Abstract
Mut L homolog-1 (MLH1) is a key DNA mismatch repair protein which participates in the sensitivity to DNA damaging agents. However, its role in the radiosensitivity of tumor cells is less well characterized. In this study, we investigated the role of MLH1 in cellular responses to ionizing radiation (IR) and explored the signaling molecules involved. The isogenic pair of MLH1 proficient (MLH1+) and deficient (MLH1–) human colorectal cancer HCT116 cells was exposed to IR for 24 h at the dose of 3 cGy. The clonogenic survival was examined by the colony formation assay. Cell cycle distribution was analyzed with flow cytometry. Changes in the protein level of MLH1, DNA damage marker γH2AX, and protein kinase A catalytic subunit (PRKAC), a common target for anti-tumor drugs, were examined with Western blotting. The results showed that the HCT116 (MLH1+) cells demonstrated increased radio-resistance with increased S population, decreased G2 population, a low level of γH2AX, a reduced ratio of phosphorylated PRKACαβ to total PRKAC, and an elevated level of total PRKAC and phosphorylated PRKACβII following IR compared with the HCT116 (MLH1–) cells. Importantly, silencing PRKAC in HCT116 (MLH1+) cells increased the cellular radiosensitivity. In conclusion, MLH1 may increase cellular resistance to IR by activating PRKAC. Our finding is the first to demonstrate the important role of PRKAC in MLH1-mediated radiosensitivity, suggesting that PRKAC has potential as a biomarker and a therapeutic target for increasing radio-sensitization.
Keywords: HCT116 cells, ionizing radiation, Mut L homolog-1, protein kinase A catalytic subunit, radiosensitivity
Impact statement
Mut L homolog-1 (MLH1)-mediated DNA mismatch repair has biological significance following ionizing radiation (IR), underscoring the need to determine the biological significance of MLH1 on the radiosensitivity of tumor cells responding to IR. MLH1-proficient human colorectal cancer HCT116 cells (MLH+) demonstrated increased radio-resistance with decreased G2 population, a low level of γH2AX protein, a reduced ratio of phosphorylated PRKACαβ to total PRKAC, and an elevated level of total PRKAC and phosphorylated PRKACβII following IR compared with the MLH1-deficient HCT116 cells. Importantly, silencing PRKAC in HCT116 (MLH1+) cells increased the cellular radiosensitivity. These results suggest that MLH1 may increase cellular resistance to IR by activating PRKAC. Our finding is the first to demonstrate the important role of PRKAC in MLH1-mediated radiosensitivity, suggesting that PRKAC has potential as a biomarker and a therapeutic target for increasing radio-sensitization.
Introduction
Cancer is a global problem and one of the main causes of death in human beings. 1 With changes in people's lifestyle and living environment, especially the consequent aging of the population, the incidence and mortality of cancer in China have been increasing year by year.2,3 Radiotherapy is one of the three main treatment methods for cancer. More than 70% of tumor patients need radiotherapy at different stages of treatment. 4 However, the radiosensitivity of tumors varies considerably among individuals, leading to wide differences in radiotherapy efficiency. 4 Although some radiosensitive tumors can be treated reliably by radiotherapy, most tumors have low sensitivity to radiotherapy. Due to the limited tolerance of normal tissues around the tumor to radiation, the tumor dose is limited, and this eventually leads to tumor recurrence. 5 Therefore, improving the radiotherapy sensitivity of tumor cells and avoiding or reducing the damage of normal tissues using a radiotherapy sensitizer, targeted therapy, concurrent chemotherapy, and other means are important ways to reduce tumor recurrence. 6
Ionizing radiation (IR) produces many types of DNA damage and the efficiency of a cell to repair DNA damage will affect cellular response to IR. 7 DNA mismatch repair (MMR) is a highly conserved DNA repair pathway for maintaining genomic integrity, and it is involved in the repair of the IR–induced DNA damages. 8 The link between MMR-deficiency and hereditary nonpolyposis colorectal cancer is well documented. Many other cancers can also arise from sporadic MMR gene mutations.8–12 Mut L homolog-1 (MLH1) is a key MMR protein participating in the repair of endogenous and exogenous mispairs in the daughter strands during S phase.13–15 Accumulating data suggest that MMR proteins may be involved in the DNA damage response to IR, but the status of how MLH1 affects the cellular responses to IR remains controversial. Yan et al. implicated a role for MLH1-proficiency in the hypersensitive response observed in HCT116 cells to prolonged low-dose rate IR. 16 Conflicting data provided by Davis et al. showed that MLH1-deficient HCT116 cells had decreased survival after IR compared with MLH1-proficient HCT116 cells. 17 Flanagan et al. also demonstrated that MLH1-deficient HCT116 cells were unable to repair drug-induced DNA mismatches and were more easily radiosensitized than MLH1-proficient HCT116 cells. 18 Therefore, MLH1-mediated MMR has biological significance following IR, underscoring the need to determine the biological significance of MLH1 on the radiosensitivity of tumor cells responding to IR.
In this study, we expressed MLH1 in MLH1-deficient HCT116 cells and evaluated the radiosensitivity of the MLH1-proficient cells responding to IR by assessing the cell survival, cell cycle, and DNA damage. In addition, we also examined whether protein kinase A catalytic subunit (PRKAC), a common target for anti-tumor drugs, was involved in MLH1-mediated radiosensitivity. The findings may provide insights into the role of MLH1 in cellular responses to IR. This may be of interest in the development of molecular labelling for assessing radiosensitivity and a new strategy for radiosensitization.
Materials and methods
Cell culture and IR
Human colon carcinoma cell line HCT116 deficient in the MLH1 gene was used in this study. The HCT116 cells expressing MLH1 (MLH1+) were obtained by transfecting pcDNA3.1-MLH1 vector (iCarTAB BioMed Inc., Suzhou, China) for 24 h. The HCT116 cells transfected with pcDNA3.1 vector were HCT116 (MLH1–) cells and used as the negative control. In addition, HCT116 (MLH1+) and HCT116 (MLH1–) were transfected with PRKAC siRNA (sense: 5’-CCUGUUGUAUGAAAUGCUUTT-3’; antisense: 5’-AAGCAUUUCAUACAACAGGAC-3’) and negative control siRNA (sense: 5’- UUCUCCGAACGAGU CACGUTT-3’; antisense: 5’- ACGUGACUCGUUCGGAGA ATT-3’) for 48 h. The lipofectamine 2000 (Thermo Fisher Scientific) was used as the transfection carrier. All cell lines were grown in Dulbecco's Modified Eagle Medium (Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Thermo Fisher Scientific), 2 mmol/L glutamine (Thermo Fisher Scientific), 0.1 mmol/L nonessential amino acids (Thermo Fisher Scientific), and 100 U/ml penicillin-streptomycin (Genom, Hangzhou, Zhejiang, China) grown in a humidified incubator at 37°C with 5% CO2. For IR, all cell lines were irradiated by an iridium-192 source at the dose rate of 3 cGy for 24 h in a humidified incubator at 37°C with 5% CO2.
Western blot
Cell lysates were prepared in phosphate-buffered saline (PBS) containing 1% Triton X-100, 1× protease inhibitor cocktail (Thermo Fisher Scientific), and 1 mM phenylmethylsulfonyl fluoride (Thermo Fisher Scientific). Protein concentration was determined using a Pierce® bicinchoninic acid protein assay kit (Thermo Fisher Scientific). Aliquots of 20 μg of proteins were separated on 12% (w/v) sodium dodecyl sulphate polyacrylamide gel electrophoresis and transferred onto an Immoblion-FL Transfer polyvinylidene difluoride membrane (Millipore, Burlington, MA, USA). Subsequently, the membrane was blocked with 5% non-fat milk or bovine serum albumin (BSA, for antibody against phosphorylated protein) at room temperature for 1 h and incubated with anti-MLH1 antibody (1:1000, Cell Signaling Technology (CST), Danvers, MA, USA), anti-γH2AX antibody (1:1000, CST), anti-PRKAC antibody (1:1000, Abcam plc., Cambridge, MA, USA), anti-phosphorylated PRKACα/β antibody (1:1000, CST), and anti-phosphorylated PRKACβII (1:1000, CST) at 4°C overnight. Following this, the membrane was incubated with goat anti-rabbit or anti-mouse IgG conjugated horseradish peroxidase (1:10,000, Proteintech Group, Inc., Rosemont, IL, USA) at room temperature for 1 h, and bands on the membrane were visualized using the enhanced chemiluminescent assay (Thermo Fisher Scientific). The protein level was quantified as the gray value of target bands detected by the corresponding antibodies normalized to those detected by β-ACTIN (loading control) using Image J software (version 1.44, National Institutes of Health, MD, USA). Each experiment was repeated at least three times.
Colony formation assay
Log-phase cells were plated into a six-well cell culture plate (Thermo Fisher Scientific) at the cell numbers of about 3000 cells per well for two weeks until colony formation. The cells were washed by PBS and fixed by methanol for 20 min. After washed by PBS, the cells were stained by 0.1% crystal violet solution (Beyotime Biotechnology, Beijing, China) for 20 min. The staining solution was removed by washing with PBS, and the stained colonies were imaged and counted.
Flow cytometry analysis
The determination of the cell cycle profile was described previously. 16 An aliquot of 1 × 106 fixed cells was incubated with 50 μg/ml propidium iodide (Sigma-Aldrich, St. Louis, MO, USA) for 30 min. The stained cells were analyzed by an Attune NxT Flow Cytometer (Thermo Fisher Scientific). The flow cytometric data were analyzed with Modfit 3.0 (Verity Software, Topsham, ME, USA).
Statistical analysis
All of the statistical analyses were performed by the statistics software GraphPad Prism (version 6.02, GraphPad Software, San Diego, CA, USA). Data are expressed as the mean ± SEM. Differences between two groups were assessed using the unpaired t test, while differences among multiple groups were assessed using one-way analysis of variance and the Dunnett’s test. Statistically significant differences were determined at P < 0.05.
Results
Expressing MLH1 in HCT116 cells increases cellular resistance to radiation
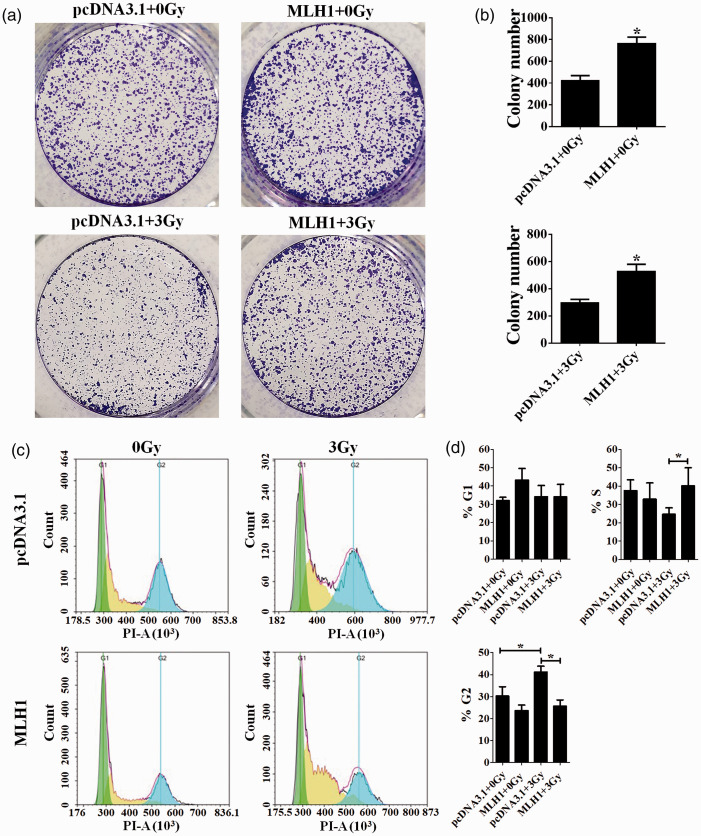
The colony formation assay was performed to examine the colony-formation ability of HCT116 (MLH1+) and HCT116 (MLH1–) cells before and after IR. A valid comparison of colony-formation ability was made by always irradiating both the cell lines simultaneously. The HCT116 (MLH1+) cells showed increased colony-formation ability compared with the HCT116 (MLH1–) cells no matter whether cells are exposed to IR or not (P < 0.05, Figure 1(a) and (b)). We next carried out a flow cytometry analysis to assess the cell cycle profile of the two HCT116 cells responding to IR at the dose of 3 Gy. The result showed that there is no significant difference in G1 cell cycle distribution between these two HCT116 cells after 3 Gy IR (P > 0.05, Figure 1(c) and (d)), while there is statistically higher percentage of S population in the HCT116 (MLH1+) cells compared with the HCT116 (MLH1–) cells after 3 Gy IR (P < 0.05, pcDNA3.1 + 3Gy vs. MLH1 + 3Gy, Figure 1(c) and (d)). 3 Gy IR induced G2-M cell cycle checkpoint arrest in the HCT116 (MLH1–) cells (pcDNA3.1 + 0Gy vs. pcDNA3.1 + 3Gy, Figure 1(c) and (d)), while the IR-induced G2-M cell cycle checkpoint arrest was relieved in the HCT116 (MLH1+) cells compared with the HCT116 (MLH1–) cells (pcDNA3.1 + 3Gy vs. MLH1 + 3Gy, Figure 1(c) and (d)).
Figure 1.
The colony-formation ability and cell cycle distribution of HCT116 cells. The human colon carcinoma cell line HCT116 deficient in the MLH1 gene was used in this study. The HCT116 cells expressing MLH1 (MLH1+) were obtained by transfecting the pcDNA3.1-MLH1 vector for 24 h. The HCT116 cells transfected with pcDNA3.1 vector were HCT116 (MLH1–) cells and used as the negative control. The colony formation assay was performed to examine the colony-formation ability of HCT116 (MLH1+) and HCT116 (MLH1–) cells before and after IR at the dose of 3 Gy (a and b). The flow cytometry analysis was performed to assess the cell cycle profile of the HCT116 (MLH1+) and HCT116 (MLH1–) cells before and after IR at the dose of 3 Gy (c). The flow cytometric data were analyzed with Modfit 3.0 (d). Each experiment was repeated at least three times. Data are expressed as the mean ± SEM. *P < 0.05. One-way analysis of variance. (A color version of this figure is available in the online journal.)
The changes of MLH1, γH2AX, PRKAC, and phosphorylated PRKAC levels in the HCT116 (MLH1+) and HCT116 (MLH1–) cells responding to IR
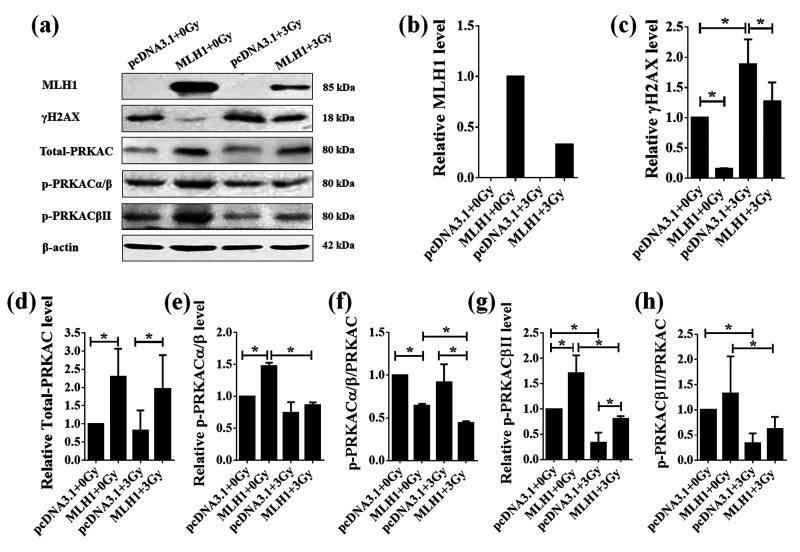
The MLH1 protein could not be detected in HCT116 (MLH1–) cells whether exposed to IR or not (Figure 2(a)), while the MLH1 protein level was decreased in the HCT116 (MLH1+) cells after a 24-h exposure to IR at the dose of 3 Gy (Figure 2(a) and (b)). Since the MLH1 protein has been implicated in DNA damage repair, we examined the level of a double strand–break (DSB) marker, γH2AX, in the HCT116 (MLH1+) and HCT116 (MLH1–) cells before or after IR by Western blot. The results showed that γH2AX protein levels were significantly increased in the HCT116 (MLH1–) and HCT116 (MLH1+) cells after a 24-h exposure to IR at the dose of 3 Gy (P <0.05, Figure 2(a) and (c)). Although the increasing rate of γH2AX in the HCT116 (MLH1+) cells was higher than that in the HCT116 (MLH1–) cells, the level of γH2AX proteins were lower in the HCT116 (MLH1+) cells than that in the HCT116 (MLH1–) cells after 3 Gy IR treatment (Figure 2(a) and (c)). Expressing MLH1 in HCT116 cells significantly increased the total PRKAC level before or after IR (P < 0.05, Figure 2(a) and (d)), but the total PRKAC level was not significantly changed in either HCT116 (MLH1+) cells or HCT116 (MLH1–) cells after a 24-h exposure to IR at the dose of 3 Gy (P > 0.05, Figure 2(a) and (d)). In addition, we also examined the level of phosphorylated PRKAC in the HCT116 (MLH1+) and HCT116 (MLH1–) cells responding to IR. The phosphorylated PRKACα/β (p-PRKACα/β) and PRKACβII (p-PRKACβII) were significantly increased, while the ratio of p-PRKACα/β to total PRKAC was significantly decreased in the HCT116 (MLH1+) cells compared with the HCT116 (MLH1–) cells (P < 0.05, Figure 2(a) and (e) to (g)). The p-PRKACα/β, the ratio of p-PRKACα/β to total PRKAC, p-PRKACβII, and the ratio of p-PRKACβII to total PRKAC were all significantly decreased in the HCT116 (MLH1+) cells after a 24-h exposure to IR at the dose of 3 Gy (P < 0.05, Figure 2(a) and (e) to (h)). The ratio of p-PRKACα/β to total PRKAC and p-PRKACβII were lower and higher, respectively, in the HCT116 (MLH1+) cells than those in the HCT116 (MLH1–) cells after a 24-h exposure to IR at the dose of 3 Gy (P < 0.05, Figure 2(a), (f), and (g)). Both p-PRKACβII and the ratio of p-PRKACβII to total PRKAC were decreased in the HCT116 (MLH1–) cells after a 24-h exposure to IR at the dose of 3 Gy (P < 0.05, Figure 2(a), (g), and (h)).
Figure 2.
The protein levels of MLH1, γH2AX, and PRKAC in the HCT116 cells before and after IR. The proteins were extracted from HCT116 (MLH1+) and HCT116 (MLH1–) cells before and after IR at the dose of 3 Gy. The protein levels of MLH1, γH2AX and total PRKAC, phosphorylated PRKACαβ (p-PRKACαβ), and phosphorylated PRKACβII (p-PRKACβII) were examined by Western blot (a) and quantified as the gray value of target bands detected by the corresponding antibodies normalized to those detected by β-actin (loading control) using Image J software (b-h). Each experiment was repeated at least three times. Data are expressed as the mean ± SEM. *P < 0.05. One-way analysis of variance.
Silencing PRKAC in the HCT116 (MLH1+) and HCT116 (MLH1–) cells
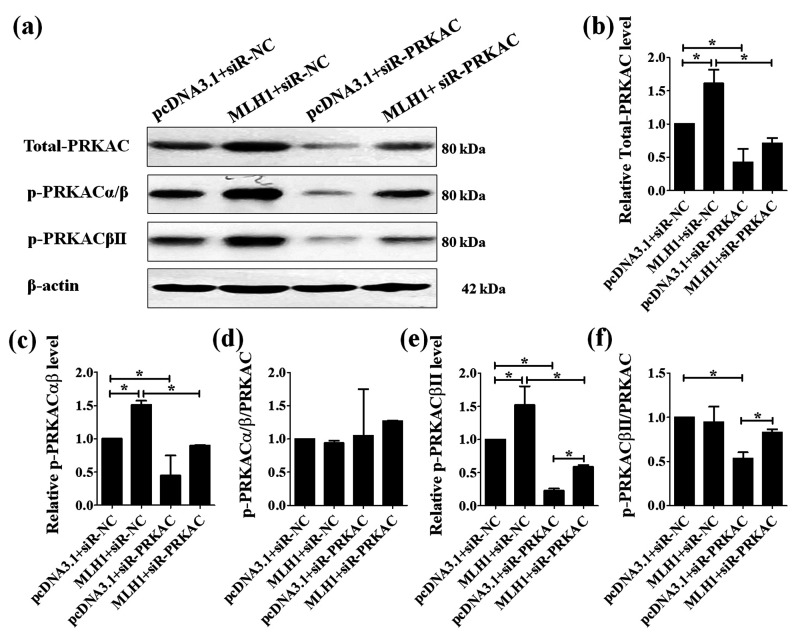
PRKAC siRNA significantly decreased the protein level of total PRKAC, p-PRKACαβ, and p-PRKACβII in both HCT116 (MLH1+) and HCT116 (MLH1–) cells (Figure 3(a) to (c) and (e)). PRKAC siRNA did not affect the ratio of p-PRKACαβ to total PRKAC in both HCT116 (MLH1+) and HCT116 (MLH1–) cells (Figure 3(a) and (d)), but it significantly lowered the ratio of p-PRKACβII to total PRKAC in the HCT116 (MLH1–) cells (Figure 3(a) and (f)). In addition, expressing MLH1 in HTC116 cells compromised the silencing effect of PRKAC siRNA on p-PRKACβII level (pcDNA3.1+siR-PRKAC vs. MLH1+siR-PRKAC, Figure 2(a), (e), and (f)).
Figure 3.
Effects of silencing PRKAC on the protein level of PRKAC in the HCT116 cells. PRKAC was silenced in HCT116 cells using PRKAC siRNA (siR-PRKAC). The negative control siRNA (siR-NC) was used. The proteins were extracted from HCT116 cells transfected with siR-PRKAC and siR-NC for 48 h. The protein levels of total PRKAC, phosphorylated PRKACαβ (p-PRKACαβ), and phosphorylated PRKACβII (p-PRKACβII) were examined by Western blot (a) and quantified as the gray value of target bands detected by the corresponding antibodies normalized to those detected by β-actin (loading control) using Image J software (b–f). Each experiment was repeated at least three times. Data are expressed as the mean ± SEM. *P < 0.05. One-way analysis of variance.
Effects of silencing PRKAC in the HCT116 (MLH1+) and HCT116 (MLH1–) cells on cellular resistance to IR
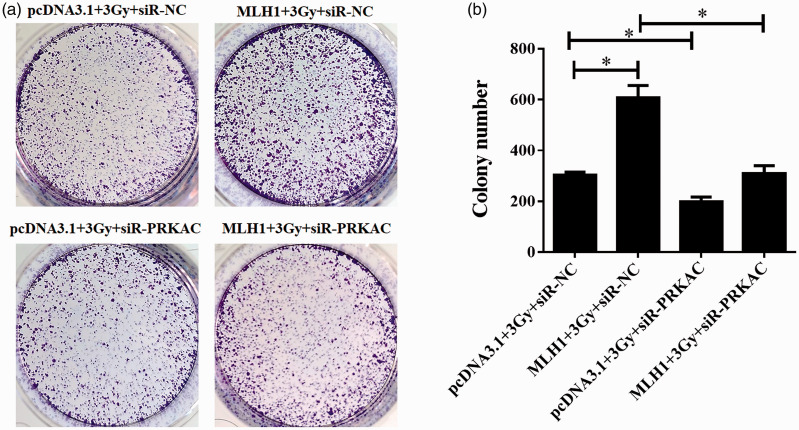
We examined whether silencing PRKAC affected the colony-formation ability in the HCT116 (MLH1+) and HCT116 (MLH1–) cells responding to IR. The HCT116 (MLH1+) cells showed a significantly increased cellular resistance to IR compared with the HCT116 (MLH1–) cells (pcDNA3.1 + 3Gy+siR-NC vs. MLH1 + 3Gy+siR-NC, P < 0.05, Figure 4(a) and (b)), while silencing PRKAC in the HCT116 (MLH1+) and HCT116 (MLH1–) cells decreased the cellular resistance to radiation (pcDNA3.1 + 3Gy+siR-NC vs. pcDNA3.1 + 3Gy+siR-PRKAC and MLH1 + 3Gy+siR-NC vs. MLH1 + 3Gy+siR-PRKAC, Figure 4(a) and (b)).
Figure 4.
Effects of silencing PRKAC on the colony-formation ability in the HCT116 cells after IR. The colony formation assay was performed to examine the colony-formation ability of HCT116 cells transfected with siR-PRKAC and siR-NC after IR at the dose of 3 Gy (a and b). Each experiment was repeated at least three times. Data are expressed as the mean ± SEM. *P < 0.05. One-way analysis of variance. (A color version of this figure is available in the online journal.)
Discussion
The MLH1 protein participates in the repair of endogenous and exogenous mispairs in the daughter strands,13,14 and it was also implicated in the response to environmentally induced DNA lesions.13,16–19 The role of MLH1 in the DNA damage response to IR has also been reported previously; however, how the status of MLH1 affects the cellular responses to IR remains unclear.16–19 In this study, we evaluated the differences in cell survival, cell cycle, and DNA damage between MLH1 proficient and deficient HTC116 cells in response to IR. MLH1-proficient HTC116 cells showed increased survival, relieved G2-M cell cycle checkpoint arrest, and a low level of γH2AX after IR compared with MLH1-deficent HTC116 cells. We also found that PRKAC was involved in MLH1-mediated radiosensitivity. The findings may provide insights into the role of MLH1 in cellular responses to IR.
MHL1 was reported to be involved in the radiotherapy sensitivity of tumor cells; however, different research groups have shown conflicting data. 8 In this study, we found that the MLH1-proficient HCT116 cells had an increased colony-formation ability after 24-h IR at the dose of 3 Gy compared with the MLH1-deficient HCT116 cells. This result supported the findings reported by Davis et al. 17 and Flanagan et al. 18 These results could be explained and understood. MLH1 is a key protein in the repair of DNA damage as a response to IR.16–19 Expressing MLH1 in HCT116 cells significantly reduced the γH2AX level before or after IR, indicating that MLH1 was involved in the repair of DNA damage no matter whether cells are exposed to IR or not. In addition, MLH1 in the HCT116 (MLH1+) cells relieved the IR induced G2-M cell cycle checkpoint arrest. These results supported the finding that expressing MLH1 in HCT116 cells increases cellular resistance to IR.
Although previous studies have investigated the cellular responses in MLH1-mediated radiosensitivity, the signaling pathway involved is not clear.16–19 Davis et al. examined the role of the P53-mediated pathway, an extensively characterized signaling pathway in response to IR, 20 in MLH1-mediated radiosensitivity, and they found that the MLH1 function was not influenced by the P53-mediated pathway. 17 In this study, we found that expressing MLH1 in HCT116 cells increased the level of total PRKAC, p-PRKACα/β, and p-PRKACβII no matter if the cells were exposed to IR. Silencing PRKAC in the HCT116 (MLH1+) and HCT116 (MLH1–) cells decreased the cellular resistance to radiation. These results suggested that PRKAC was involved in MLH1-mediated radiosensitivity. The PRKAC is the catalytic subunit of protein kinase A (PKA) which mediates cAMP-dependent cell processes such as DNA replication and cell proliferation. 21 Human tumors, such as breast, colon, and lung cancers, overexpress PKA, and PKA overexpression has been associated with a poor prognosis. 22 PKA inhibitors have demonstrated radiosensitization of various cancer cell lines. 23 The present study is the first to demonstrate the important role of PKA in MLH1-mediated radiosensitivity. However, the mechanism underlying how MLH1 affect the protein and phosphorylation levels of PRKAC and what downstream molecular(s) and/or pathway(s) are involved in PRKAC related radiosensitization have not been explored in the study, and thus further experiments are required in the future work.
In conclusion, the present study provided the evidence that expressing MLH1 in HCT116 cells increased the cellular resistance to IR and relieved the IR-induced G2-M cell cycle checkpoint arrest after IR. In addition, our finding is the first to demonstrate the important role of PKA in MLH1-mediated radiosensitivity, suggesting that PKA has potential as a biomarker and a therapeutic target for increasing radiosensitization.
Footnotes
AUTHORS’ CONTRIBUTION: All authors participated in the design, implement of the experiment, and data analysis. CLJ conceptualized the study, analyzed results, and drafted the article. YLH and LF conducted the experiments. YQB did statistical analysis. CLJ, YLH, and JHL wrote the manuscript. YZ did literature search. MQF edited the article. JGL and CLJ engaged in the experiment design and instructed students.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (No. 81760547).
ORCID iD: Chunling Jiang https://orcid.org/0000-0002-1214-5325
References
- 1.Vineis P, Wild CP. Global cancer patterns: causes and prevention. Lancet 2014; 383:549–57 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66:115–32 [DOI] [PubMed] [Google Scholar]
- 3.Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen W, Li X, Wang L, Wang L, Liu Y, Liu J, Zhang M, Qi J, Yu S, Afshin A, Gakidou E, Glenn S, Krish VS, Miller-Petrie MK, Mountjoy-Venning WC, Mullany EC, Redford SB, Liu H, Naghavi M, Hay SI, Wang L, Murray CJL, Liang X. Mortality, morbidity, and risk factors in China and its provinces, 1990–2017: a systematic analysis for the global burden of disease study 2017. Lancet 2019; 394:1145–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall EJ, Cox JD. Physical and biologic basis of radiation therapy. In: Cox JD, Ang KK. (eds) Radiation oncology. 9th ed. Philadelphia: Mosby, 2010, Chapter 1, pp. 3–49 [Google Scholar]
- 5.Sharda N, Yang C-R, Kinsella T, Boothman D. Radiation resistance. In: Bertino JR. (ed.) Encyclopedia of cancer. 2nd ed. New York: Academic Press, 2002, pp. 1–11 [Google Scholar]
- 6.Linkous AG, Yazlovitskaya EM. Novel radiosensitizing anticancer therapeutics. Anticancer Res 2012; 32:2487–99 [PubMed] [Google Scholar]
- 7.Santivasi WL, Xia F. Ionizing radiation-induced DNA damage, response, and repair. Antioxid Redox Signal 2014; 21:251–9 [DOI] [PubMed] [Google Scholar]
- 8.Martin LM, Marples B, Coffey M, Lawler M, Lynch TH, Hollywood D, Marignol L. DNA mismatch repair and the DNA damage response to ionizing radiation: making sense of apparently conflicting data. Cancer Treat Rev 2010; 36:518–27 [DOI] [PubMed] [Google Scholar]
- 9.Umar A, Kunkel TA. DNA-replication fidelity, mismatch repair and genome instability in cancer cells. Eur J Biochem 1996; 238:297–307 [DOI] [PubMed] [Google Scholar]
- 10.Marra G, Jiricny J. DNA mismatch repair and colon cancer. Adv Exp Med Biol 2005; 570:85–123 [DOI] [PubMed] [Google Scholar]
- 11.Jackson T, Ahmed MAH, Seth R, Jackson D, Ilyas M. MLH1 function is context dependent in colorectal cancers. J Clin Pathol 2011; 64:141–5 [DOI] [PubMed] [Google Scholar]
- 12.Rashid S, Freitas MO, Cucchi D, Bridge G, Yao Z, Gay L, Williams M, Wang J, Suraweera N, Silver A, McDonald SAC, Chelala C, Szabadkai G, Martin SA. MLH1 deficiency leads to deregulated mitochondrial metabolism. Cell Death Dis 2019; 10:795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellison AR, Lofing J, Bitter GA. Human MutL homolog (MLH1) function in DNA mismatch repair: a prospective screen for missense mutations in the ATPase domain. Nucleic Acids Res 2004; 32:5321–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blasi MF, Ventura I, Aquilina G, Degan P, Bertario L, Bassi C, Radice P, Bignami M. A human cell-based assay to evaluate the effects of alterations in the MLH1 mismatch repair gene. Cancer Res 2006; 66:9036–44 [DOI] [PubMed] [Google Scholar]
- 15.Kawashima N, Yoshida H, Miwa M, Fujiwara K. MLH1 is a prognostic biomarker for serous ovarian cancer treated with platinum- and taxane-based chemotherapy. Anticancer Res 2019; 39:5505–13 [DOI] [PubMed] [Google Scholar]
- 16.Yan T, Seo Y, Kinsella TJ. Differential cellular responses to prolonged LDR-IR in MLH1-proficient and MLH1-deficient colorectal cancer HCT116 cells. Clin Cancer Res 2009; 15:6912–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis TW, Wilson-Van Patten C, Meyers M, Kunugi KA, Cuthill S, Reznikoff C, Garces C, Boland CR, Kinsella TJ, Fishel R, Boothman DA. Defective expression of the DNA mismatch repair protein, MLH1, alters G2-M cell cycle checkpoint arrest following ionizing radiation. Cancer Res 1998; 58:767–78 [PubMed] [Google Scholar]
- 18.Flanagan SA, Krokosky CM, Mannava S, Nikiforov MA, Shewach DS. MLH1 deficiency enhances radiosensitization with 5-fluorodeoxyuridine by increasing DNA mismatches. Mol Pharmacol 2008; 74:863–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan T, Schupp JE, Hwang H-S, Wagner MW, Berry SE, Strickfaden S, Veigl ML, Sedwick WD, Boothman DA, Kinsella TJ. Loss of DNA mismatch repair imparts defective cdc2 signaling and G2 arrest responses without altering survival after ionizing radiation. Cancer Res 2001; 61:8290–7 [PubMed] [Google Scholar]
- 20.Mirzayans R, Andrais B, Scott A, Wang YW, Murray D. Ionizing radiation-induced responses in human cells with differing TP53 status. Int J Mol Sci 2013; 14:22409–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turnham RE, Scott JD. Protein kinase a catalytic subunit isoform PRKACA; history, function and physiology. Gene 2016; 577:101–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neary CL, Nesterova M, Cho YS, Cheadle C, Becker KG, Cho-Chung YS. Protein kinase a isozyme switching: eliciting differential cAMP signaling and tumor reversion. Oncogene 2004; 23:8847–56 [DOI] [PubMed] [Google Scholar]
- 23.Chin C, Bae JH, Kim MJ, Hwang JY, Kim SJ, Yoon MS, Lee MK, Kim DW, Chung BS, Kang CD, Kim SH. Radiosensitization by targeting radioresistance-related genes with protein kinase a inhibitor in radioresistant cancer cells. Exp Mol Med 2005; 37:608–18 [DOI] [PubMed] [Google Scholar]