Abstract
The importance of mitogen-activated protein kinase (MAPK) pathway signaling in regulating microglia-mediated neuroinflammation in Alzheimer’s disease (AD) remains unclear. We examined the role of MAPK signaling in microglia using a preclinical model of AD pathology and quantitative proteomics studies of postmortem human brains. In multiplex immunoassay analyses of MAPK phosphoproteins in acutely isolated microglia and brain tissue from 5xFAD mice, we found phosphorylated extracellular signal-regulated kinase (ERK) was the most strongly upregulated phosphoprotein within the MAPK pathway in acutely isolated microglia, but not whole-brain tissue from 5xFAD mice. The importance of ERK signaling in primary microglia cultures was next investigated using transcriptomic profiling and functional assays of amyloid-β and neuronal phagocytosis, which confirmed that ERK is a critical regulator of IFNγ-mediated pro-inflammatory activation of microglia, although it was also partly important for constitutive microglial functions. Phospho-ERK was an upstream regulator of disease-associated microglial gene expression (Trem2, Tyrobp), as well as several human AD risk genes (Bin1, Cd33, Trem2, Cnn2), indicative of the importance of microglial ERK signaling in AD pathology. Quantitative proteomic analyses of post-mortem human brain showed that ERK1 and ERK2 were the only MAPK proteins with increased protein expression and positive associations with neuropathological grade. In a human brain phosphoproteomic study, we found evidence for increased flux through the ERK signaling pathway in AD. Overall, our analyses strongly suggest that ERK phosphorylation, particularly in microglia in mouse models, is a regulator of pro-inflammatory immune responses in AD pathogenesis.
Keywords: Alzheimer’s disease, ERK, microglia, neuroinflammation, proteomics, RRID:AB_331646, RRID:AB_354872, RRID:AB_394489, RRID:AB_396772, RRID:CVCL_0470, RRID:IMSR_JAX:000664, RRID:MMRRC_034840-JAX, RRID:SCR_002798, RRID:SCR_002865, RRID:SCR_003420, RRID:SCR_017386
1 |. INTRODUCTION
Microglia, the primary immune cells of the brain, are the key enactors of neuroinflammation in Alzheimer’s disease (AD) (Hickman et al., 2018; Lambert et al., 2013), orchestrating both beneficial and detrimental mechanisms in context-dependent manners (Bachiller et al., 2018; Dubbelaar et al., 2018; Tay et al., 2017). Detrimental consequences of microglia are mediated by pro-inflammatory mechanisms, including reactive oxygen species, cytokines (IL1β, TNFα, IL6), phagocytosis of synapses and dendrites, and impaired phagocytic clearance of pathological proteins. Microglia can also promote pathogenesis via indirect mechanisms involving astrocytes or oligodendroglia, which then impact neuronal health and survival (Block et al., 2007; Geraghty et al., 2019; Hagemeyer et al., 2017; Liddelow et al., 2017; Sellgren et al., 2019). Defining critical regulators of homeostatic, disease-associated, and pro-inflammatory microglial responses in AD (Rangaraju, Dammer, Raza, Gao, et al., 2018) can lead to neuro-immunomodulatory therapies that selectively inhibit pro-inflammatory responses without impacting homeostatic functions.
Distinct microglial immunological profiles are likely to be determined by upstream signaling events that are engaged by cytokines, pathological proteins, and oxidative stress in the setting of neurodegeneration. Signaling pathways, such as the mitogen-activated protein kinase (MAPK) family of proteins, are engaged very early in immune cells (Cargnello & Roux, 2011). MAPKs include over 60 proteins broadly categorized into three families, namely the extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and p38 cascades (Yates et al., 2017), which are initiated via activation of a cell surface receptor, triggering serial phosphorylation events by serine/threonine protein kinases, ultimately activating or suppressing transcription factors (TFs; Arkun & Yasemi, 2018; Santos et al., 2007). ERK signaling is initiated by receptor tyrosine kinases (RTKs) that are typically receptors for growth factors. JNK signaling, which is activated by cellular stress, is associated with increased formation of amyloid-β (Aβ) plaques and neurofibrillary tangles, as well as neuron apoptosis, in AD (Yarza et al., 2016). p38 is activated by inflammatory stimuli and cytokines (Lee & Kim, 2017). There is also significant cross talk between these three cascades, particularly JNK and p38 (Zhang & Liu, 2002). While these signaling events are typically transient and often necessary for homeostasis, constitutive functions, and proliferation, sustained activation may lead to dysfunction and chronic inflammatory activation in microglia (Kim et al., 2004). However, the importance of MAPK signaling in regulating microglia-mediated neuroinflammation in AD remains unclear.
We investigate the role of MAPK signaling using phosphoprotein signaling studies of acutely isolated microglia and whole-brain tissues from 5xFAD mice to reveal microglia-specific ERK activation in AD pathology. In neuroinflammatory transcriptomic profiling of primary mouse microglia, we show that ERK signaling is a critical regulator of pro-inflammatory microglial activation in response to interferon γ (IFNγ), in addition to being an upstream regulator of several disease-associated microglial genes and genetic risk factors for late-onset AD. We further interrogated human postmortem mass spectrometry data from control and AD brains and provide evidence for increased ERK activation. Our integrated approach synthesizes data from mouse microglia and whole post-mortem human brains and provides novel insights into the role of ERK signaling in AD.
2 |. MATERIALS AND METHODS
2.1 |. Animals
All mouse experiments were performed in accordance with policies and procedures described in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee at Emory University. C57BL/6J (RRID:IMSR_JAX:000664) and 5xFAD mice (RRID:MMRRC_034840-JAX) were bred and housed in a 12/12 hr light/dark cycle.
2.2 |. Mouse microglial isolation and in-vitro studies
Adult microglia were isolated from the brains of wild-type and age-matched 5xFAD mice (age 6–7 mo, n = 7/group including three males and four females) using Percoll density centrifugation (35%/70%), followed by enrichment of CD11b+ microglia by magnetic-activated cell sorting (Miltenyi Biotec) and frozen on dry ice for Luminex phosphoprotein signaling studies (Rangaraju, Dammer, Raza, Gao, et al., 2018).
Primary microglial cultures were prepared from P0–3 postnatal mouse brains as previously described (Gao et al., 2019). Briefly, brain tissue was harvested from C57BL/6J mice (P0 to P3) and digested with Trypsin for 15 min. Trypsinization was halted by the addition of Dulbecco’s modified Eagle medium: Nutrient Mixture F-12 (DMEM-F12) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin-glutamine. Once filtered using a 40-μm nylon mesh strainer, the resulting cell suspension was positively selected for CD11b+ cells using mini-MACS columns. The cells, almost entirely consisting of microglia, were seeded into poly-L-lysine-coated wells and cultured in a humidified CO2 incubator at 37°C. Following 24 hr of incubation, half of the medium was replaced with fresh medium, and 1 μM SCH772984, a selective small molecule inhibitor of ERK 1/2 (IC50 for ERK1: 4 nM, IC50 for ERK2: 1 nM), was added to the culture (Morris et al., 2013). After 1 hr of preincubation, cells were stimulated with 1 ng/ml IFNγ (Sigma) and then incubated for 24 hr, after which cells were trypsinized and harvested for downstream analyses.
2.3 |. Luminex phosphoprotein analyses of acutely isolated microglia from mouse brain
Collected microglia were lysed in Bio-Plex lysis buffer (Bio-Rad) with the addition of one Complete Mini (Roche) protease inhibitor per 5 ml of buffer. Cells were lysed for 10 min on an end-over-end rotator, then centrifuged at 13 kRPM for 10 min with supernatant collected and stored at −80°C. Protein yield was measured using Pierce BCA Protein Assay kit (ThermoFisher Scientific), and 0.1 μg of total protein from each sample was added to the Luminex assay. Phosphosignaling was quantified using the MAPK/SAPK Signaling 10-Plex Multiplex assay (Millipore Sigma, 48–660MAG). Phosphorylation sites for the MAPK/SAPK as follows: ATF2 (Thr71), ERK (Thr185/Tyr187), HSP27* (Ser78), JNK (Thr183/Tyr185), c-Jun (Ser73), MEK1 (Ser222), MSK1 (Ser212), p38 (Thr180/Tyr182), p53* (Ser15), and STAT1 (Tyr707). Analytes marked with an asterisk have not been reported to be mouse-reactive and were removed from the analysis. The assay was read out using a MAGPIX instrument (Luminex, Austin, TX, USA).
2.4 |. NanoString transcriptomic profiling of microglia and statistical analyses
NanoString transcriptomic profiling of mRNA isolated from primary mouse microglia was performed as previously published (Gao et al., 2019). Primary microglia were treated with the ERK inhibitor in the presence or absence of IFNγ. The cells were washed in 1x diethyl pyrocarbonate–phosphate-buffered saline and lysed in TRIzol (Invitrogen). RNA was extracted using the nCounter Low RNA Input Kit, and a quality control assessment was performed to determine concentration and integrity of RNA (RIN scores >8). One hundred nanograms of RNA per sample was run using the nCounter Mouse Neuroinflammation Panel (770 genes), and gene expression data were imported into nSolver Analysis Software (Version 4.0, RRID:SCR_003420). After background thresholding, genes for which >50% of samples exceeded this threshold (n = 678) were analyzed. Expression levels were normalized to positive controls and then to 12 housekeeping genes and analyzed by ANOVA (two-tailed) and Tukey’s post hoc tests for groupwise differences. Differentially expressed transcripts (p < 0.05) were included in K-means clustering analysis (Morpheus: (https://software.broadinstitute.org/morpheus, RRID:SCR_017386; Metric: one minus Pearson correlation with 1,000 maxim iterations). We applied a combination of hierarchical clustering and K-means clustering approaches to identify the ideal number of clusters that best described groupwise differences and distinct expression patterns based on ERK inhibitor and IFNγ exposures. We first explored the data using hierarchical clustering analysis (one minus Pearson correlation metric, average linkage method), which revealed that at least four, up to six distinct clusters of genes exist in the data set. We then performed K-means clustering using a range of potential cluster solutions (range 2–8) and identified 5 as the optimal cluster size. Gene Ontology (GO) enrichment analysis was performed on the resulting clusters using GO-Elite (version 1.2.5), to identify GO, KEGG pathway, and Wikipathway terms enriched in lists of genes as compared to a reference list of genes (Zambon et al., 2012). We also used t-weighted stochastic neighbor embedding (t-SNE) to visualize the clusters of genes in two dimensions and determine whether known homeostatic microglial and disease-associated microglial (DAM) genes mapped to specific gene clusters identified in our data set. We used existing reference transcriptomes and network analyses to obtain lists of homeostatic and DAM genes (Keren-Shaul et al., 2017) and lists of genes specific to modules of co-expressed microglial genes previously identified using network analysis (Rangaraju, Dammer, Raza, Gao, et al., 2018).
2.5 |. Flow cytometry studies
Flow cytometric assays of microglial viability, Aβ phagocytosis, and microglia-mediated neuronal phagocytosis were performed using in-vitro primary mouse microglia. To assess the viability of microglia under the experimental conditions, primary microglia were treated with the ERK inhibitor and/or IFNγ as above. After 24 hr, the cells were washed in PBS and trypsinized (15 min 37°C), and cells were washed and incubated with LIVE/DEAD indicator (30 min). After washing (2% normal horse serum and PBS), PE-Cy7 anti-mouse CD45 mAb was added (dark, 30 min), after which the cells were washed in PBS. Compensation, gating, live/dead exclusion, and data analysis for flow cytometry were performed as published (Rangaraju, Dammer, Raza, Gao, et al., 2018).
The effect of ERK inhibition and inflammatory stimulation on microglial phagocytosis of fluorescent fibrillar amyloid-β42 (fAβ42) was determined by flow cytometry as previously described (Gao et al., 2019; Rangaraju, Raza, et al., 2018). Primary microglia treated with the ERK inhibitor and/or IFNγ were incubated with 2 μM of fAβ42–488 for 1 hr (37°C) and then subsequently labeled with anti-CD45, and phagocytosis was assessed by observing the second peak of fluorescence which represents actin-dependent Aβ phagocytosis (Rangaraju, Dammer, Raza, Gao, et al., 2018).
To measure microglia-mediated neuronal phagocytosis, we performed microglia-neuron coculture studies using mouse N2a cells (RRID:CVCL_0470) stably transduced to express GFP (Lenti-Ubc-GFP/FUGW, Addgene#14883) and primary P0–3 mouse microglia and used flow cytometry to detect microglial uptake of GFP+ neuronal material. GFP-N2a cells were washed twice with medium and allowed to grow to 70%–80% confluence. Fifty thousand N2a cells were seeded into 12-well plates, and 24 hr later, the medium was replaced with DMEM with 1% FBS and 10 μM retinoic acid to induce differentiation for 4 days (Namsi et al., 2018). Meanwhile, primary microglia were cultured in the presence or absence of IFNγ and treated with the ERK inhibitor (or DMSO control) as described above. After 4 days of N2a neuronal differentiation, fresh medium with 1% FBS replaced the differentiation medium, and 100,000 microglia were seeded into the differentiated N2a culture. Cocultures were incubated for 24 hr and then trypsinized for 2 min at 37°C for antibody staining. The cells were washed sequentially with 1% FBS and PBS and labeled with LIVE/DEAD indicator and anti-CD45 (PC-Cy7). Microglial phagocytosis of neuronal cells was assessed by determining GFP positivity in live microglia via flow cytometry with appropriate negative controls (microglia without neurons). Viability of CD45+ microglia and N2a-GFP+ cells was assessed.
Flow cytometry for phospho-ERK1/2 was performed in acutely isolated brain mononuclear cells derived from wild-type and 5xFAD mice. The cells were labeled with fluorophore-conjugated mAbs against CD11b (APC-Cy7) and CD45 (PE-Cy7), then fixed with fixation buffer for 10 min at RT and then permeabilized with cold methanol (30 min, 4°C, dark) and blocked (Fc-block Biolegend Cat #553142) (30 min). Cells were incubated with primary anti-phospho-ERK1/2 (1:100) or anti-ERK (1:20) (30 min) followed by secondary antibody (Goat anti-rabbit-FITC, 1:500, 30 min) followed by flow cytometry. Appropriate negative (unstained), secondary-alone and fluorescence-minus-one controls were performed for all experiments involving unconjugated primary antibodies. Single microglia that expressed CD11b and intermediate CD45 were analyzed, and phospho-ERK1/2 was compared between wild-type and 5xFAD microglia.
2.6 |. Quantitative reverse transcriptase PCR (qRT-PCR)
qRT-PCR was performed using RNA isolated from primary microglia (Gao et al., 2019) cultured in conditions mirroring the NanoString studies. RNA was isolated from microglia, and RNA concentration and purity were determined using the NanoDrop 2000 spectrophotometer. cDNA was synthesized, and qRT-PCR was performed on the 7500 Fast Real-time PCR System (Applied Biosystems), TaqMan PCR master mix (Applied Biosystems), and gene-specific TaqMan probes (Applied Biosystems) against Apoe (Mm01307193_g1), Tnf (Mm01210732_g1), Trem2 (Mm04209424_g1), Tyrobp (Mm00449152_m1), Spp1 (Mm00436767_m1), and Gapdh (Mm99999915_g1). All primer sets were run in duplicate, and three biological replicates were performed per condition. Gene expression was normalized to the housekeeping gene, Gapdh, and relative gene expression was calculated using the 2-ΔΔCt method.
2.7 |. Human postmortem brain quantitative proteomics data
Two existing postmortem brain proteomic data sets were used for analyses. The first is a published data set in which dorsolateral prefrontal cortex (age-matched nondisease controls, AD cases, Parkinson’s disease (PD), and both AD and PD, n = 10/group) was sampled for tandem mass tag mass spectrometry (TMT-MS) (Ping et al., 2018). The second data set is a phosphoproteomic TMT-MS data set of nondisease controls, asymptomatic AD cases, and AD cases with dementia (n = 6/group). Methodology used for phosphopeptide enrichment by immobilized metal affinity chromatography (IMAC) and isobaric tandem mass tag mass spectrometry has been previously published (Dai et al., 2018; Dammer et al., 2015). Total protein (data set 1) and phosphopeptide data (data set 2) pertaining to MAPKs were used for analyses. Spearman’s rank correlation co-efficient was calculated in SPSS to assess the relationship between MAPK1 and MAPK3 protein expression levels with BRAAK stage. The clinical and demographic data for these study cohorts are presented in Table S1.
2.8 |. Materials
ERK1/2 inhibitor SCH772984 was obtained from Selleckchem (S7101). The following reagents were used for studies: CD45-PE-Cy7 Rat mAb (BD Biosciences, #552848, RRID:AB_394489), LIVE/DEAD Fixable Blue Stain (Invitrogen, L34961), CD11b-APC-Cy7 Rat mAb (BD Biosciences, #557657, RRID:AB_396772), phospho-ERK1/2 Rabbit pAb (Cell Signaling Technology, #9101S, RRID:AB_331646), and ERK1/2 Rabbit pAb (R&D Systems, AF1576, RRID:AB_354872). Details about antibodies used are summarized in Table 1.
TABLE 1.
Antibody list and details
| Name | Host | Vendor and RRID | Immunizing agent | Dilution | Class |
|---|---|---|---|---|---|
| CD45 (flow cytometry) | Rat | BD Biosciences, #552848, RRID:AB_394489 | Mouse thymus and spleen | 1:100 | Monoclonal |
| CD11b (flow cytometry) | Rat | BD Biosciences, #557657, RRID:AB_396772 | Mouse spleen | 1:100 | Monoclonal |
| Phospho-ERK1/2 (flow cytometry) | Rabbit | Cell Signaling Technology, #9101S, RRID:AB_331646 | Residues on human ERK1/2 surrounding Thr202/Tyr204 | 1:100 | Polyclonal |
| ERK1/2 (flow cytometry) | Rabbit | R&D Systems, AF1576, RRID:AB_354872 | E. coli-derived recombinant human ERK1/2 | 1:20 | Polyclonal |
2.9 |. Statistical considerations
GraphPad Prism (Ver. 5, RRID:SCR_002798) and SPSS (Ver. 26, RRID:SCR_002865) were used to create graphs and for statistical analyses. Error bars in all bar graphs represent the standard deviation (SD). We determined sample size for Luminex studies of CD11b+ MACS-purified microglia (n = 7–9/group) based on the following parameters: expected difference between means of 25%, a standard deviation of 15%, alpha level 5%, and power of 80%. We included male and female mice across all groups. All other experiments did not have a power calculation. For experiments with >2 groups, one-way ANOVA was performed as previously described in R (Ver 4.0.2) (R Core Team, 2020; Seyfried et al., 2017) to detect differences across groups, and post hoc pairwise comparisons were performed using post hoc Tukey’s test (p < 0.05 considered significant for all experiments). Clustering analyses for visualization of NanoString data were performed using Morpheus (https://software.broadinstitute.org/morpheus) as described above.
3 |. RESULTS
3.1 |. Phosphorylated ERK and p38 MAPK are increased in microglia in 5xFAD mice
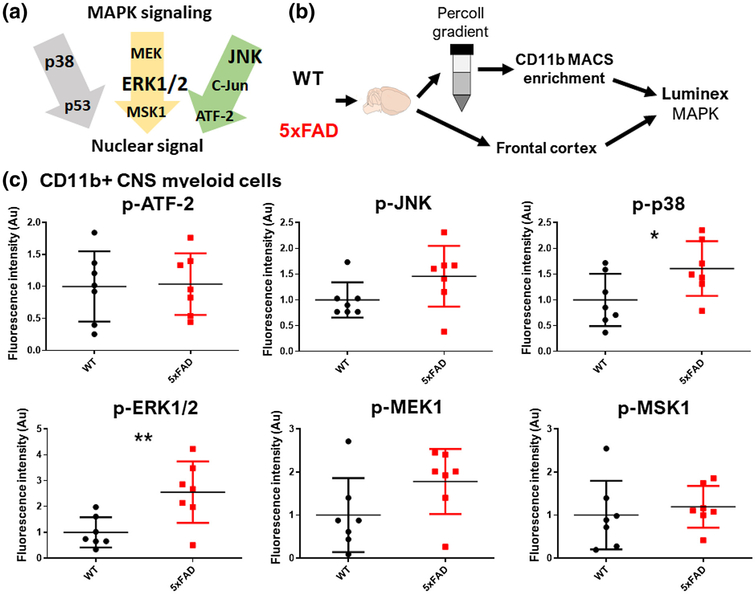
MAPKs are comprised of three subfamilies of signaling proteins, namely ERK, JNK, and p38 pathways (Figure 1a). To determine whether MAPK signaling is chronically upregulated in microglia in a mouse model of AD pathology, we acutely isolated CD11b+ brain myeloid cells from adult (6- to 7-mo-old) wild-type and age/sex-matched 5xFAD mice that exhibit accelerated Aβ pathology in the brain. CD11b+ brain myeloid cells are comprised primarily (>95%) of microglia and a small proportion of central nervous system (CNS)-infiltrating monocytes/macrophages with >95% viability (Rangaraju, Dammer, Raza, Gao, et al., 2018). We broadly interrogated phosphosignaling within MAPK pathways in microglia by assaying phosphorylation of nine proteins within the three MAPK pathways (Figure 1b). We simultaneously quantified signaling within whole tissue samples obtained from the frontal cortex where Aβ pathology is abundant in 5xFAD mice. Compared to microglia isolated from WT mice, 5xFAD mouse microglia exhibited increased activation of ERK signaling, indicated by increased phospho-ERK1/2 (p-ERK1/2) (two-tailed independent sample t-test, t(12) = 2.98, p = 0.009) and trend toward increased levels of phospho-MEK1 (p-MEK1) (two-tailed independent sample t-test, t(12) = 1.8, p = 0.090), an upstream ERK activator. Increased p38 MAPK activation was also observed, indicated by higher phospho-p38 (p-p38 levels) in 5xFAD microglia (two-tailed independent sample t-test, t(12) = 2.15, p = 0.048) (Figure 1c). Unlike in microglia, we did not observe any differences in MAPK signaling in whole-brain tissue (Figure S1). These findings provide evidence for increased activation of MAPKs, particularly ERK1/2, in microglia in 5xFAD mice.
FIGURE 1.
Increased activation of ERK1/2 and p38 mitogen-activated protein kinase (MAPK) signaling in microglia in the 5xFAD mouse Alzheimer’s disease model. (a) Schematic summarizing key MAPK signaling pathways including ERK1/2, p38, and c-Jun N-terminal kinase (JNK) signaling cascades, the three families of MAPKs, all of which result in the activation of transcription factors which regulate gene regulation. (b) Experimental outline for Luminex studies of acutely isolated CD11b+ CNS myeloid cells and frontal cortex brain samples isolated from age-matched WT and 5xFAD mice (n = 7–8 mice/group). CD11b+ cells were isolated by Percoll density centrifugation followed by CD11b enrichment using MACS columns. *p < 0.05, **p < 0.01, ***p < 0.005. (c) Summary of phospho-protein signaling data for phosphoERK1/2 (p-ERK1/2), p-p38, and p-JNK signaling cascades in CD11b+ CNS myeloid cells from WT and 5xFAD mice. Mean and SD of the expression of phospho (p)-proteins, as represented by the intensity of fluorescence, are represented. *p < 0.05, **p < 0.01, ***p < 0.005
3.2 |. MAPK protein expression is unaltered in 5xFAD microglia
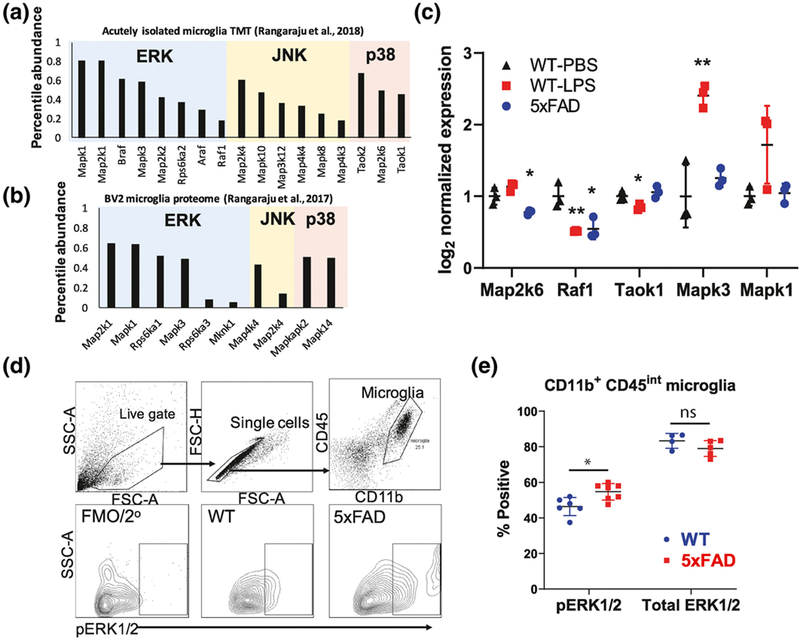
To ascertain the expression levels of MAPKs at the total protein level in mouse microglia, we assessed the relative abundance of total MAPK proteins (vs. non-MAPK proteins) in recently published proteomic data sets obtained from purified mouse microglia, as well as BV2 mouse microglia (Rangaraju, Dammer, Raza, Rathakrishnan, et al., 2018; Rangaraju, Raza, et al., 2018). In total, 60 genes/proteins have been classified as belonging to the family of MAPK cascades (Table S2) (Yates et al., 2017). Of these, 17 MAPK proteins (total protein levels) were identified in acutely isolated CD11b+ microglia from 6-mo-old mice (pooled analysis of wild-type, 5xFAD, and systemic LPS-treated wild-type mice) (Figure 2a), while 10 MAPK proteins were identified in BV2 microglia (Figure 2b). Interestingly, relative abundances of ERK proteins were highest in both acutely isolated mouse microglia (Figure 2a), as well as in BV2 microglia (Figure 2b) compared to other MAPKs. Compared to wild-type microglia, 5xFAD microglia showed lower total protein levels of RAF1 (F(2,6) = 12.7, n = 3, p = 0.007, post hoc p = 0.03) and MAP2K6 (F(2,6) = 15.9, n = 3, p = 0.004, post hoc p = 0.033), while microglia isolated from LPS-treated mice showed increased total levels of two ERK proteins, namely MAPK1 (F(2,6) = 4.5, n = 3, p = 0.061, post hoc p = 0.080) and MAPK3 (F(2,6) = 21.8, n = 3, p = 0.002, post hoc p = 0.006), and lower levels of TAOK1 (F(2,6) = 8.1, n = 3, p = 0.020, post hoc p = 0.033) and RAF1 (post hoc p = 0.009) (Figure 2c). While our Luminex studies showed alterations in levels of activated ERK phosphoproteins in 5xFAD microglia, proteomic studies show that total protein levels of ERK1 and ERK2 were not altered in 5xFAD microglia. Consistent with this, we performed flow cytometric studies of acutely isolated brain myeloid cells (Figure 2d), confirming that intracellular p-ERK1/2 levels were increased in 5xFAD microglia as compared to wild-type microglia (two-tailed independent sample t-test, t(10) = 3.16, p = 0.010), whereas total ERK1/2 was equal between the groups (Figure 2d,e).
FIGURE 2.
Confirmation of increased ERK1/2 activation in 5xFAD microglia. (a,b) Bar plot showing the relative abundance of 17 mitogen-activated protein kinase (MAPK) proteins as compared to non-MAPK proteins in microglia from WT, 5xFAD, and lipopolysaccharide (LPS)-treated WT mice (a) and BV2 microglia (b) using published proteomic data sets. (c) Differentially expressed MAPK proteins identified in PBS-treated WT, 5xFAD, and LPS-treated WT mice. Error bars represent SD. Proteins differentially expressed across groups (ANOVA p < 0.05) and meeting statistical significance (post hoc Tukey’s HSD p < 0.05) as compared to the WT-PBS group are indicated. *p < 0.05, **p < 0.01, ***p < 0.005. (d) Intracellular flow cytometric analysis of acutely isolated CNS myeloid cells for p-ERK1/2 labeling. Gating strategy used is shown (top). (e) Quantitative analyses of flow cytometric studies of p-ERK1/2 and total ERK1/2 expression in WT and 5xFAD CD11b+ CD45int microglia. *p < 0.05, **p < 0.01, ***p < 0.005
In summary, our findings demonstrate that MAPK signaling proteins belonging to the ERK cascade are highly abundant in microglia and that microglia in an AD mouse model exhibit upregulation of ERK phospho-signaling, supporting critical roles for ERK signaling in regulating neuroinflammation in AD.
3.3 |. In-vitro analysis reveals that ERK signaling is a critical regulator of pro-inflammatory microglial activation
ERK signaling may be an important regulator of pro-inflammatory microglial activation (Gao et al., 2019). Based on our results showing increased ERK signaling in 5xFAD microglia and the relevance of ERK to human AD pathology, we next hypothesized that ERK is a key regulator of DAM gene expression, particularly pertaining to pro-inflammatory responses that can be detrimental in AD. We exposed mouse primary microglia to a selective ERK1/2 inhibitor (SCH772984, 1 μM) in the presence or absence of IFNγ (1 ng/ml) and performed viability studies and neuroinflammatory transcriptomic profiling (770 genes). For our in-vitro studies, we selected IFNγ as a canonical immune stimulus to mirror pro-inflammatory activation of microglia and induce M1-like characteristics. IFNγ, produced by glia and lymphocytes, is also known to regulate the gene expression of several AD-related genes and regulate Aβ and tau pathology in mouse models of AD pathology (Mastrangelo et al., 2009).
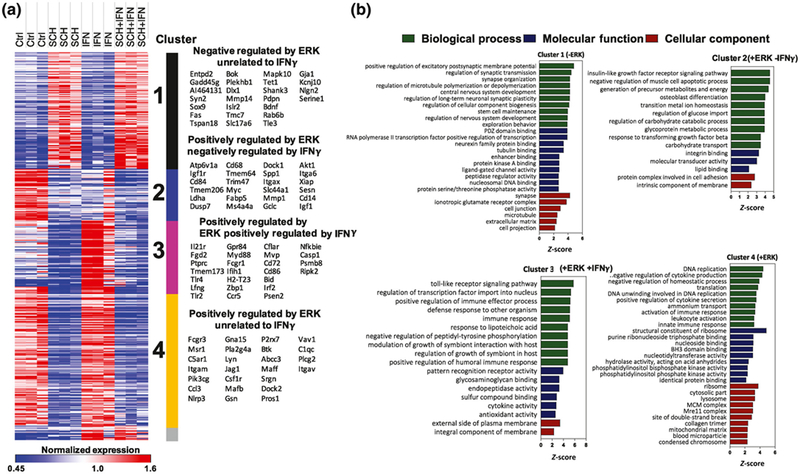
Overall, ERK inhibition minimally impacted cell health and viability under our experimental conditions (Figure S2). Of the 770 genes in the NanoString panel, 678 were included in the final analysis (Table S3), and ANOVA identified 465 genes with differential expression across all four groups (436 genes FDR adjusted p < 0.05). K-means clustering of these differentially expressed genes revealed four main clusters of genes (Figure 3a, Table S4). Cluster 1 consisted of genes negatively regulated by ERK under both unstimulated and IFNγ-stimulated conditions. Clusters 2–4 were all positively regulated by ERK signaling. Cluster 2 was suppressed by IFNγ, Cluster 3 was upregulated by IFNγ, and Cluster 4 was not affected by IFNγ. The most statistically significant and representative genes for each cluster are shown in Figure 3a. A smaller cluster of 17 genes, Cluster 5, contained genes upregulated by IFNγ independent of ERK; several genes in this cluster align with existing literature as being positively regulated by IFNγ (e.g., Stat1 and Irf1) (Zhou et al., 2015). Despite being biologically meaningful, we did not draw attention to this cluster because it was not conserved across multiple runs of K-means analysis and confidence in this cluster is low given its small size.
FIGURE 3.
Transcriptomic profiling of microglia reveals distinct clusters of genes regulated by ERK1/2. (a) Heat map showing K-means clustering analysis of 465 genes with differential expression across four treatment groups, as identified by one-way ANOVA, using NanoString gene expression data. Cluster 1: inhibited by ERK; Cluster 2: activated by ERK and inhibited by IFNγ; Cluster 3: activated by ERK and IFNγ; Cluster 4: inhibited by ERK. (b) Gene ontology (GO) analysis depicting enrichment of GO terms in genes belonging to each cluster.
To obtain a functional understanding of the clustered genes, we next performed GO enrichment analyses (GO-Elite) using all included genes in the neuroinflammation panel as the reference. Cluster 1 genes were enriched for neuronal/synaptic and glutamatergic transcripts with generally low abundance. Cluster 2 was enriched for genes involved in IGF1 receptor signaling, negative regulation of apoptosis, cell differentiation and energy metabolism, and integrin binding. Cluster 3 was enriched with genes involved in pro-inflammatory signaling and immune processes, cytokine activity, and TF activity in the nucleus. Cluster 4 was enriched with genes involved in DNA replication, cell division, cytokine secretion, ribosomal machinery, and purine metabolism (Figure 3b). Additional KEGG and Wikipathway enriched terms are shown in Table S5. Since ERK signaling regulates TF activity, we used bioinformatics approaches (Metacore, Thompson Reuters) to identify TFs downstream of ERK activation that are likely to regulate gene expression in Cluster 2, 3, and 4 genes (Table S6). PU.1, c-Jun, Bcl-6, SP3, EGR1, and SP1 were top-predicted TF regulators of Cluster 2 genes, consistent with known importance of PU.1, c-Jun, and SP1 in microglial development and survival and the enrichment of homeostatic genes within Cluster 2 (Rustenhoven et al., 2018; Smith et al., 2013). Similarly, Cluster 4 genes were predicted to be regulated primarily by PU.1, Bcl-6, SP1, and c-Myc, again consistent with the enrichment of homeostatic and constitutive gene ontologies in this cluster. In contrast, Stat1, NFκB, Bcl-6, IRF1, and PU.1 were predicted to regulate Cluster 3 genes that were enriched in pro-inflammatory genes. These agree with previous publications that demonstrated critical roles for Stat1, NFκB, IRF1 (along with IRF8), and PU.1 in microglial activation (Rustenhoven et al., 2018; Smith et al., 2013).
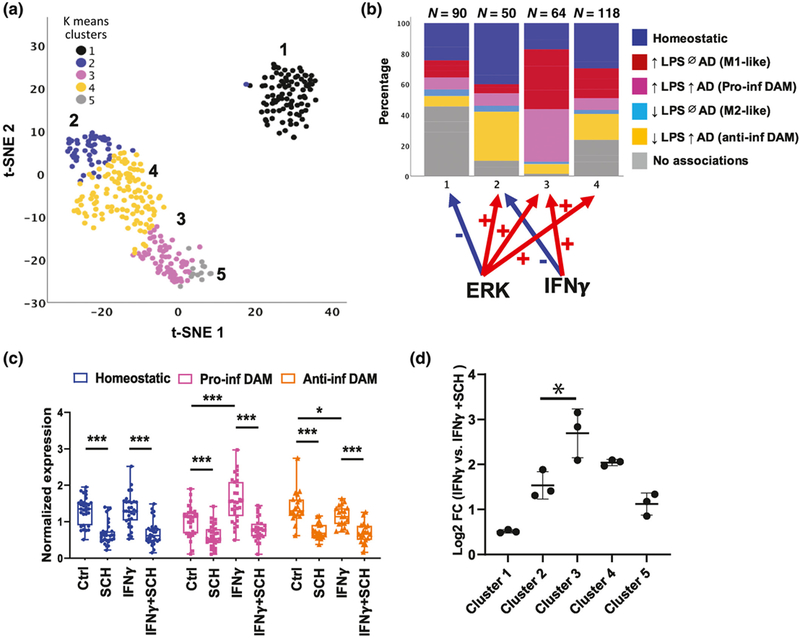
To determine the enrichment patterns of homeostatic, pro-inflammatory DAM, and anti-inflammatory DAM gene expression within each cluster, we mapped our previously published lists of homeostatic, pro-inflammatory DAM, anti-inflammatory DAM, and M1-like and M2-like gene lists (Rangaraju, Dammer, Raza, Gao, et al., 2018) to a 2D t-SNE representation of the clusters (Figure 4a). We observed that Clusters 2 and 4 were enriched with homeostatic and anti-inflammatory DAM genes, while Cluster 3 was enriched with pro-inflammatory DAM genes and M1-like genes (Figure 4b). ERK was found to positively regulate several canonical DAM genes, including Ccl3, Ch25h,Spp1, Igf1, and Itgax (Figure S3) and pro-inflammatory DAM genes, includingCcl2, Irf1, Tnf, Slamf9, and Cd69 (Figure S4). We also calculated a synthetic eigenvalue to represent overall expression of homeostatic, anti-inflammatory DAM, and pro-inflammatory DAM profiles across each condition using previously identified hub genes (Kme > 0.75) present in our NanoString data set (Dai et al., 2018; Gao et al., 2019). We confirmed that inhibition of ERK suppresses homeostatic, anti-inflammatory, and pro-inflammatory DAM gene expression, while IFNγ increases pro-inflammatory DAM and suppresses anti-inflammatory DAM gene expression (Figure 4c). A summary of the direction of regulation of each cluster by IFNγ and ERK activity is shown in Figure 4b (Homeostatic [F(3,8) = 29, n = 3, p < 0.001], Pro-inflammatory DAM [F(3,8) = 29.5, n = 3, p < 0.001], Anti-inflammatory DAM [F(3,8) = 22.4, n = 3, p < 0.001). We also performed qRT-PCR validation studies using primary mouse microglia to confirm ERK-mediated regulation of several DAM genes (Apoe [F(3,8) = 25.79, n = 3, p = 0.002, SCH772984 vs. control post hoc p = 0.015], Trem2 [F(3,8) = 4.79, n = 3, p = 0.034], Tyrobp [F(3,8) = 8.45, n = 3, p = 0.007, SCH772984 vs. Control post hoc p = 0.037],Spp1 [F(3,8) = 80.52, n = 3, p < 0.001, SCH772984 vs. Control post hoc p < 0.001]) identified in Cluster 2 and a pro-inflammatory gene, Tnf (F(3,8) = 5.20, n = 3, p = 0.028), identified in Cluster 3 (Figure S5). Although ERK was found to positively regulate several clusters of microglial genes (Clusters 2, 3, and 4), we found that the magnitude of positive regulation by ERK was highest for Cluster 3 (Figure 4d) (F(3,8) = 23.1, n = 3, p < 0.001, post hoc p-value for Cluster 3 vs. 2 = 0.030), which is enriched in pro-inflammatory DAM genes. Therefore, we hypothesized that ERK-dependent pathways disproportionately impact pro-inflammatory DAM gene expression over homeostatic and anti-inflammatory DAM gene expression. To test this hypothesis, we assessed the effects of lower concentrations of the ERK inhibitor on expression of representative homeostatic, pro-inflammatory DAM, and anti-inflammatory DAM genes. We found that ERK inhibition at dose ranges between 4 and 50 nM inhibited gene expression of Ptgs2, a pro-inflammatory DAM gene (IFNγ-stimulated conditions: F(3,10)=9.09, n = 3, p = 0.002, IFNγ+SCH772984 50 nM vs. IFNγ p < 0.001) without impacting homeostatic (Tgfbr1, Bin1) or anti-inflammatory DAM genes (Spp1, Tyrobp) (Figure S6).
FIGURE 4.
ERK1/2 positively regulates homeostatic, anti-inflammatory disease-associated microglia (DAM), and pro-inflammatory DAM gene expression in microglia. (a) t-SNE plots showing clusters of genes based on NanoString expression data. Color codes indicate K-means clusters identified in Figure 3. Clusters 2, 3, and 4 are positively regulated by ERK while Cluster 1 is negatively regulated by ERK. (b) Distribution of homeostatic, pro-inflammatory DAM, anti-inflammatory DAM, M1-like, and M2-like microglial genes in each cluster. (c) Expression of synthetic eigengenes of each microglial state (homeostatic, pro-inflammatory DAM, and anti-inflammatory DAM) across experimental conditions. (d) Plot showing the magnitude of change in overall gene expression level in each cluster (averaging across normalized gene expression within each cluster) between ERK inhibitor treated and nontreated conditions in the presence of IFNγ. N = 3 replicates per group (Error bars represent SD). *p < 0.05, **p < 0.01, ***p < 0.005 based on Tukey’s HSD post hoc pairwise comparisons
To summarize our findings from transcriptomic and in-vitro microglial validation studies, ERK appears to be important for homeostatic, protective, and pathological responses mediated bymicroglia. However, the critical role for ERK in regulating pro-inflammatory immune responses appears to be most evident under pro-inflammatory conditions, suggesting that promotion of neuroinflammatory disease mechanisms via microglial ERK may be context-dependent. While ERK activation in homeostatic microglia may regulate constitutive functions and mediate the effects of several trophic factors, ERK activation under disease-associated conditions, such as in AD, may regulate pro-inflammatory immune responses.
3.4 |. ERK signaling regulates microglial phagocytic function
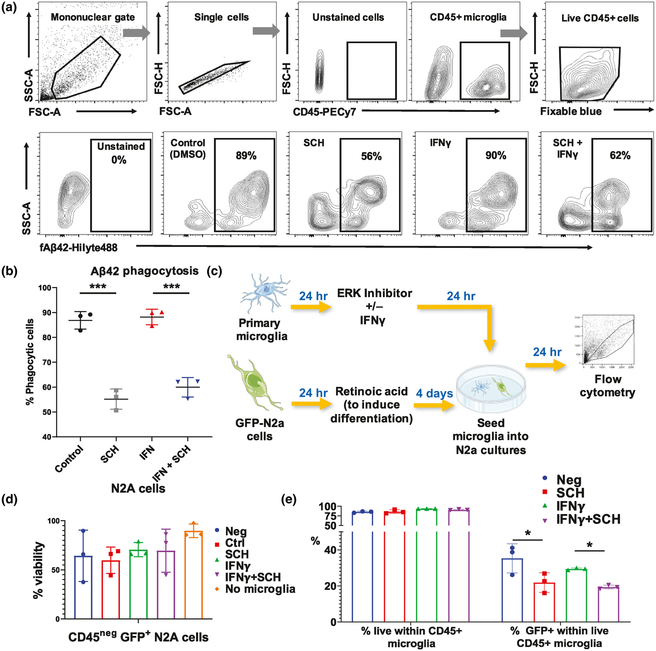
Having found that ERK signaling regulates diverse DAM-associated genes, we next asked if ERK inhibition modulates microglial phagocytic function. To test this, we used a flow cytometric assay of fluorescently labeled Aβ42 fibril phagocytosis. We observed that ERK inhibition reduced microglial phagocytosis of Aβ42 fibrils regardless of stimulation by IFNγ (Figure 5a,b, F(3,8) = 67.1, n = 3, p < 0.001, SCH772984 vs. control post hoc p < 0.001, IFNγ+SCH772984 vs. IFNγ post hoc p < 0.001, control vs. IFNγ post hoc p = 0.959 Figure S7). Since pro-inflammatory microglia can also mediate neurotoxicity via direct neuronal phagocytosis, we performed in-vitro microglia-neuron coculture studies and determined the effect of IFNγ and ERK inhibition on microglia-mediated neuronal phagocytosis. N2a cells were stably transduced to express GFP and then differentiated with retinoic acid under low serum conditions for 4 days. Primary mouse microglia, which were treated with IFNγ and/or the ERK inhibitor, were then added to differentiated N2a-GFP cells for another 24 hr, after which microglia were collected for flow cytometry to measure GFP positivity within microglia (Figure 5c). First, we observed that addition of microglia, regardless of treatment, did not significantly decrease viability in N2a cells (Figure 5d, F(4,10) = 1.36, n = 3, p = 0.314). Inhibition of ERK significantly reduced microglial phagocytosis of N2a cells without IFNγ treatment (Figure 5e, F(3,8) = 6.55, n = 3, p = 0.015, SCH772984 vs. control post hoc p = 0.017). These functional assays support a role for ERK in phagocytic activity, both for Aβ and neuronal phagocytosis.
FIGURE 5.
Roles of extracellular signal-regulated kinase (ERK) activity in regulating microglial phagocytosis of fibrillar Aβ42 and of neuronal phagocytosis. (a) Flow cytometry experimental data showing the uptake of fluorescent Aβ42 fibrils by live CD45+ microglia that had been treated with an ERK1/2 inhibitor and/or IFNγ. (b) Dotplots showing the reduction of microglial phagocytosis of Aβ42 by ERK inhibition under unstimulated and stimulated conditions in-vitro. N = 3 replicates per group. Error bars represent SD. *p < 0.05, **p < 0.01, ***p < 0.005. (c) Experimental protocol for microglia-N2a coculture study: GFP-positive N2a cells were differentiated for 4 days using retinoic acid. Meanwhile, primary microglia were isolated, and after 24 hr, were incubated with the ERK inhibitor with or without IFNγ. After incubation for 24 hr, the microglia were seeded into the differentiated N2a cultures overnight and then harvested for antibody staining and flow cytometry. This figure was created using modified images from Servier Medical Art. (d) Bar plot demonstrating the viability of N2a cells when cocultured with microglia. N = 3 replicates per group. Error bars represent SD. *p < 0.05, **p < 0.01, ***p < 0.005. (e) Bar plots showing a reduction of microglialphagocytosis of differentiated N2a cells in the ERK inhibited group. GFP+ N2A cells were differentiated and then cocultured with primary microglia that were pretreated with an ERK inhibitor with or without IFNγ. N = 3 replicates per group. Error bars represent SD. *p < 0.05, **p < 0.01, ***p < 0.005
3.5 |. GWAS and proteomic data sets reveal evidence for a pathophysiological role for ERK activation in human AD
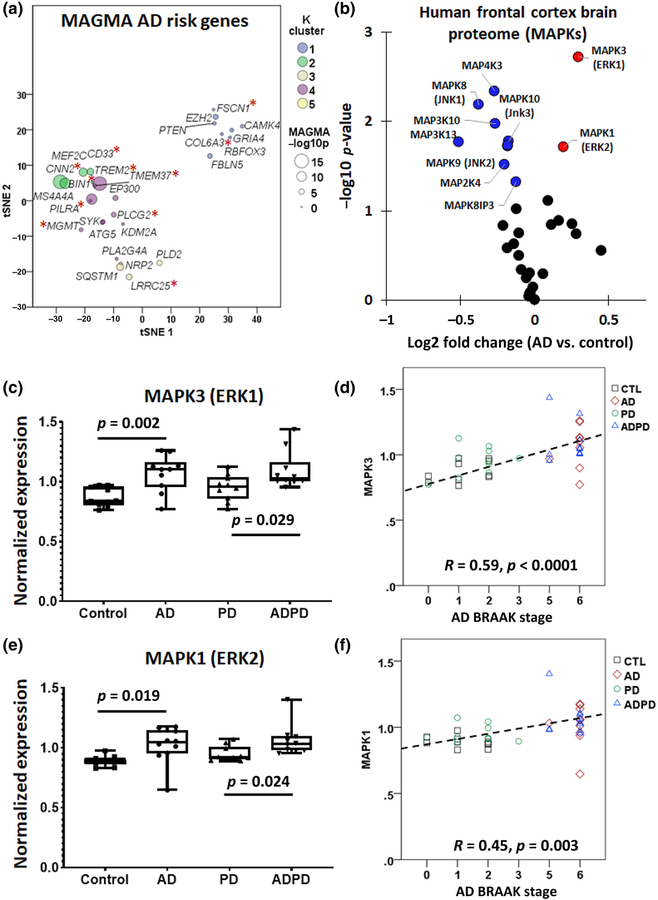
Upon finding that ERK signaling can regulate microglial function, we searched for evidence of dysregulated ERK signaling in human tissues. If ERK signaling in microglia is important to human AD pathogenesis, we predicted that ERK signaling should lie upstream of immune determinants of AD genetic risk. To test this hypothesis, we cross-referenced ERK-regulated gene clusters generated from the NanoString analysis of mouse microglia against late-onset AD risk genes identified in meta-analyses of human genome-wide association study (GWAS) studies, using an approach called multi-marker analysis of GenoMic annotation (MAGMA) (de Leeuw et al., 2015; Seyfried et al., 2017). MAGMA is a tool for gene and gene-set analysis of GWAS data that accounts for linkage disequilibrium and detects associations based on several markers, giving it distinct advantages over many existing techniques (de Leeuw et al., 2015). Of over 1,000 known AD risk genes, 43 were present in the NanoString panel and 28 demonstrated differential expression in our data set, mapping to any one of the five gene clusters (Figure 6a, Table S7). While there was no statistically significant overrepresentation of AD risk genes in any of the clusters, risk genes with highest MAGMA significance mapped to Cluster 2 (Cnn2, Ms4a4a, Mef2c, Cd33) and Cluster 4 (Bin1, Pilra, Ep300). Cluster 3 AD risk genes included Sqstm1, Lrrc25, and Nrp2. Of these 28 ERK-regulated AD risk genes, 11 genes are specifically expressed in microglia (highlighted in Figure 6a with red asterisks).
FIGURE 6.
Multi-marker analysis of GenoMic annotation (MAGMA) and TMT proteomics reveals role for extracellular signal-regulated kinase (ERK) activation in human Alzheimer’s disease (AD). (a) t-SNE plot showing late-onset AD risk genes identified in human GWAS analyses that are present in mouse microglia from the NanoString data set (*p < 0.05). (b) Differential expression analysis of human frontal cortex proteomes showing that MAPK1 (ERK2) and MAPK3 (ERK1), both shown in red, are expressed more highly in AD brains than in control brains while the opposite trend is observed in other notable MAPKs, shown in blue. (c) Box plots (median, min to max range, and individual values) showing that MAPK3 expression levels are increased in AD and AD/Parkinson’s disease (PD) brains as compared to PD or control brains (n = 10 per group). (d) Correlation between AD Braak stage and MAPK3 expression. Spearman’s R and p value are indicated. (e) Box plots (median, min to max range, and individual values) showing that MAPK1 expression levels are increased in brains with AD pathology (n = 10 per group). (f) Correlation between Braak stage and MAPK1 expression. Spearman’s R and p value are indicated
Next, to determine whether MAPK pathway protein expression and phospho-signaling is altered in human AD, we investigated two postmortem brain quantitative proteomic data sets. The clinical and demographic data for these study cohorts can be found in Table S1. The first data set was derived from 10 controls, 10 AD cases, 10 PD, and 10 with both AD and PD pathological features (Ping et al., 2018). TMT proteomics was performed on tissues obtained from the dorsolateral prefrontal cortex (Table S8) (Ping et al., 2020). In this data set, 33 MAPK proteins were identified, and differential expression analyses comparing AD with control samples revealed increased expression of MAPK1 (ERK2) and MAPK3 (ERK1) in AD, while other MAPKs including MAPK8–10 (JNK1–3) were decreased in AD brains (Figure 6b). We also found that increased levels of MAPK3 (ERK1) (F(3,36) = 6.61, n = 10, p = 0.001, post hoc AD vs. Control p = 0.002, post hoc AD-PD vs. PD p = 0.029) and MAPK1 (ERK2) (F(3,36) = 5.04, n = 10, p = 0.005, post hoc AD vs. Control p = 0.019, post hoc AD-PD vs. PD p = 0.024) were only seen in thhe brains with AD pathology, while PD brains did not exhibit increased MAPK1 or MAPK3 levels (Figure 6c–e). We also observed strong associations between AD neuropathological grade (Braak stage) and protein levels of MAPK1 (Spearman’s R = 0.56, p = 0.003) and MAPK3 (Spearman’s R = 0.63, p < 0.001) (Figure 6d,f). These findings show that ERK signaling proteins are specifically increased in AD brains. However, these total protein data do not confirm whether activation of ERK proteins is also increased.
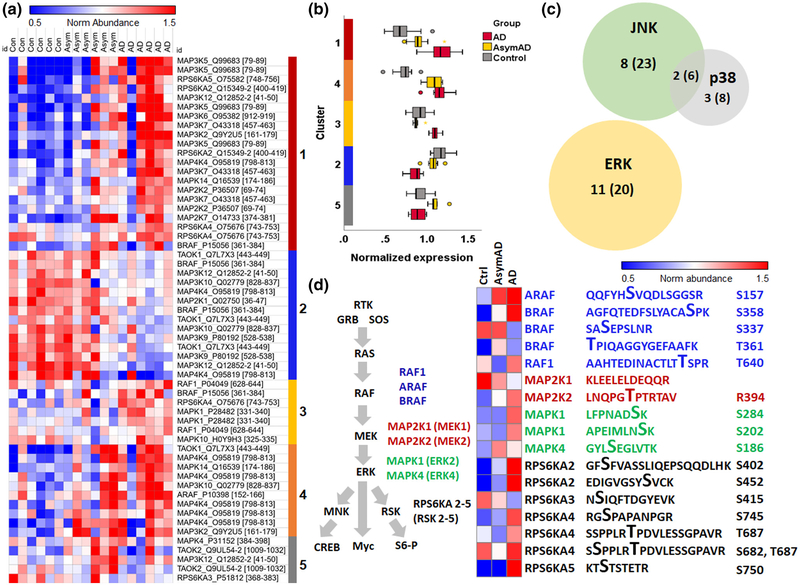
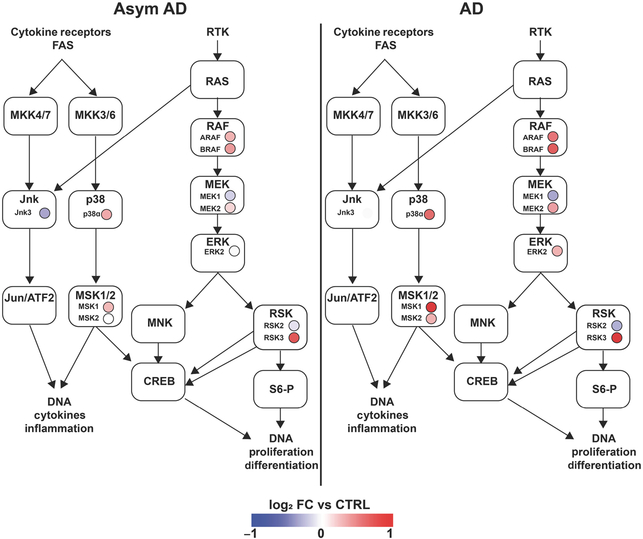
We then turned to an existing phosphoproteomic data set available on Synapse at https://www.synapse.org/#!Synapse:syn20820053/wiki/596078 in which frontal cortex samples from six controls, six asymptomatic AD pathology, and six symptomatic AD cases were used for quantitative TMT proteomics after purification of phosphopeptides on an IMAC column (Ping et al., 2020). Within this data set, we obtained expression data for phosphopeptides that mapped to any of the known 60 MAPK family reference protein/gene list. This identified 127 phosphopeptides, of which 57 peptides showed differential expression (unadjusted ANOVA p < 0.05) across the three conditions (Figure 7, Table S9). K-means clustering of these differentially expressed peptides revealed distinct clusters with unique trajectories of change across stages of AD progression (control→AsymAD→AD) (Figure 7a). Cluster 1 consisted of peptides with dose-dependent increase across the three groups, and Cluster 4 showed early and sustained increase, while Cluster 3 showed a delayed increase. Conversely, Clusters 2 showed decreased expression in clinical stages of AD pathology, although the number of these peptides was relatively small. The overall trajectories of change are presented in Figure 7b. Based on the known biological functions of each MAPK protein, we found that 20 peptides (11 gene symbols) mapped to the ERK cascade, while 23 peptides (eight gene symbols) mapped to JNK, eight peptides (three gene symbols) mapped to p38, and four peptides mapped to MAPK proteins involved in cross talk between JNK and p38 MAPK cascades (Figure 7c). To gain further insights into the roles of differentially expressed phosphopeptides in each signaling pathway, we then mapped each phosphopeptide to known MAPK signaling pathways. ERK-related peptides mapped to all the levels of the ERK signaling cascade (RAF→MEK→MAPK→RSK) (Figure 7d). Within the 20 ERK-related phosphopeptides with differential expression in this data set, we then identified key phosphosites in ERK family proteins of AD relevance. Three RSK peptides (RPS6KA2 pS434 and pS402, and RPS6KA5 pS750) and RAF peptides (BRAF dual phosphorylated pS358+pS750 and ARAF pS157) were increased at least 1.5-fold in AD compared to controls, and two RSK phosphopeptides (RPS6KA2 pS434 and pS402) were also increased 1.5-fold in the asymptomatic stage of AD (Table S9). Two phosphosites in ERK2 were also increased in AD (MAPK1/ERK2 pS284, MAPK1/ERK2 pS202). While little is known about pS284 and its impact on ERK activity, S202 in ERK2 is located at the activating lip in very close proximity to two phosphorylation sites that confer full ERK kinase activity (ERK2 T185/Y187), and a S202P mutation confers resistance to ERK inhibitors, suggesting that ERK phosphorylation at S202 may positively regulate ERK activation (Goetz et al., 2014). In addition to ERK phosphopeptides, we also found that early signaling events in the JNK and p38 cascades represented by phosphopeptides were also differentially expressed in AD brains (Table S9). To identify upstream triggers that may regulate these differentially expressed MAPK phosphopeptides in AD, we performed pathway enrichment analysis using protein analysis through evolutionary relationships and found that the majority of proteins were downstream of growth factor receptors and RTKs such as PDGF, GnRH, FGF, EGF, TGF, and VEGF receptors (Table S10) (Thomas et al., 2003). These findings using cross-sectional human postmortem brain data show unique patterns of ERK activation in asymptomatic and symptomatic stages of AD pathology, suggesting that differential flux through MAPK signaling pathways, specifically ERK, may underlie symptomatic progression and neurodegeneration. We have summarized these patterns of differential ERK signaling observed in our phosphoproteomic analyses in Figure 8. Overall, ERK signaling activation in human AD brains was consistent with our observations in the 5xFAD mouse model of Aβ pathology of AD.
FIGURE 7.
Phosphoproteomic analysis of human postmortem brain reveals extracellular signal-regulated kinase (ERK) activation in Alzheimer’s disease (AD). (a) Heat map showing k-means clustering of 57 differentially expressed phosphopeptides that mapped to a mitogen-activated protein kinase (MAPK) reference gene list, across frontal cortex samples from six non-AD control, six asymptomatic AD, and six symptomatic AD cases. (b) Box and whisker plot quantifying the trajectory of change in expression for each cluster across stages of AD progression. (c) Diagram showing the number of peptides (in parenthesis) and their respective gene symbols (outside of parenthesis) are involved in ERK, c-Jun N-terminal kinase (JNK), and p38 signaling. (d) Visual representation and k-means clustering of key phosphosites within the differentially expressed ERK-related peptides in the IMAC data set, mapping to all levels of the ERK cascade, that are relevant in AD
FIGURE 8.
Phosphoproteomics reveals progressively upregulated flux through MAPK pathway in asymptomatic Alzheimer’s disease (AD) and cognitively impaired AD cases. Colored circles represent fold-change of each detected phosphoprotein protein compared to control cases (six non-AD control, six asymptomatic AD, and six symptomatic AD cases)
4 |. DISCUSSION
Immune responses are orchestrated by early phospho-signaling events that follow activation of cell surface receptors. In turn, phospho-signaling results in regulated expression of groups of genes that together determine the immunological phenotype and function of the cell. Among the various immune signaling cascades that may regulate microglial functions, we focused on the MAPK family of signaling proteins in the 5xFAD preclinical AD model in order to identify pathologically relevant changes in MAPK signaling in microglia that may be responsible for pro-inflammatory immune activation in AD. Using Luminex analyses of phosphoproteins in acutely isolated microglia, we found that among the three main classes of MAPKs, upregulated ERK signaling was the most apparent in microglia isolated from a mouse model of AD. However, no significant changes in MAPK activation were observed in whole tissue, suggesting that ERK signaling may not be coordinated among different cell types in the brain and that ERK activation may be particularly characteristic of microglia in AD. Since ERK signaling is often transient following immune stimuli, our observation of significantly increased p-ERK1/2 levels in mouse microglia from an AD model also suggests that ERK activation may be persistently elevated in AD, potentially as a result of sustained activation of ERK via agonists of RTKs.
In support of a disease-relevant role for ERK in AD, we also found that ERK proteins are highly abundant in microglia isolated from 5xFAD mice. Moreover, in-vitro experiments using primary microglia revealed that inhibition of ERK strongly suppressed pro-inflammatory immune activation induced by IFNγ, which is an AD relevant pro-inflammatory cytokine (Wood et al., 2015; Zheng et al., 2016). We also found that ERK signaling is important for homeostatic and anti-inflammatory gene expression, as well as constitutive microglial functions including phagocytosis of Aβ and neuronal phagocytosis, which is a potentially neurotoxic microglial function during AD pathogenesis (Rajendran & Paolicelli, 2018). ERK signaling was also found to be an upstream regulator of several canonical DAM genes, including Trem2 and Tyrobp, suggesting that ERK signaling may regulate the transition of homeostatic microglia to DAM, a phenotype commonly seen in AD and other neurodegenerative diseases. However, we did not observe any role for ERK signaling in regulation of microglial Apoe expression, suggesting that the Apoe-Trem2-dependent checkpoint for homeostatic-to-DAM transition in AD (Krasemann et al., 2017) may be regulated by both ERK-independent and ERK-dependent mechanisms. Interestingly, ERK activation was also upstream of several immune AD risk genes, including Cd33,Bin1, Plcg2, Trem2, and Cnn2, emphasizing the importance of ERK signaling in regulating immune pathways responsible for AD pathogenesis. While our in-vitro findings show that ERK is a positive regulator of homeostatic, pro-inflammatory, and anti-inflammatory DAM genes in microglia, the effect of ERK inhibition on pro-inflammatory DAM gene expression after induction by IFNγ was far more profound than under unstimulated conditions, emphasizing a potential context-dependent effect of ERK-mediated gene regulation in microglia. Based on our in-vitro studies, the most robust effects of ERK inhibition seem to be the suppression of IFNγ-induced pro-inflammatory genes, many of which are detrimental in AD pathogenesis. However, there are several AD-associated genes with potentially protective and unclear roles, which are also ERK dependent. These highlight a complex role for ERK in microglial activation. Another interpretation is that the level of ERK inhibition may determine the overall protective or detrimental phenotype of microglia. In support of this idea, we found that lower levels of ERK inhibition (closer to the known EC50s for ERK1 and 2) have a more robust effect on pro-inflammatory gene expression with no impact on homeostatic or inflammatory DAM gene expression. At high doses, all three gene profiles are affected, as seen in our NanoString data, although the effect of ERK inhibition was most marked in pro-inflammatory gene expression (Cluster 3, Figures 3 and 4). While we have to explore this concept further, this raises the possibility that a lower level of ERK inhibition (modulation) may be the desired approach in vivo. One limitation of our work is that all in-vitro experiments were conducted using microglia derived from neonatal mouse pups due to challenges of keeping adult microglia alive in culture, limiting our ability to generalize the observed effects of ERK signaling to microglia from adult or aging mice.
Consistent with these important roles for ERK signaling in mouse microglia and in 5xFAD mice, we found that human AD is also characterized by increased ERK activation. Our analyses of human proteomics data, including phosphoproteomic studies, confirmed that ERK1 and ERK2 are the only two MAPKs that are increased at the total protein level in human AD brains, while other MAPKs, such as JNKs, are decreased. At the phosphoprotein level, 57 MAPK phosphopeptides were found to be differentially expressed in human AD brains, and the relative proportion of ERK proteins with differentially expressed phosphopeptides was highest compared to other MAPK proteins. A limitation of this analysis is that human postmortem proteomic data were not cell type specific, while our mouse Luminex findings confirmed microglia-specific ERK activation in 5xFAD mice. This limits our ability to determine whether ERK and other MAPK-related changes occurring in 5xFAD microglia also occur in human microglia in AD cases, and whether MAPK changes observed in the human brain proteomes are driven by microglia or by other cell types. Future human brain cell type-specific analysis may address these questions. Overall, however, our integrated analyses of signaling data obtained from preclinical 5xFAD mice and in-vitro mechanistic studies in microglia, along with confirmatory human postmortem brain proteomics findings, strongly support an important role for ERK signaling in AD pathogenesis.
From a therapeutic perspective, our results suggest that inhibition of ERK in microglia should limit pro-inflammatory disease mechanisms. They also highlight, however, the importance of ERK in homeostasis and constitutive cellular functions, such as cell division, survival, and phagocytosis. Furthermore, ERK is ubiquitously expressed in the brain in several cell types and is also important in neuronal physiological functions, including learning and memory (Hutton et al., 2017; Silingardi et al., 2011). Therefore, therapeutic inhibition of ERK signaling specifically in microglia will likely be limited by cellular nonselectivity, potentially resulting in undesired side effects. Rather than global inhibition of ERK activation, another potential strategy to attain desirable neuro-immunomodulatory effects would be to modulate microglial RTKs that primarily signal via ERK. In recent work, several RTKs were indeed identified as unique to homeostatic (CSF1R, MERTK), pro-inflammatory DAM (FLT4), and anti-inflammatory DAM (AXL, FLT3, IGF1R), among others (Fourgeaud et al., 2016; Rangaraju, Dammer, Raza, Gao, et al., 2018). If different RTKs activate ERK in distinct manners, each resulting in expression of distinct gene sets, the dynamics of ERK signaling downstream of RTKs are likely to modulate downstream gene expression, although this has yet to be explored in microglia. Strategically manipulating ERK signaling via RTKs could allow for the targeted suppression of pro-inflammatory DAM genes with known detrimental roles in AD, while sparing or even augmenting protective homeostatic and anti-inflammatory DAM genes, resulting in a net beneficial immune response. Based on our results, future studies will investigate RTK-specific MAPK signaling in microglia in AD models to guide therapeutic interventions.
A dynamic model of ERK activation in microglia also raises the intriguing possibility that short-term ERK activation regulates homeostatic and constitutive responses, while sustained and long-term ERK activation may mediate disease-associated and pro-inflammatory microglial responses in neurodegeneration. Indeed, in-vitro and in-vivo studies using real-time ERK signaling reporters in neuronal cells and T cells have shown that the immune effects of short-term ERK activation and sustained ERK activation are very different (de la Cova et al., 2017; Harvey et al., 2008; Konishi et al., 2018). The recent development of FRET-based biosensors of ERK and MAPK signaling will allow us to test mechanistic hypotheses to define the ERK signaling dynamics of diverse RTKs expressed by microglia, and then further determine the downstream effects of ERK activation via RTKs on gene expression patterns both in vitro and in vivo. These novel approaches will also facilitate the development of novel approaches to regulate ERK signaling dynamics to achieve desired neuroinflammatory microglial phenotypes.
Although we focused on ERK signaling, our Luminex studies were consistent with prior work revealing increased activation of p38 and trends toward increased JNK activation in microglia from 5xFAD mice, and human postmortem brain proteomes also suggested activation of JNK and p38 pathways, although to a lesser extent than ERK (Lee & Kim, 2017; Yarza et al., 2016). These data suggest that non-ERK MAPKs may also be relevant in regulating microglial functions in AD. Beyond these MAPKs, NFκB signaling is another key regulator of pro-inflammatory immune responses, which was not considered in our studies (Shih et al., 2015). Unlike the robust effects of ERK inhibition observed in these studies, we previously found that inhibition of JNK or NFκB in microglia had minimal effects on the expression of selected homeostatic and DAM genes or on effects of pro-inflammatory stimuli in microglia (Gao et al., 2019). Due to the limited yield of protein from acutely isolated CD11b+ microglia from adult mice, we focused our efforts on ERK, although future studies will investigate the importance of other MAPKs, specifically p38 and JNK signaling pathways, in AD. Another limitation of our work include low protein yields that limited our ability to explore regional differences in microglial ERK activation in 5xFAD mice. Lastly, a serious limitation of our study includes the fact that some in-vitro experiments were conducted with only three biological replicates per group. As a result, findings from these must be interpreted with great caution. At the time these experiments were completed and during the peer review period, additional replicates were not available and additional experiments could not be performed due to limited laboratory staffing and access due to the COVID-19 pandemic.
5 |. CONCLUSIONS
In conclusion, our integrated analysis of a preclinical model of AD pathology, in-vitro transcriptomic and functional studies in primary microglia, and human proteomic data provide novel insights into ERK activation in AD pathogenesis. Moreover, they suggest that modulation of ERK phosphorylation may be a novel avenue for immunomodulation in AD.
Supplementary Material
TABLE S8 Human frontal cortex proteomics from Alzheimer’s disease, Alzheimer’s disease/Parkinson’s disease, Parkinson’s disease, and control brains
TABLE S10 Upstream regulators of differentially expressed MAPK phosphopeptides in Alzheimer’s disease
TABLE S9 Differentially expressed ERK, JNK, and p38 phosphopeptides from human frontal cortex samples
TABLE S7 MAGMA analysis of 28 known Alzheimer’s disease risk genes differentially expressed in NanoString data set
TABLE S6 Transcription factors downstream of ERK activation likely to regulate genes in Clusters 2, 3, and 4
TABLE S5 Gene Ontology analysis of K-means clusters showing enriched GO terms, KEGG, and Wikipathways
TABLE S4 K-means clustering of 465 differentially expressed genes in NanoString data set
TABLE S3 NanoString transcriptomic data set for 678 genes with ANOVA p values
TABLE S2 List of MAPK genes/proteins cross-referenced with published proteomic data sets
TABLE S1 Clinical data for human postmortem brain samples
FIGURE S1 MAPK phospho-signaling changes in frontal cortex in WT and 5xFAD mouse brain
FIGURE S2 ERK inhibition exerts minimal impact on viability of microglia
FIGURE S3 DAM genes positively regulated by ERK signaling
FIGURE S4 Pro-inflammatory DAM genes positively regulated by ERK signaling
FIGURE S5 qRT-PCR validation confirms ERK effect of select DAM genes
FIGURE S6 qRT-PCR study showing dose-dependent regulation of pro-inflammatory DAM but not homeostatic and anti-inflammatory DAM genes by ERK inhibition in primary microglia
FIGURE S7 Flow cytometry gating strategy for live/dead cell distinction
Significance.
We report that extracellular signal-regulated kinase (ERK) activation is characteristic of activated microglia in an Aβ mouse model of Alzheimer’s disease (AD) pathology. Inhibition of ERK activation in primary microglia dampened IFNγ-induced pro-inflammatory gene expression and suppressed neuronal phagocytic activity, suggesting that ERK activity in microglia may regulate detrimental pro-inflammatory responses. Analyses of global and phosphoproteomic data from postmortem human brain indicate increased ERK activation in human AD. Our integrated analyses provide novel insights into ERK activation in microglia and support a critical ro2le for ERK signaling in AD pathogenesis.
Funding information
Georgia Clinical and Translational Science Alliance, Grant/Award Number: UL1TR002378; National Institute of Biomedical Imaging and Bioengineering, Grant/Award Number: T32-GM008433; Georgia Institute of Technology; Alzheimer’s Disease Research Center, Emory University, Grant/Award Number: P50 AG025688 and P30 AG066511; National Science Foundation CBET/EBS Program, Grant/Award Number: 1944053; National Institutes of Health, Grant/Award Number: K08-NS099474-1 and 1R01NS114130-01A1; Alzheimer’s Association, Grant/Award Number: AARG 37102; Accelerating Medicine Partnership for Alzheimer’s Disease, Grant/Award Number: U01 AG046161 and U01 AG061357
Abbreviations:
- 5xFAD
refers to Tg(APPSwFlLon,PSEN1*M146L*L286V)6799Vas/Mmjax strain on C57BL6/J background
- AD
Alzheimer’s disease
- Aβ
amyloid beta
- DAM
disease-associated microglia
- ERK
extracellular signal-regulated kinase
- GO
gene ontology
- IFNγ
interferon gamma
- JNK
c-Jun N-terminal kinase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LPS
lipopolysaccharide
- MAGMA
multi-marker analysis of GenoMic annotation
- MAPK
mitogen-activated protein kinase
- NFκB
nuclear factor kappa-light-chain-enhancer of activated B cells
- PANTHER
protein analysis through evolutionary relationships
- RTK
receptor tyrosine kinase
- TMT
tandem mass tag
Footnotes
CONFLICT OF INTEREST
The authors report no competing interests.
DECLARATION OF TRANSPARENCY
The authors, reviewers and editors affirm that in accordance to the policies set by the Journal of Neuroscience Research, this manuscript presents an accurate and transparent account of the study being reported and that all critical details describing the methods and results are present.
Additional supporting information may be found online in the Supporting Information section.
Transparent Peer Review Report
Transparent Science Questionnaire for Authors
DATA AVAILABILITY STATEMENT
Raw data from both proteomic data sets are publicly available (Data set 1: http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD007160; Data set 2: deposited at https://www.synapse.org/#!Synapse:syn20820053/wiki/596078). Supplementary data files pertinent to this manuscript are also provided within the manuscript.
REFERENCES
- Arkun Y, & Yasemi M (2018). Dynamics and control of the ERK signaling pathway: Sensitivity, bistability, and oscillations. PLoS ONE, 13(4), e0195513. 10.1371/journal.pone.0195513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachiller S, Jimenez-Ferrer I, Paulus A, Yang Y, Swanberg M, Deierborg T, & Boza-Serrano A (2018). Microglia in neurological diseases: A road map to brain-disease dependent-inflammatory response. Frontiers in Cellular Neuroscience, 12, 488. 10.3389/fncel.2018.00488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block ML, Zecca L, & Hong JS (2007). Microglia-mediated neurotoxicity: Uncovering the molecular mechanisms. Nature Reviews Neuroscience, 8(1), 57–69. 10.1038/nrn2038 [DOI] [PubMed] [Google Scholar]
- Cargnello M, & Roux PP (2011). Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiology and Molecular Biology Reviews, 75(1), 50–83. 10.1128/MMBR.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Johnson ECB, Dammer EB, Duong DM, Gearing M, Lah JJ, Levey AI, Wingo TS, & Seyfried NT (2018). Effects of APOE genotype on brain proteomic network and cell type changes in Alzheimer’s disease. Frontiers in Molecular Neuroscience, 11, 454. 10.3389/fnmol.2018.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer EB, Lee AK, Duong DM, Gearing M, Lah JJ, Levey AI, & Seyfried NT (2015). Quantitative phosphoproteomics of Alzheimer’s disease reveals cross-talk between kinases and small heat shock proteins. Proteomics, 15(2–3), 508–519. 10.1002/pmic.201400189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cova C, Townley R, Regot S, & Greenwald I (2017). A real-time biosensor for ERK activity reveals signaling dynamics during C. elegans cell fate specification. Developmental Cell, 42(5), 542–553.e544. 10.1016/j.devcel.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, & Posthuma D (2015). MAGMA: Generalized gene-set analysis of GWAS data. PLOS Computational Biology, 11(4), e1004219. 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubbelaar ML, Kracht L, Eggen BJL, & Boddeke E (2018). The kaleidoscope of microglial phenotypes. Frontiers in Immunology, 9, 1753. 10.3389/fimmu.2018.01753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourgeaud L, Través PG, Tufail Y, Leal-Bailey H, Lew ED, Burrola PG, Callaway P, Zagórska A, Rothlin CV, Nimmerjahn A, & Lemke G (2016). TAM receptors regulate multiple features of microglial physiology. Nature, 532(7598), 240–244. 10.1038/nature17630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, Jernigan J, Raza SA, Dammer EB, Xiao H, Seyfried NT, Levey AI, & Rangaraju S (2019). Transcriptional regulation of homeostatic and disease-associated-microglial genes by IRF1, LXRβ, and CEBPα. Glia, 67(10), 1958–1975. 10.1002/glia.23678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraghty AC, Gibson EM, Ghanem RA, Greene JJ, Ocampo A, Goldstein AK, Ni L, Yang T, Marton RM, Paşca SP, Greenberg ME, Longo FM, & Monje M (2019). Loss of adaptive myelination contributes to methotrexate chemotherapy-related cognitive impairment. Neuron, 103(2), 250–265.e258. 10.1016/j.neuron.2019.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz EM, Ghandi M, Treacy DJ, Wagle N, & Garraway LA (2014). ERK mutations confer resistance to mitogen-activated protein kinase pathway inhibitors. Cancer Research, 74(23), 7079–7089. 10.1158/0008-5472.Can-14-2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeyer N, Hanft K-M, Akriditou M-A, Unger N, Park ES, Stanley ER, Staszewski O, Dimou L, & Prinz M (2017). Microglia contribute to normal myelinogenesis and to oligodendrocyte progenitor maintenance during adulthood. Acta Neuropathologica, 134(3), 441–458. 10.1007/s00401-017-1747-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CD, Ehrhardt AG, Cellurale C, Zhong H, Yasuda R, Davis RJ, & Svoboda K (2008). A genetically encoded fluorescent sensor of ERK activity. Proceedings of the National Academy of Sciences of the United States of America, 105(49), 19264–19269. 10.1073/pnas.0804598105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman S, Izzy S, Sen P, Morsett L, & El Khoury J (2018). Microglia in neurodegeneration. Nature Neuroscience, 21(10), 1359–1369. 10.1038/s41593-018-0242-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutton SR, Otis JM, Kim EM, Lamsal Y, Stuber GD, & Snider WD (2017). ERK/MAPK signaling is required for pathway-specific striatal motor functions. Journal of Neuroscience, 37(34), 8102–8115. 10.1523/jneurosci.0473-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, & Amit I (2017). A unique microglia type associated with restricting development of Alzheimer’s disease. Cell, 169(7), 1276–1290.e1217. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kim SH, Smith CJ, & Van Eldik LJ (2004). Importance of MAPK pathways for microglial pro-inflammatory cytokine IL-1 beta production. Neurobiology of Aging, 25(4), 431–439. 10.1016/S0197-4580(03)00126-X [DOI] [PubMed] [Google Scholar]
- Konishi Y, Terai K, Furuta Y, Kiyonari H, Abe T, Ueda Y, Kinashi T, Hamazaki Y, Takaori-Kondo A, & Matsuda M (2018). Live-cell FRET imaging reveals a role of extracellular signal-regulated kinase activity dynamics in thymocyte motility. iScience, 10, 98–113. 10.1016/j.isci.2018.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, Greco DJ, Smith ST, Tweet G, Humulock Z, Zrzavy T, Conde-Sanroman P, Gacias M, Weng Z, Chen H, … Butovsky O (2017). The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity, 47(3), 566–581. e569. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert J-C, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, … Amouyel P (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature Genetics, 45(12), 1452–1458. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JK, & Kim N-J (2017). Recent advances in the inhibition of p38 MAPK as a potential strategy for the treatment of Alzheimer’s disease. Molecules, 22(8), 1287. 10.3390/molecules22081287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung W-S, Peterson TC, Wilton DK, Frouin A, Napier BA, Panicker N, Kumar M, Buckwalter MS, Rowitch DH, Dawson VL, Dawson TM, … Barres BA (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature, 541(7638), 481–487. 10.1038/nature21029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo MA, Sudol KL, Narrow WC, & Bowers WJ (2009). Interferon-{gamma} differentially affects Alzheimer’s disease pathologies and induces neurogenesis in triple transgenic-AD mice. American Journal of Pathology, 175(5), 2076–2088. 10.2353/ajpath.2009.090059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, Carr D, Deng Y, Jin W, Black S, Long B, Liu J, DiNunzio E, Windsor W, Zhang R, Zhao S, Angagaw MH, Pinheiro EM, Desai J, … Samatar AA (2013). Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discovery, 3(7), 742–750. 10.1158/2159-8290.Cd-13-0070 [DOI] [PubMed] [Google Scholar]
- Namsi A, Nury T, Hamdouni H, Yammine A, Vejux A, Vervandier-Fasseur D, Latruffe N, Masmoudi-Kouki O, & Lizard G (2018). Induction of neuronal differentiation of murine N2a cells by two polyphenols present in the mediterranean diet mimicking neurotrophins activities: Resveratrol and apigenin. Diseases, 6(3), 67. 10.3390/diseases6030067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping L, Duong DM, Yin L, Gearing M, Lah JJ, Levey AI, & Seyfried NT (2018). Global quantitative analysis of the human brain proteome in Alzheimer’s and Parkinson’s disease. Scientific Data, 5, 180036. 10.1038/sdata.2018.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping L, Kundinger SR, Duong DM, Yin L, Gearing M, Lah JJ, Levey AI, & Seyfried NT (2020). Global quantitative analysis of the human brain proteome and phosphoproteome in Alzheimer’s disease. Scientific Data, 7(1), 315. 10.1038/s41597-020-00650-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.r-project.org/ [Google Scholar]
- Rajendran L, & Paolicelli RC (2018). Microglia-mediated synapse loss in Alzheimer’s disease. Journal of Neuroscience, 38(12), 2911–2919. 10.1523/jneurosci.1136-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju S, Dammer EB, Raza SA, Gao T, Xiao H, Betarbet R, Duong DM, Webster JA, Hales CM, Lah JJ, Levey AI, & Seyfried NT (2018). Quantitative proteomics of acutely-isolated mouse microglia identifies novel immune Alzheimer’s disease-related proteins. Molecular Neurodegeneration, 13(1), 34. 10.1186/s13024-018-0266-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju S, Dammer EB, Raza SA, Rathakrishnan P, Xiao H, Gao T, Duong DM, Pennington MW, Lah JJ, Seyfried NT, & Levey AI (2018). Identification and therapeutic modulation of a pro-inflammatory subset of disease-associated-microglia in Alzheimer’s disease. Molecular Neurodegeneration, 13(1), 24. 10.1186/s13024-018-0254-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju S, Raza SA, Li NXA, Betarbet R, Dammer EB, Duong D, Lah JJ, Seyfried NT, & Levey AI (2018). Differential phagocytic properties of CD45(low) microglia and CD45(high) brain mononuclear phagocytes-activation and age-related effects. Frontiers in Immunology, 9, 405. 10.3389/fimmu.2018.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustenhoven J, Smith AM, Smyth LC, Jansson D, Scotter EL, Swanson MEV, Aalderink M, Coppieters N, Narayan P, Handley R, Overall C, Park TIH, Schweder P, Heppner P, Curtis MA, Faull RLM, & Dragunow M (2018). PU.1 regulates Alzheimer’s disease-associated genes in primary human microglia. Molecular Neurodegeneration, 13(1), 44. 10.1186/s13024-018-0277-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SD, Verveer PJ, & Bastiaens PI (2007). Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nature Cell Biology, 9(3), 324–330. 10.1038/ncb1543 [DOI] [PubMed] [Google Scholar]
- Sellgren CM, Gracias J, Watmuff B, Biag JD, Thanos JM, Whittredge PB, Fu T, Worringer K, Brown HE, Wang J, Kaykas A, Karmacharya R, Goold CP, Sheridan SD, & Perlis RH (2019). Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nature Neuroscience, 22(3), 374–385. 10.1038/s41593-018-0334-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfried NT, Dammer EB, Swarup V, Nandakumar D, Duong DM, Yin L, Deng Q, Nguyen T, Hales CM, Wingo T, Glass J, Gearing M, Thambisetty M, Troncoso JC, Geschwind DH, Lah JJ, & Levey AI (2017). A multi-network approach identifies protein-specific co-expression in asymptomatic and symptomatic Alzheimer’s disease. Cell Systems, 4(1), 60–72.e64. 10.1016/j.cels.2016.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih R-H, Wang C-Y, & Yang C-M (2015). NF-kappaB signaling pathways in neurological inflammation: A mini review. Frontiersin Molecular Neuroscience, 8, 77. 10.3389/fnmol.2015.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silingardi D, Angelucci A, De Pasquale R, Borsotti M, Squitieri G, Brambilla R, Putignano E, Pizzorusso T, & Berardi N (2011). ERK pathway activation bidirectionally affects visual recognition memory and synaptic plasticity in the perirhinal cortex. Frontiers in Behavioral Neuroscience, 5, 84. 10.3389/fnbeh.2011.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AM, Gibbons HM, Oldfield RL, Bergin PM, Mee EW, Faull RLM, & Dragunow M (2013). The transcription factor PU.1 is critical for viability and function of human brain microglia. Glia, 61(6), 929–942. 10.1002/glia.22486 [DOI] [PubMed] [Google Scholar]
- Tay TL, Mai D, Dautzenberg J, Fernández-Klett F, Lin G, Sagar, Datta M, Drougard A, Stempfl T, Ardura-Fabregat A, Staszewski O, Margineanu A, Sporbert A, Steinmetz LM, Pospisilik JA, Jung S, Priller J, Grün D, Ronneberger O, & Prinz M (2017). A new fate mapping system reveals context-dependent random or clonal expansion of microglia. Nature Neuroscience, 20(6), 793–803. 10.1038/nn.4547 [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, & Narechania A (2003). PANTHER: A library of protein families and subfamilies indexed by function. Genome Research, 13(9), 2129–2141. 10.1101/gr.772403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood LB, Winslow AR, Proctor EA, McGuone D, Mordes DA, Frosch MP, Hyman BT, Lauffenburger DA, & Haigis KM (2015). Identification of neurotoxic cytokines by profiling Alzheimer’s disease tissues and neuron culture viability screening. Scientific Reports, 5, 16622. 10.1038/srep16622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarza R, Vela S, Solas M, & Ramirez MJ (2016). c-Jun N-terminal kinase (JNK) signaling as a therapeutic target for Alzheimer’s disease. Frontiers in Pharmacology, 6, 321. 10.3389/fphar.2015.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates B, Braschi B, Gray KA, Seal RL, Tweedie S, & Bruford EA (2017). Genenames.org: The HGNC and VGNC resources in 2017. Nucleic Acids Research, 45(D1), D619–D625. 10.1093/nar/gkw1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon AC, Gaj S, Ho I, Hanspers K, Vranizan K, Evelo CT, Conklin BR, Pico AR, & Salomonis N (2012). GO-Elite: A flexible solution for pathway and ontology over-representation. Bioinformatics, 28(16), 2209–2210. 10.1093/bioinformatics/bts366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, & Liu HT (2002). MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Research, 12(1), 9–18. 10.1038/sj.cr.7290105 [DOI] [PubMed] [Google Scholar]
- Zheng C, Zhou X-W, & Wang J-Z (2016). The dual roles of cytokines in Alzheimer’s disease: Update on interleukins, TNF-α, TGF-β and IFN-γ. Translational Neurodegeneration, 5(1), 7. 10.1186/s40035-016-0054-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zoller T, Krieglstein K, & Spittau B (2015). TGFbeta1 inhibits IFNgamma-mediated microglia activation and protects mDA neurons from IFNgamma-driven neurotoxicity. Journal of Neurochemistry, 134(1), 125–134. 10.1111/jnc.13111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
TABLE S8 Human frontal cortex proteomics from Alzheimer’s disease, Alzheimer’s disease/Parkinson’s disease, Parkinson’s disease, and control brains
TABLE S10 Upstream regulators of differentially expressed MAPK phosphopeptides in Alzheimer’s disease
TABLE S9 Differentially expressed ERK, JNK, and p38 phosphopeptides from human frontal cortex samples
TABLE S7 MAGMA analysis of 28 known Alzheimer’s disease risk genes differentially expressed in NanoString data set
TABLE S6 Transcription factors downstream of ERK activation likely to regulate genes in Clusters 2, 3, and 4
TABLE S5 Gene Ontology analysis of K-means clusters showing enriched GO terms, KEGG, and Wikipathways
TABLE S4 K-means clustering of 465 differentially expressed genes in NanoString data set
TABLE S3 NanoString transcriptomic data set for 678 genes with ANOVA p values
TABLE S2 List of MAPK genes/proteins cross-referenced with published proteomic data sets
TABLE S1 Clinical data for human postmortem brain samples
FIGURE S1 MAPK phospho-signaling changes in frontal cortex in WT and 5xFAD mouse brain
FIGURE S2 ERK inhibition exerts minimal impact on viability of microglia
FIGURE S3 DAM genes positively regulated by ERK signaling
FIGURE S4 Pro-inflammatory DAM genes positively regulated by ERK signaling
FIGURE S5 qRT-PCR validation confirms ERK effect of select DAM genes
FIGURE S6 qRT-PCR study showing dose-dependent regulation of pro-inflammatory DAM but not homeostatic and anti-inflammatory DAM genes by ERK inhibition in primary microglia
FIGURE S7 Flow cytometry gating strategy for live/dead cell distinction
Data Availability Statement
Raw data from both proteomic data sets are publicly available (Data set 1: http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD007160; Data set 2: deposited at https://www.synapse.org/#!Synapse:syn20820053/wiki/596078). Supplementary data files pertinent to this manuscript are also provided within the manuscript.