Abstract
CDK11, a member of the cyclin-dependent kinase family, has been implicated in a diverse array of functions including transcription, RNA processing, sister chromatid cohesion, spindle assembly, centriole duplication and apoptosis. Despite its involvement in many essential functions, little is known about the requirements for CDK11 and its partner Cyclin L in a developing multicellular organism. Here we investigate the function of CDK11 and Cyclin L during development of the nematode Caenorhabditis elegans. Worms express two CDK11 proteins encoded by distinct loci: CDK-11.1 is essential for normal male and female fertility and is broadly expressed in the nuclei of somatic and germ line cells, while CDK-11.2 is nonessential and is enriched in hermaphrodite germ line nuclei beginning in mid pachytene. Hermaphrodites lacking CDK-11.1 develop normally but possess fewer mature sperm and oocytes and do not fully activate the RAS-ERK pathway that is required for oocyte production in response to environmental cues. Most of the sperm and eggs that are produced in cdk-11.1 null animals appear to complete development normally but fail to engage in sperm-oocyte signaling suggesting that CDK-11.1 is needed at multiple points in gametogenesis. Finally, we find that CDK-11.1 and CDK-11.2 function redundantly during embryonic and postembryonic development and likely do so in association with Cyclin L. Our results thus define multiple requirements for CDK-11-Cyclin L during animal development.
Keywords: CDK11, Cyclin L, Gametogenesis, RAS-ERK, C. elegans
1. Introduction
Cyclin-dependent kinases, or CDKs, are a family of serine/threonine kinases that function as master regulators of the cell cycle and integrate the transcriptional state of the cell with intracellular and extracellular signals (Malumbres, 2014). A defining feature of a CDK is that the enzymatic activity of the kinase is dependent upon its physical association with a specific cyclin regulatory partner. Humans possess 20 CDKs that can be divided among eight subfamilies (Cao et al., 2014). While most, and perhaps all, CDKs influence both the cell cycle and transcriptional state of the cell, they are often divided into two groups. Three of the families (CDK1/2/3, CDK4/6, and CDK5) are known primarily as cell cycle regulators while five (CDK7, CDK20, CDK8/19, CDK10/11, and CDK9/12/13) are known to be involved in regulating transcription and other aspects of gene expression. Given their key roles in cell cycle and transcriptional regulation, it is not surprising that CDKs are often found to be misregulated in human diseases including cancer (Whittaker et al., 2017).
CDK11 is an intriguing member of the CDK family as it has been shown to function in coordinating transcription and mRNA processing, and independently in regulating mitosis and apoptosis (Zhou et al., 2016). In humans, two highly similar genes (CDC2L1 and CDC2L2) encode essentially identical CKD11 proteins (CDK11B and CDK11A). Three structurally and functionally distinct CDK11 isoforms are known. Transcription of the complete mRNA yields the largest isoform, CDK11p110. CDK11p110 is expressed throughout the cell cycle, is localized to the nucleus, and has been implicated in regulating transcriptional initiation, splicing and 3’ end processing (Choi et al., 2014; Drogat et al., 2012; Hu et al., 2003; Loyer et al., 2008; Pak et al., 2015; Trembley et al., 2002, 2003; Valente et al., 2009). The shorter mitosis-specific CDK11p58 isoform is produced via translation from an internal ribosome entry site (Cornelis et al., 2000) and functions in centrosome maturation, centriole duplication, bipolar spindle formation, chromatid cohesion and cytokinesis (Barna et al., 2008; Franck et al., 2011; Hu et al., 2007; Petretti et al., 2006; Rakkaa et al., 2014; Wilker et al., 2007; Yokoyama et al., 2008). Finally, the apoptosis-specific isoform CDK11p46 is produced by cleavage of CDK11p110 and CDK11p58 by caspases 1 and 3 (Ariza et al., 1999; Beyaert et al., 1997; Lahti et al., 1995). When ectopically expressed, CDK11p46 induces apoptosis (Chen et al., 2006; Lahti et al., 1995; Shi et al., 2003).
CDK11 is mainly regulated via association with Cyclin L. In humans, the CCNL1 and CCNL2 genes encode 6 Cyclin L isoforms. As all three CDK11 isoforms have been shown to bind multiple Cyclin L isoforms, there apparently exist numerous distinct CDK11-Cyclin L complexes (Loyer et al., 2008). CDK11p110 in association with the alpha and beta isoforms of Cyclin L1/L2 localize to splicing compartments, physically associate with splicing factors, and regulate splice site selection (Loyer et al., 2008). In fission yeast, a complex of CDK11-Cyclin L promotes assembly of the Mediator complex, a general regulator of RNA polymerase II, to control expression of a subset of genes (Drogat et al., 2012). In addition to Cyclin L, CDK11p58 has been shown to bind Cyclin D3 specifically during G2/M phase of the cell cycle (Zhang et al., 2002). Further, knockdown of Cyclin D3 has been shown to result in a G2/M delay and polyploidy (Chen et al., 2011). Together these findings suggest that distinct CDK11-Cyclin complexes might have specific functions, but the full spectrum of biological processes in which CDK11 participates is currently unknown.
To date, almost all studies on CDK11 have focused on single-cell organisms or on tissue culture cells. In contrast, the role of CDK11 within the context of a multicellular organism and during animal development has not been explored in detail. Notably, while CDK11 is known to be required for early embryogenesis in mice, knockout of the sole mouse cdc2l gene results in proliferative defects during early embryonic divisions and developmental arrest before 4 days postcoitus, precluding analysis of its roles at later stages of development (Li et al., 2004). In the current study, we utilize a combination of RNAi and strong loss-of-function alleles to characterize the role of CDK-11 and Cyclin L during development of C. elegans. We find that both CDK11 and Cyclin L are essential for embryonic and postembryonic development. Further, we find that in mature adults CDK11 activity is required for the production of oocytes and sperm and, at least in part, functions by promoting the activity of the RAS-ERK pathway.
2. Materials and methods
2.1. Worm strains and maintenance
Unless otherwise noted, all worm strains were grown at 20 °C on MYOB plates seeded with E. coli OP50. All worm strains used in this study and their genotypes are listed in Table S1.
2.2. BLAST analysis and protein phylogeny
To identify C. elegans CDK11 and Cyclin L orthologs, we performed BLAST searches using human CDK11A (NP_076916.2), CDK11B (P21127.3) or Cyclin L (NP_064703.1) protein sequences as queries in NCBI's genome database (ncbi.nlm.nih.gov) and the Wormbase website (wormbase.org). The kinase domains of the 26 proteins in Supplemental Fig. 1 were aligned using CLC Sequence Viewer 6 (CLC bio). We removed areas of low amino acid conservation using GBlocks 0.91b (Castresana, 2000; Talavera and Castresana, 2007). The final alignment contained 182 amino acid positions broken into seven blocks. An unrooted, neighbor-joining tree was produced using MegAlign (DNASTAR, Inc.).
2.3. Quantitation of brood size, embryonic viability, and larval lethality
Brood sizes were determined by placing individual L4 hermaphrodites on 35-mm MYOB plates and allowing them to produce self-progeny at 16, 20, or 25 °C. Hermaphrodites were moved to fresh plates every 24 h and brood sizes determined by counting all viable and nonviable progeny produced either over the course of 48 h or until embryo production ceased. Embryonic viability was measured as previously described (Miller et al., 2016). Larval lethality was measured by placing individual young adult hermaphrodites onto fresh plates and allowing them to lay embryos for 6.5 h. The hermaphrodites were then removed and the embryos immediately counted. Unhatched embryos were tallied the next day and approximately 42–46 h later, adult worms were counted. Larval lethality was calculated as the percentage of live hatchlings that failed to reach adulthood.
2.4. Mating tests
For matings where brood size was quantified, four males were allowed to mate with either hermaphrodites or fog-2(q71) females overnight at 20 °C. Hermaphrodites or females were then moved to new 35-mm MYOB plates every 24 h for a specified number of days or until no more progeny were produced. All progeny, live and dead, were counted 16–20 h after removal of the parent. To test for sperm defects in cdk-11.1(tm5495) mutants, L4 mutant males, or wild-type control males, were picked to a freshly seeded plate and incubated overnight at 20 °C in the absence of a mating partner. The next day crosses were established with four healthy active males and one fog-2(q71) female. Matings were carried out for one additional day at 20 °C before females were singled to fresh plates at 20 °C and embryonic viability scored as described above. To score for mating efficiency, the same routine was employed except that before mating, males were transferred to a 1.5 ml microcentrifuge tube containing 50 μl of 70 mM SYTO17 (ThermoFisher Scientific, Inc) in TBS (10 mM Tris, 1 mM EDTA, 5 mM NaCl, pH 8). After incubation in the dark at 25 °C for 3 h, the males were allowed to recover on a fresh MYOB plate. Matings were then initiated as above. The next day, each female was anaesthetized in M9 buffer (85.6 mM NaCl, 42.3 mM Na2HPO4, 22 mM KH2PO4, 1 mM MgSO4) containing 20 mM sodium azide, mounted on 3% agar pads under a coverslip and examined by DIC and epi-fluorescence microscopy using a rhodamine filter set. To test for oocyte defects in cdk-11.1(tm5495) hermaphrodites, we performed the aforementioned procedure using four WT males to one cdk-11.1(tm5495) hermaphrodite.
2.5. RNAi experiments
Unless otherwise noted, RNAi experiments were carried out at 25 °C using a feeding protocol as previously described (Kamath et al., 2003; Timmons and Fire, 1998). L4 larvae were picked onto MYOB plates containing 50 μg/ml carbenicillin and 1 mM IPTG and seeded with dsRNA-expressing E. coli HT115(DE3) cells. The plates were then incubated for 16–24 h. As a negative control, we performed RNAi against the non-essential gene smd-1.
2.6. DAPI staining of intact germ lines
Worms were picked into a 2 μl drop of a 1:1 mixture of egg whites and M9 buffer on a SuperFrost microscope slide (Fisher Scientific). Using a pick, each worm was drawn out radially until it was isolated in a small droplet. The slides were immersed in Carnoy's fixative (60% ethanol, 30% chloroform, 10% glacial acetic acid) at 4 °C overnight or for up to 1 week. Worms were then rehydrated in five sequential steps: 1) 90% ethanol/10% water, 2) 70% ethanol/30% water, 3) 50% ethanol/50% PBS, 4) 25% ethanol /75% PBS, and 5) 100% PBS for two minutes each. Specimens were mounted in Vectashield with DAPI (Vector Labs) and stored at − 30 °C until imaged.
2.7. Production of antibodies, immunoblotting, and immunofluorescence
Antibodies specific for CDK-11.2 were prepared by YenZym Antibodies LLC (San Francisco, CA). Rabbits were immunized with a peptide whose sequence corresponded to amino acids 626–642 of CDK-11.2: CPENRRKNRLEALLADEE. Sera was then collected from rabbits exhibiting a strong response and subjected to affinity-purification.
Immunoblotting and indirect immunofluorescence staining were performed essentially as described (Miller et al., 2016; Peel et al., 2012). For immunoblotting, the following antibodies were used at 1:500–1:1000 dilutions: anti-CDK-11.2, anti-GFP (Roche), and antialpha-tubulin DM1A (Millipore Sigma). Anti-mouse and anti-rabbit IRDye secondary antibodies (LI-COR Biosciences, Inc.) were used at a dilution of 1:10,000–14,000. For indirect immunofluorescence, the following primary antibodies were used at a dilution of 1:400–1:1000: anti-di-phosphorylated MPK-1 (MilliporeSigma), anti-phospho-histone-H3 (MilliporeSigma), anti-NOP1 (Encore Biotechnology, Inc), and anti-CDK-11.2. Anti-mouse and anti-rabbit Alexa Fluor secondary antibodies (Thermo Fisher Scientific, Inc.) were used at a dilution of 1:500–1:1000.
2.8. DIC and fluorescence microscopy
To visualize germ lines by DIC, worms were placed in a drop of M9 buffer containing 20 mM sodium azide on a 3% agar pad atop a microscope slide. A cover slip was placed over the worms and the edges sealed with molten petroleum jelly. DIC imaging (Song et al., 2008) and spinning disk confocal imaging (Miller et al., 2016) were performed as previously described. For quantitative fluorescence intensity measurements, all imaging parameters including laser intensity, camera sensitivity, exposure time, and binning, were kept constant between control and experimental samples. For late pachytene dpMPK-1 staining, we measured fluorescence intensity in a region of interest (ROI) near the pachytene/diplotene border, and for diakinesis, we measured fluorescence intensity in an ROI centered on the nuclei of the most distal one or two oocytes.
2.9. Construction and injection of MosSCI vectors
The 5’ and 3’ flanking regions of cdk-11.1 and cdk-11.2, and the cdk-11.1 open reading frame were amplified using primers containing the appropriate site-specific recombination (att) sequences for MultiSite Gateway® three fragment construction (Thermo Fisher Scientific, Inc.). The coding sequences for superfolder gfp and histone H2B (his-58) were fused using the Gibson Assembly Cloning Kit (New England BioLabs, Inc.) and contained appropriate terminal att sites. DNA oligomer sequences can be found in Table S2. PCR products were purified with the QIAquick PCR Purification Kit (Qiagen) and inserted into the appropriate DONR vector via the BP reaction. Transgenes were assembled using the LR reaction to combine a 5’ flanking region, orf, and 3’ flanking region into either the chromosome I (pCFJ210) or chromosome II (pCFJ150) destination vector (Frokjaer-Jensen et al., 2012). MosSCI transgenesis was performed as previously described (Frokjaer-Jensen, 2015).
2.10. CRISPR genome editing
For genome editing, we utilized two alternative versions of the co-CRISPR technology (Arribere et al., 2014). We initially utilized a previously described protocol (Miller et al., 2016) that involved micro-injection of expression vectors encoding gRNAs and Cas9 protein into the germ lines of adult hermaphrodites. We later employed a more efficient protocol based on that of Paix et al. (2015) that involved germ-line injection of crRNAs, tracrRNA, and purified recombinant Cas9 protein. A typical injection mix contained either 8 μg of a target crRNA (single-locus editing) or 6 μg of each target crRNA (dual-loci editing), 3.2 μg of dpy-10 crRNA, 20 μg tracrRNA, 275 ng of dpy-10 repair template, either up to 500 ng/μl of target PCR repair template or 2.2 μg of target oligonucleotide repair template, and 50 μg of Cas9 protein in a final volume of 20 μl. All crRNAs and tracrRNA were synthesized by GE Healthcare Dharmacon, Inc. For repair templates, we used single stranded oligomers synthesized by Integrated DNA Technologies, Inc. Sequences of gRNAs, crRNAs and repair templates can be found in Table S3. Screening was performed as in Miller et al. (2016) and all genomic alterations were verified by Sanger Sequencing.
2.11. spe-9 cellular complementation test
spe-9(eb19) hermaphrodites were place alone, or with males (4 males: 1 hermaphrodites) on 35 mm MYOB plates seeded with OP50 E. coli for 48 h. Males were then removed, and 48 h later the progeny were counted.
2.12. Reagent and data availability
Raw data and all antibodies, plasmids, worm strains, and other reagents that are not commercially available will be shared upon request. A key resource table listing such items accompanies this manuscript.
3. Results
3.1. The two C. elegans CDK-11 paralogs are expressed in distinct but overlapping patterns
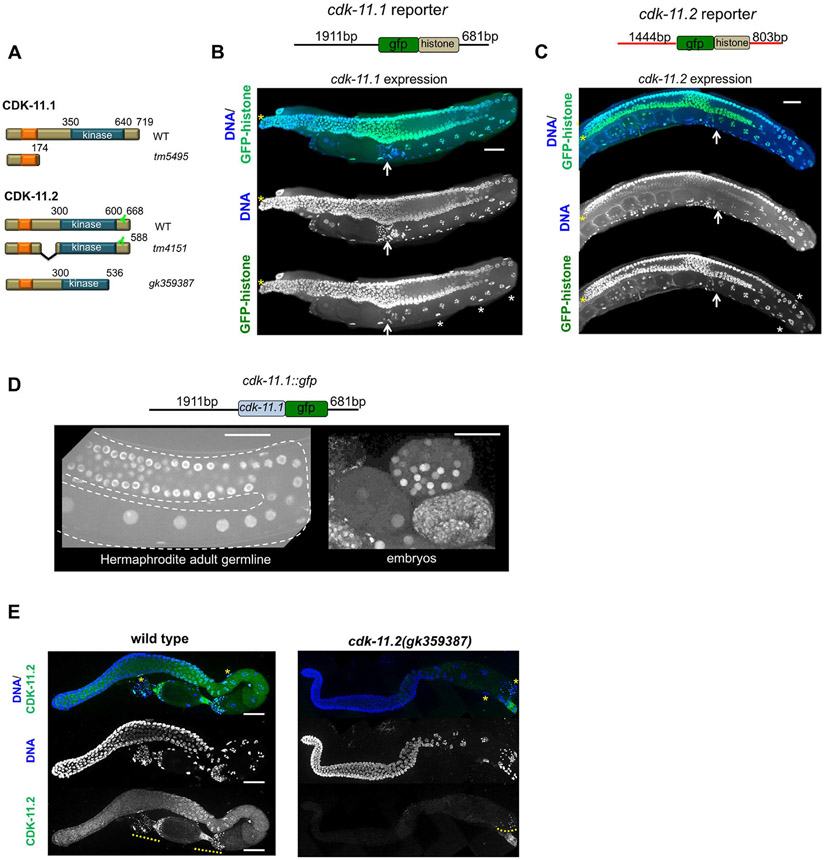
Using human CDK11 protein sequences as query, we searched the C. elegans proteome (www.wormbase.org) for worm homologs. The BLAST results revealed a large number of confirmed and previously annotated proteins believed to be CDKs. The top two hits were encoded by two uncharacterized genes, B0495.2 and ZC504.3. The most significant matches other than the top two, were genes previously identified as CDKs. B0495.2 and ZC504.3 encode proteins of 719 and 668 amino acids respectively, with each containing a single kinase domain in the C-terminus (Fig. 1A). Both proteins also contain a N-terminal RE (arginine/glutamic acid) domain (Fig. 1A, orange box); this domain is typical of CDK11 proteins and has been implicated in RNA processing (Zhou et al., 2016). We refer to these proteins as CDK-11.1 (B0495.2), and CDK-11.2 (ZC504.3). We confirmed the identities of CDK- 11.1 and CDK-11.2 using phylogenetic analysis of the amino acid sequences of CDKs from several different species: H. sapiens, M. musculus, X. laevis, D. melanogaster, S. cerevisiae and C. elegans. The two worm CDK-11 homologs grouped with the other CDK11 proteins with high confidence (Fig. S1A). Within the kinase domain, CDK-11.1 is 68% identical to CDK-11.2 and 66% identical to human CDK11A and CDK11B (Fig. S1B). CDK-11.2 is somewhat more divergent, sharing only 56% amino acid identity to each of the human CDK11 proteins. Throughout this paper, we refer to individual worm CDK-11 proteins by name (i.e. CDK-11.1 or CDK-11.2), both worm proteins collectively as CDK-11, and vertebrate orthologs as CDK11.
Fig. 1.
CDK-11.1 and CDK-11.2 are broadly expressed nuclear proteins with distinct germ line expression patterns. (A) Domain architecture of wild-type and mutant CDK-11.1 and CDK-11.2 proteins. Both proteins possess a C-terminal kinase domain and a N-terminal arginine- and glutamic acid-rich RE domain (orange box). Additionally, each protein contains a putative nuclear localization sequence within the RE domain (not shown). The position of the epitope recognized by the anti-CDK-11.2 antibody is indicated. (B) The structure of the cdk-11.1 reporter is shown at the top: the 5’ and 3’ flanking regions of cdk-11.1 were used to drive the expression of GFP::histone. GFP::histone is expressed throughout the entire germ line and in the sheath cells of the somatic gonad (white asterisks). The distal tip is marked by a yellow asterisk and the proximally located spermatheca by a white arrow. (C) The structure of the cdk-11.2 reporter is shown at the top: the 5’ and 3’ flanking regions of cdk-11.2 were used to drive the expression of GFP::histone. Expression of GFP::histone is observed throughout the germ line and somatic gonad. (D) The structure of the transgene expressing a CDK-11.1::GFP fusion protein. The fusion protein accumulates in nuclei throughout the germ line and in embryos. (E) Wild-type and cdk-11.2(gk359387) germ lines stained with anti-CDK-11.2 antibody and DAPI. CDK-11.2 is detected in the proximal germ line and somatic sheath cells (yellow asterisks). This staining is absent in the cdk-11.2(gk359387) mutant that lacks the epitope recognized by the antibody (see panel 1A). Note that the anti-CDK-11.2 antibody nonspecifically recognizes sperm (yellow dotted line). Scale bars are 25 μm in all images.
To determine where the CDK-11 proteins are produced, we created reporter constructs using the 5’ and 3’ flanking regions of cdk-11.1 and cdk-11.2 to drive the expression of a superfolder gfp::his-58 (GFP::histone) reporter gene (Fig. 1, B and C). The expression patterns of both transgenes appeared identical: GFP-histone was broadly expressed in both the soma and germ line. Within the germ line, GFP-histone labeled all nuclei from the most distal region through the most proximal region. In addition, the transgenes were expressed in the somatic gonad including sheath cells (Fig. 1, B and C), demonstrating that both cdk-11.1 and cdk-11.2 genes are broadly transcribed and translated in the worm.
Next, to determine the tissue distribution and subcellular location of the endogenous proteins, we set out to raise antisera specific for each CDK-11 protein. We immunized rabbits with a peptide whose amino acid sequence was unique to each protein; unfortunately, we were not able to obtain useful antibodies for CDK-11.1. Therefore to determine the distribution of CDK-11.1, we constructed a strain expressing a CDK-11.1::GFP fusion protein under control of the cdk-11.1 5’ and 3’-flanking regions (Fig. 1D). We then used MosSCI transgenesis (Frokjaer-Jensen, 2015) to integrate a single copy of this transgene at a specific site on chromosome I. The resulting transgenic line expressed a single protein species with an apparent molecular weight of approximately 140 kDa (Fig. S2A). While the size of CDK-11.1::GFP appears significantly larger than expected based on molecular weight, an anomalously slow electrophoretic mobility is characteristic of vertebrate CDK11 proteins. Taking into account that GFP is ~30 kDa, we surmise that CDK-11.1 is migrating as a 110 kDa protein, consistent with electrophoretic behavior of its vertebrate orthologs. We found that the CDK-11.1::GFP fusion protein accumulates in nuclei and does not appear to be enriched in any other cellular compartment. The nuclear-enriched CDK-11.1::GFP protein was detected broadly in both the soma, including somatic gonadal sheath cells, and the germ line (Fig. 1D). Within the hermaphrodite germ line, the protein was detected in all nuclei from the most distal mitotic region to the most proximal oocyte. The only cells in which we failed to detect the CDK-11.1::GFP fusion protein were sperm which lack a membrane-bound nucleus. Nuclear-localized CDK-11.1::GFP was also observed in embryos where it is ubiquitously expressed during all stages of embryogenesis (Fig. 1D). However, CDK-11.1::GFP did appear to be expressed at lower levels during the first few cell cycles than at later points in embryonic development. Thus, in worms, CDK-11.1 exists as a p110 form that is broadly localized to nuclei.
We next used the anti-CDK-11.2 antibody to explore its cellular and subcellular distribution within the germ line. Unlike the even germ line distribution of CDK-11.1::GFP, CDK-11.2 protein levels vary throughout the germ line (Figs. 1E and S2B and S2C). CDK-11.2 was found in nuclei and cytoplasm; both nuclear and cytoplasmic levels are relatively low in the mitotic region, peak in late pachytene, and then gradually decrease as oocytes mature (Fig. S2C). CDK-11.2 was also found in the sheath cell nuclei (Fig. 1E-asterisks) and in the nuclei and cytoplasm of embryos where levels steadily increase as embryos develop (Fig. S2B and S2C). Interestingly, the subcellular distribution of CDK-11.2 appears to be developmental regulated. In the mitotic region, CDK-11.2 is more concentrated in the cytoplasm than nuclei but as cells enter late pachytene, the protein becomes enriched in nuclei (Fig. S2B and S2C). In oocytes, CDK-11.2 is more evenly distributed between cytoplasm and nuclei, and in embryos the protein is initially evenly distributed between nuclei and cytoplasm but gradually becomes more concentrated in nuclei as embryos develop. This nuclear enrichment is first evident in four- and six-cell stage embryos, and increases during the ensuing cell divisions (Fig. S2C). Importantly, the antibody appears specific for CDK-11.2 as nuclear and cytoplasmic staining is absent in animals expressing a truncated form of CDK-11.2 that lacks the epitope recognized by the antibody (Fig. 1E), and in animals harboring a complete deletion of the cdk-11.2 gene (Fig. 5A). While we found that the anti-CDK-11.2 antibody also stains sperm, this staining persists in animals lacking the epitope, and thus represents nonspecific cross-reactivity (Fig. 1E).
Fig. 5.
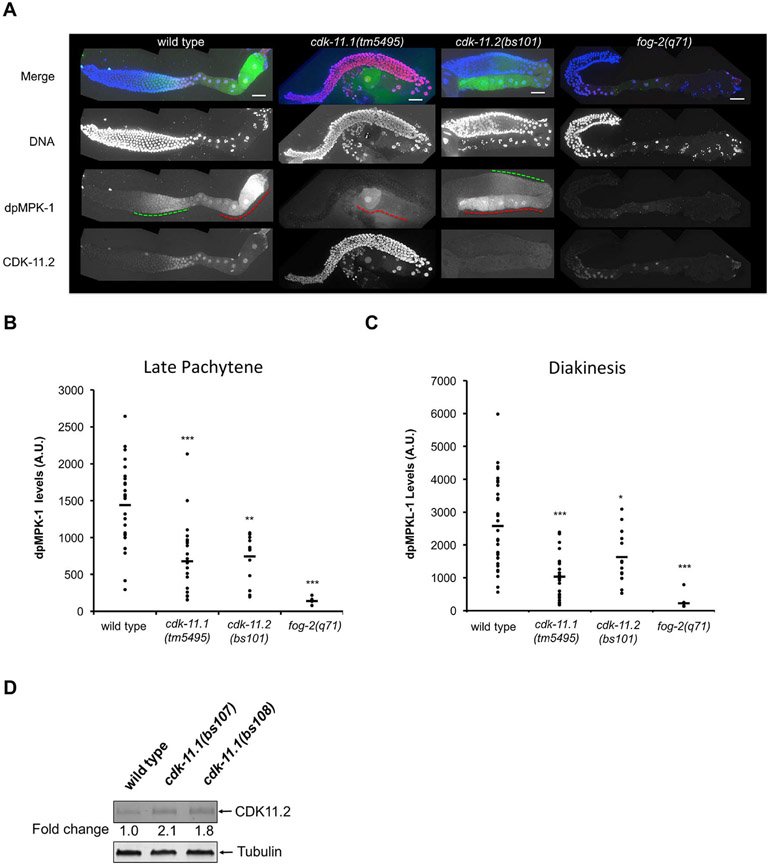
CDK-11.1 is required for full activation of the RAS-ERK pathway. (A) Dissected germ lines stained for DNA (blue), activated ERK (dpMPK-1, green), and CDK-11.2 (red). In wild-type germ lines, dpMPK-1 levels peak in late pachytene (green dashed line) and again in diakinesis (red dashed line), The fog-2(q71) germ lines serve as a negative control where very little or no dpMPK-1 staining is observed. cdk-11.1(tm5495) mutant hermaphrodites display decreased levels of dpMPK-1 in both late pachytene and diakinesis, while cdk-11.2(bs101) animals exhibit a similar reduction in the level of dpMPK-1 in late pachytene but a milder reduction during diakinesis. Note that cdk-11.1(tm5495) mutants show an increase in the level of CDK-11.2 throughout the entire germ line when compared to wild-type germ lines. Scale bars are 25 μm in all images. B and C) Quantitation of dpMPK-1 levels (plotted as arbitrary units, A.U.) during late pachytene and diakinesis in wild-type, cdk-11.1(tm5495), cdk-11.2(bs101) and fog-2(q71) germ lines. Black bars represent the mean. A one-tailed t-test was used to determine if mutants exhibit a significantly different dpMPK-1 level relative to the wild type (*p < 0.05,**p < 0.0001, ***p < 0.00001). The number of gonads scored (late pachytene/diakinesis) = 29/31 for wild type, 22/23 for cdk-11.1(tm5495), 14/13 for cdk-11.2(bs101), and 8/10 for fog-2(q71). (D) Quantitative immunoblot demonstrating that loss of CDK-11.1 leads to an increase in expression of CDK-11.2. Whole worm lysates were prepared and the equivalent of thirty adult animals were loaded in each lane.
By immunoblotting, the anti-CDK-11.2 antibody also proved to be specific. While we detected a single protein with an apparent molecular weight of ~ 110 kDa in mixed-stage wild-type worm lysates, we did not detect a band in lysates prepared from two CRISPR-derived lines: one in which the cdk-11.2 gene had been completely deleted by CRISPR genome editing (cdk-11.2(bs101)) and the other containing an early nonsense mutation (cdk-11.2(bs104)) (Fig. S2D). Furthermore, we detected a protein with a somewhat smaller apparent molecular weight (~ 85 kDa) in lysates prepared from the cdk-11.2(tm4151) mutant which possesses an internal deletion. We conclude that worms produce a p110 form of CDK-11.2 that has a dynamic expression pattern during germ line and embryonic development.
3.2. cdk-11.1 is required for normal fecundity
To determine what roles the two CDK-11 proteins may play in the worm, we set out to document phenotypes that arise from a loss of either or both paralogs. We obtained a deletion allele of cdk-11.1 from the National BioResource Project-Japan; the cdk-11.1(tm5495) mutation deletes 181-bp that spans exons 5 and 6 (Fig. S1C). We prepared and sequenced cDNA from this strain and found that the mutant transcript is predicted to encode a truncated protein consisting of the first 174 amino acids but lacking the protein kinase domain (Fig. 1A). Hermaphrodites homozygous for cdk-11.1(tm5495) are viable with normal morphology but exhibit low fecundity. To quantify the severity of this defect we determined brood size in the mutant. As compared to the wild type, the brood size of cdk-11(tm5495) hermaphrodites is drastically reduced at all temperatures measured (Fig. 2A-C). To confirm that the defects observed in the cdk-11.1(tm5495) mutant resulted solely from a loss of CDK-11.1, we used CRISPR-Cas9 genome editing to precisely delete the entire cdk-11 open reading frame and obtained two independent alleles bs107 and bs108 (Fig. S1C). Unfortunately, the CRISPR-Cas9 co-conversion marker dpy-10 is tightly linked to the cdk-11.1 gene, and thus both of our knockout lines exhibit a Dpy phenotype which made cytological analysis of germline defects difficult. However, similarly to cdk-11.1(tm5495) animals, both dpy-10(bs126) cdk-11.1(bs107) and dpy-10(bs141) cdk-11.1(bs108) hermaphrodites are viable but produced only a few progeny (Fig. 2D and data not shown). Furthermore, the bs107 and bs108 alleles failed to complement the fecundity defect of cdk-11.1(tm5495) (Fig. 2E), demonstrating that this defect is due to a loss of CDK-11.1 and not some extraneous mutation present in the deletion strain. Consistent with this, a single integrated copy of the cdk-11.1::gfp transgene shown in Fig. 1D, largely rescued the fecundity defects of cdk-11.1(bs107) homozygotes (Fig. 2F). Finally, the fecundity defects of cdk-11.1(tm5495) and cdk-11.1(bs107) proved to be recessive as heterozygous hermaphrodites possess normal brood sizes (Fig. 1A and B). We conclude that CDK-11.1 is not essential for viability or development, but is required for normal fecundity.
Fig. 2.
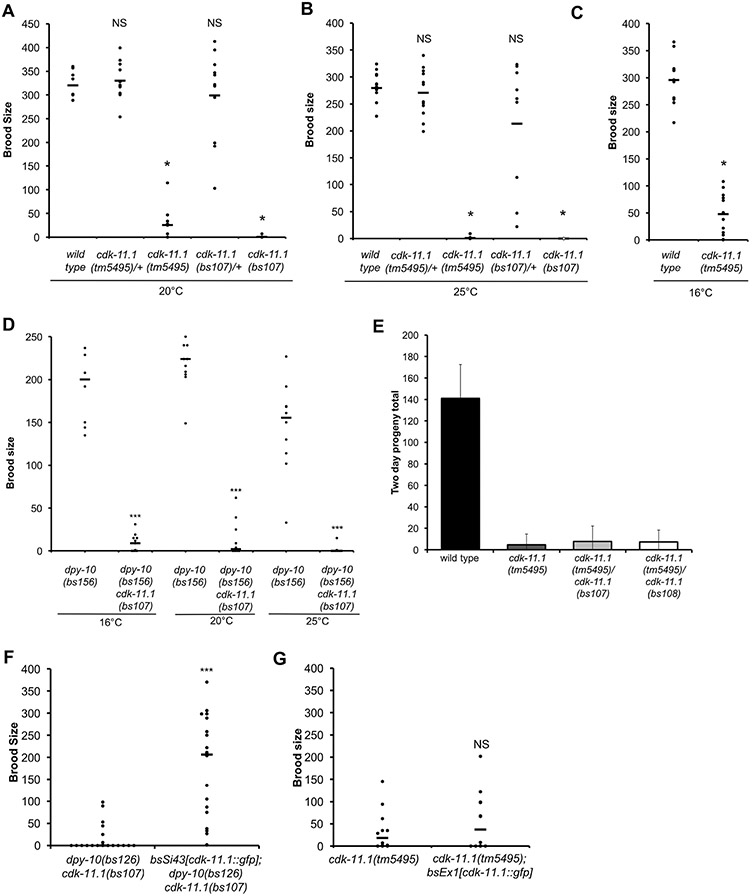
Loss of CDK-11.1 results in low fecundity. (A-C) Brood sizes of wild-type and cdk-11.1 mutant hermaphrodites at various temperatures. (D) Brood sizes of dpy-10(bs156), dpy-10(bs126) cdk-11.1(bs107), and dpy-10(bs141) cdk-11.1(bs108) hermaphrodites at various temperatures. (E) Complementation tests showing that the null alleles cdk-11.1(bs107) and cdk-11.1(bs108) fail to complement the cdk-11.1(tm5495) mutation for the fecundity defect. (F) The integrated cdk-11.1::gfp transgene (bsSi.43) rescues the fecundity defect of cdk-11.1(bs107) hermaphrodites. (G) The same cdk-11.1::gfp transgene does not rescue the fecundity defect when expressed from an extrachromosomal array (bsEx1). (ns = not significant, * p < 0.01, *** p < 0.001).
We next wanted to know if normal fecundity relies on expression of CDK-11.1 in the germ line. The integrated copy of the cdk-11.1::gfp transgene is expressed in both the soma and germ line (Fig. 1D) and rescues the fecundity defect of cdk-11.1(tm5495) hermaphrodites (Fig. 2F). To express the cdk-11.1::gfp transgene primarily in the soma, we created an extrachromosomal array carrying the same cdk-11.1::gfp transgene; extrachromosomal arrays are subject to germ line silencing, and are typically expressed poorly if at all in the germ line (Kelly et al., 1997). As expected, we detected GFP expression in the nuclei of somatic cells, including the somatic gonad, but not in the germ line (data not shown). Interestingly, animals carrying the array did not exhibit rescue of the fecundity defect (Fig. 2G). We conclude that normal fecundity requires expression of CDK-11.1 in the germ line.
3.3. cdk-11.2 is not essential for viability and fecundity
We next turned our attention to cdk-11.2. To determine the consequences of a complete loss of cdk-11.2, we used CRISPR-Cas9 genome editing to completely remove the cdk-11.2 open reading frame (Fig. S1C). Animals homozygous for the cdk-11.2(bs101) deletion allele were viable and appeared healthy in all respects. At 16 °C, 20 °C and 25 °C, cdk-11.2(bs101) hermaphrodites produced a normal brood size and lacked an embryonic lethal phenotype (Fig. S3A and S3B) revealing that despite its strong conservation, cdk-11.2 is not essential for viability or fecundity. Thus, while C. elegans possess two well-conserved cdk-11 genes, loss of only cdk-11.1 affects the reproductive capacity of worms.
3.4. CDK-11.1 functions in both egg and sperm production
The drastic reduction in the brood size of cdk-11.1(tm5495) animals could be caused by a defect in oogenesis, spermatogenesis, or both. To determine which of these possibilities was correct, we carried out a set of genetic crosses. To test for an oocyte defect, we mated wild-type males to cdk-11.1(tm5495) hermaphrodites and then measured the resulting brood sizes. Such crosses did not increase the brood sizes of cdk-11.1(tm5495) hermaphrodites (Table 1). To confirm that mating took place, we labeled male sperm with the DNA dye SYTO-17 prior to setting up crosses. Males were allowed access to hermaphrodites for approximately 24 h after which hermaphrodites were examined for the presence of SYTO-17 positive sperm within the spermatheca. We found that approximately 80% of the mutant hermaphrodites had mated, an efficiency similar to that observed in wild-type control crosses (Fig. S4A). Interestingly, in 89% of the mated hermaphrodites, sperm did not properly target to the spermatheca and were found in the uterus (Fig. S4A). This phenotype suggests that mutant oocytes are defective in production of the positional cue that sperm use to locate the oocytes (Edmonds et al., 2010; Kubagawa et al., 2006) Together, these results reveal that loss of CDK-11.1 results in the production of defective oocytes that lack the ability to signal sperm.
Table 1.
Genetics of the cdk-11.1(tm5495) fecundity defect.
| Matings | Progeny produced (SEM) | n |
|---|---|---|
| cdk-11.1(tm5495) ♀ | 7.8 (3.6) | 17 |
| WT ♂ x cdk-11.1(tm5495) ♀ | 13.4 (4.5) | 25 |
| fog-2(q71) ♀ | 0.0 (0) | 17 |
| WT ♂ x fog-2(q71) ♀ | 485.9 (65.4) | 15 |
| cdk-11.1(tm5495)/+♂ x fog-2(q71) ♀ | 317.8 (57.8) | 23 |
| cdk-11.1(tm5495) ♂ x fog-2(q71) ♀ | 0.7 (0.6) | 20 |
We next set out to determine if the cdk-11.1(tm5495) mutation also impacts male gametes. We therefore set up crosses between cdk-11.1(tm5495); him-8(e1489) males and fog-2(q71) females. The him (high incidence of males) mutation was included to ensure a reliable source of males. Both him-8(e1489) and cdk-11.1(tm5495)/+; him-8(e1489) males sired hundreds of progeny. In contrast, cdk-11.1(tm5495); him-8(e1489) males sired only a few offspring (Table 1). Examination of mating efficiency using SYTO-17-stained males revealed that cdk-11.1(tm5495); him-8(e1489) males successfully mated at a frequency comparable to that of him-8(e1489) control males (71 vs. 79%) (Fig. S4B). However, females mated to cdk-11.1(tm5495); him-8(e1489) males often possessed visibly fewer sperm than those mated to him-8(e1489) or cdk-11.1(tm5495)/+; him-8(e1489) males (Fig. S4B), possibly owing to a defect in sperm production (see below). Importantly, those cdk-11.1(tm5495) sperm that we detected were invariably located in the spermatheca (Fig. S4B), suggesting that at least some mutant sperm are capable of crawling and interpreting the oocyte-derived positional cue. Given that mutant males could successfully mate with wild-type females but not sire progeny, the cdk-11.1(tm5495) mutation also affects sperm function.
That cdk-11.1(tm5495) sperm can travel to the spermatheca but not fertilize eggs suggests that the mutants lack the ability to signal the female to ovulate and/or are defective in one or more or the critical events involved in fertilization. To determine if the sperm defects were solely due to a failure to signal the female germ line to ovulate, we carried out a type of cellular complementation assay. spe-9(eb19) hermaphrodites are sterile as their sperm lack the ability to fertilize but maintain the ability to signal to oocytes and trigger ovulation (Singson et al., 1998). We reasoned that if cdk-11.1(tm5495) sperm were only deficient in signaling but capable of fertilization, that the presence of spe-9(eb19) sperm would allow fertilization by cdk-11.1(tm5495) sperm by providing a signal for ovulation. We therefore mated males of various genotypes with spe-9(eb19) hermaphrodites and scored for the production of progeny. All unmated spe-9(eb19) hermaphrodites did not produce progeny (Table S4). In contrast, when mated to wild-type or cdk-11.1(tm5495)/+ males, all spe-9(eb19) hermaphrodites produced progeny. On the other hand, mating spe-9(eb19) hermaphrodites to cdk-11.1(tm5495) males did not appreciably increase fecundity (Table S4). Thus, cdk-11.1(tm5495) sperm not only lack the ability to signal ovulation, they also appear defective in other downstream events in fertilization.
3.5. CDK-11.1 promotes sperm and egg production
To explore the basis for the fecundity defect of the cdk-11.1(tm5495) mutant, we examined hermaphrodites by differential interference contrast (DIC) microscopy and DAPI staining. The C. elegans gonad is arranged into two elongated tubes, or arms, with a mitotic population of germ cell nuclei at the distal end, nuclei progressing through the early stages of meiotic prophase in the middle, and maturing oocytes in late prophase at the proximal end. One arm extends anteriorly and lies along the right side of the animal and the other extends posteriorly and lies along the left side. DIC imaging of wild-type gonads reveals a well-ordered arrangement of germ line nuclei that transit from mitosis distally into meiotic prophase in the central portion of the gonad. Proximally, maturing oocytes form a row of 7–9 cuboidal shaped cells that terminate next to the spermatheca (Fig. 3A and B).
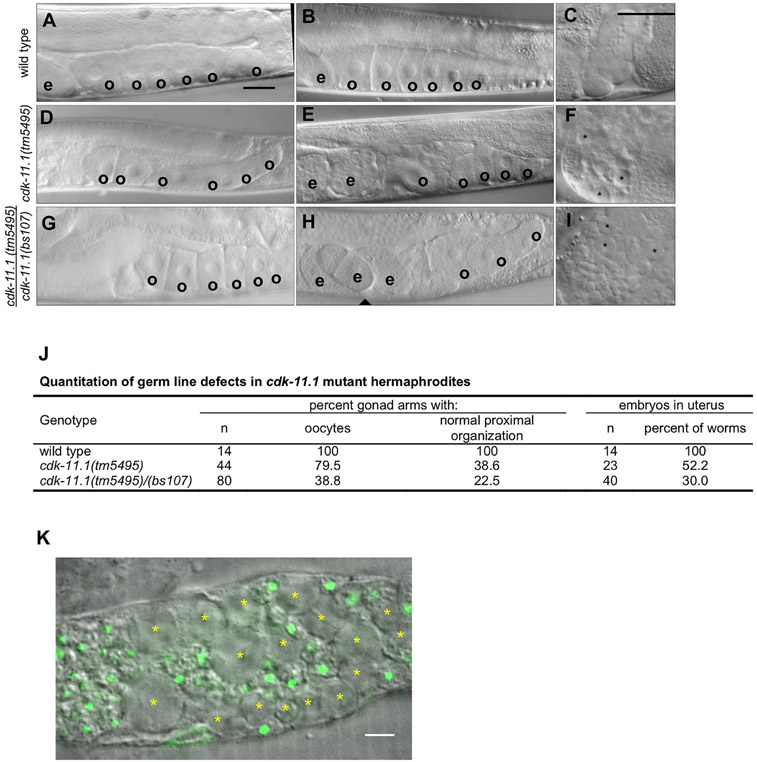
Fig. 3.
CDK-11.1 is required for normal gametogenesis. Hermaphrodites were synchronized at the L1 stage and grown for 3 days at 20 °C before analysis. (A and B) DIC images of wild-type gonads which invariably display a single row of cuboidal oocytes (o) in the proximal end of the germ line, and embryos (e) in the uterus. (C) An image of a wild-type spermatheca. Contained within are a population of sperm with uniform size. (D and E) DIC images of cdk-11.1(tm5495) hermaphrodite gonads. Although some appear normal (panel D) others display fewer oocytes that are less uniform in size and with a disorganized arrangement in the proximal germ line (panel E). (F) An image of a cdk-11.1(tm5495) spermatheca containing some normal looking sperm and some larger cells (asterisks). (G and H) DIC images of cdk-11.1(tm5495)/(bs107) hermaphrodite gonads. Some cdk-11.1(tm5495)/(bs107) trans-heterozygotes display normal-looking oocytes (panel G) while others possess fewer oocytes whose shape, size, and arrangement vary (panel H). (I) An image of a cdk-11.1(tm5495)/(bs107) spermatheca containing normal-sized sperm and some abnormally large cells (asterisks). (J) Quantitation of oocyte defects in various strains. (K) DAPI-stained spermatheca from a cdk-11.1(tm5495) hermaphrodite. Asterisks mark residual bodies which lack DNA (green). Scale bars are 25 μm (panels A–I) and 5 μm (panel K).
Examination of cdk-11.1(tm5495) hermaphrodites revealed that most possess two well-developed gonad arms of normal size. In some of the mutant animals however, one or both arms appeared somewhat shorter or did not lie entirely along one side of the animal. By DIC, the distal germ cell nuclei in the mitotic and pachytene regions look largely normal. Most cdk-11.1(tm5495) hermaphrodites produce oocytes although their number and appearance vary from animal to animal (Fig. 3D and E). We quantified this phenotype and found that 20% of gonad arms lack oocytes altogether, while in another 41%, the oocytes are reduced in number and/or appear smaller and are arranged in a disorganized manner in the proximal gonad (Fig. 3E and J). We also noticed that while cdk-11.1(tm5495) hermaphrodites possess sperm, there are often some large cells present in the spermatheca (Fig. 3F, asterisks). Unlike sperm, these large cells lack DNA (Fig. 3K) and thus are most probably residual bodies, or cell remnants that are produced during spermatogenesis. During the transition to adulthood, residual bodies are normally removed via engulfment by somatic gonadal sheath cells, and as a result, residual bodies are absent in wild-type adult hermaphrodites (Huang et al., 2012). Thus, the cdk-11.1(tm5495) mutation somehow perturbs the mechanism that removes residual bodies.
To confirm that phenotypes observed in the cdk-11.1(tm5495) mutant are due to a loss of CDK-11.1 activity, we also imaged gonads from cdk-11.1(tm5495/bs107) trans-heterozygotes and found a similar but more severe phenotype; approximately 60% of the gonad arms lack oocytes while only about 23% possess normal looking oocytes (Fig. 3G, H, and J). Similarly to cdk-11.1(tm5495) homozygotes, cdk-11(tm5495/bs107) trans-heterozygotes retain residual bodies in the spermatheca (compare Fig. 3F and I). Our data therefore argue that the fecundity and germ line defects are due to a loss of CDK-11.1 activity and that CDK-11.1 plays an important role in the production of both female and male germ cells.
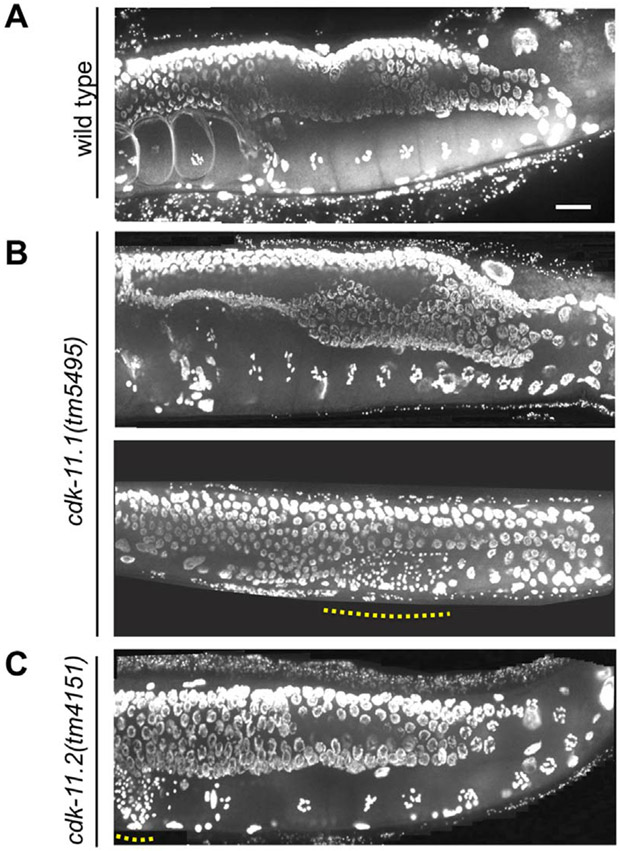
We also examined wild-type and mutant germ lines by DAPI staining. Consistent with DIC imaging, the distal regions of cdk-11(tm5495) hermaphrodite germ lines look similar to their wild-type counterparts (Fig. 4A and B). Some cdk-11(tm5495) germ lines look completely normal and contain a single row of oocytes with highly condensed chromatin (Fig. 4B, top); these oocytes always contain the normal complement of six bivalents. In other cases, cdk-11(tm4595) germ lines lack oocytes despite more distal regions appearing completely normal (Fig. 4B, bottom). Interestingly, some of the hermaphrodite germ lines that lack oocytes contain sperm in the most proximal regions. Overall, we could detect sperm in 92% of cdk-11.1(tm5495) hermaphrodites but in most cases these animals possess far fewer sperm than wild-type counterparts and the sperm are often found outside the spermatheca, either in the uterus or proximal germ line. Similarly, most cdk-11.1(tm5495) and cdk-11.1(tm5495/bs107) males contain fewer sperm than their wild-type counterparts (Fig. S5). Furthermore, 27.6% of cdk-11.1(tm5495) males (n = 29) and 23.1% of cdk-11.1(tm5495/bs107) males (n = 13) lack any detectable sperm. Finally, we examined cdk-11.2(tm4151) germ lines by DAPI staining and found that they invariably appear normal (Fig. 4C). In summary, our analysis indicates that CDK-11.1 plays a similar role in the female and male germ lines, and that in the absence of CDK-11.1 the production of both egg and sperm is severely diminished.
Fig. 4.
Germ-line defects in cdk-11.1(tm5495) hermaphrodites are restricted to the proximal end of the gonad. Animals were plated at 20 °C at the L1 stage, grown for 3 days, then fixed and stained with DAPI. (A) wild type (B) cdk-11.1(tm5495). Dotted line highlights region containing sperm nuclei. (C) cdk-11.2(tm4151). Scale bar is 25 μm.
3.6. The RAS-ERK pathway is not fully activated in cdk-11.1 and cdk-11.2 mutants
In C. elegans, the RAS-ERK pathway has been shown to regulate many aspects of oocyte development, including progression of cells through pachytene, membrane organization and morphogenesis, cell growth, and oocyte meiotic maturation and ovulation (Church et al., 1995; Lee et al., 2007). ERK activity peaks twice within the hermaphrodite germ line: in pachytene where it promotes the transit of cells through meiotic prophase in response to nutrient availability and in diakinesis where it activates oocyte meiotic maturation and ovulation in response to a sperm-derived signal (Arur et al., 2009; Lee et al., 2007; Lopez et al., 2013; Miller et al., 2001).
As show in Fig. 3, cdk-11.1(tm5495) hermaphrodites often exhibit a disorganized proximal germ line with small oocytes, one of the phenotypes observed upon partial loss of mpk-1 (ERK) activity (Lee et al., 2007). Furthermore, CDK-11.2 expression peaks within the regions where RAS-ERK signaling is active (Arur et al., 2009; Lee et al., 2007; Lopez et al., 2013; Miller et al., 2001). Thus, we wondered if the cdk-11 genes might contribute in some way to activation of the RAS-ERK pathway. We therefore stained wild-type and mutant germ lines for activated ERK (e.g. di-phosphorylated or dpMPK1). As previously reported for wild-type germ lines (Arur et al., 2009; Lee et al., 2007; Lopez et al., 2013; Miller et al., 2001), high levels of dpMPK-1 were observed in mid to late pachytene, and in diakinesis (Fig. 5A). Lower, but detectable levels of dpMPK-1 were observed in the intervening region (diplotene). Strikingly, cdk-11.1(tm5495) animals display a strong reduction in the levels of activated ERK in both pachytene and diakinesis (Fig. 5A-C), suggesting that CDK-11.1 might have a role in activation of the RAS-ERK pathway in the germ line. However, as illustrated by the low levels of dpMPK-1 staining in the female strain fog-2(q71) (Fig. 5A-C), the absence of functional sperm can also lead to loss of dpMPK-1 staining. Thus, the reduction in dpMPK-1 levels in cdk-11.1(tm5495) hermaphrodites could reflect a direct role for CDK-11.1 in this process or else might be due to an indirect effect such as a defect in sperm signaling.
As noted above, CDK-11.2 expression and ERK activation both peak within the same region of the germ line (Fig. 5A). Although cdk-11.1 null animals exhibit normal fecundity, we decided to investigate if the loss of CDK-11.2 had any effect on ERK activation. To our surprise, we detected a highly statistically significant reduction in the intensity of dpMPK-1 staining during pachytene and a smaller and less significant reduction during diakinesis in cdk-11.2(bs101) hermaphrodites (Fig. 5A-C). Since cdk-11.2(bs101) mutants possess normal fecundity, it seems unlikely that the reduction in ERK activation is due to a defect in sperm signaling. Furthermore, it is important to note that although cdk-11.1(tm5495) and cdk-11.2(bs101) hermaphrodites exhibit a similar reduction in late pachytene dpMPK-1 levels, only cdk-11.1(tm5495) mutants exhibit a fecundity defect. This suggests that the reduction in activated ERK levels that we observed during late pachytene is not sufficient to account for the fecundity defect of cdk-11.1(tm5495) hermaphrodites. In summary, our data indicate that CDK-11 activity contributes directly or indirectly to activation of the RAS-ERK pathway in the germ line but that loss of CDK-11.1 results in additional defects that lead to near complete sterility.
3.7. CDK-11.1 is also required at later stages of oogenesis for fertilization competency
Our cytological analysis indicates that nearly 50% of cdk-11.1(tm5495) germ lines appear completely normal. Despite this, all cdk-11.1(tm5495) hermaphrodites have very low fecundity (Fig. 2A) and the low fecundity can’t be rescued by mating with wild-type males (Table 1). This indicates that in addition to promoting oocyte production, CDK-11.1 is involved in the late stages of oocyte development. We therefore sought to determine if cdk-11.1-deficient animals exhibit any defects in oocyte meiotic progression, and stained germ lines for DNA and meiotic progression markers such as phospho-histone H3 (pH3) and the nucleolar protein NOP1/fibrillarin. In wild-type animals, the phosphorylation of serine 10 on histone H3 (pH3) increases as oocytes mature (Hsu et al., 2000). In wild-type animals, we found that the most proximal (−1) oocyte stained positive for pH3 ~ 90% of the time, while the −3 oocyte stained only ~ 50% of the time (Fig. S6A and S6B). While some cdk-11.1(tm5495) germ lines could be somewhat challenging to score due to disorganization within the proximal region, we found that the oocytes in the mutants still followed the same general trend; the −1 oocyte stained positive for pH3 90% of the time while the −3 oocyte stained 31% of the time. NOP1/ fibrillarin, whose levels decrease as oocytes mature, also behaved similarly in wild-type and cdk-11.1(tm5495) hermaphrodites; in both control and mutant animals, roughly 80% of −3 oocytes stained positive for NOP1/fibrillarin while staining was absent in virtually all −1 oocytes (Fig. S6A and S6C). Thus, the cdk-11.1(tm5495) mutation does not appreciably affect the events associated with late meiotic progression. Our data therefore is more consistent with a role for CDK-11.1 in the functional interactions between oocytes and sperm.
3.8. CDK-11.1 and CDK-11.2 function redundantly through development and together are required for viability
Next, we explored the possibility of functional redundancy between the two cdk-11 paralogs. Interestingly, we found that in cdk-11.1(tm5495) germ lines, the domain of peak CDK-11.2 expression expanded dramatically to include all germ line nuclei, similar to the expression pattern of CDK-11.1 (Fig. 5A). Consistent with this, we found by quantitative immunoblotting that whole worm lysates prepared from cdk-11(null) strains contained twice the level of CDK-11.2 protein as that found in wild-type lysates (Fig. 5D). We reasoned that this might represent a compensatory mechanism that could mitigate the effects of a loss of CDK-11.1. We therefore sought to determine the consequences of a complete loss of CDK-11 activity in the worm and attempted to construct a cdk-11.1(tm5495); cdk-11.2(bs101) double mutant line. However, we failed to recover a viable double mutant adult following genetic crosses. We then constructed a strain homozygous for cdk-11.2(bs101) that carried the cdk-11.1(tm5495) allele over the chromosome II balancer mnC1 (genotype: cdk-11.1(tm5495)/mnc1::gfp; cdk-11.2(bs101)). One quarter of the F1 progeny of such animals should be homozygous for both gene deletions. Interestingly, we did not detect cdk-11.1(tm5495); cdk11.2(bs101) double mutants among the adult progeny, but instead found that approximately one quarter of the progeny die during early larval development (Fig. 6A). We conclude that functional redundancy exists between the two CDK-11 proteins and that CDK-11 activity is not just required for reproductive fitness but is also essential for proper organismal development and viability.
Fig. 6.
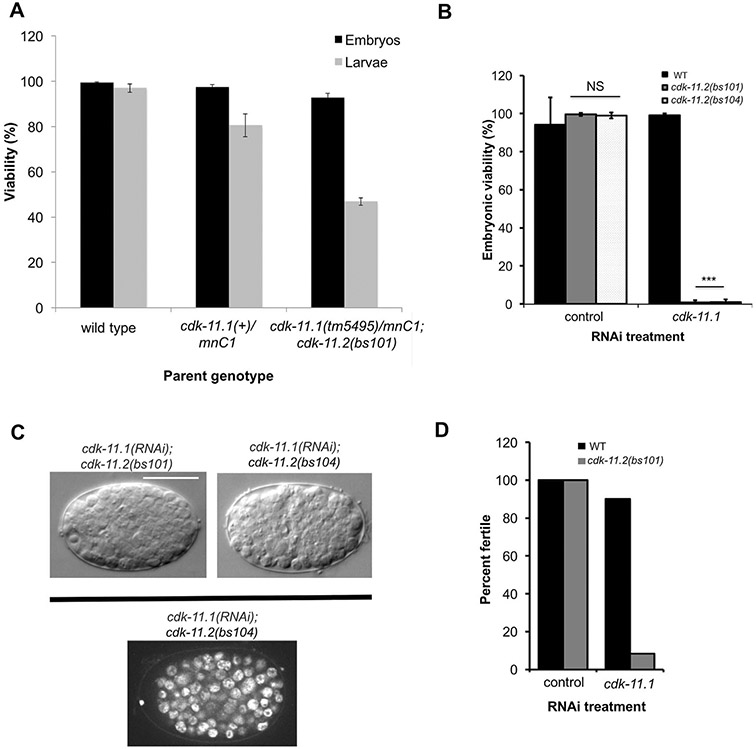
CDK-11.1 and CDK-11.2 function in a partially redundant manner. (A) The progeny of hermaphrodites of the indicated genotype were scored for embryonic and larval lethality. Note that cdk-11.1(tm5495)/mnC1; cdk-11.2(bs101) mothers produce about 25% more inviable larvae than +/mnC1 controls. (B) L4 hermaphrodites of the indicated genotypes were fed dsRNA targeting cdk-11.1 or the nonessential gene smd-1 as a negative control. Loss of embryonic viability was only observed when CDK-11.1 was depleted in animals harboring a cdk-11.2 null allele. (C) DIC Images of cdk-11.1(RNAi); cdk-11.2(null) embryos displaying the terminal phenotype (top) and an image of a DAPI-stained cdk-11.1(RNAi); cdk-11.2(bs104) embryo (bottom). Embryos arrest with approximately 130 cells but do not show any signs of gross aneuploidy, typical of mutants with cell division failures. Scale bar is 25 μm. (D) L1 hermaphrodites of the indicated genotypes were fed dsRNA targeting smd-1 or cdk-11.1. Loss of fertility was only observed when CDK-11.1 was depleted in animals harboring a cdk-11.2 null allele. The cdk-11.1(RNAi); cdk-11.2(null) animals display a nearly complete loss of fertility.
To uncover any additional redundant roles for the two CDK-11 proteins, we used RNAi to deplete CDK-11.1 at different points in the development of a cdk-11.2 null mutant. When we exposed wild-type L4 hermaphrodites to cdk-11.1 (RNAi) or cdk-11.2 null mutants to control RNAi, animals developed into fertile adults that produced viable progeny (Fig. 6B). In contrast, when cdk-11.2 null L4 hermaphrodites were exposed to cdk-11.1(RNAi), animals developed into fertile adults but produced broods almost entirely composed of dead embryos. Microscopy of the arrested embryos using DIC and DAPI imaging revealed that the arrest occurs approximately at 130 cells and that most, if not all, of the cells appeared to be arrested in interphase (Fig. 6C). Such embryos did not exhibit any evidence of cell or nuclear division failure such as enlarged cells or gross aneuploidy, suggesting that CDK-11 activity is not required for the proper operation of critical cell division machinery. We next exposed the wild type or cdk-11.2(bs101) mutants to control or cdk-11.1(RNAi) starting at the L1 stage. Neither the wild type nor cdk-11.2(bs101) mutants exhibited any obvious fertility defects following control RNAi. Similarly, treatment of wild-type animals with cdk-11.1(RNAi) only marginally affected fertility. In contrast, when exposed to cdk-11.1(RNAi) at the L1 stage, nearly all cdk-11.2(bs101) animals developed into sterile adults (Fig. 6D). For simplicity, we refer to these animals as cdk-11 L1 RNAi hermaphrodites. Our results indicate that while CDK-11.1 has the predominant germ-line role, CDK-11.2 also functions in the germ line and thus, the two CDK-11 proteins exhibit functional redundancy throughout development.
To determine what germ line processes CDK-11.2 might contribute to, we examined the germ lines of cdk-11 L1 RNAi hermaphrodites. Interestingly, while the defects present in these animals were similar to those of hermaphrodites lacking only CDK-11.1, there were some differences. Similar to cdk-11.1(tm5495)/(bs107) mutants, cdk-11 L1 RNAi animals exhibited disorganization of the cells in the proximal germ line; in these animals, oocytes and sperm were almost always present but oocytes often appeared smaller and failed to form a single row next to the spermatheca (Fig. 7A and C). However, in contrast to animals lacking cdk-11.1 only, cdk-11 L1 RNAi animals possessed oocytes in more distal regions of the germ line suggesting precocious oocyte development. These results suggest that different combinations of the two CDK-11 proteins have different effects on gametogenesis and indicate that while the two proteins might cooperate, they might on their own also possess distinct molecular activities.
Fig. 7.
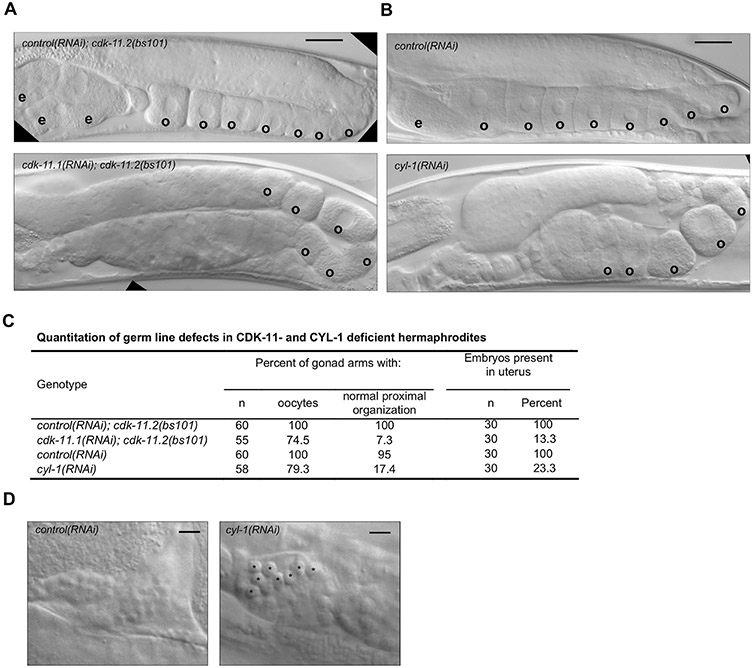
Germ line defects in cdk-11 null and cyl-1(RNAi) mutants. (A) DIC images of cdk-11.2(bs101) hermaphrodites treated with smd-1(RNAi) (top) as a negative control and cdk-11.1 (RNAi) (bottom), beginning at the L1 stage. cdk-11.2(bs101) hermaphrodites treated with smd-1 (RNAi) look normal with an ordered row of oocytes within the proximal end of the gonad. The cdk-11.1(RNAi); cdk-11.2(bs101) animals have irregularly sized and shaped oocytes which sometimes appear to form precociously. (B) Treatment of WT animals with cyl-1 RNAi from the L1 stage produces a disordered proximal germ line phenotype similar to that of cdk-11.1(RNAi); cdk-11.2(bs101) hermaphrodites. cyl-1(RNAi) animals exhibit premature cellularization in the loop region of the gonad and oocytes of abnormal shape, size and arrangement in the most proximal region. The oocytes also appear compacted, likely due to failed ovulation. (C) Quantitation of DIC phenotypes. (D) DIC images of spermathecae from smd-1(RNAi)-treated and cyl-1(RNAi)-treated animals. Note that the spermathecae of cyl-1(RNAi) animals possess unusually large cells (possibly residual bodies). Scale bars are 25 μm (panel A–B) and 5 μm (panel D).
3.9. C. elegans Cyclin-L homolog
Cyclin-dependent kinases are activated by binding to a cyclin partner and in the case of CDK-11, the cognate binding partner is Cyclin L (Berke et al., 2001; Dickinson et al., 2002; Loyer et al., 2008; Yang et al., 2004). The cyl-1 gene encodes the sole C. elegans Cyclin L homolog, a 526-amino-acid protein with a single CYCLIN box found in the N-terminal half of the protein (Fig. 8A) (Hajdu-Cronin et al., 2004). Additionally, there is a serine- and arginine-rich region near the C-terminus (not shown); this so-called RS domain is shared between some Cyclin L proteins and the SR family of splicing factors (Berke et al., 2001; Dickinson et al., 2002; Shepard and Hertel, 2009; Yang et al., 2004). C. elegans Cyclin L and the human Cyclin-L1 isoform 1 share 38% identity, with the highest conservation occurring within and near the CYCLIN box. To explore the possibility that CDK-11 function in the C. elegans germ line is Cyclin L-dependent we used CRISPR-Cas9 genome engineering to precisely delete the cyl-1 gene (Fig. 8A). Although we were able to generate an animal heterozygous for the deletion allele cyl-1(bs106), we were unable to recover cyl-1(bs106) homozygotes among its progeny, indicating that cyl-1 is an essential gene. We therefore balanced the cyl-1(bs106) allele over a dpy-11(e224) unc-76(e911) chromosome. We then analyzed the progeny of such animals and found that about a quarter of the progeny failed to develop past the L2 larval stage (Fig. 8B) and that homozygous cyl-1(bs106) animals were absent among the progeny that had developed to adulthood. Thus, CYL-1, like CDK-11, is required for larval development.
Fig. 8.
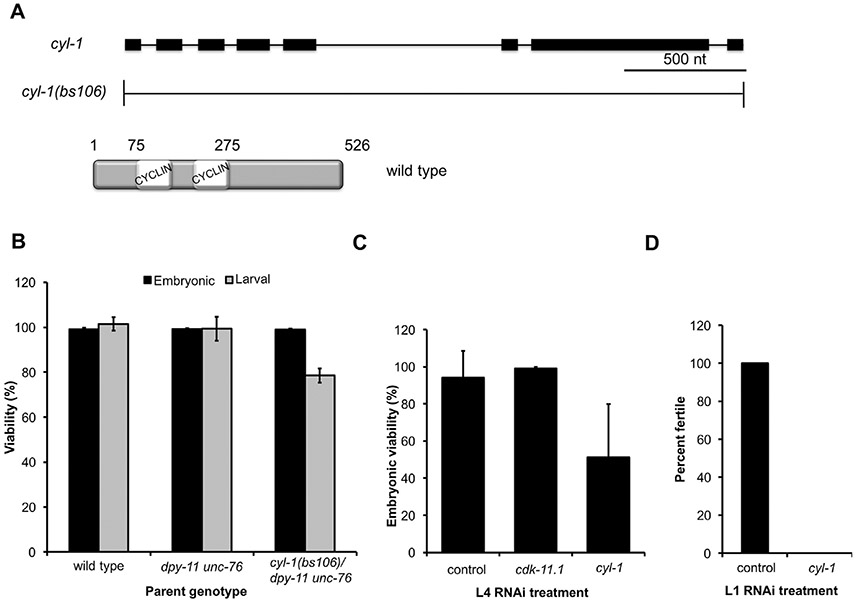
Loss of CYL-1 phenocopies the various defects of cdk-11 mutants. (A) Schematic of the cyl-1 gene and the null allele cyl-1(bs106). The product of the cyl-1 gene is a Cyclin L protein containing a single CYCLIN box composed of two cyclin folds. (B) The progeny of hermaphrodites of the indicated genotype were scored for embryonic and larval lethality. Note that the homozygous cyl-1(bs106) progeny of a heterozygous mother die during larval development. (C) Percent embryonic viability of the progeny of wild-type hermaphrodites subjected to various RNAi treatments beginning at the L4 stage. smd-1(RNAi) serves as a negative control. (D) Percent of wild-type hermaphrodites that develop to fertile adults when exposed to smd-1(RNAi) or cyl-1(RNAi) beginning at the L1 stage.
The larval arrest phenotype of cyl-1(bs106) mutants precluded our ability to determine if CYL-1 plays a role in the germ line. We therefore utilized cyl-1(RNAi) to deplete CYL-1 in animals that had developed beyond the early larval arrest point of the cyl-1(b106) deletion mutant. When we depleted cyl-1 by exposing L4 larvae to cyl-1 dsRNA-expressing bacteria, the animals developed into fertile hermaphrodites but produced many dead embryos (Fig. 8C), reminiscent of the embryonic lethal phenotype seen following inhibition of both CDK-11.1 and CDK11.2 in L4 larvae (Fig. 6B). Finally, when RNAi of cyl-1 was initiated at the L1 stage, animals developed to adulthood but exhibited a complete loss of fertility (Fig. 8D). Examination of the germ line of cyl-1(RNAi) animals revealed a phenotype similar to that of cdk-11.1 11.1; cdk-11.2 deficient animals (Fig. 7A and B). That is, while some hermaphrodites lacked oocytes altogether (~ 20%), others possessed a few oocytes that were abnormal in both size and arrangement within the proximal gonad (Fig. 7B and C). In particular, cyl-1(RNAi) resulted in the appearance of oocytes in more distal regions of the germ line, a phenotype similar to that of animals lacking both CDK-11.1 and CDK.11.2 (Fig. 7A). Further, almost half of the cyl-1(RNAi) animals contained unusually large cells in the spermatheca (Fig. 7D, black asterisks), reminiscent of the residual body elimination defect exhibited by cdk-11.1 mutants (Fig. 3F and I, black asterisk). We conclude that loss of CYL-1 phenocopies a loss of CDK-11, suggesting that as in other species, CDK-11 function in worms is dependent upon its association with a CYCLIN L partner.
4. Discussion
Prior work in vertebrate and yeast cells has shown that CDK11 functions in a multitude of roles; some of these can be classified as gene expression roles (i.e. transcriptional initiation, splicing, 3’end processing) while others can be classified as cell division roles (i.e. mitotic progression, centrosome maturation, spindle assembly). Given CDK11's diverse roles and the fact that CDK11 function has been investigated almost entirely in tissue culture cells or unicellular organisms, we were interested to explore the function of CDK11 in a developing multicellular animal. Along these lines, one prior study examined developmental defects in CDK11 knockout mice and found that knockout homozygotes arrested early in embryonic development with obvious cell division defects (Li et al., 2004). Unfortunately, this early arrest precluded the study of CDK11 function at later points in development, including postembryonic development and germ line development.
Our work demonstrates that CDK11 is essential at many points in the development of a multicellular animal. Using a combination of RNAi and gene knockouts, we have shown that CDK-11.1 and CDK-11.2 function together during embryogenesis, larval development, and within the germ line of a fully developed adult. Further, we find that both the cdk-11.1 and cdk-11.2 genes are broadly transcribed in the worm throughout development and that CDK-11.1 protein is detectable in a broad range of cells from embryos to adults. All together, these results suggest that CDK-11 may provide a basic function required in a broad-range of cells for their proper functioning.
One question that arises from our work concerns the predominant role of CDK-11.1 in the germ line. The cdk-11.1 gene is located on an autosome while its paralog cdk-11.2 is X linked. There exists in C. elegans a number of autosomal/X-linked duplicated gene pairs. Functional analysis of several of these gene pairs has indicated that they have tissue-specific roles, with the autosomal copy providing an essential germ line function and the activity of the X-linked copy biased toward the soma (Maciejowski et al., 2005). The lack of an essential germ-line role for the X-linked paralogs is thought to be due to X chromosome silencing that takes place within the germ line (Fong et al., 2002; Kelly et al., 2002). While our transcriptional cdk-11.2 reporter showed strong germ line expression (Fig. 1C), this reporter was integrated on chromosome II, thus liberating it from potential X-linked transcriptional repression. Nonetheless, we detected expression of endogenous CDK-11.2 throughout the hermaphrodite germ line (Fig. 1E, and S2) and uncovered evidence of a redundant germ-line role for both cdk-11 genes (Fig. 6D). Thus, while we have demonstrated that both cdk-11.1 and cdk-11.2 operate in the germ line and soma, like other autosomal/X-linked gene pairs, germ line development depends predominantly upon the autosomal copy.
When considering the various phenotypes exhibited by worms depleted of CDK-11, it appears that developing tissues are the most sensitive to a loss of this cyclin-dependent kinase. Depending on the time at which we depleted CDK-11 or Cyclin L, we could observe developmental defects during the proliferative stage of embryogenesis, during larval development, and in the mature germ line, which is the only tissue in the adult in which cells continue to develop. In contrast, although some gonad arms appeared misshapen or improperly positioned in cdk-11.1 mutants, the adult soma appears largely unaffected by a loss of CDK-11.1. Thus, perhaps not surprisingly, CDK-11 activity appears most important in those cells that are traversing the cell cycle but is less important, and possibly dispensable, in most terminally differentiated cells.
In vertebrates, CDK11 has been implicated in several essential cell division roles. Since dividing cells in the worm also show a requirement for CDK-11, it might seem that C. elegans CDK-11s also possess these cell division roles. However closer inspection of our data suggests that this is most likely not the case. First, cell division phenotypes were not observed in animals compromised for CDK-11.1 and/or CDK-11.2. In particular, cdk-11.1(RNAi); cdk-11.2(null) embryos arrest at the 130-cell stage but do not exhibit evidence of cell or nuclear division failure. That is, all cells are approximately equal in size with no signs of gross aneuploidy (Fig. 6C), as might be expected if defects in centrosome maturation, centriole duplication, spindle assembly or chromatid cohesion had occurred at even low to moderate frequencies. Furthermore, in contrast to the situation in cdk11-null mice embryos where blastomeres arrest in mitosis, loss of CDK-11 activity in C. elegans embryos results in an interphase arrest (Fig. 6C), suggesting the absence of mitotic defects. Likewise, loss of CDK-11.1 and/or CDK-11.2 does not appear to grossly affect the mitotic (distal) region of the germ line as the only defects we observed were limited to the late stages of oogenesis. Second, we have only detected the long p110 forms of the enzyme (Fig. S2) and not the shorter p58 and p46 forms that are responsible for most of the cell division and apoptotic roles of vertebrate and yeast CDK11s respectively (Beyaert et al., 1997; Franck et al., 2011; Hu et al., 2007; Kong et al., 2009; Lahti et al., 1995; Petretti et al., 2006; Rakkaa et al., 2014; Shi et al., 2003, 2009). Third, at a subcellular level, we detect CDK-11.1 and CDK-11.2 in the nucleus but not at the centrosome or on microtubules as observed in Human and Xenopus cells (Franck et al., 2011; Petretti et al., 2006; Yokoyama et al., 2008). Thus, in worms CDK-11 might strictly function to regulate gene expression.
Within the germ line, CDK-11 seems to be particularly important for the late stages of gametogenesis. In hermaphrodites lacking CDK-11.1, both CDK-11.1 and CDK-11.2, or Cyclin L, defects are not observed in the distal mitotic region, the transition zone, or the early stages of meiotic prophase I. Similarly, the distal region of the male germ line also does not appear to be affected by a loss of CDK-11. In the hermaphrodite germ line, loss of CDK-11 or Cyclin L affects the production and development of oocytes. Likewise, cdk-11.1 mutant males produce fewer sperm than their wild-type counterparts (Fig. S5) and the sperm that are produced are not completely functional. Thus, in both sexes, CDK-11, likely in association with Cyclin L, appears to be important for gamete production and the acquisition of fertilization competency.
Our work also shows that in cdk-11.1 mutants the RAS-ERK pathway is not fully activated. This defect might contribute to at least some of the phenotypes observed in cdk-11.1 mutants. The RAS-ERK pathway plays a central role in promoting oocyte production in response to environmental stimuli (Lopez et al., 2013; Miller et al., 2001) and is also required for proper oocyte growth, organization, and differentiation (Church et al., 1995; Lee et al., 2007). Furthermore, sperm production is nearly absent in males that lack RAS-ERK signaling (Lee et al., 2007). We observed similar phenotypes among the CDK-11- and CYL-1-deficient animals where oocytes are often absent (Fig. 3J), or of abnormal size and arrangement in the proximal gonad (Fig. 7A and B), and where sperm production is diminished (Fig. S5). One possibility is that CDK-11 plays a direct role in activating RAS-ERK signaling. This is consistent with the expression pattern of CDK-11.2, whose peak levels coincide with the region of activated ERK. However, our results do not rule out the alternative possibility that the loss of CDK-11 activity indirectly affects RAS-ERK signaling. For instance, the diminished levels of RAS-ERK signaling in the cdk-11 mutants could result from defects in the earlier stages of germ cell development that went undetected in our study. Further analysis will be needed to more clearly define the role of CDK-11 in RAS-ERK signaling.
While we do not yet understand how CDK-11 might contribute to activation of the RAS-ERK pathway, it is possible that CDK-11 together with Cyclin L regulates expression of one or more factors required for RAS-ERK signaling. This would fit with our notion that worm CDK-11 lacks cell division roles and thus only functions to regulate gene expression. This model is also consistent with our finding that CDK-11 and Cyclin L likely function together in the germ line and prior work that demonstrated a role for Cyclin L in regulating gene expression in the worm (Hajdu-Cronin et al., 2004). What genes might be regulated by CDK-11-Cyclin L is an open question, and future work aimed at identifying the relevant targets will be needed.
Finally, we have also found that CDK-11 is required for the late stages of oogenesis and spermatogenesis. About 50% of cdk-11.1(tm5495) gonads have normal-appearing oocytes but these oocytes are not capable of being fertilized by wild-type sperm. One reason for why cdk-11(tm5495) oocytes might fail to achieve fertilization competency is that they seemingly fail to properly signal the sperm. Normally, when sperm are transferred to the hermaphrodite during mating, they target to the spermatheca in response to a guidance cue emitted by the oocytes (Hoang et al., 2013; Kubagawa et al., 2006). In the absence of this guidance signal, sperm tend to remain in the uterus. Our finding that wild-type sperm do not properly target to the spermathecae of mutant hermaphrodites suggests that cdk-11.1(tm5495) oocytes are defective in the production or emission of this signal. Similarly, cdk-11.1(tm5495) sperm can crawl and target to the spermatheca but fail to achieve fertilization competency. Wild-type sperm engage in signaling of their own by releasing major sperm protein which induces oocyte maturation and ovulation (Miller et al., 2001). Thus, it is possible that cdk-11.1(tm5495) mutant sperm, like mutant oocytes, fail to properly signal their partner.
In summary, we have defined a new role for CDK-11 and Cyclin L in the production and development of gametes in C. elegans. At a molecular level, CDK-11-Cyclin L appears to contribute to oocyte (and possibly sperm) production by ensuring full activation of the ERK-RAS pathway. It is also clear from our data that CDK-11-Cyclin L is required for later aspects of gamete development. As CDK-11 is not required for cell division in worms, the C. elegans germ line should provide a less complicated system for dissecting the role of CDK-11-Cyclin L in the differentiation and development of gametes.
Supplementary Material
Acknowledgements
We would like to thank Aimee Jaramillo-Lambert for technical suggestions, Alexandre Paix and Geraldine Seydoux for sharing reagents and guidance on gene editing techniques, and Andy Golden and Swathi Arur for thoughtful comments on the manuscript. We would also like to thank Shohei Mitani and the National Bioresource Project of Japan for providing the original cdk-11 deletion alleles. Some C. elegans strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This research was supported by the Intramural Research Program of the National Institutes of Health and the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ydbio.2018.06.006.
References
- Ariza ME, Broome-Powell M, Lahti JM, Kidd VJ, Nelson MA, 1999. Fas-induced apoptosis in human malignant melanoma cell lines is associated with the activation of thep34(cdc2)-related PITSLRE protein kinases. J. Biol. Chem 274, 28505–28513. [DOI] [PubMed] [Google Scholar]
- Arribere JA, Bell RT, Fu BXH, Artiles KL, Hartman PS, Fire AZ, 2014. Efficient marker-free recovery of custom genetic modifications with CRISPR/Cas9 in Caenorhabditis elegans. Genetics 198, 837–846. 10.1534/genetics.114.169730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arur S, Ohmachi M, Nayak S, Hayes M, Miranda A, Hay A, Golden A, Schedl T, 2009. Multiple ERK substrates execute single biological processes in Caenorhabditis elegans germ-line development. Proc. Natl. Acad. Sci. USA 106, 4776–4781. 10.1073/pnas.0812285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barna M, Pusic A, Zollo O, Costa M, Kondrashov N, Rego E, Rao PH, Ruggero D, 2008. Suppression of Myc oncogenic activity by ribosomal protein haploinsufficiency. Nature 456, 971–975. 10.1038/nature07449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke JD, Sgambato V, Zhu PP, Lavoie B, Vincent M, Krause M, Hyman SE, 2001. Dopamine and glutamate induce distinct striatal splice forms of Ania-6, an RNA polymerase II-associated cyclin. Neuron 32, 277–287. [DOI] [PubMed] [Google Scholar]
- Beyaert R, Kidd VJ, Cornelis S, Van de Craen M, Denecker G, Lahti JM, Gururajan R, Vandenabeele P, Fiers W, 1997. Cleavage of PITSLRE kinases by ICE/CASP-1 and CPP32/CASP-3 during apoptosis induced by tumor necrosis factor. J. Biol. Chem 272, 11694–11697. [DOI] [PubMed] [Google Scholar]
- Cao L, Chen F, Yang X, Xu W, Xie J, Yu L, 2014. Phylogenetic analysis of CDK and cyclin proteins in premetazoan lineages. BMC Evol. Biol 14, 10. 10.1186/1471-2148-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J, 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol 17, 540–552. [DOI] [PubMed] [Google Scholar]
- Chen BB, Glasser JR, Coon TA, Mallampalli RK, 2011. FBXL2 is a ubiquitin E3 ligase subunit that triggers mitotic arrest. Cell Cycle 10, 3487–3494. 10.4161/cc.10.20.17742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Yan J, Sun Q, Yao LY, Jian YZ, Lu JQ, Gu JX, 2006. Induction of apoptosis by p110C requires mitochondrial translocation of the proapoptotic BCL-2 family member BAD. FEBS Lett. 580, 813–821. 10.1016/j.febslet.2005.12.097. [DOI] [PubMed] [Google Scholar]
- Choi H-H, Choi H-K, Jung SY, Hyle J, Kim B-J, Yoon K, Cho E-J, Youn H-D, Lahti JM, Qin J, Kim S-T, 2014. CHK2 kinase promotes pre-mRNA splicing via phosphorylating CDK11(p110). Oncogene 33, 108–115. 10.1038/onc.2012.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church DL, Guan KL, Lambie EJ, 1995. Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 121, 2525–2535. [DOI] [PubMed] [Google Scholar]
- Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R, 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol. Cell 5, 597–605. [DOI] [PubMed] [Google Scholar]
- Dickinson LA, Edgar AJ, Ehley J, Gottesfeld JM, 2002. Cyclin L is an RS domain protein involved in pre-mRNA splicing. J. Biol. Chem 277, 25465–25473. 10.1074/jbc.M202266200. [DOI] [PubMed] [Google Scholar]
- Drogat J, Migeot V, Mommaerts E, Mullier C, Dieu M, van Bakel H, Hermand D, 2012. Cdk11-CyclinL controls the assembly of the RNA polymerase II mediator complex. Cell Rep. 2, 1068–1076. 10.1016/j.celrep.2012.09.027. [DOI] [PubMed] [Google Scholar]
- Edmonds JW, Prasain JK, Dorand D, Yang Y, Hoang HD, Vibbert J, Kubagawa HM, Miller MA, 2010. Insulin/FOXO signaling regulates ovarian prostaglandins critical for reproduction. Dev. Cell 19, 858–871. 10.1016/j.devcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong Y, Bender L, Wang W, Strome S, 2002. Regulation of the different chromatin states of autosomes and X chromosomes in the germ line of C. elegans. Science 296, 2235–2238. 10.1126/science.1070790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck N, Montembault E, Rome P, Pascal A, Cremet J-Y, Giet R, 2011. CDK11(p58) is required for centriole duplication and Plk4 recruitment to mitotic centrosomes. PLoS One 6. 10.1371/journal.pone.0014600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, 2015. Transposon-assisted genetic engineering with Mos1-mediated single-copy insertion (MosSCI). Methods Mol. Biol 1327, 49–58. 10.1007/978-1-4939-2842-2_5. [DOI] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM, 2012. Improved Mos1-mediated transgenesis in C. elegans. Nat. Methods 9, 117–118. 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdu-Cronin YM, Chen WJ, Sternberg PW, 2004. The L-type cyclin CYL-1 and the heat-shock-factor HSF-1 are required for heat-shock-induced protein expression in Caenorhabditis elegans. Genetics 168, 1937–1949. 10.1534/genetics.104.028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang HD, Prasain JK, Dorand D, Miller MA, 2013. A heterogeneous mixture of F-series prostaglandins promotes sperm guidance in the Caenorhabditis elegans reproductive tract. PLoS Genet. 9, e1003271–e1003276. 10.1371/journal.pgen.1003271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Sun ZW, Li X, Reuben M, Tatchell K, Bishop DK, Grushcow JM, Brame CJ, Caldwell JA, Hunt DF, Lin R, Smith MM, Allis CD, 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102, 279–291. [DOI] [PubMed] [Google Scholar]
- Hu D, Mayeda A, Trembley JH, Lahti JM, Kidd VJ, 2003. CDK11 complexes promote pre-mRNA splicing. J. Biol. Chem 278, 8623–8629. 10.1074/jbc.M210057200. [DOI] [PubMed] [Google Scholar]
- Hu D, Valentine M, Kidd VJ, Lahti JM, 2007. CDK11(p58) is required for the maintenance of sister chromatid cohesion. J. Cell Sci 120, 2424–2434. 10.1242/jcs.007963. [DOI] [PubMed] [Google Scholar]
- Huang J, Wang H, Chen Y, Wang X, Zhang H, 2012. Residual body removal during spermatogenesis in C. elegans requires genes that mediate cell corpse clearance. Development 139, 4613–4622. 10.1242/dev.086769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J, 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237. 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Schaner CE, Dernburg AF, Lee M-H, Kim SK, Villeneuve AM, Reinke V, 2002. X-chromosome silencing in the germline of C. elegans. Development 129, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, Xu S, Montgomery MK, Fire A, 1997. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Gan H, Hao Y, Cheng C, Jiang J, Hong Y, Yang J, Zhu H, Chi Y, Yun X, Gu J, 2009. CDK11p58 phosphorylation of PAK1 Ser174 promotes DLC2 binding and roles on cell cycle progression. J. Biochem 146, 417–427. 10.1093/jb/mvp089. [DOI] [PubMed] [Google Scholar]
- Kubagawa HM, Watts JL, Corrigan C, Edmonds JW, Sztul E, Browse J, Miller MA, 2006. Oocyte signals derived from polyunsaturated fatty acids control sperm recruitment in vivo. Nat. Cell Biol 8, 1143–1148. 10.1038/ncb1476. [DOI] [PubMed] [Google Scholar]
- Lahti JM, Xiang J, Heath LS, Campana D, Kidd VJ, 1995. PITSLRE protein kinase activity is associated with apoptosis. Mol. Cell. Biol 15, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M-H, Ohmachi M, Arur S, Nayak S, Francis R, Church D, Lambie E, Schedl T, 2007. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177, 2039–2062. 10.1534/genetics.107.081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Inoue A, Lahti JM, Kidd VJ, 2004. Failure to proliferate and mitotic arrest of CDK11(p110/p58)-null mutant mice at the blastocyst stage of embryonic cell development. Mol. Cell. Biol 24, 3188–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AL III, Chen J, Joo H-J, Drake M, Shidate M, Kseib C, Arur S, 2013. DAF-2 and ERK couple nutrient availability to meiotic progression during Caenorhabditis elegans oogenesis. Dev. Cell 27, 227–240. 10.1016/j.devcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyer P, Trembley JH, Grenet JA, Busson A, Corlu A, Zhao W, Kocak M, Kidd VJ, Lahti JM, 2008. Characterization of cyclin L1 and L2 interactions with CDK11 and splicing factors: influence of cyclin L isoforms on splice site selection. J. Biol. Chem 283, 7721–7732. 10.1074/jbc.M708188200. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, Ahn JH, Cipriani PG, Killian DJ, Chaudhary AL, Lee JI, Voutev R, Johnsen RC, Baillie DL, Gunsalus KC, Fitch DHA, Hubbard EJA, 2005. Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics 169, 1997–2011. 10.1534/genetics.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, 2014. Cyclin-dependent kinases. Genome Biol. 15, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JG, Liu Y, Williams CW, Smith HE, O'Connell KF, 2016. The E2F-DP1 transcription factor complex regulates centriole duplication in Caenorhabditis elegans. G3 6, 709–720. 10.1534/g3.115.025577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D, 2001. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291, 2144–2147. 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- Paix A, Folkmann A, Rasoloson D, Seydoux G, 2015. High efficiency, homology-directed genome editing in Caenorhabditis elegans using CRISPR-Cas9 ribonucleoprotein complexes. Genetics 201, 47. 10.1534/genetics.115.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak V, Eifler TT, Jaeger S, Krogan NJ, Fujinaga K, Peterlin BM, 2015. CDK11 in TREX/THOC regulates HIV mRNA 3' end processing. Cell Host Microbe 18, 560–570. 10.1016/j.chom.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel N, Dougherty M, Goeres J, Liu Y, O'Connell KF, 2012. The C. elegans F-box proteins LIN-23 and SEL-10 antagonize centrosome duplication by regulating ZYG-1 levels. J. Cell Sci 125, 3535–3544. 10.1242/jcs.097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petretti C, Savoian M, Montembault E, Glover DM, Prigent C, Giet R, 2006. The PITSLRE/CDK11(p58) protein kinase promotes centrosome maturation and bipolar spindle formation. EMBO Rep. 7, 418–424. 10.1038/sj.embor.7400639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakkaa T, Escude C, Giet R, Magnaghi-Jaulin L, Jaulin C, 2014. CDK11(p58) kinase activity is required to protect sister chromatid cohesion at centromeres in mitosis. Chromosome Res. 22, 1–10. 10.1007/s10577-013-9400-x. [DOI] [PubMed] [Google Scholar]
- Shepard PJ, Hertel KJ, 2009. The SR protein family. Genome Biol. 10, 242. 10.1186/gb-2009-10-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Feng Y, Goulet A-C, Vaillancourt RR, Sachs NA, Hershey JW, Nelson MA, 2003. The p34cdc2-related cyclin-dependent kinase 11 interacts with the p47 subunit of eukaryotic initiation factor 3 during apoptosis. J. Biol. Chem 278, 5062–5071. 10.1074/jbc.M206427200. [DOI] [PubMed] [Google Scholar]
- Shi J, Hershey JWB, Nelson MA, 2009. Phosphorylation of the eukaryotic initiation factor 3f by cyclin-dependent kinase 11 during apoptosis. FEBS Lett. 583, 971–977. 10.1016/j.febslet.2009.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singson A, Mercer KB, L'Hernault SW, 1998. The C. elegans spe-9 gene encodes a sperm transmembrane protein that contains EGF-like repeats and is required for fertilization. Cell 93, 71–79. [DOI] [PubMed] [Google Scholar]
- Song MH, Aravind L, Muller-Reichert T, O'Connell KF, 2008. The conserved protein SZY-20 opposes the Plk4-related kinase ZYG-1 to limit centrosome size. Dev. Cell 15, 901–912. 10.1016/j.devcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J, 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol 56, 564–577. 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A, 1998. Specific interference by ingested dsRNA. (854–854)Nature 395. 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Trembley JH, Hu D, Hsu L-C, Yeung C-Y, Slaughter C, Lahti JM, Kidd VJ, 2002. PITSLRE p110 protein kinases associate with transcription complexes and affect their activity. J. Biol. Chem 277, 2589–2596. 10.1074/jbc.M109755200. [DOI] [PubMed] [Google Scholar]
- Trembley JH, Hu D, Slaughter CA, Lahti JM, Kidd VJ, 2003. Casein kinase 2 interacts with cyclin-dependent kinase 11 (CDK11) in vivo and phosphorylates both the RNA polymerase II carboxyl-terminal domain and CDK11 in vitro. J. Biol. Chem 278, 2265–2270. 10.1074/jbc.M207518200. [DOI] [PubMed] [Google Scholar]
- Valente ST, Gilmartin GM, Venkatarama K, Arriagada G, Goff SP, 2009. HIV-1 mRNA 3' end processing is distinctively regulated by eIF3f, CDK11, and splice factor 9G8. Mol. Cell 36, 279–289. 10.1016/j.molcel.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker SR, Mallinger A, Workman P, Clarke PA, 2017. Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacol. Ther. 10.1016/j.pharmthera.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker EW, van Vugt MATM, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, Yaffe MB, 2007. 14–3-3 sigma controls mitotic translation to facilitate cytokinesis. Nature 446, 329–332. 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- Yang L, Li N, Wang C, Yu Y, Yuan L, Zhang M, Cao X, 2004. Cyclin L2, a novel RNA polymerase II-associated cyclin, is involved in pre-mRNA splicing and induces apoptosis of human hepatocellular carcinoma cells. J. Biol. Chem 279, 11639–11648. 10.1074/jbc.M312895200. [DOI] [PubMed] [Google Scholar]
- Yokoyama H, Gruss OJ, Rybina S, Caudron M, Schelder M, Wilm M, Mattaj IW, Karsenti E, 2008. Cdk11 is a RanGTP-dependent microtubule stabilization factor that regulates spindle assembly rate. J. Cell Biol 180, 867–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SW, Cai MM, Zhang S, Xu SL, Chen S, Chen XN, Chen C, Gu JX, 2002. Interaction of p58(PITSLRE), a G(2)/M-specific protein kinase, with cyclin D3. J. Biol. Chem 277, 35314–35322. 10.1074/jbc.M202179200. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Shen JK, Hornicek FJ, Kan Q, Duan Z, 2016. The emerging roles and therapeutic potential of cyclin-dependent kinase 11 (CDK11) in human cancer. Oncotarget 7, 40846–40859. 10.18632/oncotarget.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.