2D, and S1). As previously observed, both MZ1 and dBET1 activity was dependent on the proteasome and inhibition of PROTAC activity with either 50 μM JQ1, the parental BRD4 ligand, or VHL1 and lenalidomide, effectively prevented the PROTACs from degrading BRD4 (Figure 2E).
Figure 2.
PROTAC activity is dependent on the cell line context
Four cell lines were treated with up to 3 μM PROTAC for 4 h and the resulting lysates were separated by capillary electrophoresis with immunodetection of BRD4 and actin proteins, (A and B), and subjected to BRD4 quantification by sandwich ELISA, (C–E).
(A) dBET1-treated lysates are shown; the activity of the dBET1 PROTAC varied greatly across the four cell lines, with little activity in a lung cancer cell line, A549.
(B) MZ1 treated cell lysates are shown, activity of the PROTAC varied but all cell lines allowed efficient degradation of BRD4 protein.
(C and D) Immunodetection of BRD4 protein with an MSD sandwich ELISA protocol recapitulated the protein levels quantified by capillary electrophoresis.
(E) Lysates were collected from A375 cells treated with a dose titration of either MZ1 or dBET1 and competing BRD4 and ligase receptor ligands. BRD4 protein was quantified by MSD ELISA. On mechanism, ligase dependent, activity of the PROTACs was demonstrated through competition with either BET ligand JQ1, VHL ligand compound #10, or CRBN ligand lenalidomide. Dose titration curves are derived from n = 2 independent experiments, error bars represent standard error of the mean (SEM).
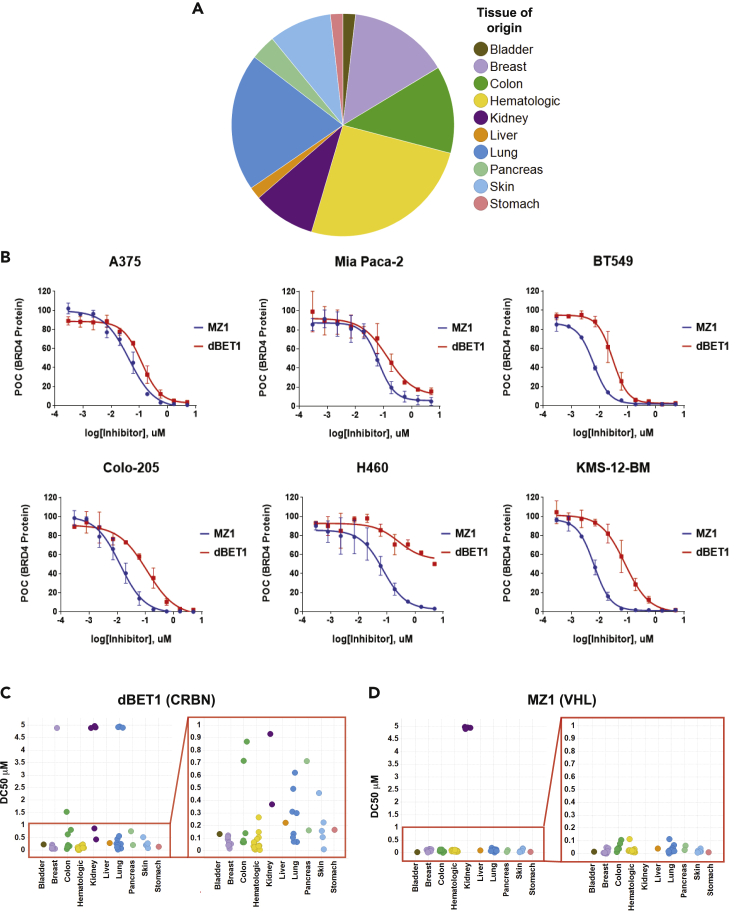
To understand the utility of CRL2VHL and CRL4CRBN across multiple cancer subtypes, we assembled a panel of 56 cell lines commonly used in research laboratories for the in vitro evaluation of small molecule inhibitors. Ten tissues of origin were represented, and a greater proportion of colon, lung, breast, and hematologic cell lines was included (Figure 3A). Each of the cell lines in our panel was represented in the CCLE (Cancer Cell Line Encyclopedia) database with retrievable gene expression and DNA sequencing data. We treated the 56 cell lines with MZ1 and dBET1 for 4 h with a 10-point dose titration. The 4-h treatment time was determined to be the optimum time to capture the breadth of activity across the panel (Figure S2). Levels of BRD4 were evaluated via MSD assay and the IC50 at the inflection point of the curve and the DC50, the concentration at which 50% of BRD4 protein was degraded, as well as the maximum activity were extracted from the data (Table 1). The variability within groups of similar cell lines was compared (Table 1, Figures 3B–3D). As observed with the initial four cell lines, the activity of dBET1 was more variable across the panel compared to the activity of MZ1, suggesting that CRBN-dependent ligase activity may be more variable. Renal cell carcinoma lines uniformly lacked VHL activity, but all other lines showed similar levels of BRD4 degradation by MZ1. Reduced dBET1 activity was seen in multiple cancer types, with at least one example of nearly complete absence of CRBN-dependent BRD4 degradation in lung, colon, and kidney cell lines, suggesting that lack of CRBN activity may be a common occurrence. Notably, hematological cancer cell lines showed consistently high activity (Figure 3C and Table 1); thus, it appears that the CRL4CRBN ligase is most active in cell lines corresponding to cancers that express high levels of the CRBN protein and where lenalidomide has significant pharmacological activity (Fischer et al., 2014).
Figure 3.
dBET1 activity varies across a cell line panel representing 10 cancer subtypes and MZ1 is inactive in renal cell carcinoma cell lines
(A) Pie chart showing tissues of origin and distribution for the 56 cell lines included in the panel.
(B) Example 10-point dose-response curves showing BRD4 protein quantified by MSD ELISA after 4-h treatment with either MZ1 or dBET1. Dose titration curves are derived from n = 2 independent experiments, error bars represent standard error of the mean (SEM).
(C and D) DC50 values calculated from dose-response curves for all 56 cell lines for both dBET1 and MZ1 are plotted by tissue of origin. Zoom in on 0–1 μM shows greater variation in dBET1 activity (C) across the cell line panel among cell lines within this range compared to MZ1 activity plotted in (D).
Table 1.
Summary of dBET1 and MZ1 activity after treatment of 56 cell lines
| Cell Line | Tissue of Origin | MZ1 IC50 (μM) | MZ1 DC50 (μM) | MZ1 max % | dBET1 IC50 (μM) | dBET1 DC50 (μM) | dBET1 max % |
|---|---|---|---|---|---|---|---|
| BT549 | Breast | 0.006, n = 2 | 0.005, n = 2 | 1.50, n = 2 | 0.030, n = 2 | 0.029, n = 2 | 2.50, n = 2 |
| CAL51 | Breast | 0.019, n = 2 | 0.015, n = 2 | 3.00, n = 2 | 0.086, n = 2 | 0.100, n = 2 | 7.00, n = 2 |
| HCC 1954 | Breast | 0.0185, n = 2 | 0.022, n = 2 | 10.19, n = 2 | 0.042, n = 2 | 0.055, n = 2 | 11.02, n = 2 |
| HCC1569 | Breast | 0.0155, n = 2 | 0.011, n = 1 | 2.00, n = 2 | 0.101, n = 1 | 0.098, n = 1 | 2.00, n = 2 |
| HCC1806 | Breast | 0.020, n = 2 | 0.024, n = 2 | 4.50, n = 2 | >5, n = 2 | >5, n = 2 | 92.00, n = 2 |
| MCF7 | Breast | 0.033, n = 2 | na | 0.50, n = 2 | 0.067, n = 2 | 0.065, n = 2 | 15.50, n = 2 |
| MDA-MB-157 | Breast | 0.013, n = 1 | 0.012, n = 1 | 4.00, n = 1 | 0.020, n = 1 | 0.019, n = 1 | 6.00, n = 1 |
| ZR 75-1 | Breast | 0.009, n = 2 | 0.006, n = 2 | 2.89, n = 2 | 0.115, n = 2 | 0.118, n = 2 | 10.15, n = 2 |
| DAUDI | Heme | 0.011, n = 2 | 0.013, n = 2 | 1.50, n = 2 | 0.069, n = 2 | 0.075, n = 2 | 3.00, n = 2 |
| HT | Heme | 0.018, n = 2 | 0.016, n = 2 | 2.00, n = 2 | 0.018, n = 2 | 0.022, n = 2 | 2.50, n = 2 |
| JURKAT | Heme | 0.021, n = 2 | 0.022, n = 2 | 2.93, n = 2 | 0.109, n = 2 | 0.132, n = 2 | 4.46, n = 2 |
| JVM3 | Heme | 0.013, n = 1 | na | 0.013, n = 1 | 0.077, n = 1 | 0.070, n = 1 | 0.08, n = 1 |
| KG1 | Heme | 0.1425, n = 2 | 0.113, n = 2 | 2.45, n = 2 | 0.308, n = 2 | 0.280, n = 2 | 5.01, n = 2 |
| KMS-12-BM | Heme | 0.006, n = 2 | 0.006, n = 2 | 1.50, n = 2 | 0.082, n = 2 | 0.085, n = 2 | 1.50, n = 2 |
| MOLM13 | Heme | 0.006, n = 2 | 0.013, n = 2 | 15.03, n = 2 | 0.030, n = 2 | 0.049, n = 2 | 16.66, n = 2 |
| MOLT4 | Heme | 0.021, n = 2 | 0.019, n = 2 | 0.50, n = 2 | 0.115, n = 2 | 0.076, n = 2 | 2.00, n = 2 |
| MV411 | Heme | 0.00075, n = 2 | 0.00076, n = 2 | 2.10, n = 2 | 0.010, n = 2 | 0.009, n = 2 | 2.40, n = 2 |
| NAMALWA | Heme | 0.00433, n = 2 | 0.00519, n = 2 | 3.72, n = 2 | 0.015, n = 2 | 0.015, n = 2 | 3.58, n = 2 |
| OPM-2 | Heme | 0.00463, n = 2 | 0.00412, n = 2 | 5.05, n = 2 | 0.020, n = 2 | 0.021, n = 2 | 5.37, n = 2 |
| PL-21 | Heme | 0.0120, n = 2 | 0.0070, n = 2 | 5.92, n = 2 | 0.047, n = 2 | 0.029, n = 2 | 5.94, n = 2 |
| RL | Heme | 0.0077, n = 2 | na | 3.00, n = 2 | 0.033, n = 2 | 0.020, n = 2 | 2.00, n = 2 |
| U266B1 | Heme | 0.031, n = 2 | 0.0340, n = 2 | 10.90, n = 2 | 0.021, n = 2 | 0.022, n = 2 | 8.31, n = 2 |
| 293T | KIdney | 0.0146, n = 2 | 0.013, n = 2 | 2.27, n = 2 | 0.045, n = 2 | 0.047, n = 2 | 2.43, n = 2 |
| 769-P | KIdney | >5, n = 2 | >5, n = 2 | 108.93, n = 2 | 0.310, n = 2 | 0.353, n = 2 | 9.12, n = 2 |
| 786–0 | KIdney | >5, n = 2 | >5, n = 2 | 98.00, n = 2 | 0.366, n = 2 | >5, n = 1 | 54.50, n = 2 |
| A498 | KIdney | >5, n = 2 | >5, n = 2 | 128.00, n = 2 | 2.30, n = 2 | >5, n = 1 | 55, n = 2 |
| A704 | KIdney | >5, n = 2 | >5, n = 2 | 100.76, n = 2 | >5, n = 2 | >5, n = 2 | 59.86, n = 2 |
| CAKI-2 | KIdney | >5, n = 2 | >5, n = 2 | 100.59, n = 2 | 0.681, n = 2 | 0.945, n = 2 | 20.46, n = 2 |
| COLO205 | Large Intestine | 0.0132, n = 2 | 0.0129, n = 2 | 0.00, n = 2 | 0.118, n = 2 | 0.074, n = 2 | 2.00, n = 2 |
| H508 | Large Intestine | 0.0605, n = 2 | 0.0935, n = 2 | 25.24, n = 2 | 0.373, n = 2 | 1.63, n = 2 | 30.18, n = 2 |
| HCT116 | Large Intestine | 0.0395, n = 2 | 0.0333, n = 2 | 3.00, n = 2 | 0.308, n = 2 | 0.886, n = 2 | 45.50, n = 2 |
| HCT15 | Large Intestine | 0.114, n = 2 | 0.122, n = 2 | 7.07, n = 2 | 0.758, n = 2 | 0.740, n = 2 | 22.75, n = 2 |
| HT29 | Large Intestine | 0.014, n = 2 | 0.013, n = 2 | 0.50, n = 2 | 0.130, n = 2 | 0.136, n = 2 | 8.50, n = 2 |
| SW1417 | Large Intestine | 0.021, n = 2 | 0.017, n = 2 | 0.50, n = 2 | 0.068, n = 2 | 0.090, n = 2 | 5.00, n = 2 |
| SW48 | Large Intestine | 0.012, n = 2 | 0.011, n = 2 | 6.94, n = 2 | 0.055, n = 2 | 0.067, n = 2 | 7.51, n = 2 |
| HEPG2 | Liver | 0.023, n = 2 | 0.026, n = 2 | 3.94, n = 2 | 0.147, n = 2 | 0.208, n = 2 | 11.36, n = 2 |
| A427 | Lung | 0.012, n = 2 | 0.013, n = 2 | 3.59, n = 2 | 0.202, n = 2 | 0.507, n = 2 | 35.85, n = 2 |
| A549 | Lung | 0.093, n = 2 | 0.093, n = 2 | 3.52, n = 2 | 0.588, n = 2 | 0.636, n = 2 | 10.63, n = 2 |
| H1568 | Lung | 0.0056, n = 2 | 0.0051, n = 2 | 4.85, n = 2 | 0.055, n = 2 | 0.069, n = 2 | 13.57, n = 2 |
| H358 | Lung | 0.0067, n = 2 | 0.0064, n = 2 | 2.41, n = 2 | 0.224, n = 2 | 0.313, n = 2 | 21.27, n = 2 |
| H441 | Lung | 0.0170, n = 2 | 0.0150, n = 2 | 4.44, n = 2 | 0.078, n = 2 | 0.087, n = 2 | 7.63, n = 2 |
| HCC 15 | Lung | 0.0032, n = 2 | 0.0033, n = 2 | 2.80, n = 2 | 0.429, n = 2 | >5, n = 2 | 75.73, n = 2 |
| NCIH1299 | Lung | 0.0215, n = 2 | 0.020, n = 2 | 0.00, n = 2 | 0.220, n = 2 | 0.325, n = 2 | 17.00, n = 2 |
| NCIH23 | Lung | 0.0110, n = 2 | 0.0105, n = 2 | 4.00, n = 2 | >5, n = 2 | >5, n = 2 | 55.50, n = 2 |
| NCIH460 | Lung | 0.069, n = 2 | 0.0485, n = 2 | 3.00, n = 2 | 0.158, n = 2 | >5, n = 2 | 56.50, n = 2 |
| NCIH661 | Lung | 0.0060, n = 2 | 0.0055, n = 2 | 0.00, n = 2 | 0.102, n = 2 | 0.119, n = 2 | 2.50, n = 2 |
| NCIH838 | Lung | 0.0175, n = 2 | 0.0185, n = 2 | 0.00, n = 2 | 0.099, n = 2 | 0.105, n = 2 | 2.00, n = 2 |
| Mia PaCa-2 | Pancreas | 0.0665, n = 2 | 0.0507, n = 2 | 5.00, n = 2 | 0.1435, n = 2 | 0.152, n = 2 | 15.50, n = 2 |
| SW1990 | Pancreas | 0.0230, n = 2 | 0.0245, n = 2 | 7.06, n = 2 | 0.404, n = 2 | 0.705, n = 2 | 24.08, n = 2 |
| A101D | Skin | 0.0133, n = 2 | 0.0136, n = 2 | 2.71, n = 2 | 0.105, n = 2 | 0.145, n = 2 | 15.29, n = 2 |
| A2058 | Skin | 0.0052, n = 2 | 0.0026, n = 2 | 2.03, n = 2 | 0.190, n = 2 | 0.446, n = 2 | 26.96, n = 2 |
| A375 | Skin | 0.0455, n = 2 | 0.0410, n = 2 | 0.50, n = 2 | 0.123, n = 2 | 0.092, n = 2 | 3.50, n = 2 |
| RPMI 7951 | Skin | 0.0020, n = 2 | 0.0019, n = 2 | 2.26, n = 2 | 0.019, n = 2 | 0.022, n = 2 | 5.75, n = 2 |
| SK MEL 28 | Skin | 0.0128, n = 2 | 0.0113, n = 2 | 2.84, n = 2 | 0.179, n = 2 | 0.215, n = 2 | 16.45, n = 2 |
| AGS | Stomach | 0.0020, n = 2 | 0.0019, n = 2 | 2.28, n = 2 | 0.099, n = 2 | 0.177, n = 2 | 23.12, n = 2 |
| UM UC-3 | Urinary Track | 0.0175, n = 2 | 0.0110, n = 2 | 2.64, n = 2 | 0.126, n = 2 | 0.110, n = 2 | 6.34, n = 2 |
The concentration at the inflection point (IC50) and the concentration at 50% degradation (DC50) were calculated with maximum degradation (max%) from 10-point dose-response curves for dBET1 and MZ1 after 4 h of treatment.
DC50 is frequently used as an indicator of relative PROTAC activity, but DC50 values generated at a fixed timepoint reflect both the concentration at which the substrate-PROTAC-ligase ternary complex is formed and the rate of ubiquitination and degradation. To understand how measured DC50 values are affected by the rate of BRD4 degradation, we probed a small number of cell lines over a time course with multiple concentrations of MZ1. In this experiment, the dose-response curves for MZ1 for each cell line never quite reached a minimum DC50, so that the longer cells were treated the lower the DC50 values became. For H838, dose-response curves resulted in DC50s between 29 and 11 nM between 2 and 8 h after treatment (Figure S2) and an observed max activity of 98%–99%. To see how PROTAC concentration affects the rate of degradation, we plotted the same data in reverse, tracking the decrease in BRD4 protein over time to calculate the time that is required to degrade 50% of BRD4 protein (T50%) for each concentration of MZ1. From these plots, we observed that T50% decreased with increasing concentration of the PROTAC until reaching an apparent plateau 3-fold above the DC50. This indicated that for each cell line, the minimal T50% measurements reflected the maximum rate at which BRD4 could be degraded. This analysis also showed that high concentrations (>3 μM) of PROTAC can lead to lower activity, an effect that is caused by excess PROTAC-inhibiting ternary complex formation by binding target and ligase separately. This “hook effect” has been described in detail by previous studies (Riching et al., 2018; Roy et al., 2019). It is worth noting that the magnitude of the “hook effect” is a characteristic of each PROTAC. If protein-protein and protein-drug interaction lead to ternary complexes that exhibit cooperativity then the concentration of PROTAC that is required to diminish activity of the PROTAC is increased and then the hook effect is only observed at very high concentrations or with very short treatment.
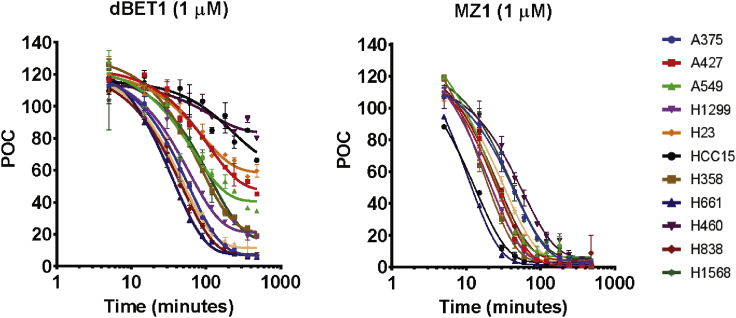
We expanded the time-course analysis to additional cell lines with MZ1 and dBET1 dosed at a single concentration to provide additional insight into the lung cancer cell line panel (Figure 4 and Table 2). We chose a fixed dose of 1 μM as it is sufficiently above the mean DC50 for the entire panel but below concentrations where the confounding hook effect sometimes appeared. Inspired in part by the work of Riching et al., we used dose-response time-course experiments on a limited number of cell lines to further validate our choice of the 1 μM dose, and found that our measurements in cancer cell lines were similar to their measurements in HEK293 cells despite our use of a protein ELISA-like assay compared to their use of a live cell imaging assay (Figure S2) (Riching et al., 2018). Consistent with our previous measurements, T50% varied greatly across the lung cancer panel for the dBET1 PROTAC. H661 and H838 cells showed the fastest dBET1-induced degradation of BRD4 (T50% = 50 and 57 min), while there was almost no activity in H23 cells (T50% > 720 min). Three of the twelve cell lines failed to degrade 50% of BRD4 protein in 24 h. This is in stark contrast to measurements with MZ1, where the shortest T50% was 14 min (H661), and the longest recorded was 49 min (H460). Although degradation of BRD4 in H460 was the slowest among the cell lines tested, and this cell line lacked dBET1 activity, low CRBN activity did not always coincide with less robust VHL PROTAC activity. In fact, some cell lines such as HCC15 and A427 lacked dBET1 activity yet were among the most active degraders of BRD4 when the MZ1 VHL-BRD4 PROTAC was employed. This suggests that low CRBN-dependent ligase activity specifically limited dBET1-dependent degradation.
Figure 4.
dBET1 and MZ1 activity time course across a cell line panel representing 10 cancer subtypes
11 lung cancer cell lines and A375 were treated with 1 μM dBET1 or MZ1 for up to 720 min. Time-course plots of MSD data from each cell line are shown. These plots were used to calculate T50% and max % degradation for the two compounds in each cell line. Time course curves are derived from n = 2 independent experiments, error bars represent standard error of the mean (SEM).
Table 2.
T 50% values and max % degradation of BRD4 after MZ1 or dBET1 treatment
| Cell line | MZ1 t 50%, min | MZ1 Max %, POC | dBET1 t 50%, min | dBET1 max %, POC |
|---|---|---|---|---|
| A375 | 42 ± 1, n = 2 | 2 ± 1, n = 2 | 60 ± 1, n = 2 | 6 ± 0, n = 2 |
| NCIH23 | 22 ± 2, n = 2 | 1 ± 0, n = 2 | >720, n = 2 | 54 ± 3, n = 2 |
| NCIH460 | 49 ± 5, n = 2 | 4 ± 1, n = 2 | >720, n = 2 | 77 ± 4, n = 2 |
| HCC15 | 15 ± 1, n = 2 | 1 ± 0, n = 2 | >720, n = 2 | 63 ± 4, n = 2 |
| A427 | 27 ± 0, n = 2 | 0 ± 0, n = 2 | 389 ± 26, n = 2 | 48 ± 3, n = 2 |
| A549 | 40 ± 13, n = 2 | 4 ± 1, n = 2 | 161 ± 11, n = 2 | 35 ± 1, n = 2 |
| NCIH1299 | 23 ± 1, n = 2 | 2 ± 1, n = 2 | 85 ± 10, n = 2 | 20 ± 2, n = 2 |
| NCIH1568 | 42 ± 2, n = 2 | 3 ± 0, n = 2 | 137 ± 46, n = 2 | 22 ± 2, n = 2 |
| NCIH358 | 21 ± 1, n = 2 | 1 ± 1, n = 2 | 95 ± 35, n = 2 | 24 ± 5, n = 2 |
| NCIH441 | 35 ± 2, n = 2 | 3 ± 1, n = 2 | 64 ± 12, n = 2 | 9 ± 2, n = 2 |
| NCIH661 | 14 ± 2, n = 2 | 0 ± 0, n = 2 | 50 ± 10, n = 2 | 8 ± 2, n = 2 |
| NCIH838 | 31 ± 2, n = 2 | 2 ± 1, n = 2 | 57 ± 9, n = 2 | 8 ± 2, n = 2 |
DC50 values shown were calculated from 10-point dose-response curves are shown with T50% and max % degradation for dBET1 and MZ1 time course treatment of each cell line.
To understand the dramatic differences in PROTAC activity seen across our cell line panel, we split the cell lines into top and bottom quartiles and looked for genes whose expression correlates with PROTAC activity for both dBET1 and MZ1 (Tables S2 and S3). Top genes positively associated with dBET1 activity are enriched in mRNA processing (e.g. HNRNPUL1, PRPF19, and RBM6), protein deubiquitinating (e.g. ATXN3, BAP1,USP4, and USP19), and protein catabolic processes (e.g. PRPF19, RNF146, RNF123, and VHL). Although the interplay between deubiquitinating and ubiquitination pathways could affect the ubiquitin-mediated proteasomal degradation, none of the above genes are immediately connected to CRBN ligase function based on current knowledge and some are likely reflecting the existing genetics and expression patterns present in the cancer cell lines in the panel. The presence of VHL in the dBET1 signature is an example of this effect; most of the renal cell carcinoma (RCC) cell lines that we tested had both low dBET1 activity and low MZ1 activity and all of them are VHL mutants. Similar to our findings for dBET1, a genome-wide search for genes that positively correlated with MZ1 activity identified many genes involved in RNA processing and transcription factors unrelated to VHL ligase function. These findings spurred us to take a different approach (Akhoondi et al., 2007, 2010; Malyukova et al., 2007).
To more thoroughly search for genes that are related to ubiquitin-mediated degradation, we narrowed our analysis to focus on curated protein ubiquitination genes identified through Ingenuity Pathway Analysis (QIAGEN Inc, https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) (Kramer et al., 2014) (Tables S4 and S5) and then further narrowed our focus on known factors directly linked to the function of the CRBN and VHL ligases (Tables 3 and 4). In these two analyses, the expression of CRBN and its associated factor FBXO7 are positively associated with dBET1 activity and the expression of VHL is positively associated with MZ1 activity. Interestingly, the expression of VHL-associated factors TCEB1, TCEB2, and E3 regulators (UBE2H and RBX1) was negatively associated with MZ1 activity. We confirmed positive associations with a Wilcoxon analysis comparing the quartiles, binning the most active degraders of BRD4 vs the least active for each of the two PROTACs. Again, low dBET1 activity is associated with low CRBN expression and high activity is associated with high CRBN expression with a high degree of certainty, p = 0.0048 (Figure 5A). The same is true when activity is compared to CRBN copy number for dBET1, low activity correlates with low CRBN copy number, suggesting that copy number loss is a significant driver of low CRBN activity in these cell lines (Figure 5A). The same analysis for MZ1 is more challenging due to the narrow range of DC50 values across the panel. Despite this complicating factor, breaking the panel into quartiles still results in a statistically significant correlation between VHL expression and MZ1 activity, p = 0.028, and to a lesser extent, VHL copy number and MZ1 activity, p = 0.059 (Figure 5B). If just the renal cell carcinoma cell lines that are annotated as mutant or copy loss at the VHL loci are compared to the rest of the panel, an intact VHL gene clearly drives VHL activity. If this same group is removed from the 56-cell line panel, then no significant predictor of MZ1 activity emerges from the dataset.
Table 3.
Analysis of the correlation of known CRBN-associated partners and regulators with dBET1 DC50
| Gene | Category | Correlation coefficient | P. value |
|---|---|---|---|
| CRBN | CRBN and partner | −0.494871713 | 0.00543 |
| FBXO7 | CRBN and partner | −0.419334578 | 0.02107 |
| UBE2D3 | CRBN and partner | −0.32612564 | 0.07861 |
| ARIH1 | E3 Ligase Regulator | −0.312511312 | 0.09269 |
| DDB1 | CRBN and partner | −0.302250886 | 0.10451 |
| COPS5 | E3 Ligase Regulator | −0.291339995 | 0.11828 |
| UBE2H | E3 Ligase Regulator | 0.255081944 | 0.1737 |
| CUL4A | CRBN and partner | −0.199830715 | 0.28972 |
| UBE2G1 | CRBN and partner | −0.098374884 | 0.60503 |
| DCUN1D1 | E3 Ligase Regulator | −0.077538506 | 0.68381 |
| CUL4B | CRBN and partner | −0.018342434 | 0.92336 |
| NAE1 | E3 Ligase Regulator | −0.006346605 | 0.97345 |
| RBX1 | E3 Ligase Regulator | −0.001237036 | 0.99482 |
Correlation analysis results of known CRBN E3 machinery partners and E3 ligases regulators with dBET1 DC50 were shown as correlation coefficient and p value of the correlation test. A negative correlation coefficient represents a positive association with dBET1 activity and vice versa.
Table 4.
Analysis of the correlation of known VHL-associated partners and regulators with MZ1 DC50
| Gene | Category | Correlation coefficient | p-value |
|---|---|---|---|
| VHL | VHL and partner | −0.637964271 | 0.000149 |
| UBE2H | E3 Ligase Regulator | 0.528277695 | 0.002693 |
| TCEB1 | VHL and partner | 0.465946632 | 0.009457 |
| TCEB2 | VHL and partner | 0.446163111 | 0.013463 |
| RBX1 | E3 Ligase Regulator | 0.387889062 | 0.034175 |
| ARIH1 | E3 Ligase Regulator | −0.31035643 | 0.095084 |
| NAE1 | E3 Ligase Regulator | −0.275538037 | 0.140547 |
| DCUN1D1 | E3 Ligase Regulator | 0.263944329 | 0.15872 |
| CUL2 | VHL and partner | 0.122842043 | 0.517826 |
| COPS5 | E3 Ligase Regulator | 0.012943911 | 0.945876 |
Correlation analysis results of known VHL E3 machinery partners and E3 ligases regulators with MZ1 DC50 were shown as correlation coefficient and p value of the correlation test. A negative correlation coefficient represents a positive association with MZ1 activity and vice versa.
Figure 5.
Correlation of PROTAC activity with CRBN and VHL RNA expression, DNA copy number, and protein level
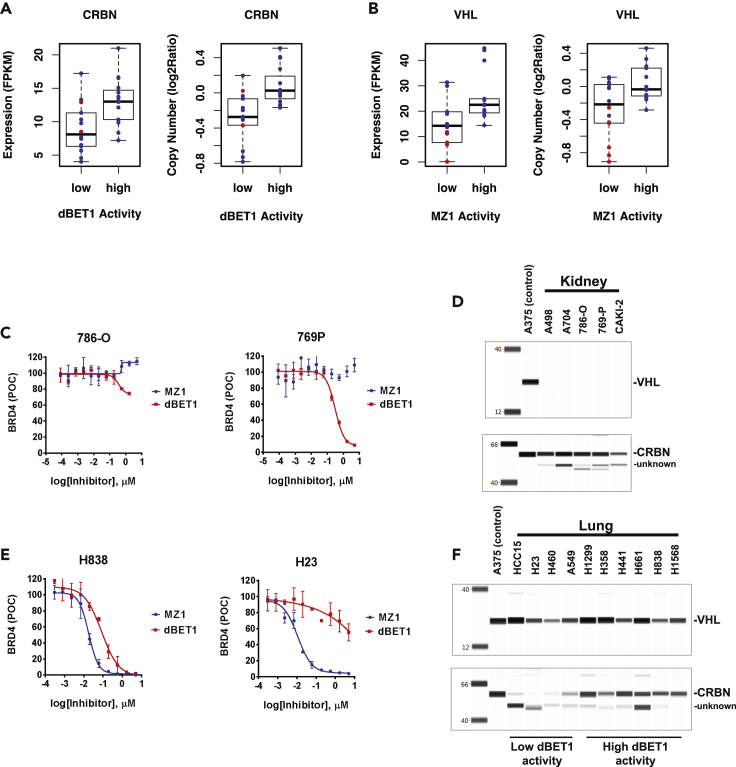
(A and B) DC50 values from the cell line panel for each compound were used to group the cell lines into the bottom (low) and top (high) quartiles. Low and high quartiles were plotted against ligase mRNA expression or copy number, p values from unpaired two-samples, two-sided Wilcoxon rank sum tests of the groups were calculated. (A) dBET1 activity correlates significantly with CRBN copy number, p = 0.00058, and mRNA expression, p = 0.0048. Cell lines with non-synonymous mutations of CRBN were marked in red. (B) MZ1 activity correlates with VHL RNA expression, p = 0.028 but not VHL copy number, p = 0.059. Cell lines with non-synonymous mutations of VHL were marked in red.
(C) Dose-response curves from representative kidney-derived cancer cell lines are shown; 786-O is devoid of both VHL and CRBN activity, 769P is lacking VHL activity. Dose titration curves are derived from n = 2 independent experiments, error bars represent standard error of the mean (SEM).
(D) Lysates from untreated cells were separated by capillary electrophoresis, VHL and CRBN proteins were immune-detected. Each of the five kidney-derived cancer cell lines is lacking VHL protein, all of the cell lines express appreciable CRBN protein.
(E) Dose-response curves from two representative lung cancer cell lines, H23 lacks dBET1-CRBN-associated activity. Dose titration curves are derived from n = 2 independent experiments, error bars represent standard error of the mean (SEM).
(F) Lung-derived cancer cell lines with low CRBN activity have low or no CRBN protein, all lung cancer cell lines express VHL protein.
Differences in RNA expression of VHL and CRBN are frequently reflected in the expression levels of the corresponding proteins; however, this does not hold true for all cases. For instance, A549 has unusually low CRBN protein, 29% of our standard cell line A375, but an FPKM of 9.77 compared to 12.55 for A375 (Figure S3 and Table S6). For some cell lines, such as the 10-cell line lung cancer panel, CRBN protein expression correlates with PROTAC activity. The four cell lines with the lowest levels of dBET1 activity, HCC15, H460, H23, and A549 also had low or nearly undetectable CRBN protein relative to A375 (Figures 5C–5F). Interestingly, for these lines, only HCC15 is indicated to have especially low CRBN mRNA, with FPKM of 4.01. For renal cell lines, where VHL-dependent MZ1 activity was clearly lacking, VHL protein was absent, as predicted by frequent copy loss and inactivating point mutations in VHL (Table S7). Surprisingly, several cell lines, A498, A704, and 786-O that displayed very low dBET1 activity appeared to have average levels of CRBN protein. To further understand the origin of the low dBET1 activity in these cell lines, we investigated the expression and mutation status of other subunits of the CRL4CRBN ligase complex, including the adaptor protein DDB1; again, neither mutation nor low mRNA levels of these components explains the lack of dBET1 activity. It seems possible that some more general effect on UPS activity might be at play in the renal cancer cell lines, though to date we have not identified a specific factor.
Our observation that CRBN activity is heterogeneous in the cell line panel and that reduced activity frequently coincides with low expression of CRBN mRNA and protein suggests that tumor-specific suppression of ligase receptor genes could seriously limit the ability of CRBN engaging PROTACs to induce degradation of therapeutic targets in some cancer subtypes. To better understand this possible complication, we retrieved the expression pattern of each ligase from the publicly available GTEX body map database and plotted this data to highlight tissue-specific expression and variability of expression within each tissue type. These data indicate that both ligases are broadly expressed, with minor increased or decreased expression levels in specific tissues and very little variability of expression within each tissue subset (Figure S4A). High expression of both ligases occurs in blood and lymphatic tissues, consistent with their roles in lineage specification and hematopoiesis. CRBN expression is higher than VHL overall, with a median FPKM of 17 and peak expression in spinal cord, brain, and muscle tissues exceeding FPKMs greater than 25. The median FPKM value for VHL was 8 with increased expression in blood and lymphoid tissue. VHL expression is relatively low in both blood vessels and heart with FPKMs below 6.
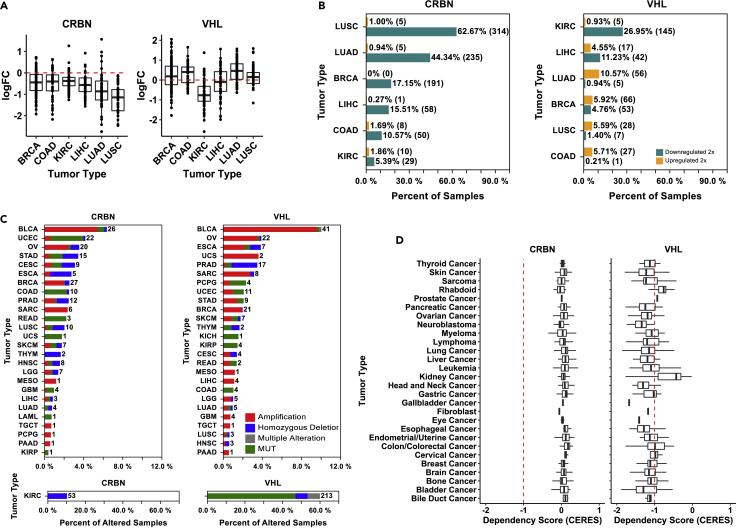
Of greatest importance to our work is the expression pattern of VHL and CRBN in human tumors. For this analysis, we retrieved RNA-Seq expression and DNA alteration data from available OmicSoft TCGA data sets and graphed these by TCGA-designated tumor type (Figure S4B). The resulting expression profiles confirmed some of the observations that we made with our cancer cell line panel. Squamous cell carcinoma of the lung (LUSC) has relatively low expression of CRBN suggesting that these lung cancers may have reduced CRBN activity, mirroring the cell line panels. Renal cell carcinoma (KIRC) showed relatively low expression of VHL in agreement with a high frequency of loss of function mutations and low expression of VHL. To better understand the significance of these expression differences, we retrieved data from the TCGA databases for matched tumor and normal tissue samples to determine if expression is downregulated in the cancer cell. Plotting this data as fold change of tumor expression relative to the matched normal revealed an overall trend of reduced CRBN expression in the tumor (Figure 6A). This contrasted with VHL, where expression was often higher in the tumor than in the matched normal. To probe these findings further, we also plotted the frequency of 2-fold changes in tumor vs normal tissue. Most dramatically, CRBN expression is frequently 2-fold lower in both LUAD and LUSC samples, 44% and 63% respectively, suggesting that loss or suppression of CRBN activity might be a common occurrence in these cancers (Figure 6B). This finding was unexpected; therefore, we probed the DNA landscape of VHL and CRBN more completely, enumerating the frequency and type of mutation across a handful of cancer subtypes (Figure 6C), the frequency with which CRBN copy number loss or gene disruptive alteration in renal cell carcinoma samples was notable. 53 of 532 samples were identified as having a homozygous deletion of the CRBN gene. The role of CRBN as a tumor suppressor in these cancers has not been previously reported. Though occurring with much less frequency, LUSC and LUAD samples can also carry either deletion or non-synonymous mutations of the CRBN gene, 2.0% and 0.7% respectively.
Figure 6.
Genomic and expression profiles of VHL and CRBN in normal tissue and cancer
(A) Boxplot showing the distribution of log transformed fold change of the expression of CRBN (left) and VHL (right) in cancer tissue compared to its paired non-cancer control tissue. Only cancer types with at least 50 non-cancer control tissue samples were included.
(B) The percentage of samples showed more than 2-fold increasing (orange) or decreasing (green) in CRBN (left) or VHL (right) level in each of the six representative cancer types.
(C) Bar graph showing percentage of samples harboring CRBN (left) or VHL (right) mutations in each tumor type in TCGA, the number of samples showing mutations is labeled in the right side of each bar. Different mutation types are color labeled, with red representing amplification, blue representing homozygous deletion, green representing non-synonymous mutations, and gray representing a mixture of the above type of mutations.
(D) Genome-scale CRISPR-Cas9 essentiality screen results for genes in across different cancer cell lines performed by the Broad Institute were characterized by dependency score (CERES) to reflect the functional importance of genes in certain cancer types. Boxplot summarized the distribution of CERES score of CRBN (left) and VHL (right) in cell lines of representative tumor types. A lower score means that a gene is more likely to be essential for the cancer cell line survival and proliferation. A score of −1 corresponds to the median of all common essential genes, used as a cutoff indicator here.
The potential roles for CRBN and VHL in tumorigenesis prompted us to explore whether cancer cell lines are dependent on either CRBN or VHL expression. The data to answer this question are readily available through the BROAD Achilles project, where greater than 500 cell lines have been treated with the Avana CRISPR library that covers 17634 genes including CRBN and VHL. CRISPR sequences-directed targeting genes that are required for normal proliferation are counter selected through multiple rounds of passaging and their underrepresentation in the final pool of passaged cells is determined by statistical methods that correct for copy number variation within the cell line panel (Meyers et al., 2017). The resulting CERES score places each gene on a sliding scale, where −1 or below is indicative of strong dependency on the given gene, 0 suggests a lack of selective pressure, and a score of +1 or greater indicates that a strong growth advantage is conferred by deletion of the gene. When we examined the Broad Avana 19Q2 dataset for both CRBN and VHL, the difference between these two genes was readily apparent (Figure 6D). The mean CERES score across multiple cancer types is at or above 0 for CRBN, suggesting that most of these cell lines are not dependent on CRBN activity under normal culturing conditions. VHL, however, had a mean CERES score of less than −1 for most cancer subtypes suggesting a strong dependency on VHL expression and activity. The noteworthy exceptions were the kidney cell lines, many of which are VHL mutants; the mean CERES score of this group was −0.4. Thus, the CRISPR dependency screens have potentially identified a critical limitation of the CRBN ligase and its use in PROTAC therapeutics. The data suggest that most tumors, like the cancer cell lines, do not require CRBN activity and can survive elimination of CRL4CRBN to suppress the activity of IMiD-derived PROTACs. This hypothesis is corroborated by the frequent downregulation of CRBN in cancer cell lines and tumors. In contrast, these data also suggest that VHL is less likely to suffer similar loss of expression after PROTAC treatment given the strong dependency of most cell lines on an intact VHL gene and the infrequent loss of VHL expression observed in TCGA tumor samples.
Discussion
PROTACs have been employed to degrade a diverse collection of target proteins including kinases, hormone receptors, and membrane-bound growth factor receptors (Burslem et al., 2018; Kang et al., 2018; Salami et al., 2018; Sun et al., 2018). These PROTACs are useful tools to probe basic biology, and some derivatives will be developed as therapeutics (Burslem and Crews, 2020). However, full realization of the promise of PROTACs will depend on the development of ligase binders that have superior biochemical and cellular activity, as well as ligands that bind new ligases with disease-specific activity. Developing effective PROTACs is an immense effort as the evolution of proof of concept molecules into potent rapid degraders requires the simultaneous optimization of two ligands, linker, and linking chemistry (Zoppi et al., 2019). In addition, extensive resources are committed to elucidate and exploit potential protein-protein interactions that stabilize the ligase-substrate complex, including the generation of biophysical and structural biology data that guides chemistry. Therefore ligase and target choice must be considered carefully before embarking on a full-scale campaign to make novel PROTACs (Hughes and Ciulli, 2017; Roy et al., 2019). Ottis et al. described an experimental approach to evaluate ligase activity using a halo-tagged ligase substrate receptor protein and a generic PROTAC that fuses the halo binder to a BRD4 ligand (Ottis et al., 2017). This approach allows for a direct comparison of ligases without engineering entirely new PROTACs. Although the halo-fusion proteins might differ from the native protein, identifying ligases that are flexible and robust enough to promote rapid ubiquitination of a generic substrate is a powerful exercise. This represents one part of a strategy to prospectively evaluate ligases prior to embarking on a novel receptor ligand discovery campaign and synthesizing proof of concept molecules. An understanding of the activity of a potential PROTAC ligase in different disease and tissue settings represents a complementary strategy.
As the first candidate molecules that degrade androgen receptor and estrogen receptor have entered clinical testing in prostate and breast cancer, there is growing hope that PROTACs will become a powerful new modality for the treatment of cancer. To understand whether genomics could be used to predict ligase activity within cancers, we chose to assess the activity of CRBN and VHL across a cell line panel using two BRD4 targeting PROTACs, MZ1 and dBET1. We profiled their activity in 56 cell lines representing 10 cancer subtypes and mined genomic datasets for these lines to establish predictive bio-markers of their activity.
It is important to note that MZ1 and dBET1 are very different molecules; strong evidence for ternary complex cooperativity has been described for MZ1 but not for dBET1. These factors certainly affect the PROTACs response to changes in the cellular levels of either ubiquitin ligases or substrate proteins (Gadd et al., 2017; Roy et al., 2019). To avoid pitfalls that may arise from these confounding factors as well as differences in the physiochemical properties of the two PROTACs, we avoid directly comparing the performance of MZ1 to that of dBET1, and instead focus on the performance of MZ1 and dBET1 across the cell line panel.
The resulting data reveal that VHL and CRBN are not equivalent, and each has tumor-type-specific limitations. Although both genes are widely expressed in normal tissues and expressed in most cancers, their utility within given subtypes of cancer varies greatly. CRBN appears to be frequently inactivated in lung and colon cell lines and its expression is downregulated in tumors of similar origin. VHL appears to more effectively promote degradation of target proteins across a larger number of cancer types. Recent publications by Zhang et al. and Ottis et al. explored the development of drug resistance in cells treated with either CRBN-based or VHL-based BRD4 degraders (Zhang et al., 2019; Ottis et al., 2019). Zhang et al. report that downregulation of CRBN through point mutation and copy loss promotes resistance to sustained knockdown of BRD proteins in cells dependent on BRD4. Although the authors did not see a similar downregulation of VHL, they did identify mutations in the CUL2 protein (Zhang et al., 2019). The work of Ottis et al. identified components of the VHL ligase complex, including COP9 protein, as important modulator of ubiquitin ligase activity (Ottis et al., 2019). Both papers expertly demonstrate the power of CRISPR-mediated screens to identify the machinery necessary for ubiquitin ligase function. Both works support our conclusion that downregulation of the ligase machinery will likely limit the utility of CRBN and VHL PROTACs. It is important to note that most of the genes identified by Zhang and Ottis were not found to correlate with PROTAC activity in our study. We surmise that CRISPR screens exhaustively probe for dependency to uncover the subunits that are necessary for ligase activity, but those genes may not represent frequent targets for mutation in cancers or those mutations may be underrepresented in our cancer cell lines.
The number of ligase substrate adapter proteins that have been exploited for the PROTAC modality continues to grow. PROTACs making use of MDM2, CIAP, XIAP, VHL, CRBN, KEAP1, RNF4, RNF114, and DCAF16 have been extensively explored in published works (Paiva and Crews, 2019). Of those in addition to VHL and CRBN, only RNF4 looks similar to VHL with respect to both its broad expression and cell line dependency (Figures S4 and S5, 7). RNF4 is expressed at high levels across the GTEX body map, and it is overexpressed with higher frequency in tumors compared to matched normal tissues in the subset of cancers we probed. This data combined with infrequent loss of function mutation or deletion in tumors suggest RNF4 could have robust activity across many cancer subtypes. A known oncogene, MDM2, also exhibits focal dependency in cancer cell lines and is frequently amplified and overexpressed, suggesting that MDM2-based PROTACs might have greater activity in tumors than in normal tissues. Unlike VHL, RNF4, and MDM2, the genomic profile of KEAP1 suggests that this ligase would be a risky choice for building lung cancer targeting PROTACs (Cancer Genome Atlas Research, 2012, 2014). Lung tumors frequently carry inactivating mutations in KEAP1, and it is considered a tumor suppressor in both squamous and adenocarcinoma subtypes. Finally, Achilles dependency data for RNF114 result in a dependency score near 0, but its expression is not frequently suppressed in tumors, and it is not frequently mutated. RNF114 is overexpressed in some tumor types compared to matched normal, and its overexpression in colon tumors is mirrored by frequent amplification of the gene in tumor samples. Thus, RNF114 might be especially useful for cancer targeting PROTACs and future evaluation of proof of concept PROTACs in our cell line panel would be informative.
Figure 7.
Comparison of genomic features for seven PROTAC Ub-ligases
(A) Boxplot showing the distribution of log transformed fold change of the expression of seven Ub-ligases in cancer tissue compared to its paired non-cancer control tissue. Only cancer types with at least 50 non-cancer control tissue samples were included.
(B) DNA alternation landscape of seven Ub-ligases across cancer types in TCGA. Bar graph showing percentage of samples harboring each Ub-ligase mutations across tumor types, the number of samples altered are labeled in the right side of each bar. Different mutation types are color labeled, with red representing amplification, blue representing homozygous deletion, green representing non-synonymous mutations, and gray representing a mixture of the above type of mutations.
(C) Genome-scale CRISPR-Cas9 essentiality screen results for genes in across different cancer cell lines performed by the Broad Institute were characterized by dependency score (CERES) to reflect the functional importance of genes in certain cancer types. Boxplots summarized the distribution of CERES score of each ligase receptor in cell lines of representative tumor types. A lower score means that a gene is more likely to be essential for the cancer cell line survival and proliferation. A score of −1 corresponds to the median of all common essential genes, used as a cutoff indicator here.
It is impossible to predict whether new ubiquitin ligase receptor proteins will be suitable for a PROTAC strategy without generating proof of concept molecules and testing them in relevant models. However, our results suggest that genomic data from cancer cell lines and tumors might provide a reasonable method to prioritize the hundreds of potential PROTAC ligases. Promising ligases can then be screened with BRD4 proof of concept PROTACs in a cell line panel to establish their utility before embarking on larger campaigns to identify therapeutic molecules. Ligases like CRBN that have low or no activity in some cell lines and are beset by frequent mutation and decreasing expression in cancer cell lines and tissues are risky unless there is clear evidence for expression and activity in a setting that is of overwhelming interest. Our brief look into the genomic data of published PROTAC ligases suggests that cancer-specific expression, broad expression, and infrequent inactivation are reasonable filters for the selection of new PROTAC ligases that have therapeutic utility.
Limitations of the study
Human cancer cell lines are related to but not copies of the tumors that they originate from. Testing the activity of potential anticancer drugs in cell lines can provide valuable insight into some of the genetic changes that alter their activity but the depth of the effect that those genetic changes cause and the frequency with which they occur may differ greatly from their occurrence in human tumors. Testing the activity of dBET1 and MZ1 activity in patient-derived tumor samples to confirm some of the findings reported in this study would be a valuable extension of our effort.
In this work, we go to great lengths to understand the activity of dBET1 or MZ1 across the cell line panel. Though most of our conclusions are derived from data that represent the activity of a single PROTAC across the cell line panel and the characteristics of the cell lines in the panel, at times we may inadvertently compare the activities of dBET1 and MZ1 to one another. It is worth noting that these molecules have very different chemical properties that may affect their ability to promote degradation of BRD4 under our in vitro assay conditions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Sulfo-Tag Anti-Mouse (Goat) | MSD | Cat# R32AC-5; RRID:AB_2783819 |
| MSD Capture Ab, Rabbit mAb & Gel Electrophoresis | Cell Signaling Tech. | Cat# 13440 lot 3–5; RRID:AB_2687578 |
| MSD Detection Ab, Mouse mAb | Atlas Antibodies | Cat# AMAb90841 lots 03087, 02741, & 02739; RRID:AB_2665685 |
| ANTI CRBN ANTIBODY Gel Electrophoresis | Sigma Millipore | Cat# HPA045910 lot BD117086; RRID:AB_10960409 |
| ANTI VHL ANTIBODY Electrophoresis | Cell Signaling Tech. | Cat# 68547 lot 1; RRID:AB_2716279 |
| Anti-Rabbit-HRP Electrophoresis | Cell Signaling Tech. | Cat# 7074; RRID:AB_2099233 |
| Critical commercial assays | ||
| ANTIBODY DETCTION Gel Electrophoresis | PROTEIN SIMPLE | Cat# DM-001 |
| 12–230 kDa Separation Module, 8 x 25 capillary cartridges Gel Electrophoresis | PROTEIN SIMPLE | Cat# SM-W004 |
| Experimental models: Cell lines | ||
| BT-549 | ATCC | HTB-122; RRID:CVCL_1092 |
| CAL-51 | DSMZ | ACC 302; RRID:CVCL_1110 |
| HCC 1954 | ATCC | CRL-2338; RRID:CVCL_1259 |
| HCC 1569 | ATCC | CRL-2330; RRID:CVCL_1255 |
| HCC 1806 | ATCC | CRL-2335; RRID:CVCL_1258 |
| MCF-7 | ATCC | HTB-22; RRID:CVCL_0031 |
| MDA-MB-157 | ATCC | HTB-24; RRID:CVCL_0618 |
| ZR 75–1 | ATCC | CRL-1500; RRID:CVCL_0588 |
| DAUDI | ATCC | CCL-213; RRID:CVCL_0008 |
| HT | ATCC | CRL-2260; RRID:CVCL_1290 |
| JURKAT E6.1 | ATCC | TIB-152 ;RRID:CVCL_0367 |
| JVM-3 | DSMZ | ACC 18; RRID:CVCL_1320 |
| KG-1 | ATCC | CCL-246; RRID:CVCL_0374 |
| KMS-12-BM | DSMZ | ACC 551; RRID:CVCL_1334 |
| MOLM-13 | DSMZ | ACC 554; RRID:CVCL_2119 |
| MOLT-4 | ATCC | CRL-1582; RRID:CVCL_0013 |
| MV4-11 | ATCC | CRL-9591; RRID:CVCL_0064 |
| NAMALWA | ATCC | CRL-1432; RRID:CVCL_0067 |
| OPM-2 | DSMZ | ACC 50; RRID:CVCL_1625 |
| PL-21 | DSMZ | ACC 536; RRID:CVCL_2161 |
| RL | ATCC | CRL-2261; RRID:CVCL_1660 |
| U266B1 | ATCC | TIB-196; RRID:CVCL_0566 |
| HEK293 T (293 T) | ATCC | CRL-3216; RRID:CVCL_0063 |
| 769-P | ATCC | CRL-1933; RRID:CVCL_1050 |
| 786-O | ATCC | CRL-1932; RRID:CVCL_1051 |
| A-498 | ATCC | HTB-44; RRID:CVCL_1056 |
| A-704 | ATCC | HTB-45; RRID:CVCL_1065 |
| CAKI-2 | ATCC | HTB-47; RRID:CVCL_0235 |
| COLO 205 | ATCC | CCL-222; RRID:CVCL_0218 |
| NCI H508 (H508) | ATCC | CCL-253; RRID:CVCL_1564 |
| HCT 116 | ATCC | CCL-247; RRID:CVCL_0291 |
| HCT 15 | ATCC | CCL-225; RRID:CVCL_0292 |
| HT-29 | ATCC | HTB-38; RRID:CVCL_0320 |
| SW1417 | ATCC | CCL-238; RRID:CVCL_1717 |
| SW48 | ATCC | CCL-231; RRID:CVCL_1724 |
| HEP-G2 | ATCC | HB-8065; RRID:CVCL_0027 |
| A-427 | ATCC | HTB-53; RRID:CVCL_1055 |
| A-549 | ATCC | CCL-185; RRID:CVCL_0023 |
| NCI-H1568 (H1568) | ATCC | CRL-5876; RRID:CVCL_1476 |
| NCI-H358 (H358) | ATCC | CRL-5807; RRID:CVCL_1559 |
| NCI-H441 (H441) | ATCC | HTB-174; RRID:CVCL_1561 |
| HCC 15 | DSMZ | ACC 496; RRID:CVCL_2057 |
| NCI-H1299 (H1299) | ATCC | CRL-5803; RRID:CVCL_0060 |
| NCI-H23 (H23) | ATCC | CRL-5800; RRID:CVCL_1547 |
| NCI-H460 (H460) | ATCC | HTB-177; RRID:CVCL_0459 |
| NCI-H661 (H661) | ATCC | HTB-183; RRID:CVCL_1577 |
| NCI-H838 (H838) | ATCC | CRL-5844; RRID:CVCL_1594 |
| Mia PaCa-2 | ATCC | CRL-1420; RRID:CVCL_0428 |
| SW1990 | ATCC | CRL-2172; RRID:CVCL_1723 |
| A101-D | ATCC | CRL-7898; RRID:CVCL_1057 |
| A2058 | ATCC | CRL-11147; RRID:CVCL_1059 |
| A-375 | ATCC | CRL-1619; RRID:CVCL_0132 |
| RPMI 7951 | ATCC | HTB-66; RRID:CVCL_1666 |
| SK-MEL-28 | ATCC | HTB-72; RRID:CVCL_0526 |
| AGS | ATCC | CRL-1739; RRID:CVCL_0139 |
| UM UC-3 | ATCC | CRL-1749; RRID:CVCL_1783 |
| Software and algorithms | ||
| Graph Pad Prism (version 7) | Graphpad Software | https://www.graphpad.com/ |
| VWORKS (12.0.1279) | Agilent Technologies | https://www.agilent.com/ |
| MSD Workbench (4.0.12 thru 4.0.12.1) | MSD | https://www.mesoscale.com/ |
| COMPASS FOR SW (version 4) | Simple Western Software | https://proteinsimple.com/ |
| SOFTMAX PRO (version 6) | MOLECULAR DEVICES | Cat# SMP6 |
| OmicSoft Suite software (version10.0.1.81) | QIAGEN | https://digitalinsights.qiagen.com/products-overview/discovery-insights-portfolio/analysis-and-visualization/qiagen-omicsoft-suite/ |
| IPA | QIAGEN | https://www.qiagen.com/us/products/discovery-and-translational-research/next-generation-sequencing/informatics-and-data/interpretation-content-databases/ingenuity-pathway-analysis/ |
| R (v3.6.0) | The R Project for Statistical Computing | https://www.r-project.org/ |
| ggplot2 (v3.2.0) | CRAN | https://cran.r-project.org/web/packages/ggplot2/index.html |
| ggpubr (v0.2.1) | CRAN | https://cran.r-project.org/web/packages/ggpubr/index.html |
| Other | ||
| JESS INSTRUMENT | PROTEIN SIMPLE | 004–650 |
| WES INSTRUMENT | PROTEIN SIMPLE | 004–600 |
| MSD Sector S Imager 600MM READER | MSD | IC1AA-0 |
| MSD Sector Imager 6000 | MSD | I10AA-0 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dane Mohl (dmohl@amgen.com).
Materials availability
All materials and cell lines used in this study are commercially available and detailed in the key resources table.
Any transfer of materials may require a materials transfer agreement.
Method details
Cell culture
A panel of 56 human cell lines was procured from the American Type Culture Collection (ATCC) or Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures (DSMZ), propagated in RPMI-1640 medium (1X), containing 10% heat inactivated fetal bovine serum and 1X penicillin–streptomycin and grown in a humidified incubator at 37 °C and 5% CO2. Cells were maintained for no more than 20 passages. The complete panel of 56 cell lines were submitted to ATCC for cell authentication. To ensure that the data we generated would be comparable with that of other research groups, short tandem repeat (STR) profiling of each of the cell lines was conducted to confirm its identity. All cell lines in our cell line bank were exact matches to the ATCC reference map, except for HCC 15 and H1299, which were greater than 90% matched to the ATCC reference map (Table S1).
BRD4 PROTAC compound treatment
For 96 well dose titration studies cells were seeded overnight as follows: Adherent cells (40,000 cells/90 μL) and suspension cells (80,000 cells/90 μL). Cells were treated with 10 μL of 10X compounds for 4 h in a humidified incubator at 37 °C and 5% CO2. Final compound concentration was 5 μM with 1:3 serial dilution. Two independent experiments were run per cell line. For HCC 1569, JVM3, and MDA MB 157, one of the two repeats was removed when the data did not pass our QC requirements. Lysates from cells were generated in a 96 well plate by removing media/compound and adding 60 μL of 1X Complete Tris MSD Lysis Buffer (150 mM NaCl, 20 mM Tris, pH 7.5, 1 mM EDTA, 1 mM EGTA, and 1% Triton X-100) supplemented with 1 tablet of Roche Phospho stop (Roche, 04906837001) and Complete (Roche, 04693116001) protease inhibitors per 10 mls of 1X Tris MSD Lysis Buffer (MSD, R60TX-2). After addition of lysis buffer 96 well plates were transferred to −80°C freezer until day of MSD assay.
A panel of 4 cells lines was used to evaluate BRD4 protein degradation with two reference PROTACs in parallel studies using WES protein blots and the MSD platform for comparison. Each cell line was plated in two 6 well dishes. One plate per treatment group with each of our two reference PROTACs. Adherent cells (500,000 cells/3 mls) and suspension cells (1,500,000 cells/3 mL) were seeded overnight. Cells were treated with 333 μL of 10X compounds for 4 h in a humidified incubator at 37 °C and 5% CO2. Final compound concentration was 3 μM starting conc with a five point 1:3 serial dilution series and DMSO control. A minimum of two independent experiments were run per cell line. Lysates from cells were generated in a 6 well plate by removing media/compound and adding 100 μL of 1X Complete Tris MSD Lysis Buffer (as described previously). All lysates were transferred to −80°C freezer for storage until protein quantitation was ready to be initiated.
Protein quantification
For the panel of 4 cell lines evaluated by WES and MSD platforms. Protein quantitation was performed using the Pierce BCA Protein Assay Kit (Cat 23225) using the Microplate Procedure per the manufacturer’s instructions. Samples were diluted 1:5 in lysis buffer and plated against BCA kit standards in 10 μL volume. The BCA assay colorimetric reaction was evaluated using the Molecular Devices M5 instrument using the BCA Protein assay protocol programmed into instrument. Standard curves had R2 values that ranged from R2 = 0.913–0.998 determined using SoftmaxPro (Molecular Devices, Version 6) software. Lysates were found to be in the linear range and were diluted 1 μg/μL for the BRD4 MSD assay and were run neat for the Wes protein analysis.
WES/JESS simple western
Cell lysates were analyzed with the WES/Jess Simple Western System according to the manufacturer’s instructions using 12–230 kDa Separation Modules with 25 Capillary Cartridges (ProteinSimple, SM-W004) and Anti-Rabbit Detection Module (DM-001). Prepared lysates at 2X working concentration (1 mg/mL) were diluted 1:1 with 0.1X Sample Buffer and combined with 5X Fluorescent Master Mix in a 1:5 ratio to make final lysate concentrations of 0.4 mg/mL 1.2 μg protein per sample, antibody diluent, primary antibodies in antibody diluent, HRP-conjugated secondary antibodies and chemiluminescent substrate were pipetted into the Separation Module. Instrument default settings were used: stacking and separation at 375 V for 25 min; blocking reagent for 5 min, primary and secondary antibody both for 30 min; Luminol/peroxide chemiluminescence detection for ∼15 min. The resulting electropherograms were inspected to check whether automatic peak detection required any manual correction using Compass for SW (Protein Simple, Version 4) software. The primary antibodies used were anti-BRD4 (Cell Signaling, 13440 lot5, 1:25), anti-CRBN (Sigma, HPA045910 lot BD117086, 1:25), and anti-VHL (Cell Signaling, 68547 lot 1, 1:25). Secondary antibodies used were Anti-Rabbit-HRP (Cell Signaling, 7074, 1:50) for CRBN and BRD4 detection and Anti-Rabbit-HRP secondary from the ProteinSimple Anti-Rabbit Detection Module (DM-001) for VHL detection. ProteinSimple Anti-Rabbit Detection Module (DM-001) also contained the following reagents used for all gel capillary electrophoresis set up: Antibody Diluent 2, Luminol-S chemiluminescence detection, and Peroxide chemiluminescence detection.
BRD4 MSD assay
The BRD4 MSD assay was a custom assay and multi array 96-well standard bind plates (MSD, L15XA-3) were solution coated by adding 50 μL/well of 1.47 μg/mL Cell Signaling Technologies BRD4 (CST, 13440 lot 3–5) capture antibody in PBS without Ca2+ or Mg2+ (Wurz et al., 2018). The plates were sealed and incubated overnight at 4 °C with no shaking for a minimum of 16 h. After the capture antibody incubation, the plates were washed per MSDs recommendation with three washes at 150 μL per wash using a BioTek 406 plate washer (BioTek, 406PSUB3) with 1X MSD Tris Wash Buffer (MSD, R61TX-1). All subsequent washes were performed in the same manner. After wash, all plates were blocked using 150 μL/well of 3% Blocker A (MSD, R93BA-4) in 1X MSD Tris Wash Buffer for 1 h at room temperature with shaking at 700 rpm on microplate shaker. The plates were then washed 3x for with 1X MSD Wash Buffer and 25 μL of lysates were added to each well. Control wells received MSD 1X Complete Tris Lysis buffer (previously described) alone as a background control. Plates were incubated 1 h at room temperature with shaking at 700 rpm on microplate shaker.
Following lysate incubation plates were washed 3x with in 1X MSD Tris Wash Buffer. Detection antibody Atlas BRD4 (Atlas, AMAb90841 lots 03087, 02741, & 02739) was added to each well at 25 μL at a concentration of 2 μg/mL in 0.6% Blocker A diluted in 1X MSD Tris Wash Buffer. Detection antibody was incubated for 1 h at room temperature with shaking at 700 rpm on microplate shaker. The plates were then washed 3x for with 1X MSD Wash Buffer and 25 μL of goat anti-mouse sulfotag antibody (MSD, R32AC-5) added to each well at a concentration of 1 μg/mL in 0.6% Blocker A diluted in 1X MSD Tris Wash Buffer. Plates were incubated for 1 h at room temperature with shaking at 700 rpm on microplate shaker. The plates were then washed 3x for with 1X MSD Wash Buffer and 150 μL of 1X MSD READ Buffer (MSD, R92TC-3) was added to each well. Plates were immediately read on MSD 6000 Sector Imager or previous version MSD Sector S Imager 600MM reader. Both instruments used MSD Workbench (4.0.12 thru 4.0.12.1) software to capture data and plate images.
Quantification and statistical analysis
MSD assays
For MSD assays, MSD signals were subtracted with the average background signal from the wells with lysis buffer alone. Normalized MSD values for individual compound treated wells were acquired by dividing assay signal with signal from vehicle control wells (Cells + 0.1% DMSO) and multiplying by 100. These data were graphed as POC with Grouped Plot Summary in Prism data-fitting software (Graphpad Prism 7, San Diego, CA, USA). The MSD data from select four cell line panel were directly compared to lysates that were run in parallel in the WES western blot platform.
Genomic expression and DNA alteration analysis
Tissue and cancer expression and genomic data were processed and obtained using OmicSoft (Cary, NC) Array Studio software platform. mRNA expression and DNA alteration distribution data of each ligase receptor were extracted from OmicSoft Body Map release GTEX_B38 and OncoLand release TCGA_B38 (data generated by the TCGA Research Network: https://www.cancer.gov/tcga) (Consortium, 2013). Quantile Strip plots were created to provide a compact visualization of expression of ligases across tissues and cancer types. The expression level was expressed as fragments per kilobase of transcript per million mapped reads (FPKM). The FPKM values in both GTEx and TCGA datasets were generated using OmicSoft’s implementation of the RSEM algorithm and normalized using upper-quartile normalization (Li and Dewey, 2011; Mortazavi et al., 2008; Robinson and Oshlack, 2010). Samples with shared annotation were sorted by ascending expression and compressed into each cell, with assigned color indicating the different expression value grouping into qualitative expression categories. Cancer versus normal gene expression comparison was done by comparing the mRNA expression of paired primary tumor and non-tumor tissue in selected cancer types in TCGA data sets. The percentage of patients showed at least 2-fold increased or decreased expression of genes in tumor versus normal in each of the selected cancer types were summarized by OmicSoft mRNA quantification comparison. DNA alteration distribution was summarized as the percentage of altered samples shown in each type of the alteration (amplification, homozygous deletion, non-synonymous mutation or combination of multiple alteration) and alteration OmicPrint was generated to comparatively visualize genomic alteration events in different ligases across TCGA cancer types by heatmap. Pearson correlation analyses between PROTAC activity and gene expression were performed in R (v3.6.0) and ingenuity pathway analysis were done on the significantly correlated genes by p value cutoff of 0.01 through the use of IPA (QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuitypathway-analysis) (Kramer et al., 2014). Cancer dependency comparison, Avana CRISPR library screening data (Achilles_gene_effect.csv Public 19 Q3) from Broad Institute Project Achilles Project was obtained from DepMap (Cancer Dependency MAP, https://depmap.org/portal/download/), which includes CRISPR screening results for all our interested ligases in 625 cell lines. Reported CERES scores (Meyers et al., 2017), which were used as indicative measurement of cancer dependency for each gene in corresponding cell line, of representative ligases were summarized in boxplot for each of 29 primary disease types. Corresponding CCLE sample info, mRNA expression, DNA copy number and DNA mutation data were obtained from DepMap portal Public 19 Q3 (Meyers et al., 2017; Ghandi et al., 2019). Copy number and mRNA expression of CRBN and VHL in dBET1 and MZ1 low and high activity group were visualized as boxplots with ggplot2 (v3.2.0) R package and unpaired two-samples, two-sided Wilcoxon rank sum tests were performed with ggpubr (v0.2.1) R package.
Acknowledgments
X.L., I.A., K.D., K.S., O.H., J.R.L., and D.M. would like to thank Ryan Wurz and his team for providing both MZ1 and dBET1. We would also like to thank Song Li for input and guidance during the preparation of this manuscript and Christine Sastri for her assay expertise and input at the inception of this work.
Author contributions
I.A., K.D., and K.S. designed and executed cell assays to characterize the activity of dBET1 and MZ1 across the cell line panel. X.L. and O.H. analyzed dBET1 and MZ1 activity data and probed publicly available gene expression and mutation data to identify predictive markers. X.L. and O.H. analyzed genomic and dependency data from publicly available datasets to identify promising E3 ligase substrate receptor proteins for future PROTAC development. D.M. and J.R.L. conceived the cell line screen project and provided insight and guidance throughout the work. D.M., I.A., and X.L. prepared the manuscript with contribution from all authors.
Declaration of interests
X.L., I.A., K.D., K.S., O.H., J.R.L., and D.M. were employees and stock holders of Amgen at the time of this report.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103985.
Supplemental information
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.Software used in the preparation of this manuscript is described with version number in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.
References
- Akhoondi S., Lindstrom L., Widschwendter M., Corcoran M., Bergh J., Spruck C., Grander D., Sangfelt O. Inactivation of FBXW7/hCDC4-beta expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer. Breast Cancer Res. 2010;12:R105. doi: 10.1186/bcr2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhoondi S., Sun D., von der lehr N., Apostolidou S., Klotz K., Maljukova A., Cepeda D., Fiegl H., Dafou D., Marth C., et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- Alqahtani A., Choucair K., Ashraf M., Hammouda D.M., Alloghbi A., Khan T., Senzer N., Nemunaitis J. Bromodomain and extra-terminal motif inhibitors: a review of preclinical and clinical advances in cancer therapy. Future Sci. OA. 2019;5:FSO372. doi: 10.4155/fsoa-2018-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem G.M., Crews C.M. Proteolysis-targeting chimeras as therapeutics and tools for biological discovery. Cell. 2020;181:102–114. doi: 10.1016/j.cell.2019.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burslem G.M., Smith B.E., Lai A.C., Jaime-Figueroa S., Mcquaid D.C., Bondeson D.P., Toure M., Dong H., Qian Y., Wang J., et al. The advantages of targeted protein degradation over inhibition: an RTK case study. Cell Chem. Biol. 2018;25:67–77.e3. doi: 10.1016/j.chembiol.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research, N Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra R., Sadok A., Collins I. A critical evaluation of the approaches to targeted protein degradation for drug discovery. Drug Discov. Today Technol. 2019;31:5–13. doi: 10.1016/j.ddtec.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium G.T. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer E.S., Bohm K., Lydeard J.R., Yang H., Stadler M.B., Cavadini S., Nagel J., Serluca F., Acker V., Lingaraju G.M., et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadd M.S., Testa A., Lucas X., Chan K.H., Chen W., Lamont D.J., Zengerle M., Ciulli A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi A.K., Kang J., Havens C.G., Conklin T., Ning Y., Wu L., Ito T., Ando H., Waldman M.F., Thakurta A., et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.) Br. J. Haematol. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., Mcdonald E.R., 3rd, Barretina J., Gelfand E.T., Bielski C.M., Li H., et al. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019;569:503–508. doi: 10.1038/s41586-019-1186-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T., Goralski M., Gaskill N., Capota E., Kim J., Ting T.C., Xie Y., Williams N.S., Nijhawan D. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science. 2017;356:eaal3755. doi: 10.1126/science.aal3755. [DOI] [PubMed] [Google Scholar]
- Hansen J.D., Condroski K., Correa M., Muller G., Man H.W., Ruchelman A., Zhang W., Vocanson F., Crea T., Liu W., et al. Protein degradation via CRL4(CRBN) ubiquitin ligase: discovery and structure-activity relationships of novel glutarimide analogs that promote degradation of Aiolos and/or GSPT1. J. Med. Chem. 2018;61:492–503. doi: 10.1021/acs.jmedchem.6b01911. [DOI] [PubMed] [Google Scholar]
- Hughes S.J., Ciulli A. Molecular recognition of ternary complexes: a new dimension in the structure-guided design of chemical degraders. Essays Biochem. 2017;61:505–516. doi: 10.1042/EBC20170041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishoey M., Chorn S., Singh N., Jaeger M.G., Brand M., Paulk J., Bauer S., Erb M.A., Parapatics K., Muller A.C., et al. Translation termination factor GSPT1 is a phenotypically relevant off-target of heterobifunctional phthalimide degraders. ACS Chem. Biol. 2018;13:553–560. doi: 10.1021/acschembio.7b00969. [DOI] [PubMed] [Google Scholar]
- Kang C.H., Lee D.H., Lee C.O., Du Ha J., Park C.H., Hwang J.Y. Induced protein degradation of anaplastic lymphoma kinase (ALK) by proteolysis targeting chimera (PROTAC) Biochem. Biophys. Res. Commun. 2018;505:542–547. doi: 10.1016/j.bbrc.2018.09.169. [DOI] [PubMed] [Google Scholar]
- Kramer A., Green J., Pollard J., JR., Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronke J., Fink E.C., Hollenbach P.W., Macbeth K.J., Hurst S.N., Udeshi N.D., Chamberlain P.P., Mani D.R., Man H.W., Gandhi A.K., et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature. 2015;523:183–188. doi: 10.1038/nature14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronke J., Udeshi N.D., Narla A., Grauman P., Hurst S.N., Mcconkey M., Svinkina T., Heckl D., Comer E., Li X., et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Girona A., Mendy D., Ito T., Miller K., Gandhi A.K., Kang J., Karasawa S., Carmel G., Jackson P., Abbasian M., et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G., Middleton R.E., Sun H., Naniong M., Ott C.J., Mitsiades C.S., Wong K.K., Bradner J.E., Kaelin W.G. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyukova A., Dohda T., von der Lehr N., Akhoondi S., Corcoran M., Heyman M., Spruck C., Grander D., Lendahl U., Sangfelt O. The tumor suppressor gene hCDC4 is frequently mutated in human T-cell acute lymphoblastic leukemia with functional consequences for Notch signaling. Cancer Res. 2007;67:5611–5616. doi: 10.1158/0008-5472.CAN-06-4381. [DOI] [PubMed] [Google Scholar]
- Matyskiela M.E., Lu G., ITO T., Pagarigan B., Lu C.C., Miller K., Fang W., Wang N.Y., Nguyen D., Houston J., et al. A novel cereblon modulator recruits GSPT1 to the CRL4(CRBN) ubiquitin ligase. Nature. 2016;535:252–257. doi: 10.1038/nature18611. [DOI] [PubMed] [Google Scholar]
- Meyers R.M., Bryan J.G., McFarland J.M., Weir B.A., Sizemore A.E., Xu H., Dharia N.V., Montgomery P.G., Cowley G.S., Pantel S., et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 2017;49:1779–1784. doi: 10.1038/ng.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., Mccue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Ottis P., Palladino C., Thienger P., Britschgi A., Heichinger C., Berrera M., Julien-Laferriere A., Roudnicky F., Kam-Thong T., Bischoff J.R., et al. Cellular resistance mechanisms to targeted protein degradation converge toward impairment of the engaged ubiquitin transfer pathway. ACS Chem. Biol. 2019;14:2215–2223. doi: 10.1021/acschembio.9b00525. [DOI] [PubMed] [Google Scholar]
- Ottis P., Toure M., Cromm P.M., Ko E., Gustafson J.L., Crews C.M. Assessing different E3 ligases for small molecule induced protein ubiquitination and degradation. ACS Chem. Biol. 2017;12:2570–2578. doi: 10.1021/acschembio.7b00485. [DOI] [PubMed] [Google Scholar]
- Paiva S.L., Crews C.M. Targeted protein degradation: elements of PROTAC design. Curr. Opin. Chem. Biol. 2019;50:111–119. doi: 10.1016/j.cbpa.2019.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel H.K., Bihani T. Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharmacol. Ther. 2018;186:1–24. doi: 10.1016/j.pharmthera.2017.12.012. [DOI] [PubMed] [Google Scholar]
- Petzold G., Fischer E.S., Thoma N.H. Structural basis of lenalidomide-induced CK1alpha degradation by the CRL4(CRBN) ubiquitin ligase. Nature. 2016;532:127–130. doi: 10.1038/nature16979. [DOI] [PubMed] [Google Scholar]
- Riching K.M., Mahan S., Corona C.R., Mcdougall M., Vasta J.D., Robers M.B., Urh M., Daniels D.L. Quantitative live-cell kinetic degradation and mechanistic profiling of PROTAC mode of action. ACS Chem. Biol. 2018;13:2758–2770. doi: 10.1021/acschembio.8b00692. [DOI] [PubMed] [Google Scholar]
- Robertson J.F. ICI 182,780 (Fulvestrant)--the first oestrogen receptor down-regulator--current clinical data. Br. J. Cancer. 2001;85:11–14. doi: 10.1054/bjoc.2001.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M.J., Winkler S., Hughes S.J., Whitworth C., Galant M., Farnaby W., Rumpel K., Ciulli A. SPR-measured dissociation kinetics of PROTAC ternary complexes influence target degradation rate. ACS Chem. Biol. 2019;14:361–368. doi: 10.1021/acschembio.9b00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K.M., Kim K.B., Kumagai A., Mercurio F., Crews C.M., Deshaies R.J. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc. Natl. Acad. Sci. U S A. 2001;98:8554–8559. doi: 10.1073/pnas.141230798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K.M., Kim K.B., Verma R., Ransick A., Stein B., Crews C.M., Deshaies R.J. Development of Protacs to target cancer-promoting proteins for ubiquitination and degradation. Mol. Cell Proteomics. 2003;2:1350–1358. doi: 10.1074/mcp.T300009-MCP200. [DOI] [PubMed] [Google Scholar]
- Salami J., Alabi S., Willard R.R., Vitale N.J., Wang J., Dong H., Jin M., Mcdonnell D.P., Crew A.P., Neklesa T.K., Crews C.M. Androgen receptor degradation by the proteolysis-targeting chimera ARCC-4 outperforms enzalutamide in cellular models of prostate cancer drug resistance. Commun. Biol. 2018;1:100. doi: 10.1038/s42003-018-0105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneekloth A.R., Pucheault M., Tae H.S., Crews C.M. Targeted intracellular protein degradation induced by a small molecule: en route to chemical proteomics. Bioorg. Med. Chem. Lett. 2008;18:5904–5908. doi: 10.1016/j.bmcl.2008.07.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneekloth J.S., JR., Fonseca F.N., Koldobskiy M., Mandal A., Deshaies R., Sakamoto K., Crews C.M. Chemical genetic control of protein levels: selective in vivo targeted degradation. J. Am. Chem. Soc. 2004;126:3748–3754. doi: 10.1021/ja039025z. [DOI] [PubMed] [Google Scholar]
- Sun Y., Zhao X., Ding N., Gao H., Wu Y., Yang Y., Zhao M., Hwang J., Song Y., Liu W., Rao Y. PROTAC-induced BTK degradation as a novel therapy for mutated BTK C481S induced ibrutinib-resistant B-cell malignancies. Cell Res. 2018;28:779–781. doi: 10.1038/s41422-018-0055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T., Minoshima Y., Sagane K., Sugi N.H., Mitsuhashi K.O., Yamamoto N., Kamiyama H., Takahashi K., Kotake Y., Uesugi M., et al. Selective degradation of splicing factor CAPERalpha by anticancer sulfonamides. Nat. Chem. Biol. 2017;13:675–680. doi: 10.1038/nchembio.2363. [DOI] [PubMed] [Google Scholar]
- Winter G.E., Buckley D.L., Paulk J., Roberts J.M., Souza A., Dhe-Paganon S., Bradner J.E. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurz R.P., Dellamaggiore K., Dou H., Javier N., Lo M.C., Mccarter J.D., Mohl D., Sastri C., Lipford J.R., Cee V.J. A "click chemistry platform" for the rapid synthesis of bispecific molecules for inducing protein degradation. J. Med. Chem. 2018;61:453–461. doi: 10.1021/acs.jmedchem.6b01781. [DOI] [PubMed] [Google Scholar]
- Zengerle M., Chan K.H., Ciulli A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 2015;10:1770–1777. doi: 10.1021/acschembio.5b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Riley-Gillis B., Vijay P., Shen Y. Acquired resistance to BET-PROTACs (Proteolysis-Targeting chimeras) caused by genomic alterations in core components of E3 ligase complexes. Mol. Cancer Ther. 2019;18:1302–1311. doi: 10.1158/1535-7163.MCT-18-1129. [DOI] [PubMed] [Google Scholar]
- Zhang X., Lee H.C., Shirazi F., Baladandayuthapani V., Lin H., Kuiatse I., Wang H., Jones R.J., Berkova Z., Singh R.K., et al. Protein targeting chimeric molecules specific for bromodomain and extra-terminal motif family proteins are active against pre-clinical models of multiple myeloma. Leukemia. 2018;32:2224–2239. doi: 10.1038/s41375-018-0044-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.X., Braggio E., Shi C.X., Bruins L.A., Schmidt J.E., Van Wier S., Chang X.B., Bjorklund C.C., Fonseca R., Bergsagel P.L., et al. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood. 2011;118:4771–4779. doi: 10.1182/blood-2011-05-356063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoppi V., Hughes S.J., Maniaci C., Testa A., Gmaschitz T., Wieshofer C., Koegl M., Riching K.M., Daniels D.L., Spallarossa A., Ciulli A. Iterative design and optimization of initially inactive proteolysis targeting chimeras (PROTACs) identify VZ185 as a potent, fast, and selective von Hippel-lindau (VHL) based dual degrader probe of BRD9 and BRD7. J. Med. Chem. 2019;62:699–726. doi: 10.1021/acs.jmedchem.8b01413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.Software used in the preparation of this manuscript is described with version number in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the Lead Contact upon request.