Jiang et al. leveraged KRAS-induced iron addiction in cancer cells to design a clever drug delivery approach to enable selective inhibition of KRAS signaling in mutant KRAS tumors but not in normal tissues, offering a new strategy for treating this largely incurable disease.
Abstract
How to specifically target oncogenic KRAS-driven cancers while sparing normal tissues remains an unmet need in cancer therapy. In this issue of JEM, Jiang et al. (2022. J. Exp. Med. https://doi.org/10.1084/jem.20210739) leveraged KRAS-induced iron addiction in cancer cells to design a clever drug delivery approach to enable selective inhibition of KRAS signaling in mutant KRAS tumors but not in normal tissues, offering a new strategy for treating this largely incurable disease.
Oncogenic KRAS mutations, which account for 85% of all RAS mutations, are common in human cancers, including colon cancer, lung cancer, and, most notably, pancreatic ductal adenocarcinomas (PDACs; >90%; Simanshu et al., 2017; Mustachio et al., 2021). Wild-type KRAS, a small G protein, cycles between its guanosine diphosphate–bound inactive and guanosine triphosphate–bound active states; in contrast, oncogenic mutant KRAS is locked in its guanosine triphosphate–bound active form, which allows oncogenic KRAS to constitutively drive pro-proliferative and pro-survival signaling through its downstream effectors, such as the RAF-MEK-MAPK cascade, independent of upstream stimuli, leading to tumor development (Simanshu et al., 2017; Kerk et al., 2021). Given its pivotal roles in tumor biology, targeting KRAS in cancer therapy, either directly or through its downstream effectors, has garnered considerable interest in cancer research communities (Ryan and Corcoran, 2018). However, several challenges remain in targeting KRAS and its effector pathways: (i) KRAS was previously considered an undruggable target because of a lack of drug-binding pockets outside its nucleotide-binding pocket (Goody et al., 1991; Ryan and Corcoran, 2018); (ii) although recently developed agents that selectively target specific KRAS mutant forms (e.g., KRASG12C) can indeed inhibit KRAS function, this strategy is effective in only a small subset of mutant KRAS cancers and, even more concerningly, wild-type KRAS can also promote tumor development by sustaining its downstream effector signaling (Lito et al., 2016; Ryan and Corcoran, 2018); and (iii) targeting pivotal downstream effectors of KRAS, such as blocking the RAF-MEK-MAPK pathway by using MEK inhibitors, is a promising approach, but its antitumor activity is hampered by dose-limiting toxicities (Zhao and Adjei, 2014; Saunders et al., 2020). Therefore, it is imperative to establish optimal therapeutic strategies that specifically inhibit KRAS or its downstream effectors in tumors without affecting KRAS signaling in nonmalignant tissues.

Insights from Guang Lei and Boyi Gan.
Iron uptake, storage, and utilization are strictly controlled in normal cells, but are generally dysregulated in cancer cells (Lelièvre et al., 2020). Oncogenic KRAS is known to “rewire” cellular metabolism, including iron metabolism, in cancer cells to meet their increased demands for energy and biosynthesis (Kerk et al., 2021). In this issue of JEM, Jiang et al. (2022) uncovered oncogenic KRAS-induced addiction to iron metabolism in cancer cells and exploited this “ferroaddiction” in cancer therapy aiming to improve therapeutic windows for targeting KRAS-driven cancers (Jiang et al., 2022). The authors began their study by analyzing expression levels of genes involved in iron metabolism in multiple cancer types using The Cancer Genome Atlas databases, and they found that the cancer types with the most active iron metabolism were RAS pathway–driven tumors, such as PDAC. Using positron emission tomography, the authors showed that PDAC metastases exhibited elevated levels of ferric iron (Fe3+) radiotracer signals relative to normal tissues in a patient with PDAC. Likewise, they observed higher radiotracer signals for both Fe3+ and ferrous iron (Fe2+) in PDAC xenografts with mutant KRAS than in breast cancer xenografts with wild-type KRAS. The authors confirmed that PDAC cell lines generally have higher intracellular Fe2+ levels than do breast cancer cell lines or nonmalignant cell lines (which harbor wild-type KRAS). These findings suggest that elevated iron uptake and its reduction to labile Fe2+ seem to be common features in oncogenic KRAS-driven PDACs.
Using cell lines in which KRASG12D (a KRAS mutant that frequently occurs in PDACs) can be turned on or off, the authors showed that KRASG12D expression increased intracellular Fe2+ levels, whereas extinguishing KRASG12D expression had the opposite effect. To further study how KRASG12D induces intracellular Fe2+ accumulation, Jiang et al. (2022) compared the expression levels of several regulators involved in iron metabolism in cells with or without KRASG12D. Although the mutations in another RAS isoform, HRAS, have been shown to expand the intracellular Fe2+ pool by inhibiting ferritin expression (Kakhlon et al., 2002), the authors found that KRASG12D did not seem to affect ferritin expression, but rather induced the expression of STEAP3 and repressed the expression of ferroportin. STEAP3 is a lysosomal ferrireductase that is capable of reducing Fe3+ to Fe2+, whereas ferroportin is an iron exporter. Therefore, oncogenic KRAS regulation of STEAP3 and ferroportin expression would presumably promote Fe3+ reduction to Fe2+ and suppress iron efflux, which together contribute to the increased intracellular labile iron pools observed in oncogenic KRAS-activated cells.
The RAF-MEK-MAPK pathway is the best-characterized downstream effector of RAS, and inhibiting this kinase cascade by using MEK inhibitors has been validated as a promising strategy for treating mutant KRAS cancers (Ryan and Corcoran, 2018). However, the dosage of MEK inhibitors required to achieve a therapeutic benefit in mutant KRAS tumors are usually close to or in excess of the clinically tolerable doses for normal tissues, as the pathway flux in cancer cells is generally higher than that in normal cells. As a result, MEK inhibitors have had only limited clinical benefit, partly because of their severe toxicities in normal tissues (including skin, eye, and gut), which compromise the dose intensity that can be attained in tumors (Saunders et al., 2020). Jiang et al. (2022) tackled this challenge by exploiting the ferroaddiction of mutant KRAS cancers they identified. Specifically, they used 1,2,4-trioxolane (TRX) chemistry to synthesize a novel ferrous iron–activatable drug conjugate (FeADC), in which the Food and Drug Administration–approved MEK inhibitor cobimetinib (COBI) was conjugated to the Fe2+-targeting TRX moiety to yield TRX-COBI; the COBI would be released from the TRX-COBI in cells by reacting with Fe2+. The authors reasoned that, because KRASG12D cancer cells generally have higher intracellular Fe2+ levels, a FeADC-targeting MEK would be more efficiently activated by the higher levels of Fe2+ in KRASG12D cancer cells than that in KRAS wild-type normal cells, leading to selective inhibition of MEK in KRASG12D cancer cells or tumors while sparing normal cells or tissues. In support of this innovative idea, PDAC cell lines were indeed identified as being the most sensitive to TRX-COBI among 750 tested cancer cell lines of a variety of cancer types.
The authors further tested their idea in vivo by using both xenograft and autochthonous oncogenic KRAS-driven tumor models. They found that the FeADC-targeting MEK demonstrated tumor suppressive effects comparable to those of the corresponding MEK inhibitor, and most importantly, that the FeADC minimized the systemic on-target, off-tumor toxicities induced by MEK inhibition, presumably because the MEK inhibitor in the FeADC remained largely inactive and consequently failed to suppress MEK in normal tissues. To speed translation of their findings to the clinic, the authors went one step further. Although inhibiting the RAF-MEK-MAPK pathway is pivotal for treating mutant KRAS cancers, activation of other KRAS downstream pathways in mutant KRAS tumors has limited the clinical efficacy of MEK inhibitors given as monotherapy (Ryan and Corcoran, 2018). SHP2 inhibition has been shown to sensitize mutant KRAS cancers to MEK inhibitors (Ruess et al., 2018), although toxicities in normal tissues compromise the effective dose and clinical benefit of such combinations. Jiang et al. (2022) demonstrated that, although a SHP2 inhibitor had pronounced adverse effects in normal tissues when it was combined with a MEK inhibitor, it showed significant tumor suppressive activity and, most importantly, was well tolerated when combined with the FeADC. Therefore, the FeADC in combination with other therapies seems to be an effective therapeutic strategy for mutant KRAS cancers.
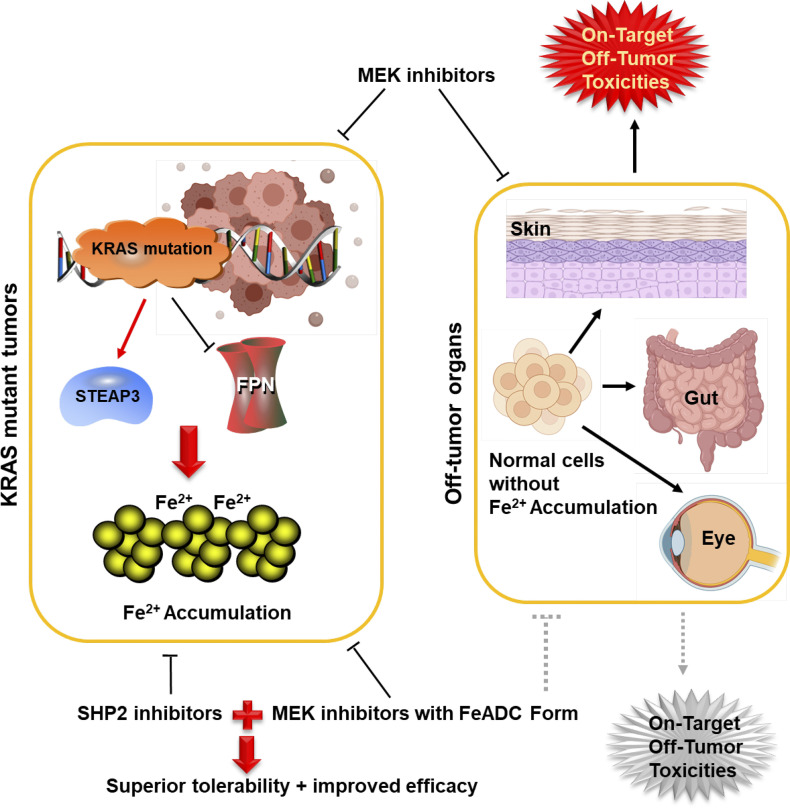
Model depicting how FeADC that targets MEK enables selective inhibition of KRAS signaling in mutant KRAS tumors while sparing normal tissues. MEK is an important downstream effector of KRAS, and MEK inhibitors have been considered a promising strategy for treating mutant KRAS cancers. However, MEK inhibition also triggers severe on-target, off-tumor toxicities in normal tissues (e.g., skin, eye, and gut), limiting its clinical benefit. Cancers with oncogenic KRAS mutations generally have high intracellular Fe2+ levels, likely due to their increased expression of STEAP3 and decreased expression of ferroportin (FPN). Consequently, such tumors can be selectively targeted by MEK inhibitors with FeADC form, which can be efficiently activated by the enriched Fe2+ in mutant KRAS tumors, but remain largely inactive and consequently fail to repress MEK in normal tissues, thereby minimizing on-target, off-tumor toxicities by MEK inhibition. Further combining the FeADC-targeting MEK with SHP2 inhibitors can trigger significant tumor suppression with superior tolerability in oncogenic KRAS-driven cancers.
Collectively, this study revealed that Fe2+ accumulation is a metabolic feature of oncogenic KRAS-driven cancers and that the FeADC is active mainly in Fe2+-enriched mutant KRAS tumors but does not impair normal tissues. The authors also proposed the novel concept that oncogenic KRAS-induced ferroaddiction provides a potential therapeutic window for targeting mutant KRAS cancers by decoupling MEK inhibition in normal tissues and tumors based on the status of bioavailable Fe2+ pool. It will be important to further explore this concept in other cancers that may have other metabolic addictions. The underlying mechanisms by which the FeADC suppresses tumor growth remain to be investigated. Because Fenton chemistry is known to be involved in FeADC activation and in an iron-dependent cell death mechanism known as ferroptosis (Dixon et al., 2012; Spangler et al., 2016), the authors considered whether their FeADC could induce ferroptosis. However, the TRX-COBI–mediated cell killing effect was not suppressed by a ferroptosis inhibitor, suggesting that ferroptosis is not involved in this context. The exact cell death mechanisms induced by the TRX-COBI require additional study. Finally, further investigations are needed to refine the FeDAC design, exploit its combination with other therapies (such as immunotherapy and radiotherapy), and eventually design clinical trials to test this innovative therapeutic strategy in cancer patients with KRAS mutations.
Acknowledgments
We thank Christine F. Wogan, MS, ELS, from MD Anderson’s Division of Radiation Oncology, for editing the manuscript.
References
- Dixon, S.J., et al. 2012. Cell. 10.1016/j.cell.2012.03.042 [DOI] [Google Scholar]
- Goody, R.S., et al. 1991. Trends Biochem. Sci. 10.1016/0968-0004(91)90134-h [DOI] [PubMed] [Google Scholar]
- Jiang, H., et al. 2022. J. Exp. Med. 10.1084/jem.20210739 [DOI] [Google Scholar]
- Kakhlon, O., et al. 2002. Biochem. J. 10.1042/0264-6021:3630431 [DOI] [Google Scholar]
- Kerk, S.A., et al. 2021. Nat. Rev. Cancer. 10.1038/s41568-021-00375-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelièvre, P., et al. 2020. Cancers. 10.3390/cancers12123524 [DOI] [Google Scholar]
- Lito, P., et al. 2016. Science. 10.1126/science.aad6204 [DOI] [Google Scholar]
- Mustachio, L.M., et al. 2021. Cancers. 10.3390/cancers13061204 [DOI] [Google Scholar]
- Ruess, D.A., et al. 2018. Nat. Med. 10.1038/s41591-018-0024-8 [DOI] [Google Scholar]
- Ryan, M.B., and Corcoran R.B.. 2018. Nat. Rev. Clin. Oncol. 10.1038/s41571-018-0105-0 [DOI] [PubMed] [Google Scholar]
- Saunders, I.M., et al. 2020. Oncologist. 10.1634/theoncologist.2019-0606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simanshu, D.K., et al. 2017. Cell. 10.1016/j.cell.2017.06.009 [DOI] [Google Scholar]
- Spangler, B., et al. 2016. J. Med. Chem. 10.1021/acs.jmedchem.6b01470 [DOI] [Google Scholar]
- Zhao, Y., and Adjei A.A.. 2014. Nat. Rev. Clin. Oncol. 10.1038/nrclinonc.2014.83 [DOI] [PubMed] [Google Scholar]